Significance
The human genome is “littered” with remnants of ancient retrovirus infections that invaded the germ line of our ancestors. Only one of these may still be proliferating, named HERV-K HML-2 (HK2). Not all humans have the same HK2 viruses in their genomes. Here we show that one specific uncommon HK2, which lies close to a gene involved in dopaminergic activity in the brain, is more frequently found in drug addicts and thus is significantly associated with addiction. We experimentally show that HK2 can manipulate nearby genes. Our study provides strong evidence that uncommon HK2 can be responsible for unappreciated pathogenic burden, and thus underlines the health importance of exploring the phenotypic roles of young, insertionally polymorphic HK2 integrations in human populations.
Keywords: HERV-K HML-2, endogenous retrovirus, RASGRF2, persons who inject drugs, addiction
Abstract
HERV-K HML-2 (HK2) has been proliferating in the germ line of humans at least as recently as 250,000 years ago, with some integrations that remain polymorphic in the modern human population. One of the solitary HK2 LTR polymorphic integrations lies between exons 17 and 18 of RASGRF2, a gene that affects dopaminergic activity and is thus related to addiction. Here we show that this antisense HK2 integration (namely RASGRF2-int) is found more frequently in persons who inject drugs compared with the general population. In a Greek HIV-1–positive population (n = 202), we found RASGRF2-int 2.5 times (14 versus 6%) more frequently in patients infected through i.v. drug use compared with other transmission route controls (P = 0.03). Independently, in a United Kingdom-based hepatitis C virus-positive population (n = 184), we found RASGRF2-int 3.6 times (34 versus 9.5%) more frequently in patients infected during chronic drug abuse compared with controls (P < 0.001). We then tested whether RASGRF2-int could be mechanistically responsible for this association by modulating transcription of RASGRF2. We show that the CRISPR/Cas9-mediated insertion of HK2 in HEK293 cells in the exact RASGRF2 intronic position found in the population resulted in significant transcriptional and phenotypic changes. We also explored mechanistic features of other intronic HK2 integrations and show that HK2 LTRs can be responsible for generation of cis-natural antisense transcripts, which could interfere with the transcription of nearby genes. Our findings suggest that RASGRF2-int is a strong candidate for dopaminergic manipulation, and emphasize the importance of accurate mapping of neglected HERV polymorphisms in human genomic studies.
The human genome is littered with retroviral elements as a result of ancient retroviral infections in the germ line of our primate ancestors. Some of these retroviral invasions proliferated successfully by continuously reintegrating in their host genomes (1). Proliferation success of endogenous retroviruses (ERVs), measured by the relative abundance within different hosts, depends on a complex interplay of factors, including viral life cycle (2) and host life history (3). Furthermore, hosts have multiple diverse control mechanisms that inactivate ERVs in place. Some of these mechanisms that reduce potentially deleterious retroviral activity include the accumulation of inactivating mutations, internal recombination events between their long terminal repeats (LTRs), host-derived restriction factors, and transcriptional silencing (4).
While pathogenic ERVs have been described throughout the animal kingdom, none have been definitively linked with harmful effects in humans. Furthermore, ERV germ-line proliferation in humans and other great apes is lower compared with other primates (5), with the majority of human endogenous retroviruses (HERVs) being defective and fixed within the population. Perhaps retroviral proliferation poses a disproportionate burden for humans compared with other mammals. Although most HERVs ceased proliferating millions of years ago, HERV-K (HML-2) (referred to in the text as HK2) continued proliferating in the germ line of our ancestors after the human–chimpanzee divergence ∼5 to 6 Mya (1, 6), with some integrations still being polymorphic in the population (7, 8). As a consequence, some HK2 proviruses are exceptionally well preserved, express viral proteins, and produce viral particles (9). HK2 integrations tend to be found near genes, with an antisense bias (10), suggesting that antisense integrations are less likely to impose a pathogenic burden (11, 12). Some evidence suggests that intronic HK2 integrations modulate transcription of nearby genes (13), but their phenotypic role remains largely unknown. Intronic HK2, as well as other intronic transposable elements, have been proposed to have contributed to host evolution by modulating transcription of the surrounding genes (14, 15). Moreover, some evidence suggests that intronic integrations of transposable elements could reshape the transcriptome upon reactivation during cancer development (16), although there is no evidence that HK2 reactivation could result in gene modulation in cancer development (17).
One of the polymorphic HK2 integrations is found between exons 17 and 18 of RASGRF2 (hereafter RASGRF2-int), a gene involved in signaling pathways. RASGRF2 has at least two domains and is expressed in T cells, the heart, and the brain. The deregulation of the gene could thus be involved in complex phenotypes (or disease syndromes) involving more than one system (the brain and immune system). Double knockout mice mutants of Rasgrf2tm1Esn displayed no significant pathological phenotype with respect to growth and development (18), but experiments into its role in T-cell signaling responses showed diminished immune responses (19); the importance of the deficient immune response has not been explored in humans.
Stronger evidence exists for the brain-related phenotype of RASGRF2 in humans; a genome-wide association study on alcohol addiction provided a potential hit for the SNP rs26907 that is located between exons 3 and 4 (20). This SNP was subsequently shown to modulate addictive behavior (21) due to modifications of noradrenergic and serotonergic responses (22). Young carriers of rs26907 were more likely to have alcohol-induced reinforcement and show enhanced reward-related dopaminergic activity in functional MRI experiments (20). Crucially, the rodent model of addiction is simple, reproducible, and predictive of addiction in humans (23), with double knockout RASGRF2(−/−) mice being remarkably resistant to addiction (21).
Here we show that RASGRF2-int is significantly associated with drug addiction in two independent, genetically distinct human populations. We also provide evidence in support of a mechanistic interaction between HK2 integration and RASGRF2 transcription, suggesting that the observed association is likely to be causal.
Results
RASGRF2-Int Is Associated with Drug Addiction.
The most well-established phenotype of RASGRF2 is linked with addiction. RASGRF2 is expressed most intensively in the brain, but also in the heart, lungs, and T cells (24, 25). Thus, we initially hypothesized that if the proviral HK2 modulated RASGRF2 in humans, it would be found at higher frequency among individuals with well-defined strong addictive behavior such as persons who inject drugs (PWIDs). We tested this by using PCR to screen the genomic DNA of 202 fully anonymized Greece-based HIV-positive individuals with blinded samples for the presence or absence of this integration (Fig. 1 A–D). The cohort consisted of 102 PWIDs (reported i.v. drug use within the past 6 mo as the likely transmission route) and 100 controls (infected by other transmission routes). In this population, we did not test for potential confounding variables including sex and age among groups. The power of our approach is 80% for recovering fourfold higher frequency in the PWIDs versus the expected 4% (7) in the general population. We found that the integration was present in 14 PWIDs vs. 6 non-PWIDs (P = 0.03, χ2, one-sided test) in the form of a solo-LTR, suggesting a more than twofold higher frequency in populations with long-term addictive behavior. Individuals who carried the integration were heterozygotic as determined by another PCR, specific for the preintegration site, which also served as an internal control to confirm the absence of the integration in other individuals.
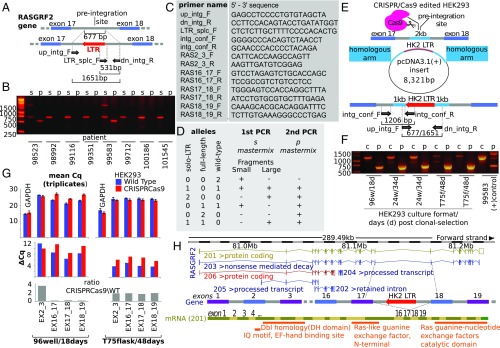
Fig. 1.
(A) LTR integration screening—PCR design. Primer mapping and product sizes relative to the wild type (Top) and the HK2-LTR (red) integrated allele (Bottom). Exonic and intronic regions are in blue and gray, respectively. (B) Integration screening results of eight (p1 to p8) random patients: Primers LTR_splc_F and dn_intg_R were used in “p,” while primers up_intg_F and dn_intg_R were used in “s” mastermix. Patients 98523 and 99583 are positive (heterozygous) for the integration. (C) Primer sequences used for the LTR integration screening and the confirmation of the editing in HEK293 cells and for the expression assessment of the RASGRF2 exonic junctions. (D) Tabular index for the genotypic interpretation of the PCR products in B. (E) CRISPR/Cas9 editing of the HEK293 cell line to incorporate RASGRF2-int. pcDNA3.1(+) plasmid containing the LTR sequence flanked by 1-kb preintegration site homologous arms (light blue) was used for the homology directed repair (HDR) insertion mechanism. Primers intg_conf_F (mastermix “c”), which avoid the false-positive amplification of the HDR plasmid, and ingr_conf_R (mapping the 5′ LTR splice site) were used in conjunction with primers up_intg_F and dn_intg_R to confirm integration/genotyping of the cell line. (F) Post CRISPR/Cas9 clonal selection and screening/genotyping for the RASGRF2-int HEK293 cells. The gel is annotated according to the scale of the cells in culture over the days posttransfection: 96w/18d (96-well plate, 18 d), 24w/34d (24-well plate, 34 d), and T75f/48d (T75 flask, 48 d). The aneuploidy of the cell line selects for the wild-type alleles versus the edited ones. (G) Differential expression of exons (triplicates of the same clone) upon editing of HEK293 cells: SYBR Green qPCR was used to evaluate the expression levels of exon–exon junctions 2–3, 16–17, 17–18, and 18–19. Error bars represent 95% confidence intervals. The expression of the exons around the integration is reduced by more than >70% (P = 0.01, P = 0.047, and P = 0.002 for 16–17, 17–18, and 18–19, respectively, t test), while the expression of exons 2–3 is increased by more than threefold (P = 0.01, t test) compared with the wild-type HEK293 cells, 18 d posttransfection. The expression levels of the exons revert to normal after 48 d posttransfection. (H) RASGRF2 gene and alternative transcripts. Positioning of HK2 solo-LTR integration, between exons 17 and 18 of the main, 201, transcript.
We further examined whether the association would be observed in an independent, genetically distinct population. Thus, we used a United Kingdom-based population of individuals with chronic hepatitis C virus (HCV) infection and tested the frequency of RASGRF2-int with respect to the route of transmission. Here, we posed a stricter criterion on the history of addictive behavior, as we selected PWIDs who had injected within the previous 6 mo from the sampling date and reported having their first injection at least 2 y before sampling (thus establishing long-term addictive behavior). The control population consisted of subjects who had been infected with HCV through bleeding disorders and matched the PWID population by age (±10 y), gender, and ethnicity. We found RASGRF2-int in 34 out of 100 PWIDs compared with 8 from the 84 controls, revealing a 3.5-fold higher frequency in populations with long-term addictive behavior (P < 0.001, χ2 test, two-sided test) compared with matched controls. We further tested potential confounding of gender, age, alcohol use, and smoking in multivariate models and found that the association of RASGRF2-int with drug addiction remained significant (P < 0.001) while no other parameters were found to be significantly associated with RASGRF2-int. A pooled analysis of the Greece- and United Kingdom-based cohorts further supported the strong significance for the association of RASGRF2-int with PWID (23.8% in 202 PWIDs versus 7.6% in 184 control patients; P < 0.0001, χ2 test, two-sided test).
Artificial Insertion of the HK2 LTR Can Modulate RASGRF2 Transcription.
We then explored whether the above-found associations were due to a causal relationship between RASGRF2-int and addiction. Our primary hypothesis is that RASGRF2-int is modulating transcription of RASGRF2 (Fig. 1), as some intronic ERVs have been shown to modulate transcription in mice (12, 26, 27). RASGRF2-int, like the majority of HK2 intronic integrations of the human genome, is antisense compared with RASGRF2 (10). Intronic ERVs in mice are mostly antisense and the majority of them do not disrupt normal gene transcription (26), while a minority of antisense intronic mouse ERVs have been shown to disrupt transcription (26, 28). The alternative explanation for the above-found associations is that RASGRF2-int is a proxy of a genetically linked (yet unknown to us) polymorphism of RASGRF2, which bears the true causal effect.
To test our primary hypothesis, we used the CRISPR/Cas9 approach (Fig. 1E) to introduce the LTR in the same position observed in the human population within the HEK293 cell line, derived from kidney cells but known to have a neuronal transcriptome (29). We performed PCR and Sanger sequencing of the integration and preintegration sites to show that the integration was heterozygous and that there was no off-target editing of the preintegration site. Eighteen days after clonal selection, we evaluated the transcription levels of RASGRF2 exons. We detected a significant modification of the normal transcription of RASGRF2 (Fig. 1G). More specifically, transcription and splicing of the early exons were significantly increased by more than five times while transcription and splicing of the surrounding exons were significantly diminished by ∼70% compared with the wild type (see Fig. 1G legend for detailed statistics). In a previous study, down-regulation of RASGRF2 by 70% with RNAi resulted in a marked decrease of the RASGRF2 protein (30), suggesting that our observed down-regulation can be also significant at the translational level. Crucially, according to ENSEMBL, two protein-coding transcripts are produced from RASGRF2 (RASGRF2-201 and RASGRF2-206; Fig. 1H), the presence of which in HEK293 cells we confirmed by analyzing publicly available RNA-sequencing (RNA-seq) datasets. RASGRF2-201 includes all of the exons, while RASGRF2-206 includes only the first 10 exons, suggesting that our findings could potentially be explained through down-regulation of RASGRF2-201 and up-regulation of RASGRF2-206. This hypothesis, however, needs to be explored with a stabilized cell-line model which will allow in-depth study of the potential underlying mechanism.
RASGRF2 has at least two independent domains responsible for signaling activities through the guanine exchange factor (GEF), one for Ras and one for Rac1 (Fig. 1H). The exons with “diminished” expression are proxies for Ras-GEF activity, and their disruption should result in a decelerated rate of cell division. We therefore expected a fitness cost for the edited cells compared with wild type. Indeed, genome editing produced a slightly diminished survival-under-stress phenotype for the population of the edited cells; 3 out of 48 wells seeded with the clonally expanded edited cell line survived de novo clonal selection in nonenriched media compared with 13 out of 48 of the wild type, suggesting a selective disadvantage of the cells harboring the integration (P < 0.002, binomial test). Furthermore, chromosome 5 of HEK293 cells (where RASGRF2 lies) is aneuploid, the copy number of which fluctuates during passages (31). We found that within 30 d during serial passaging, the cells were losing the edited allele (Fig. 1), suggesting a selective disadvantage of the allele carrying the integration and a recovery of the normal phenotype (Fig. 1H). Remarkably, the modulated transcription profile of RASGRF2 exons was restored in the cells, which eventually lost the RASGRF2-int allele. The concurrent transcriptional recovery following the loss of the integration also supported that the observed fitness cost was indeed due to the RASGRF2 editing by the knocking down of the Ras-GEF activity and not due to potential off-target effects.
Unfortunately, the fitness disadvantage resulted in destabilizing the HEK293 clone of RASGRF2-int; we attempted to stabilize the clone twice, but on both occasions loss of the clone as described above was observed (suggesting also that the fitness cost experiment is reproducible). We then attempted to edit eHAP1 cells, a haploid cell line which in theory should allow us to stabilize edited clones if the disruption caused by the integration is not deleterious when in the homozygotic state. On the other hand, if homozygosity has a significant deleterious cost, this would result in failure to expand the edited clone. After screening 92 potential clones, we obtained nine candidates, none of which survived upon expansion (see also SI Appendix). This prevented us from obtaining sufficient quantity of the edited clone that would allow us to perform direct detailed in-depth transcriptomic and proteomic analyses of RASGRF2-int. Although multiple mechanisms have been described for disruption of nearby genes by intronic integrations for other classes of human transposable elements (32), only a few studies have suggested a potential importance of intronic antisense HERVs (13), while the role of polymorphic intronic HERVs is largely unknown. To indirectly explore potential mechanisms of transcriptional modulation that could be involved in the observed in vivo and in vitro phenotypic changes, we then studied intronic transcriptional dynamics of HK2 integrations in a cell line known to highly express HK2.
Intronic HK2 Integrations Can Initiate Transcription.
Previous studies showed that at least 50% of HK2 LTRs can initiate transcription (33). To provide in-depth analysis of HK2 intronic transcription, we examined RNA-seq data from teratocarcinoma NCCIT cells known for their embryonic stem-cell identity and elevated HK2 transcription (34). To avoid potential bias as a consequence of the presence/absence of polymorphic integrations, we characterized the HK2 integration sites of NCCIT cells using a target enrichment sequencing approach (SI Appendix). We marked 10 differences compared with the hg19 reference genome, 4 integrations existing in hg19 that were missing and 6 integrations absent from hg19 integrations that were present. Based on the HK2 integration sites in NCCIT cells, we analyzed the HK2-related transcriptome in previously published RNA-seq datasets derived from this cell line (35) (Fig. 2). HK2 LTRs have enhancer and promoter signals, and therefore we focused on RNA reads spanning the integration sites (LTR–host junction) of HK2 to evaluate the potential of transcriptional modulatory activity of these integrations. The transcriptional activity of retroviral LTRs is known to be bidirectional (36), with promoter activity being mainly unidirectional (whether the promoter is on the sense or antisense strand), while enhancer activity can be bidirectional or 2D (i.e., acting on promoters found at both sense and antisense DNA strands simultaneously). We categorized the HK2–host junctions as LTR-start and LTR-end junctions and considered that LTR-start RNAs reflect antisense-5′ (A5) transcriptional activity while LTR-end RNAs reflect sense-3′ (S3) transcriptional activity of the LTR (Fig. 2A). We note that we only measured reads spanning the LTR–host junctions, that is, we only count “chimeric” reads that are partially HK2 LTR (edge), partially host sequence. These reads, unless HK2 integrations were produced as a result of duplication events, should be unique and, thus, less likely to suffer from mapping issues (e.g., due to the highly repetitive nature of HK2 LTRs or when the host flanking sequence is a highly repetitive region itself).
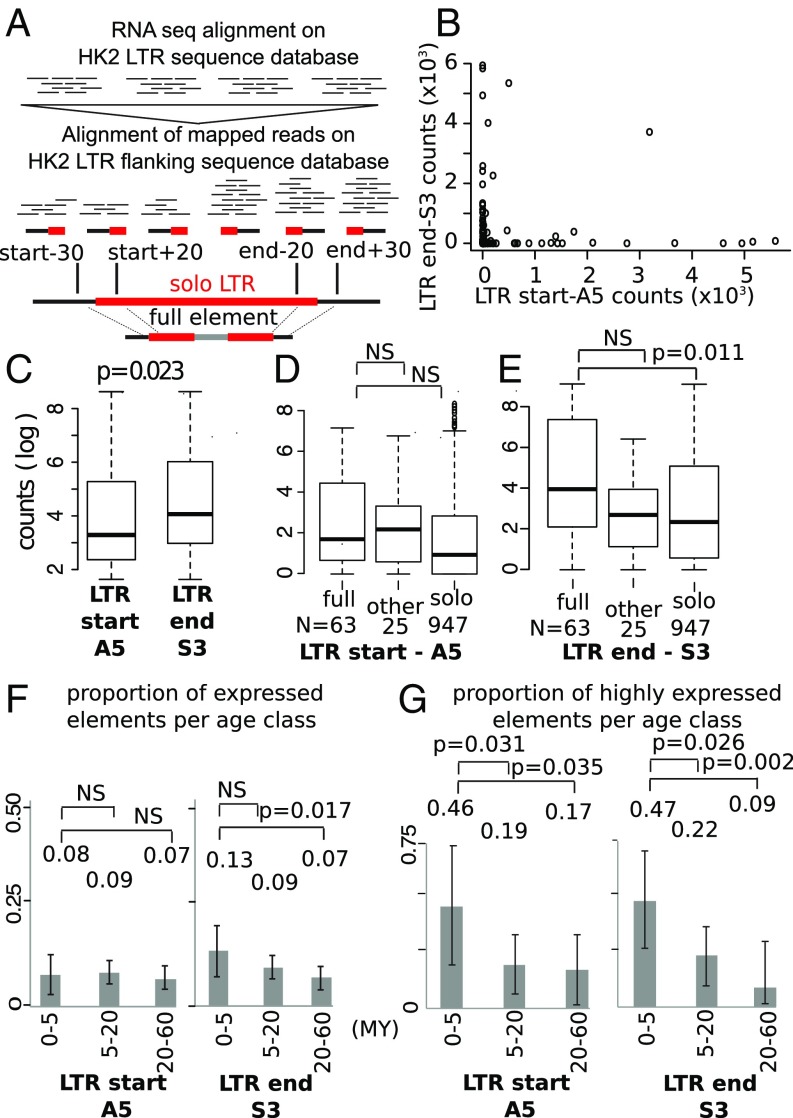
Fig. 2.
(A) Schematic representation of the pipeline used for the estimation of the expression at the edges of the HK2 integrations in NCCIT cells. LTR (in red) and full-length HK2 elements were used to build a sequence database. RNA-seq HK2-filtered reads were locally realigned against an HK2–host junction database. (B) A scatterplot of counts spanning LTR-start vs. LTR-end edges. (C–E) (Log) Boxplots of mean counts vs. type of element, over LTR-start and LTR-end edges. (F) Active (>5 reads) vs. not active (0 to 5 reads) elements, in combination with their age. (G) The magnitude of activity (more/less than 500 reads) in combination with the age of the elements, over LTR-start and LTR-end edges. NS, not statistically significant. Error bars indicate 95% confidence intervals.
There was a clear distinction in the abundance of the two classes of RNAs (i.e., those spanning the 5′ or 3′ ends of the LTR–host junction), suggesting that strong transcription is initiated within the LTRs and is due to the enhancer/promoter roles (Fig. 2B). The LTR-end RNA abundance contrast observed in the highly transcribed integrations suggests a clear dichotomy of the S3 and A5 activities (Fig. 2B), and that strong bidirectional transcriptional activity is not commonly observed in HK2 LTRs. The only exception is a polymorphic integration at chromosome 8 (position 91,696,259), which either suggests bidirectional transcriptional activity or, most likely, is part of the long noncoding RNA NONHSAT127661 that runs through this sequence. It is, consequently, not initiated within and by the LTR. Based on the biology of LTRs, we expected that the S3 activity would be more potent and, indeed, we find that the 3′ RNAs are more abundant than 5′ RNAs (P = 0.023).
We then looked at the evolutionary dynamics of the transcriptional activity of the HK2 integration sites. If the ability of the HK2 integration sites to induce RNA transcription were random, then transcription should be eventually diminished in older integrations as a result of random deleterious mutations or local accumulation of repressive epigenetic marks. In effect, we would see the reduction of transcriptionally active integrations as we move back in time (i.e., more recently integrated proviruses should be more transcriptionally active than older proviruses). Based on this model and due to the more potent S3 activity, we would normally expect that the A5 activity would be more rapidly reduced. In striking contrast, we see that the S3 activities have been significantly reduced through time (P = 0.017, one-sided, Z test; Fig. 2F) (i.e., older integrations have less activity compared with younger integrations), while the A5 has remained resistant to silencing with ∼8% of the LTRs remaining active (P ∼ 0.3, one-sided, Z test; Fig. 2F) even after 30 My. We also observe, for both the A5 and S3 activities, that, within nonsilenced elements, activity is higher for younger (integrated within the last 5 My) rather than older elements (Fig. 2G).
We then explored whether such a transcriptional dichotomy occurs within introns, which could disrupt transcription through multiple mechanisms including transcriptional collision and RNA interference by cis-naturally occurring antisense transcripts (cis-NATs) (13, 37–39). HK2 is integrated intronically or within 2 kb of 359 genes (Dataset S1), some of which are important for immunity (CD4, NFKB2, APOBEC3C) and signaling (DEK, RASGRF2). We found a clear transcriptional dichotomy in 23 genes, most notably CD4, NFKB2, and DEK, suggesting that HK2 LTRs can be triggered to initiate strong transcription within introns, which could in effect modify the functional expression of the respective genes.
Finally, to reproduce the potential of the HK2 LTR for initiating transcription, we used a plasmid reporter gene system to examine whether the HK2 LTR can act as a promoter in both the sense and antisense directions. After successfully transfecting two cell lines (HeLa and Huh7), we were able to see that both directions initiated transcription, although the antisense direction was more potent than the sense in both cell lines. The finding agrees with the previously reported bidirectional promoter activity of HERV-K LTRs (40) and further supports our hypothesis for a disruption and modulation mechanism of RASGRF2-int, either through cis-NAT interference or transcriptional collision.
Discussion
The Potential for Transcriptional Modulation by Intronic HK2.
Thirty-three percent of polymorphic HK2 integrations are within or near genes (within 2 kb), of which 60% are intronic (8). Retroviral integrations preferentially occur near genes, because euchromatic (41) areas are more easily accessible to retroviral integrase. Perhaps intronic integrations are more likely to survive within the hypomethylated introns of genes (42) where retroviral transcription is allowed, rather than within heavily methylated areas of the genome. These intronic integrations exhibit an antisense bias with respect to their directionality compared with the nearby host genes (43), suggesting stronger negative selection on sense-oriented insertions, possibly due to their interference with transcription signals (44). Indeed, at the time of integration, the reconstituted HK2 (HERVKcon) ancestral virus does not exhibit directional bias in vitro with respect to genes (45), and younger integrations seem to have less bias than older elements, although this is not statistically significant (Dataset S1). It has thus been hypothesized that antisense intronic solo-LTRs have no or minimal pathogenicity (10).
Based on the RNA-seq analysis and our functional experiment with the RASGRF2 integration, we suggest that HK2 antisense integrations can modify the human transcriptome. We thus suggest a model for transcriptional activity of HK2 elements where, at the time of integration or soon after, novel proviruses are either silenced or remain active but an adjustment in their regulatory activity results in intermediate transcription over time.
Genetics and in Vitro and in Vivo Phenotypes of RASGRF2-Int.
In accordance with the well-established role of RASGRF2 with addiction, we observed a statistically significant association of RASGRF2-int with i.v. drug use in two independent genetically distinct populations. PWIDs are three times more likely to carry the RASGRF2-int allele than non-PWID controls. Larger studies are required to solidify our findings in PWIDs and potentially with other addiction phenotypes. A reward-related phenotype such as addictive behavior is conditionally pathogenic and, in theory, might even promote beneficial behavior under different circumstances (46). We find that RASGRF2-int had a slightly higher frequency in the United Kingdom-based population compared with the Greece-based population, although the study design does not allow for a safe comparison between populations. There are, however, multiple sources of evidence that the frequency of RASGRF2-int in the human population varies in time and space.
RASGRF2-int has been found in both full genomes of Neanderthal and Denisovan, suggesting that the time of integration was older than diversification from modern humans and most importantly at a much higher frequency in the archaic hominin lineage (47). On the other hand it is rare (∼5%) in modern human populations, with no homozygotic individual recovered in our study, since RASGRF2-int was heterozygous in every person tested. This could be due to either our sample size not being sufficiently large or because the homozygous state is deleterious for the host and thus under strong negative selection. If the homozygous state is indeed deleterious while the heterozygous state provides beneficial traits to the host, the prolonged existence of RASGRF2-int in human populations could be explained by balancing selection. In this case, the reward trait of RASGRF2 could have had a much more significant effect in archaic hominin populations compared with modern humans. Interestingly, in line with our PCR screening of controls, RASGRF2-int is found in ∼5% of the human population of non-African origin, but has recently been reported to be absent among East Asian populations (8). Larger studies will, thus, also clarify the possible existence of homozygotes of RASGRF2-int, determine their potential phenotype, and allow us to test for the hypothesis of balancing selection.
Accordingly, our CRISPR/Cas9-edited cells showed a transcriptional modification of RASGRF2 even though HK2 integration was only in one of the alleles. We would typically expect the wild-type allele to sufficiently express the gene product, but the intensity of the effect suggests that the “edited” allele has a trans effect on “unedited” wild-type transcripts. This can be explained through RNA interference of the overhanging non-LTR homologous segment of the resulting NAT, but we cannot exclude that transcription collision also operates in the edited allele. We then showed that RASGRF2-int had a fitness cost in cell-culture conditions. We should, however, note that RASGRF2-int fitness cost was nonlethal in heterozygotic cells, as the cells were able to survive and proliferate though at a slower rate. Based on our experiments, we cannot, however, exclude that the fitness cost of RASGRF2-int in the homozygotic state is more deleterious; thus, disentangling the in vivo phenotype of RASGRF2-int is challenging.
The function of RASGRF2 has been well-studied in vivo and there were clear predictions to be tested with an epidemiological association study. Based on our cell-line experiments, we suggest that RASGRF2-int leads to enhancement of dopaminergic activity through higher expression of the first exons of RASGRF2, which then results in increased potential for addiction. Our study provides the rationale to explore a wider effect of RASGRF2-int in the dopaminergic activity through independent studies on the frequency of RASGRF2-int in persons with drug use and other addictive behaviors, as well as functional MRI experiments of the RASGRF2-int carriers.
Mechanism and Pathogenicity of HERV-K HML-2 Transcriptional Rewiring.
We suggest that intronic HERVs can trigger transcriptional collision and/or RNA interference through cis-NATs (48). Under this mechanistic model, the implicated genes, even if disrupted, still have ongoing transcription which can be sufficiently functional. When intronic HK2 transcription becomes up-regulated, the interrupted genes will have a modified transcriptional profile; whether the resulting phenotypes are beneficial or pathogenic will depend on the protein domains upstream and downstream of the integration, the directionality of the integration, and the environmental exposure. It is possible that other unknown potentially pathogenic mechanisms can operate as a result of HK2 activity; an intriguing hypothesis would be that HK2 integrations can modify transcription through remodeling of chromatin structure, as has been recently demonstrated for human T-lymphotropic virus (49). It needs, however, to be noted that although there is a well-established link between mRNA levels and protein translation in the case of RASGRF2 (30), this cannot be generalized to other genes; depending on the characteristics of the translated protein (e.g., half-life), a disruption at the mRNA level might have less dramatic effects (50). Our findings thus point towards a conditionally pathogenic role for HK2 integrations through rewiring of the transcriptome; although disease associations and HK2 integrations have been reported, causality has not been sufficiently supported.
Our study demonstrates that at least a percentage of genomic contributions to disease and phenotypes can be missed as a result of inaccurate mapping of polymorphisms related to repetitive elements such as HERVs and other retrotransposons. Developing algorithms and technologies that will accurately recover this difficult part of the human genome and transcriptome will be important to further establish links between HERVs and human phenotypes.
Materials and Methods
Patient Samples.
Ethical approval for the use of Greek-population DNA samples (PWIDs and controls) was obtained from the Research Ethics Committee of the University of Athens. The DNA samples of the United Kingdom-based population were obtained from HCV Research UK. Ethics approval for HCV Research UK was given by National Health Services Research Ethics Committee East Midlands-Derby 1 (Research Ethics Committee reference 11/EM/0314). Informed consent was obtained at the time of collection.
Wet Laboratory.
The wet laboratory procedures are described in detail in SI Appendix and include the design of target enrichment and high-throughput sequencing, procedures used to culture cells, design and protocols used for genome engineering (CRISPR/Cas9 design and execution), PCR screening for RASGRF2-int patient samples and qPCR on cell lines, and assessment of the HK2 LTR promoter in plasmids.
Bioinformatics.
Characterization of the HK2 integrome of NCCIT cells.
NCCIT cells were sequenced using a customized target-enrichment method (Wet Laboratory) to capture HK2 LTRs with flanking host sequences. These genomic fragments prepared in specially optimized large insert sizes (800 to 1,200 bp) and 2 × 300-bp paired-end sequencing resulted in reads that were mapped using Novoalign (www.novocraft.com/products/novoalign/) allowing for soft clipping. Reads were then overlapped with a previous list of known HK2 integration sites to mark presence/absence (8).
RNA-Seq.
Previously published RNA-seq data (50-bp-long reads) from NCCIT cells (51) were filtered after local mapping alignment [Bowtie2 default settings (52)] on an HK2 LTR sequence database. The partially mapped reads were then locally mapped against the HK2 host–junction sequence database after extracting 50-bp fragments at the edges of each element (30 nt before and 20 nt after the start coordinate or 20 nt before and 30 nt after the end coordinate of each element). We made a comprehensive list of NCCIT-integrated elements in the datasets derived from our HK2 target-enrichment NCCIT libraries. We estimated the mean read counts by calculation of the mean read depth across each 50-bp fragment. The mean counts were then adjusted to match the orientation of each element; for example, the 3′ counts were considered to be “LTR-start” counts for negative-oriented elements.
Supplementary Material
Acknowledgments
We thank Paul Klenerman for critical reading of the manuscript. We wish to acknowledge the role of the HCV Research UK Biobank (Award C0365) in collecting and making available samples and data for this publication. The study has been supported by Medical Research Council UK (Project MR/K010565/1). J.M. was supported by an MRC Award (MC_UU_12014/1). A.K. was funded by the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in Genbank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. MH626561–MH626576).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811940115/-/DCSupplemental.
References
- 1.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 2.Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. Env-less endogenous retroviruses are genomic superspreaders. Proc Natl Acad Sci USA. 2012;109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katzourakis A, et al. Larger mammalian body size leads to lower retroviral activity. PLoS Pathog. 2014;10:e1004214. doi: 10.1371/journal.ppat.1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 5.Magiorkinis G, Blanco-Melo D, Belshaw R. The decline of human endogenous retroviruses: Extinction and survival. Retrovirology. 2015;12:8. doi: 10.1186/s12977-015-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol. 2014;88:9529–9537. doi: 10.1128/JVI.00919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wildschutte JH, et al. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci USA. 2016;113:E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Löwer R, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady T, et al. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 2009;23:633–642. doi: 10.1101/gad.1762309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Burgess SM. Integration target site selection for retroviruses and transposable elements. Cell Mol Life Sci. 2004;61:2588–2596. doi: 10.1007/s00018-004-4206-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Romanish MT, Mager DL. Distributions of transposable elements reveal hazardous zones in mammalian introns. PLoS Comput Biol. 2011;7:e1002046. doi: 10.1371/journal.pcbi.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J Virol. 2009;83:6098–6105. doi: 10.1128/JVI.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sverdlov ED. Retroviruses and Primate Genome Evolution. Landes Bioscience; Georgetown, TX: 2005. [Google Scholar]
- 15.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montesion M, Bhardwaj N, Williams ZH, Kuperwasser C, Coffin JM. Mechanisms of HERV-K (HML-2) transcription during human mammary epithelial cell transformation. J Virol. 2017;92:e01258-17. doi: 10.1128/JVI.01258-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Medarde A, et al. Targeted disruption of Ras-Grf2 shows its dispensability for mouse growth and development. Mol Cell Biol. 2002;22:2498–2504. doi: 10.1128/MCB.22.8.2498-2504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz S, Santos E, Bustelo XR. RasGRF2, a guanosine nucleotide exchange factor for Ras GTPases, participates in T-cell signaling responses. Mol Cell Biol. 2007;27:8127–8142. doi: 10.1128/MCB.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108:7119–7124, and erratum (2011) 108:9316. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stacey D, et al. IMAGEN Consortium RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc Natl Acad Sci USA. 2012;109:21128–21133. doi: 10.1073/pnas.1211844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fasano S, et al. Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) controls activation of extracellular signal-regulated kinase (ERK) signaling in the striatum and long-term behavioral responses to cocaine. Biol Psychiatry. 2009;66:758–768. doi: 10.1016/j.biopsych.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barak S, Carnicella S, Yowell QV, Ron D. Glial cell line-derived neurotrophic factor reverses alcohol-induced allostasis of the mesolimbic dopaminergic system: Implications for alcohol reward and seeking. J Neurosci. 2011;31:9885–9894. doi: 10.1523/JNEUROSCI.1750-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anborgh PH, et al. Ras-specific exchange factor GRF: Oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol Cell Biol. 1999;19:4611–4622. doi: 10.1128/mcb.19.7.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutchman M, et al. Dematin interacts with the Ras-guanine nucleotide exchange factor Ras-GRF2 and modulates mitogen-activated protein kinase pathways. Eur J Biochem. 2002;269:638–649. doi: 10.1046/j.0014-2956.2001.02694.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Babaian A, Gagnier L, Mager DL. Visualized computational predictions of transcriptional effects by intronic endogenous retroviruses. PLoS One. 2013;8:e71971. doi: 10.1371/journal.pone.0071971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maksakova IA, et al. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 2016;7:24. doi: 10.1186/s13100-016-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 30.Ma X, et al. βArrestin1 regulates the guanine nucleotide exchange factor RasGRF2 expression and the small GTPase Rac-mediated formation of membrane protrusion and cell motility. J Biol Chem. 2014;289:13638–13650. doi: 10.1074/jbc.M113.511360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YC, et al. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun. 2014;5:4767. doi: 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaer K, Speek M. Retroelements in human disease. Gene. 2013;518:231–241. doi: 10.1016/j.gene.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol. 2006;80:10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boller K, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 35.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arpin-André C, Laverdure S, Barbeau B, Gross A, Mesnard JM. Construction of a reporter vector for analysis of bidirectional transcriptional activity of retrovirus LTR. Plasmid. 2014;74:45–51. doi: 10.1016/j.plasmid.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmichael GG. Antisense starts making more sense. Nat Biotechnol. 2003;21:371–372. doi: 10.1038/nbt0403-371. [DOI] [PubMed] [Google Scholar]
- 39.Zinad HS, Natasya I, Werner A. Natural antisense transcripts at the interface between host genome and mobile genetic elements. Front Microbiol. 2017;8:2292. doi: 10.3389/fmicb.2017.02292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domansky AN, et al. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 2000;472:191–195. doi: 10.1016/s0014-5793(00)01460-5. [DOI] [PubMed] [Google Scholar]
- 41.Schröder ARW, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 42.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 43.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Lagemaat LN, Medstrand P, Mager DL. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 2006;7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushman F, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 46.Spanagel R, Weiss F. The dopamine hypothesis of reward: Past and current status. Trends Neurosci. 1999;22:521–527. doi: 10.1016/s0166-2236(99)01447-2. [DOI] [PubMed] [Google Scholar]
- 47.Agoni L, Golden A, Guha C, Lenz J. Neandertal and Denisovan retroviruses. Curr Biol. 2012;22:R437–R438. doi: 10.1016/j.cub.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 48.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 49.Satou Y, et al. The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc Natl Acad Sci USA. 2016;113:3054–3059. doi: 10.1073/pnas.1423199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 51.Jung I, et al. H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 2012;22:1026–1035. doi: 10.1101/gr.120634.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.