Significance
HIV creates a persistent reservoir, which is largely resistant to current antiretroviral treatment aimed at inhibiting HIV replication. CD4+ T memory lymphocytes, key components of this reservoir, are generally refractory to infection, but stimulation with γc-chain cytokines, such as IL-15, renders these cells more susceptible to HIV. We found that, by inducing cell cycle entry of CD4+ T cells, JAK1 is a key mediator responsible for counteracting the antiviral activity of SAM domain and HD domain-containing protein 1 (SAMHD1). Pharmacological inhibition of these kinases resulted in restoration of SAMHD1 in CD4+ T cells. Protecting these cells during the critical IL-15 surge observed during primary infection has the potential to limit reservoir establishment.
Keywords: HIV, SAMHD1, IL-15, CD4 T memory stem cells
Abstract
HIV integrates into the host genome to create a persistent viral reservoir. Stimulation of CD4+ memory T lymphocytes with common γc-chain cytokines renders these cells more susceptible to HIV infection, making them a key component of the reservoir itself. IL-15 is up-regulated during primary HIV infection, a time when the HIV reservoir established. Therefore, we investigated the molecular and cellular impact of IL-15 on CD4+ T-cell infection. We found that IL-15 stimulation induces SAM domain and HD domain-containing protein 1 (SAMHD1) phosphorylation due to cell cycle entry, relieving an early block to infection. Perturbation of the pathways downstream of IL-15 receptor (IL-15R) indicated that SAMHD1 phosphorylation after IL-15 stimulation is JAK dependent. Treating CD4+ T cells with Ruxolitinib, an inhibitor of JAK1 and JAK2, effectively blocked IL-15–induced SAMHD1 phosphorylation and protected CD4+ T cells from HIV infection. Using high-resolution single-cell immune profiling using mass cytometry by TOF (CyTOF), we found that IL-15 stimulation altered the composition of CD4+ T-cell memory populations by increasing proliferation of memory CD4+ T cells, including CD4+ T memory stem cells (TSCM). IL-15–stimulated CD4+ TSCM, harboring phosphorylated SAMHD1, were preferentially infected. We propose that IL-15 plays a pivotal role in creating a self-renewing, persistent HIV reservoir by facilitating infection of CD4+ T cells with stem cell-like properties. Time-limited interventions with JAK1 inhibitors, such as Ruxolitinib, should prevent the inactivation of the endogenous restriction factor SAMHD1 and protect this long-lived CD4+ T-memory cell population from HIV infection.
Lifelong persistence of HIV-1 prevents HIV/AIDS disease cure. Within days of infection, a latent reservoir develops and persists, despite effective highly active antiretroviral therapy (HAART) (1). While CD4+ T lymphocytes are the primary target of HIV, they also contribute to establishing and maintaining the latent reservoir (2). The activation status of the CD4+ cell determines susceptibility to infection and reactivation from latency (3–5). Resting CD4+ T memory cells constitute the majority of the latently infected cell pool (4), although quiescent CD4+ T cells are resistant to HIV infection due to several blocks in the viral replication cycle (3, 4). One early block to infection in memory CD4+ T cells is mediated by SAM domain and HD domain-containing protein 1 (SAMHD1) (6–8). First described as a restriction factor in myeloid cells, SAMHD1 also blocks HIV infection in CD4+ T lymphocytes (6–10). SAMHD1 targets HIV at the level of reverse transcription by decreasing cellular dNTPs levels to prevent efficient synthesis of viral cDNA and/or by degrading viral cDNA (9, 10). T-cell receptor engagement leads to cell activation, proliferation, and SAMHD1 phosphorylation, creating a cellular environment, which is highly supportive of productive infection (11–13). The antiviral activity of SAMHD1 is abrogated when a specific threonine (at position 592) is phosphorylated by the cyclin-dependent kinase 1 (CDK1)/CyclinA2 complex (11–13). CDK1 is a serine/threonine kinase that regulates cell cycle progression (14).
Common γ-chain (γc) cytokines (IL-2, IL-7, IL-15) are critical for T-cell homeostasis and expansion (15). IL-7 and IL-15 levels increase during HIV infection (16, 17). Moreover, IL-15 is specifically up-regulated during the acute phase of HIV/simian immunodeficiency virus (SIV) infection (17, 18). Increased IL-15 plasma levels also correlate with high viremia and inflammation (19). Recent studies showed that IL-7 increased infection of CD4+ T cells by inducing SAMHD1 phosphorylation, which was inhibited by the Src inhibitor Desatinib (20–22). However, the impact of IL-15 on CD4+ T-memory cell populations remains undefined.
IL-15 supports the homeostasis of natural killer cells, memory CD8+ T cells, and memory CD4+ T cells (23–26). Type I IFN is a potent inducer of IL-15 production (27), but this cytokine also arises in antigen-presenting cells (e.g., dendritic cells, monocytes, and macrophages) on pathogenic stimuli (23, 24). While IL-15 may exert positive effects on survival and proliferation of HIV-specific CD8+ T cells (28–30), its administration in models of acute SIV infection increases the viral set point and accelerates disease progression by rendering CD4+ T memory cells more susceptible to infection (31, 32).
CD4+ T memory stem cells (TSCM) are the least differentiated of the human memory T-cell subsets and represent 1–4% of total circulating T cells. Importantly, CD4+ TSCM retain stem cell-like properties and undergo a developmental program with progression toward more differentiated CD4+ T-cell types (26, 33). CD4+ TSCM support productive infection with CCR5 and CXCR4 tropic viruses, probably due to relatively low levels of the restriction factors SAMHD1, APOBEC3G, and Trim5α (34–36). In HIV-infected patients receiving HAART, CD4+ TSCM contain replication-competent proviruses and contribute to the total HIV reservoir over time (34, 37). The percentage of infected CD4+ TSCM positively correlates with pathogenicity in HIV-infected individuals and in rhesus macaques infected with pathogenic SIV (38, 39). Since the HIV reservoir is established very early during acute infection (17, 18), we hypothesized that the increased IL-15 levels observed during this critical time window temporarily alter memory CD4+ T-cell properties, facilitating infection and reservoir establishment in a cell population generally regarded as resistant to HIV infection.
Results
IL-15 Increases HIV Infection of CD4+ T Cells by Phosphorylating SAMHD1.
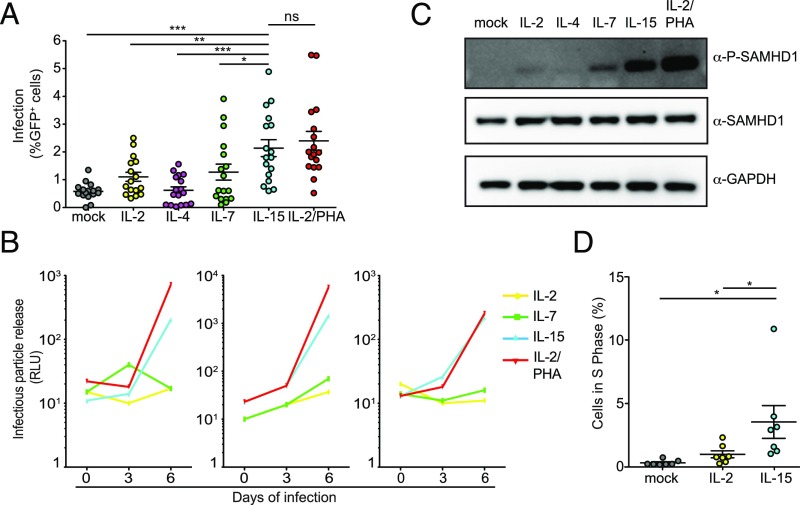
To investigate the impact of γc-cytokines on HIV infection, we stimulated primary human CD4+ T cells with equimolar amounts of IL-2, IL-4, IL-7, IL-15, and IL-2/phytohemagglutinin (PHA) (positive control) or left them untreated as baseline control before infecting them with HIV-GFP. Five days postinfection, we observed that all treatments, except IL-4, increased the number of productively infected cells as previously reported (40). IL-15 was the most efficient of the γc-cytokines with respect to increasing the susceptibility of CD4+ T cells to HIV infection (Fig. 1A and SI Appendix, Fig. S1 A and B). We confirmed these results by infecting IL-2–, IL-7–, IL-15–, and IL-2/PHA–stimulated cells from three different donors with the transmitter founder virus HIV CHO40. We found that HIV CHO40 replicates more efficiently in cells stimulated with IL-15 than with IL-7 or IL-2 (Fig. 1B). Although we observed cellular activation (CD25, CD69 expression) in IL-15–stimulated cells (SI Appendix, Fig. S1 C and D), this was significantly lower than with IL-2/PHA treatment while infection was comparable, indicating that activation alone was not sufficient to achieve the level of infection observed.
Fig. 1.
IL-15 stimulation of CD4+ T memory cells increases susceptibility to infection and induces SAMHD1 phosphorylation. (A) CD4+ T cells from 17 donors were stimulated as indicated and infected with HIV R7/3 GFP. Flow cytometry assayed the efficiency of infection 5 d postinfection. *P < 0.05 using a two-tailed paired Student’s t test; **P < 0.01 using a two-tailed paired Student’s t test; ***P < 0.001 using a two-tailed paired Student’s t test; ns, not significant. (B) Primary blood lymphocytes from three different healthy donors stimulated as indicated were infected with HIV CHO40.C/2625. Clarified supernatants of infected CD4+ T cells were collected every 3 d. Infectious particle release was determined using TZM-bl reporter cells. Infectivity is expressed in relative light units (RLUs). (C) Levels of phosphorylated and total SAMHD1 assayed by Western blotting of CD4+ T-cell lysates stimulated for 96 h with 1 nM indicated cytokines. A representative membrane is shown (n = 3 donors). (D) Percentage of CD4+ T cells in the S phase based on the FACS analysis of BrdU vs. 7AAD incorporation after 3 d in culture with the indicated ILs is shown. *P < 0.05 using a two-tailed paired Student’s t test.
The antiviral activity of SAMHD1 is regulated by phosphorylation at residue T592: SAMHD1 fails to target incoming HIV in activated CD4+ T cells, because it is mostly phosphorylated in these cells (9–12). Previous reports indicated that stimulation with IL-2, IL-7, and CD3/CD28 induced different levels of SAMHD1 phosphorylation, but IL-15 was not included in these studies (11, 12, 21). Our experiments indicate that IL-15 is more effective than IL-7 in regard to inducing phosphorylation at residue T592 (Fig. 1C). Note that the levels of total SAMHD1 remain comparable across the different stimuli or mock control cells, while phosphorylated SAM domain and HD domain-containing protein 1 (P-SAMHD1) is only observed in IL-2–, IL-7–, IL-15–, and PHA/IL-2–stimulated CD4+ T cells.
Because of the association of SAMHD1 phosphorylation with cell cycle progression (11, 12, 41), we investigated if IL-15 treatment promotes exit from G0 state and progression in the cell cycle. BrdU and 7-aminoactinomycin D (7AAD) incorporation revealed that the percentage of IL-15–treated CD4+ T lymphocytes in S phase is higher than IL-2– or mock-treated cells (Fig. 1D and SI Appendix, Fig. S1 E and F). Taken together, we found that IL-15 induces cell cycle entry, which in turn, is the cause of SAMHD1 phosphorylation. Moreover, we observed that IL-15 was superior to IL-7 in inducing SAMHD1 phosphorylation and increasing susceptibility to HIV infection in primary human CD4+ T cells.
SAMHD1 Phosphorylation by IL-15 Is JAK Dependent.
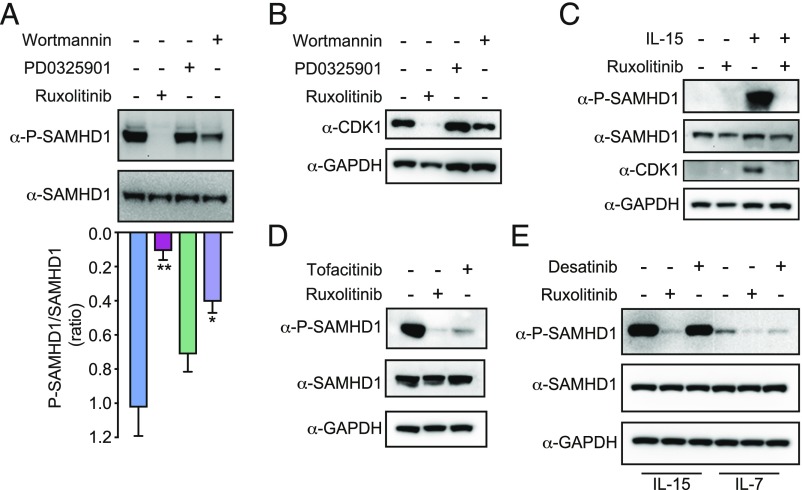
We next aimed to identify the signaling pathways that initiated IL-15–induced SAMHD1 phosphorylation. The IL-15 receptor consists of three subunits (alpha, beta, and gamma), with the alpha subunit being specific for IL-15 (23, 24). Activation of the IL-15 receptor initiates a signaling cascade that includes the activation of JAK1/2, the PI3K, and the Mitogen-activated protein kinase 1/2 (MEK1/2) (42). These combined signals induce gene transcription for antiapoptotic and proliferative processes (42). We systematically probed these three distinct signaling cascades using inhibitors: Ruxolitinib to block JAK1, Wortmannin to block PI3K, and PD0325901 to inhibit MEK1/2 (SI Appendix, Fig. S2A). We stimulated CD4+ T cells with IL-15 followed by treatment with one of these three kinase inhibitors. Only inhibition of the JAK pathway potently blocked SAMHD1 phosphorylation (Fig. 2A). Wortmannin had a modest, intermediate effect, while PD0325901 had no sizeable impact on SAMHD1 phosphorylation (Fig. 2A). We ascertained the efficacy of each of these kinase inhibitors at the chosen concentration by measuring phosphorylation of their known substrate (STAT3, ERK1/2, and Akt) (SI Appendix, Fig. S2B). Ruxolitinib also decreased the degree of CD4+ T-cell activation as assayed by CD25 and CD69 expression in both IL-2 and IL-15 conditions, with no impact on cell viability (SI Appendix, Fig. S2 C–E). Abolishing SAMHD1 phosphorylation with Ruxolitinib correlated with the absence of CDK1 in IL-15–treated CD4+ T-cell lysates (Fig. 2B). Unstimulated primary CD4+ T cells do not express CDK1, and therefore, Ruxolitinib blocks IL-15–induced CDK1 expression (Fig. 2C). To confirm the observation that inhibiting the JAK pathway blocks SAMHD1 phosphorylation, we measured IL-15–induced P-SAMHD1 levels after treatment with another JAK inhibitor, Tofacitinib. We observed that Tofacitinib also decreased the levels of SAMHD1 phosphorylation (Fig. 2D).
Fig. 2.
IL-15–induced SAMHD1 phosphorylation is JAK dependent. (A) IL-15–stimulated CD4+ T cells were treated with Wortmannin (200 nM), PD0325901 (20 nM), and Ruxolitinib (50 nM). Levels of phosphorylated and total SAMHD1 in cell lysates were assayed by Western blot and quantified by densitometric analysis. Data represent means ± SEM of the P-SAMHD1/SAMHD1 ratio. A two-tailed paired Student’s t test was applied (n = 3 donors), *P < 0.05, **P < 0.01. (B) IL-15–stimulated CD4+ T cells were treated as in A. The total levels of CDK1 were quantified in cell lysates. (C) Levels of phosphorylated and total SAMHD1, CDK1, and GAPDH were determined by Western blot analysis in CD4+ T cells cultured with or without IL-15 in the presence or absence of Ruxolitinib. (D) Lysates from IL-15–stimulated CD4+ T cells were treated with Ruxolitinib or Tofacitinib (80 nM) and probed for P-SAMHD1, SAMHD1, and GAPDH levels. (E) CD4+ T cells were stimulated with equimolar amounts of IL-15 or IL-7 and treated with Desatinib (75 nM) or Ruxolitinib (50 nM). Levels of phosphorylated and total SAMHD1 from cell lysates were analyzed by Western blot.
Since IL-7–induced phosphorylation of SAMHD1 in CD4+ T cells is blocked by Desatinib, a Bcr-Abl tyrosine kinase and Src family tyrosine kinase inhibitor (21, 22), we next tested whether IL-15–induced phosphorylation of SAMHD1 was similarly affected by this drug. In contrast to Ruxolitinib, we found that Desatinib failed to prevent IL-15–induced phosphorylation of SAMHD1 in primary CD4+ T cells (Fig. 2E). IL-7–induced SAMHD1 phosphorylation was reduced by Desatinib and was almost completely abolished by Ruxolitinib. We conclude that IL-15–induced SAMHD1 phosphorylation in primary CD4+ T cells is primarily JAK dependent, as it can be blocked by different JAK inhibitors (Ruxolitinib and Tofacitinib) but not by Desatinib. Thus, JAK activation by IL-7 and IL-15 triggers distinct downstream pathways to phosphorylate SAMHD1. Furthermore, SAMHD1 phosphorylation after IL-15 treatment is detected 48 h poststimulation and peaks at 72 h, suggesting that this process requires changes in JAK-induced gene expression and cell cycle entry (SI Appendix, Fig. S2F).
Inhibition of JAK Impairs HIV Infection in IL-15–Stimulated CD4+ T Cells at the Step of Reverse Transcription.
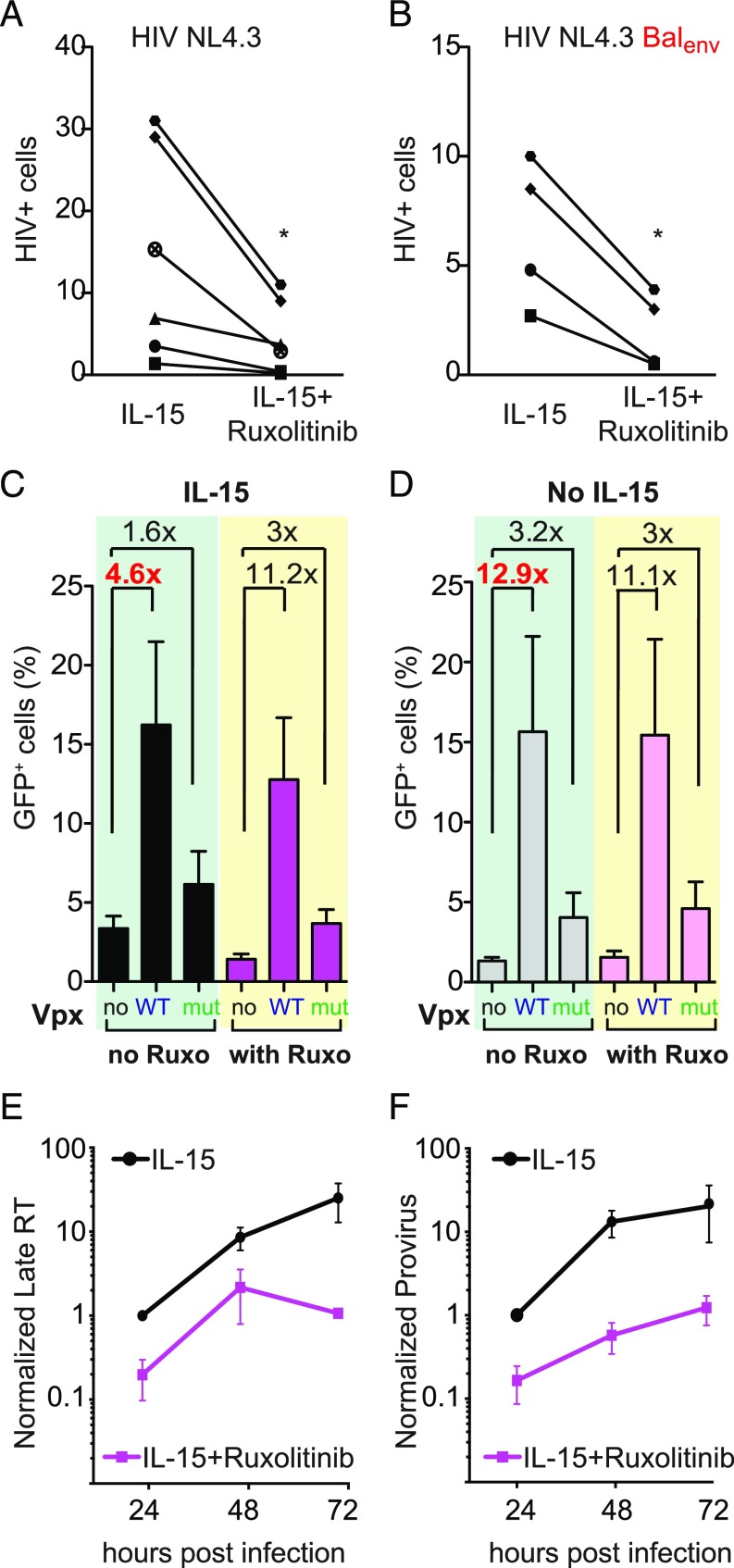
Next, we assessed whether pharmacological inhibition of JAK limits HIV infection. We infected primary CD4+ T cells treated with and without Ruxolitinib with CXCR4-tropic (HIV-NL4.3) and CCR5-tropic HIV (HIV-NL4.3/BALenv). Ruxolitinib induced, on average, a 4.8-fold reduction (range: 1.9- to 8.8-fold) in infection levels regardless of coreceptor usage (Fig. 3 A and B; a representative flow cytometry plot is shown in SI Appendix, Fig. S3A). Ruxolitinib also decreased IL-2 enhancement of susceptibility to infection in CD4+ T cells, while the infection of mock-treated CD4+ T cells remained unchanged (SI Appendix, Fig. S3B). This is not surprising, since IL-2 and IL-15 receptors share two subunits and because both signal through JAK. Moreover, Ruxolitinib blocked SAMHD1 phosphorylation in both IL-15– as well as IL-2–stimulated CD4+ T cells (SI Appendix, Fig. S3C).
Fig. 3.
Ruxolitinib impairs CD4+ T-cell infection at reverse transcription. (A and B) IL-15–stimulated CD4+ T cells treated with and without Ruxolitinib (50 nM) were infected with HIV NL4.3 or HIV NL4.3BalEnv. FACS determined the number of infected cells 5 d postinfection using intracellular Gag p24 staining (n = 4–6 donors), *P < 0.05. (C and D) CD4+ T cells stimulated as indicated were treated or not with Ruxolitinib and infected with HIV*GFP carrying SIV Vpx WT, SIV Vpx mutant Q76A, or the control plasmid pcDNA (no Vpx). The number of GFP expression cells was determined by flow cytometry. Data represent means ± SEM (n = 5 donors). (E) LRTs were quantified using qPCR in infected IL-15–stimulated CD4+ T cells treated with and without Ruxolitinib. Infections were followed for 3 d. LRT levels in IL-15–treated cells at 24 h were set to one. Values represent means ± SEM from three independent experiments/donors. (F) Levels of integrated viral DNA were quantified from infected CD4+ T cells treated as indicated. Integrated viral DNA levels in IL-15–treated cells at 24 h were set to one. Data represent means ± SEM (n = 3 donors).
We next probed the mechanism underlying the observed antiviral activity of Ruxolitinib by specifically removing SAMHD1 from the cells using SIVmac Vpx (6, 7, 10). To do that, we exploited an HIV GFP reporter virus modified to incorporate Vpx (HIV*GFP-Vpx) or a mutant form of Vpx (Q76A) that does not induce SAMHD1 degradation (6, 7, 10). We first cultured primary CD4+ T cells with or without IL-15 followed by treatment with or without Ruxolitinib and infection with one of the following three viruses: HIV*GFP (no Vpx), HIV*GFP-Vpx (Vpx WT), or HIV*GFP-Vpx Q76A (Vpx mutant).
In IL-15–treated cells, HIV*GFP-Vpx and HIV*GFP-Vpx Q76A resulted in 4.6- and 1.6-fold increases of infection, respectively, compared with HIV*GFP (Fig. 3C). In the presence of Ruxolitinib, these differences are further enhanced to 11.2-fold for the Vpx WT virus and to 3-fold for the Vpx mutant virus. Thus, in HIV Vpx-infected cells, Ruxolitinib lost its ability to reduce the levels of infection in IL-15–stimulated cells due to the Vpx-mediated elimination of SAMHD1 (Fig. 3C). Of note, a recent report suggests that both SIV Vpx WT and SIV Vpx Q76A also degrade TASOR, a subunit of the HUSH repressor complex, which could influence HIV-1 transcription downstream of SAMHD1 (43).
Control infection experiments with unstimulated CD4+ T cells (Fig. 3D) revealed that the three viruses reach infection levels comparable with those observed in Ruxolitinib-treated IL-15–stimulated cells. Thus, Ruxolitinib does not modulate susceptibility to infection in nonstimulated CD4+ T cells (compare Fig. 3C with Fig. 3D). Of note, HIV-Vpx Q76A resulted in a modest increase of infection (1.6- to 3.2-fold increase compared with no Vpx) as previously shown (9, 10). This partial activity likely reflects the fact that Vpx Q76A still binds SAMHD1 and thus, impairs its oligomerization (44). Taken together, these data indicate that the presence of SAMHD1 is necessary for the ability of Ruxolitinib to reduce infection.
We next sought to determine which step in the viral lifecycle is targeted by JAK inhibition. We quantified late reverse transcripts (LRTs) and proviruses in IL-15–stimulated CD4+ T cells infected in the presence and absence of Ruxolitinib. We observed that Ruxolitinib significantly reduced the levels of both LRTs (Fig. 3E) and integrated viral DNA (Fig. 3F), suggesting that the mechanism by which Ruxolitinib limits HIV infection involves viral reverse transcription (as the absence of an additive effect between Vpx and Ruxolitinib excludes an influence on entry). These data also provide mechanistic insight into how JAK inhibitors block HIV-1 replication (45).
Based on these findings, we speculated that IL-15 and Ruxolitinib might also influence the susceptibility of primary human macrophages to HIV infection. To test this hypothesis, we differentiated macrophages from isolated CD14+ cells, cultured them with or without IL-15, and treated them with or without Ruxolitinib for 72 h before infection with an HIV-Renilla Luciferase R5Env. We did not observe any effect of IL-15 or Ruxolitinib on HIV infection levels in human macrophages (SI Appendix, Fig. S4A). To determine whether IL-15 and the JAK pathway play different roles in SAMHD1 phosphorylation in macrophages compared with CD4+ T cells, we analyzed the levels of P-SAMHD1, SAMHD1, and CDK1 in macrophages after IL-15 and Ruxolitinib treatment. In contrast to our findings in primary CD4+ T cells, IL-15 treatment of macrophages did not increase P-SAMHD1 levels, and JAK inhibition had no effect on SAMHD1 phosphorylation in these cells. IL-15 and Ruxolitinib do not modulate CDK1 expression in macrophages to the same degree as in CD4+ T cells, which explains their lack of impact on macrophages infection (SI Appendix, Fig. S4B). Taken together, Ruxolitinib inhibits HIV infection specifically in primary CD4+ T cells by acting at a step in the viral lifecycle that maps to reverse transcription.
IL-15 Increases Proliferation of CD4+ TSCM and Their Proportion in Infected CD4+ T Cells.
Our data show that, at the molecular level, IL-15 increases the susceptibility of CD4+ T cells to HIV infection in an SAMHD1-dependent manner. We considered next whether IL-15 differentially altered the features and the susceptibility to infection of specific CD4+ T-cell subsets relevant for HIV pathogenesis [naïve cells, central memory cells (TCM), effector memory cells (TEM), and TSCM]. Therefore, we analyzed the effect of IL-15 stimulation on the immunophenotype of infected and uninfected CD4+ T cells at the single-cell level using multiplex mass cytometry by TOF (CyTOF). Our CyTOF assay performs the equivalent of a 19-parameter (13 membrane markers, 4 intracellular markers, and 2 markers for live gating) “FACS analysis” in a single tube (46, 47). We also incorporated a CD45-based barcoding system to multiplex four samples in the same CyTOF run and enhance reproducibility across our conditions (48) (SI Appendix, Fig. S5C). We stimulated CD4+ T cells from two different donors with IL-2 or IL-15 for 3 d and infected them with replication-competent HIV NL4.3 or no virus as mock control. Five days later, we analyzed cells by multiplex CyTOF and plotted the data using the dot plot display of Cytobank (36, 38) (SI Appendix, Fig. S5 D and E).
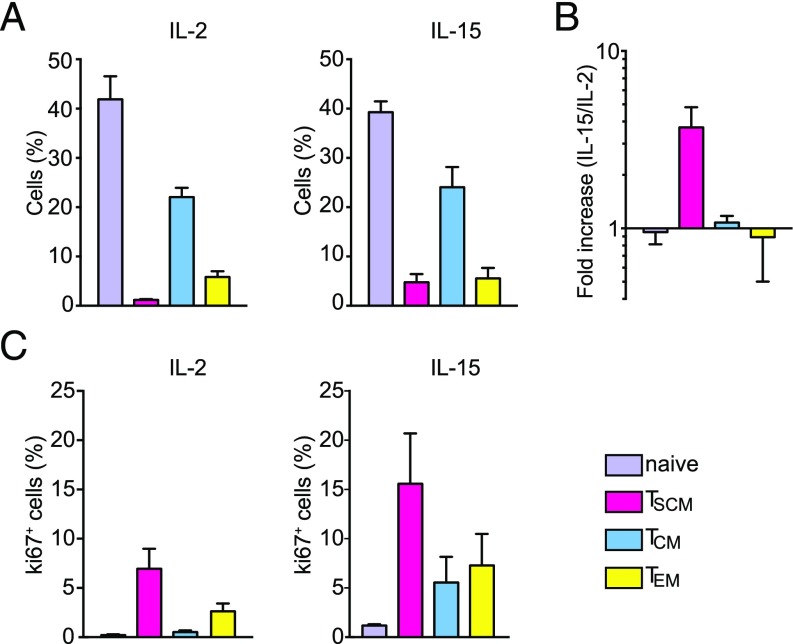
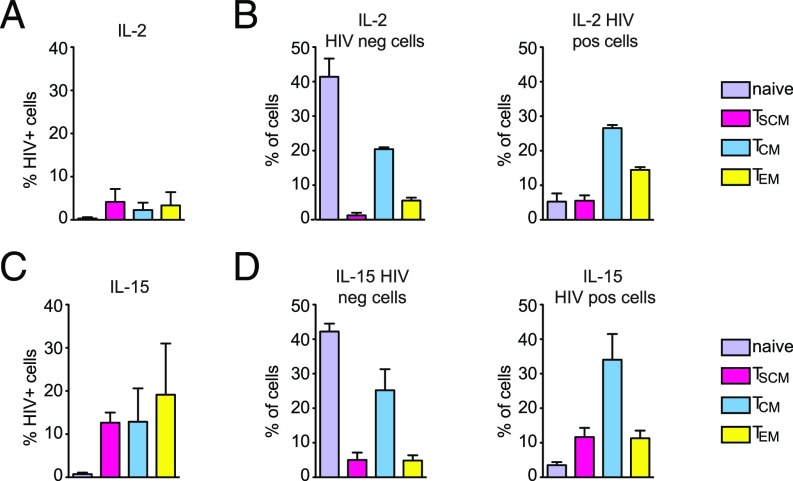
First, we analyzed the effect of IL-2 and IL-15 treatment on mock-infected CD4+ T cells. Consistent with previous reports, in the presence of IL-2, we found a small CD4+ TSCM population (3.1–6.4%) compared with other memory populations (TCM: 14.5–20%; TEM: 3.5–7%; naïve cells: 41–37%) (Fig. 4A, Left). We observed that IL-15 increased the percentage of CD4+ TSCM by 3.8-fold compared with IL-2 treatment (Fig. 4 A, Right and B). CD4+ TSCM display a high proliferative capacity as measured by Ki67 expression, and the percentage of TSCM positive for Ki67 increased in the presence of IL-15 (Fig. 4C).
Fig. 4.
IL-15 expands the CD4+ TSCM population in the absence of infection. (A) Bar graphs show the distribution of CD4+ T-cell subsets in PBMCs from two different donors after 8 d of IL-2 (Left) or IL-15 (Right) stimulation as measured by CyTOF (±SEM). (B) Fold change in the percentage of different CD4+ T-cell subsets after IL-15 treatment compared with IL-2 (n = 2 donors; ±SEM). The percentage of IL-2–stimulated CD4+ T cells was set to one. (C) The percentage of Ki67+ cells in the different CD4+ T-cell subsets present in uninfected samples was measured by CyTOF (average of two donors; ±SEM).
Second, we analyzed the effects of IL-15 on HIV infection. By gating the productively infected lymphocytes (Gag p24+) (SI Appendix, Fig. S5A), we confirmed that IL-15 stimulation increased the susceptibility of CD4+ T cells to infection compared with IL-2 treatment (SI Appendix, Fig. S5B). When treated with IL-2, CD4+ TSCM were infected as efficiently as the other memory CD4+ T cells, while naïve CD4+ T cells remained largely refractory to infection (Fig. 5A). The proportion of CD4+ T-cell subsets changed upon infection (compare Fig. 5B, Left to Right). In the presence of IL-15, susceptibility to infection increased in all CD4+ T-cell subsets (Fig. 5C). The percentage of CD4+ TSCM further increased within the productively infected cells, as CD4+ TSCM represented 8.9% of the total infected CD4+ T cells in donor 1 and 14.3% in donor 2, likely due to increased proliferation (Fig. 4C). Therefore, the memory CD4+ T-cell pool is higher in infected IL-15–treated samples, while the percentage of naïve CD4+ T cells decreased over an order of magnitude (Fig. 5D). In conclusion, CyTOF proved a powerful method to dissect the dynamics of CD4+ T-cell infection and to provide a detailed representation of cell cycle deregulation associated with productive HIV infection. Our data indicate that CD4+ TSCM are highly vulnerable to HIV infection and display the highest proliferation capacity among memory CD4+ T lymphocytes.
Fig. 5.
TSCM compartment increases in the infected CD4+ T cells. (A) The percentage of IL-2–stimulated, productively infected (Gag-p24+) CD4+ T-cell subsets from two different donors was determined by CyTOF. (B) Bar graphs show the distribution of the different CD4+ T-cell subsets in the uninfected cell population (HIV neg; Left) and in the productively infected cell population (HIV pos; Right) stimulated with IL-2 (n = 2 donors; ±SEM). (C) The percentage of IL-15–stimulated, productively HIV-infected (Gag-p24+) CD4+ T-cell subsets from two different donors was determined by CyTOF. (D) Bar graphs show the distribution of the different CD4+ T-cell subsets in the uninfected cell population (HIV neg; Left) and in the productively infected cell population (HIV pos; Right) stimulated with IL-15 (n = 2 donors; ±SEM).
SAMHD1 Is Phosphorylated in CD4+ TSCM.
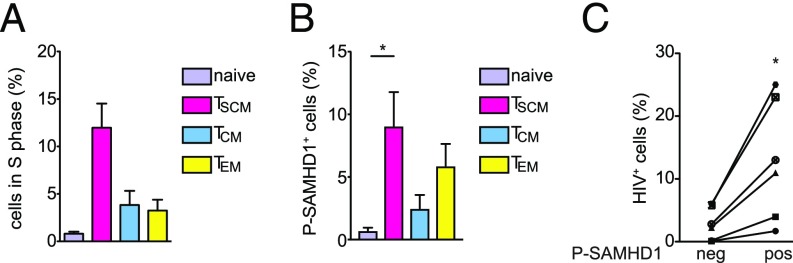
Given the inherent high proliferation capacity of CD4+ TSCM described in the CyTOF immune profiling (Fig. 4C), we hypothesized that SAMHD1 was present in its phosphorylated, nonantiviral form in CD4+ TSCM. As CDK1 is mainly expressed during the S phase of cell cycle, we first determined the percentage of CD4+ T cells in the S phase using iododeoxyuridine (IdU) IdU incorporation determined in the CyTOF experiments (SI Appendix, Fig. S6 shows the gating strategy). We observed that the CD4+ TSCM subset has the highest percentage of cells in S phase compared with the other T-cell subpopulations (Fig. 6A). We directly measured the quantity of P-SAMHD1 at the single-cell level in the different CD4+ T-cell subsets by FACS. Coupling an intracellular staining for P-SAMHD1 with membrane staining, we quantified the amount of P-SAMHD1 within the different CD4+ T-cell subsets. We first confirmed the specificity of P-SAMHD1 staining by detecting the increased positivity of CD4+ T cells for P-SAMHD1 after IL-2/PHA treatment compared with untreated and IL-2 only-treated cells (SI Appendix, Fig. S7A).
Fig. 6.
CD4+ TSCM express P-SAMHD1. (A) Bar graph shows the percentage of cells in S phase in the different CD4+ T-cell subsets as measured by IdU incorporation by CyTOF (n = 2 donors; ±SEM). (B) Quantification of P-SAMHD1 in the different CD4+ T-cell subsets using intracellular FACS (n = 8 donors; ± SEM). (C) The difference in the percentages of HIV-infected CD4+ T cells expressing the P-SAMHD1 (pos) and the CD4+ T cells that do not express P-SAMHD1 (neg) is shown. Experiments were done using CD4+ T cells from six different donors. *P < 0.05 using a two-tailed paired Student’s t test.
We cultured CD4+ T cells for 3 d in IL-15 and then separated the cells in CD45RO− and CD45RO+ subpopulations using magnetic microbeads. Subsequently, we performed membrane staining with CCR7 and CD95 and intracellular staining with anti–P-SAMHD1 antibody (SI Appendix, Fig. S7 B and C) and analyzed the cells by FACS. We observed that the percentage of CD4+ T cells containing P-SAMHD1 was highest in CD4+ TSCM followed by CD4+ TEM, CD4+ TCM, and naïve CD4+ T cells (Fig. 6B). These findings indicate that the susceptibility of CD4+ TSCM to infection in the presence of IL-15 likely derives from the presence of P-SAMHD1. Therefore, we investigated the susceptibility of cells expressing the phosphorylated form of SAMHD1 compared with CD4+ T cells that do not contain the phosphorylated form of SAMHD1. We infected IL-15–stimulated CD4+ T cells with HIV-NL4.3. Five days postinfection, we performed intracellular staining for both HIV p24 and P-SAMHD1. We observed that a higher percentage of CD4+ T cells expressing P-SAMHD1 was productively infected (Gag p24+) compared with cells negative for P-SAMHD1 (Fig. 6C). Overall, these data confirm that the presence of the phosphorylated form of SAMHD1, which lacks its antiviral activity, is strongly associated with the susceptibility to HIV infection in primary CD4+ T cells.
Discussion
Here, we show that IL-15, a γc-cytokine specifically up-regulated during acute HIV infection, increases the susceptibility of primary human CD4+ cells to HIV infection in an SAMHD1-dependent manner. Our systematic analysis of upstream signaling pathways identified the JAK1 pathway as responsible for IL-15–induced SAMHD1 phosphorylation (Fig. 2A). Treatment of CD4+ T cells with Ruxolitinib, a Food and Drug Administration-approved JAK1/2 inhibitor, blocked IL-15–induced SAMHD1 phosphorylation.
After IL-15 treatment, higher percentages of CD4+ T cells were in S phase compared with untreated cells and IL-2–treated cells (Fig. 1D). These data suggest that IL-15 induces cell cycle entry more efficiently than IL-2. CDK1, a kinase expressed mainly in S phase, is required for SAMHD1 phosphorylation (11, 12). We found that only primary CD4+ T cells stimulated with IL-15 do express CDK1, while unstimulated CD4+ T cells do not express CDK1 (Fig. 2C). This process is completely reversed by the JAK inhibitor Ruxolitinib, which abolishes CDK1 expression in IL-15–treated cells, (Fig. 2 B and C). In the context of infection, Ruxolitinib reduced HIV reverse transcription products, thereby limiting the number of productive infection events (Fig. 3 D and E).
We also observed that JAK inhibition impairs the general activation status of T cells without affecting their viability (SI Appendix, Fig. S2 C and D). Ruxolitinib action on cell activation, SAMHD1 phosphorylation, and HIV susceptibility was not limited to IL-15–treated cells. Ruxolitinib treatment decreased both activation levels and HIV susceptibility of IL-2–treated cells (SI Appendix, Figs. S2E and S3 B and C). Ruxolitinib also blocked both IL-15– and IL-7–induced SAMHD1 phosphorylation, while Desatinib, a pan-SRC kinase inhibitor, only inhibited IL-7–induced SAMHD1 phosphorylation (20–22) but not IL-15–induced SAMHD1 phosphorylation (Fig. 2D). This result is not surprising, since all three receptors (IL-2, IL-7, and IL-15) signal through JAK1 and JAK3. In addition, our data suggest that IL-7 and IL-15 initially activate JAK by binding to their respective receptors, but after this initial shared signal, the pathways responsible for IL-7– and IL-15–induced SAMHD1 phosphorylation diverge. Future studies will dissect the precise upstream components in each pathway that control SAMHD1 phosphorylation in CD4+ T cells and investigate whether activation of the JAK pathway exerts positive effects on other steps in the HIV lifecycle [e.g., LTR-driven transcription (49)].
Ruxolitinib treatment limits infection only in IL-15–stimulated lymphocytes, but baseline infection levels in resting CD4+ T cells remain unchanged. The presence of Vpx in the virion rescued HIV infection in IL-15– and Ruxolitinib-treated cells, suggesting that the presence of SAMHD1 is necessary for the antiviral activity of Ruxolitinib (Fig. 3D). Previous reports using immortalized cell lines and activated CD4+ T cells have shown that Ruxolitinib interferes with HIV replication and reactivation from latency (45, 49, 50). Our studies support that Ruxolitinib exerts additional antiviral activity that maps to SAMHD1. This SAMHD1-based antiviral activity of Ruxolitinib is limited to primary CD4+ T cells and would not occur in T-cell lines given the high dNTP pools inherent to these cells (10). A recent report suggests that SIV Vpx degrades not only SAMHD1 but also, TASOR, a subunit of the HUSH repressor complex in activated CD4+ T cells (43). Thus, we cannot exclude that some of the infectivity observed with HIV*GFP-Vpx and HIV*GFP-Vpx Q76A is due to counteraction of the HUSH repressor complex (43). Future studies are need to determine the impact of this repressor complex on infection of IL-15–stimulated primary CD4+ T memory cells, such as the ones used in our study.
We also investigated the possible role of IL-15 and Ruxolitinib in human macrophages. Macrophages can secrete IL-15, which may also directly regulate macrophage function (51, 52). However, IL-15 does not increase expression of CDK1 or modulate SAMHD1 phosphorylation in macrophages. Therefore, IL-15 does not enhance HIV infection in this cell type (SI Appendix, Fig. S4), suggesting that IL-15 exerts different functions in a cell type-dependent manner. Macrophages are terminally differentiated cells, and their susceptibility to infection has been linked to a G1-like phase, in which cell cycle-associated proteins are expressed without cell cycle progression (53, 54). It is possible that IL-15 signaling in macrophages is not linked to cell proliferation and does not impact the percentage of these G1-like macrophages. However, we cannot rule out that macrophages differentiated with other stimuli (GM-CSF or M-CSF) might be responsive to IL-15. GM-CSF–derived macrophages are more resistant to HIV infection than M-CSF–derived macrophages due to cyclin D2 expression (54). In our experimental settings, we differentiated macrophages in the presence of human serum without adding any other growth factors (55). Taken together, our investigations into the molecular effects of IL-15 in the context of HIV infection of CD4+ T-cell populations suggest that IL-15 stimulation augments the cellular susceptibility of CD4+ T cells by eliminating the protective effects provided by SAMHD1, which is prevented by Ruxolitinib.
Multidimensional immune profiling of CD4+ T cells by CyTOF showed that expansion of the CD4+ TSCM subset in both infected and uninfected populations further amplifies the proviral effect of IL-15 (Figs. 4 and 5). Emerging evidence suggests that CD4+ TSCM play a key role in HIV and SIV pathogenesis (34, 37–39). In HIV-infected patients, CD4+ TSCM contain replication-competent proviruses (34), and the percentage of infected TSCM is linked to pathogenicity (38, 39). Our high-resolution maps of primary human CD4+ T cells exposed to IL-15 with and without HIV infection generated by CyTOF captured around 86% of all CD4+ T cells. Since some CD4+ T-cell subsets, such as CD4+ T-regulatory and CD4+ T-terminal effector cells, were not identified using the antibodies that we selected for this analysis, our findings remain limited to the four main subsets of primary CD4+ T cells (naïve cells, TCM, TEM, and TSCM).
We also observed that CD4+ TSCM have the highest proliferation capacity of all CD4+ T subsets, a phenotype that was further enhanced by IL-15 (Ki67 expression) (Fig. 6C). This pronounced renewal capacity would allow these special cells, if infected, to propagate infection by cellular proliferation (56). Clonal expansion of infected cells could arise from a few CD4+ TSCM infected during the early phase of HIV infection, which subsequently seed the persistent HIV reservoir by proliferation and differentiation (57, 58).
Since the antiviral activity of SAMHD1 is influenced by the cell cycle status, we wondered whether TSCM display such a high susceptibility to HIV infection, because they are proliferating and express a phosphorylated, nonantiviral form of SAMHD1. We found a higher percentage of TSCM in S phase compared with the other analyzed subsets (Fig. 6A). In addition, in the presence of IL-15, the CD4+ TSCM subset expressed more P-SAMHD1 (Fig. 6B). The importance of this posttranslational modification in regulating HIV susceptibility was confirmed by our analysis of CD4+ T-cell infection levels in cells positive for P-SAMHD1 compared with cells that do not express P-SAMHD1. We did observe that expression of the nonantiviral form of SAMHD1 strongly correlates with infection susceptibility in CD4+ T cells (Fig. 6C).
Based on our findings, we propose a model in which IL-15 produced during primary HIV infection helps establish the persistent reservoir in two ways. First, it induces cell cycle entry and thereby, phosphorylation of SAMHD1. Second, it increases the proliferation of CD4+ TSCM. Since CD4+ TSCM persist for prolonged periods and give rise to different lineages of progeny cells, we hypothesize that HIV exploits the stemness of CD4+ TSCM to create a self-renewing, persistent reservoir, which bypasses the need for de novo infections. Timely intervention with Ruxolitinib or other JAK inhibitors may prevent IL-15–induced phosphorylation of SAMHD1, which will preserve its natural antiviral activity and constrain the establishment of the persistent HIV reservoir.
Experimental Procedures
Cell Isolation and Cell Culture.
HEK 293T and TZM-bl cells were maintained in DMEM supplemented with 10% FBS (Gibco) and penicillin/streptomycin (Sigma) at 37 °C in a 5% CO2 humidified incubator. TZM-bl was obtained from John C. Kappes, Department of Medicine, University of Alabama at Birmingham, Birmingham, AL, Xiaoyun Wu, State Key Laboratory of Virology and the Modern Virology Research Center, College of Life Sciences, Wuhan University, Wuhan, PR China, and Tranzyme Inc. through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH (59). Testing for mycoplasma was carried out using the MycoAlert Mycoplasma Detection kit (Lonza).
CD4+ T cells were purified from peripheral blood mononuclear cells (PBMCs) or peripheral blood lymphocytes obtained from anonymous healthy blood donors (New York Blood Center). Ficoll (Ficoll Hystopaque; Sigma) density centrifugation was performed as per the manufacturer’s instructions, and CD4+ cells were negatively selected using magnetic beads (CD4+ T-cell isolation kit I; Miltenyi Biotec). CD4+ T cells were cultured in RPMI 1640 supplemented with 10% FBS (Gibco), 100 IU penicillin, 100 µg/mL streptomycin, 0.1 M Hepes, 2 mM l-glutamine, and/or recombinant human IL-2 (NIH AIDS Reagent Program), IL-15, IL-7, or IL-4 (R&D) as indicated. Cells were maintained at 37 °C in a 5% CO2 humidified incubator. CD45RA and CD45RO populations were isolated using CD45RO MicroBeads (Miltenyi Biotec) as per the manufacturer’s instructions.
CD14+ cells were isolated from PBMCs using an MACS CD14 isolation kit (Miltenyi Biotec). CD14+ cells were differentiated into macrophages by culturing the cells in RPMI supplemented with 10% human serum for 6 d as previously described (55).
Production of Viral Stocks.
pBR HIV NL4.3 nef-IRES-Renilla Δenv was previously described (60, 61), HIV R7/3 GFP was a gift of Cecilia Cheng Mayer, Aaron Diamond AIDS Research Center, The Rockefeller University, New York (62), and HIV NL4.3 was obtained from the AIDS Research and Reference Reagent Program (63). Transmitter founder molecular clone HIV pCH040.c/2625 was a gift of Beatrice H. Hahn, Departments of Microbiology and Medicine, University of Pennsylvania, Philadelphia (64).
Viral stocks were generated by transfection of HEK 293T with polyethylenimine (Polysciences). Two days after transfection, culture supernatants were collected, clarified at 441 × g for 5 min, and filtered (0.45 μm). pHIV*GFP and pcDNA3.1Vpx SIVmac239-Myc (WT and Q76A) were gifts of Oliver Fackler, Infectious Disease Research, Integrative Virology, University Hospital Heidelberg, Heidelberg (10, 65). HIV*GFP with and without Vpx was generated by cotransfection of pHIV*GFP with pcDNA3.1Vpx SIVmac239-Myc WT, pcDNA3.1Vpx SIVmac239-Myc Q76A, or pcDNA3.1 in a 2:1 ratio. Viruses were purified on a 6% Optiprep cushion (Sigma) by centrifugation at 14,000 × g for 6 h. Viral titers were determined by infecting TZM-bl reporter with triplicate serial dilutions of the viral stocks as previously described (66).
HIV Infection Experiments.
Primary CD4+ T cells were stimulated with ILs for 72 h or with 1 μg/mL phytohemagglutinin-P (Sigma) for 48 h before infection.
Drugs were added at the time of stimulation at the following concentrations: Ruxolitinib, 50 nM; Desatinib, 75 nM; Wortmannin, 200 nM; PD0325901, 20 nM (all from Sigma). Infections with HIV R7/3-GFP, HIV NL4.3, HIV CHO40.C/2625, HIV-Renilla Luciferase R5Env, and HIV-NL4.3/BALenv were performed overnight in the presence of polybrene (2 μg/mL), and fresh media were replaced. The number of infected cells was determined by measuring GFP or intracellular HIV p24 by FACS at different time points after infection (ranging from 48 h to 7 d depending on the experiments). Renilla luciferase values were measured using the Renilla Luciferase Assay System kit (Promega).
Infections with HIV*GFP with and without Vpx were performed as previously described (10, 65). Briefly, CD4+ T cells were spinoculated at 115 × g for 2 h with equal amounts of viruses. Media were replaced after 4 h. The multiplicity of infection used for infection varied between 0.1 and 0.05. The number of GFP-expressing cells was analyzed 48 h postinfection by FACS.
Immunoblotting.
CD4+ T cells (between 106 and 3 × 106) were lysed in RIPA buffer supplemented with complete protease inhibitor (Roche). Proteins were separated on 10% SDS-polyacrylamide gels (Invitrogen). The following antibodies were used: anti-SAMHD1 (12361; Cell Signaling), anti-GAPDH (32233; Santa Cruz), anti-STAT3 (sc-482; Santa Cruz), antiphospho-STAT3 (9145; Cell Signaling), anti-STAT5 (9358; Cell Signaling), antiphospho-STAT5 (9351; Cell Signaling), anti-Erk1/2 (4696; Cell Signaling), antiphospho-Erk1/2 (4370; Cell Signaling), anti-Akt (2920; Cell Signaling), antiphospho-Akt (4060; Cell Signaling), and anti-CDK1 (Cdc2; 9116; Cell Signaling). The anti–P-SAMHD1 was previously described (11).
Flow Cytometry Analysis.
Between 105 and 106 CD4+ T cells were stained and analyzed on an LSRII (BD Biosciences) FACS machine or Guava EasyCyte Flow Cytometer (Millipore). An average of 105 cells were acquired per sample, and data were analyzed using FlowJo software. The following antibodies were used: CD25-PerCP (1:100), anti–CD95-Apc-Cy7 (1:200), anti–CCR7-FITC (1:50), anti–CD45RA-PerCP-Cy5 (1:50; Biolegend), and CD69-PE (1:100) (BD).
For the HIV p24 intracellular staining, cells were fixed and permeabilized with the BD fix and perm kit and stained with Kc-57-RD1 (1:100; Beckam-Coulter). For P-SAMHD1 intracellular staining, CD4+ T cells were fixed in formaldehyde (4%), permeabilized in cold 90% methanol (30 min at −20 °C), and stained with anti–P-SAMHD1 (1:200) followed by anti-rabbit Alexa647 (1:500; Thermo Fisher Scientific).
When optimizing the intracellular P-SAMHD1 FACS assay, cell viability was determined using the LIVE/DEAD Fixable Dead Cell Stain (Thermo Fisher Scientific). We noted an increased positivity for the LIVE/DEAD Fixable Dead Cell Stain after permeabilization of the cells with cold 90% methanol, resulting in an overestimation of the number of dead cells. In all of the experiments, viability was, thus, determined using forward vs. side scatter (FSC vs. SSC) gating.
Quantification of Viral DNA.
Viral stocks were pretreated for 1 h at room temperature with DnaseI (10 U/mL; BioLabs) and Benzonase (50 U/mL; Millipore) before infection to remove background coming from plasmid DNA.
Cellular DNA was isolated from infected CD4+ T cells at the indicated time points postinfection using the QiAmp DNA Blood Mini kit (Qiagen); 100 ng of total DNA was used to quantify LRTs and integrated proviral DNA as previously described using real time PCR (BioRad) (67–69). Of note, we used the same amount of DNA as input for both LRT and Alu-PCR and in addition, normalized each sample to an internal control of total cellular DNA (RNAseP gene) using the delta cycle threshold (CT) method. For each donor, these data were then expressed as fold change over LRT or provirus levels detected in IL-15–treated cells at 24 h (set to one).
CyTOF.
Ten million infected and uninfected CD4+ T cells were stained with IdU to determine the amount of DNA and Rh103 as a viability dye for 30 min at 37 °C. Cells were washed in FACS buffer (2% FBS in PBS), stained with CD45 antibody for barcoding (CD45_Pr141Di, CD45_Ho165Di, CD45_Tm169Di, CD45_In115Di), and pooled. Cells were washed with FACS buffer and resuspended in the membrane antibody mixture containing CD45RA_Nd143Di, CCR5_Nd144Di, CD4_Nd145Di, CD8_Nd146Di, CD45RO_Sm149Di, CD62L_Eu152Di, CD3_Sm154Di, CD27_Gd155Di, CCR7_Tb159Di, CD95_Dy164Di, CD122_Er170Di, and CXCR4_Yb173Di. After 15 min, cells were washed in FACS buffer, and Cisplatin was added for 5 min. Lymphocytes were fixed and permeabilized with Cytofix/Cytoperm solution (BD), and they were stained with KC57-FITC (Beckam-Coulter) for 20 min. Cells were washed with Perm Wash Buffer (BD) and stained with anti–FITC-160_Gd. Cells were washed and resuspended in Fix solution (2% paraformaldehyde) and iridium intercalator. Between 200,000 and 600,000 CD4+ T cells were acquired per individual sample on the CyTOF2 mass cytometer (Fludigm).
On acquisition, gating was done manually using the dot plot display on Cytobank. The gating strategy for the CD45RO+ memory compartment consisted of CD45RA− gating followed by gating on CD27 and CCR7 to discriminate between TCM and TEM (70). To distinguish naïve cells from TSCM, we used a CD95 gate on CD45RA+, CCR7+ and CD27+ as previously described (36, 38). IdU incorporation was used to determine cells in S phase (71).
Statistical Analysis.
Statistical tools in GraphPad Prism (version 7) were used to perform all of the statistical analysis. All results are given as mean ± SEM. Statistical analyses were performed using a two-tailed Student t test. Differences with P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Adeeb Rahman and the Human Immune Monitoring Core at Icahn School of Medicine at Mount Sinai for expertise and assistance. We also thank Dabeiba Bernal for processing of PBMCs, Shane Curran and Bipin Bathia for their assistance on the Guava EasyCyte Flow Cytometer, and Life Science Editors for editing assistance. L.C.F.M. is supported by NIH/National Institute of General Medical Sciences Grants R01 GM113886 and R01 GM113886-03S1. B.L. acknowledges partial support from Australian Government National Health and Medical Research Council Grant APP1086178 and the Ward–Coleman estate for endowing the Ward–Coleman Chairs at the Icahn School of Medicine at Mount Sinai. This research was supported by NIH/National Institute of Allergy and Infectious Diseases Grants U19 AI1186101 (to A.F.-S.), R01 AI07345 (to A.F.-S.), AI125173 (to V.S.), R01 AI136916 (to V.S.), and R01 AI120998 (to V.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806695115/-/DCSupplemental.
References
- 1.Chun TW, et al. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: What, where and how to target them. Nat Rev Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 3.Zack JA, Kim SG, Vatakis DN. HIV restriction in quiescent CD4+ T cells. Retrovirology. 2013;10:37. doi: 10.1186/1742-4690-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan X, Baldauf HM, Keppler OT, Fackler OT. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 2013;23:876–885. doi: 10.1038/cr.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shan L, Siliciano RF. From reactivation of latent HIV-1 to elimination of the latent reservoir: The presence of multiple barriers to viral eradication. BioEssays. 2013;35:544–552. doi: 10.1002/bies.201200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descours B, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldauf HM, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White TE, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol. 2013;87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benanti JA. Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity. Curr Genet. 2016;62:271–276. doi: 10.1007/s00294-015-0535-5. [DOI] [PubMed] [Google Scholar]
- 15.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Napolitano LA, et al. Increased production of IL-7 accompanies HIV-1-mediated T-cell depletion: Implications for T-cell homeostasis. Nat Med. 2001;7:73–79. doi: 10.1038/83381. [DOI] [PubMed] [Google Scholar]
- 17.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberly MD, et al. Increased IL-15 production is associated with higher susceptibility of memory CD4 T cells to simian immunodeficiency virus during acute infection. J Immunol. 2009;182:1439–1448. doi: 10.4049/jimmunol.182.3.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, et al. Interleukin-15 (IL-15) strongly correlates with increasing HIV-1 viremia and markers of inflammation. PLoS One. 2016;11:e0167091. doi: 10.1371/journal.pone.0167091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das J, et al. 2-aminothiazole as a novel kinase inhibitor template. Structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1- piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J Med Chem. 2006;49:6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- 21.Coiras M, et al. IL-7 induces SAMHD1 phosphorylation in CD4+ T lymphocytes, improving early steps of HIV-1 life cycle. Cell Rep. 2016;14:2100–2107. doi: 10.1016/j.celrep.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermejo M, et al. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol. 2016;106:30–45. doi: 10.1016/j.bcp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Verbist KC, Klonowski KD. Functions of IL-15 in anti-viral immunity: Multiplicity and variety. Cytokine. 2012;59:467–478. doi: 10.1016/j.cyto.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodolce JP, Burkett PR, Koka RM, Boone DL, Ma A. Regulation of lymphoid homeostasis by interleukin-15. Cytokine Growth Factor Rev. 2002;13:429–439. doi: 10.1016/s1359-6101(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 25.Cieri N, et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood. 2013;121:573–584. doi: 10.1182/blood-2012-05-431718. [DOI] [PubMed] [Google Scholar]
- 26.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattei F, Schiavoni G, Belardelli F, Tough DF. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J Immunol. 2001;167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 28.Kanai T, Thomas EK, Yasutomi Y, Letvin NL. IL-15 stimulates the expansion of AIDS virus-specific CTL. J Immunol. 1996;157:3681–3687. [PubMed] [Google Scholar]
- 29.Mueller YM, et al. IL-15 enhances survival and function of HIV-specific CD8+ T cells. Blood. 2003;101:1024–1029. doi: 10.1182/blood-2002-07-1957. [DOI] [PubMed] [Google Scholar]
- 30.Petrovas C, et al. HIV-specific CD8+ T cells exhibit markedly reduced levels of Bcl-2 and Bcl-xL. J Immunol. 2004;172:4444–4453. doi: 10.4049/jimmunol.172.7.4444. [DOI] [PubMed] [Google Scholar]
- 31.Mueller YM, et al. IL-15 treatment during acute simian immunodeficiency virus (SIV) infection increases viral set point and accelerates disease progression despite the induction of stronger SIV-specific CD8+ T cell responses. J Immunol. 2008;180:350–360. doi: 10.4049/jimmunol.180.1.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okoye A, et al. Profound CD4+/CCR5+ T cell expansion is induced by CD8+ lymphocyte depletion but does not account for accelerated SIV pathogenesis. J Exp Med. 2009;206:1575–1588. doi: 10.1084/jem.20090356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. T memory stem cells in health and disease. Nat Med. 2017;23:18–27. doi: 10.1038/nm.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzon MJ, et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cashin K, et al. Differences in coreceptor specificity contribute to alternative tropism of HIV-1 subtype C for CD4(+) T-cell subsets, including stem cell memory T-cells. Retrovirology. 2014;11:97. doi: 10.1186/s12977-014-0097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabler CO, et al. CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J Virol. 2014;88:4976–4986. doi: 10.1128/JVI.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaafoura S, et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T Cells. Nat Commun. 2014;5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klatt NR, et al. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog. 2014;10:e1004345. doi: 10.1371/journal.ppat.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartwright EK, et al. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J Immunol. 2014;192:4666–4673. doi: 10.4049/jimmunol.1303193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J Exp Med. 1999;189:1735–1746. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pauls E, et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 42.Budagian V, Bulanova E, Paus R, Bulfone-Paus S. IL-15/IL-15 receptor biology: A guided tour through an expanding universe. Cytokine Growth Factor Rev. 2006;17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Chougui G, et al. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat Microbiol. 2018;3:891–897. doi: 10.1038/s41564-018-0179-6. [DOI] [PubMed] [Google Scholar]
- 44.Reinhard C, Bottinelli D, Kim B, Luban J. Vpx rescue of HIV-1 from the antiviral state in mature dendritic cells is independent of the intracellular deoxynucleotide concentration. Retrovirology. 2014;11:12. doi: 10.1186/1742-4690-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavegnano C, et al. Novel mechanisms to inhibit HIV reservoir seeding using Jak inhibitors. PLoS Pathog. 2017;13:e1006740. doi: 10.1371/journal.ppat.1006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung RK, Utz PJ. Screening: CyTOF-the next generation of cell detection. Nat Rev Rheumatol. 2011;7:502–503. doi: 10.1038/nrrheum.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai L, Ong R, Li J, Albani S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytometry A. 2015;87:369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavegnano C, et al. Ruxolitinib and tofacitinib are potent and selective inhibitors of HIV-1 replication and virus reactivation in vitro. Antimicrob Agents Chemother. 2014;58:1977–1986. doi: 10.1128/AAC.02496-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spivak AM, et al. Janus kinase inhibition suppresses PKC-induced cytokine release without affecting HIV-1 latency reversal ex vivo. Retrovirology. 2016;13:88. doi: 10.1186/s12977-016-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alleva DG, Kaser SB, Monroy MA, Fenton MJ, Beller DI. IL-15 functions as a potent autocrine regulator of macrophage proinflammatory cytokine production: Evidence for differential receptor subunit utilization associated with stimulation or inhibition. J Immunol. 1997;159:2941–2951. [PubMed] [Google Scholar]
- 52.Rückert R, et al. Interleukin-15 stimulates macrophages to activate CD4+ T cells: A role in the pathogenesis of rheumatoid arthritis? Immunology. 2009;126:63–73. doi: 10.1111/j.1365-2567.2008.02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mlcochova P, et al. A G1-like state allows HIV-1 to bypass SAMHD1 restriction in macrophages. EMBO J. 2017;36:604–616. doi: 10.15252/embj.201696025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badia R, et al. The G1/S specific cyclin D2 is a regulator of HIV-1 restriction in non-proliferating cells. PLoS Pathog. 2016;12:e1005829. doi: 10.1371/journal.ppat.1005829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown A, Wang X, Sawai E, Cheng-Mayer C. Activation of the PAK-related kinase by human immunodeficiency virus type 1 Nef in primary human peripheral blood lymphocytes and macrophages leads to phosphorylation of a PIX-p95 complex. J Virol. 1999;73:9899–9907. doi: 10.1128/jvi.73.12.9899-9907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med. 2017;214:959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol. 2009;83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manganaro L, et al. HIV Vpu interferes with NF-κB activity but not with interferon regulatory factor 3. J Virol. 2015;89:9781–9790. doi: 10.1128/JVI.01596-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Münch J, et al. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol. 2007;81:13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakrabarti LA, Ivanovic T, Cheng-Mayer C. Properties of the surface envelope glycoprotein associated with virulence of simian-human immunodeficiency virus SHIV(SF33A) molecular clones. J Virol. 2002;76:1588–1599. doi: 10.1128/JVI.76.4.1588-1599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baldauf HM, et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc Natl Acad Sci USA. 2017;114:2729–2734. doi: 10.1073/pnas.1613635114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ooms M, Majdak S, Seibert CW, Harari A, Simon V. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J Virol. 2010;84:7961–7969. doi: 10.1128/JVI.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 68.Manganaro L, et al. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat Med. 2010;16:329–333. doi: 10.1038/nm.2102. [DOI] [PubMed] [Google Scholar]
- 69.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flynn JK, et al. Quantifying susceptibility of CD4+ stem memory T-cells to infection by laboratory adapted and clinical HIV-1 strains. Viruses. 2014;6:709–726. doi: 10.3390/v6020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.