Significance
Human fungal pathogens resistant to conventional therapeutics pose a major threat to global human health. Thus, there is an urgent need to discover new antifungal drugs that act via novel mechanisms of action. Here, we show that commercial herbicides that inhibit acetohydroxyacid synthase (AHAS) have potent and broad-spectrum antifungal activity in vitro and that chlorimuron ethyl, a member of the sulfonylurea herbicide family, has antifungal activity in a mouse model. Thus, this study shows that AHAS inhibitors have strong potential to be developed into potent antifungal therapeutic agents.
Keywords: acetohydroxyacid synthase, Candida albicans, antifungal drug, crystal structure, herbicide
Abstract
The increased prevalence of drug-resistant human pathogenic fungal diseases poses a major threat to global human health. Thus, new drugs are urgently required to combat these infections. Here, we demonstrate that acetohydroxyacid synthase (AHAS), the first enzyme in the branched-chain amino acid biosynthesis pathway, is a promising new target for antifungal drug discovery. First, we show that several AHAS inhibitors developed as commercial herbicides are powerful accumulative inhibitors of Candida albicans AHAS (Ki values as low as 800 pM) and have determined high-resolution crystal structures of this enzyme in complex with several of these herbicides. In addition, we have demonstrated that chlorimuron ethyl (CE), a member of the sulfonylurea herbicide family, has potent antifungal activity against five different Candida species and Cryptococcus neoformans (with minimum inhibitory concentration, 50% values as low as 7 nM). Furthermore, in these assays, we have shown CE and itraconazole (a P450 inhibitor) can act synergistically to further improve potency. Finally, we show in Candida albicans-infected mice that CE is highly effective in clearing pathogenic fungal burden in the lungs, liver, and spleen, thus reducing overall mortality rates. Therefore, in view of their low toxicity to human cells, AHAS inhibitors represent a new class of antifungal drug candidates.
The number of diagnosed pathogenic fungal infections continues to rise significantly each year, with an estimated 1 billion people worldwide now at risk (1) and ∼2 million people per annum succumbing to these infections (2), a number that exceeds the annual death toll for tuberculosis or malaria (3). Despite this, major advances in the search for novel antifungal treatments have been a rarity (4). Individuals commonly affected by fungal infections (e.g., candidiasis, cryptococcosis, aspergillosis, or Pneumocystis pneumonia) are transplant recipients, the immunocompromised suffering HIV, or patients receiving immunosuppressant therapy. However, existing antifungal therapeutics are limited to just a few classes of drugs (e.g., polyenes, azoles, echinocandins, and fluorinated pyrimidine analogs), and these are costly to produce. Furthermore, treatment using these medications requires high doses to be taken over a prolonged time, which can result in serious side effects and lack of compliance (5, 6). In addition to these factors, several clinically relevant pathogenic fungi, including Aspergillus fumigatus, Candida glabrata, Candida krusei, and Cryptococcus spp., are intrinsically resistant to some first-line antifungals (4, 5, 7). Moreover, acquired resistance to all these classes of antifungal drugs has been extensively reported (8–11), and the prevalence of multidrug-resistant strains of pathogenic fungi is continuing to rise (4, 12). Thus, it is clear that new drugs are urgently needed to combat this major threat to human health.
A potential target for the development of new antifungal drugs is acetohydroxyacid synthase (AHAS; EC 2.2.1.6), the first enzyme in the branched-chain amino acid (BCAA) biosynthesis pathway. BCAAs are synthesized by a common pathway in plants and microorganisms; however, this pathway is absent in animals, making it an especially attractive drug target (13). It has previously been shown that AHAS activity is crucial for survival of Candida albicans and Cryptococcus neoformans in vitro, and BCAA auxotrophic strains of these two fungal pathogens are avirulent in vivo (14, 15). These findings suggest AHAS inhibitors could be useful as antifungal agents.
It has been 30 y since the first AHAS inhibitors were introduced as commercial herbicides, with five families of compounds [i.e., sulfonylureas (SUs), imidazolinones (IMIs), pyrimidinyl-benzoates (PBs), triazolopyrimidines (TPs), and sulfonylamino-carbonyl-triazolinones (SCTs)] still in use today (16), which is a testament to their robustness as biocidal agents. Further enhancing their credentials as drug candidates, these compounds display low toxicity in mammals (LD50 in rat >5 g/kg), which is expected since animals do not possess the BCAA pathway (13, 17). Studies on AHAS inhibitors as antifungals have been sporadic and inconclusive. Chlorimuron ethyl (CE) (Fig. 1A), an SU herbicide, has previously been shown to inhibit the growth of C. albicans in culture with a minimum inhibitory concentration, 50% (MIC50) of 2 μM (18), and several noncommercial TPs have activity against C. albicans, C. neoformans, and A. fumigatus with MIC50 values in the range of 1–4 μg/mL (19). However, beyond these reports, a comprehensive study into the suitability of the AHAS-inhibiting herbicides as antifungal agents has yet to be undertaken.
Fig. 1.
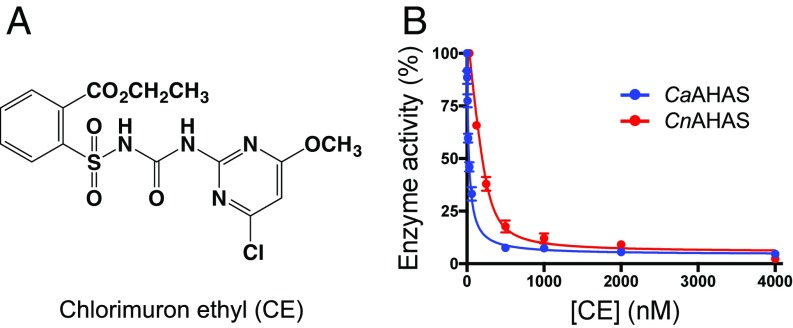
Inhibition of CaAHAS and CnAHAS by CE. (A) Chemical structure of CE. (B) Percentage of enzyme activity vs. inhibitor concentration. The solid line represents the best fit to the data using the equation for weak to medium binding (Eq. 1; CnAHAS) or for tight-binding (Eq. 2; CaAHAS). Error bars represent the SD of the mean (SEM) (n = 3).
Results
CaAHAS and CnAHAS Inhibition by Commercial Herbicides.
Representatives of the five families of herbicides that target AHAS (SI Appendix, Fig. S1) were tested as inhibitors of C. albicans (Ca) AHAS and C. neoformans (Cn) AHAS. Members of the SU and TP families are the most potent inhibitors (Table 1), with CE having a Ki of 24.26 nM for CaAHAS and 119.7 nM for CnAHAS (Fig. 1B), metosulam (MT) a Ki of 800 pM for CaAHAS, and cloransulam methyl a Ki of 35.3 nM for CnAHAS. Members of the SCT, PB, and IMI families inhibit CaAHAS, but with Ki values only in the micromolar range, while the SCTs and PBs inhibit CnAHAS, with Ki values in the millimolar range (Table 1). CnAHAS is partially resistant to SU herbicides, including bensulfuron methyl (BSM), ethoxysulfuron (ES), and sulfometuron methyl (SM). This lower inhibitory activity appears to be due to the P188A and A191L substitutions in the CnAHAS herbicide binding pocket compared with CaAHAS (discussed below) (SI Appendix, Fig. S2A). Two noncommercial SUs derived from the chemical structure of CE, iodomuron ethyl (IE) and iodomuron methyl (IM) (18) (SI Appendix, Fig. S1), were also tested as inhibitors of the fungal enzymes. Compared with CE, these compounds show a two- to fourfold reduction in the Ki values against CaAHAS; however, this improvement in binding affinity is not extended to CnAHAS (Table 1).
Table 1.
Inhibition constants and kinetic rate constants of accumulative inhibition of commercial herbicides for fungal AHASs
| Inhibitor | C. albicans | C. neoformans | ||||||
| Ki, nM | kiapp, min−1 | k3, min−1 | kiapp/k3 | Ki, nM | kiapp, min−1 | k3, min−1 | kiapp/k3 | |
| SU | ||||||||
| IE | 6.3 ± 0.7 | 0.33 ± 0.05 | 3.3 × 10−11 | 10 × 1010 | 110.5 ± 10.8 | 0.17 ± 0.02 | 0.023 | 7.4 |
| IM | 12.7 ± 1.1 | 1.10 ± 0.15 | 0.020 | 55.0 | 472.7 ± 25.5 | 0.17 ± 0.03 | 0.056 | 3.0 |
| CE | 24.3 ± 2.5 | 1.00 ± 0.32 | 0.045 | 22.2 | 119.7 ± 4.2 | 0.73 ± 0.03 | 0.022 | 33.0 |
| BSM | 29.9 ± 2.7 | 1.82 ± 0.25 | 0.064 | 28.4 | NI | — | — | — |
| ES | 69.3 ± 6.9 | 1.69 ± 0.20 | 0.082 | 20.6 | NI | — | — | — |
| CS | 570 ± 59 | NP | NP | — | 769 ± 89 × 103 | NP | NP | — |
| SM | 630 ± 51 | 0.67 ± 0.14 | 0.081 | 8.3 | NI | — | — | — |
| TP | ||||||||
| MT | 0.8 ± 0.1 | 3.39 ± 0.69 | 0.023 | 147.4 | 65.4 ± 4.9 | 1.14 ± 0.06 | 0.029 | 39.3 |
| DS | 1.7 ± 0.8 | 2.31 ± 0.20 | 0.071 | 32.5 | 901.0 ± 72.9 | 0.13 ± 0.02 | 0.007 | 18.6 |
| PS | 2.4 ± 0.3 | 15.26 ± 3.42 | 0.074 | 206.2 | 331.1 ± 20.0 | 2.18 ± 0.11 | 0.037 | 58.9 |
| CSM | 4.6 ± 1.2 | 3.86 ± 0.54 | 0.052 | 74.2 | 35.3 ± 3.7 | 0.94 ± 0.06 | 0.018 | 52.2 |
| FS | 10.1 ± 1.7 | 5.97 ± 0.40 | 0.068 | 87.8 | 590 ± 41.0 | 0.67 ± 0.03 | 0.025 | 26.8 |
| PYS | 212 ± 18 | 2.26 ± 0.10 | 0.067 | 33.7 | 320 ± 29 × 103 | 3.99 ± 0.60 | 5.3 × 10−10 | 7.5 × 109 |
| FT | 773 ± 62 | 3.78 ± 0.27 | 0.068 | 55.6 | 137 ± 9 × 103 | 2.00 ± 0.59 | 1.5 × 10−10 | 1.4 × 1010 |
| SCT | ||||||||
| PC | 484 ± 37 | 1.83 ± 0.28 | 0.080 | 22.9 | 9.4 ± 1.6 × 103 | 0.19 ± 0.04 | 3.7 × 10−11 | 5.1 × 109 |
| TCM | 28 ± 6 × 103 | 21.3 ± 4.4 | 0.109 | 195.0 | 1.2 ± 0.2 × 106 | NP | NP | — |
| PB | ||||||||
| BS | 2.6 ± 0.3 × 103 | NP | NP | — | 5.8 ± 0.6 × 106 | NP | NP | — |
| PTB | 403 ± 37 × 103 | NP | NP | — | 765 ± 65 × 103 | NP | NP | — |
| IMI | ||||||||
| IQ | 716 ± 61 × 103 | NP | NP | — | NI | — | — | — |
| IT | 4.3 ± 0.7 × 106 | NP | NP | — | NI | — | — | — |
BS, bispyribac; CS, chlorsulfuron; CSM, cloransulam methyl; DS, diclosulam; FS, florasulam; IQ, imazaquin; IT, imazethapyr; NI, no inhibition detected; NP, no reversible accumulative inhibition present; PS, penoxsulam; PTB, pyrithiobac; TCM, thiencarbazone methyl.
Recently, we demonstrated AHAS inhibition by commercial herbicides fits a model of time-dependent accumulative inhibition from which first-order rate constants of enzyme inactivation (kiapp) and enzyme recovery (k3) can be determined (20). Here, we found that the five families of herbicides show different levels of reversible accumulative inhibition for CaAHAS and CnAHAS (Table 1 and SI Appendix, Fig. S3). The kiapp and k3 values were determined by curve fitting to Eq. 3 (20) and show that three families of herbicides (i.e., SUs, TPs, and SCTs) are highly effective in promoting the inactivation of CaAHAS (kiapp values from 0.33 to 21.26 min−1). Despite the high level of conservation observed in the herbicide-binding site of CnAHAS and CaAHAS (SI Appendix, Fig. S2A), the former enzyme is less prone to undergo inactivation by commercial herbicides with kiapp values in the range of 0.13–3.99 min−1. k3 values for CaAHAS in the presence of IE, and for CnAHAS in the presence of pyroxsulam (PYS), flumetsulam (FT), or propoxycarbazone (PC) are so low that the inactivation evoked by these herbicides is virtually irreversible (Table 1). This implies that, when kiapp >> k3, the enzyme’s ability to recover from herbicide-induced inactivation determines the potency of accumulative inhibition (kiapp/k3).
CaAHAS Overall Structure.
Crystal structures of the catalytic subunit of uninhibited CaAHAS and CaAHAS in complex with eight different herbicidal inhibitors were determined at resolutions ranging from 1.81 to 2.97 Å (SI Appendix, Table S1). CaAHAS is homotetrameric (Fig. 2A) consistent with Arabidopsis thaliana (At) AHAS but different from Saccharomyces cerevisiae (Sc) AHAS, which is homodimeric (SI Appendix, Fig. S4A). The overall fold for the polypeptide of uninhibited CaAHAS closely resembles that of uninhibited ScAHAS (SI Appendix, Fig. S4B) but differs significantly in the capping region [“mobile loop” (Q576–Y591) and “C-terminal arm” (P646–Y683)] (Fig. 2B). In uninhibited ScAHAS the majority of residues in these two regions are not observed in the electron density maps or adopt a different fold (e.g., an antiparallel β-sheet from V583 to H597 is observed in ScAHAS that changes to a helix when herbicide binds) (21) (SI Appendix, Fig. S4B). In this respect, CaAHAS more closely resembles that of uninhibited AtAHAS (rmsd = 0.699 Å when 428 Cα of a single polypeptide are superposed) that has a folded capping region and preformed binding pocket for the herbicide (22) (SI Appendix, Fig. S4B).
Fig. 2.
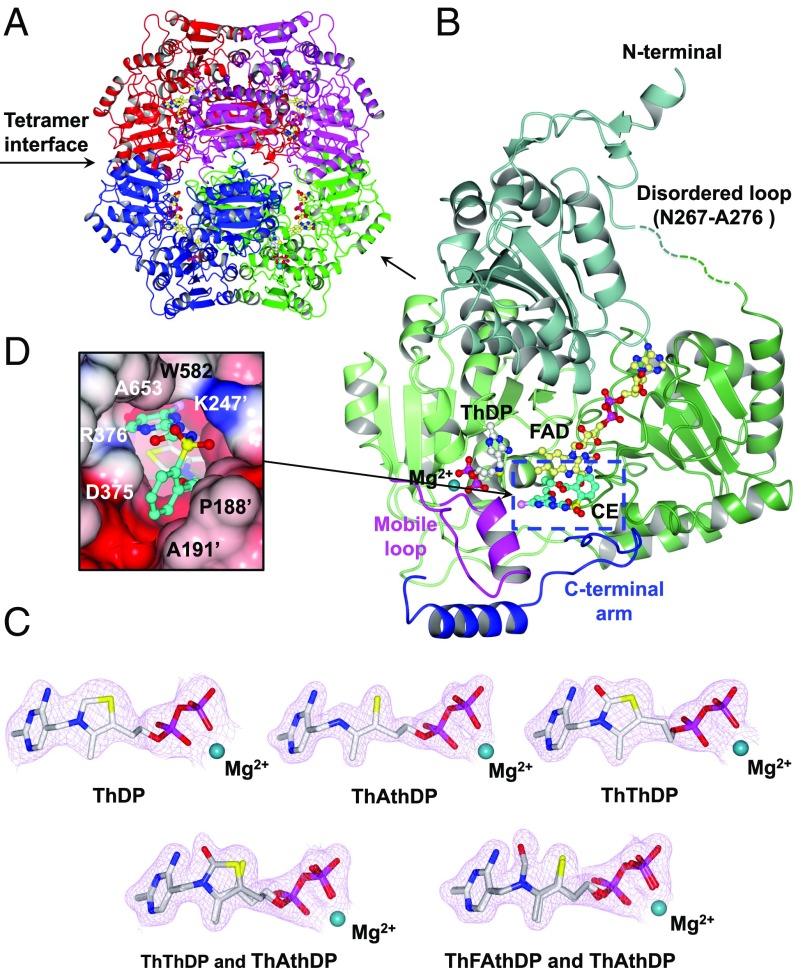
CaAHAS structure. (A) Overall fold of the uninhibited CaAHAS tetramer in cartoon representation. The cofactors ThDP (white) and FAD (yellow) are drawn as ball and sticks models, and the magnesium ion is depicted as a dark cyan sphere. (B) Cartoon representation of a single subunit of CaAHAS in complex with CE. The three domains: α (76–272), β (273–454), and γ (468–639) are colored in different shades of green. The two elements of the capping region, the “mobile loop” (Q576-Y591) and “C-terminal arm” (P646-Y683), which are key for trapping the herbicide in its binding pocket, are colored magenta and blue, respectively. The carbon atoms in CE are colored cyan. A green broken line represents the N267–A276 region, which is disordered in all structures. (C) 2Fo − Fc electron density maps (contoured at ≥2.0 σ) for ThDP, ThAthDP, ThThDP, ThThDP/ThAthDP, and ThFAthDP/ThAthDP in the uninhibited enzyme (2.9-Å resolution), the IE (1.8-Å resolution), PC (2.4-Å resolution), MT (2.1-Å resolution), and CE (2.1-Å resolution) complexes, respectively. (D) Connolly surface showing CE (cyan) inserted into the substrate access channel thereby blocking the active site, where ThDP (white) is located (enclosed in B with broken lines). Nitrogen, blue; oxygen, red; sulfur, yellow; potassium, magenta; and chlorine, pink.
In the uninhibited CaAHAS structure the entire polypeptide is well-resolved, except for the regions between I192–Q198 and N267–A276. Importantly, I192–Q198 is in the vicinity of the active site, forming part of the substrate access channel and herbicide-binding site (SI Appendix, Fig. S5A). Upon herbicide binding, these residues are locked in position by direct contacts to the herbicides (SI Appendix, Fig. S5B). The N267–A276 region is remote from the active site and herbicide-binding site and not likely to influence the binding of substrates, cofactors, or inhibitors (Fig. 2B).
The electron densities for FAD, thiamine diphosphate (ThDP), and magnesium ion (Mg2+) and for the herbicides are all unequivocal, noting that in all of the enzyme-inhibitor complexes the chemical structure of ThDP is chemically modified (Fig. 2C). When the SCT, PC, binds ThDP is modified to thiamine thiazolone diphosphate (ThThDP), and when the SUs, BSM, IE, and IM, or the TP, PS, bind the cofactor is altered to thiamine aminoethenethiol diphosphate (ThAthDP). However, the electron densities in the CaAHAS–SM and the CaAHAS–MT complexes suggest both ThThDP and ThAthDP are present. In contrast, the crystallographic data of the CaAHAS–CE complex suggests that ThAthDP along with an intermediate product of degradation of ThDP, thiamine formamidoethenethiol diphosphate (ThFAthDP), are present. The assignment of these chemical modifications was confirmed by MS of the cofactor isolated from a solution of enzyme incubated with inhibitor (SI Appendix, Fig. S6). Herbicide (e.g., PB, SCT, and TP families)-induced modifications to ThDP have also been observed for AtAHAS (23, 24), and these are the result of oxidative reactions that promote the formation of peracetate in the active site (24) and the subsequent alteration of ThDP (Fig. 3). Importantly, ThThDP has been attributed to have antimetabolic activity (ThDP antagonist) (25) and reactive oxygen species, such as peracetate, inhibit C. albicans growth via NADH depletion (26). These factors, in addition to ThDP’s being replenished at a metabolic cost to cells, could be major contributors to biocidal activity.
Fig. 3.
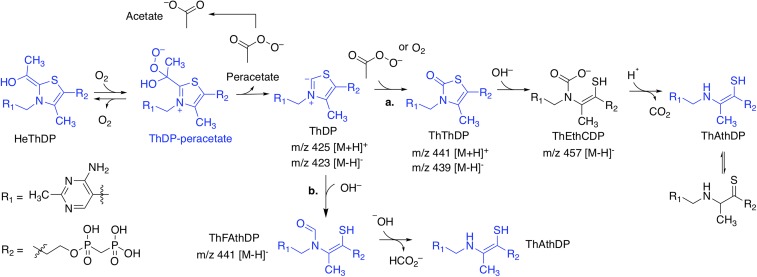
Proposed mechanism for AHAS modification of ThDP. Our kinetic, structural, and MS data suggest that, upon herbicide binding, ThDP is primarily altered into two analogs: ThThDP or ThAthDP. Turnover conditions and molecular oxygen [which binds AHAS nearby ThDP (59)] are required for this to occur (23, 24), implying that the presence of the hydroxyethyl (He) ThDP intermediate in the active site is required for either herbicide binding and/or the generation of peracetate, a product of the oxygenase activity of AHAS (60). From these results, the following mechanism has been developed. In pathway a, free peracetate or molecular oxygen (61, 62) (both trapped in the active site by the herbicide) (24) react with the C2 carbene of ThDP, producing ThThDP. The thiazolone ring is then hydrolyzed by hydroxide and ring-opened, leading to intermediate thiamine ethenethiolcarbamate diphosphate (ThEthCDP), which undergoes decarboxylation of the carbamic acid group to give ThAthDP (63). Alternatively, in pathway b, ThAthDP is produced by a parallel mechanism that involves direct nucleophilic ring opening of the thiazolium ring of ThDP, producing formamidoethenethiol diphosphate (ThFAthDP) (62, 64). Hydroxide then induces cleavage of the N–CHO bond to give ThAthDP (65). The ThDP related molecules depicted in blue have been observed in the crystal structures of AHAS in complex with the herbicide, except for intact ThDP, which is observed in the crystal structure of uninhibited CaAHAS.
CaAHAS Structures in Complex with Herbicide.
To understand the molecular basis of herbicide inhibition of CaAHAS, we cocrystallized this enzyme in complex with five SUs, CE, BSM, SM, IE, and IM; two TPs, MT and PS; and the SCT, PC (SI Appendix, Table S1). The overall fold of these structures is similar to that of the uninhibited enzyme, with rmsd values between 0.145 and 0.271 Å when all Cα atoms are superposed, but in these structures the I192–Q198 region, which is involved in herbicide binding (SI Appendix, Fig. S5), is now fully resolved.
Herbicide Binding Mode to CaAHAS.
All eight herbicides are located in a binding pocket (Figs. 2D and 4) that is situated above the active site and thus operate by preventing the substrate, pyruvate, access to ThDP (SI Appendix, Fig. S5A). This pocket is formed by 12 residues and FAD (SI Appendix, Fig. S7) and is the putative binding site of quinones, which inhibit AHAS activity via FAD oxidation (27). Key to the binding of the three families of herbicides are the contacts formed with P188, F187, K247, M350, R376, M578, and W582 (Fig. 4 and SI Appendix, Fig. S8). Except for P188, all these residues are highly conserved and located in the deepest part of the herbicide-binding site (SI Appendix, Fig. S2B). K247 and R376 contribute with up to five hydrogen bonds when the SUs or the SCT, PC, bind (Fig. 4 A and C), whereas between four to six hydrogen bonds are formed with the TPs (Fig. 4B). In addition, the guanidinium group of R376 forms a π cation interaction with the aromatic ring of the herbicide. The remaining residues of the herbicide-binding site form a large hydrophobic surface (SI Appendix, Fig. S7) that accounts for 30–50 noncovalent interactions. Of these, probably the most important are the π staking interaction formed between the side chain of W582 and the heterocyclic ring of the herbicide and the carbon–π interaction with P188, both of which anchor the herbicide in the binding pocket. A more detailed analysis of the binding mode of each family of herbicides is available as SI Appendix.
Fig. 4.
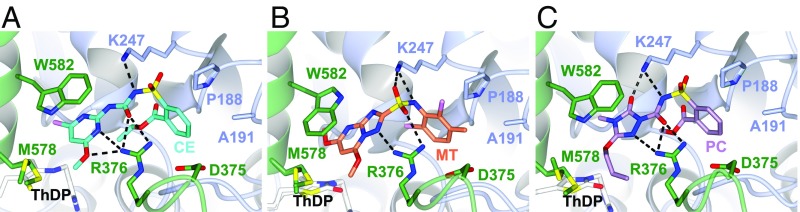
SU, TP, and SCT binding sites in CaAHAS. (A) CE, (B) MT, and (C) PC. The herbicides and key residues for herbicide binding are depicted in stick models, whereas the polypeptide is shown in ribbon display with each subunit colored differently. Hydrogen bonds formed between the herbicides and the polypeptide are presented as dashed lines.
An overlay of the CE complex with an example of the other herbicide complexes shows that the three different families partially overlap their modes of binding to CaAHAS (SI Appendix, Fig. S9). The aromatic rings, and the sulfonyl and carbonyl groups of the SUs and the SCT, PC, are the only components of these two families that adopt a similar position and conformation. In contrast, the TPs have a different orientation, with the aromatic ring overlapping with the ester substituent of the SUs or SCTs and the heterocyclic and linker moieties inserted deepest in the herbicide-binding site.
Conformational Changes in CaAHAS upon Herbicide Binding.
A comparison of uninhibited CaAHAS and the CaAHAS-inhibitor complexes shows that, in the uninhibited enzyme, R376 and W582 protrude toward the herbicide-binding cavity and occupy the location taken by the heterocyclic ring of all three families of herbicides (SI Appendix, Fig. S9). This implies that these residues would block herbicide binding. However, the side chains of R376 and W582 change their conformation so as to make numerous contacts with the herbicides. The adjustments of W582 are the most important for herbicide binding because these allow the π stacking interaction that locks the herbicides into the binding pocket. Other residues in the herbicide-binding site, including P188, A191, K247, and D375, undergo less pronounced side-chain and backbone adjustments to accommodate the herbicides (SI Appendix, Fig. S9). Compared with AtAHAS (22, 23) the herbicide-binding site in CaAHAS is much more rigid and ready to receive inhibitors with only minor conformational changes, a feature of importance for the rational design of alternative CaAHAS inhibitors.
Effect of P188A and A191L Substitutions in CnAHAS.
To understand why some SU herbicides are potent inhibitors of CaAHAS but fail to inhibit CnAHAS we performed in silico mutation and energy minimization of P188A and A191L on the structures of uninhibited CaAHAS and for the CE and MT complexes. These models show that in the P188A substitution key nonpolar interactions (including CH–π interactions; see SI Appendix, Fig. S7) formed between P188 and the aromatic ring of the SUs are no longer present due to the smaller alanine side chain (SI Appendix, Fig. S2C). However, the side chain of leucine in the A191L substitution protrudes toward the binding cavity, reducing its volume; therefore, conformational adjustments are required to accept the herbicide (SI Appendix, Fig. S2C). Although extra contacts may be formed in the presence of this substitution, the overall effect of the P188A and A191L substitutions is negative as witnessed by the increase in the Ki value of CE and resistance in the presence of SM, BSM, and ES. In contrast, the Ki values of the TPs are less affected by these substitutions (Table 1). In the light of these results, this appears to be because these herbicides rely less on the interactions formed with P188 than on the additional contacts that may occur with the A191L substitution (SI Appendix, Fig. S2D).
Fungal Growth Inhibition by AHAS-Inhibiting Herbicides.
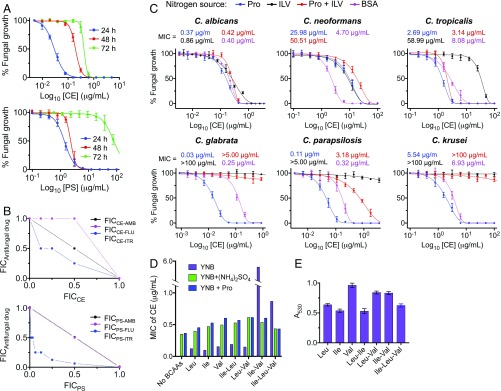
The five classes of commercial herbicide that target AHAS were assessed for their ability to prevent the growth of five Candida species (C. albicans, Candida parapsilosis, C. glabrata, Candida tropicalis, and C. krusei), C. neoformans, and S. cerevisiae in culture (SI Appendix, Table S2). CE was found to display broad-spectrum antifungal activity, being highly effective in preventing the growth of C. albicans, C. parapsilosis, C. glabrata, and S. cerevisiae with MIC values as low as 0.03 μg/mL (72.32 nM), 0.003 μg/mL (7.23 nM), 0.005 μg/mL (12.05 nM), and 0.008 μg/mL (19.29 nM), respectively (Table 2), while other herbicides, including MT, are also effective at preventing growth, but not with the same level of broad-spectrum activity (SI Appendix, Table S2). Independent of the strain tested, some herbicides reduced growth for 24 h, whereas CE, among others, maintained potency for up to 72 h (Fig. 5A and SI Appendix, Table S2). CE has a minimum fungicidal concentration (MFC) for C. albicans that is four times MIC, and thus is considered fungicidal rather than fungistatic (Table 2).
Table 2.
MIC and MFC of CE for fungal pathogen growth in cell culture
| MIC50 | MIC* | MFC/MIC | |||
| Fungal species | μg/mL | nM | μg/mL | nM | |
| Ca | 0.030 | 72.32 | 0.070 | 168.7 | 4 |
| Cg | 0.005 | 12.05 | 0.010 | 24.1 | >8 |
| Cp | 0.003 | 7.23 | 0.004 | 9.6 | 8 |
| Ct | 0.550 | 1.33 × 103 | 1.450 | 3.5 × 103 | 8 |
| Ck | 0.430 | 1.04 × 103 | 1.960 | 4.7 × 103 | >8 |
| Cn | 2.750 | 6.63 × 103 | 7.980 | 19.2 × 103 | >8 |
| Sc† | 0.008 | 19.29 | 0.02 | 48.2 | 2–4 |
MIC50 and MIC values determined after 24-h culture.
The MIC values for fluconazole were 0.31 μg/mL (C. albicans, Ca), >100 μg/mL (C. glabrata, Cg), 4.66 μg/mL (C. parapsilosis, Cp), 6.22 μg/mL (C. tropicalis, Ct), 25.81 μg/mL (C. krusei, Ck), 15.34 μg/mL (C. neoformans, Cn), and 17.03 μg/mL (S. cerevisiae, Sc).
MIC50 and MIC values determined after 48-h culture.
Fig. 5.
Fungal growth inhibition by herbicide. (A) Percentage of C. albicans cell growth vs. log10 [CE] or [PS] after 24, 48, and 72 h. The solid lines represent the best fit to the data using the modified Gompertz function (57). (B) Isobolograms showing the interactions between CE (Top) or PS (Bottom) and three front-line antifungal drugs against C. albicans growth. The FICCE are defined as the MIC of CE in combination with an antifungal drug (itraconazole, fluconazole, or amphotericin B) divided by the MIC of CE alone. A straight line represents an additive interaction (CE–amphotericin B combination), a concave curve indicates synergy (CE–itraconazole combination), and a convex curve shows antagonism (CE–fluconazole interaction). The FIC values for PS and each antifungal drug were determined in the same way. AmB, amphotericin B; Flu, fluconazole; Itr, itraconazole. (C) Fungal growth inhibition by CE in presence of different nitrogen sources: (i) 10 mM proline (blue lines); (ii) 2.5 mM proline, 2.5 mM leucine, 2.5 mM valine, and 2.5 mM isoleucine (red lines); (iii) 3.3 mM leucine, 3.3 mM valine, and 3.3 mM isoleucine (black lines); and (iv) 4 g/L BSA (magenta lines). The MIC values were determined at 48-h incubation using the modified Gompertz function. Note that the substitution of ammonium sulfate for proline, as the nitrogen source, does not produce a significant change in the MIC values reported in SI Appendix, Table S2. (D) Inhibition of C. albicans growth by CE in presence different combinations of BCAAs. YNB media was supplemented with 5 mM valine, 5 mM leucine, and/or 5 mM isoleucine in the presence or absence of 10 mM ammonium sulfate or 10 mM proline. (E) C. albicans growth in the presence of different combinations of BCAAs (5 mM each) and in the absence of herbicide. Error bars represent the SD of the mean (SEM) (n = 3).
Antifungal Activity of CE Is Enhanced by Itraconazole.
To test if the current first-line antifungal compounds, fluconazole, itraconazole, or amphotericin B, can influence the activity of CE in reducing the growth of C. albicans, a grid experiment was performed. This showed that CE in combination with itraconazole had a synergistic effect, resulting in a fractional inhibitory concentration (FIC) value of 0.63 (Fig. 5B, Top). In addition, an additive interaction was observed when CE and amphotericin B were combined (FIC = 1). In contrast, the combination of CE with fluconazole shows an antagonistic interaction (FIC = 1.5). It is worth noting that fluconazole (a fungistatic agent) is an antagonist of other antifungal drugs, for example amphotericin B (a fungicidal) (28), an interaction usually observed when the drugs have different mechanisms of action.
A method that plants adopt to detoxify AHAS-inhibiting herbicides is to use P450 enzymes. This is achieved by adding or removing functional groups, thereby preventing binding to the enzyme (29, 30). Azoles are potent inhibitors of the fungal cytochrome P450 lanosterol 14-α-demethylase (CYP51A1). They also have activity toward human P450 enzymes (CYP3A4 and CYP2C9), increasing the serum concentration of several drugs (31). Therefore, we hypothesize itraconazole may inhibit CE degradation via P450s, thus maintaining the concentration of the compound in the cell for a longer time period. This implies existing fungal P450s can play a role in reducing the antifungal activity of those herbicides that are susceptible to modification. To answer this question, a synergy grid experiment was performed using PS, a TP that has a low MIC value for C. albicans growth of 6.41 μg/mL after 48 h, but which loses activity after 72 h (MIC = 185.5 μg/mL) (Fig. 5A), in combination with fluconazole, itraconazole, or amphotericin B. The results show that only itraconazole enhances the longevity of antifungal activity of PS (synergistic effect, FIC = 0.31) (Fig. 5B, Bottom), resulting in a >15-fold reduction in MIC value for PS after 72 h. However, adding fresh PS alone after 24, 48, and 72 h to concentrations equal to those used in the microdilution tests or additive self-drug combination of PS did not produce an improvement in the MIC values. These data suggest that itraconazole could be coadministered to minimize the effect of herbicide metabolism.
Effect of BCAA Supplementation on the Antifungal Activity of CE.
Next, we investigated whether BCAA supplementation affects the efficacy of CE in reducing the growth of five Candida species and C. neoformans in culture media. Compared with the control [yeast nitrogen base (YNB) broth supplemented with 10 mM proline as nitrogen source], the addition of BCAAs (10 mM; >15-fold the concentration found in human blood) as sole nitrogen source produces a shift in the MICs of CE for all five Candida strains. C. neoformans is unable to grow under these conditions (15) (Fig. 5C). The effect of BCAAs on C. albicans growth in the presence of CE is significantly lower (2.3-fold increase of MIC) compared with that of the other Candida strains (>20-fold increase of MIC). The inclusion of proline along with BCAAs partially restores the antifungal activity of CE against C. albicans, C. parapsilosis, and C. tropicalis (Fig. 5C). In this situation, C. neoformans growth was viable (15) with a twofold increase in the MIC of CE. Given that serum albumin could be a potential source of BCAAs during fungal infections, we next investigated the effect of BSA (4 g/L) on the performance of CE. These results showed that only C. glabrata and C. parapsilosis can partially bypass AHAS inhibition through scavenging BCAAs from BSA (8.3- and 2.9-fold increase of MIC, respectively). Remarkably, the MIC of CE against C. neoformans was reduced a further 5.5-fold compared with the control under this assay condition (Fig. 5C).
To identify the requirements of these fungal pathogens to circumvent AHAS inhibition in the presence of BCAAs, we next assessed the growth of C. albicans in YNB broth with leucine, valine, isoleucine, proline, and/or ammonium sulfate as the only nitrogen source or in combinations, and in the presence of different concentrations of CE (Fig. 5D). These experiments showed that the addition of 10 mM valine, leucine, or isoleucine as the sole nitrogen source does not affect the efficacy of CE in preventing C. albicans growth. Indeed, the MICs of CE determined under these conditions are lower compared with the MIC determined using YNB media supplemented with 10 mM ammonium sulfate or proline. However, the combination of valine and isoleucine suppresses significantly the antifungal activity of CE (MIC = 5.6 μg/mL), while this effect is nullified by addition of leucine, proline, or ammonium sulfate (Fig. 5D). Amino acid transport is highly regulated by the nitrogen catabolite repression or the Ssy1p-Ptr3p-Ssy5p systems, the intracellular and extracellular amino acid ratio, and the presence of nitrogenated molecules in the environment (32, 33). Ammonium diminishes BCAA transport via Gap1 (Gap2 in C. albicans) and Bap2 repression (34, 35), a mechanism which explains why ammonium sulfate restores the antifungal activity of CE in the presence of BCAAs. In agreement with this result, 100 μg/mL eugenol, an inhibitor of BCAA permeases (36), synergizes with CE (FIC 0.38; 48-h MICCE = 0.125 μg/mL) against C. albicans when BCAAs are present in the growth media (SI Appendix, Fig. S10). However, while it has been shown that proline derepresses BCAA transport (34), here we observed that it also diminishes the ability of C. albicans to bypass AHAS inhibition by taking up BCAAs from the media. In addition, we observed when leucine, valine, and isoleucine are added in combination in the media, growth is reduced (Fig. 5E). This could be due to reciprocal competition as the three BCAAs try to use the same method of transport into the cell (37) (Fig. 5E). It has been shown that absence of valine is highly detrimental for C. albicans AHAS (ilv2Δ) mutant growth (14). Here, we show valine increases the growth of C. albicans and that both leucine and isoleucine compete with valine, as seen by a 15% reduction in fungal growth when these are in combination (Leu–Val or Ile–Val). Isoleucine also inhibits growth in presence of leucine and the combination of the three BCAAs yields 35% less growth compared with that observed when valine is the nitrogen source (Fig. 5E). These results suggest that, even though C. albicans is able to scavenge BCAAs from the environment and, to some extent, bypass AHAS inhibition by herbicide to support growth, the intricate interactions between substrates and amino acids limit the absorption of BCAAs and therefore add further support for the concept of using AHAS-inhibiting herbicides to treat human fungal infections.
Antifungal Activity of CE in Vivo.
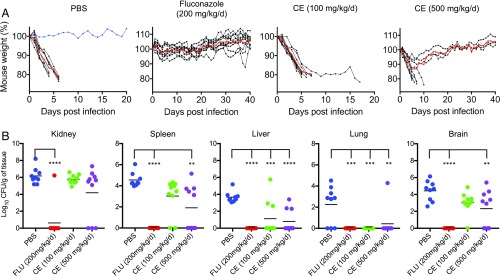
We next assessed if CE could be an effective antifungal agent in mice. First, we performed a toxicity study where CE was injected i.p. twice per day and showed that by this procedure over a 7-d period doses as high as 500 mg⋅kg−1⋅d−1 were tolerated by the mice, with no adverse effects. To assess the efficacy of CE we infected groups of 10 mice via tail vein with C. albicans (strain SC5314; 24-h MIC of CE = 0.05 μg/mL) and tested their ability to survive given different doses of CE (100 and 500 mg⋅kg−1⋅d−1) delivered in two IP injections, each including half of the daily dose of CE. PBS and fluconazole (200 mg/kg/d) were included as negative and positive controls and delivered in the same way as CE. Weight loss is a gold-standard symptom indicative of disseminated fungal infection (38) and we used this reference to assess the severity of the infection. With this premise, only one mouse in the PBS group did not show signs consistent with the disease (Fig. 6A) and therefore was not considered in the final statistical analysis, while all mice receiving CE or fluconazole showed signs consistent with the disease (i.e., weight loss) (Fig. 6A). Compared with the control group receiving PBS, the results show that administration of CE prolongs the life of the infected mice in a concentration-dependent manner (Fig. 6A). Organ fungal burden analysis of kidneys, spleen, liver, lungs, and brain showed that all mice that survived until the end of the experiment, except one individual receiving fluconazole, had cleared the infection (Fig. 6B). In addition, CE significantly decreased fungal load in the spleen, liver, lungs, and brain compared with those mice without receiving antifungal treatment, suggesting that the herbicide is able to reach its target via an adequate distribution of the herbicide in these organs. Importantly, CE is effective in clearing infection from the lungs and liver, even at low concentrations (Fig. 6B). Previous studies have shown that the SUs are readily absorbed following oral administration (17, 39); therefore, here we additionally show that i.p. injection is also a viable route for drug administration. The SUs, including CE, and most AHAS-inhibiting herbicides show a pH-dependent solubility in aqueous solutions, with these compounds becoming insoluble at low pH. We therefore hypothesize the weaker effect of CE in reducing the fungal burden in the kidneys could be due to precipitation of the compound in urine (40).
Fig. 6.
Survival of BALB/c mice infected i.v. with C. albicans. (A) Percentage of mouse weight postinfection. Mice (n = 10 per group) received PBS, fluconazole (200 mg⋅kg−1⋅d−1), CE (100 mg⋅kg−1⋅d−1), or CE (500 mg⋅kg−1⋅d−1) via i.p. injection over a 7-d period after i.v. infection with C. albicans (Methods). The weight of the mice was recorded every day for the duration of the experiment (40 d), and mice were killed by CO2 asphyxiation when they showed ≥20% weight loss or signs consistent with disseminated candidiasis. Each mouse showed weight loss for at least four consecutive days postinfection. Black lines represent each individual within groups, red lines the mean of each group, and a blue line represents an outlier in the PBS group that did not show signs of infection (i.e., weight loss). (B) Organ burden analysis calculated in colony forming units (CFU) per gram of tissue. One-way ANOVA test was performed to determine statistical significance (kidney: F = 17.82, P < 0.0001; spleen: F = 18.26, P < 0.0001; liver: F = 14.87, P < 0.0001; lung: F = 8.93, P = 0.0002; and brain: F = 17.7, P < 0.0001; degrees of freedom = 3 for treatments and 35 for residuals), followed by Tukey test (α = 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001).
Discussion
Since the discovery that AHAS is essential for virulence and survival of C. albicans and C. neoformans cells in vitro and in vivo (14, 15), this enzyme has been proposed as a potential target for developing antifungal compounds. Preliminary studies have shown that inhibitors targeting AHAS have antifungal activity against C. albicans, C. neoformans, and A. fumigatus in culture (18, 19). Here, we have performed a comprehensive study to identify drug lead candidates with potent antifungal activity within the five families of AHAS-inhibiting herbicides. Our kinetic assays concluded that CE and several other AHAS inhibitors of the SU and TP families show potent inhibition of CaAHAS and CnAHAS that also correlates with strong antifungal activity. The ability of the herbicides to induce accumulative inhibition via ThDP alteration and FAD oxidation (23, 24) is an important determinant in how effective these compounds are in biological assays, so should be a key factor in consideration of the design of AHAS inhibitors to act as antifungal drugs. Importantly, CE stands out over all commercial herbicides tested in this study because it possesses a broad spectrum of antifungal activity against five different Candida species and C. neoformans and is fungicidal against C. albicans in vitro. Our cell-based studies suggest several fungal pathogens have the ability to detoxify some AHAS-inhibiting herbicides via P450s. It is worth noting that CE and the other commercial AHAS inhibitors assayed here were developed to possess crop vs. weed selectivity via herbicide detoxification. O-dealkylation of the methoxy substituents at the heterocyclic ring via P450s is an example of how plants detoxify AHAS inhibitors by affecting their performance (16), suggesting a similar mechanism could occur in fungi. CE has a chlorine atom attached to the heterocyclic ring, a unique feature among the commercial herbicides tested (SI Appendix, Fig. S1). Accordingly, CE is effective in reducing the growth of a number of fungal pathogens over a period of 72 h, indicative that this herbicide is less affected by detoxification. In this context, we have shown itraconazole enhances the antifungal activity of AHAS inhibitors, suggesting there is potential to exploit/develop specific fungal P450 inhibitors to further maximize the performance of CE and other herbicides that target AHAS.
The broad-spectrum antifungal activity of CE is a result of the high conservation of the AHAS herbicide-binding site across fungi (SI Appendix, Fig. S2). Of those residues forming direct contacts with the herbicides, only four have been changed during evolution and are located at the entrance of the binding cavity (SI Appendix, Fig. S2B). The A653G substitution observed in ScAHAS and C. parapsilosis AHAS has little effect, since ScAHAS (41) and S. cerevisiae and C. parapsilosis cells are highly susceptible to AHAS inhibitors. However, C. neoformans is resistant to the SU herbicide, SM (15). Modeling suggests this is in part due to the P188A and A191L substitutions in CnAHAS (SI Appendix, Fig. S2 C and D). Based on these findings it was advocated AHAS inhibitors might not be suitable for treating infections caused by this pathogen. While we have confirmed CnAHAS exhibits a natural insensitivity to several AHAS-inhibiting herbicides of the SU (including SM, BSM, and ES), SCT, and IMI families, we also observed that CE (a SU) and MT and CSM (TPs) inhibit this enzyme at the low-nanomolar level. Models of the P188A and A191L substitutions in CaAHAS and our kinetic data show that CE is effective in inhibiting CnAHAS because it possesses a longer ethyl ester substituent at the aromatic ring and this contributes to anchoring the herbicide in its binding cavity even in presence of these two unfavorable substitutions (SI Appendix, Fig. S2C). The TPs, including MT and CSM, are potent inhibitors of CnAHAS because this family of herbicides form fewer contacts with P188, their linker has more flexibility (SI Appendix, Fig. S2D), and these locate the aromatic ring deeper in the herbicide-binding site compared with the SUs (SI Appendix, Fig. S9). For future drug design efforts, a focus should be placed on developing inhibitors that bind only to the highly conserved residues in the AHAS binding site. This should reduce the propensity for resistance to develop.
Previous studies point out the possibility that fungal pathogens might scavenge BCAA from the hosts’ serum and bypass AHAS inhibition by herbicides (15, 19). Here, we show the antifungal activity of CE against C. albicans, C. parapsilosis, C. tropicalis, and C. neoformans is not affected by BCAA supplementation, which is a product of the complex regulation of BCAA absorption by BCAAs themselves and by other molecules that could serve as nitrogen source (e.g., ammonium ions or proline). Further supporting our observation, there is evidence during protein synthesis inhibition, which is the latter effect of blocking BCAA biosynthesis by herbicide, endogenous amino acids accumulate inhibiting amino acid uptake below detection limits (42). Conclusively, CE is effective in reducing fungal burden in the spleen, liver, lungs, and brain of mice systemically infected with C. albicans, confirming that the hosts’ BCAAs absorbed by this pathogen during infection cannot completely bypass AHAS inhibition by CE. Therefore, we identify the SU herbicides as having great potential to be developed as a novel class of antifungal drugs to treat human fungal pathogenic infections.
Conclusions
Here, AHAS-inhibiting commercial herbicides, developed to efficiently kill weeds over crops, were assessed as antifungal agents. This is possible by taking advantage of the fact that the sequences of AHAS are highly conserved across both kingdoms and the activity of this enzyme is essential for the survival of plants and microorganisms. The absence of this enzyme in mammals and the low toxicity of AHAS inhibitors in humans (17) make them an appealing class of compound to be optimized into therapeutic agents to treat infections due to microbial pathogens.
The kinetics of inhibition of CaAHAS and CnAHAS by representatives of the five families of commercial herbicides were determined and these served to identify several potent accumulative inhibitors of these enzymes. CaAHAS inhibition by these herbicides is different from that observed when they bind to ScAHAS. In ScAHAS much of the polypeptide has to fold around these compounds to optimize binding. However, in CaAHAS no major conformational changes occur. This is a key finding because it allows the rational structure-based design of inhibitors based on the native structure of CaAHAS. Previously, we have demonstrated AHAS inhibitors are able to take full advantage of the high reactivity of ThDP and to induce reactions that lead to the ultimate inactivation of this cofactor (23, 24). Here, the crystallographic data and MS allowed us to describe the mechanism by which the herbicides convert ThDP into ThThDP and ThAthDP.
Our cell-based assays provide unique insights into the pathogens’ response to the potent antifungal activity of several commercial herbicides that target AHAS and the potential use of P450 inhibitors to further enhance antifungal activity and to diminish the prospects of the development of resistance. More importantly, CE possesses antifungal activity in mice infected with C. albicans, which illustrates CE is a drug lead suitable for further development and that AHAS inhibitors have great promise to treat human pathogenic fungal infections.
Methods
Cloning of CnAHAS and CaAHAS.
The ORF for the catalytic subunit of the CnAHAS gene was amplified from C. neoformans cDNA. The first 101 aa corresponding to the mitochondrial transit peptide were deleted using the forward primer 5′-AAAGGATCCATGCCCACTAATATGCC-3′, which contains a BamHI site at the N terminus to facilitate its cloning into pET30a(+) vector. The reverse primer was 5′-TCTGCGGCCGCTTACTCAGAGCCGTTCCG-3′ and contains a NotI restriction site after the stop codon. This construct therefore encodes the first 50 residues of the vector, including a hexahistidine tag, followed by the CnAHAS polypeptide from M102 to E718. Cloning of the CaAHAS catalytic subunit into pET30a(+) vector was reported previously (18). The CaAHAS and CnAHAS constructs were then transformed into Escherichia coli strain Rosetta BL21(DE3) and BL21(DE3)pLysS, respectively, for protein expression.
Protein Expression and Purification.
Expression and purification of the catalytic subunits of CaAHAS and CnAHAS was carried out as reported previously for AtAHAS (23). The enzymes were stored in aliquots of 30 μL at −70 °C. Protein concentration was measured using the Direct Detect system (Merck Millipore).
AHAS Assay.
For all assays, unless specifically stated, the standard reaction buffer contained 200 mM potassium phosphate (pH 7.2), 100 mM sodium pyruvate, 10 mM MgCl2, 1 mM ThDP, and 10 μM FAD and assays were conducted at 30 °C. Measurement of Ki values was carried out in triplicate using a colorimetric method (43) and partial anaerobic conditions as described previously (23). These values were then calculated by nonlinear regression using Eq. 1 [weak to medium binding (44)] or Eq. 2 [tight binding (18)]:
| [1] |
| [2] |
For accumulative inhibition assays, 1 μM enzyme was assayed in triplicates using a continuous method (45). The reaction buffer for these assays contained 50 mM pyruvate. The inhibitors were added 15 min after the enzyme so that inhibition could be measured when maximum activity of the enzyme was attained. The reaction was then monitored for a further 35 min. The data were fitted using Eq. 3 (20) for the calculation of kiapp and k3 involved in accumulative inhibition:
| [3] |
For BS, CS, DS, FS, FT, IM, PC, PS, PTB, PYS, SM, and TCM, their relatively high Ki values were taken into account for the estimation of the value F (free enzyme/enzyme.inhibitor complex ratio). The effective concentration of enzyme.inhibitor was calculated using Eq. 4 (inhibition constant formula), the known initial concentrations of enzyme and inhibitor, and the Ki value of Table 1:
| [4] |
Protein Crystallization and Structure Determination.
Crystallization trials were performed using the hanging-drop vapor-diffusion method as described previously (23). The protein solution included freshly thawed protein (30 mg/mL), 5 mM MgCl2, 1 mM ThDP, 1 mM FAD, 5 mM DTT, 3 mM sodium pyruvate, and, when appropriate, 1 mM herbicide. The hanging drops consisted of protein and well solution in a 2:1 ratio. For uninhibited CaAHAS and the CaAHAS–MT complex, the well solution contained 1 M Na/K tartrate, 0.1 M 2-(N-cyclohexylamino)ethanesulfonic acid, and 0.2 M Na2SO4. The pH of the solution ranged from 9.4 to 9.8. Crystals for the remaining CaAHAS-herbicide complexes were obtained in similar way, with the only difference being the well solution contained (NH4)2SO4 instead of Na2SO4. The crystallization experiments were kept at 290 K and protected from light until the crystals reached their maximum size. For cryocooling the crystals were transferred to a drop containing well solution, 35% vol/vol ethylene glycol, cofactors, and inhibitor (when appropriate), mounted in cryoloops and frozen in liquid nitrogen.
The datasets of the uninhibited CaAHAS and the CaAHAS–herbicide complexes were collected at the Australian Synchrotron. The datasets were indexed, integrated, and scaled using XDS (46) and Aimless (47). The crystal structure of uninhibited CaAHAS was solved by molecular replacement with PHASER (48) using ScAHAS in complex with chlorsulfuron (Protein Data Bank ID code 1T9B) as the starting model. Model building was carried out using Coot 0.8.1 (49) and refinement was with PHENIX 1.9-1692 (50). The crystal structures of CaAHAS in complex with herbicides were then solved by the same method using the uninhibited CaAHAS as the starting model. Figures were generated with CCP4mg (51).
Protein Sequence Alignment Protein–Ligand Interaction Analysis.
The amino acid sequences for the AHAS catalytic subunits of C. albicans (XP_721571.1), C. glabrata (XP_448375.1), C. parapsilosis (CCE44821.1), C. tropicalis (XP_002548114.1), C. krusei (KGK39551.1), C. neoformans (AAR29084.1), A. fumigatus (EAL92550.1), and S. cerevisiae (NP_013826.1) were obtained from GenBank and sequence alignment was performed with Clustal Omega (52). Interactions with each herbicide in CaAHAS were analyzed using Arpeggio (53).
CnAHAS Herbicide-Binding Site Model.
The P188A and A191L substitutions present in CnAHAS were modeled in the 3D coordinates of uninhibited CaAHAS and the CaAHAS-CE/MT complexes using Coot 0.8.1 (49) and structure optimization was carried out with Yasara (54).
MS.
Samples for MS analysis of ThDP were prepared as described previously (23). One microliter of each sample was then injected onto an Agilent InfinityLab Poroshell 120 EC-C18 column (2.1 × 50 mm). Liquid chromatography gradient was 0–98% B over 7 min, run at 100 μL/min, with the column oven set to 45 °C. Eluted samples were sprayed using a heated electrospray ionization source and analyzed on an Orbitrap Elite mass spectrometer. Source parameters included capillary temperature = 320 °C, source heater temperature = 200 °C, sheath gas = 25 L/min, auxiliary gas = 8 L/min, source voltage = 4.5 kV, and s-lens = 60%. Samples were analyzed with the Orbitrap analyzer set to 30,000 resolution.
Cell-Based Studies.
Antifungal susceptibility testing was performed using the broth microdilution method according to a standard protocol (55). All experiments were performed in 96-well plates using YNB broth without amino acids and ammonium sulfate (BD Difco) supplemented with 2% glucose, 100 mM (NH4)2SO4, and 50 mM Hepes. The pH of the media was adjusted to 7.2 to ensure inhibitor solubility and stability (56). The herbicides were dissolved in DMSO and then serially diluted in 96-well plates using YNB. The herbicide solutions were used fresh and kept at −20 °C for no more than 1 wk. The latter was particularly important to guarantee accurate MIC measurements, since some of the herbicide tested had degraded when stored in DMSO for prolonged periods (>1 wk). The well-established antifungal drugs fluconazole and itraconazole were used as controls. All studies were conducted using C. albicans ATCC 90028, C. glabrata ATCC 90030, C. parapsilosis ATCC 22019, C. tropicalis ATCC 750, C. krusei ATCC 6258, C. neoformans ATCC 90113, and S. cerevisiae Sigma 1278b. Growth was determined by measuring the OD530 every 24 h using a Molecular Devices Spectramax 250 microplate reader (Marshall Scientific). The MIC50 and MIC values were determined using the modified Gompertz function (57). All experiments were conducted in triplicate, with each performed on a different day. The MFC was determined as described previously (58).
The FIC index of CE and PS in combination with fluconazole, itraconazole, or amphotericin B against C. albicans growth was determined using a synergy grid experiment in a microdilution format as described above. The FIC values for CE (FICCE) were defined as the MIC of CE in combination with an antifungal drug (itraconazole, fluconazole, or amphotericin B) divided by the MIC of CE alone. The FIC values for PS and each antifungal drug were determined in the same way. The FIC index for each drug combination was calculated as FICherbicide + FICantifungal drug. Synergism is defined as FIC index values <1, antagonism by FIC index values >1, and additivity by FIC index values = 1. All experiments were conducted in triplicate.
The effect of BCCAs and BSA on the antifungal activity of CE was measured in triplicate using broth microdilution in a 96-well plate format as described above, with the only difference being (NH4)2SO4 was replaced by (i) 10 mM proline; (ii) 2.5 mM proline in addition to different combinations of BCAAs: 2.5 mM leucine, 2.5 mM valine, and/or 2.5 mM isoleucine; (iii) 3.3 mM of each BCAA as sole nitrogen source; or (iv) 4 g/L BSA as sole nitrogen source. Inhibition of C. albicans growth by CE in the presence different combinations of BCAAs was measured using YNB broth supplemented with 5 mM valine, 5 mM leucine, and/or 5 mM isoleucine in the presence or absence of 10 mM ammonium sulfate or 10 mM proline.
Mouse Studies.
A mouse model study of disseminated C. albicans infection was conducted following the requirements of animal welfare in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes by the National Health and Medical Research Council, and after approval by the Molecular Biosciences Animal Ethics Committee (AEC) of The University of Queensland (AEC approval no. SCMB/175/17).
For toxicity assays of CE in mice, 25 6-wk-old BALB/c mice received 0.1-mL PBS injections via the tail vein using a 1-mL tuberculine syringe and a 27-gauge, 0.5-inch needle. Subsequently, five mice received one of two different doses of CE (100 or 500 mg⋅kg−1⋅d−1) delivered by two i.p. injections (morning and evening), each containing half of the daily dose dissolved in 100 μL PBS (adjusted to pH 8). Fluconazole (200 mg/kg/d) and PBS alone were used as controls and delivered in the same way as CE. All injections were applied using a 1-mL tuberculine syringe and a 27-gauge, 0.5-inch needle. Weight and aspect of the mice were examined on day 1 and every day after for 2 wk, at which point all mice were killed via CO2 inhalation. Kidneys, spleen, liver, lungs, and brain were collected to record organ weight and aspect. Mice were provided with food and water for ad libitum intake and with cardboard as environmental enrichment.
For murine infection assays, 40 6-wk-old female BALB/c mice were infected by tail-vein injection of 0.1 mL PBS containing 106 viable cells (38) using a 1-mL tuberculine syringe and a 27-gauge, 0.5-inch needle. Mice were separated in groups of 10 and treated during a 7-d period, beginning the same day of infection with two different doses of CE (100 and 500 mg⋅kg−1⋅d−1), fluconazole (200 mg⋅kg−1⋅d−1), or PBS delivered in 100-μL i.p. injections twice per day (morning and evening) as described above. A maximum of five mice were housed per individually ventilated cage and provided with food and water for ad libitum intake and cardboard. The weight and aspect of the mice were monitored daily during the duration of the experiment (40 d). Mice were killed via CO2 asphyxiation once they declined to ≥80% of preinfection body weight or when symptoms consistent with infection were obvious. The date of death was recorded as the following day. Kidneys, spleen, liver, lungs, and brain were collected and homogenized in 1 mL PBS using a TissueLyser II (Qiagen) and plated on YPD agar containing 25 μg/mL chloramphenicol, 50 μg/mL kanamycin, and 100 μg/mL ampicillin in 1:10 serial dilutions. The colony-forming units per gram of tissue were determined after 24 h incubation at 30 °C. One-way ANOVA was performed to determine statistical significance within organ fungal burden datasets, followed by the Tukey test.
The inoculum for mice infection was prepared from fresh cultures of C. albicans strain SC5314 plated on YPD agar from a glycerol stock and incubated at 30 °C for 48 h. A single colony was transferred to 10 mL YPD broth and further incubated at 30 °C with shaking (150 rpm) for 20 h. Cells were harvested by centrifugation (800 × g) for 1 min at room temperature and washed twice with sterile PBS and the cell density was adjusted to 1 × 107 cells per mL using a hemocytometer. The cell suspension was kept on ice and used immediately. Cell concentration was confirmed by plating 1:10 serial dilutions on YPD agar and incubated for 24–48 h at 30 °C.
Supplementary Material
Acknowledgments
We thank Barbara Arnts and David Herne (Australian Institute for Bioengineering and Nanotechnology–University of Queensland Biological Resources) for their assistance. This work was supported by Australian National Health and Medical Research Council Grants 1008736 and 1087713 and Natural Science Foundation of China Grant 21672114. Preliminary crystallization conditions and X-ray data were obtained at the University of Queensland Remote-Operation Crystallization and X-Ray diffraction facility. Final datasets were collected using beamlines MX1 and MX2 at the Australian Synchrotron, Clayton, Victoria.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6DEK, 6DEL, 6DEM, 6DEN, 6DEO, 6DEP, 6DEQ, 6DER, and 6DES).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809422115/-/DCSupplemental.
References
- 1.Brown GD, et al. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.GAFFI 2017 Fungal disease frequency (Global Action Fund for Fungal Infections, Geneva). Available at https://www.gaffi.org/why/fungal-disease-frequency/
- 3.Centers for Disease Control and Prevention 2018. Data and statictics for tuberculosis and malaria (CDC, Atlanta)
- 4.Denning DW, Bromley MJ. Infectious disease. How to bolster the antifungal pipeline. Science. 2015;347:1414–1416. doi: 10.1126/science.aaa6097. [DOI] [PubMed] [Google Scholar]
- 5.Nett JE, Andes DR. Antifungal agents: Spectrum of activity, pharmacology, and clinical indications. Infect Dis Clin North Am. 2016;30:51–83. doi: 10.1016/j.idc.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Souza ACO, Amaral AC. Antifungal therapy for systemic mycosis and the nanobiotechnology era: Improving efficacy, biodistribution and toxicity. Front Microbiol. 2017;8:336. doi: 10.3389/fmicb.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonardelli F, et al. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob Agents Chemother. 2016;60:5420–5426. doi: 10.1128/AAC.00905-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacCallum DM, et al. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob Agents Chemother. 2010;54:1476–1483. doi: 10.1128/AAC.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baixench MT, et al. Acquired resistance to echinocandins in Candida albicans: Case report and review. J Antimicrob Chemother. 2007;59:1076–1083. doi: 10.1093/jac/dkm095. [DOI] [PubMed] [Google Scholar]
- 10.Vandeputte P, et al. Molecular mechanisms of resistance to 5-fluorocytosine in laboratory mutants of Candida glabrata. Mycopathologia. 2011;171:11–21. doi: 10.1007/s11046-010-9342-1. [DOI] [PubMed] [Google Scholar]
- 11.Martel CM, et al. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother. 2010;54:3578–3583. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad R, Kapoor K. Multidrug resistance in yeast Candida. Int Rev Cytol. 2005;242:215–248. doi: 10.1016/S0074-7696(04)42005-1. [DOI] [PubMed] [Google Scholar]
- 13.Duggleby RG, Pang SS. Acetohydroxyacid synthase. J Biochem Mol Biol. 2000;33:1–36. [Google Scholar]
- 14.Kingsbury JM, McCusker JH. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2Delta) mutants is influenced by the carbon source and rapamycin. Microbiology. 2010;156:929–939. doi: 10.1099/mic.0.034348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 degrees C and in vivo. Microbiology. 2004;150:1547–1558. doi: 10.1099/mic.0.26928-0. [DOI] [PubMed] [Google Scholar]
- 16.Gutteridge S, et al. Acetohydroxyacid synthase inhibitors (AHAS/ALS) In: Krämer W, Schirmer U, Jeschke P, Witschel M, editors. Modern Crop Protection Compounds. Wiley-VCH; Weinheim, Germany: 2012. pp. 29–162. [Google Scholar]
- 17.Healy CE, Heydens WF, Naylor MW. Mammalian toxicology overview and human risk assessment for sulfosulfuron. Regul Toxicol Pharmacol. 2004;39:310–324. doi: 10.1016/j.yrtph.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y-T, et al. Sulfonylureas have antifungal activity and are potent inhibitors of Candida albicans acetohydroxyacid synthase. J Med Chem. 2013;56:210–219. doi: 10.1021/jm301501k. [DOI] [PubMed] [Google Scholar]
- 19.Richie DL, et al. Identification and evaluation of novel acetolactate synthase inhibitors as antifungal agents. Antimicrob Agents Chemother. 2013;57:2272–2280. doi: 10.1128/AAC.01809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonhienne T, et al. Commercial herbicides can trigger the oxidative inactivation of acetohydroxyacid synthase. Angew Chem Int Ed Engl. 2016;55:4247–4251. doi: 10.1002/anie.201511985. [DOI] [PubMed] [Google Scholar]
- 21.Pang SS, Guddat LW, Duggleby RG. Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase. J Biol Chem. 2003;278:7639–7644. doi: 10.1074/jbc.M211648200. [DOI] [PubMed] [Google Scholar]
- 22.Garcia MD, Wang J-G, Lonhienne T, Guddat LW. Crystal structure of plant acetohydroxyacid synthase, the target for several commercial herbicides. FEBS J. 2017;284:2037–2051. doi: 10.1111/febs.14102. [DOI] [PubMed] [Google Scholar]
- 23.Garcia MD, Nouwens A, Lonhienne TG, Guddat LW. Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families. Proc Natl Acad Sci USA. 2017;114:E1091–E1100. doi: 10.1073/pnas.1616142114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonhienne T, et al. Structural insights into the mechanism of inhibition of AHAS by herbicides. Proc Natl Acad Sci USA. 2018;115:E1945–E1954. doi: 10.1073/pnas.1714392115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arjunan P, et al. Structural determinants of enzyme binding affinity: The E1 component of pyruvate dehydrogenase from Escherichia coli in complex with the inhibitor thiamin thiazolone diphosphate. Biochemistry. 2004;43:2405–2411. doi: 10.1021/bi030200y. [DOI] [PubMed] [Google Scholar]
- 26.Avci P, et al. Sodium ascorbate kills Candida albicans in vitro via iron-catalyzed Fenton reaction: Importance of oxygenation and metabolism. Future Microbiol. 2016;11:1535–1547. doi: 10.2217/fmb-2016-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonhienne T, Garcia MD, Guddat LW. The role of a FAD cofactor in the regulation of acetohydroxyacid synthase by redox signaling molecules. J Biol Chem. 2017;292:5101–5109. doi: 10.1074/jbc.M116.773242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louie A, Banerjee P, Drusano GL, Shayegani M, Miller MH. Interaction between fluconazole and amphotericin B in mice with systemic infection due to fluconazole-susceptible or -resistant strains of Candida albicans. Antimicrob Agents Chemother. 1999;43:2841–2847. doi: 10.1128/aac.43.12.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonné-Pfister R, Gaudin J, Kreuz K, Ramsteiner K, Ebert E. Hydroxylation of primisulfuron by an inducible cytochrome P450-dependent monooxygenase system from maize. Pestic Biochem Physiol. 1990;37:165–173. [Google Scholar]
- 30.Brown HM. Mode of action, crop selectivity and soil relations of the sulfonylurea herbicides. Pestic Sci. 1990;29:263–281. [Google Scholar]
- 31.Katz HI. Drug interactions of the newer oral antifungal agents. Br J Dermatol. 1999;141:26–32. doi: 10.1046/j.1365-2133.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 32.Cooper TG, Sumrada RA. What is the function of nitrogen catabolite repression in Saccharomyces cerevisiae? J Bacteriol. 1983;155:623–627. doi: 10.1128/jb.155.2.623-627.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forsberg H, Ljungdahl PO. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol Cell Biol. 2001;21:814–826. doi: 10.1128/MCB.21.3.814-826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grenson M, Hou C, Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol. 1970;103:770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didion T, Grauslund M, Kielland-Brandt MC, Andersen HA. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2025–2029. doi: 10.1128/jb.178.7.2025-2029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darvishi E, Omidi M, Bushehri AAS, Golshani A, Smith ML. The antifungal eugenol perturbs dual aromatic and branched-chain amino acid permeases in the cytoplasmic membrane of yeast. PLoS One. 2013;8:e76028. doi: 10.1371/journal.pone.0076028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szmelcman S, Guggenheim K. Interference between leucine, isoleucine and valine during intestinal absorption. Biochem J. 1966;100:7–11. doi: 10.1042/bj1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacCallum DM. 2012. Mouse intravenous challenge models and applications. Host-Fungus Interactions: Methods and Protocols, Methods in Molecular Biology, eds Brand A, MacCallum DM (Humana, New York), Vol 845.
- 39.Kreisberg JF, et al. Growth inhibition of pathogenic bacteria by sulfonylurea herbicides. Antimicrob Agents Chemother. 2013;57:1513–1517. doi: 10.1128/AAC.02327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold LL, Cano M, St John MK, Healy CE, Cohen SM. Effect of sulfosulfuron on the urine and urothelium of male rats. Toxicol Pathol. 2001;29:344–352. doi: 10.1080/019262301316905309. [DOI] [PubMed] [Google Scholar]
- 41.Duggleby RG, Pang SS, Yu H, Guddat LW. Systematic characterization of mutations in yeast acetohydroxyacid synthase. Interpretation of herbicide-resistance data. Eur J Biochem. 2003;270:2895–2904. doi: 10.1046/j.1432-1033.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 42.Grenson M, Crabeel M, Wiame JM, Béchet J. Inhibition of protein synthesis and simulation of permease turnover in yeast. Biochem Biophys Res Commun. 1968;30:414–419. doi: 10.1016/0006-291x(68)90760-2. [DOI] [PubMed] [Google Scholar]
- 43.Singh BK, Stidham MA, Shaner DL. Assay of acetohydroxyacid synthase. Anal Biochem. 1988;171:173–179. doi: 10.1016/0003-2697(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 44.Chang AK, Duggleby RG. Herbicide-resistant forms of Arabidopsis thaliana acetohydroxyacid synthase: Characterization of the catalytic properties and sensitivity to inhibitors of four defined mutants. Biochem J. 1998;333:765–777. doi: 10.1042/bj3330765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss JV, Van Dyk DE, Vasta JF, Kutny RM. Purification and properties of Salmonella typhimurium acetolactate synthase isozyme II from Escherichia coli HB101/pDU9. Biochemistry. 1985;24:4952–4959. doi: 10.1021/bi00339a034. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jubb HC, et al. Arpeggio: A web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol. 2017;429:365–371. doi: 10.1016/j.jmb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krieger E, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.NCCLS 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved standard–Second edition (NCCLS, Wayne, PA)
- 56.Sarmah AK, Sabadie J. Hydrolysis of sulfonylurea herbicides in soils and aqueous solutions: A review. J Agric Food Chem. 2002;50:6253–6265. doi: 10.1021/jf025575p. [DOI] [PubMed] [Google Scholar]
- 57.Lambert RJW, Pearson J. Susceptibility testing: Accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J Appl Microbiol. 2000;88:784–790. doi: 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
- 58.Espinel-Ingroff A. In vitro fungicidal activities of voriconazole, itraconazole, and amphotericin B against opportunistic moniliaceous and dematiaceous fungi. J Clin Microbiol. 2001;39:954–958. doi: 10.1128/JCM.39.3.954-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lonhienne T, Garcia MD, Fraser JA, Williams CM, Guddat LW. The 2.0 Å X-ray structure for yeast acetohydroxyacid synthase provides new insights into its cofactor and quaternary structure requirements. PLoS One. 2017;12:e0171443. doi: 10.1371/journal.pone.0171443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abell LM, Schloss JV. Oxygenase side reactions of acetolactate synthase and other carbanion-forming enzymes. Biochemistry. 1991;30:7883–7887. doi: 10.1021/bi00246a002. [DOI] [PubMed] [Google Scholar]
- 61.Machius M, et al. A versatile conformational switch regulates reactivity in human branched-chain α-ketoacid dehydrogenase. Structure. 2006;14:287–298. doi: 10.1016/j.str.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Itoh T, Nagata K, Okada M, Ohsawa A. The reaction of 3-methylthiazolium derivatives with superoxide. Tetrahedron. 1993;49:4859–4870. [Google Scholar]
- 63.Lebegue N, Charrier G, Carato P, Yous S, Berthelot P. Synthesis of 2-(ethylsulfanyl)aniline derivatives through the unexpected ring opening of N-substituted- 2(3H)-benzothiazolones. Tetrahedron Lett. 2004;45:9509–9511. [Google Scholar]
- 64.Kluger R, Tittmann K. Thiamin diphosphate catalysis: Enzymic and nonenzymic covalent intermediates. Chem Rev. 2008;108:1797–1833. doi: 10.1021/cr068444m. [DOI] [PubMed] [Google Scholar]
- 65.Blumberger J, Ensing B, Klein ML. Formamide hydrolysis in alkaline aqueous solution: Insight from ab initio metadynamics calculations. Angew Chem Int Ed Engl. 2006;45:2893–2897. doi: 10.1002/anie.200600283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.