Significance
A central event during autophagy is the biogenesis of double-membrane vesicles called autophagosomes, which sequester various intracellular materials for degradation in lysosomes/vacuoles. Recent studies have suggested the involvement of the endoplasmic reticulum (ER) in autophagosome formation, and that pre-autophagosomal membranes contact with the ER. However, the mechanistic basis of these contacts has remained unknown. Here we describe two membrane-binding domains responsible for autophagosome formation in the autophagy-related protein Atg2, which localizes to the pre-autophagosomal membrane–ER contact sites in yeast cells. Our data suggest that the amphipathic helix in the C-terminal region of Atg2 targets the protein to pre-autophagosomal membranes, whereas the N-terminal region of the same molecule associates with the ER, tethering these membranes together to mediate membrane expansion during autophagosome formation.
Keywords: autophagy, Atg protein, membrane tether, yeast
Abstract
The biogenesis of double-membrane vesicles called autophagosomes, which sequester and transport intracellular material for degradation in lysosomes or vacuoles, is a central event in autophagy. This process requires a unique set of factors called autophagy-related (Atg) proteins. The Atg proteins assemble to organize the preautophagosomal structure (PAS), at which a cup-shaped membrane, the isolation membrane (or phagophore), forms and expands to become the autophagosome. The molecular mechanism of autophagosome biogenesis remains poorly understood. Previous studies have shown that Atg2 forms a complex with the phosphatidylinositol 3-phosphate (PI3P)-binding protein Atg18 and localizes to the PAS to initiate autophagosome biogenesis; however, the molecular function of Atg2 remains unknown. In this study, we show that Atg2 has two membrane-binding domains in the N- and C-terminal regions and acts as a membrane tether during autophagosome formation in the budding yeast Saccharomyces cerevisiae. An amphipathic helix in the C-terminal region binds to membranes and facilitates Atg18 binding to PI3P to target the Atg2-Atg18 complex to the PAS. The N-terminal region of Atg2 is also involved in the membrane binding of this protein but is dispensable for the PAS targeting of the Atg2-Atg18 complex. Our data suggest that this region associates with the endoplasmic reticulum (ER) and is responsible for the formation of the isolation membrane at the PAS. Based on these results, we propose that the Atg2-Atg18 complex tethers the PAS to the ER to initiate membrane expansion during autophagosome formation.
Autophagy is an intracellular degradation pathway highly conserved from yeast to humans (1). When autophagy is induced, a flattened membrane vesicle called the isolation membrane (or phagophore) appears, expands, and closes to form a double-membrane vesicle called the autophagosome. The autophagosome is delivered to, and fuses with, the lysosome or vacuole, where its contents are degraded. In the budding yeast Saccharomyces cerevisiae, more than 40 autophagy-related (Atg) proteins have been identified to date (2), of which 19 proteins are required for autophagosome formation. These 19 Atg proteins constitute six functional groups: (i) the Atg1 protein kinase complex, (ii) vesicles containing the membrane protein Atg9 (Atg9 vesicles), (iii) the Atg14-containing phosphatidylinositol 3-kinase (PI3K) complex, (iv) the Atg12-Atg5 conjugation system, (v) the Atg8-phosphatidylethanolamine conjugation system, and (vi) the Atg2-Atg18 complex. These proteins assemble in an ordered manner and organize the preautophagosomal structure (PAS) on the surface of the vacuolar membrane (3). Previous studies have suggested that Atg9 vesicles serve as starting material to form the isolation membrane at the PAS, followed by expansion of the isolation membrane using an as-yet unknown membrane source (4).
The Atg2-Atg18 complex localizes to the PAS in a manner dependent on PI3P, which is produced by the Atg14-containing PI3K complex (3, 5, 6). Atg18 binds to PI3P and PI3,5P2 via the conserved FRRG motif (7, 8). Atg18 forms another complex with Fab1, Vac7, Vac14, and Fig4 that is involved in the regulation of vacuolar morphology, but not of autophagy (5, 7, 9). Atg18 binding to PI3,5P2 is required for the localization of this protein to the vacuolar membrane (7, 10). The replacement of two arginine residues in the FRRG motif of Atg18 with threonine abolishes the binding of Atg18 to both PI3P and PI3,5P2 (7, 8). Cells expressing the Atg18FTTG mutant are defective in the PAS localization of the Atg2-Atg18 complex and exhibit reduced autophagic activity. Fusion of the FYVE domain, which specifically binds PI3P, to the C terminus of the Atg18FTTG mutant rescues these defects (5). These results suggest that the PI3P-binding ability of Atg18 is required for PAS localization of the Atg2-Atg18 complex, and thus autophagy.
In the absence of the Atg2-Atg18 complex, the isolation membrane is not formed, although other Atg proteins accumulate at the PAS (3). Previous studies have shown that the Atg2-Atg18 complex localizes to a few specific spots on the opening edge of the isolation membrane that lie in the vicinity of sites for COPII vesicle formation in the endoplsasmic reticulum (ER), or ER exit sites (11, 12). These findings led us to speculate that the Atg2-Atg18 complex functions at a contact between the PAS and the ER and serves to initiate the formation and expansion of the isolation membrane. How the Atg2-Atg18 complex is involved in this process remains to be clarified, however. To address this issue, it is necessary to reveal the molecular function of Atg2.
In this study, we identified two membrane-binding domains in the N- and C-terminal regions of Atg2 that are important for autophagosome formation. In vivo and in vitro analyses suggested that membrane binding in the C-terminal amphipathic helix facilitates Atg18 binding to PI3P, and thus targeting of the Atg2-Atg18 complex to the PAS. On the other hand, the N-terminal membrane-binding domain of Atg2 is suggested to associate with the ER to form the isolation membrane. We also showed that Atg2 has the ability to tether liposomes. Based on these results, we propose that Atg2, via the two membrane-binding domains, tethers the PAS to the ER to mediate membrane expansion during autophagosome formation.
Results
The C-Terminal Region of Atg2 Is Important for Its Localization to the PAS.
Although S. cerevisiae Atg2 consists of 1,592 amino acid residues, no functional domains can be predicted based on the primary sequence. Analysis of Atg2 sequences from different species revealed that conserved amino acid residues are enriched in both the N- and C-terminal regions (SI Appendix, Fig. S1A), suggesting that these regions play important roles in autophagosome formation. To identify a functional domain in the C-terminal region of Atg2, we generated a series of deletion mutants (SI Appendix, Fig. S1B) and analyzed their functions by three different methods. First, we monitored the maturation of the vacuolar aminopeptidase Ape1. Ape1 is synthesized in the cytoplasm as a proform (prApe1) that is delivered into the vacuole, where it is converted to the mature form (mApe1). This vacuolar transport of Ape1 is mediated by the cytoplasm-to-vacuole targeting pathway in vegetatively growing cells or by autophagy in cells treated with the autophagy-inducing reagent rapamycin. Both of these pathways depend on Atg2 (13). Therefore, the efficiency of Ape1 maturation indicates the functionality of Atg2 mutants.
Second, we examined the processing of the Pgk1-GFP fusion protein (14). Pgk1 is an abundant cytoplasmic protein. When cells are treated with rapamycin, Pgk1-GFP is transported along with other cytoplasmic components to the vacuoles via autophagy. Vacuolar cleavage of the fusion protein yields GFP fragments (GFP′), which are relatively resistant to vacuolar degradation. The accumulation of GFP′ correlates with the autophagic activity of the cell.
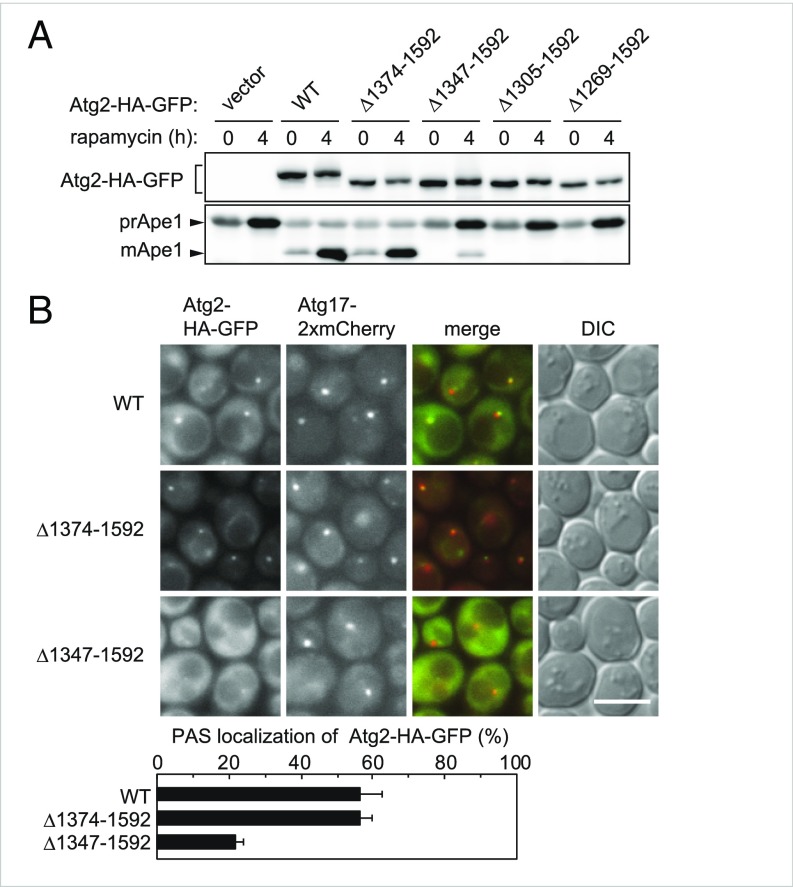
Third, we performed an alkaline phosphatase (ALP) assay (15), in which a mutant form of the vacuolar ALP Pho8 (Pho8Δ60) is expressed in the cytoplasm. When Pho8Δ60 is transported to the vacuole via autophagy, it is processed into an active form, and its enzymatic activity can be biochemically quantified. We expressed C-terminally truncated Atg2 mutants tandemly tagged with the 3×HA sequence and EGFP (Atg2-HA-GFP; hereinafter, the tag is not noted, although all the Atg2 variants expressed in yeast cells have the tag at their C termini) from centromeric plasmids in yeast cells lacking the genomic ATG2 gene (atg2Δ cells), and confirmed that similar levels of the mutant and wild-type proteins were expressed (Fig. 1A).
Fig. 1.
The C-terminal region of Atg2 is important for its PAS localization. (A) Yeast cells were grown to midlog phase, treated with rapamycin, and examined by immunoblotting using anti-HA and anti-Ape1 antibodies. mApe1, mature Ape1; prApe1, Ape1 proform. (B) Cells were treated with rapamycin for 2 h and then observed under a fluorescence microscope. DIC, differential interference contrast. (Scale bar: 5 μm.) The graph shows the PAS localization of Atg2-HA-GFP, reflected by the proportion of Atg17-2× mCherry puncta that were positive for Atg2-HA-GFP. Error bars represent SD (n = 3).
The results of all three assays led to the same conclusion: whereas Atg2 lacking residues 1374–1592 (Δ1374–1592) had autophagic activity comparable to that of the wild-type protein, further deletion of the C-terminal region (Δ1347–1592, Δ1305–1592, and Δ1269–1592) caused severe defects in autophagy (Fig. 1A and SI Appendix, Figs. S1C and S2A). Atg2 interacts with Atg18 and Atg9 (3, 16). In immunoprecipitation experiments, levels of Atg18 and Atg9 coprecipitated with Atg2 were reduced by Δ1269–1592 and Δ1305–1592, but not by Δ1347–1592 and Δ1374–1592 (SI Appendix, Fig. S1D). These results suggest that the Atg2Δ1347–1592 mutant interacts with Atg18 and Atg9 but lacks an unknown function of Atg2 that is important for autophagosome formation.
We examined the PAS localization of Atg2Δ1347–1592 by fluorescence microscopy, using ATG8-knockout cells to clearly observe the PAS localization of Atg2 (Fig. 1B) (3). Atg2Δ1374–1592 colocalized with Atg17, which served as a marker for the PAS, with a frequency similar to that of the wild-type protein. In contrast, Atg2Δ1347–1592 exhibited defective localization to the PAS. These results suggest that region 1347–1373 of Atg2 is involved in PAS targeting of the protein.
The C-Terminal Region of Atg2 Contains an Amphipathic Helix Required for Targeting to the PAS.
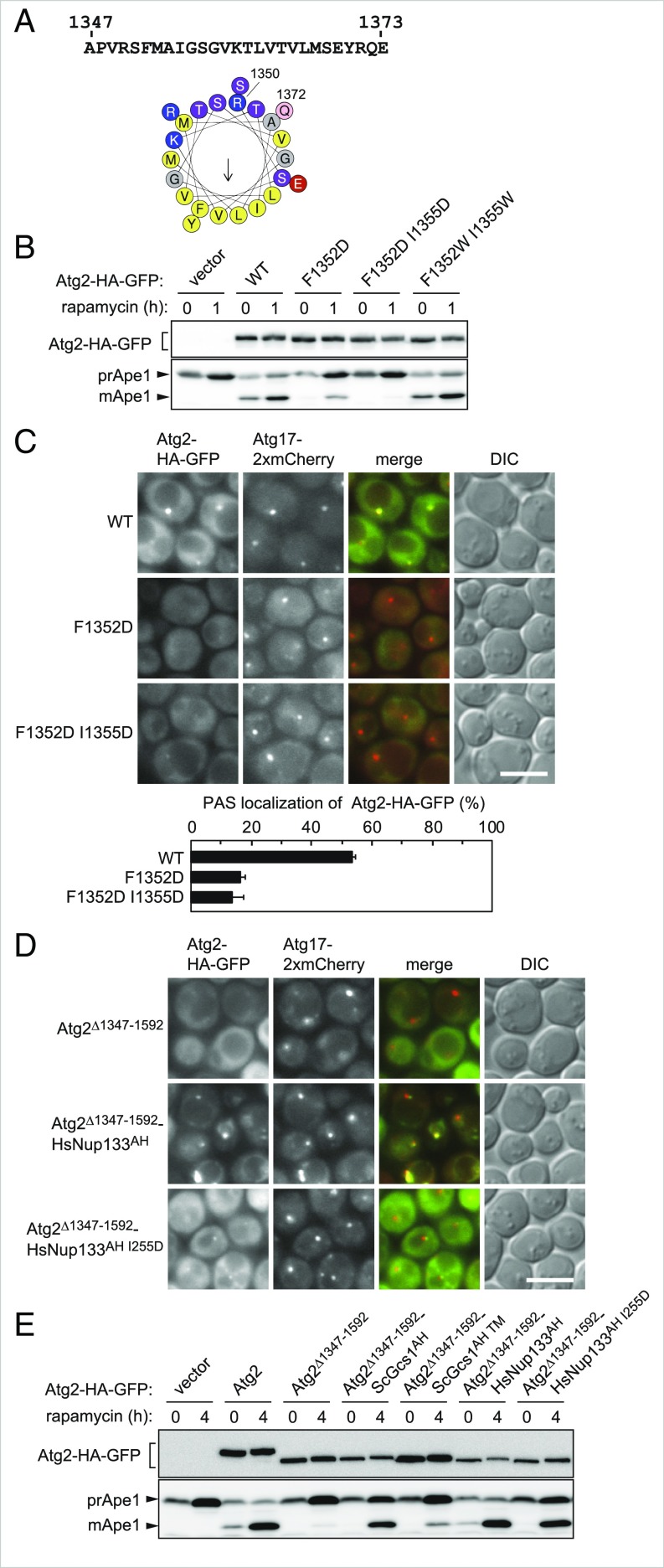
The PSIPRED method (17) predicted that amino acid residues 1350–1372 of Atg2 would adopt an α-helical configuration. A helical wheel representation of these residues showed that the two opposing surfaces of the helix are separately composed of polar and hydrophobic residues (Fig. 2A). This type of α-helix, called an amphipathic helix, can bind membranes with its hydrophobic surface buried in the hydrophobic membrane interior. To investigate the significance of this amphipathic helix in the C-terminal region of Atg2, we introduced mutations in the hydrophobic face of the helix. Although the replacement of F1352 and I1355 with another hydrophobic residue (tryptophan) did not affect the function of Atg2, as assessed by Ape1 maturation and the ALP assay, their replacement with a charged residue (aspartic acid), which could affect membrane-binding properties, caused severe defects in autophagy (Fig. 2B and SI Appendix, Fig. S2B). As with Atg2Δ1347–1592, Atg2F1352D and Atg2F1352D I1355D interacted with Atg18 and Atg9, but hardly localized to the PAS (Fig. 2C and SI Appendix, Fig. S3A).
Fig. 2.
The C-terminal region of Atg2 contains an amphipathic helix required for PAS targeting of the protein. (A) The amino acid sequence of residues 1347–1737 of Atg2 and a helical wheel depiction of residues 1350–1372, generated using Heliquest (heliquest.ipmc.cnrs.fr/). (B) The maturation of Ape1 in the Atg2 mutants for F1352 and I1355 was examined as described in Fig. 1A. (C and D) PAS localization of the Atg2-HA-GFP mutants was examined as described in Fig. 1B. (Scale bar: 5 μm.) Error bars represent SD (n = 3). (E) Ape1 maturation in the Atg2 mutants was examined as described in Fig. 1A.
To validate that region 1347–1373 acts as a membrane-binding amphipathic helix, we investigated whether a well-characterized amphipathic helix from an unrelated protein could rescue the autophagic defect in Atg2Δ1347–1592. To this end, we fused a well-characterized amphipathic helix of S. cerevisiae Gcs1 (ScGcs1235-275 or ScGcs1AH) or Homo sapiens Nup133 (HsNup133245-267 or HsNup133AH) (18, 19) to the C terminus of Atg2Δ1347–1592. In contrast to Atg2Δ1347–1592, both Atg2Δ1347–1592-ScGcs1AH and Atg2Δ1347–1592-HsNup133AH localized to the PAS (Fig. 2D and SI Appendix, Fig. S3B). In the Atg2Δ1347–1592-ScGcs1AH mutant, a significant level of Ape1 maturation was observed, but Pgk1-GFP processing and ALP activity were not detected (Fig. 2E and SI Appendix, Figs. S2C and S3C). Fusion of HsNup133AH resulted in a stronger effect: Ape1 maturation proceeded normally, and Pgk1-GFP processing and ALP activity were markedly recovered (Fig. 2E and SI Appendix, Figs. S2C and S3C). Mutations in these amphipathic helices that show decreased affinity to membranes—the triple mutations L246A, W250A, and F253A in ScGcs1AH and I255D in HsNup133AH) (18, 19)—abolished the rescue of PAS localization and autophagic activities by these helices (Fig. 2 D and E and SI Appendix, Figs. S2C and S3 B and C). These results suggest that the amphipathic helix in the C-terminal region of Atg2 functions in PAS targeting via its membrane-binding activity.
The N-Terminal Region of Atg2 Is also Responsible for Autophagosome Formation.
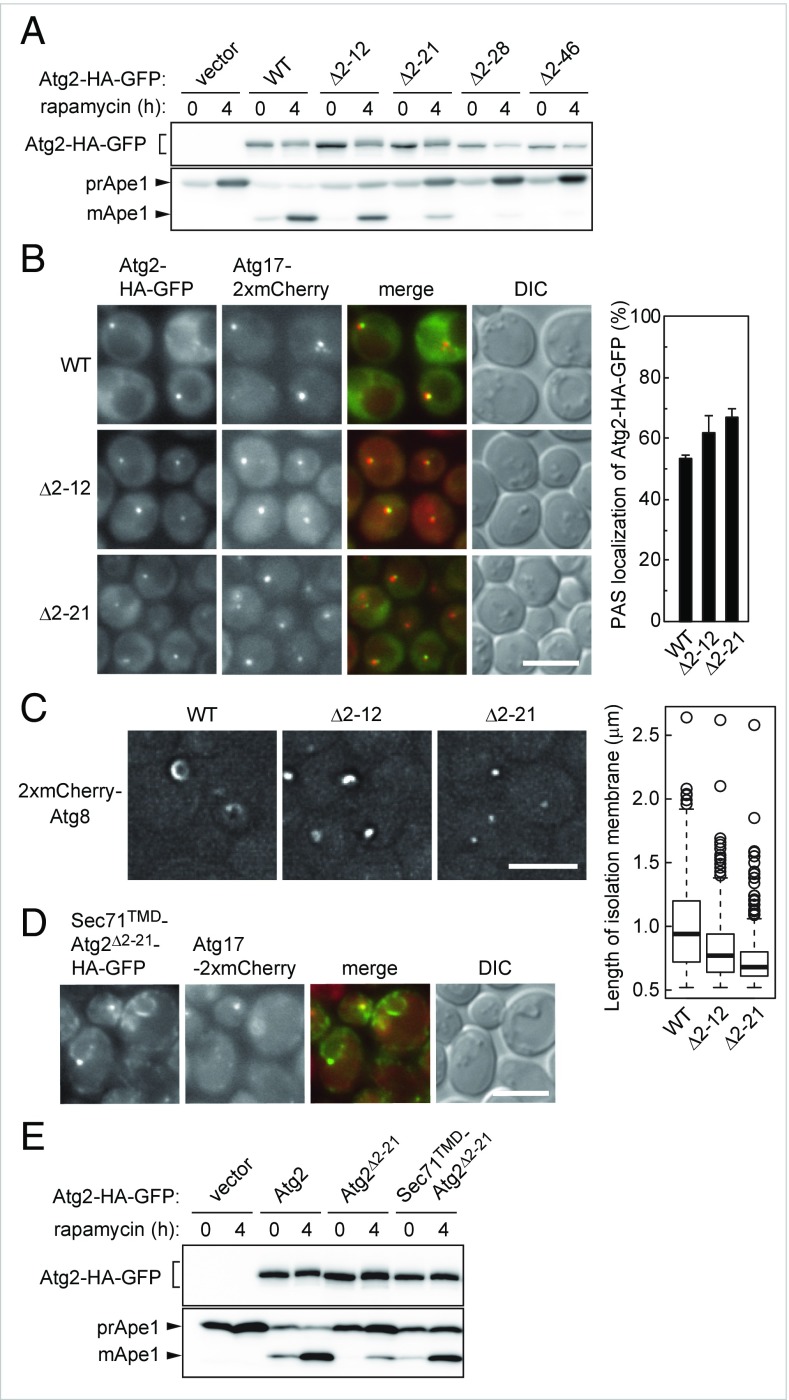
As we did for analysis of the C-terminal region, we constructed a series of N-terminally truncated mutants of Atg2. We found that deletion of the N-terminal 12 residues (Atg2Δ2–12) caused a strong defect in autophagy, which was exacerbated by further deletion of the N-terminal region (Fig. 3A and SI Appendix, Figs. S2D and S4A). Atg2Δ2–12 bound to Atg18 and Atg9 as well as the wild-type protein (SI Appendix, Fig. S4B). As more of the N-terminal region was truncated, the interactions of Atg2 with Atg18 and Atg9 were gradually diminished and were nearly completely abolished in the Atg2Δ2–46 mutant. Unlike the C-terminal mutants, Atg2Δ2–12 and Atg2Δ2–21 localized to the PAS as efficiently as wild-type Atg2 (Fig. 3B). These results suggest that the N-terminal region of Atg2 plays a crucial role in autophagosome formation exerted after its localization to the PAS. We examined the expansion of the isolation membrane in the N-terminal deletion mutants of Atg2. When Ape1 is overexpressed, it forms a giant assembly, and the isolation membrane expanding along the surface of this assembly can be observed by fluorescence microscopy under autophagy-inducing conditions as a curved structure decorated with mCherry-Atg8 (12) (Fig. 3C). We found that cells expressing Atg2Δ2–12 had a severe defect in the expansion of the isolation membrane, and that the isolation membrane barely expanded in the Atg2Δ2–21 mutant.
Fig. 3.
The N-terminal region of Atg2 is also responsible for autophagosome formation. (A) Ape1 maturation in the N-terminal deletion mutants of Atg2 were analyzed as described in Fig. 1A. (B) PAS localization of the Atg2-HA-GFP mutants was examined as described in Fig. 1B. (Scale bar: 5 μm.) Error bars represent SD (n = 3). (C) Cells overexpressing Ape1 were incubated in nitrogen-starvation medium for 1 h and observed under a fluorescence microscope. (Scale bar: 5 μm.) The graph shows the lengths of isolation membranes, measured as described in Materials and Methods. (D and E) Cells were treated with rapamycin for 2 h (D) or 4 h (E) and observed under a fluorescence microscope (D) or examined for Ape1 maturation (E). (Scale bar: 5 μm.)
Implications for the Association of the Atg2 N-Terminal Region with the ER.
The N-terminal region of Atg2 contains a number of conserved residues, including Q10-K11-R12 (SI Appendix, Fig. S4C). The Atg210-12D mutant, in which these residues were replaced with aspartic acid, was severely defective in autophagy, although it interacted with Atg18 and Atg9 and localized normally to the PAS (SI Appendix, Figs. S2E and S4 D–F). Thus, as in the case of the N-terminal truncation, the 10–12D mutation is likely to impair the function of Atg2 performed after its PAS targeting.
Wild-type Atg2 localizes to the PAS and the cytoplasm (SI Appendix, Fig. S4F). We found that when the N-terminal 46 residues of Atg2 (Atg21-46) were fused to the N terminus of GFP, the fusion protein localized to the ER (SI Appendix, Fig. S4G). The 10–12D mutation, which abolished autophagy when it was introduced into full-length Atg2, dispersed Atg21-46 into the cytoplasm (SI Appendix, Fig. S4G). These results suggest that the function of the N-terminal region of Atg2 is associated with the ER.
To further investigate the relationship between the N-terminal region of Atg2 and the ER, we fused the ER-targeting sequence (transmembrane domain) of Sec71 (Sec71TMD) to the N terminus of the N-terminally truncated, autophagy-defective mutant Atg2Δ2–21. Fluorescence microscopy confirmed that the fusion protein Sec71TMD-Atg2Δ2–21 localized to the ER in addition to the PAS (Fig. 3D). Consistent with this, centrifugation of cell lysates sedimented almost all Sec71TMD-Atg2Δ2–21, as well as the ER membrane protein Dpm1 (SI Appendix, Fig. S4H). Remarkably, fusion of Sec71TMD partially but significantly recovered autophagic activity abrogated by deletion of the N-terminal region of Atg2 (Fig. 4E and SI Appendix, Figs. S2F and S4I). These results suggest that the function of the Atg2 N-terminal region is related to its association with the ER.
Fig. 4.
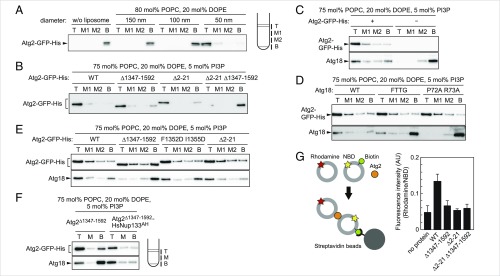
Membrane-binding and tethering functions of Atg2 and the Atg2-Atg18 complex. (A) Flotation assay with purified Atg2-GFP-His and various-sized liposomes of the indicated lipid composition were performed as described in Materials and Methods. The top (T), middle (M1 and M2), and bottom (B) fractions were collected and analyzed by immunoblotting using antibodies against Atg2 and Atg18. (B) Flotation assay using the Atg2-GFP-His mutants and liposomes with PI3P. (C) Flotation assay with Atg18 and PI3P-containing liposomes in the presence or absence of Atg2-GFP-His. (D) Flotation assay with Atg2-GFP-His, the Atg18 mutants, and PI3P liposomes. (E) Flotation assay with Atg2-GFP-His mutants complexed with Atg18 and PI3P liposomes. (F) Flotation assay with Atg2Δ1347–1592-HsNup133AH complexed with Atg18 and PI3P liposomes. After flotation, the three fractions as depicted (T, M, and B) were collected for immunoblotting analysis. (G) Liposome tethering assay. NBD liposomes containing biotinylated PE, Rho liposomes, and Atg2-GFP-His were mixed and incubated, and then NBD liposomes were collected using streptavidin beads. The graph shows rhodamine fluorescence in the collected fractions, which have been corrected with NBD fluorescence in the same fractions. Error bars represent SD (n = 3).
In Vitro Analysis of the Membrane Binding and Tethering Functions of Atg2 and the Atg2-Atg18 Complex.
Our results suggest that both the N- and C-terminal regions of Atg2 are involved in Atg2 binding to membranes. To directly test this possibility, we performed an in vitro membrane flotation assay using an OptiPrep step gradient, Atg2 purified from yeast cells (SI Appendix, Fig. S5A), and liposomes of various sizes (Fig. 4A). We showed that Atg2 bound to only the smallest liposomes (∼50 nm in diameter), suggesting that Atg2 has an affinity for membranes with high curvature. We also showed that the C- and N-terminal deletion mutants Atg2Δ1347–1592 and Atg2Δ2–21 could bind to liposomes, whereas Atg2 lacking both the N- and C-terminal regions (Atg2Δ2–21 Δ1347–1592) hardly bound (Fig. 4B). These results suggest that the N- and C-terminal regions contribute to the membrane binding of Atg2 independent of each other.
Autophagosome formation requires PI3P, which is produced by the autophagy-specific PI3K complex. Previous studies reported that Atg18 binds to PI3P in vitro, but this conclusion remains controversial (6–8, 20–22). In our flotation assay, Atg18 alone did not bind to PI3P-containing liposomes (Fig. 4C), although Atg18 did bind to liposomes containing PI3,5P2 (SI Appendix, Fig. S5B). However, Atg18 interacted with PI3P-containing liposomes in the presence of Atg2 (Fig. 4C). When a flotation assay was performed using the Atg2-Atg18 complex and liposomes without PI3P, Atg2 floated with the liposomes, but Atg18 remained in the bottom fraction (SI Appendix, Fig. S5C). This is probably because the interaction between these proteins is insufficiently stable to keep Atg18 bound to Atg2 during liposome flotation. The Atg18FTTG and Atg18P72A R73A mutants, which are unable to interact with phosphoinositides and Atg2, respectively (SI Appendix, Fig. S5 B and D), could not bind to PI3P-containing liposomes even in the presence of Atg2 (Fig. 4D). These results suggest that Atg18 binds to membranes depending on its interactions with Atg2 and PI3P.
We next investigated the membrane binding ability of Atg18 in complex with the N- or C-terminal mutants of Atg2 in the presence of PI3P. Atg18 in complex with the N-terminal deletion mutant Atg2Δ2–21 was able to bind to PI3P liposomes (Fig. 4E). In contrast, the C-terminal deletion mutant Atg2Δ1347–1592 did not allow Atg18 to associate with PI3P liposomes. The C-terminal amphipathic helix mutant Atg2F1352D I1355D was also defective in binding Atg18 to PI3P liposomes (Fig. 4E). Given that fusion of the unrelated amphipathic helix HsNup133AH could rescue autophagic defects in Atg2Δ1347–1592 (Fig. 2 D and E and SI Appendix, Fig. S2C), we investigated whether this amphipathic helix rescued Atg18 binding to PI3P liposomes. Indeed, Atg2Δ1347–1592-HsNup133AH allowed Atg18 to bind PI3P liposomes (Fig. 4F). These results suggest that the membrane association of the C-terminal region of Atg2 via the amphipathic helix is important for Atg18 binding to PI3P-containing membranes.
Finally, we performed a liposome tethering assay, in which two different liposomes were used, one containing NBD-labeled PE and biotinylated PE (NBD liposomes) and the other containing rhodamine-labeled PE (Rho liposomes). These two liposomes were mixed with Atg2, and then NBD liposomes were pulled down using streptavidin-coated beads. Tethering between NBD and Rho liposomes was evaluated by measuring the fluorescence of rhodamine in the collected fractions (Fig. 4G). In this assay, Atg2 substantially increased rhodamine fluorescence, suggesting that Atg2 has an ability to tether two membranes. Both the N- and C-terminal regions of Atg2 were important for this liposome-tethering activity (Fig. 4G). These results are consistent with the idea that the N- and C-terminal regions of Atg2 bind to different membranes and thereby tether these membranes together. We also performed this assay in the presence of Atg18 and obtained similar results (SI Appendix, Fig. S5E).
Discussion
Previous studies have shown that formation of the Atg2-Atg18 complex and production of PI3P at the PAS are prerequisites for the PAS localization of these proteins (3, 23). In this study, we identified two membrane-binding domains in Atg2, both of which are indispensable for autophagosome biogenesis. The amphipathic helix in the C-terminal region is important not only for the membrane binding of Atg2 itself, but also for Atg18 binding to PI3P-containing membranes. Consistent with these in vitro results, the amphipathic helix was required for PAS targeting of the Atg2-Atg18 complex. This finding provides a clear answer to the long-standing question of why Atg18 requires Atg2 to localize to the PAS even though Atg18 alone can bind to PI3P in vitro (3, 6–8, 22). We speculate that membrane association with the amphipathic helix of Atg2 brings Atg18 in close proximity to the membrane, facilitating its binding to PI3P. The membrane binding in the Atg2 amphipathic helix, and resultant increase in PI3P binding by Atg18, are likely to enable the PAS localization of the Atg2-Atg18 complex.
Atg9 vesicles are the membranes most likely to be bound by the Atg2-Atg18 complex at the PAS. Our in vitro assay revealed that Atg2 preferentially binds to small vesicles ∼50 nm in diameter, consistent with the size of Atg9 vesicles, reported to be 30–60 nm (4). In addition, during expansion of the isolation membrane, the Atg2-Atg18 complex localizes to the highly curved opening edge of the isolation membrane (11, 12). A previous immunoelectron microscopy study demonstrated that the isolation membrane contains PI3P (24). Based on the observation that the Atg14-containing PI3K complex is recruited to the PAS after Atg9 vesicles, PI3P also seems to be produced on these Atg9 vesicles. Moreover, the Atg2-Atg18 complex interacts directly with Atg9 (3, 16). These results suggest that the amphipathic helices of Atg2 and Atg18 act cooperatively to target the Atg2-Atg18 complex to Atg9 vesicles at the PAS, and once membrane expansion is initiated, they subsequently serve to limit localization of the complex to the opening edge of the isolation membrane. Preferential binding of Atg2 to highly curved membranes can also explain why the Atg2-Atg18 complex does not localize to the vacuolar membrane, which also contains PI3P but is relatively planar.
During the course of this study, a mammalian homolog of Atg2 was reported to have an amphipathic helix required for its localization to the isolation membrane (25). This helix likely corresponds to the amphipathic helix of yeast Atg2 identified in this study (SI Appendix, Fig. S6). In addition, mammalian Atg2 can also bind liposomes, although the regions responsible for its membrane binding remain unknown (26, 27). These observations suggest that similar mechanisms are responsible for targeting Atg2 to autophagy-related membranes in yeast and mammals.
We found that the N-terminal region of Atg2 is also important for autophagosome biogenesis. In contrast to the amphipathic helix in the C-terminal region, this region is not required for PAS targeting of the Atg2-Atg18 complex but is responsible for a subsequent step: expansion of the isolation membrane. We found that a GFP fusion of this Atg2 region localized to the ER, and that the autophagy-impairing mutation abolished this ER localization. In addition, artificial ER anchoring of the N terminus of Atg2 significantly recovered autophagic defects caused by deletion of the N-terminal region. These results suggest that the N-terminal region of Atg2 associates with the ER during its function in expansion of the isolation membrane. Previous studies demonstrated that autophagosome formation occurs in association with the ER in both mammalian and yeast cells (11, 12, 28, 29). In yeast, the Atg2-Atg18 complex forms a few puncta on the opening edge of the isolation membrane, in the vicinity of ER exit sites where COPII vesicles form (11, 12). In mammalian cells, autophagosome formation is mediated by ER subdomains, such as omegasomes and isolation membrane-associated tubules (29). We speculate that the N-terminal region of Atg2 binds to these highly curved ER-related membranes.
Based on these observations and the data obtained in this study, we propose the following model for the initiation of membrane expansion in autophagosome formation. While the amphipathic helices in the C-terminal region of Atg2 and Atg18 cooperate to bind Atg9 vesicles at the PAS, the N-terminal region of Atg2 associates with the ER. These interactions result in tethering of the PAS to the ER, leading to initiation of expansion of the isolation membrane. The Atg2-Atg18 complex may also be involved in the association between the isolation membrane edge and the ER, which continuously mediates membrane expansion. This model is supported by our in vitro results showing that Atg2 can tether two membranes (Fig. 4G); in addition, a recent study also reported that the mammalian Atg2A-WIPI4 complex has a similar function (27). It is also noteworthy that both the N- and C-terminal regions of Atg2 are homologous to Vps13, which is involved in the organization of different organelle contact sites (30, 31), consistent with the idea that Atg2 functions as a membrane tether.
The results of this study elucidate the molecular function of Atg2 and provide significant insights into the mechanism of autophagosome biogenesis. Nonetheless, some critical issues remain to be addressed. First, how does the N-terminal region of Atg2 associate with the ER, and how is this association regulated? This regulation may involve a conformational change in Atg2, which is stimulated by binding to the Atg9 vesicle or protein phosphorylation at the PAS. Another key question is how the ER participates in the expansion of the isolation membrane. One plausible possibility is that COPII vesicles fuse with the isolation membrane to expand it. If this is the case, then tethering mediated by the Atg2-Atg18 complex would spatially facilitate targeting and fusion of COPII vesicles to the PAS and isolation membrane. Further studies will clarify these issues and reveal the molecular mechanisms underlying membrane expansion during autophagosome formation.
Materials and Methods
Yeast Strains, Plasmids, and Media.
The yeast strains and plasmids used in this study are constructed as described in SI Appendix and are listed in SI Appendix, Tables S1 and S2, respectively. Media for yeast culture and for autophagy induction are also described in SI Appendix.
Immunoblotting and Immunoprecipitation.
Methods for immunoblotting and immunoprecipitation, as well as the antibodies used in these experiments, are described in SI Appendix.
Fluorescence Microscopy.
Methods for fluorescence microscopy to observe the PAS localization of Atg2-HA-GFP and the isolation membrane are described in SI Appendix.
Liposome Flotation Assay and Liposome Tethering Assay.
Proteins and liposomes used in these experiments were prepared as described in SI Appendix. Methods for liposome flotation assay and liposome tethering assay are also described in SI Appendix.
Supplementary Material
Acknowledgments
We thank the members of the H.N. and Y.O. laboratories for materials, discussions, and technical and secretarial support; the Biomaterial Analysis Center, Technical Department at Tokyo Institute of Technology for the DNA sequencing service; and Dr. Kazuhisa Nakayama for the plasmid. This work was supported in part by KAKENHI Grants-in-Aid for Scientific Research 25111003 (to H.N.), 25711005 (to H.N.), 17H01430 (to H.N.), and 23000015 (to Y.O.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Japan Science and Technology Agency CREST Grant JPMJCR13M7 (to H.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806727115/-/DCSupplemental.
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieter E, et al. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- 7.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krick R, Tolstrup J, Appelles A, Henke S, Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–4638. doi: 10.1016/j.febslet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 9.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 13.Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- 14.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6:794–797. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 15.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 16.Wang CW, et al. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 18.Zendeh-boodi Z, Yamamoto T, Sakane H, Tanaka K. Identification of a second amphipathic lipid-packing sensor-like motif that contributes to Gcs1p function in the early endosome-to-TGN pathway. J Biochem. 2013;153:573–587. doi: 10.1093/jb/mvt025. [DOI] [PubMed] [Google Scholar]
- 19.Drin G, et al. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 20.Strømhaug PE, Reggiori F, Guan J, Wang C-W, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han B-K, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopaldass N, Fauvet B, Lashuel H, Roux A, Mayer A. Membrane scission driven by the PROPPIN Atg18. EMBO J. 2017;36:3274–3291. doi: 10.15252/embj.201796859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe Y, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 25.Tamura N, et al. Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 2017;591:3819–3830. doi: 10.1002/1873-3468.12901. [DOI] [PubMed] [Google Scholar]
- 26.Zheng JX, et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy. 2017;13:1870–1883. doi: 10.1080/15548627.2017.1359381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowdhury S, et al. 2017. Structural analyses reveal that the ATG2A-WIPI4 complex functions as a membrane tether for autophagosome biogenesis. bioRxiv 180315; doi: https://doi.org/10.1101/180315.
- 28.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Wandelmer J, Ktistakis NT, Reggiori F. ERES: Sites for autophagosome biogenesis and maturation? J Cell Sci. 2015;128:185–192. doi: 10.1242/jcs.158758. [DOI] [PubMed] [Google Scholar]
- 30.Lang AB, John Peter ATAT, Walter P, Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol. 2015;210:883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J-S, et al. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol Biol Cell. 2016;27:2435–2449. doi: 10.1091/mbc.E16-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.