Significance
The neural substrate for “sleepiness,” as well as the molecular mechanisms determining daily sleep amounts, remain mysterious. Here, we found that a single amino acid, S551, of the protein kinase SIK3 has a crucial role in the regulation of sleep need and daily nonrapid-eye movement sleep amounts. Importantly, S551 in SIK3 is evolutionally conserved as a PKA phosphorylation site from nematodes and fruit flies to mice and humans. These findings implicate SIK3 in the molecular basis of homeostatic sleep/wake regulation.
Keywords: protein kinase, sleep debt, behavioral genetics, electroencephalography, electromyography
Abstract
Sleep is an evolutionally conserved behavior from vertebrates to invertebrates. The molecular mechanisms that determine daily sleep amounts and the neuronal substrates for homeostatic sleep need remain unknown. Through a large-scale forward genetic screen of sleep behaviors in mice, we previously demonstrated that the Sleepy mutant allele of the Sik3 protein kinase gene markedly increases daily nonrapid-eye movement sleep (NREMS) amounts and sleep need. The Sleepy mutation deletes the in-frame exon 13 encoding a peptide stretch encompassing S551, a known PKA recognition site in SIK3. Here, we demonstrate that single amino acid changes at SIK3 S551 (S551A and S551D) reproduce the hypersomnia phenotype of the Sleepy mutant mice. These mice exhibit increased NREMS amounts and inherently increased sleep need, the latter demonstrated by increased duration of individual NREMS episodes and higher EEG slow-wave activity during NREMS. At the molecular level, deletion or mutation at SIK3 S551 reduces PKA recognition and abolishes 14-3-3 binding. Our results suggest that the evolutionally conserved S551 of SIK3 mediates, together with PKA and 14-3-3, the intracellular signaling crucial for the regulation of daily sleep amounts and sleep need at the organismal level.
Across animal species, from invertebrates to vertebrates, sleep behavior is regulated in a homeostatic manner (1–3). A homeostatic sleep need reflects the “integral” of the quantity and quality of waking in the recent past. In mammals, the EEG spectral power in the δ-range frequency (1–4 Hz) during nonrapid-eye movement sleep (NREMS) has been considered the best marker of sleep need. In addition to NREMS δ-power density, it has been reported that mean duration of individual NREMS episodes is an additional index for sleep need in mice (4). However, the cellular/molecular mechanism for homeostatic sleep/wake regulation and the neuronal basis for sleep need remain unknown. Our large-scale forward genetic screen of the sleep/wake phenotype in ethylnitrosourea-mutagenized mice identified a splicing mutation in the Sik3 protein kinase gene, termed Sleepy mutation, which causes marked hypersomnia (increased NREMS amounts) due to an increase in inherent sleep need (5). Our recent phosphoproteomic analyses indicate that Sleepy mutant brains, as well as sleep-deprived brains, exhibit a hyperphosphorylated state of a mostly synaptic subset of neuronal proteins (6). A pan-SIK–specific kinase inhibitor reduces both the hyperphosphostate and the elevated sleep need in Sleepy mutant mice, as well as sleep-deprived WT mice. SIK3 has a serine-threonine kinase domain at the N terminus and a protein kinase A (PKA) recognition site S551 in the middle portion (7). In the Sleepy mutants, the skipping of exon 13 (ΔEx13) results in an in-frame deletion of 52 amino acids encompassing S551 (5). Thus, we hypothesized that S551 of Sik3 has an essential role for the regulation of daily sleep amounts and sleep need. To examine this hypothesis, we herein generated mutant mice with single amino acid substitutions in SIK3, S551A, and S551D, by using the CRISPR/Cas9-based genome editing. These mutants are subjected to further biochemical experiments and EEG/EMG-based sleep recordings.
Results
S551A Mutation in Sik3 Increases NREMS Amounts.
To confirm whether the genome-edited S551A and S551D mice showed intact transcription of the Sik3 mRNA, we first performed RT-PCR and quantitative PCR (qPCR) using RNA extracted from the cortex of WT, Sleepy, S551A, and S551D mice. As previously reported (5), the mRNA size of heterozygous and homozygous Sleepy mutants was altered because of exon 13 skipping (SI Appendix, Fig. S1A). In contrast, both S551A and S551D mutants showed the same transcriptional patterns of Sik3 mRNA compared with WT mice (SI Appendix, Fig. S1A). In addition, the qPCR results showed that there was no difference in Sik3 mRNA expression levels among littermate WT, S551A, and S551D mice (SI Appendix, Fig. S1B). To examine the SIK3 protein expression in S551A and S551D mutant mice, we next performed Western blotting using an antibody that recognizes the C terminus of SIK3. The anti-SIK3 antibody detected the same size and amount of SIK3 protein in both S551A and S551D mutants compared with WT littermate mice (SI Appendix, Fig. S1C). Taking these data together, we confirmed that these single amino acid changes at S551 had no appreciable effect on the transcription or translation of the Sik3 gene.
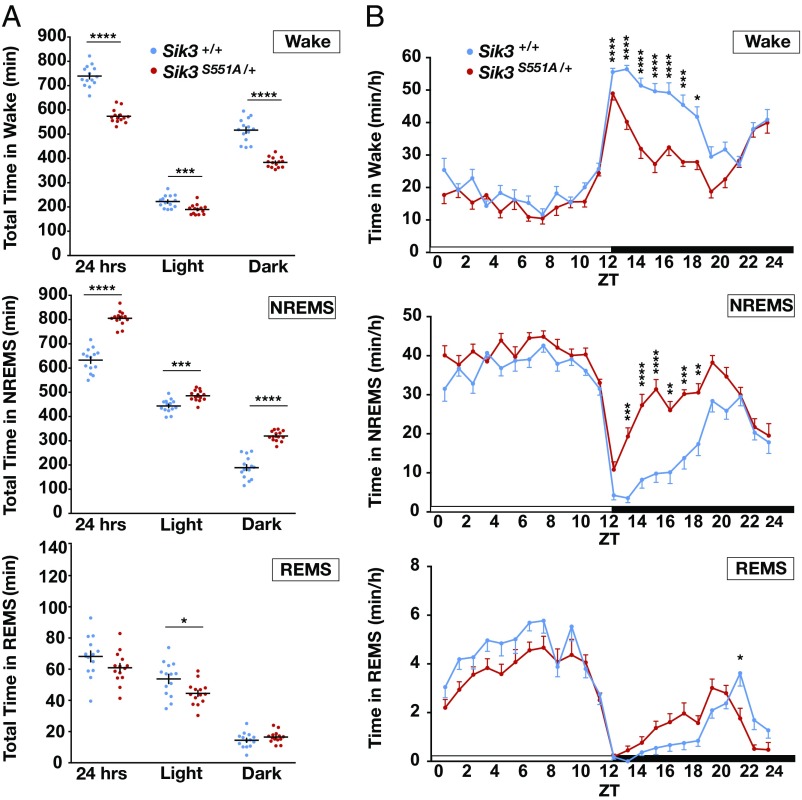
The S551A mutants exhibited a reduced wake amount and an increased NREMS amount in both the light and dark phases (Fig. 1A), with the hypersomnia phenotype more prominent in the dark phase (Fig. 1B). The opposite effect was observed for wakefulness: S551A mutants displayed decreased wakefulness in both the light and dark phases (Fig. 1A). Although there was a slight difference in total REMS amounts during light phase, there was no difference in total REMS amounts over 24 h, suggesting a NREMS-specific phenotype in S551A mutants. Analysis of the state transition frequencies among wakefulness, NREMS, and REMS over 24 h showed that S551A mutants had a reduced number of transitions from NREMS to REMS (SI Appendix, Fig. S2A). This suggests that S551A mutants tended to stay in NREM state longer than the WT did, which was consistent with the increased NREMS amount.
Fig. 1.
Sleep/wake phenotype of S551A mutant mice. (A) Total time in wakefulness, NREMS, REMS. n = 14 for each genotype. *P < 0.05, ***P < 0.001, ****P < 0.0001, two-tailed unpaired t test. (B) Circadian variation in wakefulness, NREMS and REMS. n = 14 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. Wake: the main effect of genotype, F(1, 26) = 143.8, ****P < 0.0001. NREMS: the main effect of genotype, F(1, 26) = 126, ****P < 0.0001. REMS: the main effect of genotype, F(1, 26) = 2.694, P = 0.1128. NREMS/REMS/wakefulness stages are assigned in 20-s epochs. Data are presented as the mean ± SEM.
S551A Mutation in Sik3 Increases Sleep Need.
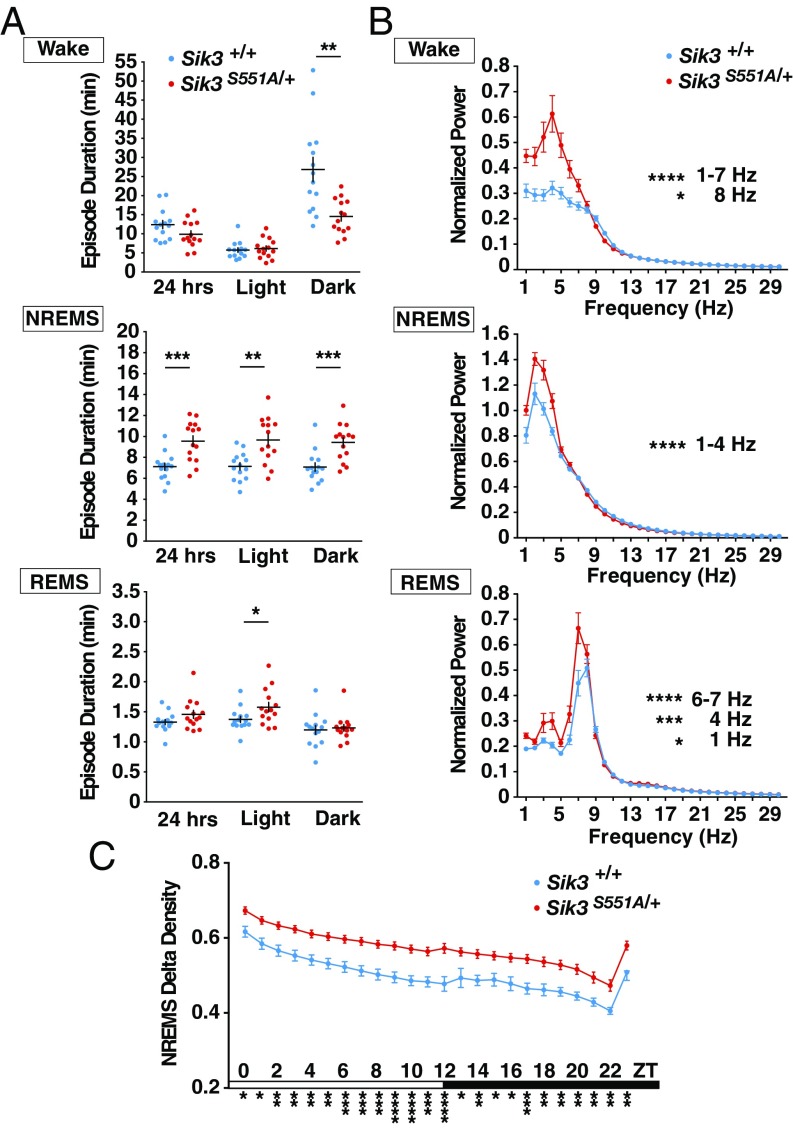
S551A mutants exhibited a longer mean duration of NREMS episodes in both the light and dark phases, but there was no change in the mean REMS episode duration (Fig. 2A). The episode duration of wakefulness was decreased in the dark phase compared with that of the WT littermates (Fig. 2A). These results were consistent with the increased total NREMS amount and decreased total wake amount in S551A mutants. The increased NREMS episode duration suggests an increased homeostatic sleep need of the S551A mutants.
Fig. 2.
Sleep/wake episode durations and EEG power spectra of S551A mutants. (A) Episode durations of wakefulness, NREMS and REMS. n = 14 for each genotype. *P < 0.05 **P < 0.01 ***P < 0.001, two-tailed unpaired t test and Mann–Whitney U test. (B) EEG power spectra in wakefulness, NREMS and REMS states. n = 14 for each genotype. *P < 0.05, ***P < 0.001, ****P < 0.0001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. Wake: the main effect of genotype, F(1, 26) = 10.83, **P = 0.0029. NREMS: the main effect of genotype, F(1, 26) = 8.521, **P = 0.0072. REMS: the main effect of genotype, F(1, 26) = 5.978, *P = 0.0216. (C) NREMS δ-power density of S551A mutants. n = 14 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. The main effect of genotype, F(1, 26) = 19.223, ***P = 0.0002. NREMS/REMS/wakefulness stages are assigned in 20-s epochs. Data are presented as the mean ± SEM.
The EEG spectral analysis of S551A mutant mice during NREMS showed elevated power in the δ-range (1–4 Hz) (Fig. 2B). The EEG δ-power during NREMS has been regarded as the best marker reflecting the ongoing level of sleep need. We further analyzed the change in the NREMS δ-density over 24 h, the ratio of EEG δ-power (1–4 Hz) to overall power (1–30 Hz). The hourly plot of NREMS δ-density revealed that the S551A mutants exhibited significantly increased NREMS δ-density over 24 h compared with that of the WT littermates (Fig. 2C), indicating that the baseline sleep need was inherently increased in the S551A mutants. Additionally, the EEG spectral analysis of S551A mutant mice during wakefulness showed increased low-frequency power (1–7 Hz) (Fig. 2B), suggesting the possible association between increased sleep need and the local sleep state in cortical regions during wakefulness (8). Thus, the increased slow-wave activity during wakefulness suggest the possibility that S551A mutants may suffer from sleepiness even during wakefulness. This may also explain the decreased episode duration of wakefulness during a dark period (Fig. 2A) when normally the mice are active, suggesting that S551A mutants are unable to normally maintain wakefulness due to sleep pressure. During REMS, the power in the 6–7 Hz range was increased in S551A mutant mice (Fig. 2B), although the significance of this finding was unclear.
Importantly, the single amino acid change S551A was sufficient to reproduce the hypersomnia phonotype of the ethylnitrosourea-induced Sleepy mutants lacking 52 amino acids (5), in terms of increased NREMS amounts [S551A heterozygotes vs. Sleepy heterozygotes: not significant (n.s.), P = 0.84; S551A WT vs. Sleepy WT: n.s., P = 0.86; one-way ANOVA followed by Tukey’s test] and lengthened NREMS episode durations (S551A vs. Sleepy: n.s., P = 0.61; S551A WT vs. Sleepy WT: n.s., P = 0.93). Thus, S551 in exon 13 of the Sik3 gene plays a crucial role in determining the daily amount of sleep and the level of sleep need.
S551D Mutation in Sik3 Increases NREMS Amounts and Sleep Need.
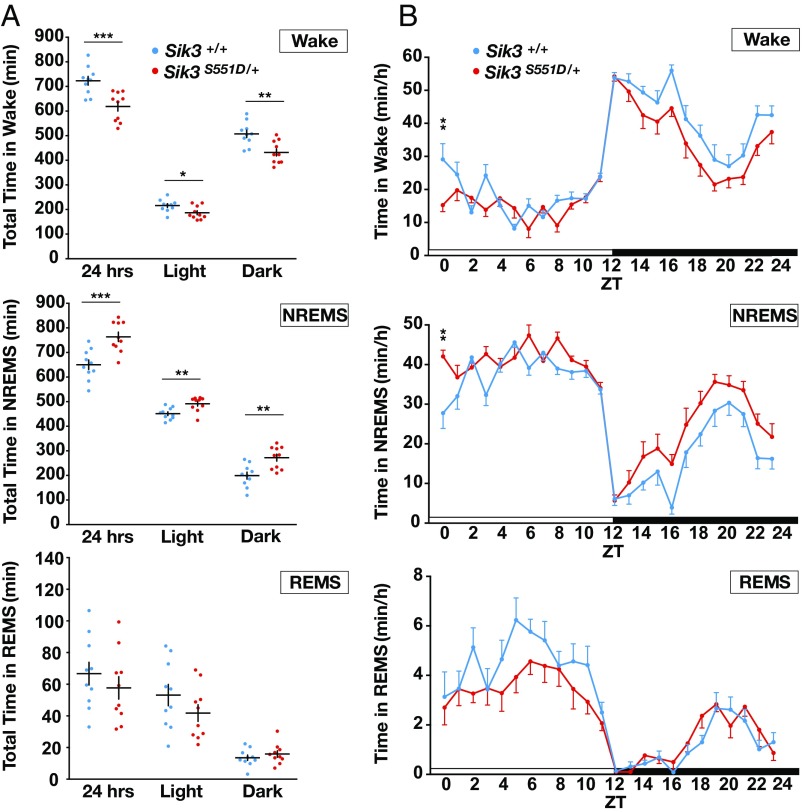
S551D mutants exhibited reduced wake amounts and increased NREMS amounts in both the light and dark phases (Fig. 3A), similar to the phenotype of S551A mutants. S551D mutants also displayed decreased wakefulness in both the light and dark phases (Fig. 3B). However, there was no difference in the total REMS amount over 24 h, indicating a NREMS-specific change. For the transitions to wakefulness, NREMS, and REMS over 24 h, S551D showed no significant changes in the transition profiles compared with WT littermate mice (SI Appendix, Fig. S2B).
Fig. 3.
Sleep/wake phenotype of S551D mutant mice. (A) Total amount in wakefulness, NREMS, REMS. n = 10 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired t test and Mann–Whitney U test. (B) Circadian variation in wakefulness, NREMS, and REMS. n = 10 for each genotype. **P < 0.01, two-way repeated measure ANOVA followed by Bonferroni’s multiple comparisons test. Wake: the main effect of genotype, F(1, 18) = 16.06, ***P < 0.001. NREMS: the main effect of genotype, F(1, 18) = 17.15, ***P < 0.001. REMS: the main effect of genotype, F(1, 18) = 0.7886, P = 0.3862. NREMS/REMS/Wakefulness stages are assigned in 20-s epochs. Data are presented as the mean ± SEM.
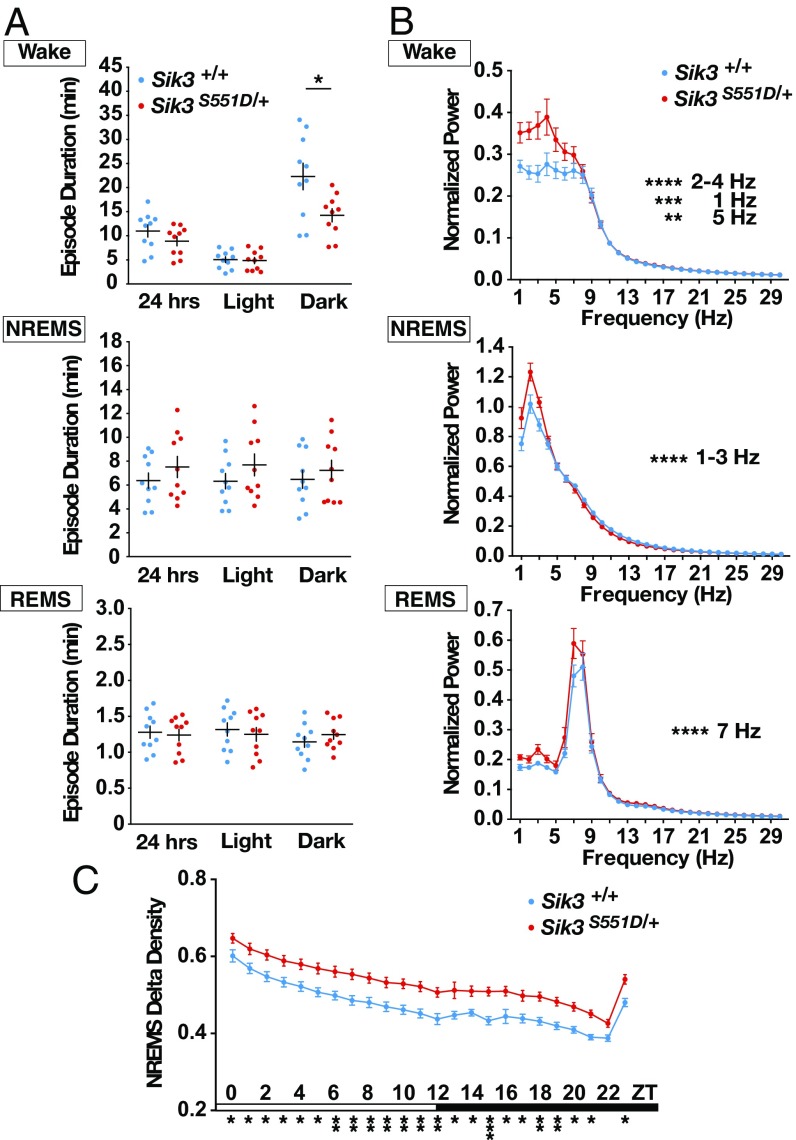
S551D mutants displayed shorter-wake episode duration in the dark phase but no change in REMS episode duration (Fig. 4A). The episode duration of NREMS tended to increase, but no statistically significant difference was observed compared with the WT littermates (Fig. 4A). The EEG spectral profile of S551D mutants during NREMS showed increased power in the low-frequency range (1–3 Hz) compared with the WT littermates (Fig. 4B). Moreover, the NREMS δ-power density of S551D mutants was significantly increased over 24 h (Fig. 4C), suggesting that the daily sleep need of S551D mutants was inherently increased. In addition, the EEG spectral analysis of S551D mutant mice during wakefulness showed increased low-frequency power (1–5 Hz) (Fig. 4B), along with the decreased episode duration of wakefulness (Fig. 4A). For REMS, the power in the 7-Hz range was significantly increased in S551D mutant mice (Fig. 4B).
Fig. 4.
Sleep/wake episode durations and EEG power spectra of S551D mutants. (A) Episode durations of wakefulness, NREMS and REMS. n = 10 for each genotype. *P < 0.05, two-tailed unpaired t test and Mann–Whitney U test. (B) EEG power spectra in wakefulness, NREMS, and REMS states. n = 10 for each genotype. **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. Wake: the main effect of genotype, F(1, 18) = 6.562, *P = 0.0196. NREMS: the main effect of genotype, F(1, 18) = 1.459, P = 0.2427. REMS: the main effect of genotype, F(1, 18) = 4.451, *P = 0.0491. (C) NREMS δ-power density of S551D mutants. n = 10 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, two-way repeated-measure ANOVA followed by Bonferroni’s multiple comparisons test. The main effect of genotype, F(1, 18) = 14.285, **P = 0.0014. NREMS/REMS/wakefulness stages are assigned in 20-s epochs. Data are presented as the mean ± SEM.
Compared with S551A mutants, S551D mutants showed similarly increased sleep amounts and sleep need, demonstrated by increased NREMS amounts over 24 h (S551A mutants vs. S551D mutants: n.s., P = 0.20; S551A WT vs. S551D WT: n.s., P = 0.83) and increased NREMS episode durations (S551A vs. S551D: n.s. P = 0.08; S551A WT vs. S551D WT: n.s., P = 0.81). Thus, although we had started with the assumption that the S551D mutation would mimic constitutively phosphorylated S551, these findings indicate that, in the context of the signaling pathway involved in sleep/wake regulation, proteins interacting with SIK3 mostly require a bona fide phospho-S551 to deliver the downstream signaling. On the other hand, S551D mutants had a less-robust phenotype of NREMS amounts during the dark phase (S551A vs. S551D: P = 0.03; S551A WT vs. S551D WT: n.s., P = 0.93), possibly suggesting that, for some SIK3-interacting proteins relevant for sleep time regulation, S551D does mimic a phosphoserine. To examine how the mutations in the S551 region affect the SIK3 signaling pathway and its binding partners, we next performed a series of biochemical analyses.
Mutations at S551 Alter the Interaction of SIK3 with PKA and 14-3-3.
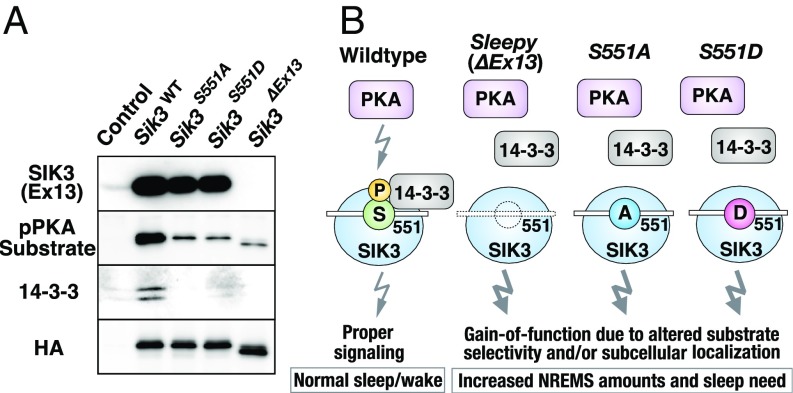
It has been reported that SIK3 is regulated by LKB1 (liver kinase B1: serine/threonine kinase 11)-mediated phosphorylation of T221, which activates the T-loop within the kinase domain (9). Phosphorylation of T221 is closely associated with the kinase activity of SIK3 (10, 11), and T221 phosphorylation is elevated in vivo when sleep need is increased by sleep deprivation (5). It has also been reported that cAMP elevation induces a PKA-dependent phosphorylation of SIK3 at T469, S551, and S674, which increases binding of the 14-3-3 protein (11). We focused on the interaction of these mutant SIK3 with PKA and 14-3-3 in subsequent analyses. We constructed HA-Sik3 (WT), HA-S551A, HA-S551D, and HA-Sleepy (ΔEx13) expression plasmids and then individually transfected them into HEK293 cells. After immunoprecipitation (IP) with anti-HA antibody, Western blotting was performed for the target molecules.
Proper expression of the SIK3 protein was confirmed by using anti-HA antibody, with HA-Sik3 (WT), HA-S551A, and HA-S551D showing the same band size and intensity (Fig. 5A). Only HA- Sleepy showed an altered band size because of the skipped exon 13, as previously reported (5). As a negative control, HEK293 cells with mock transfection were used to show blank expression. The anti-SIK3 (Ex13) antibody, which recognizes 14 amino acids in the middle of exon 13, detected the same size and amount of SIK3 protein in HA-Sik3 (WT), HA-S551A, and HA-S551D but not in HA- Sleepy.
Fig. 5.
Mutations at S551 alter the interaction of SIK3 with PKA and 14-3-3. (A) Expression plasmids for HA-Sik3 (WT), HA-S551A, HA-S551D, and HA-Sleepy were transfected to HEK293 cells, cell lysates immunoprecipitated with anti-HA antibodies, and precipitates were probed with anti-SIK3 (Ex13) antibody, anti-pPKA substrate antibody, anti–14-3-3 (pan) antibody, and anti-HA antibody. Control: no plasmid transfected. (B) The schematic diagram of molecular mechanisms for the sleep/wake phenotypes of WT, Sleepy (ΔEx13), S551A, and S551D mutants. A deletion/mutation at S551 of SIK3 result in decreased recognition from PKA and abolished binding with 14-3-3. This may alter the substrate selectivity or subcellular localization of SIK3, which leads to the increased NREMS and sleep need in Sleepy, S551A, and S551D mutant mice.
We next examined how the deletion or mutations at SIK3 S551 affect the recognition by PKA. We used a phospho-PKA–substrate antibody, which detects a phospho-Ser/Thr residue within PKA-substrate peptide motifs. Compared with HA-Sik3 (WT), the mutant lines HA-S551A, HA-S551D, and HA-Sleepy all showed similarly decreased binding to the phospho-PKA–substrate antibody (Fig. 5A). This suggests that either S551A, S551D, or Sleepy mutations reduces recognition by PKA as its substrates. The remaining band expression may be attributed to the additional PKA recognition sites of SIK3, such as T469 and S674.
Similar to the binding patterns of the phospho-PKA–substrate antibody, anti–14-3-3 antibody detected clear presence of 14-3-3 in HA-Sik3 (WT) IPs but not in HA-S551A, HA-S551D, and HA-Sleepy precipitates (Fig. 5A). These results demonstrate that either of the three mutations (S551A, S551D, or Sleepy) abolishes the binding of 14-3-3. This indicates that, as for the SIK3/14-3-3 interaction in the present context, the negatively charged S551D mutation cannot functionally mimic phosphorylated S551. Previous studies also reported similar findings regarding the interaction of 14-3-3 with other target proteins (12–14). Overall, our biochemical experiments indicate that the existence of a single phosphorylation site at the S551 position in SIK3 is crucial for recognition by PKA and interaction with 14-3-3. We presume that this signaling pathway in part mediated by PKA/SIK3/14-3-3 is required for the proper sleep/wake regulation (Fig. 5B).
Discussion
In addition to the increased daily NREMS, S551A mutants exhibited increased NREMS δ-power density and longer duration of individual NREMS episodes, two indices of homeostatic sleep need. The S551A mutation was sufficient to reproduce the phenotype of the original Sleepy (ΔEx13) mutants. Additionally, S551D mutants also exhibited prolonged NREMS amount and increased NREMS δ-power density compared with WT mice. Taken together, we found that the deletion or mutations of SIK3 S551 caused prolonged NREMS amount and increased sleep need in vivo, suggesting that the existence of S551, a PKA recognition site, is crucial for the normal sleep/wake regulation and maintenance of daily sleep need.
These findings also indicate that the S551D mutation cannot serve as a constitutive phospho-mimic for the SIK3-interacting partners that regulate sleep/wake by recognizing and binding the phospho-S551 residue. Indeed, the 14-3-3 family fulfills this criterion of such SIK3 partners (12–14). We confirmed biochemically that Sleepy, S551A, and S551D mutants showed decreased binding to the phospho-PKA–substrate antibody. This strongly suggests that the SIK3 proteins with either of the S551A, S551D, and Sleepy mutations are excluded as substrates of PKA. These mutations also abolished binding of 14-3-3 to SIK3, which corresponded to their consistent hypersomnia phenotype in vivo. We speculate that S551A, S551D, and Sleepy mutations may alter the substrate selectivity and subcellular localization of SIK3 in vivo, resulting in the hypersomnia phenotype (Fig. 5B).
A recent study reported that loss of either of the two PKA phosphorylation sites (T469 and S551) in SIK3 abolished 14-3-3 association, and rendered SIK3 insensitive to cAMP signaling in a cell-based transcription reporter assay (15). The 14-3-3 have been known to form a dimer and modulate protein–protein interactions, subcellular localization, and catalytic activity of target molecules (16–19). Because both T469 and S551 of SIK3 are the essential phospho-acceptor sites for 14-3-3 to form a dimer, a mutation or deletion at one of the two sites, S551, may cause the conformational changes that results in the abolished 14-3-3 association. Our recent interactome study also revealed that Sleepy mutation alters protein–protein interactions involving SIK3, rendering SIK3 to preferentially associate with synaptic proteins (6). These studies, taken together with the present results, raises the possibility that Sleepy, S551A, and S551D mutations may affect the interaction of SIK3 with its intrinsic binding partners and downstream targets, such as cAMP-regulated transcriptional coactivators (CRTCs) and class IIA histone deacetylases, due in part to the abolished 14-3-3 association. Because 14-3-3 isoforms have been found in the synaptic terminals, where they regulate synaptic transmission and plasticity (20–23), abolished 14-3-3 binding may also affect synapse versus soma localization of SIK3.
The SIK family of serine/threonine kinases has recently been implicated in the regulation of circadian rhythms. Through the analysis of light-regulated transcriptome in the suprachiasmatic nucleus, SIK1, another member of the SIK family, together with CRTC1, has been reported to regulate light entrainment of the circadian clock (24). Constituting a negative-feedback mechanism, SIK1 acts to suppress reentrainment of the clock to a new time zone. Another recent study reported that SIK3 also plays a role in circadian clock regulation by destabilizing PER2 protein in a phosphorylation-dependent manner (25). Together with the present findings, these reports indicate the involvement of SIK family kinases in both circadian and sleep behaviors. However, in contrast to the abnormal circadian behavior in the Sik3-null mice (25), the gain-of-function Sleepy mutation had no obvious effects on circadian rhythms (5). The presence or absence of light/dark photic cycle had no effects on the hourly NREMS amounts in the Sleepy mutant mice (SI Appendix, Fig. S3). This suggests that the dip of NREMS amounts at ZT12 (dark onset) is due to the onset of circadian behavioral activity but not photic masking effects. These data support the idea that the Sleepy allele primarily alters homeostatic but not circadian mechanisms of sleep/wake regulation.
Our findings of in vivo sleep phenotype suggest that the SIK3 S551 and its signaling pathway, including PKA and 14-3-3, may play a key role in sleep/wake regulation under normal physiological conditions (Fig. 5B). Notably, the exon 13-encoded region of SIK3, including S551, is highly conserved among both vertebrates and invertebrates, which suggests the biological importance of this PKA/SIK3/14-3-3 pathway in sleep/wake regulation. These findings provide landmark information about sleep/wake regulatory mechanisms by connecting the intracellular signaling pathway to the dynamic in vivo sleep/wake behaviors. Clinically, idiopathic hypersomnia is characterized by an insatiable need to sleep resulting in excessive daytime sleepiness, difficulty waking up, but without sleep attacks, cataplexy, or other known causes of sleepiness (26–28). Patients often present with an increased sleep need which cannot be dissipated by long naps, or by increased nightly amounts of sleep. In contrast to well-studied pathophysiology of narcolepsy, the cause of idiopathic hypersomnia remains a mystery in sleep medicine. Because our series of Sik3 mutant mice have an inherently higher sleep need despite increased sleep amounts, further research on these animal models may provide insights into the pathophysiology of idiopathic hypersomnia.
Materials and Methods
Animals.
S551A and S551D mutant mice and littermate WT mice were maintained on a C57BL/6 background. Mice were housed under a 12:12-h light/dark cycle and controlled temperature and humidity conditions. Food and water were delivered ad libitum. When the mice were under EEG/EMG recording, a water gel pack (Napa Nector, 8 oz.; System Engineering Lab Group) was used for a water source to intake water easily (29). All procedures were approved by the Institutional Animal Care and Use Committee of the University of Tsukuba.
Generation of S551A and S551D Mice by CRISPR/Cas9 System.
To construct a Cas9/single-guide RNA (sgRNA) expression vector, oligonucleotide DNAs (for both S551A/D: 5′-caccggccggagagcctcagatgg-3′ and 5′-aaacccatctgaggctctccggcc-3′) were annealed and inserted into pX330 vector (Addgene). Based on the EGxxFP system (30), the cleavage activity of the pX330-S551A/D vector was verified in cultured cells (SI Appendix, Fig. S4). Genomic DNA containing exon 13 (including S551 in the center) of the Sik3 gene was amplified and inserted into pCAG-EGxxFP to produce pCAG-EGxxFP-Sik3Ex13 using In-Fusion HD Cloning Kit (TaKaRa). The pX330-S551A/D and pCAG-EGxxFP-Sik3Ex13 were transfected into HEK293 cells with Lipofectamine LTX (ThermoFisher). The EGFP expression was evaluated 48 h after transfection. As a donor oligonucleotide to induce the amino acid change, a single-stranded 200-nucleotide DNA was synthesized (Integrated DNA Technologies), which contained a S551A or S551D coding sequence in the center and 99-nucleotide arms at the 5′ and 3′ ends. Female C57BL/6J mice were injected with pregnant mare serum gonadotropin and human CG (hCG) at a 48-h interval and were then mated with male C57BL/6J mice. The fertilized one-cell embryos were collected from the oviducts. Subsequently, 5 ng μL−1 of pX330-S551A/D vector and 10 ng μL−1 of the donor oligonucleotide were injected into the pronuclei of these one-cell embryos. The injected one-cell embryos were transferred into pseudopregnant Institute of Cancer Research mice. F0 mice were genotyped for the presence of S551A or S551D mutation in exon 13 of the Sik3 gene. F0 mice were checked for the absence of the Cas9 transgene and off-target effects. Candidate off-target sites were identified based on a complete match of S551A/D 16 bp target sequence (gagcctcagatggagg), including the PAM sequence. F0 mice were mated with the counter C57BL/6N mice to obtain F1 offspring.
qRT-PCR.
Total RNA was extracted from a tissue sample using the RNeasy Lipid Tissue Mini kit (Qiagen) and QiAzol Lysis Reagent (Qiagen). cDNA was synthesized with oligo dT primers using a PrimeScript reverse-transcriptase kit (TaKaRa). Real-time quantitative PCR reactions were performed with ViiA7 Real-Time PCR System (ThermoFisher) using SYBR GREEN PreMix Ex Taq (TaKaRa). The averages of GAPDH mRNA were used for normalization. The following PCR primers were used: Gapdh forward, 5′-agaacatcatccctgcatcc-3′; Gapdh reverse, 5′-cacattgggggtaggaacac-3′; Sik3 forward, 5′-ttgtcaatgaggaggcacac-3′; Sik3 reverse, 5′-tgttaggggccacttgtagg-3′. For the RT-PCR experiment in Fig. 4, the following PCR primers were used: Sik3 forward, 5′-catccacaggggtaactgct-3′; Sik3 reverse 5′-gaacagatctgcaggcacaa-3′.
Western Blot and IP.
Brain tissues were homogenized using a homogenizer (Polytron) in ice-cold lysis buffer [20 mM Hepes, pH 7.5, 100 mM NaCl, 10 mM Na4P2O7, 1.5% Triton X-100, 15 mM NaF, 1× PhosSTOP (Roche), 5 mM EDTA, 1× protease inhibitor (Roche)] and then centrifuged at 13,000 × g at 4 °C. The supernatants were separated by SDS/PAGE and transferred on PVDF membrane. Cultured cell lysates were prepared with ice-cold lysis buffer [1 M Hepes-NA, pH 7.5, 5 M NaCl, 0.1 M Na4P2O7, 10% Triton X-100, 0.5 M NaF, 10× PhosSTOP (Roche), 0.5 M EDTA, 25× proteases inhibitor EDTA (Roche)], rotated >15 min at 4 °C, and centrifuged at 13,000 × g at 4 °C. Western blotting was performed according to standard protocols using the corresponding antibodies (see also Antibodies, below). IP was performed for the HA-tagged SIK3 proteins [HA-Sik3 (WT), HA-S551A, HA-S551D, and HA-Sleepy] using HA tagged Protein Purification Kit (MBL 3325).
Antibodies.
For anti-SIK3 (C-terminal) antibody, a rabbit polyclonal antibody against the C-terminal 171 amino acids of mouse SIK3 was generated using custom antibody production service. For anti-SIK3 (Ex13) antibody a rabbit polyclonal antibody against 14 amino acids of mouse SIK3 Exon 13 (LHAQQLLKRPRGPS) was generated using a custom antibody production service. For antiphospho-PKA substrate (RRXS/T) (100G7E) antibody, a rabbit monoclonal antibody against pPKA substrate was used (commercially available from Cell Signaling). For anti–14-3-3 (pan) antibody, a rabbit polyclonal antibody detects all known isoforms of mammalian 14-3-3 proteins (Cell Signaling). For anti–HA-Tag (C29F4) antibody, a rabbit monoclonal antibody against HA-tag (Cell Signaling) was used. For anti–β-Tubulin (9F3) antibody, a rabbit monoclonal antibody was used as a loading marker (Cell Signaling).
Plasmid Constructs.
HA-tagged Sik3 cDNA were subcloned into pcDNA3.1 vectors using an In-Fusion HD Cloning kit (TaKaRa). The nucleotide substitutions (ΔEx13/S551A/S551D) were introduced into HA-tagged Sik3 cDNA using a KOD-Plus-Mutagenesis kit (Toyobo) and a PrimeSTAR Mutagenesis Basal Kit (TaKaRa).
Cell Culture and Transfection.
HEK293 cells were cultured in DMEM with 10% FBS and penicillin/streptomycin. For plasmid transfection, HEK293 cells were grown to 80% confluency in six-well plates. HEK293 cells were transfected with 500 ng pcDNA constructs encoding HA-Sik3 (WT), HA-Sleepy, HA-S551A, and HA-S551D (see also Plasmid Constructs, above) using FuGENE HD Transfection Regent (Promega) 1.5 μL.
EEG/EMG Electrode Implantation Surgery.
EEG/EMG electrodes with four EEG electrode pins and two EMG wires were implanted in the mice under anesthesia using isoflurane (3% for induction, 1% for maintenance) (31). The coordination of EEG electrodes was set over the frontal and occipital cortices [antero-posterior (AP): 0.5 mm, medio-lateral (ML): 1.3 mm, dorso-ventral (DV): −1.3 mm; and AP: −4.5 mm, ML: 1.3 mm, DV: −1.3 mm]. The two wires for the EMG recording were inserted in the neck muscles and subsequently attached to the skull using dental cement. All mice were allowed 7 d to recover from surgery. After recovery, for habituation to the recoding conditions, the mice were tethered to a counterbalanced arm (Instech Laboratories) that allowed free movement and exerted minimal weight for at least 3 d before the recording session. In this study, Sik3+/+, Ski3S551A/+, Sik3flag, S551A/+, and Sik3S551D/+ mice were subjected to surgery and EEG/EMG recordings.
Sleep Behavior Analysis.
EEG/EMG signals were recorded for 2 consecutive days from the onset of the light phase to examine sleep/wake behavior under baseline conditions. The recording room was kept under a 12:12-h light/dark cycle and at a constant temperature (24–25 °C). EEG/EMG data were analyzed using a MatLab (MathWorks)-based, custom semiautomated staging program that allowed the classification of NREMS, REMS, or wakefulness based on a 20-s epoch time window. Wakefulness was scored based on the criteria of the presence of fast EEG, high amplitude, and variable EMG. NREMS was staged based on high amplitude, δ- (1–4 Hz) frequency EEG and low EMG tonus. REMS was characterized by θ (6–9 Hz) -dominant EEG and EMG atonia. EEG signals were subjected to fast Fourier transform analysis from 1 to 30 Hz with a 1-Hz bin using MatLab-based custom software. The hourly δ-density during NREMS indicates the hourly averages of δ-density, which is the ratio of δ-power (1–4 Hz) to total EEG power (1–30 Hz) at each 20-s epoch or all epochs (29, 31, 32). Based on the normalization method described in Bjorness et al. (4), the EEG power in each frequency bin was normalized to the relatively constant higher frequency range (16–30 Hz).
Statistics.
Statistical analysis was performed using Graph Pad Prism 7.0 (GraphPad Software) and SigmaPlot 13.0 (Systat Software). All data were subjected to a D’Agostino–Pearson (Omnibus K2) normality test for Gaussian distribution and variance. For the dataset that showed a Gaussian distribution (P > 0.05 in normality test), we performed parametric tests such as two-tailed unpaired t test and ANOVA followed by Bonferroni’s multiple comparisons. For the dataset that failed to show a Gaussian distribution, we performed a nonparametric test, such as a Mann–Whitney u test. Significance levels in the figures are represented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Error bars in the graphs represent mean ± SEM.
Supplementary Material
Acknowledgments
We thank all M.Y. and H.F. laboratory and International Institute for Integrative Sleep Medicine (WPI-IIIS) members; and Dr. M. Lazarus and Dr. J. S. Takahashi for the critical comments and suggestions on this project. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 15J06369 (to T.H.); 26220207 and 17H06095 (to M.Y. and H.F.); and 17H04023, 16K15187, and 15H05942 (to H.F.); Japan Science and Technology Association CREST Grant JPMJCR1655 (to M.Y.); the FIRST program from JSPS (M.Y.); the Uehara and Takeda Foundations (M.Y.); and the WPI program from Japan’s Ministry of Education, Culture, Sports, Science, and Technology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810823115/-/DCSupplemental.
References
- 1.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 2.Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raizen DM, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 4.Bjorness TE, et al. An adenosine-mediated glial-neuronal circuit for homeostatic sleep. J Neurosci. 2016;36:3709–3721. doi: 10.1523/JNEUROSCI.3906-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funato H, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539:378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature. 2018;558:435–439. doi: 10.1038/s41586-018-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takemori H, Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. J Steroid Biochem Mol Biol. 2008;108:287–291. doi: 10.1016/j.jsbmb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Vyazovskiy VV, et al. Local sleep in awake rats. Nature. 2011;472:443–447. doi: 10.1038/nature10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh Y, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 11.Berggreen C, Henriksson E, Jones HA, Morrice N, Göransson O. cAMP-elevation mediated by β-adrenergic stimulation inhibits salt-inducible kinase (SIK) 3 activity in adipocytes. Cell Signal. 2012;24:1863–1871. doi: 10.1016/j.cellsig.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Yasmin L, et al. Delineation of exoenzyme S residues that mediate the interaction with 14-3-3 and its biological activity. FEBS J. 2006;273:638–646. doi: 10.1111/j.1742-4658.2005.05100.x. [DOI] [PubMed] [Google Scholar]
- 13.Ottmann C, et al. Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: From structure to pathogenesis. EMBO J. 2007;26:902–913. doi: 10.1038/sj.emboj.7601530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menon RP, et al. The importance of serine 776 in Ataxin-1 partner selection: A FRET analysis. Sci Rep. 2012;2:919. doi: 10.1038/srep00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonntag T, Vaughan JM, Montminy M. 14-3-3 proteins mediate inhibitory effects of cAMP on salt-inducible kinases (SIKs) FEBS J. 2018;285:467–480. doi: 10.1111/febs.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: A comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Yaffe MB. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 18.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin H, Rostas J, Patel Y, Aitken A. Subcellular localisation of 14-3-3 isoforms in rat brain using specific antibodies. J Neurochem. 1994;63:2259–2265. doi: 10.1046/j.1471-4159.1994.63062259.x. [DOI] [PubMed] [Google Scholar]
- 21.Moya KL, et al. Immunolocalization of the cellular prion protein in normal brain. Microsc Res Tech. 2000;50:58–65. doi: 10.1002/1097-0029(20000701)50:1<58::AID-JEMT9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Qiao H, Foote M, Graham K, Wu Y, Zhou Y. 14-3-3 proteins are required for hippocampal long-term potentiation and associative learning and memory. J Neurosci. 2014;34:4801–4808. doi: 10.1523/JNEUROSCI.4393-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg D, Holzmann C, Riess O. 14-3-3 proteins in the nervous system. Nat Rev Neurosci. 2003;4:752–762. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- 24.Jagannath A, et al. The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell. 2013;154:1100–1111. doi: 10.1016/j.cell.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayasaka N, et al. Salt-inducible kinase 3 regulates the mammalian circadian clock by destabilizing PER2 protein. eLife. 2017;6:e24779. doi: 10.7554/eLife.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billiard M, Dauvilliers Y. Idiopathic hypersomnia. Sleep Med Rev. 2001;5:349–358. doi: 10.1053/smrv.2001.0168. [DOI] [PubMed] [Google Scholar]
- 27.Ali M, Auger RR, Slocumb NL, Morgenthaler TI. Idiopathic hypersomnia: Clinical features and response to treatment. J Clin Sleep Med. 2009;5:562–568. [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson KN, Pilsworth S, Sharples LD, Smith IE, Shneerson JM. Idiopathic hypersomnia: A study of 77 cases. Sleep. 2007;30:1274–1281. doi: 10.1093/sleep/30.10.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komiya H, et al. Sleep/wake behaviors in mice during pregnancy and pregnancy-associated hypertensive mice. Sleep (Basel) March 1, 2018 doi: 10.1093/sleep/zsx209. [DOI] [PubMed] [Google Scholar]
- 30.Mashiko D, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funato H, et al. Loss of Goosecoid-like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proc Natl Acad Sci USA. 2010;107:18155–18160. doi: 10.1073/pnas.1012764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franken P, Malafosse A, Tafti M. Genetic variation in EEG activity during sleep in inbred mice. Am J Physiol. 1998;275:R1127–R1137. doi: 10.1152/ajpregu.1998.275.4.R1127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.