Abstract
Target-directed dynamic combinatorial chemistry (DCC) has emerged as a strategy for the identification of inhibitors of relevant therapeutic targets. In this contribution, we use this strategy for the identification of a high-affinity binder of a parasite target, the Trypanosoma cruzi bromodomain-containing protein TcBDF3. This protein is essential for viability of T. cruzi, the protozoan parasite that causes Chagas disease. A small dynamic library of acylhydrazones was prepared from aldehydes and acylhydrazides at neutral pH in the presence of aniline. The most amplified library member shows (a) high affinity for the template, (b) interesting antiparasitic activity against different parasite forms, and (c) low toxicity against Vero cells. In addition, parasites are rescued from the compound toxicity by TcBDF3 overexpression, suggesting that the toxicity of this compound is due to the TcBDF3 inhibition, i.e., the binding event that initially drives the molecular amplification is reproduced in the parasite, leading to selective toxicity.
Keywords: Dynamic combinatorial libraries, target-directed chemistry, Trypanosoma cruzi, bromodomain inhibitor
The identification of small-molecule modulators of protein function is a key activity in modern drug discovery and chemical biology. For targets with limited biostructural information available, random hit-identification strategies are frequently used, wherein target affinity is measured to select the best binder out of large compound libraries.1 Target binding can also be exploited for the molecular recognition of reactive small molecule building blocks and for both the chemical ligand assembly and the identification of high-affinity building block combinations, integrating in this way chemical synthesis and library screening.2 When this ligand assembly is achieved through dynamic covalent chemistry, reversibility allows proofreading to increase the yield of good binders.3,4
Dynamic combinatorial chemistry (DCC) allows the combination of building blocks through reversible reactions. Since the product distribution of these dynamic combinatorial libraries (DCLs) is dictated by the stability of the library members, it can adapt to environmental changes that affect such stability. Therefore, appropriate analysis of a DCL response to a given stimulus can give useful information about some properties of library members.5 Addition of a template molecule is a popular strategy to induce composition changes in DCLs. It has been used to discover synthetic receptors for guest templates as well as ligands for biomacromolecular templates,3 including proteins such as enzymes,6−10 lectins,11 and transporters.12,13 Although some of these proteins are potential therapeutic targets, examples of biologically active molecules resultant of template-induced molecular amplification from dynamic combinatorial libraries are very rare.6
One limitation of covalent DCC as a tool for the identification of drug discovery relevant ligands is the limited number of reversible reactions that can be carried out under biomacromolecule-compatible conditions and form bonds that are stable under physiological conditions.14 The exploitation of hydrazones in biomacromolecule-compatible DCC was precluded for more than a decade because of the required acid conditions for their exchange.12,13,15 The introduction of aniline16,17 and related amines as catalysts has paved the way toward the few examples of biomacromolecule-induced adaptation of hydrazone based DCLs.6,9,11,16,18 Another potential limitation for the exploitation of DCC in drug discovery is the unproductive ligand amplification, i.e., a concentration increase driven by noncovalent interactions with the template that do not affect the biomolecule functionality. This type of interaction cannot be detected by DCL composition analysis in the presence and in the absence of the template. It requires a parallel analysis of the effect of the DCL on the template.
Bromodomains are protein-interaction modules that bind to acetyl-lysine residues of histone and nonhistone proteins. In recent years, human bromodomains have become attractive drug targets for the treatment of a variety of diseases.19 The inhibition of eukaryotic pathogen bromodomains is considered a promising strategy to treat infections, even though it continues to be almost unexplored.20,21 Several bromodomain-containing proteins have been identified in different protozoan parasites. Since some of them are essential for viability, they represent new opportunities for the discovery of antiparasitic drugs.21−25 We have characterized three bromodomain-containing coding sequences in Trypanosoma cruzi, the protozoan parasite that causes Chagas disease.22−25TcBDF3 is a rare cytoplasmic bromodomain containing protein that interacts with acetylated α-tubulin and is involved in flagella morphogenesis and differentiation. These distinct features makes it an interesting drug target against T. cruzi.23,24
Here, we report the preparation and TcBDF3-induced adaptation of a DCL of hydrazones. The most amplified library member binds to the acetyl-lysine recognition pocket of the template and shows high cytotoxicity against different T. cruzi parasite forms. On the contrary, it shows low toxicity against Vero cells and TcBDF3 overexpressing parasites.
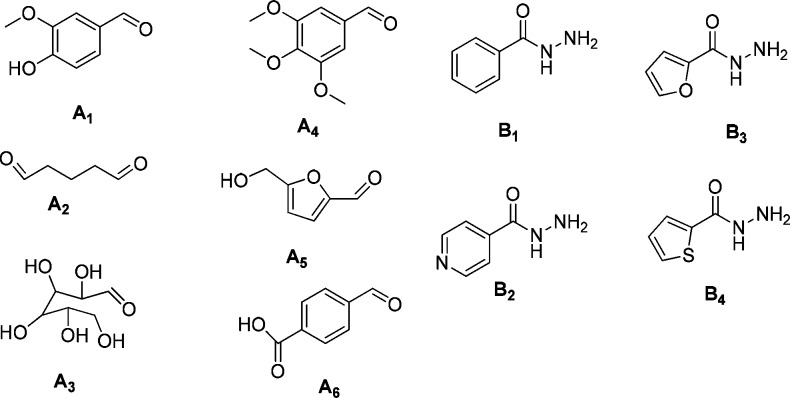
The library was prepared from a set of six aldehydes (A1–A6) and four acylhydrazides (B1–B4) (Figure 1). Their combination can produce hydrazones that include, aromatic, aliphatic, polyhydroxylated, and heterocyclic (including N, O, or S) moieties, with phenol, methoxyl, carboxyl, and alcohol substituents as potential recognition groups. One or more of these recognition groups are present in most of the reported compounds that possess affinity for different human bromodomains.26
Figure 1.
Building blocks used for DCL generation.
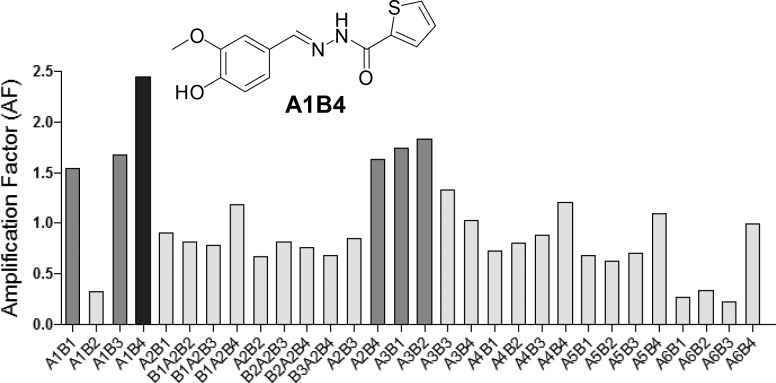
Aldehydes A1–A6 (75 μM each) and acylhydrazides B1–B4 (75 μM each) were dissolved in ammonium acetate buffer (100 mM, pH 6.5) in the presence of aniline (3.75 mM). After 10 h of reaction, the system reached a constant composition. LC–MS analysis showed the presence of 30 hydrazones with unique molecular weights (Table S1). When the dynamic library was exposed to recombinant TcBDF3 (100 μM), a shift in the composition was observed. The template-induced response of the DCL favored mainly the formation of hydrazone A1B4 that showed a 2.4-fold increase in concentration (Figure 2). Another five library members increased their concentrations to a lesser extent, between 1.5 and 1.8 times, whereas the rest of the library members either increased slightly or decreased their concentrations.
Figure 2.
Amplification factors (AFs) observed for library members in the presence of the template TcBDF3. AFs were calculated by dividing the LC–MS peak area of each compound in the templated and untemplated libraries. Black: AF higher than 2. Dark gray: AF between 1.5 and 2. Light gray: AF lower than one.
A template-induced molecular amplification in DCC is ideally driven by specific noncovalent interactions between the amplified ligand and the template. However, different factors can disrupt the correlation between amplification and affinity of the amplified compound to the template.27,28 In particular, when a biomacromolecule is used as a template to drive amplification of relatively small ligands, interactions between library members with different regions of the template are possible, and some of those interactions may not affect the biological function of the template biomolecule. To gain insight into the type of interaction between the library members and TcBDF3, the effect of the DCL on some of the properties of the template was analyzed.
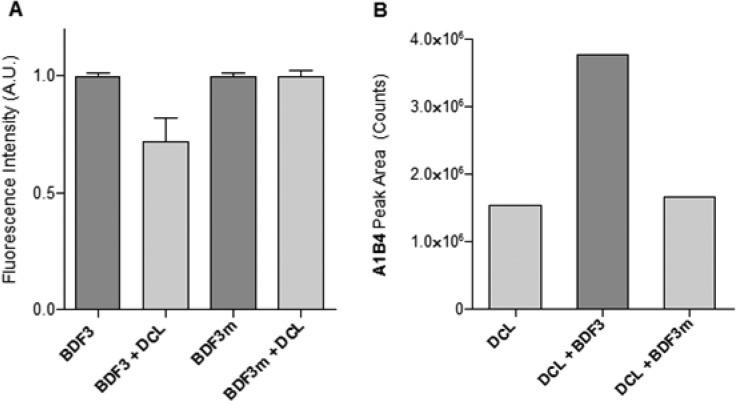
TcBDF3 possesses a tryptophan residue (W117) placed in a hydrophobic pocket that is the acetyl-lysine binding site. The intrinsic fluorescence of this aromatic residue can be affected by compounds that enter the TcBDF3 hydrophobic pocket.20,24 In the presence of the DCL, a significant decrease in the TcBDF3 maximum intrinsic fluorescence was observed (Figure 3A), suggesting that one or more library members could enter the hydrophobic pocket. In order to support this observation, the effect of the library was also measured with a mutated version of TcBDF3 (TcBDF3m) that includes two point mutations (Y123A and L130A). TcBDF3m was designed to retain its secondary structure while losing its ability to bind to its acetylated ligand in vitro.24 Neither the maximum intrinsic fluorescence of TcBDF3m was affected by the library (Figure 3A) nor the A1B4 concentration was affected by the presence of TcBDF3m (Figure 3B), suggesting a specific binding of A1B4 to the binding pocket of the wild type protein.29
Figure 3.
(A) Normalized intrinsic fluorescence of recombinant TcBDF3 or TcBDF3m (10 μM) in ammonium acetate buffer (100 mM, pH 6.5) in the absence and in the presence of DCL. (B) LC–MS peak area for A1B4 in the untemplated DCL and in the DCLs in the presence of TcBDF3 or TcBDF3m (100 μM).
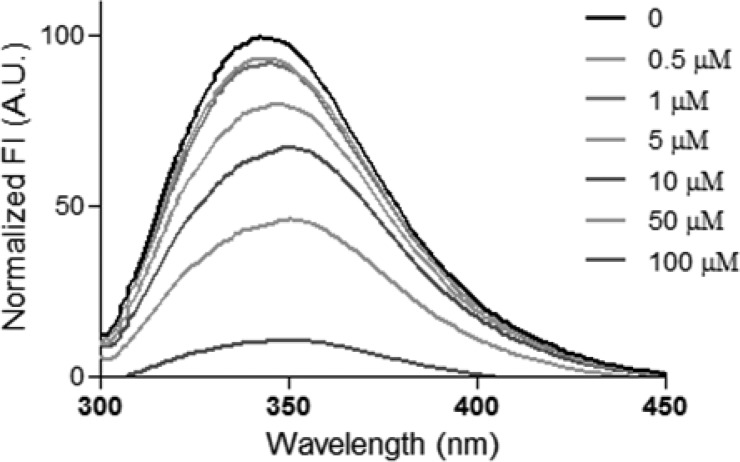
Hydrazone A1B4 was then prepared individually and its interaction, as well as the interaction of each of its component building blocks A1 and B4, with the template TcBDF3 were evaluated by fluorescence quenching and by differential scanning fluorimetry (DSF) or thermal shift. Individually, building blocks A1 and B4 (100 μM) did not produce any detectable change in fluorescence. However, the maximum intrinsic fluorescence of TcBDF3 decreased regularly in the presence of increasing concentrations of A1B4 (Figure 4). The fluorescence data were analyzed by Stern–Volmer, modified Stern–Volmer, and double logarithmic plots. The Stern–Volmer plot showed a negative deviation (toward the x-axis) as previously reported for TcBDF3 with bromodomain inhibitors.20,24 From the double logarithmic plot a dissociation constant value of 1.7 μM was obtained. In addition, binding to A1B4 significantly increased the thermal stability of TcBDF3 with ΔTm observed of 3.43 °C, whereas for TcBDF3m ΔTm was 0.4 °C, which further supports the binding of A1B4 to TcBDF3 (Figure S8).
Figure 4.
Fluorescence spectra of TcBDF3 (10 μM) alone and with increasing amounts of A1B4. λex = 295 nm. FI, fluorescence intensity; A.U., arbitrary units.
To support these results, the modeled 3D structure of TcBDF3 was used to perform docking predictions using the Swissdock server.30 The best prediction located the hydrazone A1B4 inside the hydrophobic pocket of TcBDF3 (Figure S9).
In view of the promising biophysical data and considering the novelty of TcBDF3 as a therapeutic target for Chagas disease, the effect of A1B4 on the different life cycle stages of T. cruzi was evaluated in vitro. Amastigotes are present in the mammalian host cells, whereas epimastigotes and the infective metacyclic trypomastigotes are present in the gut of the insect vector. The cytotoxicity was also studied in the Vero cell line to evaluate selectivity. A1B4 shows a trypanocidal effect on the three life cycle stages of T. cruzi with IC50 values between 13 and 23 μM (Table 1). Interestingly, its toxicity against Vero Cells was low, with an IC50 value higher than 200 μM, giving a selectivity index ≥9 depending on the parasite form.
Table 1. Cytotoxicity of A1B4 on the Different Life Cycle Stages of T. cruzi and Vero Cells.
| IC50 (μM)a,b (SIc) |
||||
|---|---|---|---|---|
| compound | epimastigotes | trypomastigotes | amastigotes | Vero cells |
| A1B4 | 23 ± 3.8 (>9) | 17.8 ± 2.29 (>11) | 13.1 ± 1.28(>15) | >200 ± 6 |
| BZNd | 18.16 ± 5.13 (2) | 27.07 ± 4.23 (1) | 3.92 ± 1.24 (8) | 30.17 ± 12 |
| NFXe | 8 ± 3 (14) | 7 ± 2 (16) | 3 ± 1.5 (38) | 115 ± 12 |
The results are averages of three separate determinations.
IC50 is the concentration required to give 50% inhibition, calculated by nonlinear regression analysis from beta-galactosidase activity at the used concentrations (0–200 μM).
Selectivity index (SI) is the ratio between IC50 on Vero cell toxicity and IC50 activity of extracellular or intracellular forms of the parasite.
MTT assay on Vero cells incubated 72 h with BZN (0–250 μM).
Values obtained from the literature.31 BZN, benznidazol; NFX, nifurtimox (reference drugs for the treatment of Chagas disease).
To gain insight into the specificity of A1B4 for TcBDF3 in vivo, the sensitivity of bromodomain-overexpressing lines was evaluated. T. cruzi epimastigotes overexpressing TcBDF3 through a tetracycline-induced plasmid were treated with A1B4, at a concentration around its IC50 value, in the absence and in the presence of tetracycline (Figure 5). Overexpression of TcBDF3 completely rescued epimastigotes from the growth inhibition produced by A1B4, suggesting that the toxicity of this compound is due to the TcBDF3 inhibition.
Figure 5.
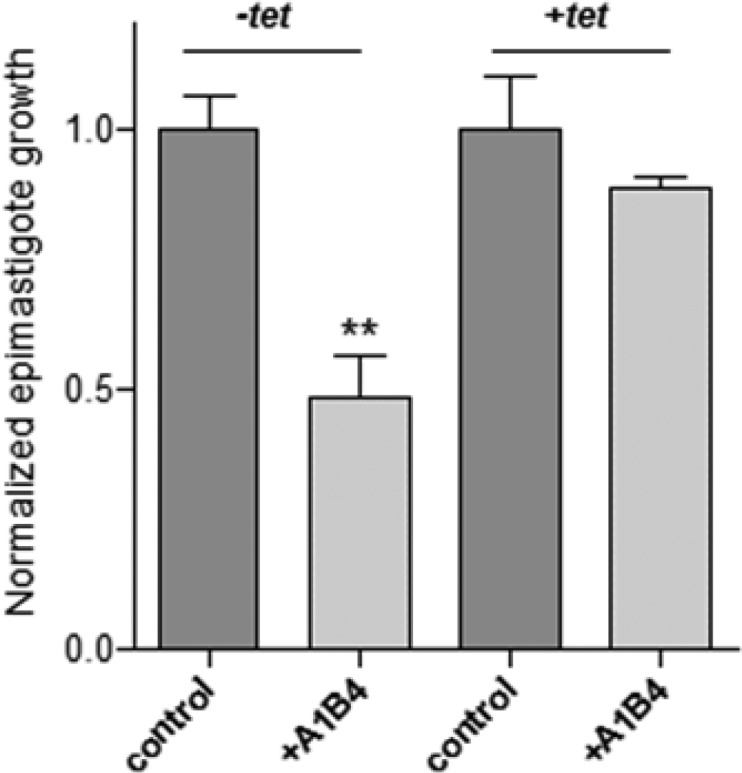
Relative growth of T. cruzi Dm28c epimastigotes transfected with pTcINDEXGW-BDF3HA, uninduced (-tet) and induced (+tet) with tetracycline (tet), untreated (control), and treated with A1B4 (25 μM). The experiment was performed in triplicate, and cell growth was determined after 72 h of culture by counting viable forms. The values obtained were normalized to the condition without compound (control). The bar graph represents the mean SD; **P < 0.05 (unpaired, two-tailed Student’s t test). In the experiment with tetracycline, the observed difference in relative growth in the absence (+tet control) and in the presence of inhibitor (+tet + A1B4) is not significant.
In order to evaluate if amplification of A1B4 is somehow related to the properties shown by this compound, a poorly amplified compound was prepared and analyzed (A4B4). Compared to A1B4, A4B4 was amplified in a lesser extent (AF = 1.21, Figure 2), produced a smaller change in the intrinsic fluorescence and thermal shift of TcBDF3 in vitro, and showed lower cytotoxicity against the parasite in vivo (Figure S12).
In summary, a hydrazone based DCL was prepared under reaction conditions that are compatible with the biologically relevant template TcBDF3. When mixed, DCL and template affected each other: the template induced changes in the relative concentration of the library members, and the library produced changes in the intrinsic fluorescence of a tryptophan residue placed in the acetyl-lysine recognition pocket of the bromodomain. Blockage of the entrance to this pocket by specific mutations deleted both effects. Binding of the most amplified library member, A1B4, to the template TcBDF3 was studied by fluorescence quenching and thermal shift observing a Kd of 1.7 μM, slightly lower than the value for the best TcBDF3 inhibitor reported.20 The binding process observed in vitro also seems to be possible in cell since the compound A1B4 shows interesting toxicity activity against different parasite forms, but it is not toxic for other eukaryotic cells, such as Vero cells, or for parasites wherein TcBDF3 has been overexpressed. Hydrazone A1B4 showed very interesting antiparasitic activity and selectivity index.
These results illustrate how biologically active small-molecules can be identified through a DCC casting approach by using mild reaction conditions, biologically relevant templates, and appropriate analysis of DCL adaptation and DCL effect on the template. In addition, the biophysical and biological properties observed for compound A1B4 give relevance to bromodomain-containing proteins as attractive targets for the discovery of selective antiparasitic molecules.
Acknowledgments
P.G. and V.A. acknowledge CONICET for their postdoctoral fellowships. The authors thank CIBION for HRMS results.
Glossary
ABBREVIATIONS
- DCC
dynamic combinatorial chemistry
- DCL
dynamic combinatorial library
- TcBDF3
Trypanosoma cruzi bromodomain
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00247.
TcBDF3 and TcBDF3m purification, library preparation, library analysis, synthesis and characterization of A1B4 and A4B4, binding experiments, and biological activities and docking prediction (PDF)
Author Contributions
§ These authors contributed equally to this work.
This work was supported by FONCYT (PICT2015–3574), CONICET (PIP 695 and 797), and Universidad Nacional de Rosario.
The authors declare no competing financial interest.
Supplementary Material
References
- Bleicher K. H.; Böhm H. J.; Müller K.; Alanine A. I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discovery 2003, 2, 369–378. 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- Jaegle M.; Wong E. L.; Tauber C.; Nawrotzky E.; Arkona C.; Rademann J. Protein-Templated Fragment Ligations—From Molecular Recognition to Drug Discovery. Angew. Chem., Int. Ed. 2017, 56, 7358–7378. 10.1002/anie.201610372. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 7464 - 7485.10.1002/ange.201610372
- Mondal M.; Hirsch A. K. H. Dynamic combinatorial chemistry: a tool to facilitate the identification of inhibitors for protein targets. Chem. Soc. Rev. 2015, 44, 2455–2488. 10.1039/C4CS00493K. [DOI] [PubMed] [Google Scholar]
- Herrmann A. Dynamic combinatorial/covalent chemistry: a tool to read, generate and modulate the bioactivity of compounds and compound mixtures. Chem. Soc. Rev. 2014, 43, 1899–1933. 10.1039/C3CS60336A. [DOI] [PubMed] [Google Scholar]
- Corbett P. T.; Leclaire J.; Vial L.; West K. R.; Wietor J. L.; Sanders J. K. M.; Otto S. Dynamic combinatorial chemistry. Chem. Rev. 2006, 106, 3652–3711. 10.1021/cr020452p. [DOI] [PubMed] [Google Scholar]
- Fu J.; Fu H.; Dieu M.; Halloum I.; Kremer L.; Xia Y.; Pand W.; Vincent S. P. Identification of inhibitors targeting Mycobacterium tuberculosis cell wall biosynthesis via dynamic combinatorial chemistry. Chem. Commun. 2017, 53, 10632–10635. 10.1039/C7CC05251K. [DOI] [PubMed] [Google Scholar]
- Kanfar N.; Tanc M.; Dumy P.; Supuran C. T.; Ulrich S.; Winum J.-Y. Effective Access to Multivalent Inhibitors of Carbonic Anhydrases Promoted by Peptide Bioconjugation. Chem. - Eur. J. 2017, 23, 6788–6794. 10.1002/chem.201700241. [DOI] [PubMed] [Google Scholar]
- Soubhye J.; Gelbcke M.; Van Antwerpen P.; Dufrasne F.; Boufadi M. Y.; Nève J.; Furtmüller P. G.; Obinger C.; Zouaoui Boudjeltia K.; Meyer F. From Dynamic Combinatorial Chemistry to in Vivo Evaluation of Reversible and Irreversible Myeloperoxidase Inhibitors. ACS Med. Chem. Lett. 2017, 8, 206–210. 10.1021/acsmedchemlett.6b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M.; Radeva N.; Fanlo-Virgos H.; Otto S.; Klebe G.; Hirsch A. K. H. Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry. Angew. Chem., Int. Ed. 2016, 55, 9422–9426. 10.1002/anie.201603074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q.-Q.; Sicking W.; Ehlers M.; Schmuck C. Discovery of potent inhibitors of human β-tryptase from pre-equilibrated dynamic combinatorial libraries. Chem. Sci. 2015, 6, 1792–1800. 10.1039/C4SC02943G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei P.; Pang L.; Silbermann M.; Eris D.; Mehlethaler T.; Schwardt O.; Ernst B. Target-directed Dynamic Combinatorial Chemistry: A Study on Potentials and Pitfalls as Exemplified on a Bacterial Target. Chem. - Eur. J. 2017, 23, 11570–11577. 10.1002/chem.201701601. [DOI] [PubMed] [Google Scholar]
- Monjas L.; Swier L. J. Y. M.; Setyawati I.; Slotboom D. J.; Hirsch A. K. H. Dynamic Combinatorial Chemistry to Identify Binders of ThiT, an S-Component of the Energy-Coupling Factor Transporter for Thiamine. ChemMedChem 2017, 12, 1693–1696. 10.1002/cmdc.201700440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern F. T.; Wanner K. T. Generation and screening of oxime libraries addressing the neuronal GABA transporter GAT1. ChemMedChem 2015, 10, 396–410. 10.1002/cmdc.201402376. [DOI] [PubMed] [Google Scholar]
- Miller B. J. Dynamic covalent chemistry: Catalysing dynamic libraries. Nat. Chem. 2010, 2, 433–434. 10.1038/nchem.659. [DOI] [PubMed] [Google Scholar]
- Hydrazone based libraries were pre-equilibrated and neutralized, stopping the exchange, before addition of the biomolecular template.
- Bhat T.; Caniard A. M.; Luksch T.; Brenk R.; Campopiano D. J.; Greaney M. F. Nucleophilic catalysis of acylhydrazone equilibration for protein-directed dynamic covalent chemistry. Nat. Chem. 2010, 2, 490–497. 10.1038/nchem.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen A.; Dirksen S.; Hackeng T. M.; Dawson P. E. Nucleophilic Catalysis of Hydrazone Formation and Transimination: Implications for Dynamic Covalent Chemistry. J. Am. Chem. Soc. 2006, 128, 15602–15603. 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- Clipson A. J.; Bhat V. T.; McNae I.; Caniard A. M.; Campopiano D. J.; Greaney M. F. Bivalent Enzyme Inhibitors Discovered Using Dynamic Covalent Chemistry’ Chemistry - A European Journal. Chem. - Eur. J. 2012, 18, 10562–10570. 10.1002/chem.201201507. [DOI] [PubMed] [Google Scholar]
- Hewings D. S.; Rooney T. P. C.; Jennings L. E.; Hay D. A.; Schofield C. J.; Brennan P. E.; Knapp S.; Conway S. J. Progress in the Development and Application of Small Molecule Inhibitors of Bromodomain–Acetyl-lysine Interactions. J. Med. Chem. 2012, 55, 9393–9413. 10.1021/jm300915b. [DOI] [PubMed] [Google Scholar]
- Ramallo I. A.; Alonso V. L.; Rua F.; Serra E.; Furlan R. L. E. A Bioactive Trypanosoma cruzi Bromodomain Inhibitor from Chemically Engineered Extracts. ACS Comb. Sci. 2018, 20, 220–228. 10.1021/acscombsci.7b00172. [DOI] [PubMed] [Google Scholar]
- Jeffers V.; Yang C.; Huang S.; Sullivan W. J. Jr. Bromodomains in Protozoan Parasites: Evolution, Function, and Opportunities for Drug Development. Microbiol. Mol. Biol. Rev. 2017, 81, e00047–16. 10.1128/MMBR.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanova G. V.; Nardelli S. C.; Cribb P.; Magdaleno A.; Silber A. M.; Motta M. C. M.; Schenkman S.; Serra E. Trypanosoma cruzi bromodomain factor 2 (BDF2) binds to acetylated histones and is accumulated after UV irradiation. Int. J. Parasitol. 2009, 39, 665–673. 10.1016/j.ijpara.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Alonso V. L.; Villanova G. V.; Ritagliati C.; Machado Motta M. C.; Cribb P.; Serra E. C. Trypanosoma cruzi Bromodomain Factor 3 Binds Acetylated α-Tubulin and Concentrates in the Flagellum during Metacyclogenesis. Eukaryotic Cell 2014, 6, 822–831. 10.1128/EC.00341-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso V. L.; Ritagliati C.; Cribb P.; Cricco J. A.; Serra E. C. Overexpression of bromodomain factor 3 in Trypanosoma cruzi (TcBDF3) affects differentiation of the parasite and protects it against bromodomain inhibitors. FEBS J. 2016, 283, 2051–2066. 10.1111/febs.13719. [DOI] [PubMed] [Google Scholar]
- Ritagliati C.; Villanova G. V.; Alonso V. L.; Zuma A. A.; Cribb P.; Machado Motta M. C.; Serra E. C. Glycosomal bromodomain factor 1 from Trypanosoma cruzi enhances trypomastigote cell infection and intracellular amastigote growth. Biochem. J. 2016, 1, 73–85. 10.1042/BJ20150986. [DOI] [PubMed] [Google Scholar]
- Ferri E.; Petosa C.; McKenna C. E. Bromodomains: Structure, function and pharmacology of inhibition. Biochem. Pharmacol. 2016, 106, 1–18. 10.1016/j.bcp.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Corbett P. T.; Sanders J. K. M.; Otto S. Exploring the relation between amplification and binding in dynamic combinatorial libraries of macrocyclic synthetic receptors in water. Chem. - Eur. J. 2008, 14, 2153–2166. 10.1002/chem.200701413. [DOI] [PubMed] [Google Scholar]
- Severin K. The Advantage of Being Virtual—Target-Induced Adaptation and Selection in Dynamic Combinatorial Libraries. Chem. - Eur. J. 2004, 10, 2565–2580. 10.1002/chem.200305660. [DOI] [PubMed] [Google Scholar]
- The observed selectivity in amplification and binding for A1B4 with TcBDF3 or TcBDF3m and the following biological results are particularly important taking into consideration that the Aggregation Advisor indicates that this compound is similar to a compound that can form aggregates under certain conditions.Irwin J. J.; Duan D.; Torosyan H.; Doak A. K.; Ziebart K. T.; Sterling T.; Tumanian G.; Shoichet B. K. An Aggregation Advisor for Ligand Discovery. J. Med. Chem. 2015, 58, 7076–7087. 10.1021/acs.jmedchem.5b01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A.; Zoete V.; Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270. 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Martínez C.; Correa-Basurto J.; Nieto-Meneses R.; Márquez-Navarro A.; Aguilar-Suárez R.; Montero-Cortes M. D.; Nogueda-Torres B.; Suárez-Contreras E.; Galindo-Sevilla N.; Rojas-Rojas Á. Design, synthesis and biological evaluation of quinazoline derivatives as anti-trypanosomatid and anti-plasmodial agents. Eur. J. Med. Chem. 2015, 96, 296–307. 10.1016/j.ejmech.2015.04.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.