Significance
Understanding the structure and function of the human brain at a cellular level is a fundamental aim of neuroscience. Tremendous progress has been made in recent years based on different in vivo and ex vivo approaches, including major advances in brain MRI. However, uncertainties remain in determining how brain MRI measurements relate to the brain’s underlying cellular composition. In this paper we use a recently developed MRI technique, quantitative gradient recalled echo (qGRE), and information on gene profiles in the human brain available from the Allen Human Brain Atlas. We demonstrate that qGRE and related MRI techniques can be used to probe the underlying cellular composition of the human brain in vivo.
Keywords: functional MRI, microstructural MRI, Allen Human Brain Atlas, human cortex cellular composition, R2t*
Abstract
fMRI revolutionized neuroscience by allowing in vivo real-time detection of human brain activity. While the nature of the fMRI signal is understood as resulting from variations in the MRI signal due to brain-activity-induced changes in the blood oxygenation level (BOLD effect), these variations constitute a very minor part of a baseline MRI signal. Hence, the fundamental (and not addressed) questions are how underlying brain cellular composition defines this baseline MRI signal and how a baseline MRI signal relates to fMRI. Herein we investigate these questions by using a multimodality approach that includes quantitative gradient recalled echo (qGRE), volumetric and functional connectivity MRI, and gene expression data from the Allen Human Brain Atlas. We demonstrate that in vivo measurement of the major baseline component of a GRE signal decay rate parameter (R2t*) provides a unique genetic perspective into the cellular constituents of the human cortex and serves as a previously unidentified link between cortical tissue composition and fMRI signal. Data show that areas of the brain cortex characterized by higher R2t* have high neuronal density and have stronger functional connections to other brain areas. Interestingly, these areas have a relatively smaller concentration of synapses and glial cells, suggesting that myelinated cortical axons are likely key cortical structures that contribute to functional connectivity. Given these associations, R2t* is expected to be a useful signal in assessing microstructural changes in the human brain during development and aging in health and disease.
Understanding the structure and function of the human brain at a cellular level is a fundamental aim of neuroscience. Most of our knowledge on brain microstructure comes from numerous ex vivo studies originating with Ramón y Cajal (1). Tremendous progress in ex vivo analysis of the human brain has been made with multiple efforts, including the recent development of the Allen Human Brain Atlas (AHBA), which provides a comprehensive view of gene expression profiles across the human brain (2).
MRI similarly revolutionized neuroscience by providing researchers with tools for in vivo study of the human brain (3). Given these complementary advances, an emerging trend in neuroscience is to combine MRI measurements with information available from the AHBA (4, 5) to explore the relationship between gene expression profiles, which in part reflect the cellular architecture of the brain, and MRI metrics. Pioneering studies (4, 5) have provided important information on the relationships between MRI-defined functional brain connectivity obtained from analysis of data available from the Human Connectome Project (HCP) (6) and networks of coexpressing genes.
The vast majority of fMRI-based functional brain studies are based on exploring variations in the gradient recalled echo (GRE) MRI signal. These variations are due to a specific element of brain activity [i.e., infraslow activity (7)] that induces changes in the blood oxygenation level—the so-called dynamic BOLD (blood-oxygen-level-dependent) effect (8–11). The GRE signal that is measured at the GRE echo time (TE) after excitation by the rf pulse is usually expressed in terms of the R2* (=1/T2*) signal relaxation:
| [1] |
The GRE signal relaxation rate constant R2* has two tissue-specific contributions, R2′ and R2t*, and is also affected by adverse effects of background gradients (B0 field inhomogeneities). The R2′ part of R2* is related to the GRE signal loss due to the presence of paramagnetic deoxygenated blood in veins and the prevenous part of the capillary bed (8). The R2t* relaxation rate constant depends solely on the cellular environment of water molecules (the main source of the MRI signal), thus providing information on tissue microstructure (12, 13). The recently introduced quantitative GRE (qGRE) method for separation of these two contributions (12) demonstrates that the R2′ of the baseline BOLD effect (i.e., corresponding to the intrinsic, activity-independent blood oxygenation level of about 60%) contributes less than 20% to the total R2*. Brain-activity-related changes in blood oxygenation level (dynamic BOLD) cause changes in R2′ and consequent changes in the GRE signal. This effect is usually quite small—R2* changes by about 1–3%. In the brain tissue outside of large veins, the majority of R2* is not represented by the portion used for fMRI and is rather largely driven by R2t*. Hence, the fundamental (and still not addressed) questions are how the underlying brain microstructure at the cellular level defines the major part of R2* (R2t*) and how R2t* affects changes in the GRE signal during brain activity.
Herein we combine MRI measurements with gene expression information available from AHBA to explore the relationship between human brain gene networks and quantitative metrics of GRE signal related to tissue microstructure and functioning. This allows us to identify the genetic underpinnings of the qGRE signal in human cerebral cortex. We found that three modules of correlated gene networks identified by weighted gene correlation network analysis (WGCNA) strongly correlate with microstructural qGRE metrics, allowing quantitative estimation of cellular contributions to the GRE signal. These modules represent two major types of cortical cells: neurons (two modules) and glial cells. The data also show that regions of the brain with higher R2t* “govern” brain functional activity—they have higher concentration of neuronal processes and produce stronger functional connections with other brain regions. Since MRI is a broadly available noninvasive tool, we anticipate that our findings will open new opportunities for interpreting and applying MRI to the in vivo study of tissue cellular microstructure and functioning in health and disease.
Results
Microstructural and Macrostructural MRI.
We used in vivo brain data obtained from 26 healthy volunteers (age range 23–76 y, 9 males, 17 females) with a Siemens 3T Trio MRI scanner. While the GRE signal decay is usually expressed in terms of the total transverse decay rate constant R2* in Eq. 1, the innovative qGRE method of data analysis (12) allows disentanglement of tissue-cellular-specific (R2t*) and BOLD-related contributions to the total GRE MRI signal decay:
| [2] |
In this equation, S(0) is the GRE signal intensity amplitude, TE is the gradient echo time, function F(TE) accounts for the adverse effects of macroscopic magnetic field inhomogeneities [herein accounted for by using a voxel spread function (VSF) method (14)], and function accounts for the BOLD contributions to the GRE signal decay (12, 15). Whereas in the literature the term BOLD is often attributed to the dynamic, activity-induced changes in the MRI signal, in this paper we adopt a more broad BOLD definition that also includes the effect of the baseline blood oxygenation level on the GRE signal (8, 15). The BOLD model (15) was previously validated in phantoms (16), in silico (17), and in vivo small animal (18) experiments. Details of qGRE data analysis are presented in Materials and Methods.
For all volunteers, we also obtained macrostructural measurements with a magnetization-prepared rapid gradient echo (MPRAGE) (19) sequence to calculate cortical thickness.
WGCNA Analysis of Gene Networks.
To allow comparison with MRI, we summarized gene expression data obtained from six separate AHBA datasets into 68 Desikan–Killiany (20) cortical regions using FreeSurfer (Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital) and analyzed them using WGCNA software and a statistical approach similar to that proposed in ref. 4 (schematically shown in Fig. 1 and described in Materials and Methods). By using all six AHBA datasets, this analysis generated eight consensus gene expression networks (common to all six AHBA datasets) characterized by modules with assigned genes: Black (n = 462), Blue (n = 1,536), Brown (n = 628), Green-yellow (n = 474), Pink (n = 1,115), Purple (n = 1,794), Salmon (n = 978), and Yellow (n = 1,099). All unassigned genes were put into the Gray (n = 12,652) module. Each module is characterized by a module eigengene (ME) that reflects the most common spatial pattern of expression for that module of genes. The resulting cluster dendrogram for the genes and their module assignment is shown in Fig. 2A. The detailed gene assignments are listed in Dataset S1. The stability of this procedure was tested for which results are presented in SI Appendix, Figs. S1–S4.
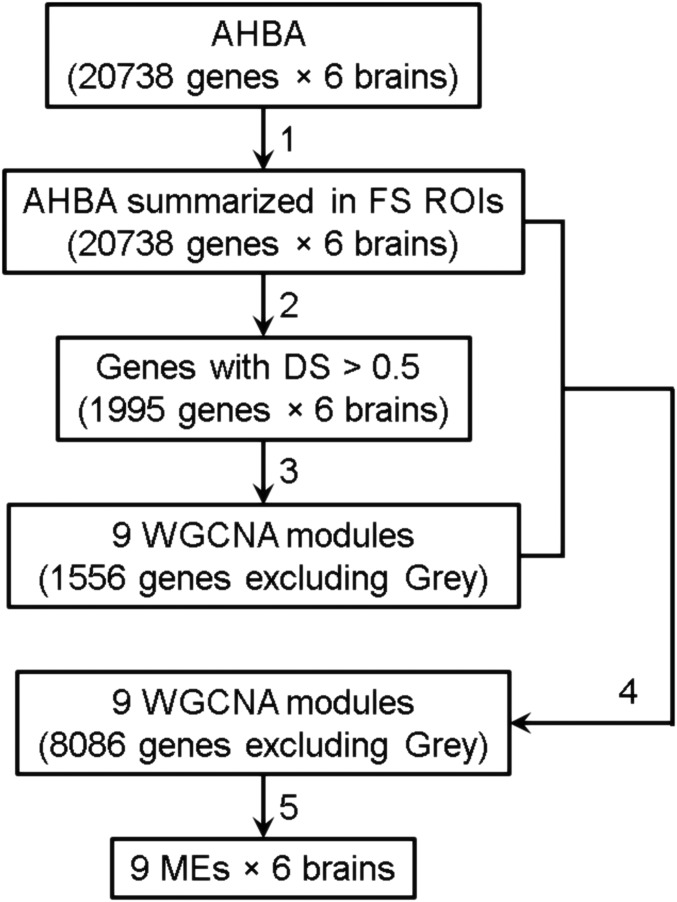
Fig. 1.
Flowchart of the WGCNA analysis. Step 1: Gene expression profiles of six AHBA datasets (each with 20,738 genes) are summarized in FreeSurfer (FS) ROIs. Step 2: DS (4) of each gene expression profile across six AHBA datasets is calculated, resulting in 1,995 genes with DS > 0.5. Step 3: Genes with DS > 0.5 are used as input to the WGCNA analysis, which generated eight consensus networks, i.e., modules containing 1,556 genes. All other genes are assigned to the Gray module. Step 4: Expression profiles of 20,738 genes are correlated with the eight MEs and genes with a correlation coefficient bigger than 0.5 are assigned to the corresponding module. This resulted in 8,086 assigned genes. Step 5: MEs of the nine new modules are calculated.
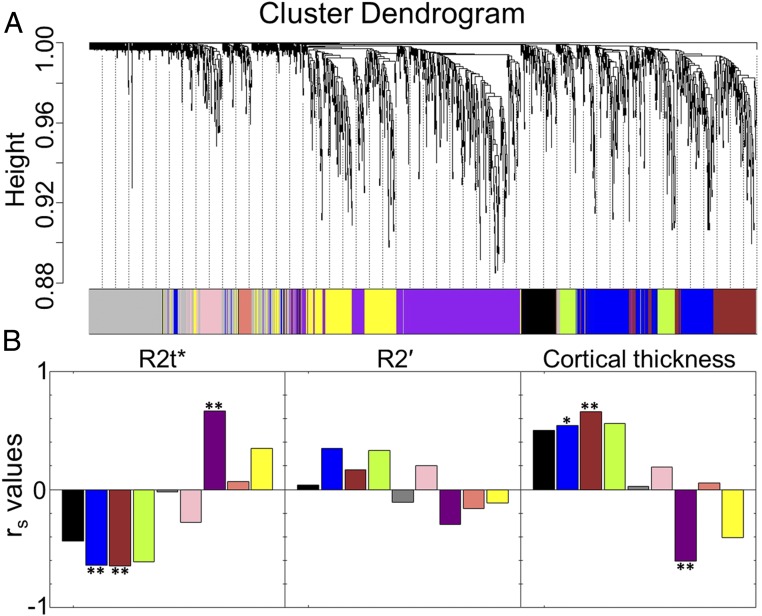
Fig. 2.
Gene coexpression networks and correlations with MRI metrics. (A) Cluster dendrogram showing gene distribution among nine consensus networks (color-coded modules) as generated by WGCNA of 1,995 genes that had similar expression patterns across all six AHBA datasets. The dendrogram is a visual representation of the gene correlation data. The vertical axis refers to a distance measure between genes or gene clusters. Branches of the dendrogram group together densely interconnected, highly coexpressed genes. (B) Correlations between MEs (from step 5 in Fig. 1) and MRI microstructural (R2t*), BOLD (R2′), and macrostructural (cortical thickness) metrics. The significant correlations are marked by ** (averaged Spearman |rs| > 0.6 and P < 0.05 for each of the six AHBA datasets) and * (averaged 0.5 < |rs| < 0.6 and P < 0.05 for each of the six AHBA datasets). We use the Spearman correlation analysis to reduce the role of outliers. All P values were adjusted for multiple comparisons using the FDR method with the Benjamini–Hochberg procedure.
Correlations Between Gene Networks and MRI Metrics.
We used only those genes that have conserved expression profiles across all six AHBA brain data with high differential stability (DS) as described in ref. 4. Averaged Spearman correlation coefficients, rs, were calculated for all nine modules using six AHBA datasets and six age-matched in vivo MRI data (each dataset was averaged from a group of four or five healthy volunteers with ages matched to the AHBA datasets). As shown in Fig. 2B, only eigengenes of three modules showed significant correlations (with averaged | rs | > 0.6 and P < 0.05 across all six AHBA datasets) with MRI (R2t* and cortical thickness). Among them, module eigengene ME-Purple showed positive correlation for R2t* and negative correlation with cortical thickness. Module eigengenes ME-Blue and ME-Brown negatively correlated with R2t* and positively correlated with cortical thickness. Those significantly correlated modules were selected for further analysis.
Cellular/Subcellular Affiliation of MRI-Essential Gene Networks.
To understand the tissue substrate underlying the gene modules that are correlated with R2t*, we employed a ToppGene tool (21) for gene enrichment analysis. To test stability of the enrichment analysis we also used DAVID Bioinformatics Resources (ref. 22; https://david.ncifcrf.gov/) with different background choices (see details in Materials and Methods). As shown in Datasets S3–S8, the selection of background has only a minor effect on the enrichment analysis. The results for cellular affiliation (Coexpression Atlas in ToppGene) shown in Fig. 3A demonstrate that genes in modules Purple and Brown are mainly related to neurons while genes in module Blue are mainly related to glial [including astrocytes, microglia, and oligodendrocyte progenitor cells (OPCs)] and endothelial cells.
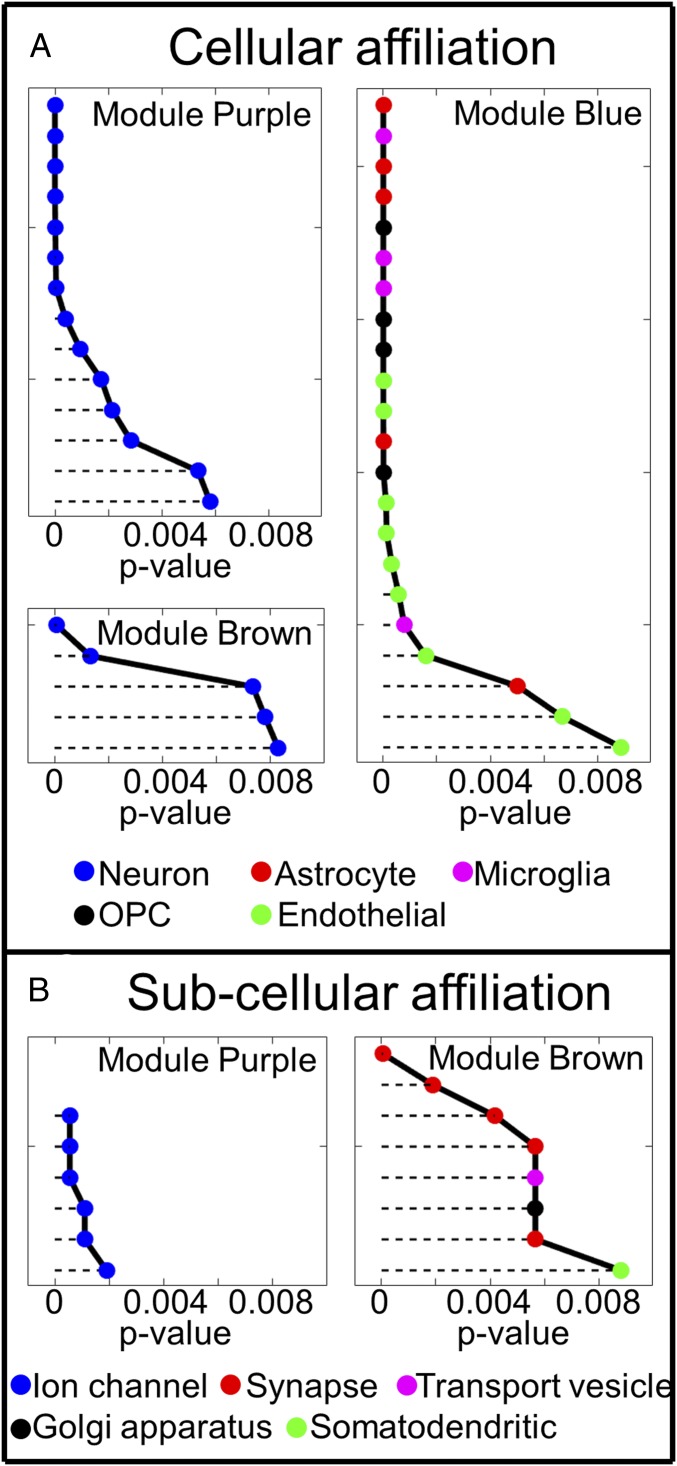
Fig. 3.
Analysis of cellular (A) and subcellular (B) gene affiliation using ToppGene. Each point in this figure represents a predefined cluster of genes that have different cellular (Coexpression Atlas in ToppGene) and subcellular (GO Cellular Component in ToppGene) affiliation in ToppGene. Different clusters associated with different cell types are marked by different colors, as shown at the bottom. Detailed cluster names are listed in Dataset S2. Results show that genes in modules Purple and Brown are affiliated with different parts of the neuron (voltage-gated ion channels in Purple, and mostly synapsis in Brown), while genes in the Blue module are mostly affiliated with glial (astrocytes, microglia, and OPCs) and endothelial cells. All P values were adjusted for multiple comparison using the FDR method with the Benjamini–Hochberg procedure. Only those clusters with P < 0.01 are shown in the figure. Lower P values correspond to higher gene enrichment profiles.
Fig. 3A shows that both the Purple and the Brown modules are associated with neurons. However, these two modules correlate with R2t* with opposite signs. Subcellular [Gene Ontology (GO) Cellular Component in ToppGene] affiliation shown in Fig. 3B demonstrates that genes in module Purple are mainly associated with ion channels that are primarily distributed along neuronal processes—axons (myelinated and not myelinated) and dendrites (23) that typically occupy over 85% of the space comprised by neurons (24). Module Purple also contains gene NeuN, which is a commonly used neuronal marker (25). Noting these findings, the Purple module likely largely reflects a transcriptional correlate for major parts of the neuron—cell bodies and neuronal processes and will be treated in this paper as a representative of neuronal contribution to brain tissue cellular composition.
Conversely, genes in module Brown are mostly associated with the synaptic substructure of neurons. Genes in module Blue are associated with glial cells and also endothelial cells forming blood vessel walls. However, it is well known that blood vessels occupy a small portion of the cortical tissue, with the total blood volume fraction ranging from 2 to 5% (26). Hence, it is reasonable to assign module Blue to glial cells, keeping in mind that glial cells and endothelial cells have highly correlated genetic profiles across the brain.
To quantitatively characterize contributions of neurons, synapses, and glial cells to the MRI metrics we introduce the gene expression indices Yneuron, Ysynapse, and Yglia (instead of module eigengenes ME-Purple, ME-Brown, and ME-Blue, correspondingly) that characterize normalized levels of their corresponding eigengene expressions:
| [3] |
In this equation we use an inverse Fisher transform for ME but shifted corresponding values so that values of the expression index Y vary between 0 (lowest expression level) and 1 (highest expression value).
Spatial Variation of MRI Metrics and Gene Networks.
Mean values of R2t*, R2′, and cortical thickness from all 26 healthy volunteers were summarized in FreeSurfer regions of interest (ROIs) for comparison with the gene expression indices. The surface maps are shown in Fig. 4 along with the surface maps of gene expression indices Y for three modules (Purple/neuron, Brown/synapse, and Blue/glia). Fig. 4 also shows boxplots of MRI metrics and gene expression for cingulate, frontal, temporal, parietal, and occipital cortices.
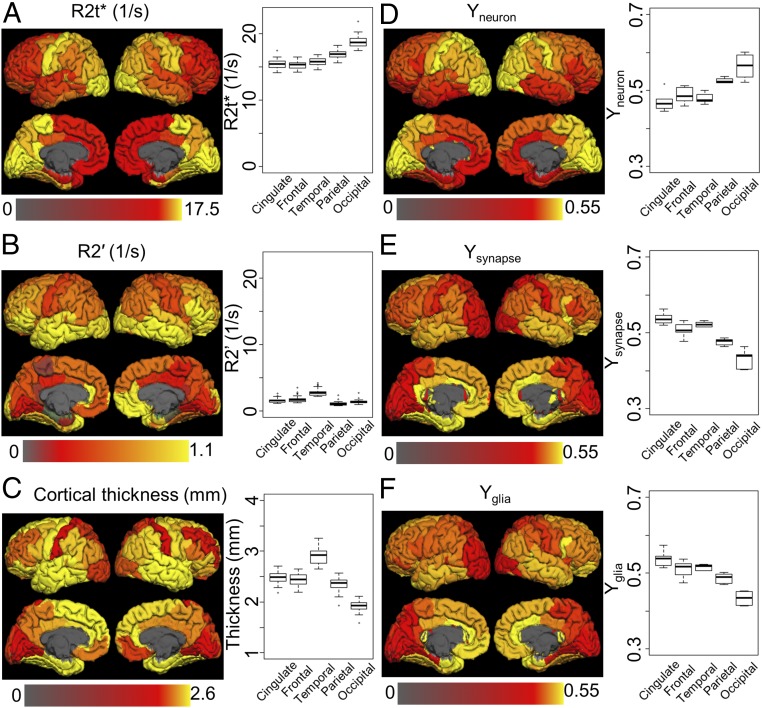
Fig. 4.
Cortical distribution of MRI metrics (A, R2t*; B, R2′; C, cortical thickness) and gene expression indices (D, Yneuron; E, Ysynapse; F, Yglia). Maps represent values averaged across all corresponding datasets (26 for MRI metrics and 6 for gene modules) in FreeSurfer ROIs projected onto the brain surface. Box plots represent median values and variation across the same datasets for five cortical regions (cingulate, frontal, temporal, parietal, and occipital). Surface maps and box plots show significant resemblance between distribution of R2t* and neuron index (Yneuron), suggesting that neurons provide significant contribution to the brain tissue-specific R2t* relaxation. Detailed statistical analysis is provided in Table 1. Data also show a significantly smaller contribution of the BOLD effect (R2′) to the total relaxation (R2* = R2t* + R2′) of the GRE signal.
Contribution of Neurons, Synapses, and Glial Cells to Microstructructural and Macrostructural MRI Metrics.
The analysis above shows that modules representing neurons, glial cells (astrocyte, microglia, and OPCs), and synapses strongly correlate (positively or negatively) with R2t* and cortical thickness. To determine the relative weightings of the three gene modules to MRI metrics, we performed a multiple linear regression analysis of the relationships between MRI metrics (R2t* and cortical thickness) and expression indices Yneuron, Yglia, and Ysynapse. The results are shown in Table 1.
Table 1.
Multiple linear regression analysis of the contributions of Yneuron, Ysynapse, and Yglia to R2t* and cortical thickness models
| R2t* (1/s) | Cortical thickness, mm | |||
| Variable | Parameter | Error | Parameter | Error |
| Intercept | 16.1 | 0.7 | 2.2 | 0.2 |
| Yneuron | 10.0 | 0.9 | −2.1 | 0.2 |
| Ysynapse | 1.4 | 0.3 | ||
| Yglia | −10.3 | 0.9 | 1.3 | 0.3 |
| R2 | 0.61 | 0.58 | ||
The P values for all shown parameters are smaller than 0.0001. Ysynapse is not shown in the R2t* model because its contribution is not significant (P = 0.4), although it contributes significantly to the cortical thickness model. Note that all indices Y are dimensionless.
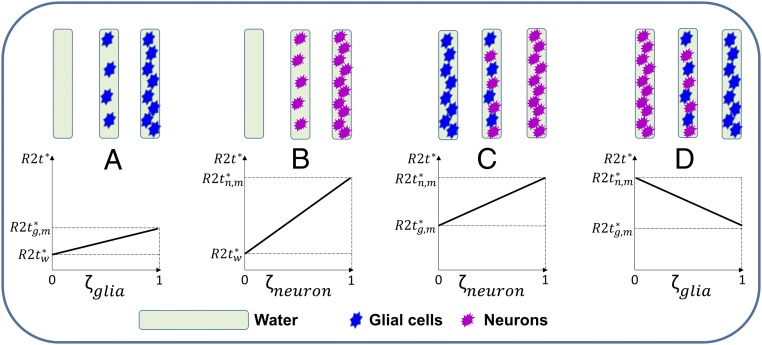
Correlation analysis in Table 1 shows relationships between the MRI and gene metrics across brain regions. Disparate signs of cellular contributions to R2t* and cortical thickness models can be explained by the fixed brain tissue “packing capacity”—higher concentration of neurons would allow less space for glial cells, and vice versa (Fig. 5).
Fig. 5.
Schematic relationship between R2t* and cellular volume fraction ζ for different combinations of media and cells. In the top row boxes represent the background media (water) filled with different concentrations of cells (glia and neurons). Media and cellular types are marked as indicated at the bottom of the figure. The bottom row shows R2t* dependence on cellular volume fraction for different combinations of media and cells. (A) Glial cells in water: R2t* increases with increasing glial cells volume fraction ζglia. (B) Neurons in water: R2t* increases with increasing neuronal cells volume fraction ζneuron. (C) Neurons and glial cells fill the whole volume with gradual increase of neuronal volume fraction ζneuron: R2t* increases as neurons replace glial cells. (D) Glial cells and neurons fill the whole volume with gradual increase of glial cells volume fraction ζglia: R2t* decreases as glial cells replace neurons. , , and represent the R2t* values when the whole volume is occupied by water molecules, glia cells, and neurons, respectively. Importantly, the negative correlation in D is due to the neurons with higher relaxivity r2t*neuron are replaced by glial cells with lower relaxivity r2t*glia.
The data also show that across all ROIs (Yneuron + Yglia) = 1 ± 0.085 (mean ± STD). This suggests that Yneuron and Yglia can be considered as surrogates for volume fractions ζ of neurons and glial cells. By substituting Yglia = 1 − Yneuron in the R2t* model in Table 1 we arrive at a simple equation expressing R2t* in terms of neuronal volume fraction ζneuron in the brain:
| [4] |
In this equation, R2t* is measured in seconds−1, while ζneuron is dimensionless.
It is important to note that Eq. 4 is derived for brain tissue composed of neurons and glial cells and it cannot be treated as a general relationship between R2t* and neuronal density for an arbitrary medium containing neurons. However, data in Table 1 provide information that allows separation of neuronal and glial contributions to R2t*. Indeed, in the hypothetical case of tissue with pure neurons (Yneuron = 1, Yglia = 0), R2t* would be = 26.1 s−1, while in the opposite case of pure glial tissue (Yneuron = 0, Yglia = 1), = 5.8 s−1. These results allow introducing individual relaxivities r2t* (relaxation rates per unit cellular volume fraction ζ) of neurons and glial cells with respect to arbitrary media with a given R2t*:
| [5] |
In fact, Eq. 4 is a particular case of Eq. 5 corresponding to a mixture of neuronal and glial cells (increased ζneurons corresponds to decreased ζglia as neurons replace glial cells). The R2t* dependencies on different combination of cells and media are illustrated in Fig. 5.
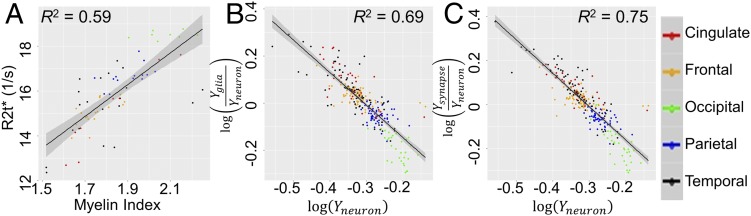
The major role of neurons in affecting R2t* can be explained mostly by a contribution from myelinated axons that dominate in module Purple (Fig. 3 and Discussion). This is further demonstrated by a significant correlation between R2t* and the myelin index [defined as a ratio of T1 and T2 weighted images (27)] that we generated using data from the HCP (Materials and Methods), as shown in Fig. 6A.
Fig. 6.
Correlation of R2t* with myelin index and correlations between neuronal, glial, and synaptical indices. (A) Correlation between R2t* (seconds−1) and myelin index (dimensionless). Myelin index [defined as a ratio of T1 and T2 weighted images (26)] values were generated using data from the HCP averaged across 1,018 healthy subjects. Regional R2t* values were calculated by averaging results from 12 healthy volunteers, ages 34 ± 7.6 y, approximately matching HCP age group. Each point represents a FreeSurfer region. (B and C) Scatter plots of relative (to neurons) concentrations of glial cells (Yglia/Yneuron) and synapses (Ysynapse/Yneuron) vs. (Yneuron) for all six AHBA datasets. Each point represents the logarithm of gene expression in one FreeSurfer ROI in one AHBA dataset. In all panels blocks of FreeSurfer regions are marked by different colors: cingulate, red; frontal, orange; occipital, green; parietal, blue; and temporal, black. The shaded areas in the panels represent the 95% confidence bands.
Eq. 4 can be very useful for understanding neuronal contributions to the qGRE metrics in the brain. It represents a relationship between R2t* and neuronal volume fraction with a caveat that neuronal features (cell body, axonal, dendritic, and synaptic substructures) in different brain regions can be quite different. This is illustrated in Fig. 6 B and C, showing that the ratios of Yglia/Yneuron and Ysynapse/Yneuron are negatively correlated with Yneuron, that is, the brain regions with higher concentration of neurons (larger Yneuron) have lower concentration of glial cells relative to neurons (smaller Yglia/Yneuron) and lower concentration of synapses relative to neurons (smaller Ysynapse/Yneuron). These results are in agreement with direct histological measurements (28, 29).
The equations for the curves shown in Fig. 6 are
| [6] |
In these equations, R2t* is measured in seconds−1, while all other variables [including myelin index MI (27)] are dimensionless.
Relationships Between Cortical Tissue Microstructure and Functional Activity.
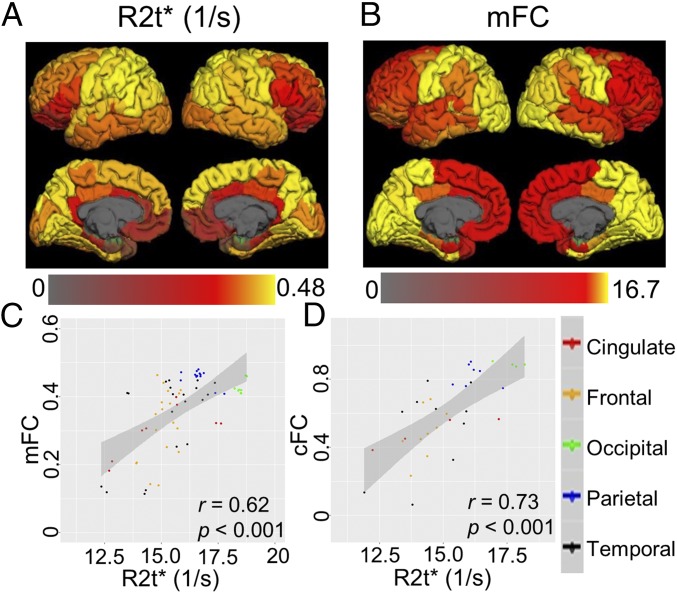
To establish relationships of cortical tissue microstructure (defined by R2t*) with functional activity, we used functional connectivity data from the HCP. Fig. 7 shows very strong associations between the cytostructure of cortical brain regions defined by cortical R2t* and functional connectivity between brain regions. The data show that regions with higher R2t* have stronger functional connections with other brain regions.
Fig. 7.
Relationships between cortical tissue microstructure and functional connectivity. (A) Cortical R2t* map projected on the brain surface. Regional R2t* values were calculated by averaging from 12 healthy volunteers, ages 34 ± 7.6 y, approximately matching HCP age group. (B) Mean functional connectivity (mFC) map representing mean connectivity between a given cortical FreeSurfer ROI and all other ROIs in the cortex (Materials and Methods) projected on the brain surface. Data were generated from HCP averaged across 1,017 healthy subjects with age 28.5 ± 4.2 y. The bottom shows correlations of cortical R2t* with mFC (C) and cFC (FC of contralateral FreeSurfer ROIs in different hemispheres) (D). Each point represents a cortical FreeSurfer region and affiliation with selected blocks of FreeSurfer regions is marked by different colors: cingulate, red; frontal, orange; occipital, green; parietal, blue; and temporal, black. The shaded areas in each panel represent 95% confidence bands.
Discussion
The ability to obtain high-quality information on brain structure and functioning in vivo with MRI has contributed significantly to our understanding of the human brain and has offered new insights into multiple disease processes. However, due to the relatively low resolution of in vivo MRI (usually millimeter-sized voxels) it is challenging to connect MRI measurements to underlying cellular composition. Hence, one of the most important questions in this regard is how to relate MRI neuroimaging measurements to the underlying microstructural anatomy, biology, and neurophysiology of brain cells (3).
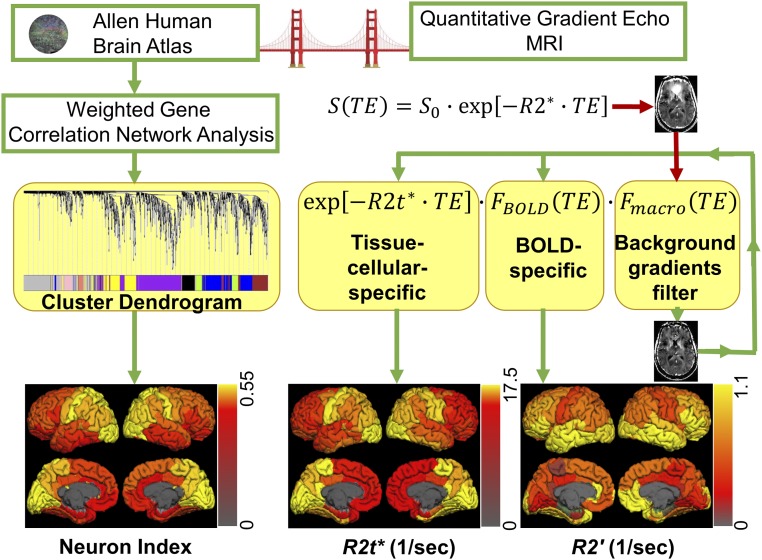
In this study we used AHBA data to establish genetic and cellular fingerprints of the GRE MRI signal that is widely used to study brain responses to stimulated and resting-state functional activity. A schematic structure of our approach is presented in Fig. 8. While the overwhelming majority of studies rely on analysis of stimulus-induced or resting-state-induced fluctuations of GRE signal, these fluctuations account for only a few percent above the baseline GRE signal. Herein, we focus on the nature of the baseline GRE signal and its relation to brain microstructure and function.
Fig. 8.
Schematic summary of the analysis performed in this study that provided a “bridge” linking brain genetic and cellular information with MRI measurements. The genetic analysis is summarized in the left side of the figure: Gene expression data were obtained from the AHBA and analyzed using WGCNA. This procedure generated eight modules characterized by eight MEs shown by different colors in the cluster dendrogram. We found that one of the modules (Purple) was related to neurons and we used a corresponding eigengene to calculate “neuron index” as indicated in Eq. 3. The analyses performed on MRI data are summarized in the right side of the figure: qGRE MRI was used to measure tissue cellular-specific (R2t*) and BOLD-related (R2′) relaxation rate constants. Instead of using traditional monoexponential model, we used a more sophisticated approach that allowed elimination of artifacts related to background field gradients and separate measurements of R2t* and R2′. All genetic and MRI results were summarized in FreeSurfer ROIs and projected onto the brain surface as shown at the bottom. Neuron index was found to be significantly correlated with R2t*, indicating potential use of R2t* as an in vivo neuron index.
By comparing gene networks generated by weighted correlation network analysis with MRI metrics, we identified three modules, Purple (associated with neurons), Blue (associated with glia), and Brown (associated with synapsis), that were significantly associated with microstructural (R2t*) and macrostructural (cortical thickness) MRI metrics. Our results suggest that regional variation in R2t* relates to variation in neuronal and glial cellular composition, as indicated by the gene expression data. Importantly, higher R2t* reflects tissue containing more neurons with myelinated axons and relatively lower numbers of glial cells and synapses. It is possible that R2t* represents more “mature” cortical tissue, for example with more myelination and less immature synapses, which is also supported by our prior report indicating a gradual rise in R2t* with age in all cortical regions (13).
There are a number of reasons to suggest that the positive correlation between R2t* and the Purple module of genes related to voltage-gated channels likely reflects a contribution from myelin-covered axons. Indeed, the majority of voltage-gated channels are located along axons that are covered by myelin sheaths (23) and are enriched near the nodes of Ranvier (30). Thus, higher expression of voltage-gated-channel-related genes might indicate more myelin content. Also, module Purple includes gene NRG1, which has been shown to regulate myelin sheath thickness (31)—reduced NRG1 expression in mouse brain can cause hypomyelination and reduced nerve conduction velocity. All these properties point to myelin as a main neuron-related “relaxation agent” affecting the R2t* relaxation rate constant of the GRE signal. This conclusion is also supported by a strong correlation between R2t* and the T1/T2 evaluated myelin index as shown in Fig. 6A. Conversely, our data (Fig. 6) show that relatively unmyelinated cortex has more synapses and glia (relative to neurons), which is in agreement with direct histology (28, 29). From a biophysical perspective, myelin also represents a cellular substructure with high relaxivity for water. Indeed, the tissue-cellular-specific (R2t*) MRI relaxation parameter depends on the environment of water molecules (the main source of MRI signal): higher concentrations of proteins, lipids, and other constituents of biological tissue and cellular constituents (sources of MRI signal relaxation) leading to higher relaxation rate constants. While the presence of high concentrations of macromolecules (relaxation agents) is a necessary condition for high R2t*, access of water molecules to these macromolecules is another important condition. From this perspective, myelin’s intermixed structure of water and lipid layers represents a significantly increased opportunity for water–macromolecule interaction.
In this paper we have introduced normalized quantitative module-related gene expression indices (Y, Eq. 3) that allowed derivation of quantitative relationships between R2t* measurements and cortical tissue cellular contents (Table 1 and Eqs. 4 and 6). It should be noted that glial cells and synapses contribute to relaxation effects governing R2t*, but the reason for the negative correlations (Table 1) is due to the relatively larger effects of myelinated axons compared with these other cellular structures (Fig. 5). Equations we derived account for this effect and allow evaluation of relative tissue cellular composition based on qGRE measurements: Using the relationship in Eq. 4 between R2t* and Yneuron we can first estimate the neuronal content. The corresponding fractions of glial cells and synaptic structures can then be estimated from Eq. 6. Thus, R2t* reflects unique cortical tissue characteristics that can be used in vivo to explore cortical structure and changes over time in health and disease in the human brain.
Cortical thickness is one of the most ubiquitous measures of MRI. Our data also show negative correlation between neuronal density and cortical thickness, which is also in agreement with previous studies (32–34).
Data in Fig. 7 demonstrate significant positive correlations between tissue R2t* and the strength of functional connectivity defined by resting-state fMRI. Data show that the brain regions with higher R2t* provide a platform for stronger connections with other brain regions and potentially can be considered as hubs for such connections. Herein we have demonstrated this concept by providing relationships between R2t*, mFC (mean regional strength of functional connectivity), and cFC (the strength of functional connectivity between symmetric contralateral brain regions in different hemispheres). These functional relationships may be related to the relationship between R2t* and underlying tissue cellular content—brain regions with higher R2t* have higher concentration of neurons (higher Yneuron), which are mainly responsible for structural brain connectivity. Interestingly enough, brain areas with a higher concentration of neurons have relatively lower concentration of synapses and glial cells (see negative correlation in Fig. 4). This suggests that a high concentration of neurons (especially myelinated axons) significantly contributes to brain functional connectivity, providing the structural support for signal transmission. Importantly, changes in cortical cellular composition due to development, aging, and/or disease states can be identified by qGRE R2t* measurements and thereby probed as potential causes of changes in functional connectivity.
Data in Fig. 4 A and B show that the BOLD-related part of the GRE signal relaxation rate parameter (R2′) constitutes only a small part of the tissue-cellular-related relaxation rate parameter (R2t*). About 40% of oxygen delivered to the brain by the blood vessel network is consumed by the brain, but only minor changes in oxygen consumption happen during stimulus-induced changes in functional brain activity (35). The rest is used to maintain the existing brain cytoarchitecture and default brain activity (36). Interestingly, this small change in oxygen consumption, and hence energy consumption, vs. baseline oxygen consumption resembles a small ratio between changes in the R2* GRE signal during brain activity and the baseline cellular-specific part of R2* (R2t*) defined by the brain basic cytoarchitecture. From this perspective, it would be interesting to investigate a potential relationship between baseline oxygen consumption and R2t*-defined brain cellular composition.
R2t* is a quantitative tissue-specific measure that has been successfully applied in studying different diseases in the central nervous system, including multiple sclerosis (37), Alzheimer disease (38), schizophrenia, and bipolar disorder (39). Our findings relating R2t* to tissue cellular content provide the potential for new insights on the results of these studies and corroborate potential use of R2t* as a biomarker of changes in the tissue cellular content. Indeed, findings of reduced cortical R2t* in Alzheimer disease (38) and multiple sclerosis (37) are consistent with the loss of neurons. Importantly, this loss was detected even in the cortical tissue spared of atrophy.
In conclusion, understanding the structure and function of the human cortex is a prominent goal of modern neuroscience. The analysis provided in this study offers a unique genetic perspective on the cellular contributions to the brain MRI signal in humans. This became possible by combining multimodality MRI data that include qGRE MRI, structural MRI, and resting-state fMRI with gene expression data from the AHBA. We demonstrated that the R2t* metric of qGRE signal provides an in vivo window into quantitative evaluation of the cellular components of cortical tissue. Data show that areas of the brain characterized by higher R2t* have high neuronal density and have stronger functional connections to other brain areas. Interestingly, these areas have a relatively smaller concentration of synapses and glial cells, suggesting that myelinated cortical axons are likely key cortical structures that contribute to functional connectivity. Given these associations, R2t* is likely to be a useful signal in assessing microstructural changes in the brain during development, aging, and in health or disease.
Materials and Methods
WGCNA Analysis of AHBA Data.
We used data from all six human brains from the AHBA, namely H0351.2001, H0351.2002, H0351.1009, H0351.1012, H0351.1015, and H0351.1016 (for details see human.brain-map.org/mri_viewers/data). The AHBA provides a comprehensive view of the relationship between brain anatomy and gene expression (2). Because in AHBA some genes have more than one probe, the first step is to choose a single probe to represent a gene. This was completed by using the “collapseRows” function in WGCNA. Then, genes without entrez IDs were removed from the analysis. These steps reduce the original 58,692 measurements to 20,738 measurements. To simplify the complexity of the AHBA data and allow comparison with MRI R2t* measurements, the original AHBA data were summarized using the standard cortical atlas (20) built into FreeSurfer. A two-step procedure was employed to complete this task. (i) Segmentations were obtained by running FreeSurfer on each of the six AHBA MRI data. Tissue samples were assigned to 68 cortical regions based on their coordinates in their own MRI space. However, some tissue cortical samples might fall outside the FreeSurfer cortical ROIs; these samples were assigned to the adjacent FreeSurfer cortical ROI. (ii) For each FreeSurfer ROI and each brain, median values were calculated from the gene expression profiles that were located inside the ROI. Since the six AHBA datasets have different numbers of anatomical samples, this procedure resulted in gene expression profiles assigned in 35–66 FreeSurfer cortical ROIs for different AHBA datasets. These summarized gene expression datasets were used to construct a consensus network. We used a general framework of WGCNA similar to that previously described (40). To use all six AHBA datasets and improve the accuracy and power of the analysis, we only chose those genes that had similar expression patterns across all six brains (4). Specifically, pairwise correlations using Pearson method were calculated between gene expression profiles of all six AHBA brains for each gene. Then, averaged correlation coefficients (so-called DS) were calculated and only those genes with DS > 0.5 (n = 1,995) were used in WGCNA analysis. This DS value was used as a compromise between distinguishing detailed cellular affiliation of different gene modules and the ability of using all six Allen brain datasets for analysis. Different DS thresholds were also tested and results are shown in SI Appendix, Fig. S1. The value DS = 0.5 is also similar to previously used in ref. 4. A consensus network was constructed from these 1,995 genes using a previously established framework (40) (soft-thresholding power = 14, reference percentile to rescale different datasets = 0.95, deepSplit = 4, cutHeight = 0.999, minClusterSize = 30, and pamRespectsDendro = FALSE). This procedure resulted in identifying nine modules represented by nine MEs.
The next step was to reassign each of original 20,738 genes from AHBA to the module to which it is most highly correlated. Pearson correlations were calculated between each gene expression profile and each ME in each brain. As a result, 8,086 genes were reassigned to WGCNA modules based on their mean (across six brains) correlation coefficients (≥0.5) with MEs. Different thresholds were also tested and results are shown in SI Appendix, Figs. S2 and S3. All genes with mean correlation coefficients <0.5 against all modules were considered unassigned. MEs were recalculated using the new modules. After this step, all nongray modules preserved more than 70% (Black, 87.0%; Blue, 75.2%; Brown, 86.2%; Green-yellow, 89.6%; Pink, 90.1%; Purple, 88.2%; Salmon, 97.3%; and Yellow, 79.1%) of the original genes assigned in the first step. These numbers are comparable with a previous study (4), demonstrating the stability of these modules.
Genome Annotation.
The ToppGene portal (21) was used to access an extensive list of databases and to calculate P values from hypergeometric tests and corrected for multiple comparison. In this study, we used cellular component in GO annotations and coexpression atlas developed in Barres’s laboratory (41, 42). We used default background settings in ToppGene, which includes 19061 genes in the “Cellular Component” category and 23956 genes in the “Coexpression Atlas” category. To test the sensitivity of the enrichment analysis with different background settings, we performed gene enrichment analysis using DAVID Bioinformatics Resources (ref. 22; https://david.ncifcrf.gov/) with the default background and a background that contains genes in the AHBA datasets. As shown in Datasets S3–S8, the selection of background has only minor effect on the enrichment analysis.
MRI.
For qGRE and structural analysis we used previously published (13) brain image data collected from 26 healthy volunteers (age range 23–76 y) using Siemens 3T Trio MRI scanner and a 32-channel phased-array head coil. Studies were conducted with approval of the local IRB of Washington University. All volunteers provided informed consent. All data were obtained using a 3D version of the GRE sequence with 10 gradient echoes followed by a navigator echo (43) used to reduce image artifacts induced by physiological fluctuations during the scan. Sequence parameters were flip angle FA = 30°, voxel size of 1 × 1 × 2 mm3, first echo time TE1 = 4 ms, echo spacing ΔTE = 4 ms (monopolar read-out gradients), repetition time TR = 50 ms, and the total imaging time for each acquisition was around 10 min. Standard clinical MPRAGE (19) images with a resolution of 1 × 1 × 1 mm3 were also collected for brain segmentation purposes. The total acquisition time of MPRAGE is about 6 min.
qGRE Data Analysis.
Raw k-space data were read into MATLAB (The MathWorks, Inc.) for processing. After applying Fourier transform to the k-space, data from different channels were combined for each voxel in a single data set S(TE) using a previously developed strategy (44, 45):
| [7] |
where the sum is taken over all M rf channels (m), TE is the gradient echo time (TE1 is the time of the first gradient echo), denotes complex conjugate of S, and λm are the rf channel-specific weighting parameters defined by the noise amplitudes (rms). We omit index corresponding to voxel position for clarity. This algorithm allows for the optimal estimation of model parameters and also removes the initial phase incoherence between channels (44, 45). A Hanning filter was also applied to further decrease noise and reduce Gibbs ringing artifacts. Maps of model parameters were obtained by fitting theoretical model in Eq. 2 to experimental data, Eq. 7, on a voxel-by-voxel basis.
The function in Eq. 2 describes GRE signal decay due to the presence of the blood vessel network with deoxygenated blood (veins and adjacent to them part of capillaries) – the baseline BOLD effect, and the function F(TE) in Eq. 2 describes effect of macroscopic magnetic field inhomogeneities. In this study, we use a VSF method (14) for calculating F(TE) and an expression from ref. 12:
| [8] |
where ζ is the deoxygenated cerebral blood volume fraction and δω is the characteristic frequency determined by the susceptibility difference between deoxygenated blood and surrounding tissue (15):
| [9] |
In this equation, (46) is the susceptibility difference between fully oxygenated and fully deoxygenated red blood cells, Yb is the blood oxygenation level (with corresponding to fully deoxygenated and to fully oxygenated red blood cells), Hct is the blood hematocrit, and γ is the gyromagnetic ratio. Function was defined in ref. 15 and can also be expressed in terms of a generalized hypergeometric function (47):
| [10] |
The function has a characteristic quadratic behavior at short TE and then becomes linear (15). The nonlinearity of is very important for our method because it allows separation of BOLD and cellular (R2t*) contributions to the GRE signal decay (16). Eq. 2 with definitions in Eqs. 8–10 is used to fit to experimental data and extract model parameters R2t*, ζ, and δω. R2′ is then calculated as
| [11] |
Signal amplitude and global frequency shifts are also evaluated from this fitting routine (we are using complex data). The image of the signal amplitudes represents a qGRE T1-weighted (T1W) image and is used herein for coregistration with MPRAGE images. The global frequency map is used to calculate function F(TE) in the framework of the VSF method (14). Eq. 9 potentially allows evaluation of blood oxygenation level, Yb, but would require additional information on blood hematocrit level that was not measured in this study.
As we already mentioned, the BOLD model (15) was previously validated in phantoms (16), in silico (17), and in vivo in small-animal (18) experiments. Model restrictions and the accuracy of model parameters estimations are discussed in great details in refs. 12, 13, and 15.
Note that conventionally in a GRE experiment the total relaxation rate constant is considered to be a sum of two components: relaxation rate constant representing the part of MR signal decay that cannot be reversed by a refocusing 180° rf pulse and relaxation rate constant representing the part of the signal decay that can be reversed by a refocusing 180° rf pulse (usually attributed to BOLD effect in a static dephasing regime): . However, such a consideration would only be valid for a homogeneous single component tissue. For a brain tissue composed of a multitude of cells and cell types, the part of remaining after the subtraction of the BOLD effect can still have contributions from magnetic susceptibility effects resulting from the presence of different cells and cell-building components. Hence, in our consideration we call it (13).
Image Segmentation.
FreeSurfer software (Laboratory for Computational Neuroimaging, Martinos Center for Biomedical Imaging) is used to generate brain segmentations based on MPRAGE images. MPRAGE images are registered to qGRE T1W images using FMRIB’s linear image registration tool (48, 49) in FSL and the transformation matrices of the registration are generated. These matrices are applied to the brain segmentations from FreeSurfer and transformed to the space of qGRE T1W images. Since qGRE T1W and R2t* images are generated from the same single MRI scan they are naturally coregistered. Hence, segmentations of qGRE T1W images are naturally coregistered with R2t* maps. Cortical thickness was also obtained from FreeSurfer. To minimize contamination of signals from CSF, CSF masks were generated based on the qGRE T1W images using FSL and only those voxels with R2t* bigger than 3 s−1 were used to calculate median R2t* values.
Correlations Between Gene Expression Profiles and R2t*.
R2t* median values were calculated in each FreeSurfer ROI for each healthy volunteer and averaged across several age-matched subjects corresponding to the age of each AHBA dataset. This was done to minimize known effects of R2t* changes with age (13). We first selected the healthy volunteer who had the closest age to an AHBA dataset. Then, we included two younger and two older neighbors, which results in a total number of five healthy volunteers, to calculate averaged R2t* values. Due to the limited availability of younger subjects, we only used four healthy volunteer to match AHBA dataset H0351.2001. The Spearman method was used to correlate MEs with R2t*. False discovery rate (FDR) was used to correct for multiple comparisons.
To address possible contamination of cortical R2t* values due to partial volume effect of adjacent white matter (WM), we also calculated the correlation between R2t* in the subcortical WM regions and gene expression profiles in the adjacent cortex. The results showed significantly weaker correlations compared with cortical regions, thus supporting our conclusion regarding contribution of cortical neurons to the correlation we found between cortical R2t* and gene expression profiles.
Stability.
To demonstrate the stability of the correlations between MEs and MRI (R2t* and cortical thickness), we carried out a permutation experiment. By randomly permuting R2t* and thickness values between ROIs, new correlation coefficients (r values) and P values were recorded. After a simulation of 10,000 steps, the distribution of r values is shown in SI Appendix, Fig. S4. The r values without permutation are also marked in the figure using solid blue lines. The 99% percentile of the distribution is marked using dashed red lines. The stability of the WGCNA analysis was also tested using different thresholds. The results are shown in SI Appendix, Figs. S1–S3.
Functional Connectivity and Myelin Index Data Analysis.
We used the HCP1200 dataset (March 1, 2017 data release) of adults aged 22–35 y (50). We selected only subjects that had all four (15 min each) fully preprocessed resting state (rfMRI) scans (1,017 subjects). HCP preprocessing pipelines used by HCP team (50) to analyze the data (see ref. 51 for the detailed description) before release of the HCP1200 dataset included three structural pipelines (PreFreeSurfer, FreeSurfer, and PostFreeSurfer) and two functional pipelines (fMRIVolume and fMRISurface) (51, 52). We used data after an initial gentle nonrigid surface registration based on folding patterns (MSMSulc) was performed. This supplanted the FreeSurfer folding-based registration previously used (52), because it achieved slightly better initial alignment of functionally corresponding regions than Freesurfer’s algorithm while inducing much lower local distortions (53). This registration, together with the FNIRT nonlinear registration, was used to bring an initial version of the data [rfMRI, myelin maps, and individually defined FreeSurfer regions based on Desikan–Killiany Atlas (20)] into standard grayordinates space (32k standard mesh for each hemisphere’s cortical surface at 2 mm average vertex spacing and 2 mm isotropic MNI-space voxels for the subcortical volume data). For rfMRI runs, the ICA+FIX pipeline (54–57) was also used to remove spatially specific temporally structured artifacts (see ref. 51 for the detailed description). Because the HCP rfMRI data were collected in four runs, the following steps were performed by using HCP tools (wb_command): (i) demean and normalize the individual time series (subtracting the mean of each grayordinate’s time course, multiplying by the bias field, and dividing by the variance normalization map); (ii) temporally concatenating the individual subject dense time-series data (1,200 time points × four runs); and (iii) for each subject, time series were extracted from cortical individually defined 68 FreeSurfer regions based on the Desikan–Killiany Atlas (20). Data for 1,017 subjects were concatenated and correlation analysis was performed in MATLAB (The MathWorks, Inc.) by using Pearson correlation coefficient.
Mean FC map (mFC) was calculated as follows:
| [12] |
where is the mean correlation coefficient in region n with all other regions, is the correlation coefficient between region n and region m, and is the volume fraction of FreeSurfer defined region n.
Myelin index maps were generated by using the ratio of T1w/T2w images and were normalized for residual transmit field inhomogeneity (see ref. 51 for the detailed description). For each subject, the myelin index was extracted from cortical individually defined 68 FreeSurfer regions based on the Desikan–Killiany Atlas (20). Then, individual myelin indexes were averaged in each FreeSurfer ROI across all subjects.
Supplementary Material
Acknowledgments
We thank the reviewers for their thoughtful comments and suggestions. J.W. and D.A.Y. thank Biao Xiang for useful discussions. This work was supported by NIH Grant R01 AG054513.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808121115/-/DCSupplemental.
References
- 1.Garcia AM. Recollections of my life. Santiago Ramon y Cajal, E. Horne Craigie, Juan Cano. Q Rev Biol. 1967;42:468. [Google Scholar]
- 2.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raichle ME, Mintun MA. Brain work and brain imaging. Annu Rev Neurosci. 2006;29:449–476. doi: 10.1146/annurev.neuro.29.051605.112819. [DOI] [PubMed] [Google Scholar]
- 4.Hawrylycz M, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832–1844. doi: 10.1038/nn.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richiardi J, et al. IMAGEN consortium BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasser MF, et al. The Human Connectome Project’s neuroimaging approach. Nat Neurosci. 2016;19:1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra A, et al. Spontaneous infra-slow brain activity has unique spatiotemporal dynamics and laminar structure. Neuron. 2018;98:297–305.E6. doi: 10.1016/j.neuron.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci USA. 1990;87:9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa S, et al. Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci USA. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandettini PA, Wong EC, Hinks RS, Tikofsky RS, Hyde JS. Time course EPI of human brain function during task activation. Magn Reson Med. 1992;25:390–397. doi: 10.1002/mrm.1910250220. [DOI] [PubMed] [Google Scholar]
- 11.Kwong KK, et al. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci USA. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrich X, Yablonskiy DA. Separation of cellular and BOLD contributions to T2* signal relaxation. Magn Reson Med. 2016;75:606–615. doi: 10.1002/mrm.25610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Wen J, Cross AH, Yablonskiy DA. On the relationship between cellular and hemodynamic properties of the human brain cortex throughout adult lifespan. Neuroimage. 2016;133:417–429. doi: 10.1016/j.neuroimage.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yablonskiy DA, Sukstanskii AL, Luo J, Wang X. Voxel spread function method for correction of magnetic field inhomogeneity effects in quantitative gradient-echo-based MRI. Magn Reson Med. 2013;70:1283–1292. doi: 10.1002/mrm.24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yablonskiy DA, Haacke EM. Theory of NMR signal behavior in magnetically inhomogeneous tissues: The static dephasing regime. Magn Reson Med. 1994;32:749–763. doi: 10.1002/mrm.1910320610. [DOI] [PubMed] [Google Scholar]
- 16.Yablonskiy DA. Quantitation of intrinsic magnetic susceptibility-related effects in a tissue matrix. Phantom study. Magn Reson Med. 1998;39:417–428. doi: 10.1002/mrm.1910390312. [DOI] [PubMed] [Google Scholar]
- 17.Dickson JD, et al. Quantitative phenomenological model of the BOLD contrast mechanism. J Magn Reson. 2011;212:17–25. doi: 10.1016/j.jmr.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Zhu M, Yablonskiy DA. Validation of oxygen extraction fraction measurement by qBOLD technique. Magn Reson Med. 2008;60:882–888. doi: 10.1002/mrm.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mugler JP, 3rd, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15:152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- 20.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao X, et al. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trimmer JS, Rhodes KJ. Localization of voltage-gated ion channels in mammalian brain. Annu Rev Physiol. 2004;66:477–519. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- 24.Karbowski J. Cortical composition hierarchy driven by spine proportion economical maximization or wire volume minimization. PLoS Comput Biol. 2015;11:e1004532. doi: 10.1371/journal.pcbi.1004532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 26.Martin WR, Powers WJ, Raichle ME. Cerebral blood volume measured with inhaled C15O and positron emission tomography. J Cereb Blood Flow Metab. 1987;7:421–426. doi: 10.1038/jcbfm.1987.85. [DOI] [PubMed] [Google Scholar]
- 27.Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herculano-Houzel S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 2014;62:1377–1391. doi: 10.1002/glia.22683. [DOI] [PubMed] [Google Scholar]
- 29.Cullen DK, Gilroy ME, Irons HR, Laplaca MC. Synapse-to-neuron ratio is inversely related to neuronal density in mature neuronal cultures. Brain Res. 2010;1359:44–55. doi: 10.1016/j.brainres.2010.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasband MN, Trimmer JS. Developmental clustering of ion channels at and near the node of Ranvier. Dev Biol. 2001;236:5–16. doi: 10.1006/dbio.2001.0326. [DOI] [PubMed] [Google Scholar]
- 31.Michailov GV, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 32.Wagstyl K, Ronan L, Goodyer IM, Fletcher PC. Cortical thickness gradients in structural hierarchies. Neuroimage. 2015;111:241–250. doi: 10.1016/j.neuroimage.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.la Fougère C, et al. Where in-vivo imaging meets cytoarchitectonics: The relationship between cortical thickness and neuronal density measured with high-resolution [18F]flumazenil-PET. Neuroimage. 2011;56:951–960. doi: 10.1016/j.neuroimage.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Cahalane DJ, Charvet CJ, Finlay BL. Systematic, balancing gradients in neuron density and number across the primate isocortex. Front Neuroanat. 2012;6:28. doi: 10.3389/fnana.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox PT, Raichle ME. Focal physiological uncoupling of cerebral blood flow and oxidative metabolism during somatosensory stimulation in human subjects. Proc Natl Acad Sci USA. 1986;83:1140–1144. doi: 10.1073/pnas.83.4.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raichle ME, Gusnard DA. Appraising the brain’s energy budget. Proc Natl Acad Sci USA. 2002;99:10237–10239. doi: 10.1073/pnas.172399499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen J, Yablonskiy DA, Salter A, Cross AH. Limbic system damage in MS: MRI assessment and correlations with clinical testing. PLoS One. 2017;12:e0187915. doi: 10.1371/journal.pone.0187915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. In vivo detection of microstructural correlates of brain pathology in preclinical and early Alzheimer disease with magnetic resonance imaging. Neuroimage. 2017;148:296–304. doi: 10.1016/j.neuroimage.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mamah D, et al. Subcomponents of brain T2* relaxation in schizophrenia, bipolar disorder and siblings: A gradient echo plural contrast imaging (GEPCI) study. Schizophr Res. 2015;169:36–45. doi: 10.1016/j.schres.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen J, Cross AH, Yablonskiy DA. On the role of physiological fluctuations in quantitative gradient echo MRI: Implications for GEPCI, QSM, and SWI. Magn Reson Med. 2015;73:195–203. doi: 10.1002/mrm.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo J, Jagadeesan BD, Cross AH, Yablonskiy DA. Gradient echo plural contrast imaging–Signal model and derived contrasts: T2*, T1, phase, SWI, T1f, FST2*and T2*-SWI. Neuroimage. 2012;60:1073–1082. doi: 10.1016/j.neuroimage.2012.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quirk JD, Sukstanskii AL, Bretthorst GL, Yablonskiy DA. Optimal decay rate constant estimates from phased array data utilizing joint Bayesian analysis. J Magn Reson. 2009;198:49–56. doi: 10.1016/j.jmr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spees WM, Yablonskiy DA, Oswood MC, Ackerman JJ. Water proton MR properties of human blood at 1.5 Tesla: Magnetic susceptibility, T(1), T(2), T*(2), and non-Lorentzian signal behavior. Magn Reson Med. 2001;45:533–542. doi: 10.1002/mrm.1072. [DOI] [PubMed] [Google Scholar]
- 47.Yablonskiy DA, Sukstanskii AL, He X. Blood oxygenation level-dependent (BOLD)-based techniques for the quantification of brain hemodynamic and metabolic properties–Theoretical models and experimental approaches. NMR Biomed. 2013;26:963–986. doi: 10.1002/nbm.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 49.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 50.Van Essen DC, et al. WU-Minn HCP Consortium The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glasser MF, et al. WU-Minn HCP Consortium The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson EC, et al. MSM: A new flexible framework for multimodal surface matching. Neuroimage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffanti L, et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salimi-Khorshidi G, et al. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith SM, et al. WU-Minn HCP Consortium Resting-state fMRI in the Human Connectome Project. Neuroimage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.