Significance
Gloeobacter violaceus ligand-gated ion channel (GLIC) is a proton-gated, cation-selective channel, a Cys-loop family member of the pentameric ligand-gated ion channels (pLGICs). In a pursuit to provide evidence of function-related conformational changes of a pLGIC in membrane, at ambient temperature and pressure and of the very same unlabeled molecules, we used high-speed atomic force microscopy (HS-AFM) combined with a buffer exchange system to visualize reversible changes of the supramolecular arrangement and conformations of GLIC during pH changes: a structural titration experiment. Reference-free classification of the molecules assigns conformations to states during the pH titration. We find large conformational changes with an unexpected closed-state structure with tightened extracellular domains. Furthermore, we provide evidence for the short-term existence of asymmetric channels at early stages of activation.
Keywords: HS-AFM, conformational change, GLIC, Cys-loop, ion channel
Abstract
Gloeobacter violaceus ligand-gated ion channel (GLIC), a proton-gated, cation-selective channel, is a prokaryotic homolog of the pentameric Cys-loop receptor ligand-gated ion channel family. Despite large changes in ion conductance, small conformational changes were detected in X-ray structures of detergent-solubilized GLIC at pH 4 (active/desensitized state) and pH 7 (closed state). Here, we used high-speed atomic force microscopy (HS-AFM) combined with a buffer exchange system to perform structural titration experiments to visualize GLIC gating at the single-molecule level under native conditions. Reference-free 2D classification revealed channels in multiple conformational states during pH gating. We find changes of protein–protein interactions so far elusive and conformational dynamics much larger than previously assumed. Asymmetric pentamers populate early stages of activation, which provides evidence for an intermediate preactivated state.
The Cys-loop pentameric ligand-gated ion channel (pLGIC) superfamily includes acetylcholine receptors (AchRs), the serotonin receptor 3A and zinc-activated channels that allow passage of cations, and channels such as GABAA receptors and glycine receptors that funnel anions; they are of utmost importance in synaptic transmission (1–5). Upon ligand binding, the channels undergo a conformational change into an activated state that allows ions to flow down their concentration gradient, thereby raising membrane potential and stimulating muscle contraction or neuronal action potentials (6–9). The first solved structure of the pLGIC family was AchR from Torpedo marmorata, which is a heteropentamer formed by α, γ, α, β, and δ subunits (10).
Structural analysis of prokaryotic pLGIC homologs, such as the proton-activated, cation-selective Gloeobacter violaceus ligand-gated ion channel (GLIC), which are homopentamers, allowed detailed structural insights (11–14): The extracellular domain (ECD) of each subunit consists of 10 β-strands, while the transmembrane domain (TMD) consists of four α-helices (M1 to M4), where M2 lines the central pore and M4 is membrane-exposed. Structural comparison between the closed (pH 7) (11, 13) and active/desensitized (pH 4) (12) structures suggested a gating mechanism and conformational changes in both the ECD and TMD domains (SI Appendix, Fig. S1). However, these changes, collected on detergent-solubilized proteins embedded in a 3D-crystal lattice, may not reflect the full dynamic range of the channel during gating. Indeed, EPR spectroscopy detected larger conformational changes at the extracellular end of M2 during conversion from the closed state to the desensitized state (15). Moreover, helix M4 displayed large outward movements of at least 4 Å away from the central axis, in lipid-sensitive gating (16).
In summary, direct evidence of function-related conformational changes of any pLGIC in membrane and under native conditions is of utmost need to contextualize the increasing number of pLGIC structures and relate them to functional states.
Results
High-speed atomic force microscopy (HS-AFM) (17) is a unique tool for the investigation of membrane protein dynamics in a native-like environment (i.e., in membrane), in physiological buffer, and at ambient temperature and pressure, with ∼1-nm lateral, ∼0.1-nm vertical, and ∼100-ms temporal resolution (6–9). Furthermore, the tip–sample interaction of HS-AFM is minimized by the use of small cantilever oscillation amplitude (∼1 nm), minimized amplitude damping, and optimized feedback operation (17, 18), allowing us here to study GLIC single molecules over extended experimental durations up to 1 h. GLIC was purified and reconstituted at a low lipid-to-protein ratio, yielding densely packed vesicles with a diameter up to 500 nm (SI Appendix, Fig. S2). We do not actively control the insertion orientation of the channels during reconstitution, but think that the molecular architecture with hydrophobic α-helical TMD and hydrophilic β-sheet ECD drives the channels to all reconstitute in vesicles in an outside-out manner. To prepare for direct structural titration experiments, the HS-AFM fluid chamber was coupled over a tubing system to a high-precision, buffer-exchange pumping device equipped with syringes of pH 3.4 and pH 7.5 buffers (8, 19) (SI Appendix, Fig. S3).
HS-AFM Structural Titration Experiments.
The HS-AFM fluid cell is preequilibrated with 150 μL of pH 3.4 buffer that is slowly (flow rates: 5 μL⋅min−1, 10 μL⋅min−1, and 15 μL⋅min−1) exchanged with pH 7.5 buffer (Fig. 1A). Using multiple syringes, the buffer exchange conditions can be reversed, returning the fluid cell to pH 3.4 (Fig. 1A, dark gray traces). Throughout the pH titration, the HS-AFM continuously monitors the structure and dynamics of the membrane-embedded channels (Fig. 1A, Inset).
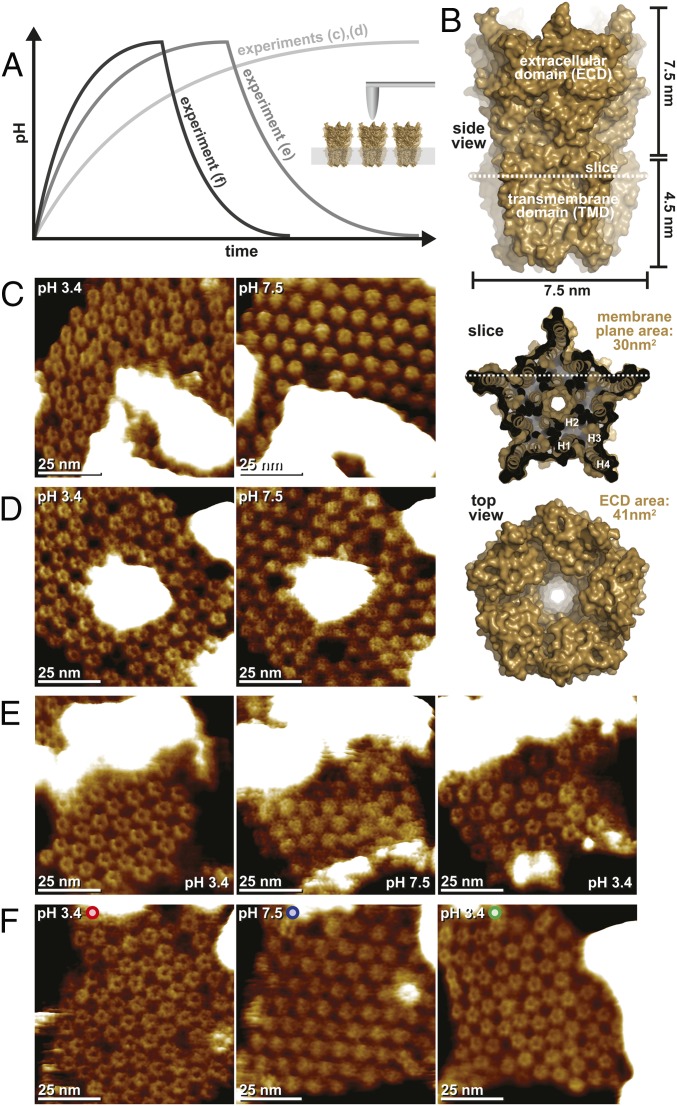
Fig. 1.
HS-AFM structural titration experiments with GLIC channels. (A) Schematic representation of the experimental procedure. (Inset) During HS-AFM imaging of membrane-embedded GLIC channels, the pH is gradually changed in the HS-AFM fluid chamber volume (150 μL) through a buffer exchange microfluidic device. The flow rate is adjusted to 5 μL⋅min−1 (experiments shown in C and D), 10 μL⋅min−1 (experiment shown in E), and 15 μL⋅min−1 (experiment shown in F). In the experiments shown in E and F, the pH was reversed to observe reversibility of assembly and structural changes. (B) Analysis of the GLIC architecture. A side view (Top), membrane slice close to the extracellular surface (Middle), and top view (Bottom) are shown. The dimensions of the molecule are indicated. High-resolution images (time averages over 5 s) of the initial pH 3.4 state (C and D, Left; open channel) and pH 7.5 state (C and D, Right; closed channel), as well as the reversed pH 3.4 state (E and F, Right; open channel) are shown.
For a better understanding of the HS-AFM results, we must first consider the general molecular features of GLIC (Fig. 1B). Viewed from the side, GLIC has an overall height of ∼12 nm, the TMD spans ∼4.5 nm in the membrane, and the ECD protrudes ∼7.5 nm (Fig. 1B, Top). In the membrane plane, the protein TMDs reach a maximum width at the level of Phe312, where the channels are star-shaped, with a 7.5-nm diameter (Fig. 1B, Middle). When viewed from the extracellular face, the ECDs have the same overall dimensions but form a regular pentagon exposing a central cavity (Fig. 1B, Bottom). On the intracellular face, the protein exposes only small loops close to the membrane surface. Thus, the sidedness of the molecule can unambiguously be assigned, and all further analysis concerns the protein viewed on the ECDs.
In all experiments, the GLIC HS-AFM topography compared favorably with the high-resolution structure in the active/desensitized state (pH 3.4). GLIC appeared as pentagons consisting of five subunits with a top-ring diameter of ∼3 nm and a central cavity into which the HS-AFM tip could penetrate ∼1.5 nm (Fig. 1 C–F, Left). While we imaged these molecules at a frame rate of 1 s−1 at submolecular resolution, the pH 7.5 buffer slowly titrated into the fluid chamber (and an equal volume of buffer was extracted from it; SI Appendix, Fig. S3). During this process, the channels underwent stunning dynamics (Movies S1 and S2): Molecules wiggle around their position; the ECDs narrow to the central axis; lateral diffusion increases; and, finally, the channels stabilize with apparently tightened ECDs, which only rarely allow the detection of a central cavity, in a hexagonal lattice (Fig. 1 C–F, Center). Such experiments at a buffer exchange flow rate of 5 μL⋅min−1 are very challenging as about 3,000 images are acquired over a period of almost 1 h (Fig. 1 C and D). Thus, we considered, that the observed rearrangements and conformational changes might be influenced by the tip–sample interaction over the extended experimental time. To exclude this, we designed a more complex experiment where a third syringe allows for a second titration back to the initial pH 3.4 buffer (SI Appendix, Fig. S3). As a result, the open-ECD conformation was recovered (Fig. 1 E and F, Right, and Movie S3) and, when using a buffer exchange flow rate of 15 μL⋅min−1, full reversibility of the process could be observed while continuously monitoring the same ∼70 molecules (Fig. 1F and Movie S4). The full reversibility of the assembly and structural changes shows that observations are not due to tip–sample or sample–surface interactions, but are intrinsic properties of the protein under investigation.
The Supramolecular Structure of GLIC in the Open and Closed Conformations.
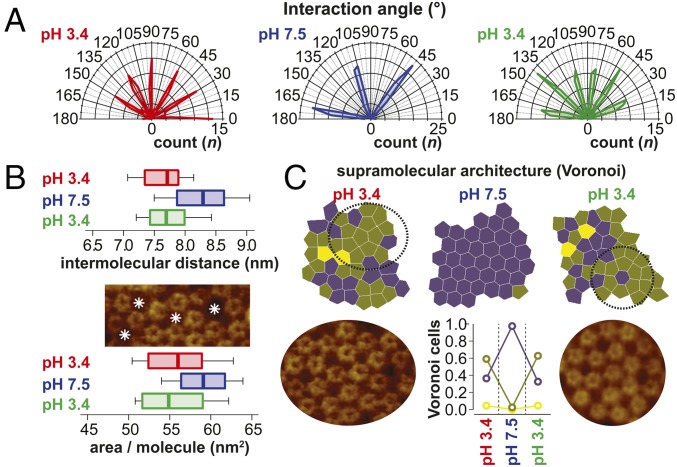
Visual inspection readily indicated important changes of the supramolecular assembly of the GLIC channels depending on the pH conditions, and hence their functional states (Fig. 1 C–F). To gain quantitative insights into these changes, we performed thorough nearest neighbor analysis. Plotting the intermolecular connection angles, we found that an apparently nonordered (we will come back to this) assembly with six dominant angular arrangements at pH 3.4 converges into a hexagonal packing with three “lattice” angles with a 60° difference at pH 7.5, a process that is fully reversible (Fig. 2A). It cannot be excluded that the pH-related rearrangements are influenced by the bilayer, but changes of membrane physics as a function of pH are minor and the abundance of ions in solution should shield charges on the headgroups. Plotting the nearest neighbor distances, we found that GLIC at pH 3.4 is significantly narrower than at pH 7.5 (Fig. 2B, Top). Indeed, at pH 3.4, the molecules space at ∼7.5 nm (in excellent agreement with the diameter of the active/desensitized conformation X-ray structure; Fig. 1B), while they are significantly larger, ∼8.4 nm, at pH 7.5. The active/desensitized conformation distance distribution displays some shorter interactions, <∼7.5 nm, certainly revealing molecules where subunits intercalate between subunits of the neighbor (Fig. 1B, Middle). In contrast, the closed conformation intermolecular distances are always >∼7.5 nm, indicating that the molecular diameter has overall increased. Next, we studied the average area occupied per molecule (Fig. 2B, Bottom and Materials and Methods). In the active/desensitized conformation, each GLIC occupies ∼56 nm2 of membrane, while they occupy ∼59 nm2 in the closed conformation. This corresponds to an ∼54% and ∼73% packing density in the membrane plane TMD (30 nm2) and on the ECD (41 nm2) level, respectively (Fig. 1B), for the active/desensitized conformation. In agreement, empty membrane areas are detected between the molecules (Fig. 2B, Middle). For the closed conformation, the protein density amounts to ∼51% and ∼69% in the membrane plane and on the ECD level, respectively. Thus, the closed conformation would, given it had the structure as described by X-ray crystallography, be even slightly less densely packed than the active/desensitized state, despite the fact that a hexagonal arrangement typically allows the densest packing of globally circular objects. This area occupation per molecule analysis indicates that we are looking at a system with a protein density and arrangement that are very similar to AchR in the postsynaptic membrane (20). Finally, we analyzed the architecture of the supramolecular structure. For this, we plotted the Voronoi cells, depicting the space that can be attributed to each molecule (Materials and Methods), and colored them according to the numbers of neighbors (Fig. 2C, Top). We found that while essentially all molecules have six neighbors in the closed conformation at pH 7.5, in the active/desensitized conformation at pH 3.4 in the beginning and the end of the structural titration experiment, only ∼40% had six neighbors and ∼60% had five neighbors (Fig. 2C, Bottom Center). The changes of the supramolecular assembly are a result of important rearrangements and lateral diffusion. On the one hand, this highlights the nativeness of the sample, where molecules have the freedom to diffuse and rearrange, and thus sample–surface interactions are minor. On the other hand, the high mobility of the molecules at pH 7.5 diminished the imaging resolution and the single subunits of the channels are not resolved in these conditions.
Fig. 2.
Analysis of the supramolecular structure of GLIC channels. (A) Polar coordinates plot of the intermolecular connections in Fig. 1F. At pH 7.5, the molecules engage in an ordered hexagonal packing with only three angles with 60° difference populated. At pH 3.4 (beginning and end of the experiment), the interaction angles populate at least six angles, indicating dense packing or a more complex order. (B) Box-and-whisker plots (0, 25, 50, 75, and 100 percentiles) of the intermolecular distances (Top) and area per molecule (Bottom) in Fig. 1F. Empty membrane areas are detected between molecules in pH 3.4 conditions as indicated by asterisks (Middle). (C, Top) Voronoi cell geometry analysis of molecules in the membrane in Fig. 1F. Molecules with five neighbors dominate (∼0.6) over molecules with six neighbors (∼0.4) in the pH 3.4 state. (C, Bottom Center) In the pH 7.5 state, essentially all molecules (∼1.0) have six neighbors. Intricate high-order supramolecular structures are found in the pH 3.4 state (Bottom Left and Right) (SI Appendix, Supplementary Information Text and Fig. S4).
Given that pLGICs are pentamers, the high percentage of channels in the active/desensitized state having five neighbors attracted our interest as a possible indication of specific protein–protein interactions. Close inspection of the Voronoi cell distributions indicated the existence of higher order patterns in these apparently randomly distributed molecules; indeed, analysis of the corresponding supramolecular architecture of the molecules in these regions revealed intricate ordered assemblies of up to 20 pentamers (Fig. 2C, Bottom Left and Right). Given that fivefold symmetry does not match any of the planar 2D symmetry space groups (21), probably the reason for the very moderate success of 2D-crystal growth of this family of membrane proteins (22), the fact of finding highly ordered supramolecular structures with conserved, regular protein–protein interactions shows that the active/desensitized state is favorably and specifically interacting with neighboring molecules (SI Appendix, Supplementary Information Text and Fig. S4). These interactions may have influence on channel function through intermolecular cooperativity (Discussion).
Structural Titration: GLIC Conformations as a Function of pH.
Next, we set out to analyze the structural changes during gating on the single-molecule level. For this, we adapted a method developed for electron microscopy (EM) that allowed unbiased assignment of molecular structures to environmental conditions (23) (SI Appendix, Fig. S5). In brief, an HS-AFM movie (Fig. 1F and Movie S4) was split into individual images, and the molecules in the images were automatically “picked.” This resulted in a total of about 120,000 molecules, with about 40 molecules in each of the 3,056 image frames. To characterize the conformational state of each particle, the particles were individually extracted from the images and sorted into five classes using a reference-free 2D classification algorithm in RELION (24) (SI Appendix, Fig. S5). Finally, each molecule was reassigned to the original movie frame from which it had been extracted and the relative abundance of molecules of each class was quantified. For this last step, the counts of molecules in bins of 100 movie frames were pooled to gather meaningful numbers of molecules (e.g., if a class populates a fraction of particles of 0.3, this corresponds to about 0.3⋅100⋅40 = 1,200 molecules). The entire processing was independently repeated several times to characterize the robustness of the classification algorithm to assign particles to particular states. In the HS-AFM experiments, the channels are embedded in a lipid bilayer that is deposited on atomically flat mica. Thus, the channels are confined to the membrane plane and their fivefold axis is perpendicular to it. While it cannot be excluded that the channels could wiggle a little in the membrane, they are rather densely packed, further stabilizing their vertical orientation. Thus, for 2D classification, we have indeed assumed that we look at “top views” only of the GLIC channels.
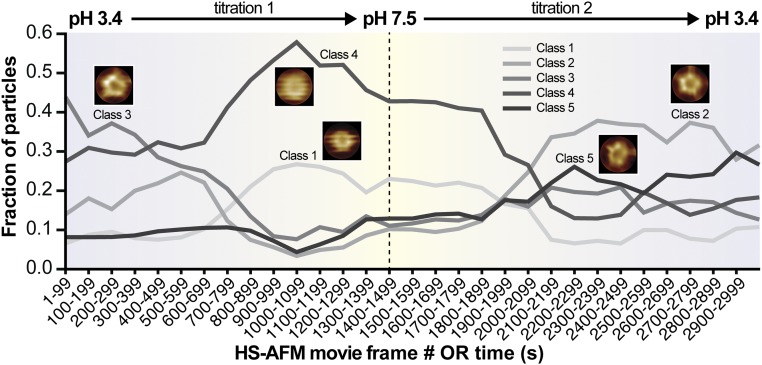
In the HS-AFM structural titration experiment, we aim at assigning conformational states to environmental conditions that correspond to relevant functional states (Fig. 3). It is furthermore important to note that we are looking at the same channels over time and under varying (reversible) conditions. We find in the beginning of the experiment, at pH 3.4, and at the end of the experiment, also at pH 3.4, two similar classes 2 and 3 dominating the conformational space. This result corroborates the reversibility of the experiment, as also assessed on the supramolecular level (Fig. 2C). Classes 2 and 3 depict a molecule in which all five protomer ECDs are well resolved, with a top-ring diameter of 2.8 ± 0.5 nm surrounding a central cavity. In principle, classes 2 and 3 should be the same, but unbiased classification separated them into two similar classes, with class 3 being slightly distorted likely due to piezo creep at very early stages of the experiment. Imaged at pH 3.4, we assign these molecules to the active/desensitized state. As the pH increased to ∼pH 6, in the vicinity of frame 700, class 4 emerges and dominates the conformational space. Another similar class 1 copopulates the higher pH regime, which reaches pH 7.5 in the vicinity of frame 1,500 (center of Fig. 3) and reaches out to about frame 2,100. Both classes 1 and 4 describe a rather featureless molecule reporting increased flexibility of the ECDs at physiological pH. While class 4 dominates the conformational space at pH 7.5, it is present throughout the movie; class 1 is almost exclusively found around pH 7.5 and displays narrowed ECDs to a top-ring diameter of ∼1 nm almost entirely constricting the access to the central cavity. Imaged at pH 7.5, we assign class 1 to the functionally closed state. As the pH crosses again at ∼pH 6 toward lower pH, the active/desensitized-state class 2 emerges and prevails. Interestingly, in this reactivation regime around frame 2,300, a class 5 with an ∼0.28 particle fraction [this class was always present in the background (fraction of particles = ∼0.1)] ranks second, and this class depicts a pentameric but rather asymmetric molecule (Discussion). The possibility that the HS-AFM tip interaction and image drift might be at the origin of this asymmetry must be considered, but seems unlikely because class 5 populates in the same movie frames that are dominated by the most symmetric class 2 molecules (right side of Fig. 3).
Fig. 3.
Structural titration of GLIC channels. Abundance of GLIC channel conformations as a function of HS-AFM movie frame acquisition number or time. During HS-AFM operation, the pH in the fluid chamber was gradually exchanged from pH 3.4 to pH 7.5 and back to pH 3.4. At the beginning and end of the movie, under active/desensitized-state conditions at pH 3.4, the very similar class 3 and class 2 molecules are most abundant. This structure depicts the ECDs as a flower-shaped pentamer surrounding a central cavity. In the middle of the movie, under closed-state conditions at pH 7.5, class 1 and class 4 molecules dominate, both revealing narrowed ECDs. An asymmetric but well resolved class 5 exists throughout the experiment, but peaks to almost 0.3 upon reexposure to low activating pH. OR, frame number or time, given that the images were acquired at 1 frame per second.
Each frame in these structural titration experiments is taken at a rate of 1 s−1. However, typical images comprise 250 scan lines; thus, each scan line is acquired in just 4 ms. Each individual GLIC channel will be contoured in ∼10 consecutive scan lines; thus, each molecule that we see and later analyze existed for just ∼40 ms in the conformation in which it is “captured” in the images. Thus, while we document the existence of a transient state in an average from frames recorded at 1 s per frame (during an experimental period of about 200 s, in frames 2,200–2,400), we have to recall that each molecule that contributes to this class was only there for just ∼40 ms. Thus, the slow titration experiment can potentially detect short-lived transient states.
This real-time structural titration experiment of the same individual molecules exposed to varying environmental conditions opens novel possibilities for structure function assignments in channel research.
Discussion
Using HS-AFM combined with a buffer exchange system (8), we present reversible supramolecular rearrangements and conformational changes of the proton-gated ion channel GLIC during pH changes. HS-AFM has the considerable advantage of imaging the channels directly in membrane at ∼60% membrane density, noncrystallized, and at ambient temperature and pressure in a physiological environment.
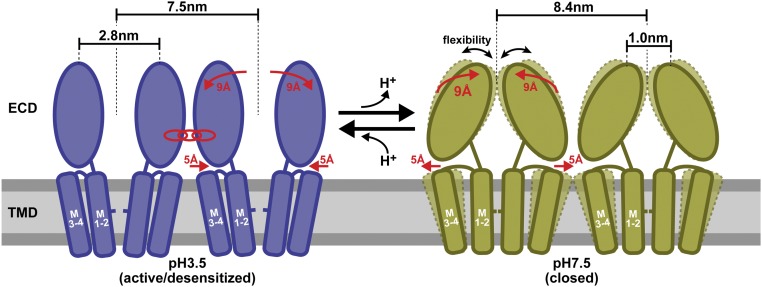
The direct HS-AFM structural titration describes the following scenario of GLIC supramolecular and conformational changes during pH gating (Fig. 4): At acidic pH, previously shown to induce channel opening (14), GLIC has a total diameter of 7.5 nm and open ECDs with a top-ring diameter of 2.8 nm (left side of Fig. 4), in excellent agreement with the active/desensitized-state X-ray structure (12). In this state, GLIC engages into favorable protein–protein contacts, as reflected by its tendency to have five nearest neighbors forming regular and predictable patterns in an ∼60% packed membrane (Fig. 2C and SI Appendix, Fig. S4). The membrane packing observed here is strongly reminiscent of the packing of AchR in the postsynaptic membrane (20, 25). It might thus be considered, although very difficult to study, that individual channels can influence the opening transition of their neighbors. Such an intermolecular cooperativity would enhance the ensemble channels’ response, for example, upon synaptic stimulation in mammalians. In support of such intermolecular interactions, cooperative activation has been shown in sodium channels in fast action potential onset (26), as evidenced for AMPA and NMDA receptors as a mechanism in learning (27) and even for oocyte-expressed P2X2 receptors (28). Most importantly, it has been evidenced for the related AchR (29). The potential physiological impact of such intermolecular potentiation is currently discussed (30).
Fig. 4.
Schematic representation of pH-dependent conformational changes in GLIC channels. At acidic pH in the activated/desensitized state (blue), the ECDs exhibit a 2.8-nm top-ring diameter and the channels engage in specific intermolecular interactions, while at physiological pH in the closed state (green), the ECDs collapse toward the channel fivefold axis and exhibit increased conformational flexibility. This transition is associated with an increase in the overall diameter of the pentamer by about 1 nm (from 7.5 to 8.4 nm), which concerns either the bottom part of the ECDs or the top part of the TMDs as schematically represented. The membrane plane is schematized by a horizontal rectangle (gray). The transient occurrence of asymmetric conformations between these two major states is evidenced.
Upon buffer titration toward physiological pH (i.e., the closed state) (13, 31), GLICs adopt an enlarged diameter of 8.4 nm and the top of the ECDs forms a contracted conformation. These changes occur when the titration experiment crosses roughly the pH 6 range (left side of Fig. 3). These conformational changes have so far never been observed and are opposite to current structural information on GLIC and pLGICs in general (11, 12, 32). Indeed, the structure of the closed state observed herein by HS-AFM is distinct from the closed-state structure solved by crystallography (SI Appendix, Fig. S1), indicating that larger conformational changes may occur during gating than were resolved by X-ray crystallography of 3D crystals. We lack a thorough explanation of the differences compared with the closed-state GLIC X-ray structure, but note that the channels in the 3D crystals make extensive crystal contacts, possibly hampering conformational rearrangements (SI Appendix, Fig. S6). It is notable, however, that the ECD–ECD interactions in the closed-state X-ray structure seem to be relatively weak, as documented by a high variability of the ECDs found among the four pentamers of the asymmetric unit cell. Thus, these ECDs might adopt a different conformation and/or fluctuate in solution without 3D-crystal contacts (13), in line with the increased mobility and flexibility of the channels observed here. It cannot be excluded that the HS-AFM imaging mechanism amplifies positional freedom of flexible protein parts in the closed state, but the fact that the topography of the active/desensitized state at the beginning and end of the two titrations favorably compares with the active/desensitized-state X-ray structure is strong evidence that the molecules suffer no damage during the entire experiments. Double electron–electron resonance has shown a widening of the peripheral transmembrane helix M4 in the desensitized state (15, 16), indicative that the transmembrane parts may also undergo larger movements than X-ray structures show, and potentially comparable to the diameter enlargement (7.5 to 8.4 nm) found in our experiments. Finally, in activating conditions, a qualitatively similar conformation to the one we detect for the GLIC closed state (right side of Fig. 4) has been described for the ivermectin-bound GluCl, where, as a result of rigid body movement, narrowing of the ECDs and enlargement of the TMs at the extracellular face were detected (32). Thus, similar conformational dynamics have been observed, but rather for opposite functional states. Together with the structural changes, the channels transit a period of increased lateral diffusion that results in a structural rearrangement where channels form a hexagonal lattice at physiological pH. The fact that the pentameric molecules arrange in a hexagonal packing indicates that the closed-state intermolecular interactions are nonspecific, in contrast to the active/desensitized state.
Titrating the GLIC channels back to acidic pH, to trigger channel opening again, we found perfect state reversibility in terms of both supramolecular architecture and molecular conformation. While we cannot “see into the interior” of the channel with HS-AFM, we hypothesize that the recovery of the 7.5-nm-wide state with expanded ECDs, matching the active/desensitized-state X-ray structure (12), is a manifestation of the channel opening. Interestingly, reference-free particle classification shows that asymmetric proteins populate the transition around pH 6 when titrating from physiological to acidic pH. Thus, we must consider a more complex state transition model that implies more states than just active/desensitized and closed states at pH 3.4 and pH 7.5, respectively (SI Appendix, Fig. S7): All channels should begin the titration experiments in the desensitized state because of prolonged exposure to acidic pH. As the pH increases during the first titration, channels reach the closed state. During the reverse titration to low pH, intermediate states may be found until the open state is reached, which is only short-lived, however, and quickly followed by adoption of the desensitized state. We thus hypothesize that the asymmetric GLIC during the titration from physiological to low (activating) pH could represent partially protonated channels where only a subset of the five subunits has undergone a conformational change, while the other subunits have not. In support of a more complex-state transition scheme, a preactive conformation has been evidenced using tryptophane/tyrosine fluorescence quenching assays (33).
In this context, one can note the recent structural description of another pentameric ion channel, CorA, that displayed a completely asymmetric state in the open, ligand-free state (34). It can thus be conceived that oligomeric ion channels generally can adopt asymmetric structures during gating, at least intermediately when the individual subunits are in structural transition.
We show that HS-AFM dynamic imaging during controlled buffer titration provides the envelopes of conformational changes of a Cys-loop pLGIC in a native-like environment, elucidating so far unidentified conformations and structural transitions. Furthermore, working at the “many-molecule level,” we provide evidence that the different conformational states also have different ways to interact with their molecular environment, potentially amplifying gating.
Materials and Methods
Protein Reconstitution.
GLIC was purified as described (14). In brief, pure (as judged by SDS/PAGE) protein sample at 0.44 mg/mL in 20 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, 2 mM Tris(2-carboxyethyl)phosphine (TCEP), and 0.02% dodecyl-maltoside (DDM) was mixed with a 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)/1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS) 8:1:1 lipid mixture or Escherichia coli lipids at a lipid-to-protein ratio of ∼1 (wt/wt) and complemented with a buffer containing 10 mM Hepes (pH 8.0), 200 mM KCl, 2 mM NaN3, and 0.025% DDM or 10 mM Na-acetate (pH 4.5), 200 mM KCl, 2 mM NaN3, and 0.025% DDM to a final protein concentration of 0.3 mg/mL The protein/lipid/detergent mixture was allowed to equilibrate for 2 h, and biobeads were then added for detergent removal overnight at room temperature. Progress of reconstitution was followed by negative-stain EM. Once intact densely packed vesicles were detected by EM, the biobeads were removed from the sample.
Sample Preparation.
Two microliters of reconstituted GLIC vesicles were injected onto a 1-mm-diameter freshly cleaved mica HS-AFM sample support covered with adsorption buffer [10 mM citric acid (pH 3.4), 200 mM KCl, 500 mM NaCl, and 5 mM MgCl2]. The vesicles were allowed to adsorb for 15 min, followed by ×10 washing with imaging buffer 10 mM citric acid (pH 3.4), 200 mM KCl, and 500 mM NaCl.
HS-AFM.
All images in this study were taken using an HS-AFM (SS-NEX; RIBM) microscope operated in amplitude modulation mode using optimized scan and feedback parameters. Short cantilevers (NanoWorld) with a nominal spring constant of 0.15 N/m, resonance frequency of 0.7 MHz, and quality factor of ∼1 in buffer were used. Typical image acquisition parameters were as follows: image size = 80 × 80 nm, full-color scale = 10 nm, frame size = 200 × 200 pixels, image acquisition rate = 1 frame per second, free amplitude = 1 nm, and setpoint amplitude = 0.9 nm. A constant pressure and constant flow pump (Harvard Instruments) was connected over silicon tubes to the fluid cell pool of the cantilever holder of the HS-AFM microscope for injecting and extracting buffers during HS-AFM observation. The two buffers that were exchanged during HS-AFM observation were 20 mM citric acid-NaOH (pH 3.4) and 100 mM KCl and 20 mM Hepes-HCl (pH 7.8) and 100 mM KCl. A tiny air bubble in the injecting buffer tube at the interface to the fluid cell initially separated the injecting buffer from the fluid cell pool and prevented fluid mixing into the fluid cell before active buffer exchange. The buffer exchange flow rate was constant and fixed to 5 μL⋅min−1, 10 μL⋅min−1, or 15 μL⋅min−1.
Buffer Exchange System.
For single-titration experiments, two silicon tubes, “buffer in” and “buffer out,” were attached to the fluid cell, which has a volume of 150 μL; injection and extraction were performed by two syringes fixed on the pumping system. The starting buffer 1 in the fluid cell contained 10 mM citric acid, 200 mM KCl, and 500 mM NaCl (pH 3.4) (SI Appendix, Fig. S3, blue), followed by gradual exchange with buffer 2 [10 mM Hepes, 200 mM KCl, and 500 mM NaCl (pH 7.8)] (SI Appendix, Fig. S3, red) at a constant flow rate of 5 μL⋅min−1. For reverse titration experiments, a second parallel injection syringe containing buffer 3 [10 mM citric acid, 200 mM KCl, and 500 mM NaCl (pH 3.4)] (SI Appendix, Fig. S3, green), the same buffer as buffer 1 (blue), was injected into the fluid cell following adjustment of a switch in the tubing system.
HS-AFM Movie Treatment.
HS-AFM movies were X–Y drift-corrected using the ImageJ plug-ins “template matching” and “slice alignment.” Image flattening was achieved by means of three-point plane fitting, followed by a median (0 order) horizontal line-by-line leveling. These steps were performed by in-house software routines developed in MATLAB (MathWorks).
Analysis of the Supramolecular Structure.
The analysis of the GLIC supramolecular architecture was performed in ImageJ (https://imagej.nih.gov/ij/). To assess the intermolecular angles and distances, the area per molecule, and the number of neighbors of each molecule of the GLIC channels in the membrane, we first picked the center X–Y coordinates of each molecule through cross-correlation searches and entered them into a peak list (ROI Manager in ImageJ). To calculate angles and distances, the Delaunay triangulation (Delaunay/Voronoi plug-in in ImageJ) was calculated from the coordinate peak lists. Delaunay triangulation draws lines connecting point coordinates in a way such that no other point can lie in between the two points that are connected. We term the lengths of the triangulation vectors “intermolecular distance” and use it as a characteristic of how close two neighboring molecules can approach each other. The direction of the triangulation vectors, between 0° and 180°, is used to assess the “interaction angle” between molecules (Fig. 2 A and B). Related to Delaunay triangulation, Voronoi tessellation (Delaunay/Voronoi plug-in in ImageJ) of the coordinate peak list was calculated. Borders of Voronoi cells are drawn in the middle and perpendicular to the connection between neighboring points (perpendicular to the Delaunay vectors), and thus delineate fields extending precisely to the middles between the analyzed point and all neighboring points. This process results in a network of fields with distinct area and geometry from which the number of neighboring fields is counted (Fig. 2 B and C).
Reference-Free Particle Classification and Analysis of Conformational Changes.
HS-AFM movies were disjointed into individual 1-s frames in TIFF (.tiff) format using ImageJ and converted to MRC (.mrc) format using tif2mrc (21). To process HS-AFM images using RELION (35), which was designed for analyzing electron micrographs, all contrast transfer function estimations and corrections were turned off. Approximately 2,000 GLIC particles were interactively selected from the images, extracted, and used for reference-free 2D classification in RELION. To minimize bias in particle selection, the interactive particle selection was performed on images selected throughout the movie. Following 2D classification, low-pass-filtered templates corresponding to active/desensitized and closed states were used for autopicking in RELION, generating 171,246 particles. Manual inspection of the images revealed that many of the selected particles corresponded to high-contrast noise in the images. To eliminate these false-positive results, all 171,246 particles were extracted and used as the input for reference-free 2D classification. Thirty-seven of the 50 classes from this classification corresponded to false-positive findings and were eliminated, resulting in 120,019 particles. Manual inspection of the images following false-positive removal indicated that few true particles were eliminated by this procedure and that most images contained ∼40 particles. Using the 120,019 particles as input, a second round of reference-free 2D classifications was performed specifying 2, 3, 5, 10, 20, and 50 output classes. Inspection of the resulting classes indicated that five classes yielded the most robust class separation. To assess the influence of pH on class distribution, the fraction of particles assigned to each of the five classes was determined for each of the 3,056 HS-AFM images. Due to the relatively low number of particles per image, bins of 100 frames were combined. To compare the robustness of the particle sorting, 2D classification was repeated three additional times and compared. The resulting class averages were similar, as were the fractions of particles that comprised the classes as a function of pH.
Supplementary Material
Acknowledgments
We thank Mohamed Chami and Henning Stahlberg for assistance with the sample preparation. This work was funded by the Agence Nationale de la Recherche (ANR) Labex Information Flow and Organization at the Membrane (INFORM) (Grant ANR-11-LABX-0054) and the ANR Initiative d’excellence Aix-Marseille (A*MIDEX) program (Grant ANR-11-IDEX-0001-02), by the European Research Council (Grant 310080, MEM-STRUCT-AFM) (to S.S.), and by the Josie Robertson Investigators Program and the Searle Scholars Program (to R.K.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 10198.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805621115/-/DCSupplemental.
References
- 1.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 2.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du J, Lü W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 6.Casuso I, et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat Nanotechnol. 2012;7:525–529. doi: 10.1038/nnano.2012.109. [DOI] [PubMed] [Google Scholar]
- 7.Chiaruttini N, et al. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyagi A, Chipot C, Rangl M, Scheuring S. High-speed atomic force microscopy shows that annexin V stabilizes membranes on the second timescale. Nat Nanotechnol. 2016;11:783–790. doi: 10.1038/nnano.2016.89. [DOI] [PubMed] [Google Scholar]
- 9.Ruan Y, et al. Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc Natl Acad Sci USA. 2017;114:1584–1588. doi: 10.1073/pnas.1616413114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 12.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 13.Sauguet L, et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci USA. 2014;111:966–971. doi: 10.1073/pnas.1314997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocquet N, et al. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445:116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 15.Velisetty P, Chalamalasetti SV, Chakrapani S. Structural basis for allosteric coupling at the membrane-protein interface in Gloeobacter violaceus ligand-gated ion channel (GLIC) J Biol Chem. 2014;289:3013–3025. doi: 10.1074/jbc.M113.523050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basak S, Schmandt N, Gicheru Y, Chakrapani S. Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel. eLife. 2017;6:e23886. doi: 10.7554/eLife.23886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ando T, Uchihashi T, Scheuring S. Filming biomolecular processes by high-speed atomic force microscopy. Chem Rev. 2014;114:3120–3188. doi: 10.1021/cr4003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagi A, Scheuring S. Automated force controller for amplitude modulation atomic force microscopy. Rev Sci Instrum. 2016;87:053705. doi: 10.1063/1.4950777. [DOI] [PubMed] [Google Scholar]
- 19.Rangl M, et al. Real-time visualization of conformational changes within single MloK1 cyclic nucleotide-modulated channels. Nat Commun. 2016;7:12789. doi: 10.1038/ncomms12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuser JE, Salpeter SR. Organization of acetylcholine receptors in quick-frozen, deep-etched, and rotary-replicated Torpedo postsynaptic membrane. J Cell Biol. 1979;82:150–173. doi: 10.1083/jcb.82.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 22.Rheinberger J, et al. Two-dimensional crystallization of the mouse serotonin 5-HT3A receptor. Micron. 2017;92:19–24. doi: 10.1016/j.micron.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Hite RK, MacKinnon R. Structural titration of Slo2.2, a Na+-dependent K+ channel. Cell. 2017;168:390–399.e11. doi: 10.1016/j.cell.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheres SHW. RELION: Implementation of a bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuber B, Unwin N. Structure and superorganization of acetylcholine receptor-rapsyn complexes. Proc Natl Acad Sci USA. 2013;110:10622–10627. doi: 10.1073/pnas.1301277110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naundorf B, Wolf F, Volgushev M. Unique features of action potential initiation in cortical neurons. Nature. 2006;440:1060–1063. doi: 10.1038/nature04610. [DOI] [PubMed] [Google Scholar]
- 27.Malenka RC. The long-term potential of LTP. Nat Rev Neurosci. 2003;4:923–926. doi: 10.1038/nrn1258. [DOI] [PubMed] [Google Scholar]
- 28.Ding S, Sachs F. Evidence for non-independent gating of P2X2 receptors expressed in Xenopus oocytes. BMC Neurosci. 2002;3:17. doi: 10.1186/1471-2202-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeramian E, Trautmann A, Claverie P. Acetylcholine receptors are not functionally independent. Biophys J. 1986;50:253–263. doi: 10.1016/S0006-3495(86)83459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi KH. Cooperative gating between ion channels. Gen Physiol Biophys. 2014;33:1–12. doi: 10.4149/gpb_2013076. [DOI] [PubMed] [Google Scholar]
- 31.Prevost MS, et al. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol. 2012;19:642–649. doi: 10.1038/nsmb.2307. [DOI] [PubMed] [Google Scholar]
- 32.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menny A, et al. Identification of a pre-active conformation of a pentameric channel receptor. eLife. 2017;6:e23955. doi: 10.7554/eLife.23955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthies D, et al. Cryo-EM structures of the magnesium channel CorA reveal symmetry break upon gating. Cell. 2016;164:747–756. doi: 10.1016/j.cell.2015.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.