Ryanodine receptors (RyRs) are large intracellular Ca2+ channels that provide the molecular basis of the process termed Ca2+-induced Ca2+ release (1). Ca2+ signaling through RyRs has been shown to be critical for skeletal, cardiac, and smooth muscle physiology (2) as well as for neurons (3) and secretory cells like pancreatic beta cells. RyRs were first seen in the electron microscope as ∼30-nm-size particles (4), and it had been known from EM studies that they form clusters, typically in the narrow “junctional” space between sarcolemma and sarcoplasmic reticulum (SR) membranes. Due to the inherent positive feedback of Ca2+-induced Ca2+ release, the size and shape of RyR clusters are expected to affect their functional activity. Indeed, detailed mathematical modeling suggests that RyR clustering is important for determining the excitability of RyR-mediated Ca2+ release (5, 6) and could, in principle, also affect the amount of Ca2+ released in microscopic release events termed Ca2+ sparks (7). The experimental study of RyR clustering has received a boost with the advent of optical superresolution microscopy, which has made the assessment of RyR cluster size more accessible by using essentially standard immunolabeling protocols. When first applied in cardiac muscle cells, this approach revealed a broad, approximately exponential RyR cluster size distribution (8), which had previously not been fully recognized. In PNAS, Pritchard et al. (9) describe changes in type 2 RyR (RyR2) clustering in smooth muscle of cerebral arteries in the mdx mouse model of Duchenne muscular dystrophy. The study is important because Pritchard et al. (9) identify these nanoscale receptor clustering changes as the likely source of cerebral muscular dysfunction in the mdx mouse.
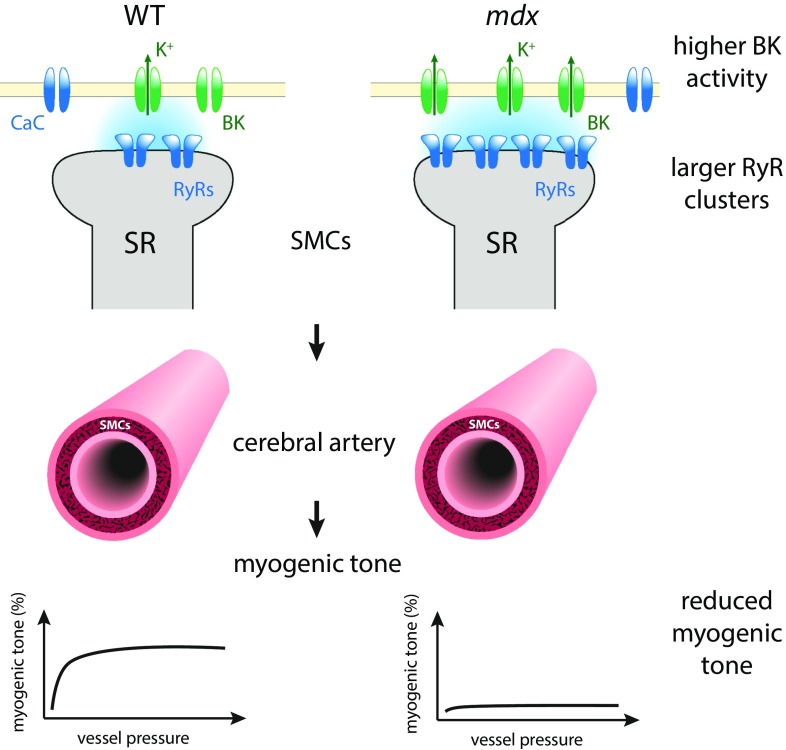
Different from striated muscle in which RyR-mediated Ca2+ release mainly contributes to the cell-wide Ca2+ transient to activate contraction, in cerebral smooth muscle, the functional role of RyR2 clusters involves the close proximity to Ca2+-activated K+ (BK) channels in the opposing sarcolemma (Fig. 1). Microscopic Ca2+ release events from RyR clusters, seen in the confocal microscope as Ca2+ sparks, stimulate large transient K+ outward currents that hyperpolarize the cell membrane (10). Pritchard et al. (9) find that, in smooth muscle cells (SMCs) from mdx mice, the mean projected area of RyR2 clusters—a proxy for RyR2 cluster size (the number of RyR2s in a cluster)—was increased in superresolution images of RyR2 labeling compared with control SMCs. Functionally, these larger RyR2 clusters resulted in more frequent Ca2+ sparks, and these had a larger signal mass (a measure of the amount of Ca2+ released). Consistent with the increased frequency and size of Ca2+ sparks, spontaneous Ca2+-activated K+ channel activity was elevated in SMCs from mdx mice. The physiological role of SMCs involves the constriction and dilation of blood vessels to regulate blood flow. The physiological consequences of the increased K+ channel activity were explored by Pritchard et al. (9) by investigating pressure responses in cerebral arteries from mdx mice. Cerebral pial arteries from mdx mice exhibited a reduced myogenic tone—that is, a change in the SMC-mediated response of arteries to increased pressure, which normally involves vasoconstriction at elevated pressures. This response was blunted in SMCs from mdx mice but could be restored to a degree via partial RyR2 blockade with tetracaine. This is consistent with a crucial role of RyR2 cluster size in causing this defect, as partial RyR2 blockade reduces the effective size of clusters.
Fig. 1.
Pritchard et al. (9) find larger RyR2 clusters in cerebral SMCs from mdx mice. These enlarged RyR2 clusters are associated with more frequent (and slightly larger) Ca2+ sparks, resulting in increased spontaneous BK activity, which tends to hyperpolarize the cell membrane. The physiological correlate of these changes is a reduced myogenic tone in cerebral arteries from mdx mice—that is, a reduced vasoconstriction response to pressurizing these vessels. Also shown are Ca2+ channels (CaC) in the sarcolemmal membrane.
Few prior studies, generally conducted with cardiac muscle cells, have linked changes in RyR clustering to physiological changes. Macquaide et al. (11) observed a reduced distance between RyR clusters (but no change in mean cluster size) in persistent atrial fibrillation. This was correlated with higher Ca2+ spark frequency and altered Ca2+ spark time course. Munro et al. (12) investigated an overexpression model of the cardiac junction protein junctophilin 2 (JPH2), which was associated with increased RyR cluster size, but functionally, this resulted in lower Ca2+ spark frequency and smaller sparks despite a larger SR load (a measure of the Ca2+ content of the SR).
It is exciting that the study by Pritchard et al. (9) provides further support for the idea that the details of RyR2 clustering at the nanoscale affect cell- and tissue-wide physiology. The emerging field of nanophysiology aims to make this connection explicit by developing robust assays of protein nanoscale distribution and associated functional cell biophysics and physiology. It should be noted that the assays that we use to measure protein clustering are still relatively new and involve a complex set of optical and chemical processes to obtain and then quantitatively analyze superresolution images. Current assays often make simplifying assumptions in analyzing and interpreting the data, such as using 2D analysis of a structure that is inherently 3D or assuming a protein density that is not directly measured. The latter assumption is typically required because the single-molecule localizations do not map one-to-one to receptor counts; the methodologies of photoactivated localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), ground-state depletion microscopy followed by individual molecule return (GDSIM), and related localization microscopy are not per se quantitative [nor are alternative stimulated emission depletion (STED)-based superresolution measurements (11, 13)]. However, they can be made more quantitative with careful calibration measurements (14, 15), and the recent DNA-based point accumulation for imaging in nanoscale topography (DNA-PAINT) approach directly supports a quantitative analysis modality (16, 17). Invariably, one needs to work quite a bit harder to make superresolution imaging fully quantitative; however, careful calibration will increase the confidence in new findings relating to RyR cluster size. In any case, it will be an important task for future work to further test and, where necessary, improve the robustness of superresolution-based clustering assays, ruling out potential artifacts that may arise from technical aspects of the broad and continuously growing range of superresolution approaches now available.
The consequences of changes in RyR clustering for functional behavior are not always easy to predict. On first sight, it may seem obvious that larger RyR clusters give rise to larger Ca2+ release events, and Pritchard et al. (9) indeed observe increased spark signal mass. However, the relationship between Ca2+ spark properties (frequency and size) and RyR cluster size is, in general, complicated by local ultrastructure, volume, and Ca2+ content of the terminal SR components, as well as any changes that may affect RyR gating and open probabilities. For example,in a recent study aiming to correlate RyR cluster size and spark amplitude in cardiac muscle (18), the authors observed a rapidly saturating flat relationship between the two quantities. In the JPH2 overexpression model, larger RyR clusters were associated with smaller and less frequent sparks (12), presumably because of a net inhibitory
It is exciting that the study by Pritchard et al. provides further support for the idea that the details of RyR2 clustering at the nanoscale affect cell- and tissue-wide physiology.
effect of a larger fraction of RyRs associated with JPH2. As a bottom line, the relationship between cluster size and functional release will have to be probed in each study and may vary widely across models and cell types. The use of mathematical modeling to formulate detailed mechanistic hypotheses for the underlying complex biophysical scenarios will remain essential, as are experimental data beyond RyR cluster size—for example, for terminal SR volumes, structural geometries, and, as in Pritchard et al. (9), data on effector molecules such as the BK channel.
Given the interesting findings by Pritchard et al. (9), two questions naturally arise: what mechanisms drive changes in RyR2 clustering and what molecular signaling cascades are responsible for the alterations observed in the mdx mouse? How lack of dystrophin, ultimately the source of alterations in the mdx mouse, triggers these changes is not clear, as Pritchard et al. (9) rightly state. They raise two possibilities for mechanisms downstream of lack of dystrophin: (i) increased RyR2 cross-linking as a result of oxidative modifications and (ii) a microtubule-dependent mechanism in the face of microtubule network disorganization in the mdx mouse. In general, at this stage, we have very little direct evidence of how molecular RyR clustering is controlled [although see the study by Asghari et al. (19), which identifies an effect of phosphorylation on RyR orientation in clusters, but not cluster size per se], and by necessity, we currently mostly have to resort to speculation. This gap will need to be closed if we are to make progress in achieving a deeper mechanistic understanding and ultimately obtain the insight needed for targeted manipulation of RyR clustering properties. Closely related is the question of how dynamic RyR cluster arrangements are. After all, current studies of clustering have mostly used fixed preparations. Some first information on the dynamic nature of clusters has recently been obtained using diffraction-limited imaging (20); corresponding superresolution imaging studies capable of resolving RyR cluster outlines in live cells are as-yet outstanding but may be on the horizon (21).
The remarkable advances in light microscopy and related imaging modalities over recent years are now enabling receptor studies at the molecular scale in intact cells and tissues. There is a distinct prospect that we will unravel currently unknown mechanisms of modulating cell biology via changes in protein clustering in health and disease, which could lead to novel treatment strategies. The study by Pritchard et al. (9) is an important step in this direction.
Acknowledgments
This research was supported by the Engineering and Physical Sciences Research Council of the United Kingdom (Grant EP/N008235/1) and Biotechnology and Biological Sciences Research Council Grants BB/P026508/1 and BB/R022127/1.
Footnotes
The author declares no conflict of interest.
See companion article on page E9745.
References
- 1.Fabiato A. Time and calcium dependence of activation and inactivation of calcium-induced release of calcium from the sarcoplasmic reticulum of a skinned canine cardiac Purkinje cell. J Gen Physiol. 1985;85:247–289. doi: 10.1085/jgp.85.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franzini-Armstrong C, Protasi F. Ryanodine receptors of striated muscles: A complex channel capable of multiple interactions. Physiol Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 3.Manita S, Ross WN. Synaptic activation and membrane potential changes modulate the frequency of spontaneous elementary Ca2+ release events in the dendrites of pyramidal neurons. J Neurosci. 2009;29:7833–7845. doi: 10.1523/JNEUROSCI.0573-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzini-Armstrong C. Studies of the triad I. Structure of the junction in frog twitch fibers. J Cell Biol. 1970;47:488–499. doi: 10.1083/jcb.47.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannell MB, Soeller C. Numerical analysis of ryanodine receptor activation by L-type channel activity in the cardiac muscle diad. Biophys J. 1997;73:112–122. doi: 10.1016/S0006-3495(97)78052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker MA, et al. Superresolution modeling of calcium release in the heart. Biophys J. 2014;107:3018–3029. doi: 10.1016/j.bpj.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: Elementary events underlying excitation–contraction coupling in heart muscle. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 8.Baddeley D, et al. Optical single-channel resolution imaging of the ryanodine receptor distribution in rat cardiac myocytes. Proc Natl Acad Sci USA. 2009;106:22275–22280. doi: 10.1073/pnas.0908971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard HAT, Pires PW, Yamasaki E, Thakore P, Earley S. Nanoscale remodeling of ryanodine receptor cluster size underlies cerebral microvascular dysfunction in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2018;115:E9745–E9752. doi: 10.1073/pnas.1804593115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaggar JH, Porter VA, Lederer WJ, Nelson MT. Calcium sparks in smooth muscle. Am J Physiol Cell Physiol. 2000;278:C235–C256. doi: 10.1152/ajpcell.2000.278.2.C235. [DOI] [PubMed] [Google Scholar]
- 11.Macquaide N, et al. Ryanodine receptor cluster fragmentation and redistribution in persistent atrial fibrillation enhance calcium release. Cardiovasc Res. 2015;108:387–398. doi: 10.1093/cvr/cvv231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro ML, et al. Junctophilin-2 in the nanoscale organisation and functional signalling of ryanodine receptor clusters in cardiomyocytes. J Cell Sci. 2016;129:4388–4398. doi: 10.1242/jcs.196873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner E, et al. Stimulated emission depletion live-cell super-resolution imaging shows proliferative remodeling of T-tubule membrane structures after myocardial infarction. Circ Res. 2012;111:402–414. doi: 10.1161/CIRCRESAHA.112.274530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puchner EM, Walter JM, Kasper R, Huang B, Lim WA. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc Natl Acad Sci USA. 2013;110:16015–16020. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanacchi FC, et al. A DNA origami platform for quantifying protein copy number in super-resolution. Nat Methods. 2017;14:789–792. doi: 10.1038/nmeth.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jungmann R, et al. Quantitative super-resolution imaging with qPAINT. Nat Methods. 2016;13:439–442. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayasinghe I, et al. True molecular scale visualization of variable clustering properties of ryanodine receptors. Cell Reports. 2018;22:557–567. doi: 10.1016/j.celrep.2017.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galice S, Xie Y, Yang Y, Sato D, Bers DM. Size matters: Ryanodine receptor cluster size affects arrhythmogenic sarcoplasmic reticulum calcium release. J Am Heart Assoc. 2018;7:e008724. doi: 10.1161/JAHA.118.008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asghari P, et al. Nonuniform and variable arrangements of ryanodine receptors within mammalian ventricular couplons. Circ Res. 2014;115:252–262. doi: 10.1161/CIRCRESAHA.115.303897. [DOI] [PubMed] [Google Scholar]
- 20.Hiess F, et al. Dynamic and irregular distribution of RyR2 clusters in the periphery of live ventricular myocytes. Biophys J. 2018;114:343–354. doi: 10.1016/j.bpj.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Y, Manfra O, Li J, Shen X, Louch WE. Live cell PALM techniques for super resolution imaging of murine cardiac myocytes. Biophys J. 2018;114:549a. [Google Scholar]