Abstract
The osteogenic differentiation of mesenchymal stem cells (MSCs) is governed by multiple mechanisms. Growing evidence indicates that ubiquitin‐dependent protein degradation is critical for the differentiation of MSCs and bone formation; however, the function of ubiquitin‐specific proteases, the largest subfamily of deubiquitylases, remains unclear. Here, we identify USP34 as a previously unknown regulator of osteogenesis. The expression of USP34 in human MSCs increases after osteogenic induction while depletion of USP34 inhibits osteogenic differentiation. Conditional knockout of Usp34 from MSCs or pre‐osteoblasts leads to low bone mass in mice. Deletion of Usp34 also blunts BMP2‐induced responses and impairs bone regeneration. Mechanically, we demonstrate that USP34 stabilizes both Smad1 and RUNX2 and that depletion of Smurf1 restores the osteogenic potential of Usp34‐deficient MSCs in vitro. Taken together, our data indicate that USP34 is required for osteogenic differentiation and bone formation.
Keywords: bone formation, mesenchymal stem cells, osteogenic differentiation, ubiquitin‐specific protease 34
Subject Categories: Development & Differentiation; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Introduction
Mesenchymal stem cells (MSCs) are heterogeneous progenitors which self‐renew and differentiate into multiple lineages of mesenchymal tissues, including bone, fat, cartilage, tendon, and muscle (Caplan, 1991; Pittenger et al, 1999; Sacchetti et al, 2007; Bianco et al, 2008; Mendez‐Ferrer et al, 2010). They have been identified and isolated from various tissues such as bone marrow, adipose tissue, umbilical cord blood, and dental tissues (Pittenger et al, 1999; Lee et al, 2004; Schaffler & Buchler, 2007; Sharpe, 2016). The intrinsic properties of MSCs make them an attractive candidate for clinical regenerative therapies (Caplan, 2007; Grayson et al, 2015; Malhotra et al, 2016).
The skeleton undergoes continuous remodeling with resorption by osteoclasts and formation by osteoblasts which are derived from MSCs (Zaidi, 2007; Crane & Cao, 2014). The process of MSC osteogenic differentiation begins with osteoprogenitor commitment followed by differentiation to pre‐osteoblasts and eventually development to mature osteoblasts (James, 2013). Disruption of this process leads to improper bone formation and imbalanced skeletal homeostasis. Mechanically, the osteogenic differentiation of MSCs is governed by master transcriptional factors, such as runt‐related gene 2 (RUNX2) and osterix (SP7), as well as by extrinsic regulators, such as cytokines, growth factors, and systemic hormones (Frith & Genever, 2008; Raggatt & Partridge, 2010; Yuan et al, 2014). In addition, there is growing evidence indicates that ubiquitin‐dependent proteolysis is crucial for the fine tuning of MSC differentiation.
The ubiquitination system is an enzymatic cascade that adds ubiquitin chains to target proteins and thereby directs their degradation (Ciechanover, 2005). Ubiquitination is carried out by ubiquitin‐activating enzymes (E1), ubiquitin‐conjugating enzymes (E2), and ubiquitin ligases (E3), which is the most abundant group of enzymes involved in ubiquitination (Komander, 2009). The ubiquitination process is reversible through the action of deubiquitinases that remove ubiquitin marks (Komander et al, 2009). Significant progress has been made in understanding the molecular regulation of MSC differentiation by ubiquitin ligases (Severe et al, 2013; Vriend & Reiter, 2016). For example, the E3 enzyme Smurf1 mediates the degradation of RUNX2, MEKK2, and JunB, resulting in inhibition of osteoblast differentiation and bone formation (Zhao et al, 2003, 2010; Yamashita et al, 2005). However, very little is known about the role of deubiquitinases in regulating MSC commitment.
The ubiquitin‐specific protease (USP) family, which consists of nearly 60 known members, is the largest of five families of deubiquitylases. These families are organized by the architecture of their catalytic domains (Liu et al, 2016a). It has been suggested that a few USPs, such as USP1 and USP6, are involved in preserving a mesenchymal stem cell program or antagonizing osteoblast differentiation (Williams et al, 2011; Zhou et al, 2016). Here, we screen the members of USP family and identify USP34 as a previously unknown regulator of osteogenesis. Conditional knockout of Usp34 leads to low bone mass in mice, blunted responses to BMP2, and impaired bone regeneration. Mechanically, we demonstrate that USP34 stabilizes both Smad1 and RUNX2.
Results
USP34 is required for osteogenic differentiation of MSCs
To investigate the potential role of USPs in osteogenesis, we first profiled the expression of 54 known USPs in MSCs after osteogenic induction. Human bone marrow MSCs were depleted with serum overnight and then treated with a combination of 100 ng/ml BMP2 and 100 ng/ml Wnt3a, which are critical regulators for osteogenic differentiation and synergistically activate the transcription of osteogenic genes to stimulate new bone formation (Rodriguez‐Carballo et al, 2011). Quantitative reverse transcriptase‐polymerase chain reaction (RT–PCR) revealed that USP34, together with USP13, USP18, USP21, and USP53, were the top five genes induced by the treatment (Appendix Fig S1A–C).
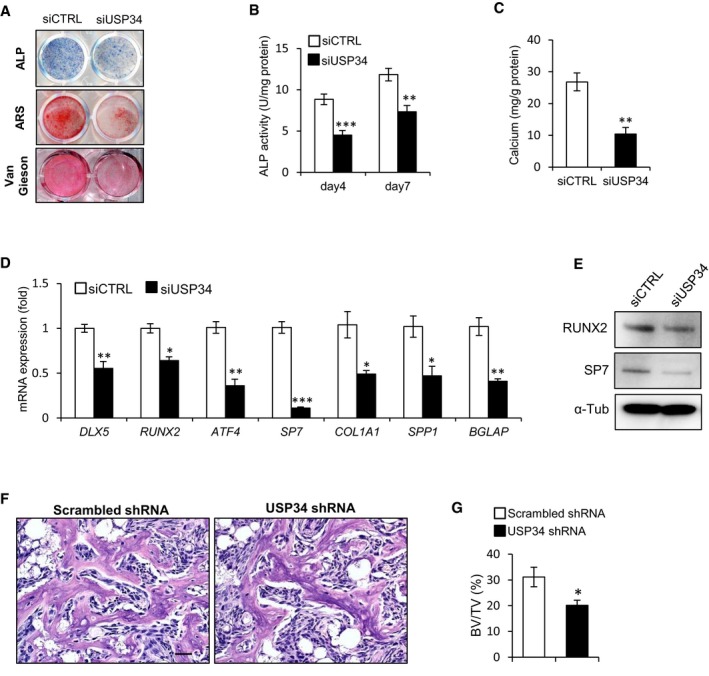
Next, we sought to investigate whether these 5 USPs are required for the osteogenic differentiation of MSCs by loss‐of‐function method using small interfering RNAs (siRNAs; Appendix Fig S2A and B). After culturing with osteogenic induction medium for 5 days, the intensities of alkaline phosphatase (ALP) and Van Gieson's staining were markedly reduced upon USP34 knockdown (Fig 1A). Moreover, USP34 depletion reduced the formation of mineralized nodules after osteogenic induction for 2 weeks (Fig 1A). These observations were confirmed by quantitative analyses of ALP activity (Fig 1B) and calcium mineralization (Fig 1C). USP34 knockdown also resulted in decreased expression of the master osteogenic transcription factors and markers, such as DLX5, RUNX2, ATF4, SP7, COL1A1, SPP1, and BGLAP (Fig 1D and E). However, depletion of USP13, USP18, USP21, or USP53 did not affect the osteogenic potential of human MSCs (Appendix Fig S2C–F).
Figure 1. USP34 is required for osteogenic differentiation of human MSCs.
-
ARepresentative images of alkaline phosphatase (ALP), alizarin Red S (ARS), and Van Gieson's staining of human MSCs.
-
B, CQuantitative analyses of the ALP activity and calcium mineralization. Results are shown as mean ± SEM; n = 6; **P < 0.01 and ***P < 0.001 by t‐test.
-
D, EQuantitative RT–PCR and Western blot analyses of the expression of osteogenic markers. Cells were cultured in osteogenic medium for 5 days. Results are shown as mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
-
FH&E staining of the MSC‐mediated ectopic bone formation. Scale bar, 50 μm.
-
GQuantitative analysis of bone volume versus total tissue volume (BV/TV). Results are shown as mean ± SEM; n = 6. *P < 0.05 by t‐test.
Source data are available online for this figure.
In addition, we generated stable knockdown human MSCs using lentiviruses expressing shRNA and implanted them with β‐TCP carriers into immunocompromised mice subcutaneously. The knockdown efficiency was confirmed by quantitative RT–PCR and Western blot (Appendix Fig S2G and H). H&E staining showed that USP34‐depleted cells formed much less bone tissue (Fig 1F) when compared with control cells that had scrambled shRNA. Quantitative measurement of newly formed bone area revealed a 35% reduction (Fig 1G).
Conditional deletion of Usp34 in MSCs leads to low bone mass
To further investigate the role of USP34 in vivo, we performed immunohistochemical staining of USP34 on mouse femoral sections and observed that USP34 is widely expressed in bone marrow cells, osteoblasts, osteocytes, and hypertrophic chondrocytes of the growth plate (Appendix Fig S3A). We also generated Prx1‐Cre;tdTomato mice and sorted the Tomato+ cells (Appendix Fig S3B). Interestingly, the expression of Usp34 was decreased in Tomato+ cells isolated from aged mice compared to those from young adults (Appendix Fig S3C and D), suggesting a potential role of USP34 in age‐related osteoporosis.
Next, we generated Usp34 floxed (Usp34 f/+) mice using CRISPR/Cas9 (Appendix Fig S4A). LoxP sites were integrated into the Usp34 allele flanking the second exon, which contains the start codons of all known Usp34 isoforms. Cre recombinase‐mediated removal of exon 2 is thus expected to cause a translation termination of Usp34. We then bred Usp34 f/+ mice with Prx1‐Cre transgenic mice to conditionally delete Usp34 from MSCs (Appendix Fig S4A and B). Prx1 is a transcription factor expressed during early limb bud mesoderm development; therefore, Prx1‐Cre targets all cells derived from limb bud mesoderm (Logan et al, 2002). The knockout efficiency was confirmed by quantitative RT–PCR and Western blot (Appendix Fig S4C and D). Prx1‐Cre;Usp34 f/f mice were viable and born at the expected Mendelian ratio. The body size and weight of Prx1‐Cre;Usp34 f/f mice were comparable to their littermate controls (Appendix Fig S4E and F).
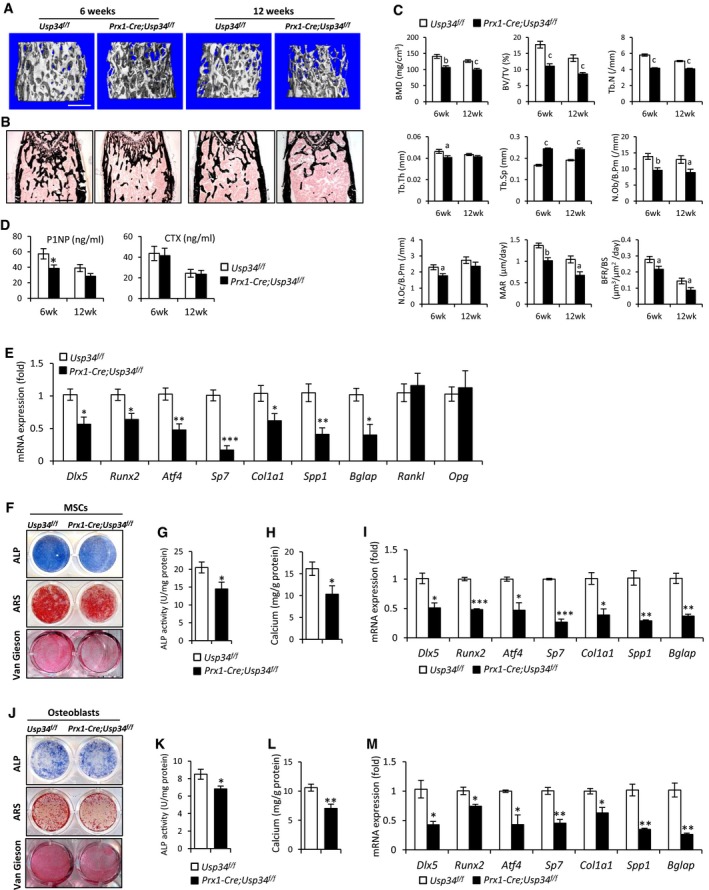
MicroCT analysis of trabecular bone from the distal femur metaphysis revealed that the bone mineral density (BMD) and bone volume (BV/TV) in Prx1‐Cre;Usp34 f/f male mice were significantly lower than those of their Usp34 f/f littermates (Fig 2A and C). Prx1‐Cre;Usp34 f/f mice also had reduced trabecular number (Tb.N), decreased trabecular thickness (Tb.Th), and increased trabecular separation (Tb.Sp) (Fig 2C). Von Kossa staining of undecalcified sections further confirmed the decrease in trabecular bone in Prx1‐Cre;Usp34 f/f mice (Fig 2B).
Figure 2. Conditional deletion of Usp34 in MSCs leads to low bone mass.
-
ARepresentative microCT images of trabecular bone from the femoral metaphysis of male Prx1‐Cre;Usp34 f/f and littermate control mice. Scale bar, 500 μm.
-
BVon Kossa staining of undecalcified sections of femurs. Scale bar, 500 μm.
-
CHistomorphometric analysis of trabecular bone from the femoral metaphysis. Results are shown as mean ± SEM; n = 8; a: P < 0.05, b: P < 0.01, and c: P < 0.001 by t‐test.
-
DSerum levels of P1NP and CTX. Results are shown as mean ± SEM; n = 8; *P < 0.05 by t‐test.
-
EQuantitative RT–PCR analyses of the gene expression of femoral bone samples. Results are shown as mean ± SEM; n = 6; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
-
FRepresentative images of ALP, ARS, and Van Gieson's staining of MSCs obtained from Prx1‐Cre;Usp34 f/f and littermate control mice.
-
G, HQuantitative analyses of the ALP activity and calcium mineralization of MSCs. Results are shown as mean ± SEM; n = 6; *P < 0.05 by t‐test.
-
IQuantitative RT–PCR analyses of the transcription of osteogenic markers. MSCs were differentiated in osteogenic medium for 5 days. Results are shown as mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
-
JRepresentative images of ALP, ARS, and Van Gieson's staining of primary calvarial osteoblasts.
-
K, LQuantitative analyses of the ALP activity and calcium mineralization of primary calvarial osteoblasts. Results are shown as mean ± SEM; n = 6; *P < 0.05 and **P < 0.01 by t‐test.
-
MQuantitative RT–PCR analyses of the transcription of osteogenic markers. Calvarial osteoblasts were differentiated in osteogenic medium for 5 days. Results are shown as mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 by t‐test.
Bone homeostasis is dependent on the coupling of bone formation and resorption (Sims & Martin, 2014). To unveil the cellular basis for decreased bone mass in Prx1‐Cre;Usp34 f/f mice, we performed the histomorphometric analysis. Osteoblast numbers (N.Ob/B.Pm) were significantly reduced, while osteoclast numbers (N.Oc/B.Pm) were unchanged (Fig 2C). Meanwhile, the mineral apposition rate (MAR) and bone formation rate (BFR/BS) in Prx1‐Cre;Usp34 f/f mice were both significantly lower than in the controls (Fig 2C), indicating a reduction in bone formation. We also performed ELISA to assess the serum markers for bone turnover. The serum levels of the bone formation marker P1NP were reduced in 6‐week‐old Prx1‐Cre;Usp34 f/f mice, whereas the levels of c‐terminal telopeptides of collagen type I (CTX), a marker for bone resorption, were not affected (Fig 2D). We excluded the potential difference between sexes by thoroughly examining the skeletal phenotype of female animals and observed a similar reduction in bone formation (Appendix Fig S5A–D).
Next, we obtained the femoral bone samples from Usp34 f/f and Prx1‐Cre;Usp34 f/f mice and performed quantitative RT–PCR analyses. The expression of osteogenic markers, such as Dlx5, Runx2, Atf4, Sp7, Col1a1, Spp1, and Bglap, were significantly decreased, while those of Rankl, Opg, Axin1, Axin2, Dkk1, and β‐catenin was unchanged (Fig 2E and Appendix Fig S6). We also isolated the bone marrow MSCs from Usp34 f/f and Prx1‐Cre;Usp34 f/f mice and compared their osteogenic potential in vitro. Prx1‐Cre;Usp34 f/f MSCs exhibited inhibited osteogenic differentiation, as evidenced by decreased ALP activity, calcium mineralization, collagen synthesis, and osteogenic marker gene expression (Fig 2F–I). Similar results were also observed in primary calvarial osteoblasts (Fig 2J–M).
Conditional deletion of Usp34 in pre‐osteoblasts impairs bone formation
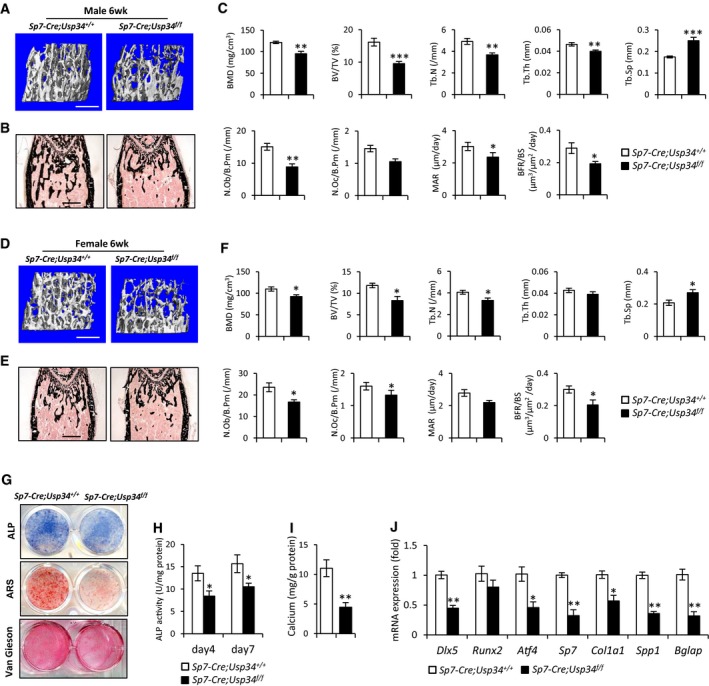
To further investigate whether deletion of Usp34 from committed osteoblast progenitors could recapitulate similar bone loss, we used Sp7‐Cre, which recombines efficiently in pre‐osteoblasts (Rodda & McMahon, 2006). Sp7‐Cre;Usp34 f/f mice exhibited normal Mendelian inheritance and growth features. MicroCT analysis revealed an impairment in trabecular bone micro‐architecture, as measured by reduced skeletal parameters of BMD, BV/TV, and Tb.N and increased Tb.Sp, in the femur metaphysis of both male and female Sp7‐Cre;Usp34 f/f mice (Fig 3A–F). In addition, the decline in osteoblast numbers and dynamic bone histomorphometry characteristics consolidated the osteopenic phenotype of mutant mice (Fig 3A–F).
Figure 3. Conditional deletion of Usp34 in pre‐osteoblasts impairs bone formation.
-
ARepresentative microCT images of trabecular bone from the femoral metaphysis of male Sp7‐Cre;Usp34 f/f and littermate control mice. Scale bar, 500 μm.
-
B, CVon Kossa staining and histomorphometric analyses of male femurs. Scale bar, 500 μm. Results are shown as mean ± SEM; n = 8; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
-
DRepresentative microCT images of trabecular bone from the femoral metaphysis of female Sp7‐Cre;Usp34 f/f and littermate control mice. Scale bar, 500 μm.
-
E, FVon Kossa staining and histomorphometric analyses of female femurs. Scale bar, 500 μm. Results are shown as mean ± SEM; n = 8; *P < 0.05 by t‐test.
-
GRepresentative images of ALP, ARS and Van Gieson's staining of MSCs obtained from Sp7‐Cre;Usp34 f/f and littermate control mice.
-
H, IQuantitative analyses of the ALP activity and calcium mineralization. Results are shown as mean ± SEM; n = 6; *P < 0.05 and **P < 0.01 by t‐test.
-
JQuantitative RT–PCR analyses of the transcription of osteogenic markers. Cells were differentiated in osteogenic medium for 5 days. Results are shown as mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 by t‐test.
Sp7‐Cre;Usp34 f/f MSCs also showed an equivalent resistance to osteogenic differentiation as seen with Prx1‐Cre‐driven Usp34‐ablated MSCs. Knockout of Usp34 inhibited ALP activity, collagen synthesis, mineralization, and expression of osteogenic marker genes in vitro (Fig 3G–J), establishing the essentiality of Usp34 as a regulator during osteogenic lineage commitment.
Loss of USP34 attenuates BMP2 signaling
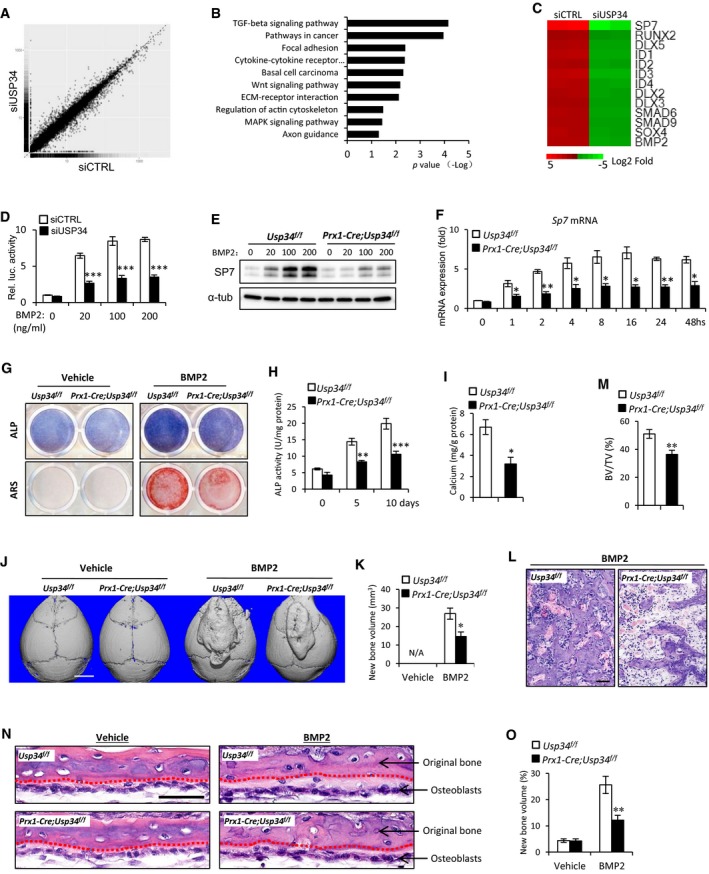
Next, we sought to elucidate the molecular mechanism for USP34 actions on osteogenesis by performing RNA‐Seq analysis. Human MSCs were treated with USP34 or control siRNAs and cultured in osteogenic induction medium for 2 days prior to RNA isolation. Depletion of USP34 from human MSCs increased the expression of 508 genes and decreased the expression of 391 genes (Fig 4A). Notably, a KEGG pathway analysis indicated that USP34 primarily affected the expression of genes associated with TGF‐β/BMP signaling pathway (Fig 4B). The well‐known target genes of BMP2, including SP7, RUNX2, ID1, ID2, and DLX5, were significantly downregulated (Fig 4C).
Figure 4. Loss of USP34 blunts BMP2‐induced responses.
-
AScatter plot of RNA‐seq expression analysis. A total of 508 genes were upregulated and 391 genes were downregulated. Human MSCs were transfected with USP34 or control siRNAs and cultured in osteogenic medium for 2 days. Two biological replicates per group.
-
BKEGG pathway analysis indicated the altered function of TGF‐β/BMP signaling pathway.
-
CHeatmap of representative genes associated with BMP2 signaling pathway.
-
DRelative activity of BRE luciferase assay. 293T cells were depleted with serum overnight followed by a treatment with BMP2 for 6 h. Results are shown as mean ± SEM; n = 3. ***P < 0.001 by t‐test.
-
EImmunoblot analysis of BMP2‐induced Sp7 expression. Prx1‐Cre;Usp34 f/f and Usp34 f/f control MSCs were starved overnight and then stimulated with 100 ng/ml BMP2 for 24 h.
-
FQuantitative RT–PCR analysis of BMP2‐induced Sp7 transcription. Cells were depleted from serum overnight before BMP2 stimulation. Results are shown as mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 by t‐test.
-
GRepresentative images of ALP and ARS staining. MSCs were cultured with normal medium supplemented with 100 ng/ml BMP2 for 10 and 21 days, respectively.
-
H, IQuantitative analyses of the ALP activity (n = 4) and calcium mineralization (n = 5). Results are shown as mean ± SEM; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
-
J, KRepresentative microCT images and quantification of newly formed bone in response to BMP2. Results are shown as mean ± SEM; n = 8. *P < 0.05 by t‐test. Scale bar, 2,500 μm.
-
LH&E staining of BMP2‐induced newly formed bone. Scale bar, 50 μm.
-
MQuantitative analysis of bone volume versus total tissue volume (BV/TV). Results are shown as mean ± SEM; n = 8; **P < 0.01 by t‐test.
-
NH&E staining of calvarial organ cultures stimulated with or without 100 ng/ml BMP2. Red dotted lines indicate the interface between original and newly formed bone. Scale bar, 50 μm.
-
OQuantitative analysis of newly formed bone volume versus original bone volume (%). Results are shown as mean ± SEM; n = 8; **P < 0.01 by t‐test.
Source data are available online for this figure.
To verify that USP34 is essential for the activation of BMP2 signaling, we then performed a BRE luciferase assay. The BRE‐Luc construct used is derived from the promoter of Id1 gene and contains the most sensitive BMP‐responsive elements (Korchynskyi & ten Dijke, 2002; Logeart‐Avramoglou et al, 2006). We observed that knockdown of USP34 in 293T cells markedly restricted BMP2‐induced responses (Fig 4D). As BMP2 induces the expression of SP7, a master transcriptional factor for osteogenic differentiation, we examined whether this induction requires USP34. Both Western blot and quantitative RT–PCR revealed that deletion of Usp34 from mouse MSCs significantly blunted the expression of Sp7 (Fig 4E and F). MSCs obtained from Prx1‐Cre;Usp34 f/f mice also showed decreased ALP activity and calcium mineralization after prolonged BMP2 treatment (Fig 4G–I).
Next, we investigated the in vivo function of USP34 in BMP2‐induced bone formation by performing calvarial injection experiments. Prx1‐Cre recombines in progenitors not only from long bone, but also in those from calvaria (Elefteriou & Yang, 2011; Xiong et al, 2011; Ouyang et al, 2014). Subcutaneous injection of BMP2 onto the calvaria induces the local recruitment and differentiation of cranial mesenchymal cells into osteoblasts, resulting in the de novo formation of cancellous bone (Mundy et al, 1999; Addison et al, 2014). In comparison with their control littermates, Prx1‐Cre;Usp34 f/f mice were markedly less responsive to BMP2 injection, with a nearly 50% reduction in the volume of newly formed bone on the calvaria (Fig 4J and K). Vehicle injections did not induce any bone formation (Fig 4J and K). Histological analysis also revealed that the newly formed bone in Prx1‐Cre;Usp34 f/f mice had larger bone marrow spaces and lower bone volume versus total volume (BV/TV) as compared to those of controls (Fig 4L and M).
Finally, we performed calvarial organ culture to assess the role of Usp34 in BMP2 signaling further up. As expected, BMP2 induced sub‐periosteal bone formation (Fig 4N and O; light red, in contrast to the red original bone). However, the bone formation in Prx1‐Cre;Usp34 f/f calvariae was significantly less than that observed in Usp34 f/f controls (Fig 4N and O). These data demonstrate that Usp34 deletion within mesenchymal cells blunts their responses to BMP2.
Deletion of Usp34 in MSCs impairs bone regeneration
Considering the indispensability of BMP2 activity in initiating fracture healing (Tsuji et al, 2006), we subsequently carried out mid‐diaphyseal femoral fractures to Prx1‐Cre;Usp34 f/f and control mice to evaluate the effect of Usp34 on bone healing. Three weeks after the fracture, microCT analysis showed that healing proceeded much more slowly in Prx1‐Cre;Usp34 f/f mice as compared to control mice (Fig 5A). There is a significant decrease in trabecular BV/TV in the callus of Prx1‐Cre;Usp34 f/f mice (Fig 5B), along with diminished Tb.N and Tb.Th, as well as elevated trabecular separation (Fig 5C–E). Histomorphometric analysis confirmed that the mineralization in callus bone at the fracture site was significantly lower in Prx1‐Cre;Usp34 f/f mice (Fig 5F and H). In contrast to the controls, Usp34‐deficient mice failed to achieve a comparable bone healing at the fracture site, with a large amount of callus cartilage still remained (Fig 5G and I).
Figure 5. Deletion of USP34 in MSCs impairs bone regeneration.
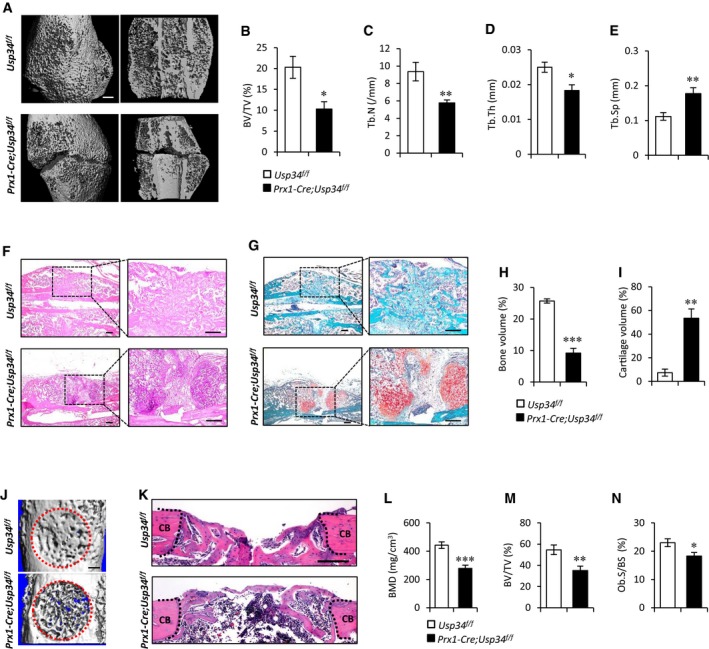
-
ARepresentative microCT 3D reconstruction and cut‐plane images of the fracture site 3 weeks after bone fracture. Scale bar, 500 μm.
-
B–EMicroCT analyses of trabecular bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp). Results are shown as mean ± SEM; n = 8; *P < 0.05 and **P < 0.01 by t‐test.
-
F, GRepresentative images of H&E (F) and Safranin O (G) staining of the fracture callus 3 weeks after bone fracture. Scale bar, 200 μm.
-
H, IQuantification of the mineralized bone callus (H) and cartilage callus (I) in the fracture site. Results are shown as mean ± SEM; n = 8; **P < 0.01 and ***P < 0.001 by t‐test.
-
JRepresentative microCT images of femoral cortical bone defects in Prx1‐Cre;Usp34 f/f and Usp34 f/f control mice. The red dotted lines indicate the position of the original defect margin. Scale bar, 500 μm.
-
KH&E staining of femoral cortical bone defects. The black dotted lines indicate the position of the original defect margin. CB, cortical bone. Scale bar, 200 μm.
-
L–NBone mineral density (BMD), bone volume (BV/TV), and osteoblast surface (Ob.S/BS) of the regenerated bone in femoral cortical gaps. Results are shown as mean ± SEM; n = 8; *P < 0.05, **P < 0.01, and ***P < 0.001 by t‐test.
For further validation, we surgically created skeletal defects by drilling holes in femoral cortical bone to examine the inhibitory effect of Usp34 deficiency on bone regeneration. MicroCT and histological analyses consistently showed that the cortical gaps in Usp34 f/f control mice were almost completely bridged after 2 weeks, while those in Prx1‐Cre;Usp34 f/f were only partially filled (Fig 5J and K). Additionally, the BMD and BV/TV of the mineralized callus of Prx1‐Cre;Usp34 f/f mice were significantly lower than their controls (Fig 5L and M). The osteoblast surfaces (Ob.S/BS) were also diminished (Fig 5N).
USP34 stabilizes Smad1 and RUNX2
BMP2 triggers osteogenic differentiation mainly through activation of Smad1/5/8 to induce the expression of downstream transcriptional factors such as DLX5, RUNX2, and SP7 (Rahman et al, 2015; Salazar et al, 2016; Wu et al, 2016). To uncover the substrates of USP34, we first ectopically expressed FLAG‐tagged Smad1 in 293T cells and revealed that it could be detected in USP34 immunoprecipitates (Fig 6A). And knockdown of USP34 significantly reduced the protein levels of FLAG‐Smad1 (Fig 6B), while increased its ubiquitination (Fig 6C), indicating a key role for USP34‐dependent deubiquitination of Smad1. We then determined whether USP34 in fact regulates the stability of the Smad1 protein by observing FLAG‐Smad1 protein levels in the presence of cycloheximide (CHX), an inhibitor of protein translation. Depleting USP34 using siRNA resulted in much faster degradation of FLAG‐Smad1 protein (Fig 6D).
Figure 6. USP34 deubiquitinates and stabilizes Smad1 and RUNX2.
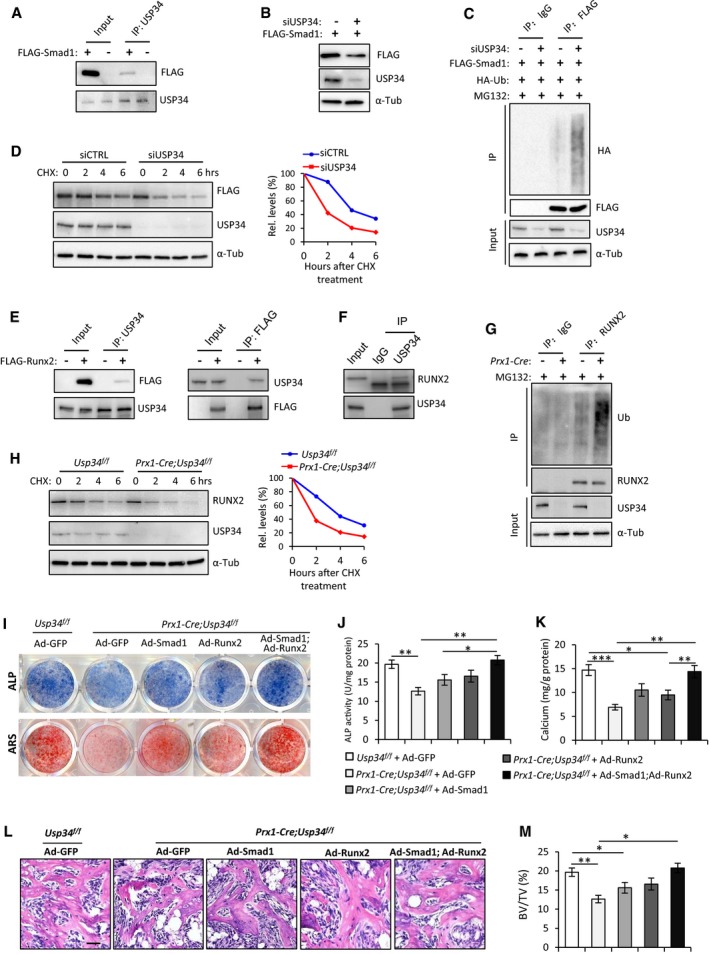
-
ACo‐immunoprecipitation of USP34 with ectopically expressed FLAG‐tagged Smad1 in 293T cells.
-
BImmunoblot analysis showing decreased FLAG‐Smad1 after USP34 knockdown.
-
CImmunoblot of Smad1‐linked polyUb. 293T cells were co‐transfected with FLAG‐Smad1 overexpression plasmid and USP34 siRNA, and treated with 10 μM MG132 for 4 h before collection.
-
DImmunoblot analysis of the degradation of FLAG‐Smad1 protein. 293T cells were co‐transfected with FLAG‐Smad1 overexpression plasmid and USP34 siRNA, and treated with 50 μg/ml cycloheximide (CHX) before collection at designated time points. Right: relative quantification of FLAG‐Smad1 protein levels at different time points.
-
ECo‐immunoprecipitation of USP34 with ectopically expressed FLAG‐tagged RUNX2 in 293T cells.
-
FCo‐immunoprecipitation of USP34 with endogenous RUNX2 in mouse MSCs.
-
GImmunoblot of RUNX2‐linked polyUb. Differentiated mouse MSCs were treated with 10 μM MG132 for 4 h before collection. Prx1‐Cre(+) indicates MSCs isolated from Prx1‐Cre;Usp34 f/f mice, and Prx1‐Cre(−) indicates those from Usp34 f/f control mice.
-
HImmunoblot analysis of the degradation of endogenous RUNX2 protein in MSCs. Prx1‐Cre;Usp34 f/f and Usp34 f/f control MSCs were stimulated with BMP2, treated with 50 μg/ml CHX, and collected at designated time points. Right: relative quantification of RUNX2 protein levels at different time points.
-
IRepresentative images of ALP and ARS staining. Usp34 f/f and Prx1‐Cre;Usp34 f/f MSCs were transduced with GFP, Smad1, Runx2, or Smad1 + Runx2 adenoviruses and then cultured in osteogenic medium for 5 and 10 days, respectively.
-
J, KQuantitative analyses of the ALP activity and calcium mineralization. Results are shown as mean ± SEM; n = 8; *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey's post hoc test.
-
L, MH&E staining and quantitative analysis of MSC‐mediated ectopic bone formation. Scale bar, 50 μm. Results are shown as mean ± SEM; n = 8; *P < 0.05 and **P < 0.01 by ANOVA with Tukey's post hoc test.
Source data are available online for this figure.
In addition, we discovered that USP34 could physically interact with RUNX2, a downstream factor of Smad1 (Fig 6E and F). And Prx1‐Cre;Usp34 f/f MSCs showed significantly increased RUNX2 ubiquitination compared to Usp34 f/f controls (Fig 6G). We then evaluated the stability of RUNX2 protein in Prx1‐Cre;Usp34 f/f MSCs and observed a more prominent degradation compared to the Usp34 f/f controls (Fig 6H). These results indicate that USP34 also acts as a bona fide deubiquitinase that stabilizes RUNX2.
Combined overexpression of Smad1 and Runx2 rescues the osteogenic potential of Usp34‐deficient MSCs
To further establish that decreased Smad1 and RUNX2 contributed to inhibited osteogenesis of Usp34‐deficient MSCs, we transduced Prx1‐Cre;Usp34 f/f MSCs with Smad1 adenoviruses (Ad‐Smad1), Runx2 adenoviruses (Ad‐Runx2), or a combination. The combination of Ad‐Smad1 and Ad‐Runx2 successfully restored the weak ALP activity and calcium mineralization in Prx1‐Cre;Usp34 f/f MSCs (Fig 6I–K), while single restoration of either Smad1 or Runx2 was insufficient to reverse the osteogenic incompetence.
In addition, we subcutaneously implanted cells that overexpressed Smad1 or/and Runx2 with β‐TCP carriers into immunocompromised mice to testify their ability to ectopically form bone tissue. In consistent with in vitro findings, histological analysis confirmed that only concurrent overexpression of Smad1 and Runx2 could fully rescue the compromised bone formation of Prx1‐Cre;Usp34 f/f cells (Fig 6L and M), demonstrating that both Smad1 and RUNX2 are critical substrates for USP34's control in osteogenic differentiation.
Depletion of Smurf1 restores the BMP2‐induced responses and osteogenic potential of Usp34‐deficient MSCs
Since Smurf1, an E3 ubiquitin ligase, regulates BMP2 signaling through directly interacting and degrading Smad1 and RUNX2 (Zhu et al, 1999; Zhao et al, 2003), we hypothesized that it couples USP34 to balance the BMP2 signaling. To this end, we knocked down both of them in 293T cells and performed BRE luciferase assay. Interestingly, knockdown of Smurf1 in Usp34‐depleted cells successfully restored BMP2‐induced luciferase activity (Fig 7A). Considering the master role of SP7 in osteogenesis, we measured the expression of Sp7 to assess the extent of recovery in BMP2‐induced response of Prx1‐Cre;Usp34 f/f MSCs upon depletion of Smurf1. Both Western blot and quantitative RT–PCR showed that the restricted expression of Sp7 in Prx1‐Cre;Usp34 f/f MSCs was rescued by Smurf1 siRNA (Fig 7B and C). Moreover, depletion of Smurf1 restored the BMP2‐induced ALP activity and calcium mineralization in Prx1‐Cre;Usp34 f/f MSCs (Fig 7D and E).
Figure 7. Depletion of Smurf1 restores the BMP2‐induced responses and osteogenic potential of Usp34‐deficient MSCs.
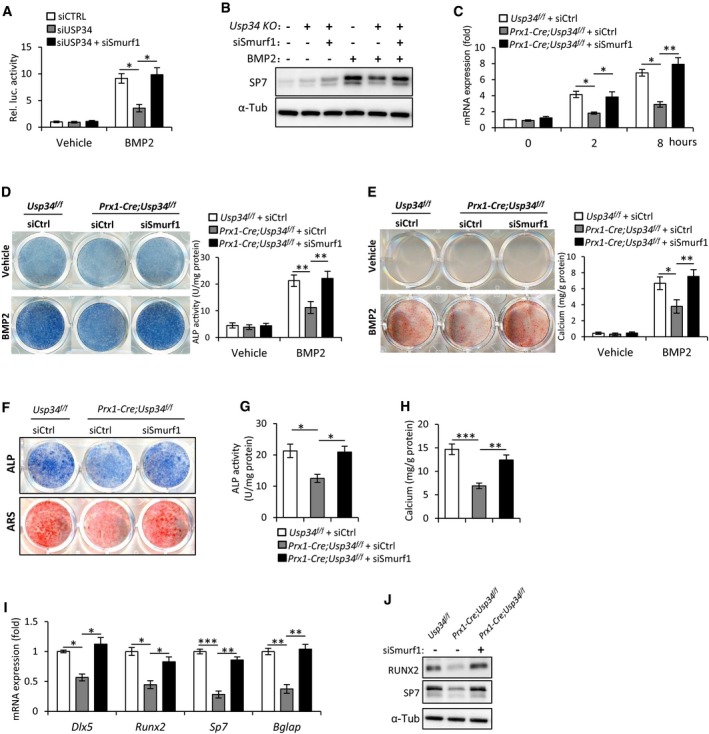
-
ARelative activity of BRE luciferase assay. 293T cells were depleted with serum overnight followed by a treatment with BMP2 for 6 h. Results are shown as mean ± SEM; n = 3. *P < 0.05 by ANOVA with Tukey's post hoc test.
-
BImmunoblot analysis of BMP2‐induced Sp7 expression. Prx1‐Cre;Usp34 f/f MSCs were treated with Smurf1 or control siRNAs, starved overnight, and then stimulated with 100 ng/ml BMP2 for 24 h.
-
CQuantitative RT–PCR analysis of BMP2‐induced Sp7 transcription. Prx1‐Cre;Usp34 f/f MSCs were treated with Smurf1 or control siRNAs, and depleted from serum overnight before BMP2 stimulation. Results are shown as mean ± SEM; n = 3; *P < 0.05 and **P < 0.01 by ANOVA with Tukey's post hoc test.
-
DALP staining and quantitative measurement of ALP activity. MSCs were cultured with normal medium supplemented with 100 ng/ml BMP2 for 5 days. Results are shown as mean ± SEM; n = 8; **P < 0.01 by ANOVA with Tukey's post hoc test.
-
ERepresentative images of ARS staining and quantification. Cells were cultured with normal medium supplemented with 100 ng/ml BMP2 for 14 days. Results are shown as mean ± SEM; n = 8; *P < 0.05 and **P < 0.01 by ANOVA with Tukey's post hoc test.
-
FRepresentative images of ALP and ARS staining. Mouse MSCs were treated with Smurf1 or control siRNAs, and cultured in osteogenic medium for 5 and 10 days, respectively.
-
G, HQuantitative analyses of the ALP activity and calcium mineralization. Results are shown as mean ± SEM; n = 8; *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey's post hoc test.
-
I, JQuantitative RT–PCR and Western blot analyses of the expression of osteogenic markers. Cells were treated with Smurf1 or control siRNAs, and cultured in osteogenic medium for 5 days. Results are shown as mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey's post hoc test.
Source data are available online for this figure.
Finally, we sought to investigate whether depletion of Smurf1 is sufficient to recover the osteogenic potential of Prx1‐Cre;Usp34 f/f MSCs in vitro. After culturing with osteogenic induction medium for 5 and 10 days, respectively, the ALP activity and calcium mineralization of Prx1‐Cre;Usp34 f/f + siSmurf1 MSCs were comparable to those of Usp34 f/f controls (Fig 7F–H). Besides, the expression of osteogenic markers, such as Dlx5, Runx2, Sp7, and Bglap, was also recovered and no longer inhibited after the treatment (Fig 7I and J), indicating that USP34 sequesters Smurf1 to promote osteogenic differentiation.
Discussion
The proper osteogenic differentiation is essential for bone homeostasis. Here, we have profiled the expression of 54 USPs in human MSCs after osteogenic induction and identified USP34 as a previously unknown regulator of osteogenesis. Our in vitro and in vivo data demonstrate that USP34 is essential for the osteogenic differentiation and bone formation. Although the expression of USP13, USP18, USP21, and USP53 was also increased after osteogenic induction, knockdown of these genes did not have a significant effect. Previous reports show that USP1 and USP6 maintain the stem‐cell‐like properties and inhibit the osteogenic commitment in osseous tumors (Ye et al, 2010; Williams et al, 2011). A recent study also suggests that USP4 antagonizes osteoblast differentiation in vitro through deubiquitination of Dishevelled (Zhou et al, 2016). These data, together with our findings, indicate that USPs may play a pivotal role in maintaining skeletal homeostasis.
In this study, we show that loss of USP34 blunts BMP2 signaling, which is indispensable for osteogenesis and bone fracture healing. It triggers osteogenic commitment of MSCs mainly through activation of Smad1/5/8 that subsequently induces the expression of transcriptional factors, such as RUNX2, MSX1, and DLX5 (Rahman et al, 2015; Salazar et al, 2016; Wu et al, 2016). To elucidate the underlying molecular mechanism, we first identified Smad1, the critical element of BMP2 signaling, as a substrate of USP34. This is consistent with the previous study which shows that USP34 interacted with Smad1/5/8 and that depletion of USP34 in HeLa cells by siRNA reduced the level of total Smad1/5/8 (Cheng et al, 2012). We also discovered that USP34 targeted the downstream transcriptional factor RUNX2. Both mRNA and protein levels of RUNX2 were decreased in the Usp34‐deficient MSCs, indicating that USP34 regulates the expression of RUNX2 not only through Smad‐dependent transcriptional activation, but also through post‐transcriptional ubiquitination. Our rescue experiments also showed that only combined overexpression, but not single overexpression of Smad1 or Runx2, was able to fully recover osteogenic potential of Usp34‐deficient MSCs, confirming USP34 targets on both Smad1 and Runx2.
The previous study reported that USP34 regulates axin stability and Wnt/β‐catenin signaling (Lui et al, 2011). Our KEGG pathway analysis of the RNA‐Seq data also suggests that Wnt signaling may be associated with USP34 activity (P = 0.007, Fig 4B). To confirm this observation, we isolated femoral bone samples from 12‐week‐old Prx1‐Cre;Usp34 f/f and Usp34 f/f mice, and compared the mRNA levels of Axin1, Axin2, Dkk1, and β‐catenin. However, no significant difference was identified (Appendix Fig S6). The potential role of Wnt signaling in USP34‐mediated bone formation needs further investigation.
Here, we report USP34 as the first documented deubiquitinase for RUNX2, a runt domain family protein that is essential for the commitment of mesenchymal cells to the osteoblast lineage. Both intramembranous and endochondral ossification are absent in Runx2 knockout mice (Komori et al, 1997; Otto et al, 1997). Previous studies have shown that RUNX2 could be degraded by ubiquitin ligases, such as Smurf1, WWP1, and CHIP (Zhao et al, 2003, 2004; Jones et al, 2006; Li et al, 2008; Shu et al, 2013). However, its deubiquitinase has not been discovered before.
Smurf1 is the first E3 ligase identified for RUNX2 ubiquitination (Zhao et al, 2003, 2004). It also mediates the degradation of Smad1 (Zhu et al, 1999). Smurf1‐deficient mice exhibit enhanced osteoblast activities and an age‐dependent increase in bone mass (Zhao et al, 2004; Yamashita et al, 2005). This phenotype is opposite to that we observed in Prx1‐Cre;Usp34 f/f and Sp7‐Cre;Usp34 f/f mice. Interestingly, we observed that depletion of Smurf1 is able to fully restore the BMP2‐induced responses and osteogenic potential of Usp34‐deficient MSCs, suggesting USP34 couples Smurf1 to orchestrate the BMP2 signaling.
The data in this study were based on loss‐of‐function experiments. We attempted overexpression of USP34 in MSCs and MC3T3‐E1 cells, but were unsuccessful due to technical difficulty. The entire ORF of USP34 is over 10 kb, which is too large. We have tried to activate transcription of endogenous USP34 using a CRISPR/Cas9 lentiviral activation particles (Santa Cruz). Unfortunately, it turned out that the efficiency for USP34 in MSCs was not sufficient. We believe the evidence presented in this study is strong but could be improved with the performance of such experiments.
In summary, we show that expression of USP34 in human MSCs increases after osteogenic induction and depletion of USP34 inhibits osteogenic differentiation. Specific deletion of Usp34 from MSCs or pre‐osteoblasts results in low bone mass and decreased osteoblast function in mice. Moreover, loss of Usp34 blunts BMP2‐induced responses and impairs bone regeneration. Mechanically, USP34 stabilizes both Smad1 and RUNX2. Our data demonstrate that USP34 is a previously unknown regulator of osteogenic differentiation and bone formation.
Materials and Methods
Reagents
Recombinant human BMP2 (#120‐02C) and Wnt3a (#315‐20) were purchased from PeproTech (Rocky Hill, NJ). The antibodies used are as follows: rabbit anti‐USP13 (A302‐762A, Bethyl), rabbit anti‐USP18 (sc‐98431, Santa Cruz), goat anti‐USP21 (sc‐79305, Santa Cruz), rabbit anti‐USP34 (A300‐824A, Bethyl), rabbit anti‐USP53 (HPA035844, Sigma), rabbit anti‐DLX5 (HPA005670, Sigma), rabbit anti‐RUNX2 (Ab23981, Abcam), rabbit anti‐SP7 (ab22552, Abcam), mouse anti‐FLAG‐M2 (F1804, Sigma), rabbit anti‐FLAG (F7425, Sigma), mouse anti‐HA (901502, BioLegend), and mouse anti‐α‐tubulin (sc‐32293, Santa Cruz). The construct for HA‐Ubiquitin (#17608) was obtained from Addgene (Cambridge, MA). The FLAG‐RUNX2 plasmid is a gift from Dr. Weiguo Zou (Institute of Biochemistry and Cell Biology, Shanghai).
Mice
Usp34 conditional knockout mice were generated by a CRISPR/Cas9‐based approach. Briefly, two sgRNAs were designed with a CRISPR design tool (http://crispr.mit.edu) to target either a region upstream or downstream of the exon 2 (Appendix Table S1). These were then screened for on‐target activity using a Universal CRISPR Activity Assay (UCA™, Biocytogen Inc, Beijing). A T7 promoter sequence was added to the Cas9 or sgRNA template by PCR amplification in vitro. A circular targeting vector containing exon 2 flanked by two LoxP sites and two homology arms was mixed with Cas9 mRNA and sgRNAs and then co‐injected into the cytoplasm of one‐cell stage fertilized C57BL6/J eggs. The injected zygotes were transferred into oviducts of Kunming pseudopregnant females to generate F0 mice. F0 mice with expected genotype confirmed by tail genomic DNA PCR and sequencing were mated with C57BL6/J mice to establish germline‐transmitted F1 founders. F1 founders were genotyped by tail genomic PCR and DNA sequencing. Southern blot examination was performed to further confirm correct genotype. Prx1‐Cre mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and mated with Usp34 f/f mice for at least three generations to generate Prx1‐Cre;Usp34 f/f conditional knockout mice. Genotypes were determined by PCR amplification of purified tail genomic DNA. Primers are listed in Appendix Table S1. Female immunocompromised nude mice were purchased from the Experimental Animal Center of Sichuan University. Mice were housed in pathogen‐free facilities under a 12‐h light and 12‐h dark cycle. All protocols were approved by the Subcommittee on Research and Animal Care (SRAC) of Sichuan University.
Cell culture
Human bone marrow‐derived mesenchymal stem cells (MSCs) were obtained from ATCC. We cultured the cells in α‐MEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), plus 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37°C with a humidified atmosphere of 5% CO2. Primary mouse MSCs were isolated from 6‐week‐old mice by flushing the bone marrow of tibiae and femurs (Song et al, 2012). Primary osteoblasts were isolated from the calvariae of 3‐day‐old mice as described previously (Yuan et al, 2012). To induce osteogenic differentiation, MSCs were seeded in 6‐well or 24‐well plates. After confluence, cells were treated with osteogenic medium containing 50 μM ascorbic acid, 10 mM β‐glycerophosphate, and 10 nM dexamethasone (all from Sigma).
Isolation of Prx1‐Cre;tdTomato + cells
Bone marrow was flushed out from femurs of young (3‐month‐old) and aged (18‐month‐old) Prx1‐Cre;tdTomato mice and then treated with red blood cell lysis buffer (Santa Cruz) for 10 min at room temperature. After rinsing with cold PBS, the nucleated marrow cells were incubated with APC‐conjugated CD45 antibody (BioLegend) and APC‐conjugated CD11b antibody (BioLegend) and were subjected to flow cytometry (Becton‐Dickinson LSRII). The cell gating was based on comparison with negative control and single‐stained controls.
siRNA knockdown, lentivirus‐mediated shRNA knockdown, and adenovirus‐mediated overexpression of MSCs
All targeted and control small interfering RNAs (siRNAs) were purchased from Santa Cruz (Dallas, Texas). Each siRNA consists of pools with three to five target‐specific 19‐ to 25‐nucleotide siRNAs designed to knock down target gene expression. For siRNA‐mediated knockdown, human MSCs or 293T cells were transfected using Lipofectamine RNAimax (Invitrogen) following the manufacturer's instructions. Knockdown efficiencies were examined by quantitative RT–PCR and Western blot 48 h after the start of transfection. For shRNA‐mediated knockdown, the USP34 shRNA expression lentiviruses were purchased from GeneCopoeia (Guangzhou, China). Stably transfected clones were selected with puromycin (Invitrogen) for about 7 days. For overexpression of Smad1 and Runx2, the adenovirus particles expressing mouse Smad1, Runx2, or GFP (control) were obtained from Cyagen (Guangzhou, China).
Quantitative RT–PCR and RNA‐Seq
We isolated the total RNA using an RNeasy mini kit (Qiagen). cDNA was prepared from 2 μg RNA using a QuantiTect reverse transcription kit (Qiagen) and analyzed with SYBR Premix Ex Taq (Takara, Dalian, China) in ABI7500 real‐time PCR system (Applied Biosystems, Foster City, CA). Relative expression was calculated using a method by normalizing with Gapdh housekeeping gene expression and presented as fold increase relative to control.
For RNA‐Seq, libraries were prepared using the Illumina TrueSeq mRNA sample preparation kit according to the manufacturer's instruction and single‐end sequenced on an Illumina HiSeq 3000 machine. The reads were aligned to hg19 genome using TopHat. The analysis of RNA‐seq data was done using Cufflinks with Refseq mRNAs to guide assembly and the cummeRbund package in R (Trapnell et al, 2012). Transcripts, which were regulated greater than twofold with significant difference, were used for KEGG pathway analysis.
Western blot and co‐immunoprecipitation
Cells were lysed in RIPA buffer (Pierce, Rockford, IL) on ice. The samples were heated at 95°C for 5 min in sample buffer containing 2% SDS and 1% 2‐mercaptoethanol, separated on 5% or 10% SDS–polyacrylamide gels, and transferred to PVDF membranes by a wet transfer apparatus (Bio‐Rad). The membranes were blotted with 5% milk for 1 h and then incubated with primary antibodies overnight, followed by incubation with a horseradish peroxidase‐conjugated IgG (Jackson ImmunoResearch, West Grove, PA). The antibody–antigen complexes were visualized with Immobilon reagents (Millipore).
For co‐immunoprecipitation, cells or primary mouse MSCs were lysed in CelLytic M buffer (Sigma) supplemented with protease inhibitor cocktail (Roche) for 20 min on ice and centrifuged at 10,000 g for 15 min at 4°C. The supernatant was incubated with the indicated antibodies at 4°C overnight, followed by incubation with a 30 μl of Dynabeads protein G (Life Technologies) for additional 1 h. Co‐precipitated proteins were washed, eluted with SDS‐loading buffer at 95°C for 5 min, and then subjected to Western blot analyses.
ALP and mineralization assays
The differentiated cells were fixed with 10% buffered formalin and incubated with 0.1 M Tris buffer (pH 9.3) containing 0.25% naphthol AS‐BI phosphate (Sigma) and 0.75% Fast Blue BB (Sigma). ALP activity was also quantified using a commercial kit according to the manufacturer's protocol (Cell Biolabs, San Diego, CA).
The mineralization assay was performed as previously described (Liu et al, 2016c). Briefly, cells were cultured in osteogenic medium for 10–14 days, fixed with 10% buffered formalin, and stained with 1% alizarin red S (pH 4.2, Sigma‐Aldrich) for 5 min. Mineralized matrix stained with alizarin red were destained with 10% cetylpyridinium chloride in 10 mM sodium phosphate (pH 7.0), and the calcium concentration was determined by absorbance measurements at 562 nm using a standard calcium curve in the same solution.
BRE luciferase assay
293T cells seeded in the 24‐well plates were transfected with USP34 or control siRNA, together with 100 ng of BRE luciferase (#45126 Addgene) and 50 ng of CMV‐beta‐galactosidase constructs using Lipofectamine 2000 (Invitrogen). After about 24 h, the cells were depleted with serum overnight followed by a treatment with 100 ng/ml BMP2 for 6 h. The luciferase and β‐galactosidase activity of total cell lysates were determined using Luc‐Screen and Galacto‐Star kits (Applied Biosystems).
Xenograft model for MSC‐mediated bone formation
Approximately 5 × 106 cells were mixed with 60 mg of pure phase beta‐tricalcium phosphate particles (SynthoGraft, Bicon, Boston, MA), which is a kind of tricalcium phosphate material that exists as crystalline polymorphs beta, and then transplanted subcutaneously under the dorsal surface of immunocompromised beige mice as described (Zhou et al, 2017). Six weeks after transplantation, the transplants were collected, fixed with 10% formalin, and decalcified with 10% EDTA. Paraffin sections were fabricated and stained with hematoxylin and eosin.
Immunohistochemical staining
The dissected femurs were fixed in 4% polyoxymethylene for 2 days and decalcified in 10% EDTA for 2 weeks before sectioning (5 μm). Slides were subjected to sodium citrate buffer at 99°C for 20 min for antigen retrieval and then incubated with mouse anti‐USP34 (1:50, sc‐100631, Santa Cruz).
MicroCT analysis and bone histomorphometry
The harvested bone tissues were fixed in 10% buffered formalin for 2 days and then stored in 70% ethanol at 4°C before being processed. MicroCT analysis was performed according to recent guidelines (Bouxsein et al, 2010) using a μCT 80 microCT system (Scanco Medical, Bassersdorf, Switzerland) with a spatial resolution of 8 μm (55 kV, 114 mA, 500 ms integration time). For the analysis of cortical bone regeneration, the volume of interest (VOI) was defined as a cylindrical area covering the initial bone defect. Bone volume (BV/TV, %) was calculated within the delimited VOI.
Processing of undecalcified bone specimens and cancellous bone histomorphometry was performed as described previously (Yuan et al, 2014). Followed by microCT scanning, femurs were dehydrated and embedded in methyl methacrylate. Five‐μm‐thick sections were prepared using a Leica RM2235 microtome and were stained by the von Kossa/nuclear fast red method. Histomorphometric measurements in the distal end of femurs were made using OsteoMeasure software (OsteoMetrics, Decatur, GA). All histomorphometric parameters were calculated and expressed according to the suggestions made by the ASBMR nomenclature committee (Dempster et al, 2013).
ELISA
Serum concentrations of P1NP and CTX were measured using ELISA kits from IDS (Fountain Hills, AZ). Animals were fasted for 4 h, and then, blood was collected by puncturing the cheek pouch.
BMP2‐induced bone formation
In vivo analysis of BMP2‐induced bone formation by calvarial injection was conducted as described (Mundy et al, 1999; Addison et al, 2014). Briefly, 7‐day‐old pups were subcutaneously injected with 2 μg recombinant BMP2 (PeproTech) or saline (vehicle control) twice a day for 5 days onto the sagittal sutures of calvariae. Animals were sacrificed 9 days after the last injection for analysis of bone formation.
For in vitro calvarial organ culture, calvariae harvested from 5‐day‐old pups were cultured in BGJb medium (Life Technologies) containing 0.1% bovine serum albumin (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco), supplemented with or without 100 ng/ml BMP2. After 7 days, calvariae were fixed in 10% buffered formalin and decalcified with 10% EDTA for 3 days. Tissue samples were then processed, paraffin‐embedded, sectioned, and stained with H&E.
Surgeries for femoral fracture and cortical defect
Twelve‐week‐old mice were anesthetized by intraperitoneal injection of a combination of ketamine (100 mg/kg) and xylazine (10 mg/kg). In addition, buprenorphine (0.05 mg/kg) was given for perioperative analgesia to minimize suffering and pain. Unilateral fractures were generated in the right femurs as described by others (Tsuji et al, 2006; Yue et al, 2016). Briefly, a metal pin (Ø = 0.45 mm) was introduced into the femoral canal through a medial parapatellar incision and arthrotomy of the knee. After closing the wound, a mid‐diaphyseal fracture was produced by using a falling weight apparatus over a three‐point bending mechanism. At 10 and 21 days post‐fracture, samples were collected for radiography, microCT scanning, and histology analysis. For femoral cortical bone defect, a 1.0 mm hole was generated using a round bur (Komet®, Germany) operating at 10,000 rpm under saline irrigation (Liu et al, 2016b,c). Samples were harvested 2 weeks after the surgery.
Statistical analysis
All values are expressed as mean ± SEM. Statistically significant differences were evaluated by two‐tailed Student's t‐test for comparison between two groups or by one‐way analysis of variance (ANOVA) followed by the Tukey's post hoc test for multiple comparisons. A P‐value of less than 0.05 was considered to be statistically significant.
Data availability
All Seq data have been deposited into NCBI database with the identifier GSE114933, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114933.
Author contributions
YG, LY, QC, and QY designed the project; YG, MW, SZ, YW, CZ, RZ, BS, LX, and WL performed experiments; YG, SZ, MW, YW, CZ, NS, JJ, and QY analyzed the data; YG, YW, QC, and QY wrote and edited the manuscript. All authors reviewed the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Dr. Weiguo Zou from Institute of Biochemistry and Cell Biology, Shanghai, for valuable advices. This work was partly supported by grants from the National Natural Science Foundation of China (NSFC 81722014, 81571001) and Sichuan Province Science and Technology Innovation Team Program (2017TD0016) to Q.Y.; by project of Innovative Research Team of Education Department of Sichuan Province (13TD0038) to L.Y.; and by grants from NSFC (81621062) and 111 Project of Ministry of Education of China to Q.C.
The EMBO Journal (2018) 37: e99398
References
- Addison WN, Fu MM, Yang HX, Lin Z, Nagano K, Gori F, Baron R (2014) Direct transcriptional repression of Zfp423 by Zfp521 mediates a bone morphogenic protein‐dependent osteoblast versus adipocyte lineage commitment switch. Mol Cell Biol 34: 3076–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Robey PG, Simmons PJ (2008) Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R (2010) Guidelines for assessment of bone microstructure in rodents using micro‐computed tomography. J Bone Miner Res 25: 1468–1486 [DOI] [PubMed] [Google Scholar]
- Caplan AI (1991) Mesenchymal stem cells. J Orthop Res 9: 641–650 [DOI] [PubMed] [Google Scholar]
- Caplan AI (2007) Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 213: 341–347 [DOI] [PubMed] [Google Scholar]
- Cheng X, Alborzinia H, Merz KH, Steinbeisser H, Mrowka R, Scholl C, Kitanovic I, Eisenbrand G, Wolfl S (2012) Indirubin derivatives modulate TGFbeta/BMP signaling at different levels and trigger ubiquitin‐mediated depletion of nonactivated R‐Smads. Chem Biol 19: 1423–1436 [DOI] [PubMed] [Google Scholar]
- Ciechanover A (2005) Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin‐proteasome system, and onto human diseases and drug targeting (Nobel lecture). Angew Chem Int Ed Engl 44: 5944–5967 [DOI] [PubMed] [Google Scholar]
- Crane JL, Cao X (2014) Bone marrow mesenchymal stem cells and TGF‐beta signaling in bone remodeling. J Clin Invest 124: 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM (2013) Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 28: 2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F, Yang X (2011) Genetic mouse models for bone studies–strengths and limitations. Bone 49: 1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith J, Genever P (2008) Transcriptional control of mesenchymal stem cell differentiation. Transfus Med Hemother 35: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Bunnell BA, Martin E, Frazier T, Hung BP, Gimble JM (2015) Stromal cells and stem cells in clinical bone regeneration. Nat Rev Endocrinol 11: 140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AW (2013) Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica (Cairo) 2013: 684736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH (2006) Regulation of adult bone mass by the zinc finger adapter protein Schnurri‐3. Science 312: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein‐specific response elements in the Id1 promoter. J Biol Chem 277: 4883–4891 [DOI] [PubMed] [Google Scholar]
- Lee OK, Kuo TK, Chen WM, Lee KD, Hsieh SL, Chen TH (2004) Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood 103: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Li X, Huang M, Zheng H, Wang Y, Ren F, Shang Y, Zhai Y, Irwin DM, Shi Y, Chen D, Chang Z (2008) CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol 181: 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, de Boeck M, van Dam H, Ten Dijke P (2016a) Regulation of the TGF‐beta pathway by deubiquitinases in cancer. Int J Biochem Cell Biol 76: 135–145 [DOI] [PubMed] [Google Scholar]
- Liu W, Kang N, Seriwatanachai D, Dong Y, Zhou L, Lin Y, Ye L, Liang X, Yuan Q (2016b) Chronic kidney disease impairs bone defect healing in rats. Sci Rep 6: 23041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhou L, Zhou C, Zhang S, Jing J, Xie L, Sun N, Duan X, Jing W, Liang X, Zhao H, Ye L, Chen Q, Yuan Q (2016c) GDF11 decreases bone mass by stimulating osteoclastogenesis and inhibiting osteoblast differentiation. Nat Commun 7: 12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ (2002) Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33: 77–80 [DOI] [PubMed] [Google Scholar]
- Logeart‐Avramoglou D, Bourguignon M, Oudina K, Ten Dijke P, Petite H (2006) An assay for the determination of biologically active bone morphogenetic proteins using cells transfected with an inhibitor of differentiation promoter‐luciferase construct. Anal Biochem 349: 78–86 [DOI] [PubMed] [Google Scholar]
- Lui TT, Lacroix C, Ahmed SM, Goldenberg SJ, Leach CA, Daulat AM, Angers S (2011) The ubiquitin‐specific protease USP34 regulates axin stability and Wnt/beta‐catenin signaling. Mol Cell Biol 31: 2053–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra S, Hu MS, Marshall CD, Leavitt T, Cheung AT, Gonzalez JG, Kaur H, Lorenz HP, Longaker MT (2016) Mesenchymal stromal cells as cell‐based therapeutics for wound healing. Stem Cells Int 2016: 4157934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez‐Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS (2010) Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy G, Garrett R, Harris S, Chan J, Chen D, Rossini G, Boyce B, Zhao M, Gutierrez G (1999) Stimulation of bone formation in vitro and in rodents by statins. Science 286: 1946–1949 [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Chen Z, Ishikawa M, Yue X, Kawanami A, Leahy P, Greenfield EM, Murakami S (2014) Prx1 and 3.2 kb Col1a1 promoters target distinct bone cell populations in transgenic mice. Bone 58: 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147 [DOI] [PubMed] [Google Scholar]
- Raggatt LJ, Partridge NC (2010) Cellular and molecular mechanisms of bone remodeling. J Biol Chem 285: 25103–25108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM (2015) TGF‐beta/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Res 3: 15005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133: 3231–3244 [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Carballo E, Ulsamer A, Susperregui AR, Manzanares‐Cespedes C, Sanchez‐Garcia E, Bartrons R, Rosa JL, Ventura F (2011) Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. J Bone Miner Res 26: 718–729 [DOI] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P (2007) Self‐renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 131: 324–336 [DOI] [PubMed] [Google Scholar]
- Salazar VS, Gamer LW, Rosen V (2016) BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol 12: 203–221 [DOI] [PubMed] [Google Scholar]
- Schaffler A, Buchler C (2007) Concise review: adipose tissue‐derived stromal cells–basic and clinical implications for novel cell‐based therapies. Stem Cells 25: 818–827 [DOI] [PubMed] [Google Scholar]
- Severe N, Dieudonne FX, Marie PJ (2013) E3 ubiquitin ligase‐mediated regulation of bone formation and tumorigenesis. Cell Death Dis 4: e463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe PT (2016) Dental mesenchymal stem cells. Development 143: 2273–2280 [DOI] [PubMed] [Google Scholar]
- Shu L, Zhang H, Boyce BF, Xing L (2013) Ubiquitin E3 ligase Wwp1 negatively regulates osteoblast function by inhibiting osteoblast differentiation and migration. J Bone Miner Res 28: 1925–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims NA, Martin TJ (2014) Coupling the activities of bone formation and resorption: a multitude of signals within the basic multicellular unit. Bonekey Rep 3: 481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Liu M, Ono N, Bringhurst FR, Kronenberg HM, Guo J (2012) Loss of wnt/beta‐catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res 27: 2344–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V (2006) BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet 38: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Vriend J, Reiter RJ (2016) Melatonin, bone regulation and the ubiquitin‐proteasome connection: a review. Life Sci 145: 152–160 [DOI] [PubMed] [Google Scholar]
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM (2011) USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 146: 918–930 [DOI] [PubMed] [Google Scholar]
- Wu M, Chen G, Li YP (2016) TGF‐beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 4: 16009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O'Brien CA (2011) Matrix‐embedded cells control osteoclast formation. Nat Med 17: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE (2005) Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Pringle LM, Lau AW, Riquelme DN, Wang H, Jiang T, Lev D, Welman A, Blobel GA, Oliveira AM, Chou MM (2010) TRE17/USP6 oncogene translocated in aneurysmal bone cyst induces matrix metalloproteinase production via activation of NF‐kappaB. Oncogene 29: 3619–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Sato T, Densmore M, Saito H, Schuler C, Erben RG, Lanske B (2012) Deletion of PTH rescues skeletal abnormalities and high osteopontin levels in Klotho‐/‐ mice. PLoS Genet 8: e1002726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Jiang Y, Zhao X, Sato T, Densmore M, Schuler C, Erben RG, McKee MD, Lanske B (2014) Increased osteopontin contributes to inhibition of bone mineralization in FGF23‐deficient mice. J Bone Miner Res 29: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ (2016) Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell 18: 782–796 [DOI] [PubMed] [Google Scholar]
- Zaidi M (2007) Skeletal remodeling in health and disease. Nat Med 13: 791–801 [DOI] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D (2003) E3 ubiquitin ligase Smurf1 mediates core‐binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem 278: 27939–27944 [DOI] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D (2004) Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo . J Biol Chem 279: 12854–12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Huang J, Guo R, Wang Y, Chen D, Xing L (2010) Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. J Bone Miner Res 25: 1246–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Li F, Fang P, Dai T, Yang B, van Dam H, Jia J, Zheng M, Zhang L (2016) Ubiquitin‐specific protease 4 antagonize osteoblast differentiation through dishevelled. J Bone Miner Res 31: 1888–1898 [DOI] [PubMed] [Google Scholar]
- Zhou CC, Xiong QC, Zhu XX, Du W, Deng P, Li XB, Jiang YZ, Zou SJ, Wang CY, Yuan Q (2017) AFF1 and AFF4 differentially regulate the osteogenic differentiation of human MSCs. Bone Res 5: 17044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400: 687–693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
All Seq data have been deposited into NCBI database with the identifier GSE114933, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114933.


