Significance
Changes in the number and/or structure of chromosomes (i.e., chromosomal rearrangements) have the potential to drive speciation. However, their accumulation in a population is considered both difficult and unpredictable, because the greatly reduced reproductive fitness of chromosomal hybrids prevents fixation of novel karyotypes. Here, we provide evidence for a mechanism that rescues fertility of chromosomal hybrids in species with holocentric chromosomes. We demonstrate that chromosomal heterozygotes of Leptidea Wood White butterflies have a reverse order of main meiotic events in which the first and most critical stage of the chromosome number reduction is replaced by the less risky stage of sister chromatid separation. This may facilitate long-term persistence of chromosomal rearrangements, which is a major prerequisite for chromosomal speciation.
Keywords: chromosomal evolution, chromosomal rearrangement, hybridization, inverted meiosis, speciation
Abstract
Chromosomal rearrangements (e.g., fusions/fissions) have the potential to drive speciation. However, their accumulation in a population is generally viewed as unlikely, because chromosomal heterozygosity should lead to meiotic problems and aneuploid gametes. Canonical meiosis involves segregation of homologous chromosomes in meiosis I and sister chromatid segregation during meiosis II. In organisms with holocentric chromosomes, which are characterized by kinetic activity distributed along almost the entire chromosome length, this order may be inverted depending on their metaphase I orientation. Here we analyzed the evolutionary role of this intrinsic versatility of holocentric chromosomes, which is not available to monocentric ones, by studying F1 to F4 hybrids between two chromosomal races of the Wood White butterfly (Leptidea sinapis), separated by at least 24 chromosomal fusions/fissions. We found that these chromosomal rearrangements resulted in multiple meiotic multivalents, and, contrary to the theoretical prediction, the hybrids displayed relatively high reproductive fitness (42% of that of the control lines) and regular behavior of meiotic chromosomes. In the hybrids, we also discovered inverted meiosis, in which the first and critical stage of chromosome number reduction was replaced by the less risky stage of sister chromatid separation. We hypothesize that the ability to invert the order of the main meiotic events facilitates proper chromosome segregation and hence rescues fertility and viability in chromosomal hybrids, potentially promoting dynamic karyotype evolution and chromosomal speciation.
Chromosomal rearrangements, i.e., alterations in the number and structure of chromosomes, attract the attention of biologists and health professionals because they are often triggers in both evolution and pathogenesis. In medicine, they cause many syndromes and heritable diseases (1, 2), and play important roles in the pathogenesis of human cancers (3, 4). In evolutionary biology, chromosomal rearrangements are known to facilitate speciation via reducing fertility of chromosomal heterozygotes or/and via suppressed recombination (5, 6). The rearrangements maintain postzygotic isolation between species by preventing merging through hybridization (7), are crucial in adaptive evolution by protecting blocks of linked genes from recombination (8, 9), and may also alter gene expression in ways not possible through point mutations (10).
Despite their importance, the question how novel chromosomal rearrangements appear, accumulate, go through the stage of heterozygosity, and become fixed in a population is still poorly resolved (11–13). Theory predicts that this process may be difficult because chromosomal heterozygosity results in the formation of multivalents in meiosis (instead of normal bivalents). This leads to segregation problems in the first meiotic division and to the production of unbalanced gametes (12, 13).
Simple theory predicts that even a single heterozygous chromosomal rearrangement, such as reciprocal translocation or chromosomal fusion, should result in 50% reduction of fertility (13). In the case of heterozygosity for multiple rearrangements, the rate of balanced gametes should decrease strongly and be as low as 1/2n, where n is the number of heterozygous rearrangements (12). In reality, the proportion of inviable gametes is often less than this expectation, because it depends on the orientation of multivalents at meiosis (13), preferential inclusion of inviable nuclei in polar bodies in females (14), and lower recombination rates in the heterogametic sex (15). However, in general, fertility decreases with increased chromosomal heterozygosity (13, 16). Nevertheless, for reasons that are often unknown, organisms can sometimes tolerate heterozygosity for multiple rearrangements (17, 18), raising questions about additional mechanisms that rescue fertility in chromosomal hybrids.
The butterfly genus Leptidea (family Pieridae) represents an excellent system to study the role of chromosomal rearrangements in speciation, because several species display notable levels of interspecific and intraspecific variability in the number of chromosomes (17, 19–21). Much of the recent karyological research on Leptidea has been triggered by the unexpected levels of cryptic diversity found within the most widespread of these species, namely the Wood White Leptidea sinapis. Until the end of the 20th century, L. sinapis was regarded as a single common Eurasian species, but research on male and female genitalia (e.g., refs. 22 and 23) coupled with behavioral (23, 24) and genetic data (25, 26) led to the unexpected discovery of a cryptic species, Leptidea reali. Even more surprisingly, recent molecular, karyological, and behavioral analyses revealed that the pair L. sinapis and L. reali actually consists of a triplet of species, L. sinapis, L. reali and Leptidea juvernica, which represents one of the most striking examples of cryptic diversity in Eurasian butterflies (19, 27). Previous research (i.e., before the discovery of L. reali and L. juvernica) reported a considerable variability of the haploid chromosome number (n) in L. sinapis (n = 28 to n = 41; see ref. 28). Given the discovery of L. reali and L. juvernica, the correct interpretation of the previous karyological data required new analyses to establish if the variation reflects intraspecific or interspecific patterns. Recent research has shown that L. reali has the diploid chromosome number (2n) ranging between 2n = 51 to 55 and L. juvernica between 2n = 76 to 91 (19, 20).
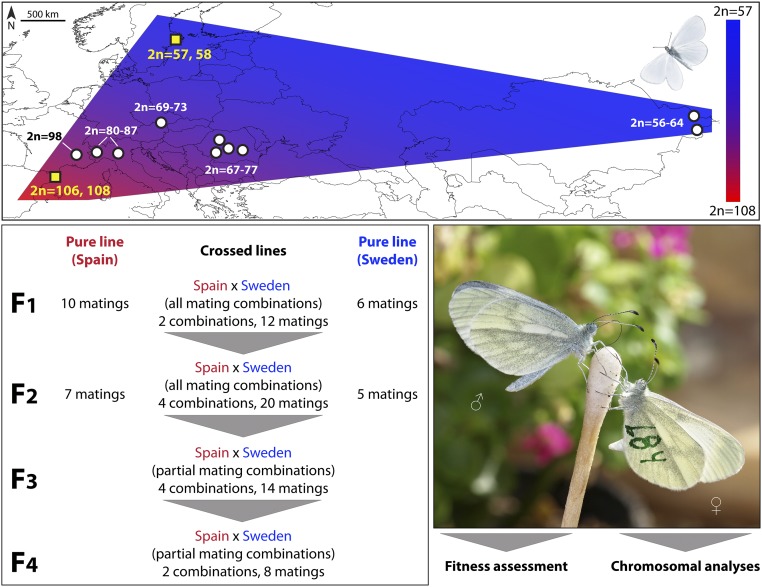
However, the most striking pattern was found in L. sinapis, which displays the widest documented intraspecific variability of the chromosome number known in eukaryotes, excluding cases of polyploidy (17). Within this species, the diploid chromosome number gradually decreases from 2n = 106, 108 in northeastern Spain to 2n = 56 in eastern Kazakhstan (17), and to 2n = 57, 58 in southeastern Sweden (this study) (Fig. 1). This likely happened due to the accumulation of chromosomal fusions/fissions, resulting in a wide cline of relatively recent origin (29). The direction of the formation of the cline is currently unclear [i.e., if fusions or fissions occurred (17)]. The intraspecific nature of this extreme level of variability in chromosome number is supported by genetic and morphological data, as well as by mating experiments (17, 27).
Fig. 1.
Chromosomal system of the Eurasian butterfly L. sinapis and general experimental plan. L. sinapis displays a wide chromosome number cline ranging from 2n = 106, 108 in Spain (17, 20) to 2n = 56 in Kazakhstan (17, 20) and 2n = 57, 58 in Sweden (data from this study). Laboratory crosses between specimens with high (Spain) and low (Sweden) chromosome numbers (yellow squares on the map) involved four generations of hybrid lines and two generations of pure lines used as controls. The progeny of successful matings were used for fitness and chromosomal analyses.
Like other Lepidoptera, L. sinapis has holocentric chromosomes (30, 31), which are characterized by kinetic activity distributed along almost the entire chromosome length (30, 32–35). Species with holocentric chromosomes occur in multiple phyla of animals and plants (30, 35, 36) and, based on the available literature, may represent as much as 20 to 30% of eukaryotic diversity.
Here, we analyzed the intriguing chromosomal system of the Eurasian butterfly L. sinapis coupled with experimental hybridization of two chromosomal races that are separated by at least 24 chromosomal fusions/fissions to (i) demonstrate an extreme case of tolerance to heterozygosity for multiple chromosomal fusions/fissions; (ii) document the inverted order of meiotic events in chromosomal hybrids (Fig. 2), a highly debated phenomenon, which until recently had only limited support (32–34); and (iii) demonstrate a link between spatial orientation of multivalents in meiosis, inverted meiosis, and viability in holocentric chromosomal hybrids.
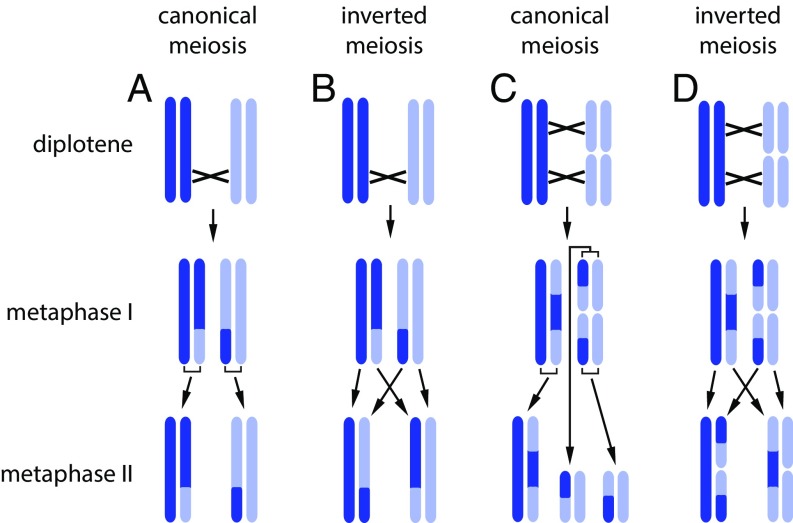
Fig. 2.
Schematic representation of canonical (A and C) and inverted (B and D) meiosis in a chromosome bivalent (A and B) and a chromosome trivalent (C and D). Chiasmata are shown by crosses. (A and C) Canonical meiosis. Following DNA replication, homologous chromosomes (shown by different colors) are separated in the first meiotic division (i.e., this division is reductional), whereas sister chromatids (shown by the same color) are separated in the second division (equational division). (B and D) Inverted meiosis. Sister chromatids are separated in the first division (equational division), and homologous chromosomes are separated in the second division (reductional division). Note that application of the concept of inverted meiosis has a limitation: If meiosis is chiasmatic, then, as a consequence of meiotic recombination, the first meiotic division can be reductional at some loci and equational at others (37–39). Nevertheless, this concept is completely applicable to (i) monocentric chromosomes for which a classification of the first meiotic division as reductional or equational can be made in terms of division at the centromere since centromere regions are not involved in recombination; (ii) achiasmatic holocentric chromosomes lacking recombination (39); (iii) structural holocentric heterozygotes, such as trivalents, in which chromosomal fusion/fission serves as a marker allowing distinction between chromosomal homologs and sister chromatids. In this case, there is a possible coherent definition of prereductional and postreductional (inverted) meiosis, depending on whether the first division segregates two and four chromatids (reductional division) or three and three chromatids (equational division), respectively (C and D); (iv) chiasmatic holocentric chromosomes, in which chromosomal homologs (but not sister chromatids) are linked after metaphase I by satellite DNA-enriched chromatin threads (32), allowing distinction between chromosomal homologs and sister chromatids; and (v) holocentric chromosomes with single (sub)terminal chiasma (Fig. 9 B, ii). The latter type of chromosomes is very common among holocentrics (38, 40, 41). Loidl (37) described how inverted meiosis may be an acceptable term for this case when nearly entire sister chromatids segregate in anaphase I due to the (sub)terminal chiasma. Note that, for simplification, the spatial structure of bivalents and trivalents is not shown.
Results
Experimental Crosses and Reproductive Fitness in F1 to F4 Hybrids.
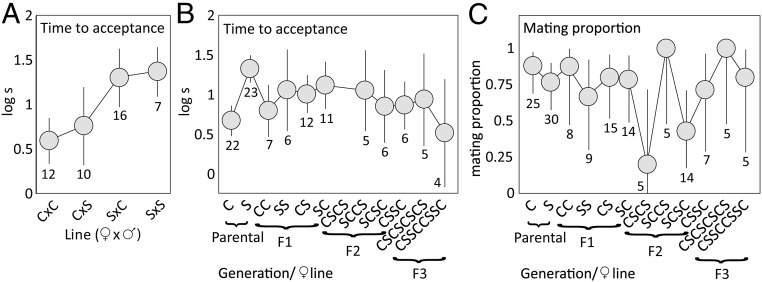
We crossed (Movie S1) chromosomal races of L. sinapis with low (2n = 57, 58 from southeastern Sweden) and high (2n = 106, 108 from Catalonia, northeastern Spain) chromosome numbers (SI Appendix, Table S1) and performed both fitness and chromosomal analyses (Fig. 1). We maintained hybrid lines for four generations (F1 to F4), and pure control lines (Sweden and Spain) for two generations (F1 and F2) (Fig. 1 and SI Appendix, SI Methods, Fig. S1, and Table S2). There was no evidence of assortative mating in the parental generation [mating proportion, generalized linear model (glm): female population χ21 = 1.07, P = 0.30; male population χ21 = 2.67, P = 0.10; female × male population χ21 = 0.20, P = 0.65], but Spanish females accepted mating faster than Swedish females (Fig. 3). Mating propensity was generally high across all pure and hybrid generations (Fig. 3).
Fig. 3.
Mean time to female mating acceptance (±95% confidence intervals CI) (A) in the parental generation where Spanish females (C) accepted mating faster than Swedish females (S) regardless of male population affiliation (linear model: female population F1,28 = 17.1, P < 0.001; male population F1,28 = 0.49, P = 0.49; female × male population F1,28 = 0.087, P = 0.77) and (B) for all female lines across the four generations of this study, where hybrid line acceptance times are distributed between acceptance times of the pure populations (female line F10,95 = 2.65, P = 0.0069). In C, the variation in mating propensity is reported for females of different lines (±95% confidence intervals CI), and a weakly significant difference among lines is largely driven by only one out of five CSCS females accepting mating (glm: χ211 = 17.1, P = 0.017).
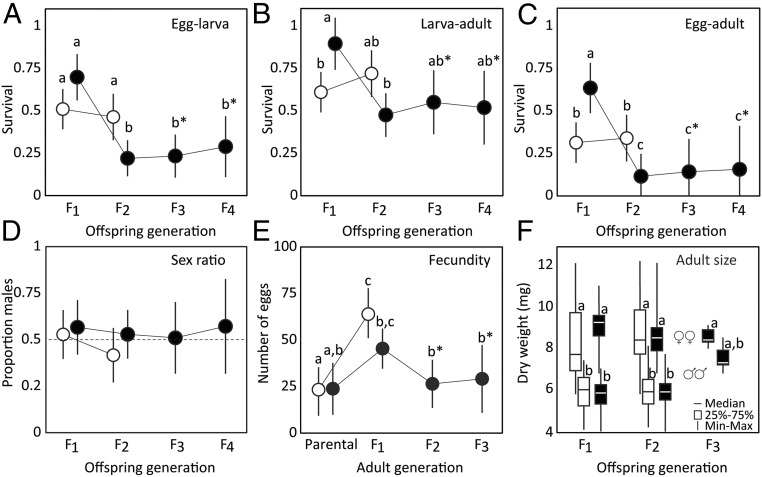
When comparing the first two generations of pure and hybrid lines, we found significant interactive effects (SI Appendix, Table S3) of cross type (pure/hybrid) and offspring generation (F1/F2) on larval fitness. Survival, measured as mean egg hatch rate (Fig. 4A), survival from larva to adult (Fig. 4B), and from egg to adult (Fig. 4C), was higher in the F1 hybrids than in the pure lines, whereas mean survival in the hybrid lines was significantly lower in the F2 generation than in the control lines (Fig. 4 A–C). There was no indication that fitness variation was influenced by a small number of sibling matings (SI Appendix, Table S4) or that a particular crossing direction or hybrid line had consistently higher fitness than other lines or crosses, with a potential exception of survival from larva to adult in generation F1, where small but significant differences were found among cross directions (SI Appendix, Fig. S2 and Table S5). The reduced hybrid fitness in the F2 generation was maintained in hybrid generations F3 and F4 (Fig. 4 A–C and SI Appendix, Table S6). The sex ratio of adults was similar across hybrid generations and pure lines, and distributed close to 1:1 (Fig. 4D and SI Appendix, Tables S3 and S6). There was no significant difference in female fecundity between pure lines and hybrids, but females of both cross types tended to lay more eggs in the F1 generation than in any other generation (Fig. 4E and SI Appendix, Table S3). Adult size (dry weight) varied between sexes (Kruskal Wallis ANOVA χ29 = 237.2, P < 0.001), but showed no significant pattern in relation to hybrid status (Fig. 4F).
Fig. 4.
Fitness measures across two generations of Swedish and Spanish pure lines (here pooled because no major differences between them were found; see SI Appendix, Fig. S2 and Table S5) (within-population crosses; open symbols), and four hybrid generations (offspring of the initial between-population crosses of Swedish and Spanish L. sinapis; filled symbols). Shown are family means (±95% confidence intervals) unless otherwise noted. These measures include (A) the egg hatch rate, (B) survival from larva to adult, (C) the overall survival (from egg to adult), (D) the sex ratio for each line and generation, (E) the female fecundity (number of eggs laid) in each generation, and (F) the dry weight of the emerged adult females (Top) and males (Bottom) of generations F1 to F3. Letters above markers indicate post hoc patterns of statistical significance [Tukey’s honest significant difference test in A–E, pairwise Mann−Whitney U tests in F; letters with asterisks (*) indicate values compared only within hybrid lines]. In A–C, note the increase in hybrid survival in F1 followed by a decrease to a consistent level of about half the pure line survival in generations F2 to F4.
Thus, contrary to the theoretical prediction (12, 13), the hybrids displayed a relatively high reproductive fitness, and their average survival from egg to adult was just below half (42%) of that of the within-population crosses.
Karyotype and Chromosome Behavior in Hybrid Males.
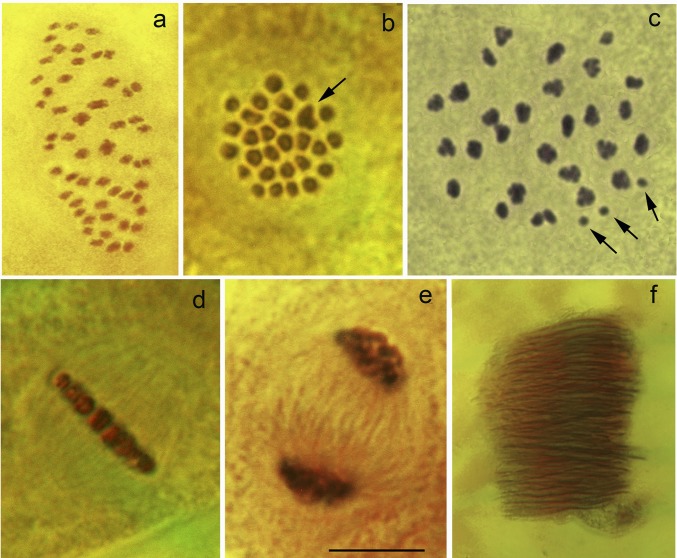
The Swedish and Spanish races are differentiated by at least 24 fixed chromosome fusions/fissions (Fig. 5 A and B). Despite this, a quite regular chromosome pairing was observed in the first meiotic division of F1 hybrid males, resulting in numerous multivalents (most likely trivalents). Thus, the total number of observed entities was close to the haploid chromosome number of the Swedish race (n = 28 to 29) (Fig. 5 C and D). However, variations in the number of chromosomal entities among meiotic metaphase I cells within a single specimen were always observed (from 29 to 33), sometimes with clear univalents indicating an imperfect meiotic pairing (Fig. 5C and SI Appendix, Table S1). In F2, F3, and F4 male hybrids, each metaphase I karyotype showed numerous multivalents and bivalents, and, sometimes, several univalents. The total number of entities (multivalents + bivalents + univalents) was very variable (from 29 to 37) (SI Appendix, Table S1).
Fig. 5.
Male meiosis and spermatogenesis in L. sinapis (stained with acetic orcein). (A) Squashed metaphase I plate in Spanish race (53 bivalents, 2n = 106). (B) Intact metaphase I plate in Swedish race (27 bivalents and one trivalent indicated by arrow, 2n = 57). (C) Squashed metaphase I plate in F1 hybrid between Spanish and Swedish races, where most of the chromosome entities are represented by multivalents (univalents are indicated by arrows). (D) Side view of an intact metaphase I plate in F1 hybrid between Spanish and Swedish races displaying a regular structure with no obvious anomalies. (E) Telophase I in F1 hybrid between Spanish and Swedish displaying no lagging chromosomes. (F), Bundle of eupyrene sperm heads in F2 hybrid between Spanish and Swedish races displaying no obvious abnormalities. (Scale bar: 10 μm in A–E and 40 μm in F.)
The first meiotic anaphase and telophase in F1, F2, F3, and F4 male hybrids appeared normal, without any lagging chromosomes (Fig. 5E). Moreover, morphologically normal bundles of eupyrene sperm were always observed in gonads (Fig. 5F), indicating that the studied specimens were fertile. These findings demonstrate that the chromosome pairing between highly rearranged chromosomes of Swedish and Spanish populations is apparently normal in F1 males. Subsequent crosses resulted in highly recombinant but genetically balanced gametes, and fertile offspring (F2, F3, and F4). These cytological patterns are consistent with the results of fitness analyses.
Inverted Meiosis.
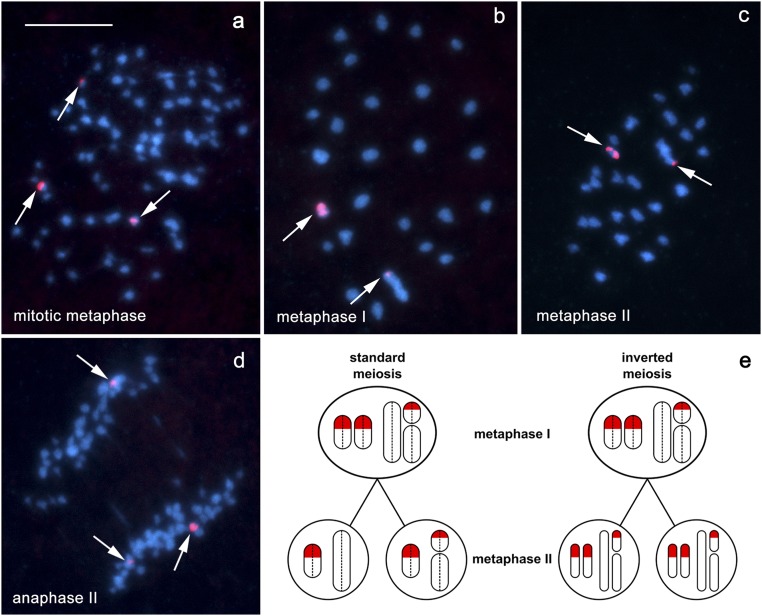
Whereas, in normal canonical meiosis, homologous chromosomes are separated during meiotic division I and sister chromatids are separated during meiosis II, we found that, in Leptidea chromosomal hybrids, this order was inverted. Two lines of evidence support the existence of inverted meiosis in our data. One is based on the analysis of asymmetrical 18S rDNA markers in metaphase I and metaphase II cells (Figs. 6 and 7). The other is based on the fact that the same numbers of chromosomal entities were observed at metaphase I and metaphase II (Table 1).
Fig. 6.
The 18S rDNA−FISH analysis of inverted meiosis in F1 hybrid males of L. sinapis. Chromosomes were stained with DAPI (blue); arrows indicate hybridization signals of the 18S rDNA probe (red). The identical distribution of rDNA signals in both the metaphase I and metaphase II is only possible in the case of inverted meiosis, i.e., when sister chromatids segregate in metaphase I and nonsister chromatids segregate in metaphase II. This pattern was observed in our study. (A) Mitotic metaphase showing hybridization signals, two large and one small, on three chromosomes. (B) Metaphase I showing one small entity (probably a bivalent) with two large signals, and one large entity (probably a multivalent) with a terminal dot-like signal. (C) Metaphase II showing one small entity with two large signals and one large entity with a terminal dot-like signal; thus, the patterns in metaphase I and metaphase II are generally the same. (D) Anaphase II showing an unequal segregation of rDNA signals; two large signals segregate to opposite poles, while a small signal segregates to one of the poles, resulting in chromosome complements (i) with one large signal and (ii) with two signals. (E) Schematic illustration of the rDNA chromosome behavior in standard and inverted meiosis. In standard meiosis, two types of metaphase II cells are expected: (i) with one large signal and (ii) with one large + one small signal. In inverted meiosis, the segregation of sister chromatids of each chromosome results in two metaphase II nuclei, each with three rDNA signals. Dotted lines separate sister chromatids in metaphase I. Note that, for simplification, meiotic recombination was not considered. (Scale bar: 10 μm.)
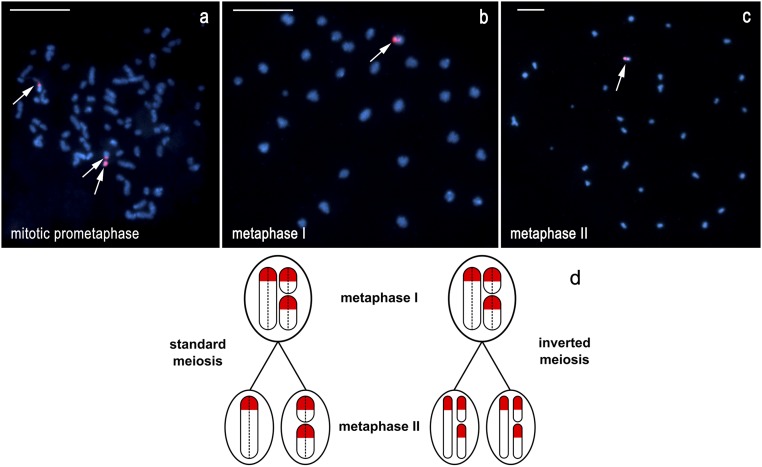
Fig. 7.
The 18S rDNA−FISH analysis of inverted meiosis in F1 hybrid males of L. sinapis. (A) Mitotic prometaphase: 73 elements, a total of three chromosomes with hybridization signals—one larger chromosome with very large interstitial signal localized in a constriction-like part of the chromosome; one middle-sized chromosome with weaker telomeric signal; and one small chromosome covered with strong signal. (B) Metaphase I: 30 elements, a total of two hybridization signals (one large terminal and one small interstitial) in the same middle-sized element. (C) Metaphase II: 30 elements, a total of two hybridization signals (one large terminal and one smaller interstitial) in the same middle-sized element. (D) Schematic illustration of inverted meiosis in F1 hybrid males of L. sinapis, based on the results of 18S rDNA−FISH. The presented case with three chromosomes, each carrying one terminal rDNA cluster (red), approximately reflects the situation shown in A–C. The three rDNA chromosomes form a trivalent in metaphase I. In standard meiosis (only one of three possible variants of segregation is shown), segregation of individual chromosomes results in two daughter (metaphase II) nuclei with one and two rDNA signals, respectively. In inverted meiosis, segregation of the sister chromatids of each chromosome results in two metaphase II nuclei, each with three rDNA signals. Dotted lines separate sister chromatids in metaphase I. Note that, for simplification, meiotic recombination was not considered. (Scale bar: 10 μm.)
Table 1.
Expected and observed number of chromosomal entities in F1 hybrids of L. sinapis in the case of canonical (prereductional) and inverted (postreductional) meiosis
| Sample ID | Observed modal number of chromosome entities in metaphase I | Expected modal number of entities in metaphase II of canonical meiosis | Expected modal number of entities in metaphase II of inverted meiosis | Observed modal number of chromosome entities in metaphase II |
| 12-Z051 | 30 | 40–42 | 30 | 30–32 |
| 12-Z054 | 30 | 40–42 | 30 | 30–33 |
| 12-Z065 | 28 | 40–42 | 28 | 28–31 |
| 12-Z066 | 29 | 40–42 | 29 | 29–32 |
| 13Y045 | 29 | 40–42 | 29 | 29 |
| 13Y057 | 29 | 40–42 | 29 | 29 |
| 13Y058 | 28 | 40–42 | 28 | 28 |
| 13Y063 | 29 | 40–42 | 29 | 29 |
Analysis of Inverted Meiosis by FISH with 18S rDNA Probe.
We found two types of F1 males with respect to the distribution and number of the 18S rDNA clusters. The first type is characterized by the presence of three hybridization signals, two large and one small, on three chromosomes in mitosis (Fig. 6A), resulting in one entity with large double signal, and one entity with small red signal in meiosis (cf. figure 3d in ref. 20). The identical distribution of rDNA signals in both meiotic metaphases (metaphase I and metaphase II), which we found (Fig. 6 B and C), is only possible in the case of inverted meiosis, i.e., when sister chromatids segregate in metaphase I and nonsister chromatids segregate in metaphase II (see scheme in Fig. 6E).
The second type of F1 male is characterized by the presence of three hybridization signals in mitosis (Fig. 7A), but only two signals were found in metaphase I, indicating that all three rDNA chromosomes formed a trivalent (Fig. 7B). This trivalent belonged to the class of middle-sized elements and included one large terminal and one small interstitial signals. A similar pattern was observed in metaphase II (Fig. 7C). The identical distribution of rDNA signals in both the metaphase I and metaphase II complements is only possible in the case of inverted meiosis, i.e., when sister chromatids segregate in metaphase I and nonsister chromatids in metaphase II (see scheme in Fig. 7D). Furthermore, two signals in the same metaphase II element are only possible if the labeled element consists of two nonsister chromatids (Fig. 7D, Left and Right).
The equal separation of rDNA signals at anaphase I and unequal separation at anaphase II (Fig. 6D) represent further evidence for inverted meiosis.
Analysis of Inverted Meiosis by Counting Chromosomal Elements in Metaphase I and Metaphase II Cells.
The Swedish (n = 28 to 29) and Spanish (n = 53 to 54) populations represent two extremes of the chromosomal variation in L. sinapis. The F1 hybrids produced 28 to 30 elements at metaphase I, which means that nearly all of these elements are trivalents. Therefore, we can formulate predictions about how many elements should be observed at metaphase I and at metaphase II in case of prereductional (also known as canonical meiosis) and in case of postreductional (inverted) meiosis. Under a prereductional scenario, we expect that every trivalent will result in 1 + 2 chromosomes (42). Thus, for 28 to 30 elements that segregate randomly, we expect that the number of chromosomes in the secondary spermatocytes will vary from 28 to 54, with an average of n = ca. 40 to 42. Under a postreductional scenario, we expect that each two-sister chromatids trivalent results in a one-sister chromatids trivalent. Thus, we expect that the number of elements in the secondary spermatocytes will be similar to the number in primary spermatocytes, i.e., n = 28 to 30 in metaphase I, and n = 28 to 30 in metaphase II. These predictions are very different and easily discriminated. The analysis of the available data (Table 1) indicates that meiosis in Leptidea F1 hybrids was indeed inverted, in line with a postreductional scenario.
Discussion
Negative effects of heterozygosity for chromosomal rearrangements are mostly observed in meiosis when malsegregation in structural hybrids results in aneuploidy, duplications, and deficiencies (13). Therefore, chromosomal rearrangements usually do not affect survival in F1 hybrids, but are expected to be detrimental to fitness in subsequent generations. This is in accordance with results of our experiment: Survival was high in F1 hybrids between chromosomal races of L. sinapis but was significantly lower in the F2, F3, and F4 (see also SI Appendix, Text 1 for discussion of the relative effects of spermatogenesis and oogenesis on reduced fitness).
However, the reproductive isolation between the L. sinapis chromosomal races was by no means complete. In fact, hybrid viability was unexpectedly high given the number of chromosomal rearrangements involved and, in F2 to F4 generations, the survival from egg to adult plateaued at a rate just below half (42%) of the fitness of the within-population crosses (Fig. 4C). To understand this phenomenon, we conducted cytological analyses of male meiosis, a crucial stage when chromosomal malsegregation and formation of unbalanced gametes takes place. The Swedish and Spanish races are differentiated by at least 24 fixed chromosome fusions/fissions (Fig. 5 A and B). Despite this pronounced difference, chromosome pairing was regular in the first meiotic division of F1 males, resulting in numerous multivalents (most likely trivalents).
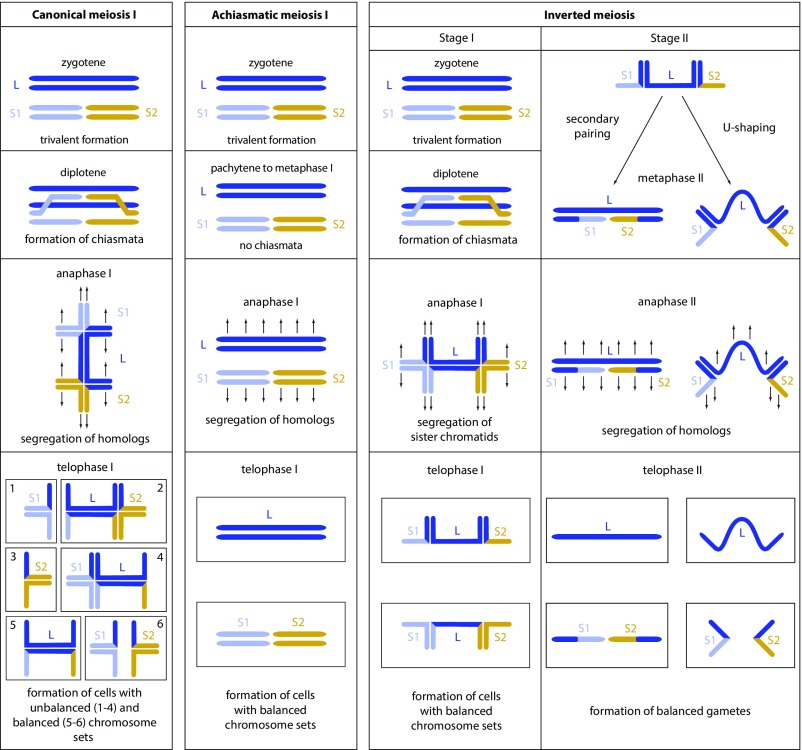
In most organisms, the high number of multivalents detected would cause serious meiotic segregation problems, resulting in complete or almost complete sterility (12, 13) (Fig. 8, Left). What mechanisms could explain how Leptidea butterflies retain substantial fertility in hybrids despite such extreme parental chromosomal differences? In Lepidoptera, females have a special organization of bivalents and multivalents that reduces unbalanced segregation. Female meiosis is achiasmatic, with the parallel position of homologs in the relatively long (not dot-shaped as in males) bivalents and multivalents, and the homologs are maintained together until segregation by a modified synaptonemal complex (43). This structure results in a parallel orientation of bivalents and multivalents in the equatorial plane during metaphase I and, consequently, in balanced segregation (Fig. 8, Middle), although shorter chromosomes exhibit a tendency to segregate more randomly (44).
Fig. 8.
Schematic representation of main peculiarities of canonical, achiasmatic, and inverted meiosis in a sample heterozygous for chromosomal fusion/fission. Only one large chromosome (L) corresponding to two small chromosomes (S1 and S2) is shown. Homologs (L, S1, and S2) are shown with different colors. Sister chromatids are shown with the same color. The scheme applies to holokinetic chromosomes, which lack a localized centromere. Arrows schematically represent microtubules that attach to the chromosomes and pull them to opposite poles. Canonical and inverted meiosis concerns males; achiasmatic meiosis is typical for lepidopteran females. In achiasmatic meiosis, the balanced segregation is ensured by the modified synaptonemal complex that maintains homologs in a parallel arrangement until metaphase I.
Surprisingly, male hybrids were still partially fertile in our experiments. We propose that this unexpectedly high fertility found in Leptidea chromosomal hybrids is facilitated by inverted meiosis (Fig. 2), a phenomenon that has been highly debated and, until recently, had very limited support (38, 39, 41, 42). In normal canonical meiosis (known also as prereductional meiosis), the first division (reductional division) involves the segregation of chromosomal homologs resulting in the reduction of chromosome number, and the second division (equational division) involves the separation of sister chromatids. Inverted meiosis (known also as postreductional meiosis) has an opposite order of these main meiotic events (37, 45). Currently, inverted meiosis has been demonstrated for some flowering plants (32, 46, 47), insects (33, 34, 48), and mites (49). Until recently, it has been believed that inverted meiosis occurs only in organisms with holokinetic chromosomes (30), which are characterized by kinetic activity distributed along almost the entire chromosomal length (30, 32–35). However, the discovery of inverted meiosis in some chromosome pairs in human female meiosis (50) indicates that this type of meiosis is more widespread than previously thought.
The application of the concept of inverted meiosis has a limitation: If meiosis is chiasmatic, then, as a consequence of meiotic recombination, the first meiotic division can be reductional at some loci and equational at others (37–39) (see Fig. 1 for explanation and justification of the term “inverted meiosis”). Despite these limitations and criticism (37–39), inverted meiosis has become a generally accepted term in recent years (32, 34, 39, 46, 48, 49).
Interestingly, meiosis can be variable among and within holocentric species. This means that bivalents and multivalents are able to undergo either prereductional or inverted (postreductional) meiosis, depending on their orientation at metaphase I (Fig. 9) (51, 52). Lepidopterans (i.e., moths and butterflies) have holocentric chromosomes (43, 44), and their male meiosis has been reported to be prereductional, based on a few studied species (53, 54). However, male meiosis was found to be inverted in chromosomal trivalents of the silkworm, Bombyx mori (42), and the butterfly Polyommatus damonides (55), while Murakami and Imai (31) demonstrated that, in B. mori, male meiosis was opportunistic (either prereductional or postreductional) with a tendency to be inverted (postreductional) in structural chromosomal heterozygotes.
Fig. 9.
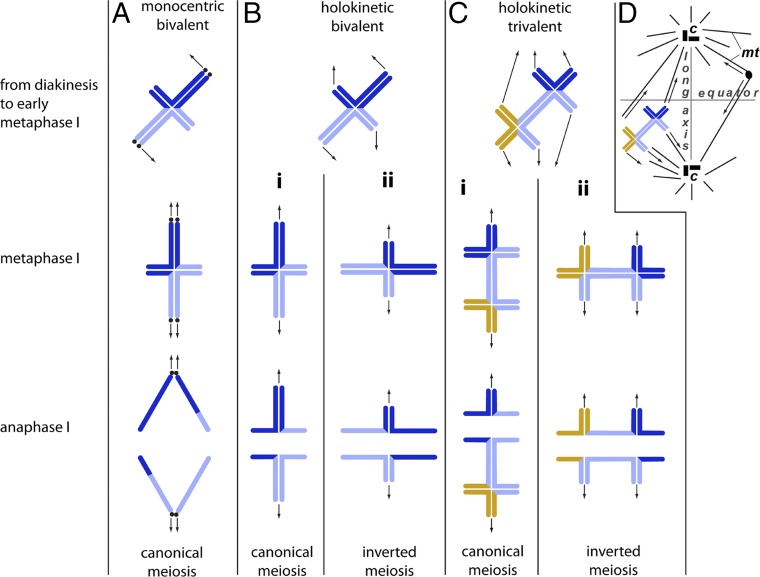
(A) Monocentric canonical meiosis and (B and C) holocentric meiosis in (B) a bivalent and (C) a trivalent. The arrows represent direction of the pulling force by microtubules. (A) In the case of monocentric chromosomes, the pulling forces by the microtubules orient the bivalent in such a way that, in metaphase I, its long axis lies in parallel to the axis of the spindle, resulting in the separation of chromosome homologs in anaphase I (i.e., in canonical meiosis). Localized centromeres are shown by black dots. (B) Holocentric meiosis in a bivalent. The pulling forces by the microtubules lead to two types of bivalent orientation in metaphase I: (i) axial orientation resulting in reductional division at the loci on the long segment and equational division at the loci on the short segment (classified as prereductional meiosis) or (ii) equatorial orientation resulting in equational division at the loci on the long segment and reductional division at the loci on the short segment (classified as postreductional inverted meiosis). (C) Holocentric meiosis in a trivalent. As in the holokinetic bivalent, the pulling forces by the microtubules may lead to two types of trivalent orientation in metaphase I, resulting in prereductional (i) or postreductional inverted (ii) meiosis. (D) Schematic representation of the spindle with the long axis (vertical line) and equator (horizontal line); mt, microtubule; c, centrosome.
The principal features of holocentric meiosis resulting in either prereductional or postreductional scenarios are explained in Fig. 9 using Lepidoptera as an example. In male meiosis of Lepidoptera (as well as in chiasmatic meiosis of other organisms with holokinetic chromosomes; see ref. 41), there is usually one subterminal or interstitial chiasma per bivalent, resulting in a cross-shaped configuration. (In rare cases, there are two subterminal chiasmata per bivalent, resulting in a ring-shaped configuration; see refs. 56–58.) Whereas, in mitosis, microtubules attach to nearly the whole surface of the chromosomes, in meiosis, the kinetic activity is usually restricted to the chromosomal ends facing the spindle poles (30, 32–35). This results in two possible orientations for each of the cross-shaped bivalents (Fig. 9B): with its long axis parallel to the spindle (“axial orientation” in ref. 40) or perpendicular to the spindle (“equatorial orientation” in ref. 40). In the former case, the first meiotic division is reductional for loci on the long segment and equational for loci on the short segment. In the latter case, it is the opposite, and, according to Loidl (37), this case can be classified as inverted meiosis.
In case of a heterozygote for chromosomal fusion/fission, there are two subterminal chiasmata per trivalent, since at least two chiasmata are required to keep the chromosomal elements together (three and more chiasmata are less likely in holokinetic organisms due to spatial restrictions; see ref. 41). Therefore, the heterozygosity for chromosomal fusion/fission results in a long chromosomal trivalent with two crosses (Fig. 8, Left, anaphase I). If this trivalent orients parallel to the spindle fibers (Fig. 9 C–I) (axial orientation), it will divide reductionally (i.e., chromosome homologs, but not sister chromatids, will segregate) and most likely unequally, resulting in aneuploid products (Fig. 8, Left and Fig. 9 C–I). However, such orientation seems unlikely in the case of a long trivalent with the kinetic activity restricted to the telomeric regions: The resultant pulling forces of the spindle fibers (see ref. 59 for explanation) will lead to equatorial-like orientation (e.g., see SI Appendix, Fig. S3 and subsequent postreductional inverted meiosis in Fig. 8, Right).
The latter has fundamental consequences for the behavior and fate of the trivalent. In canonical meiosis, chromosomal heterozygotes are expected to have segregation problems in the first meiotic anaphase, since homologous chromosome pairing is complicated by crossing over. This produces a very intricate multivalent structure and, with a high probability, results in unequal segregation of genetic material (Fig. 8, Left). In inverted meiosis, these problems are avoided because sister chromatids (but not homologs) segregate in the first anaphase, and the transmission of genetic material to metaphase II cells is thus balanced (Fig. 8, Right). The resulting metaphase II trivalents have a simpler structure compared with metaphase I trivalents, and their equal balanced segregation at anaphase II is more probable as a consequence of two described mechanisms (33, 38). First, the secondary pairing of homologs is sometimes possible and can result in the parallel orientation of multivalents in equatorial plane during metaphase II and, consequently, in balanced segregation (32, 33). Second, the metaphase II trivalents have half of their initial thickness, because every chromosome homolog consists now of only one sister chromatid, not of two as before. These thinner trivalents are more flexible and more capable of becoming U-shaped and cooriented, i.e., to orient a larger homolog toward one pole, while two smaller homologs orient toward the opposite pole (38). Such a coorientation is a prerequisite of balanced chromosome segregation (38), and was regularly observed in metaphase II in Leptidea trivalents (SI Appendix, Fig. S4). We propose that, through these mechanisms, inverted meiosis may rescue fertility in chromosomal heterozygotes to a certain degree, which provides an explanation for the fertility results we obtained and for the persistence of multiple fusions/fissions in natural populations.
However, the equatorial orientation at metaphase I with the subsequent equational division (inverted meiosis) is not an indispensable condition for proper segregation of a holocentric trivalent. For a specific sex chromosome trivalent in males of a holocentric homopteran species (Cacopsylla mali), the axial orientation at metaphase I followed by reductional division was found to be the best option for balanced segregation (38) (SI Appendix, Text 2).
The comparison between L. sinapis and C. mali indicated that sometimes axial orientation is better at meiotic metaphase I and sometimes equatorial alignment is better for subsequent trivalent segregation. Thus, there is a clear flexibility of holocentric organisms in terms of equatorial or axial orientation at metaphase I that suggests an intrinsic versatility of holocentric chromosomes in dealing with chromosomal rearrangements at meiosis. This is not an option available to organisms with monocentric chromosomes.
In Lepidoptera, some clades demonstrate an extremely high rate of chromosomal evolution (11, 60) and a tendency toward chromosomal speciation (7, 61), and researchers explained these phenomena based on the holocentric organizations of chromosomes in Lepidoptera (62). In contrast to species with monocentric chromosomes, where acentric fragments are lost during cell division, the breakage of holocentric chromosomes creates fragments with normal kinetic activity. However, we should note that kinetic activity is a necessary but insufficient condition for the survival of the chromosome fragments. To be viable, they should (i) obtain telomeres and (ii) be able to pass successfully through meiosis despite multivalent formation. The first problem is solved through the fast formation of new telomeres via a telomerase-based mechanism (62). Here we show that the second problem could be solved via the inverted order of meiotic divisions. An additional role could also be played by achiasmatic meiosis (present, for example, in females of Lepidoptera), which ensures a balanced segregation in the absence of crossing over (43). Hence, a combination of holokinetic centromere activity, fast formation of new telomeres at break points, and inverted meiosis causes a rapid karyotype evolution in species with holocentric chromosomes.
In this study, we documented a case in which hybrids between two populations of the butterfly L. sinapis, differentiated by at least 24 chromosomal fusions/fissions, retained a relatively high percentage of fertility. We also demonstrated the inverted order of meiotic events in L. sinapis males, and explained how this particular mechanism could decrease harmful effects of chromosomal rearrangements in chromosomal heterozygotes. We hypothesize that the high level of fertility in chromosomal heterozygotes made possible the gradual accumulation of chromosomal rearrangements observed in the chromosomal cline of L. sinapis (17). Thus, the studied system seems to represent a narrow time window of the very first steps of speciation driven by chromosomal change, as well as of clinal speciation—a process in which the ordered accumulation of differences along a geographic gradient results in partially reproductively isolated extremes (63), such as we found in the populations from Sweden and Spain.
Methods
Methodological aspects are described in more detail in SI Appendix, SI Methods.
Experimental Design.
Two main laboratory lines were established based on wild-caught individuals. One was representative for L. sinapis populations with high chromosome number (2n = 106, 108) (17) and included specimens originating from northeastern Spain (Montseny area, Barcelona province, Catalonia). The other laboratory line was representative for L. sinapis populations with low chromosome number (2n = 57, 58) (this study) (SI Appendix, Table S1) and included specimens originating from south central Sweden (two field sites in the vicinity of Stockholm) (Fig. 1).
Pure Spanish and Swedish laboratory lines were maintained and used as controls with respect to crossed lines between male Spanish and female Swedish and female Spanish and male Swedish L. sinapis. All possible mating combinations between Spanish and Swedish L. sinapis were performed until F2. The offspring of these pairs were bred to adults and used for further experiments (Fig. 1 and SI Appendix, Fig. S1 and Table S2). For generations F3 and F4, a subset of the potential hybrid mating combinations were performed, and the pure lines were stopped.
A number of (larval and adult) offspring from each mating combination were killed for karyological studies (SI Appendix, Table S1).
Statistical Analyses.
For a full description of statistical procedures, see SI Appendix, SI Methods. In brief, female mating propensity (yes/no) was tested in a set of generalized linear models (binomial distribution), with logit as link function. The models tested effects of male and female origin (Spain or Sweden) in the first cross, and, in later generations, compared individual cross types. The female time to mating acceptance (log transformed) was tested in a similar scheme using linear models (ANOVA II) (Fig. 3).
The survival analysis included three separate response variables: survival from egg to larva; survival from larva to adult; and the combined, total survival from egg to adult. We analyzed these survival rates, as well as the resulting sex ratio, using the survival rate/male rate (arcsine square root transformed) of each maternal family as the statistical unit. Because data on pure lines were available only for generation F1 and F2, whereas data on hybrid fitness were available also for generation F3 and F4, we first included data only from generation F1 and F2 in the models and tested the effect of cross type (pure/hybrid), generation (F1/F2), and their interaction on survival rates, sex ratio, and female fecundity (log number of eggs). Then we compared the same response variables only among the four hybrid generations (F1 to F4). Additional survival and sex ratio analyses using a generalized linear mixed-model approach (SI Appendix, SI Methods) generated qualitatively very similar results (SI Appendix, Tables S5 and S6).
Finally, we compared adult dry weights among generations (F1 to F3), sexes, and crosses (a total of 10 groups) using a nonparametric Kruskal Wallis ANOVA.
Chromosomal Analysis.
Only fresh adult males were used to analyze meiosis and to study meiotic karyotype. Testes were excised and placed into vials with freshly prepared Carnoy fixative (ethanol and glacial acetic acid, 3:1). Gonads were stored in fixative for 2 mo to 6 mo at 4 °C and then stained with 2% acetic orcein for 30 d at 20 °C. Cytogenetic analysis was conducted using a two-phase method as previously described (SI Appendix, SI Methods).
FISH with 18S rDNA Probe.
Testes of F1 hybrids were dissected and placed into freshly prepared Carnoy fixative (ethanol: and glacial acetic acid, 3:1) overnight. Then the testes were squashed on slides using the two-phase method as stated in Chromosomal Analysis, the coverslips were flicked off with a razor blade, and the slides were passed through a graded ethanol series (70%, 80%, and 100%, 1 min each). Unlabeled 18S rDNA probe was generated by PCR from Cydia pomonella (Tortricidae) genomic DNA and labeled with biotin-16-dUTP as described in SI Appendix, Fish with 18S rDNA Probe. FISH with the probe was carried out using a routine protocol.
Supplementary Material
Acknowledgments
We thank Oleg Kosterin (Institute of Cytology and Genetics, Russian Academy of Sciences) and Valentina Kuznetsova (Zoological Institute, Russian Academy of Sciences) for discussing the matter of inverted meiosis, and three anonymous reviewers for helpful comments on previous versions of the manuscript. V.A.L. was supported by Grant N 14-14-00541 from the Russian Science Foundation to the Zoological Institute of the Russian Academy of Sciences. V.D. was supported by the Wenner-Gren Foundation Sweden and by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (Project 625997). M.F. was supported by a Junior Research Grant from the Swedish Research Council. R.V. was supported by Agencia Estatal de Investigación and Fondo Europeo de Desarrollo Regional (Project CGL2016-76322-P) and by Agència de Gestió d’Ajuts Universitaris i de Recerca (Project 2014-SGR-1532). J.Š. and F.M. acknowledge support from Grants 14–22765S and 17-13713S of the Czech Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802610115/-/DCSupplemental.
References
- 1.Sumner AT. Chromosomes: Organization and Function. Wiley-Blackwell; Malden, MA: 2003. [Google Scholar]
- 2.Stone JL, et al. International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 5.Navarro A, Barton NH. Chromosomal speciation and molecular divergence–Accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- 6.Faria R, Navarro A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25:660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Kandul NP, Lukhtanov VA, Pierce NE. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution. 2007;61:546–559. doi: 10.1111/j.1558-5646.2007.00046.x. [DOI] [PubMed] [Google Scholar]
- 8.Butlin RK. Recombination and speciation. Mol Ecol. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 9.Sodeland M, et al. “Islands of divergence” in the Atlantic cod genome represent polymorphic chromosomal rearrangements. Genome Biol Evol. 2016;8:1012–1022. doi: 10.1093/gbe/evw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinjan DA, Lettice LA. Long-range gene control and genetic disease. Adv Genet. 2008;61:339–388. doi: 10.1016/S0065-2660(07)00013-2. [DOI] [PubMed] [Google Scholar]
- 11.Vershinina AO, Lukhtanov VA. Evolutionary mechanisms of runaway chromosome number change in Agrodiaetus butterflies. Sci Rep. 2017;7:8199. doi: 10.1038/s41598-017-08525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant V. Plant Speciation. Columbia Univ Press; New York: 1981. [Google Scholar]
- 13.King M. Species Evolution: The Role of Chromosomal Change. Cambridge Univ Press; Cambridge, UK: 1993. [Google Scholar]
- 14.Cortes DB, McNally KL, Mains PE, McNally FJ. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. eLife. 2015;4:e06056. doi: 10.7554/eLife.06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenormand T, Dutheil J. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castiglia R, Capanna E. Contact zone between chromosomal races of Mus musculus domesticus. 2. Fertility and segregation in laboratory-reared and wild mice heterozygous for multiple Robertsonian rearrangements. Heredity (Edinb) 2000;85:147–156. doi: 10.1046/j.1365-2540.2000.00743.x. [DOI] [PubMed] [Google Scholar]
- 17.Lukhtanov VA, Dincă V, Talavera G, Vila R. Unprecedented within-species chromosome number cline in the Wood White butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol Biol. 2011;11:109. doi: 10.1186/1471-2148-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobigny G, Britton-Davidian J, Robinson TJ. Chromosomal polymorphism in mammals: An evolutionary perspective. Biol Rev Camb Philos Soc. 2017;92:1–21. doi: 10.1111/brv.12213. [DOI] [PubMed] [Google Scholar]
- 19.Dincă V, Lukhtanov VA, Talavera G, Vila R. Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nat Commun. 2011;2:324. doi: 10.1038/ncomms1329. [DOI] [PubMed] [Google Scholar]
- 20.Šíchová J, et al. Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies. BMC Evol Biol. 2015;15:89. doi: 10.1186/s12862-015-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Šíchová J, et al. Fissions, fusions, and translocations shaped the karyotype and multiple sex chromosome constitution of the northeast-Asian wood white butterfly, Leptidea amurensis. Biol J Linn Soc Lond. 2016;118:457–471. [Google Scholar]
- 22.Réal P. Lépidoptères nouveaux principalement jurassiens. Mém Comité de Liaison Rech Ecofaunist Jura. 1988;4:1–28. [Google Scholar]
- 23.Lorković Z. Leptidea reali Reissinger, 1989 (=lorkovicii real 1988), a new European species (Lepid., Pieridae) Natura Croat. 1993;2:1–26. [Google Scholar]
- 24.Friberg M, et al. Female mate choice determines reproductive isolation between sympatric butterflies. Behav Ecol Sociobiol. 2008;62:873–886. [Google Scholar]
- 25.Martin J-F, Gilles A, Descimon H. Species concepts and sibling species: The case of Leptidea sinapis and Leptidea reali. In: C.L. Boggs, WB Watt, PR Ehrlich., editors. Butterflies: Ecology and Evolution Taking Flight. Univ Chicago Press; Chicago: 2003. pp. 459–476. [Google Scholar]
- 26.Verovnik R, Glogovčan P. Morphological and molecular evidence of a possible hybrid zone of Leptidea sinapis and L. reali (Lepidoptera: Pieridae) Eur J Entomol. 2007;104:667–674. [Google Scholar]
- 27.Dincă V, et al. Reproductive isolation and patterns of genetic differentiation in a cryptic butterfly species complex. J Evol Biol. 2013;26:2095–2106. doi: 10.1111/jeb.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorković Z. Die Chromozomenzahlen in der Spermatogenese der Tagfalter. Chromosoma. 1941;2:155–191. [Google Scholar]
- 29.Talla V, et al. Rapid increase in genome size as a consequence of transposable element hyperactivity in wood-white (Leptidea) butterflies. Genome Biol Evol. 2017;9:2491–2505. doi: 10.1093/gbe/evx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melters DP, Paliulis LV, Korf IF, Chan SWL. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- 31.Murakami A, Imai HT. Cytological evidence for holocentric chromosomes of the silkworms, Bombyx mori and B. mandarina, (Bombycidae, Lepidoptera) Chromosoma. 1974;47:167–178. doi: 10.1007/BF00331804. [DOI] [PubMed] [Google Scholar]
- 32.Heckmann S, et al. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat Commun. 2014;5:4979. doi: 10.1038/ncomms5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manicardi GC, Mandrioli M, Blackman RL. The cytogenetic architecture of the aphid genome. Biol Rev Camb Philos Soc. 2015;90:112–125. doi: 10.1111/brv.12096. [DOI] [PubMed] [Google Scholar]
- 34.Bogdanov YF. [Inverted meiosis and its place in the evolution of sexual reproduction pathways] Genetika. 2016;52:541–560. [PubMed] [Google Scholar]
- 35.Bureš P, Zedek F, Marková M. 2013. Holocentric chromosomes. Plant Genome Diversity Volume 2, Physical Structure, Behaviour and Evolution of Plant Genomes, Plant Genome Diversity, eds Leitch IJ, Greilhuber J, Doležel J, Wendel JF (Springer-Verlag, Vienna), Vol 2, pp 187–208.
- 36.Kuznetsova VG. Chromosomes of holokinetic type and their distribution among insects and other invertebrate animals. In: Scarlato OA, editor. Karyosystematics of Invertebrate Animals. Zool Inst; Leningrad: 1979. pp. 5–19. [Google Scholar]
- 37.Loidl J. Conservation and variability of meiosis across the eukaryotes. Annu Rev Genet. 2016;50:293–316. doi: 10.1146/annurev-genet-120215-035100. [DOI] [PubMed] [Google Scholar]
- 38.Nokkala S, Kuznetsova VG, Maryanska-Nadachowska A, Nokkala C. Holocentric chromosomes in meiosis. II. The modes of orientation and segregation of a trivalent. Chromosome Res. 2006;14:559–565. doi: 10.1007/s10577-006-1053-6. [DOI] [PubMed] [Google Scholar]
- 39.Viera A, Page J, Rufas JS. Inverted meiosis: The true bugs as a model to study. Genome Dyn. 2009;5:137–156. doi: 10.1159/000166639. [DOI] [PubMed] [Google Scholar]
- 40.White MJD. Animal Cytology and Evolution. Cambridge Univ Press; Cambridge, UK: 1973. [Google Scholar]
- 41.Nokkala S, Kuznetsova VG, Maryanska-Nadachowska A, Nokkala C. Holocentric chromosomes in meiosis. I. Restriction of the number of chiasmata in bivalents. Chromosome Res. 2004;12:733–739. doi: 10.1023/B:CHRO.0000045797.74375.70. [DOI] [PubMed] [Google Scholar]
- 42.Banno Y, Kawaguchi Y, Koga K, Doira H. Postreductional meiosis revealed in males of the mutant with chromosomal aberration “T(23;25)Nd” of the silkworm, Bombyx mori. J Sericol Sci Jpn. 1995;64:410–414. [Google Scholar]
- 43.Marec F. Synaptonemal complexes in insects. Int J Insect Morphol Embryol. 1996;25:205–233. [Google Scholar]
- 44.Marec F, Tothová A, Sahara K, Traut W. Meiotic pairing of sex chromosome fragments and its relation to atypical transmission of a sex-linked marker in Ephestia kuehniella (Insecta: Lepidoptera) Heredity (Edinb) 2001;87:659–671. doi: 10.1046/j.1365-2540.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 45.Lenormand T, Engelstaedter J, Johnston SE, Wijnker E, Haag CR. Evolutionary mysteries in meiosis. Phil Trans R Soc B. 2016;371:20160001. doi: 10.1098/rstb.2016.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlögelhofer P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat Commun. 2014;5:5070. doi: 10.1038/ncomms6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pazy B, Plitmann U. Unusual chromosome separation in meiosis of Cuscuta L. Genome. 1991;10:533–536. [Google Scholar]
- 48.Bongiorni S, Fiorenzo P, Pippoletti D, Prantera G. Inverted meiosis and meiotic drive in mealybugs. Chromosoma. 2004;112:331–341. doi: 10.1007/s00412-004-0278-4. [DOI] [PubMed] [Google Scholar]
- 49.Wrensch DL, Kethley J, Norton RA. Cytogenetics of holokinetic chromosomes and inverted meiosis: Keys to the evolutionary success of mites, with generalizations on eukaryotes. In: Houck MA, editor. Mites: Ecological and Evolutionary Analyses of Life-history Patterns. Springer; Dordrecht, The Netherlands: 1994. pp. 282–343. [Google Scholar]
- 50.Ottolini CS, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47:727–735. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helenius O. The mode of bivalent orientation in the Hemiptera. Hereditas. 1952;38:420–424. [Google Scholar]
- 52.Nokkala S, Nokkala C. The absence of chiasma terminalization and inverted meiosis in males and females of Myrmus miriformis Fn. (Corizidae, Heteroptera) Heredity. 1997;78:561–566. [Google Scholar]
- 53.Suomalainen E. The kinetochore and the bivalent structure in the Lepidoptera. Hereditas. 1953;39:88–96. [Google Scholar]
- 54.Marques A, Pedrosa-Harand A. Holocentromere identity: From the typical mitotic linear structure to the great plasticity of meiotic holocentromeres. Chromosoma. 2016;125:669–681. doi: 10.1007/s00412-016-0612-7. [DOI] [PubMed] [Google Scholar]
- 55.Lukhtanov VA, Dantchenko AV. A new butterfly species from south Russia revealed through chromosomal and molecular analysis of the Polyommatus (Agrodiaetus) damonides complex (Lepidoptera, Lycaenidae) Comp Cytogenet. 2017;11:769–795. doi: 10.3897/CompCytogen.v11i4.20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traut W. A study of recombination, formation of chiasmata and synaptonemal complexes in female and male meiosis of Ephestia kuehniella (Lepidoptera) Genetica. 1977;47:135–142. [Google Scholar]
- 57.Traut W, Clarke CA. Karyotype evolution by chromosome fusion in the moth genus Orgyia. Hereditas. 1997;126:77–84. [Google Scholar]
- 58.McClure M, Dutrillaux B, Dutrillaux A-M, Lukhtanov V, Elias M. Heterozygosity and chain multivalents during meiosis illustrate ongoing evolution as a result of multiple holokinetic chromosome fusions in the genus Melinaea (Lepidoptera, Nymphalidae) Cytogenet Genome Res. 2017;153:213–222. doi: 10.1159/000487107. [DOI] [PubMed] [Google Scholar]
- 59.Lukhtanov VA, Dantchenko AV. Principles of highly ordered metaphase I bivalent arrangement in spermatocytes of Agrodiaetus (Lepidoptera) Chromosome Res. 2002;10:5–20. doi: 10.1023/a:1014249607796. [DOI] [PubMed] [Google Scholar]
- 60.Lukhtanov VA. The blue butterfly Polyommatus (Plebicula) atlanticus (Lepidoptera, Lycaenidae) holds the record of the highest number of chromosomes in the non-polyploid eukaryotic organisms. Comp Cytogenet. 2015;9:683–690. doi: 10.3897/CompCytogen.v9i4.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lukhtanov VA, et al. Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature. 2005;436:385–389. doi: 10.1038/nature03704. [DOI] [PubMed] [Google Scholar]
- 62.Jankowska M, et al. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma. 2015;124:519–528. doi: 10.1007/s00412-015-0524-y. [DOI] [PubMed] [Google Scholar]
- 63.Coyne JA, Orr HA. Speciation. Sinauer Assoc; Sunderland, MA: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.