Abstract
Transforming growth factor-β (TGFβ) signaling through SMAD2/3 is an important driver of pathological fibrosis in multiple organ systems. TGFβ signaling and extracellular matrix (ECM) stiffness form an unvirtuous pathological circuit in which matrix stiffness drives activation of latent TGFβ, and TGFβ signaling then drives cellular stress and ECM synthesis. Moreover, ECM stiffness also appears to sensitize cells to exogenously activated TGFβ through unknown mechanisms. Here, using human fibroblasts, we explored the effect of ECM stiffness on a putative inner nuclear membrane protein, LEM domain–containing protein 3 (LEMD3), which is physically connected to the cell's actin cytoskeleton and inhibits TGFβ signaling. We showed that LEMD3–SMAD2/3 interactions are inversely correlated with ECM stiffness and TGFβ-driven luciferase activity and that LEMD3 expression is correlated with the mechanical response of the TGFβ-driven luciferase reporter. We found that actin polymerization but not cellular stress or LEMD3–nuclear-cytoplasmic couplings were necessary for LEMD3–SMAD2/3 interactions. Intriguingly, LEMD3 and SMAD2/3 frequently interacted in the cytosol, and we discovered LEMD3 was proteolytically cleaved into protein fragments. We confirmed that a consensus C-terminal LEMD3 fragment binds SMAD2/3 in a stiffness-dependent manner throughout the cell and is sufficient for antagonizing SMAD2/3 signaling. Using human lung biopsies, we observed that these nuclear and cytosolic interactions are also present in tissue and found that fibrotic tissues exhibit locally diminished and cytoplasmically shifted LEMD3–SMAD2/3 interactions, as noted in vitro. Our work reveals novel LEMD3 biology and stiffness-dependent regulation of TGFβ by LEMD3, providing a novel target to antagonize pathological TGFβ signaling.
Keywords: SMAD transcription factor, transforming growth factor β (TGF-β), mechanotransduction, pulmonary fibrosis, nuclear membrane, actin, Buschke-Ollendorff syndrome, LEM, LEMD3, MAN1, nuclear lamina
Introduction
Transforming growth factor β (TGFβ) 2 plays important roles in human development, wound repair, and pathology. In particular, overactivation of TGFβ signaling has been implicated as a pathological driver of fibrosis in several organ systems, including the lung, kidneys, skin, and liver (1–3). Recently, the Food and Drug Administration approved the use of pirfenidone in pulmonary fibrosis, which acts in part to antagonize TGFβ signaling. Pirfenidone has led to a slowing of disease progression, indicating that this is a promising yet not fully realized therapeutic axis for addressing fibrotic pathologies (4–7).
Previous work has shown a clear connection between extracellular matrix (ECM) stiffness and TGFβ activation. TGFβ is deposited in the ECM in an inactive state in latent TGFβ–associated protein complexes. To activate TGFβ, cells can mechanically disengage TGFβ from the latent complex through αv integrin–mediated mechanical stress, after which TGFβ acts in a paracrine fashion. Stiffer ECMs allow for more force transmission to the latent complex and potentiate the liberation of TGFβ. This allows for pathological feedback whereby TGFβ signaling drives matrix synthesis and cellular stress, and a stiffer matrix increases the free amount of TGFβ available to cells (8, 9). In addition to activating TGFβ, several in vitro studies have indicated that a stiff ECM potentiates cellular responses to exogenously activated (pre-activated) TGFβ (10–13). Here, our work focused on understanding this cellular potentiation to TGFβ to identify additional targets of possible therapeutic intervention along the TGFβ-signaling pathway (i.e. targets that specifically address matrix-driven sensitization to TGFβ and not matrix-driven activation of TGFβ).
We focused this study of rigidity-driven cellular sensitization to TGFβ on LEM domain–containing protein 3 (LEMD3). LEMD3 is an integral inner nuclear membrane protein and a compelling target for two reasons: 1) it is a known inhibitor of TGFβ signaling; 2) it is biophysically connected to the cell's cytoskeleton, which is remodeled in an ECM stiffness-dependent fashion. LEMD3 binds and inactivates the downstream transcription factor partners of both TGFβ and bone morphogenic protein (BMP), the receptor proteins' mothers against decapentaplegic (Smads) 2 and 3 and 1, 5, and 8, respectively (14–21). LEMD3's RNA recognition motif (RRM) domain, on its C-terminal end, competes with other transcription factors for binding to Smad2/3 and promotes Smad2/3 dephosphorylation and nuclear export by acting as a coordinating scaffold for protein phosphatase, Mg2+/Mn2+- dependent 1α (PPM1α) (16, 22).
In humans, heterozygous loss of LEMD3 is frequently, but not always (23), associated with the development of Buschke-Ollendorf syndrome (BOS), in which patients develop some constellation of cutaneous collagenomas and elastomas (from aberrant TGFβ signaling) and osteopoikilosis (from aberrant BMP signaling), hyperostic lesions in long bones, which can mimic osteoblastic metastatic lesions radiographically (20, 24–32). Depending on the penetrance and specific genotype, heterozygous loss of LEMD3 can also be associated with more severe ECM pathologies, such as melorheostosis, a progressive sclerosis frequently in long bones that can lead to disfigurement and joint destruction (20, 24).
In addition to its clear role in antagonizing TGFβ signaling both in vitro and in vivo, LEMD3 is retained in the inner nuclear membrane through its connection to the nuclear lamin superstructure by the activity of its N-terminal end, which contains the molecule's Lapb2/Emerin/MAN1 (LEM) domain (14, 33–37). This connection creates a direct biophysical link to the cell's actin microfilament network through the nesprin–sun linker of nucleus and cytoskeleton (LINC) complex (38–40).
Given LEMD3's role in antagonizing Smad2/3 and its biophysical connection to the cell's cytoskeleton, we hypothesized that LEMD3's inhibition of Smad2/3 would be reduced by cytoskeletal stress thereby potentiating TGFβ signaling in an ECM stiffness-dependent fashion. Here, dermal fibroblasts displayed ECM stiffness-dependent TGFβ responsiveness, and LEMD3 knockdown or overexpression modulated this stiffness responsiveness. We also showed that LEMD3's interactions with Smad2/3 were negatively regulated by ECM stiffness and were independent of TGFβ dosing. LEMD3–Smad2/3 interactions were increased by disruption of the actin cytoskeleton, but these complexes were not potentiated by disruption of the LINC complex or overexpression of the LEM domain. Unexpectedly, we discovered cytosolic interactions between LEMD3 and Smad2/3, which called into question basic assumptions about the current understanding of LEMD3's biology and its regulation of TGFβ. We identified and genetically localized N- and C-terminal LEMD3 fragments, which separated LEMD3's Smad-binding RRM and lamin-binding LEM domains. We established that these fragments were created through post-translational proteolysis by a serine protease and were differentially regulated by lamin integrity. We verified that a consensus C-terminal fragment of LEMD3 bound Smad2/3 in a stiffness-dependent fashion, antagonized Smad2/3's ability to complex with Smad4, and showed that both endogenous LEMD3 and this LEMD3 fragment bound in the cytoplasm the PPM1α protein phosphatase previously shown to be coordinated by LEMD3 to antagonize TGFβ signaling. Finally, we found correlates between our in vitro findings and human lung tissue from idiopathic pulmonary fibrosis (IPF) and non-IPF patients. Interactions between LEMD3 and Smad2/3 demonstrated greater intra-patient/total variability in frequency and were more frequent in the cytoplasm in human lung core biopsies from IPF patients relative to non-IPF tissues. Additionally, IPF tissues had unique low-frequency LEMD3–Smad2/3 interaction regions, possibly indicative of local fibrotic disease. Our work demonstrates novel aspects of LEMD3 biology as well as identifies a potential target for TGFβ antagonism relevant to fibrosis that complements current medical management.
Results
Fibroblast TGFβ responsiveness is potentiated by ECM stiffness and inhibited by LEMD3 expression
We sought to validate that fibroblasts in our system displayed stiffness-dependent TGFβ responsiveness using primary human dermal fibroblasts (HFFs) stably transfected with a Smad-driven luciferase. We characterized stiffness and morphology phenotypes associated with mechanotransduction using optical microscopy and atomic force microscopy (AFM). With increasing stiffness, fibroblasts underwent cellular compliance-matching (p = 0.0022) and increased their cell-spreading surface area (p = 0.1801) and polarization (p = 0.0263) seen in Fig. 1A and Fig. S1, respectively. Luciferase-transfected HFFs showed a sigmoidal responsiveness to recombinant human TGFβ as a function of substrate stiffness (Fig. 1B). This stiffness potentiation was dose-dependent with increasing picogram/ml doses of TGFβ appearing to shift the transition point of the sigmoid toward softer substrates (“soft-shift”, p = 0.07, Fig. 1B). Transition points in the picogram/ml dose-range occurred at stiffness over a range from ≈1–4 kPa. These cells also showed the expected TGFβ dose-response (increasing luminescence with increasing TGFβ dose) on stiff substrates (p < 0.0001, Fig. 1B). Previous studies have also shown stiffness-dependent modulation of TGFβ signaling, and our work improves the resolution of these results by more finely probing the stiffness space across a range from 0.5 to 50 kPa (10–13).
Figure 1.
LEMD3 modifies the stiffness response of fibroblasts to activated TGFβ. A, fibroblasts demonstrated mechano-sensitivity to ECM stiffness through cytoskeletal compliance matching as measured by AFM on increasingly stiff matrices (p = 0.001). B, fibroblasts stably transfected with Smad-responsive luciferase demonstrated a dose-dependent stiffness modulation of their TGFβ responsiveness. Increasing doses of TGFβ were associated with a softer sigmoidal inflection point (p = 0.07), and cells demonstrated a dose-response to TGFβ on stiff surfaces (p < 0.0001). C, LEMD3 expression was correlated with a stiffer transition point in TGFβ stiffness responsiveness (p < 0.0001) and with decreased luminescence (p = 0.04). D, cytoskeletal depolymerization but not myosin II inhibition was associated with a decreased and flattened luminescent response of fibroblasts to TGFβ. LEMD3 KD does not rescue actin-depolymerization phenotypes. Treatments with cytochalasin D did not converge to a sigmoidal model, and the data are represented as mean with standard error of the mean to convey the heterogeneity of the results. All cell stiffness/morphology phenotypes and stiffness model parameters were statistically analyzed using an ANOVA test for trends analysis. All data are represented by the mean with S.E. unless otherwise noted. Bleb, blebbistatin.
Having confirmed the TGFβ/stiffness phenotype, we tested how modulating LEMD3 alters the stiffness responsiveness of fibroblasts using Lipofectamine2000-delivered siRNA against LEMD3 (“siLEMD3”) and electroporation with pFLAG–LEMD3–V5 (LEMD3 overexpression plasmid with N-terminal FLAG epitope tag and C-terminal V5 epitope tag). Increasing LEMD3 expression (Fig. S2A) induced a decrease in maximal luminescence (p = 0.04, Fig. 1C) and a “stiff-shift” in the stiffness response's sigmoidal transition point (p < 0.0001, Fig. 1C). There was a corresponding “soft-shift” in the transition point (p < 0.0001, Fig. 1C) and an increase in maximal luminescence (p = 0.04, Fig. 1C) when LEMD3 expression was decreased through siRNA treatment (Fig. S2B). There were no significant differences in either maximal luminescence or the stiffness transition point of the sigmoid model between HFFs treated with Lipofectamine2000, electroporated control HFFs (“Neon HFF”), or HFFs treated with concentration-matched siRNA against GFP (“siGFP”).
To connect LEMD3's TGFβ antagonism seen in Fig. 1C to Smad2/3's activation, we investigated the effect of LEMD3's modulation on Smad3 phosphorylation (Fig. S3). 100 pg/ml TGFβ treatment for 90 min increased the ratio of phospho-Smad3 to total Smad3 relative to untreated HFFs (p = 0.0506, Fig. S3). Treatment with either Lipofectamine2000 or electroporation reduced the phospho-Smad3/Smad3 ratio, but treatments with 25 and 200 nm siRNA against LEMD3 increased the phospho-Smad3/Smad3 ratio relative to siRNA against GFP (p = 0.1103 and p = 0.0057 for 25 and 200 nm siRNA groups, respectively, see Fig. S3). Electroporation with pFLAG–LEMD3–V5 or a C-terminal fragment (pFLAG–LEMD3p.Δ21–669–V5 (CTF)), previously shown to be sufficient for Smad2/3 binding and de-phosphorylation (16, 18, 19, 21, 41), decreased the pSmad3/Smad3 ratio; however, as in Fig. 1C, there was no difference between electroporation controls and electroporation with any LEMD3 plasmids on tissue culture plastic. The stiffness-dependence of LEMD3's inhibition of TGFβ here is a novel finding, but the general inhibition of TGFβ signaling (14–16, 20, 21) and Smad phosphorylation (18, 19, 21) by LEMD3 (on stiff substrates) confirms findings from previous reports.
To test which components of the cell's microfilament cytoskeleton control the stiffness-response of HFFs to TGFβ, we used chemical inhibition of the structural (cytochalasin D, a globular actin (g-actin) stabilizer) and contractility machinery (blebbistatin, a myosin II inhibitor). Stiffness-dependent TGFβ responses required actin polymerization but did not require cellular contractility (Fig. 1D). Treatment with 10 μm blebbistatin (a myosin II inhibitor) did not significantly shift the transition point nor increase the maximal luminescence of cells relative to HFFs treated with TGFβ alone. Cells treated with 2 μm cytochalasin D showed a low, flat luminescent profile over increasing ECM stiffness, and the data did not converge to a sigmoidal model. Interestingly, siRNA against LEMD3 did not rescue the cytochalasin D phenotype, although the degree of LEMD3 knockdown may not have been sufficient (Fig. S2B). These data indicate that the actin cytoskeleton, but not cellular stress per se, is needed for stiffness-modulated TGFβ signaling.
LEMD3–SMAD2/3 complexes are inversely correlated to substrate stiffness and occur throughout the cell
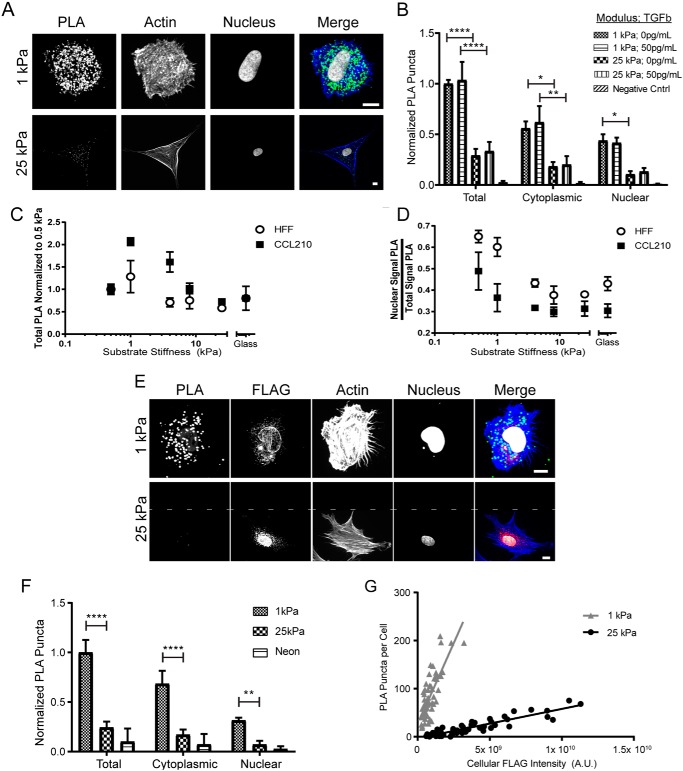
Given that LEMD3 modulated the stiffness response of fibroblasts to TGFβ, we used proximity ligation assays (PLA, methodologically reviewed in Ref. 42) to examine the stiffness dependence of LEMD3–Smad2/3 interactions on hydrogels whose stiffness is representative of physiological (1 kPa) and fibrotic (25 kPa) lung tissue in humans (43). LEMD3–Smad2/3 interactions were negatively correlated with substrate stiffness in both the cytoplasm and the nucleus (Fig. 2, A and B). This negative correlation was independent of TGFβ dosing (50 pg/ml, inside the range of stiffness-responsive doses identified in Fig. 1B), consistent with previous findings that LEMD3–Smad2/3 interactions are phosphorylation-independent (21). Although LEMD3–Smad2/3 interactions were unperturbed by TGFβ dosing, we explicitly tested the ability of LEMD3 to bind phospho-Smad2/3 (Fig. S4). We found that HFFs treated with 100 pg/ml TGFβ on glass had significantly higher LEMD3–phospho-Smad2/3 interactions than untreated fibroblasts (p = 0.0333, Fig. S4) and that there was a significant negative correlation between substrate stiffness and LEMD3–phospho-Smad2/3 interactions (p = 0.0305, Fig. S4). These data reveal the stiffness dependence of LEMD3–Smad2/3 interactions. The data also confirm previous observations that LEMD3 can bind both phosphorylated and unphosphorylated Smad2/3 (21).
Figure 2.
LEMD3–Smad2/3 interactions are inversely correlated to substrate stiffness and occur in the nucleus and cytoplasm. A, micrographs of LEMD3–Smad2/3 PLA interactions on soft (top row, 1 kPa) and stiff (bottom row, 25 kPa) matrices (PLA in green, f-actin in blue, and nucleus in white). B, quantification of PLA interactions grouped by substrate stiffness and by TGFβ dose. Total LEMD3–Smad2/3 interactions were negatively correlated to substrate stiffness (p < 0.0001 for 1 kPa versus 25 kPa for both 0 and 50 pg/ml TGFβ) but not correlated to TGFβ dose. Cytoplasmic (p = 0.0164 for 0 pg/ml TGFβ and p = 0.0087 for 50 pg/ml TGFβ) and nuclear compartment (p = 0.0428 for 0 pg/ml TGFβ and p = 0.1228 for 50 pg/ml TGFβ) interactions were also negatively correlated to substrate stiffness. C, total HFFs and CCL210s LEMD3–Smad2/3 interactions by PLA normalized to 0.5 kPa on surfaces with stiffness of 0.5, 1, 4, 8, 25 kPa and glass. Each fibroblast population showed a biphasic trend, centered around a peak of LEMD3–Smad2/3 interactions at 1 kPa. CCL210 demonstrated greater dynamic range in interaction frequency and a slower loss of interactions on stiffer substrates than HFFs. D, subcellular location of LEMD3–Smad2/3 PLA interactions in HFFs and CCL210s from C. Each cell line demonstrated a cytoplasmic shift in location with increasing substrate stiffness (p < 0.0001 and p = 0.0199 for HFFs and CCL210s, respectively; ANOVA test for trend). E, micrographs of V5–Smad2/3 PLA interactions with pFLAG–LEMD3–V5 on soft (top row) and stiff (bottom row) matrices (PLA in green, f-actin in blue, FLAG in red, and nucleus in white). Area above the dashed line in 25-kPa image indicates filler space. F, V5–Smad2/3 PLA interactions were also negatively correlated with substrate stiffness (for 1 kPa versus 25 kPa: total PLA, p < 0.0001; cytoplasmic PLA, p < 0.0001; nuclear PLA, p = 0.0018) and also occurred in the cytoplasm. G, V5–Smad2/3 PLA interactions were significantly higher on soft substrates independent of the degree of pFLAG–LEMD3–V5 expression (difference in linear regression slopes, p < 0.0001). All PLA groups were statistically compared using a two-way ANOVA with Tukey post-test unless noted. All scale bars, 10 μm. All data represented by the mean with S.E. except for G, where individual data points are plotted. *, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001.
We extended our findings by performing PLA across a finer range of stiffness with both HFFs and CCL210s, an adult pulmonary-derived fibroblast line (Fig. 2C). Both CCL210s and HFFs showed a biphasic response to stiffness with peak values at 1 kPa. CCL210s demonstrated a greater dynamic range in response to stiffness as well as a more gradual loss of PLA interactions on progressively stiffer substrates relative to HFFs. Additionally, the abrupt loss of LEMD3–Smad2/3 interactions between 1 and 4 kPa in HFFs was inversely correlated with the stiffness dependence of the sigmoidal transition point of the luciferase signal measured in Fig. 1B.
Interestingly, we found that approximately half of the PLA interactions on both substrates occurred in the cytoplasm of the cells. Across a broader range of stiffness, we found that the nuclear proportion of LEMD3–Smad2/3 interactions was inversely correlated to substrate stiffness in both HFFs (p < 0.0001) and CCL210s (p = 0.0199), as seen in Fig. 2D. Given that LEMD3 is thought to be an integral protein of the inner nuclear membrane, we validated our findings through V5–Smad2/3 PLA in cells transfected with pFLAG–LEMD3–V5 (Fig. 2, E and F). Near identical trends were observed with this independent PLA reaction: a reduction in PLA frequency overall (p < 0.0001, Fig. 2F), and reductions in both the cytoplasm (p < 0.0001, Fig. 2F) and nuclear (p = 0.0018, Fig. 2F) compartments with increasing substrate stiffness. To control for the degree of recombinant LEMD3 expressed across stiffness conditions, we found a significant difference in the linear regression between PLA puncta per cell and the per cell FLAG intensity in cells on both soft and stiff surfaces (p < 0.0001, Fig. 2G). The steeper slope observed for cells on soft substrates confirmed a higher rate of LEMD3–Smad2/3 complex formation per arbitrary unit of LEMD3 expressed (as measured by the fused FLAG epitope) relative to stiff substrates.
LEMD3–Smad2/3 complexes are inhibited by actin polymerization and not potentiated by disrupting cytoplasmic–nuclear–lamina–LEMD3 coupling
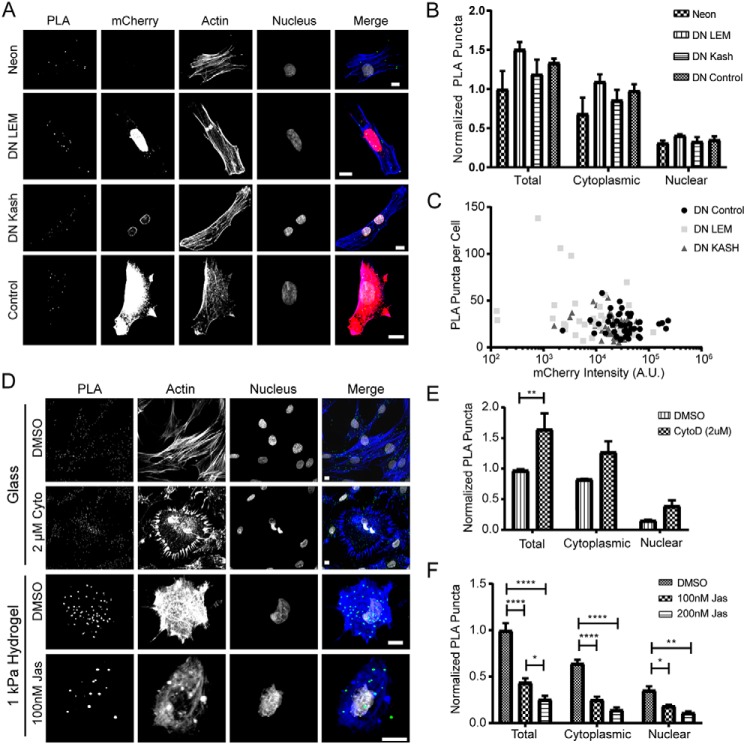
We hypothesized that the frequency of LEMD3–Smad2/3 complexes would be decreased by transmission of ECM-driven cytoskeletal tension to LEMD3 through a two-part physical linkage as follows: 1) the nesprin–sun LINC complex joining the nuclear lamina and actin cytoskeleton; and 2) through nuclear lamina–LEMD3 coupling via the LEM domain. We tested this hypothesis by disrupting the molecular linkages at either the level of the LINC complex, by expressing a previously validated dominant-negative nesprin construct, DN-Kash-mCherry (40), or by disrupting LEMD3–lamin interactions, by expressing a novel DN–LEM–mCherry constructs and assessing LEMD3–Smad2/3 complex formation by PLA. Neither DN Kash nor DN LEM expression increased the LEMD3–Smad2/3 PLA frequency relative to mCherry-only expressing fibroblasts on glass (Fig. 3, A and B). To assess whether our findings were biased by the degree of DN Kash or DN LEM expression, we correlated PLA puncta against the mCherry expression of the fusion protein per cell. None of the Pearson's coefficients (r = −0.251 and r = −0.087 for DN Kash and DN LEM, respectively, Fig. 3C) varied significantly from 0 (p = 0.0579 and p = 0.43 for DN-Kash and DN-LEM, respectively, Fig. 3C), indicating that the degree of dominant-negative protein expression was not a likely explanation for our findings. These results also corroborated findings in Fig. 1D, where cellular contractility inhibition, through blebbistatin antagonism of myosin II, did not significantly modify the TGFβ stiffness response of fibroblasts.
Figure 3.
LEMD3–Smad2/3 interactions are negatively associated with actin polymerization but not nucleus–cytoplasm coupling or LEMD3–lamin coupling. A, micrographs of LEMD3–Smad2/3 PLA in electroporated cells (top row), mCherry–DN LEM-expressing cells (2nd row), mCherry–DN Kash-expressing cells (3rd row), or mCherry only-expressing cells (bottom row), all on glass (PLA in green, f-actin in blue, mCherry in red, and nucleus in white). B, no significant differences in PLA frequency were seen in the nucleus or cytoplasm of cells expressing either DN LEM or DN Kash relative to mCherry control cells. C, no correlation between DN LEM or DN Kash expression level and PLA frequency in transfected cells. D, micrographs of LEMD3–Smad2/3 PLA in cells treated with cytochalasin D (CytoD) (top rows, g-actin stabilizer) on glass or jasplakinolide (bottom rows, f-actin stabilizer) on 1-kPa gels (PLA in green, f-actin in blue, and nucleus in white). E, cytochalasin D treatment significantly increased the total frequency (p = 0.0089) of PLA interactions per cell on glass. F, jasplakinolide (Jas) treatment significantly decreased the total frequency of LEMD3–Smad2/3 interactions in a dose-dependent fashion (p < 0.0001 for DMSO versus 100 or 200 nm jasplakinolide, p = 0.0312 for 100 nm jasplakinolide versus 200 nm jasplakinolide) and decreased the cytoplasmic (p < 0.0001 for DMSO versus 100 or 200 nm jasplakinolide) and nuclear frequencies (p = 0.002 for DMSO versus 200 nm jasplakinolide, p = 0.0466 for DMSO versus 100 nm jasplakinolide) of LEMD3–Smad2/3 interactions on 1 kPa surfaces. All groups were statistically compared using a two-way ANOVA with Tukey post-test. All scale bars are 10 μm. All data represented by the mean with S.E. except for C where individual data points are plotted. *, p ≤ 0.05; **, p ≤ 0.01; ****, p ≤ 0.0001.
Because g-actin–stabilizing agents did modify the fibroblast stiffness response (Fig. 1D), we examined the effect of actin polymerization on LEMD3–Smad2/3 complex formation by PLA. Actin polymerization was significantly negatively correlated to the frequency of LEMD3–Smad2/3 complexes (Fig. 3, D–F). Fibroblasts on glass treated with 2 μm cytochalasin D for 90 min demonstrated an increase in the total frequency of LEMD3–Smad2/3 complexes overall (p = 0.0089, Fig. 3E). The frequency of LEMD3–Smad2/3 complexes also increased in the cytosolic (p = 0.0871, Fig. 3E) and nuclear compartments (p = 0.5657, Fig. 3E). Conversely, f-actin stabilization through jasplakinolide treatment for 2 h on 1-kPa hydrogels demonstrated a dose-dependent decrease in LEMD3–Smad2/3 interactions (p < 0.0001 for vehicle/DMSO-treated cells versus both 100 and 200 nm jasplakinolide, and p = 0.0312 for 100 nm versus 200 nm jasplakinolide treatment, Fig. 3F). Significant reductions in LEMD3–Smad2/3 complex formation were observed in both the cytoplasm (p < 0.0001 for DMSO-treated cells versus both 100 and 200 nm jasplakinolide, Fig. 3F) and in the nucleus (p = 0.0466 for DMSO-treated cells versus 100 nm jasplakinolide and p = 0.002 for DMSO-treated cells versus 200 nm jasplakinolide, Fig. 3F). Considered with the results in Fig. 1A and Fig. S1, showing increased cell spreading, polarization, and stiffness as a function of substrate stiffness, these data suggested that actin polymerization in response to substrate stiffness helps coordinate the LEMD3-dependent stiffness response of cells to TGFβ.
LEMD3 fragments are generated by a serine protease and differentially regulated by the integrity of the nuclear lamina
We observed cytosolic LEMD3–Smad2/3 interactions with PLA antibody pairs against the native protein and against a full-length recombinant LEMD3 protein, expressing a V5 epitope tag. These data were supported by fractionated cell Western blottings of endogenous LEMD3, which revealed an ≈50-kDa LEMD3 fragment in the cytosol (Fig. S5B). These cytosolic LEMD3–Smad2/3 interactions seemed to be stiffness-regulated given their negative correlation with actin polymerization (Fig. 3, D and E).
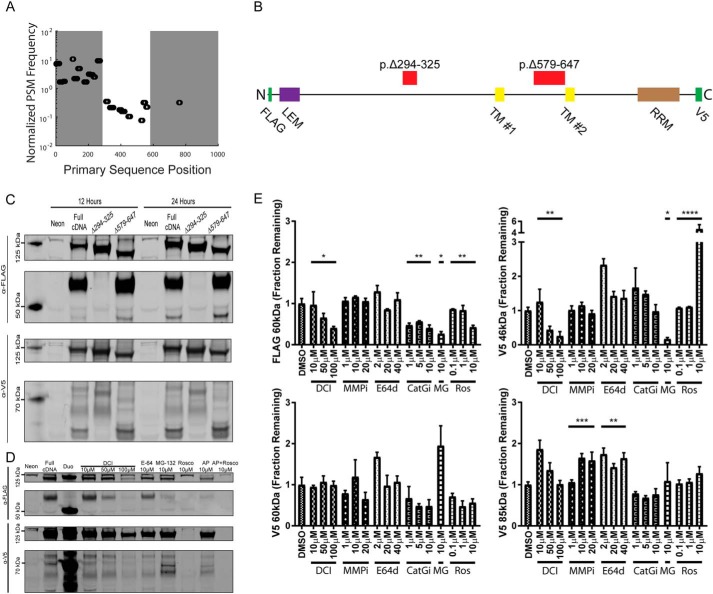
To understand how LEMD3 could interact with Smad2/3 in the cytosol, we performed MS on lysates from cells transfected with pFLAG–LEMD3–V5 constructs (Fig. 4A). Results from a FLAG-purified 60-kDa fragment showed two transitions in the peptide spectral matches to LEMD3's primary sequence, first at p.294–325 and second at p.579–647 (these protein coordinates are offset by 20 amino acids inserted with the N-terminal FLAG tag in the pFLAG–LEMD3–V5 construct from the native protein sequence). The primary sequence relationship between these two regions and other known domains of LEMD3 are shown in Fig. 4B. Deletion mutants (pFLAG–LEMD3p.Δ294–325–V5 and pFLAG–LEMD3p.Δ579–647–V5) exploring these two regions showed differential fragment presentation in Western blottings that probed either the N-terminal FLAG tag or the C-terminal V5 tag of the recombinant protein (Fig. 4C). Specifically, pFLAG–LEMD3p.Δ294-325–V5 lysates lacked the 60-kDa FLAG fragment (the FLAG fragment at <50 kDa was also missing but was not consistently found in all blots) and the 46-kDa V5 fragment. Interestingly, the 46-kDa V5 fragment was similar in size to the cytoplasmically localized fragment of LEMD3 noted earlier from endogenous lysates (Fig. S5B). The pFLAG–LEMD3p.Δ579–647–V5 lysates did not appear to change the FLAG fragment presentation but did eliminate V5 fragments at 60 and 85 kDa. These genetic data suggested that the protein is modified in at least two regions, one in the nucleoplasm (Δ294–325) and one in the peri-nuclear membrane space (Δ579–647). Moreover, all the V5/C-terminal fragments identified by our genetic mutations contained the CTF used in Fig. S3, indicating that they are sufficient for binding Smad2/3 in a stiffness-dependent fashion and binding Smad2/3 in the cytoplasm.
Figure 4.
LEMD3 is proteolytically modified by a serine protease. A, peptide spectral matches (PSMs) were normalized from a 60-kDa FLAG fragment of LEMD3 by PSM frequencies measured in full-length LEMD3. Normalized PSM frequency revealed three distinct zones: over-enriched (left shaded region)- under-enriched (unshaded region)- and absent (right shaded region). B, LEMD3 cartoon showing relative position of the two deletion mutants and known protein domains: LEM, transmembrane (TMs 1 and 2), and RRM domains, and FLAG and V5 epitope tags. C, Western blots from full-length and each deletion mutant using N-terminal FLAG tag (top two blots) and C-terminal V5 tag (bottom two blots) at 12 and 24 h after electroporation. FLAG blots consistently produced a 60-kDa fragment, whereas V5 blots produced 85-, 60-, and 46-kDa fragments. D, representative Western blots for protease and cell cycle inhibitor experiments using N-terminal FLAG tag (top two blots) and C-terminal V5 tag (bottom two blots). E, quantification of blots from D showing that 60-kDa FLAG fragment was significantly reduced relative to the full-length protein when cells were treated with DCI (p = 0.0153), MG-132 (MG) (p = 0.0238), cathepsin-G inhibitor (Cat-Gi) (p = 0.0075), or roscovitine (Ros) (p = 0.0051). V5-tagged 46-kDa fragment was similar in that DCI (p < 0.0001) and MG-132 (p = 0.0238) treatments decreased its abundance but was dissimilar in that roscovitine increased its abundance (p < 0.0001). V5-tagged 85-kDa fragment was increased with MMP inhibitor treatment (p = 0.0004) and E64D treatment (p = 0.0044). All treatment groups, except MG-132, were tested statistically using ANOVA test for trends with a correction for multiple hypotheses using a false discovery rate of α = 0.05. MG-132 was compared with DMSO treated lysates with a Mann-Whitney test and then also corrected using the false discovery rate approach above. All data in E are represented by the mean with S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Following the observation of cytoplasmic interactions with an intron-less LEMD3 cDNA construct, we hypothesized that these fragments were generated proteolytically. We used Western blottings to determine the effects of several broad-spectrum protease inhibitors against cysteine (E64 and E64d), serine (3,4-dichloroisocoumarin (DCI)), and matrix metallo-proteinases (MMP inhibitor III (MMPi)) as seen in Fig. 4, D and E. From fragments controlled by the Δ294–325 region, the 60-kDa FLAG fragment was reduced relative to the full-length transcript in cells treated with DCI (p = 0.0153, Fig. 4E), which is a known inhibitor of cathepsin G, elastase, thrombin, plasmin, factors Xa and X11a, and granzymes A, B, and H (44). This fragment was also reduced when cells were treated with MG-132 (p = 0.0238, Fig. 4E), an inhibitor of the 26S proteosome complex, an important regulator of overall protein homeostasis and particularly critical for degrading ubiquitinated proteins (45). These same trends were observed for the 46-kDa V5 fragment (DCI: p < 0.0001; MG-132: p = 0.0238, Fig. 4E), which is also controlled by the Δ294–325 region. Monash University's PROSPER (Protease Specificity Prediction Server as described in Ref. 46)) predicted only a single serine protease, cathepsin G, cleaving between p.Val-275–Leu-276 (native protein coordinates, score = 1.14) in pFLAG–LEMD3p.Δ294–325–V5's deletion region. Using a more selective cathepsin G inhibitor, we found a significant reduction in the 60-kDa FLAG fragment (p = 0.0075, Fig. 4E) but no significant trend with the 46-kDa V5 fragment. From the Δ579–647 region, the 60-kDa V5 fragment was not significantly modified by any protease inhibitor treatments explored here. Interestingly, the 85-kDa fragment's abundance was only potentiated by MMPi (p = 0.0004, Fig. 4E) and E64d (p = 0.0044, Fig. 4E) treatment. These fragment data together suggested that a serine protease, possibly cathepsin G, operates in the Δ294–325–V5 region, with a distinct mechanism of fragment generation occurring in the Δ579–647 region.
Because pFLAG–LEMD3p.Δ579–647–V5's deletion region lies in the peri-nuclear membrane space, we hypothesized that this location might only be susceptible to degradation during mitosis. Because LEMD3's localization is controlled necessarily by its association to the nuclear lamina through its LEM domain (14, 15, 35–37), we chose roscovitine, a selective inhibitor of Cdk1, which prevents cell cycle progression in part by preventing lamin phosphorylation and disassembly (47). Interestingly, roscovitine only modulated fragments associated with the nucleoplasmic site (sites controlled by pFLAG–LEMD3p.Δ294–325–V5), significantly reducing the generation of the 60-kDa FLAG fragment (p = 0.0051, Fig. 4E) and significantly increasing the generation of the 46-kDa fragment (p < 0.0001, Fig. 4E).
Finally, Fig. 2D showed that LEMD3–Smad2/3 interactions shifted toward the cytoplasm with increasing substrate stiffness. We tested whether this shift was driven by alterations in C-terminal LEMD3 fragment abundances as a function of substrate stiffness with Western blottings assaying HFFs transfected with pFLAG–LEMD3–V5 (Fig. S6, A and B). After 24 h, cells grown on 1- and 25-kPa hydrogels each had decreased proportions of full-length LEMD3 relative to fibroblasts cultured on tissue culture plastic (TC) (p = 0.0062 and p = 0.0031 for TC versus 1 kPa and TC versus 25 kPa, respectively, see Fig. S6A). Lysates from 1- and 25-kPa hydrogels had similar abundances of the 85- and 60-kDa fragments, although the 45-kDa fragment was almost twice as abundant on 25-kPa hydrogels as on 1-kPa hydrogels (p = 0.5196, Fig. S6A). We extended our findings to endogenous LEMD3 and found that full-length native LEMD3's abundance was not modulated significantly by culture on 1- or 25-kPa surfaces (Fig. S7A). LEMD3's mRNA expression level was also invariant to culture on these surfaces (Fig. S7B).
C-terminal fragment of LEMD3 binds Smad2/3 in a stiffness-dependent fashion, antagonizes Smad2/3–Smad4 complexes, and binds PPM1α
To understand the biological significance of these novel CTFs of LEMD3 in the regulation of TGFβ signaling, we used PLA to ascertain their association with Smad2/3 and their ability to antagonize TGFβ signaling. The consensus (i.e. cloned past the 2nd cleavage site so as to contain the conserved sequence from all C-terminal fragments) CTF described earlier for Fig. S3 has been shown to be sufficient for Smad2/3 binding (16, 18, 19, 21, 41) and lacks the necessary domains for nuclear localization (14, 18, 33). We repeated the recombinant PLA experiments from Fig. 2, B and F, between CTF's V5 epitope and Smad2/3 to ascertain the following: 1) whether this fragment of LEMD3 was sufficient for generating stiffness-dependent Smad2/3 interactions; 2) whether these fragments were uniquely localized to the cytoplasm; and 3) whether the CTF-Smad2/3 interactions were dependent on the phosphorylation state of Smad2/3.
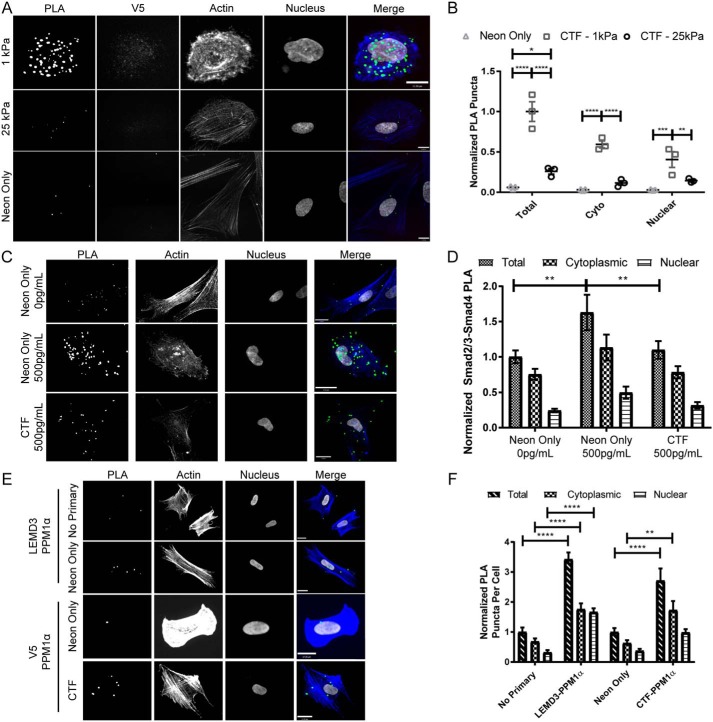
The CTF-expressing fibroblasts on 1-kPa hydrogels had significantly more Smad2/3 interactions overall (p < 0.0001, Fig. 5B), in the cytoplasm (p < 0.0001, Fig. 5B), and in the nucleus (p = 0.0082, Fig. 5B) relative to CTF-expressing HFFs on 25-kPa hydrogels. Transfected fibroblasts on 1- and 25-kPa hydrogels had significantly more overall CTF–Smad2/3 interactions than electroporation control fibroblasts (p < 0.0001 and p = 0.0227 for 1- and 25-kPa conditions, respectively, Fig. 5B). To control for degree of recombinant CTF expression, we compared the V5 signal and the PLA interactions per cell across each group. We found that fibroblasts on 1-kPa hydrogels had a steeper slope (i.e. more PLA per arbitrary unit of V5 expressed) than fibroblasts on 25-kPa hydrogels (p = 0.1476, Fig. S8). Furthermore, we found that TGFβ dosing (50 pg/ml) did not significantly vary the CTF–Smad2/3 interaction frequency on 1- or 25-kPa substrates (p = 0.98 and p = 0.99, respectively, Fig. S9).
Figure 5.
C-terminal fragments of LEMD3 bind Smad2/3 and antagonize Smad2/3–Smad4 complexes. A, representative images of V5–Smad2/3 PLA reactions imaged in fibroblasts transfected with pFLAG–LEMD3p.Δ21–669–V5 (top two rows) and electroporation control cells (bottom row, neon only) on 1-and 25-kPa hydrogel surfaces (PLA in green, f-actin in blue, V5 in red, and nucleus in white). B, PLA interactions from A between the V5 tag of a C-terminal fragment of LEMD3 (pFLAG–LEMD3p.Δ21–669–V5) and Smad2/3, normalized to the total interactions observed on 1-kPa hydrogels. HFFs on 1-kPa hydrogels have significantly more interactions overall (p < 0.0001), in the cytosol (p < 0.0001), and in the nucleus (p = 0.0082) relative to fibroblasts on 25-kPa hydrogels. All transfected cells had more PLA interactions than electroporation-only (neon) populations (p < 0.0001 and p < 0.0227 for 1- and 25-kPa hydrogels, respectively). Scale bars, 11 μm. All statistical testing done with two-way ANOVA with Tukey's post-test. C, representative images from HFFs on 1-kPa hydrogels assayed for Smad2/3–Smad4 interactions. Cells in the 2nd and 3rd rows were treated with 500 pg/ml TGFβ for 1 h before fixation, whereas cells in the 1st row were untreated. Cells in the 3rd row were electroporated with pFLAG–LEMD3p.Δ21–669–V5, and cells in the top two rows were electroporation control cells (PLA in green, f-actin in blue, and nucleus in white). All scale bars, 17 μm. D, normalized quantification of data from C. Fibroblasts treated with 500 pg/ml TGFβ and overexpressing the CTF of LEMD3 had significantly fewer Smad2/3–Smad4 complexes than untransfected cells treated with 500 pg/ml TGFβ (p = 0.0081). Untransfected TGFβ fibroblasts treated with 500 pg/ml also had significantly more Smad2/3–Smad4 complexes than untransfected fibroblasts (p = 0.001). All statistical testing done with a two-way ANOVA with Tukey's post-test. E, interactions between PPM1α and endogenous LEMD3 or its CTF assayed by PLA on glass surfaces. The 1st and 2nd rows used antibody pairs between PPM1α and endogenous LEMD3. The 3rd and 4th rows used antibody pairs between PPM1α and V5 tag on cells transfected with pFLAG–LEMD3p.Δ21–669–V5 (PLA in green, f-actin in blue, and nucleus in white). All scale bars are 17 μm. F, normalized quantification of the interaction rates between LEMD3 or its CTF by subcellular compartment. Both endogenous LEMD3 and its CTF interacted with PPM1α as demonstrated by PLA frequencies above their respective negative controls (for total interactions: p < 0.0001 for both LEMD3 versus no primary and for CTF versus neon only, respectively). Both LEMD3 and CTF transfected cells also had significantly higher PPM1α interaction rates than their respective controls in the cytoplasm (for cytoplasmic rates: p < 0.0001 and p = 0.0081 for LEMD3 and CTF pairs, respectively), but only endogenous LEMD3 had statistically significant higher interaction rate in the nucleus (for nuclear rates: p < 0.0001 and p = 0.2569 for endogenous LEMD3 and CTF-expressing fibroblasts, respectively). All statistical testing was performed by a two-way ANOVA with Sidak's post-test. All data represented by the mean with S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Unexpectedly, a significant fraction (≈40–55%, depending on stiffness) of CTF–Smad2/3 interactions took place in the nucleus (Fig. 5B). Although the N terminus of LEMD3 up to the first transmembrane domain has been shown to be necessary for faithful localization of full-length LEMD3 to the nucleus (14, 33), the CTF does have an independent ability to bind both barrier-to-autointegration factor (a nuclear chromatin protein (34)) and DNA directly (41), which may allow for enrichment of the CTF in the nucleus. Overall, these V5–Smad2/3 PLA assays with recombinant LEMD3 proteins confirmed the following: 1) the negative stiffness correlation and TGFβ independence of CTF–Smad2/3 complexes, mirroring the results observed with endogenous LEMD3 assays; and 2) the CTF was sufficient for cytoplasmic LEMD3–Smad2/3 interactions.
Having demonstrated the CTF binds Smad2/3 in a stiffness, but not TGFβ dose-dependent fashion, we assayed whether CTF overexpression could antagonize TGFβ signaling by examining the effect of CTF's expression on the abundance of Smad2/3–Smad4 complexes. Smad2/3–Smad4 complexation is a necessary step for efficient nuclear translocation and activity as a transcription factor and is sensitive to Smad2/3's phosphorylation state (48, 49). HFFs treated with 500 pg/ml TGFβ on 1-kPa hydrogels showed an increase in Smad2/3–Smad4 complexes relative to untreated HFFs (p = 0.001, Fig. 5D). Overexpression of the CTF significantly abrogated this TGFβ-driven increase in Smad2/3–Smad4 complexes (p = 0.0081, Fig. 5D). These results mirrored the finding of previous studies, which have shown that the RRM domain of LEMD3 is sufficient for Smad antagonism (16, 18, 19, 21, 41). Previous studies have shown that LEMD3 antagonizes TGFβ/Smad signaling by dephosphorylating Smads 2 and 3 in a protein phosphatase, Mg2+/Mn2+–dependent 1α (“PPM1α”)-dependent manner (16, 22). We queried whether endogenous LEMD3 and the CTF could bind PPM1α in the cytoplasm and nucleus by PLA. Both endogenous LEMD3 and the CTF bound PPM1α, demonstrated by interaction frequencies significantly above their respective negative controls (Fig. 5F, total interactions, p < 0.0001 for both endogenous LEMD3 versus a no primary control, and electroporation only/neon only control versus CTF). Both endogenous LEMD3 and the CTF bound PPM1α significantly above background in the cytoplasm (Fig. 5F, cytoplasmic interactions: p < 0.0001 for endogenous LEMD3 and p = 0.0081 for the CTF), although only endogenous LEMD3 was found to significantly bind PPM1α in the nucleus (Fig. 5F, nuclear interactions: p < 0.0001 for endogenous LEMD3 and p = 0.2569 for the CTF). These data specifically demonstrated the CTF's ability to antagonize Smad2/3 signaling and its connection to PPM1α, a known LEMD3-associated phosphatase that antagonizes TGFβ signaling.
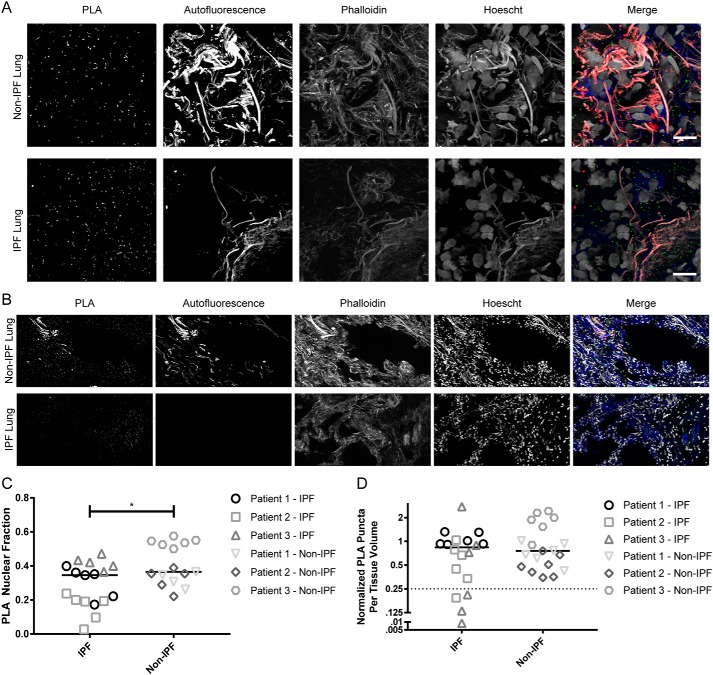
LEMD3–SMAD2/3 complexes are more cytosolic, and their frequencies are more varied in IPF biopsies than non-IPF biopsies
We sought to validate our in vitro findings in human lung core biopsies from six patients (three with and three without IPF). IPF patients had significantly more cytosolic LEMD3–Smad2/3 interactions than non-IPF patients (p = 0.0307, Fig. 6, A and C). These data are compelling because they connect the cytoplasmic PLA localization trend seen with increasing stiffness in vitro in Fig. 2D and the pattern observed in fibrosed relative to nonfibrotic human tissue ex vivo. Additionally, these data directly demonstrate that extra-nuclear LEMD3–Smad2/3 interactions occur in human lung tissue, which we also discovered in Fig. 2, indicating that extra-nuclear LEMD3 is not an artifact of in vitro culture.
Figure 6.
LEMD3–Smad2/3 PLA interactions are more cytoplasmic and more varied in frequency in IPF versus non-IPF human lung tissue. A and B, LEMD3–Smad2/3 PLA frequency imaged at high-magnification (×63) (A) and low-magnification (×20) (B) in non-IPF (top row) and IPF (bottom row) tissue (PLA in green, f-actin in blue, autofluorescence in red, and nuclei in white). C, quantification of subcellular localization of LEMD3–Smad2/3 PLA events from A showed a cytoplasmic shift in PLA interactions in IPF patients (p = 0.0307, Mann-Whitney test), mirroring the in vitro trends seen in Fig. 2D. D, quantification of total LEMD3–Smad2/3 PLA frequency from B showed a similar frequency of interactions between IPF and non-IPF patients (p = 0.5783, Mann-Whitney test). IPF tissue had a higher intra-patient variability (coefficient of variance = 67 and 30% for IPF and non-IPF patients, respectively) and more extreme dispersion overall (kurtosis = 4.765 and −0.1391 for IPF and non-IPF patients, respectively). 22% of IPF tissue areas sampled formed a unique low-interaction “tail” (<25% of the mean IPF interaction frequency, denoted by data under the dotted line), which was absent in non-IPF tissues. All scale bars are 10 μm. Bars in C and D represent grand medians.
Patients with IPF also had a slightly higher median frequency of LEMD3–Smad2/3 interactions relative to patients without IPF (Fig. 6D, p = 0.5783). However, although each non-IPF patient had a fairly consistent LEMD3–Smad2/3 interaction “set-point,” IPF patients displayed a higher degree of intra-patient heterogeneity across their sampled tissue regions (67% versus 30% average coefficient of variance for IPF and non-IPF patients, respectively). Overall, the whole IPF data set had a higher degree of kurtosis (4.765 versus −0.1391 for IPF and non-IPF patients, respectively), indicating that more of the LEMD3–Smad2/3 variance in IPF tissue comes from extreme deviations in interaction frequency. In particular, ≈22% of tissue regions in IPF patients had uniquely low LEMD3–Smad2/3 interaction rates (<25% the mean interaction frequency) not observed in non-IPF tissue. Previous micro-mechanical investigations of IPF tissue revealed a high degree of spatial heterogeneity to tissue stiffness in IPF lungs (43). Our findings were consistent with the interpretation that spatially heterogeneous regions of fibrosis in the IPF tissue create both increased variability in the LEMD3–Smad2/3 interaction rate and regions of locally lower LEMD3–Smad2/3 interactions. These data in aggregate showed that LEMD3 regulation of TGFβ through Smad2/3 is locally diminished in IPF tissue relative to non-IPF tissue and that LEMD3's role is not confined to the nucleus in human tissue.
Discussion
The medical management of fibrotic diseases, such as pulmonary fibrosis, has enjoyed recent success with the approval of pirfenidone, an “anti-fibrotic agent,” and nintedanib, an inhibitor of multiple tyrosine kinase receptors, after years of failed clinical trials with a variety of other agents, including anti-inflammatory and anti-oxidant drugs (4, 6, 50, 51). Subsequent study of the mechanisms of pirfenidone has shown that pirfenidone antagonizes fibrotic progression in part by inhibiting the synthesis of TGFβ (5, 7), which is preferentially activated in fibrotic matrices (8, 9). Although pirfenidone may successfully decrease the supply of TGFβ available in fibrotic pathology, ours and others' findings indicate that the fibrotic matrix also potentiates cellular responses (e.g. synthesis of extracellular matrix and increased cellular contractility) to remaining TGFβ (10–13). We focused on the role of LEMD3 in this matrix-driven sensitization, and a graphical summary of our working model for LEMD3–Smad2/3 stiffness-driven interactions is shown in Fig. 7. We showed in vitro that LEMD3 overexpression antagonized TGFβ-driven transcription and stiff-shifts the responsiveness of these fibroblasts to TGFβ, whereas LEMD3 knockdown by siRNA soft-shifts the mechanical response to TGFβ and potentiates TGFβ signaling. We also showed in vitro that LEMD3–Smad2/3 interactions are inhibited and cytoplasmically shifted with increasing substrate stiffness and actin polymerization. Ex vivo, we identified these same phenotypic correlates in IPF biopsies relative to non-IPF tissue; IPF tissue had a pronounced cytoplasmic shift in PLA subcellular localization; and ≈22% of IPF tissue regions had a unique reduction in LEMD3–Smad2/3 interactions relative to non-IPF tissue and a higher degree of variation in the LEMD3–Smad2/3 interaction rate across various tissue regions, both in individual patients and overall. These low interaction regions in IPF are not likely attributable to changes in overall LEMD3 abundance in pathology. Ours and others' data showed that full-length LEMD3 was not regulated by ECM stiffness at the mRNA or protein level in vitro for endogenous LEMD3 (Fig. S7) or for recombinant LEMD3 (Fig. S6) (52, 53). Moreover, IPF patients do not show evidence of altered LEMD3 transcription in either microarray or next generation sequencing of IPF and control patients (54–56).
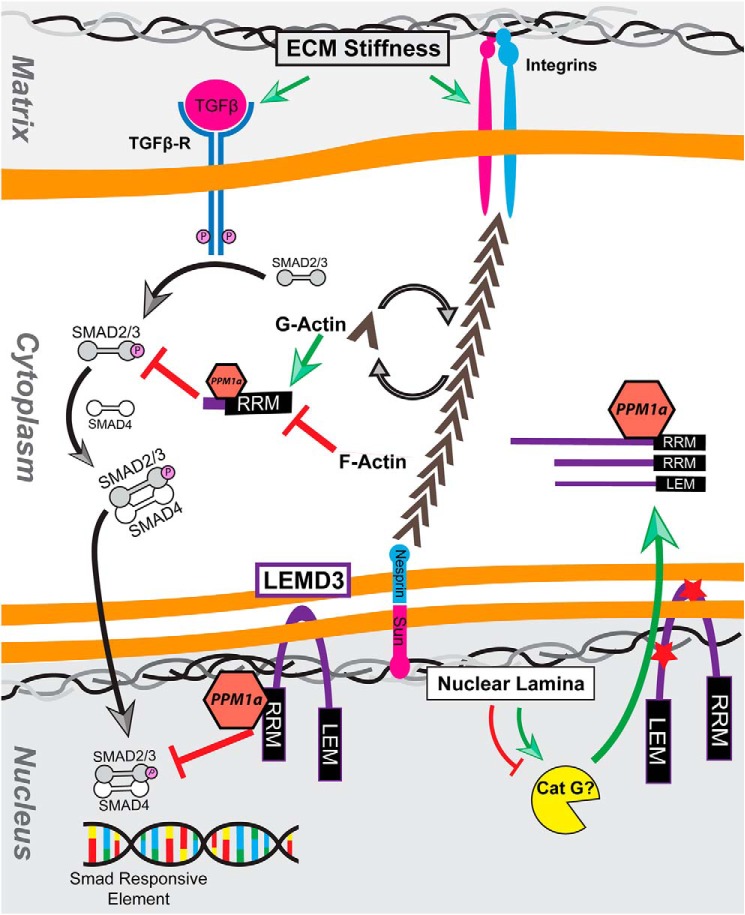
Figure 7.
Summary cartoon. Mechanical cues from the extracellular matrix potentiate TGFβ activation, which ultimately drives the phosphorylation-dependent formation of Smad2/3/4 complexes. These complexes translocate to Smad response elements in the nucleus and facilitate the transcription of TGFβ-dependent genes. LEMD3 antagonizes this TGFβ/Smad2/3 signaling in a stiffness-dependent fashion, altering the stiffness response of fibroblasts to activated TGFβ. LEMD3 achieves this by binding both Smad2/3 and PPM1α, a protein phosphatase previously shown to be coordinated by LEMD3, both in the nucleus and in the cytosol. Cytosolic LEMD3 fragments are post-translationally generated at two sites, which separate the nuclear-localizing LEM domain and the Smad2/3 interacting RRM domain. Processing at the nucleoplasmic site (bottom red star) generates LEM- and RRM-containing fragments, which are reduced in abundance by serine protease inhibitors but have differential responses to lamin phosphorylation inhibitors. Both nuclear and cytosolic LEMD3–Smad2/3 complexes are inhibited by actin polymerization, which is driven by mechanical cues from the matrix, thereby connecting ECM mechanics to the inhibition of an inhibitor of Smad2/3. Inhibitory interactions are shown with red block-end arrows, and activating signals are denoted with green arrows. Black arrows indicate translocation. Abbreviations used are as follows: Cat G, cathepsin G; RRM, RNA Recognition motif; LEM, Lap2b–Emerin–MAN1 domain; TGFβ-R, TGFβ receptor; PPM1α, protein phosphatase, Mg2+/Mn2+-dependent 1α.
Supporting our discovery of LEMD3–Smad2/3 cytoplasmic interactions, we have discovered here that LEMD3 was post-translationally cleaved at two sites, and we have implicated serine proteases (possibly cathepsin G) in the generation of C-terminal fragments that lack the necessary LEM domain for nuclear targeting (33, 34, 57). We have also shown that a C-terminal fragment of LEMD3 bound Smad2/3 throughout the cell, associated with PPM1α, and antagonized Smad2/3–Smad4 complex abundance. These results were consistent with previous studies showing that the C-terminal RRM domain is sufficient for LEMD3's antagonism of Smad2/3 (16, 18, 19, 21, 41).
However, the underlying mechanism for a cytoplasmic shift in LEMD3–Smad2/3 interactions with increasing substrate stiffness in vitro and in IPF patient ex vivo is not clear. In vivo, there is strong evidence for an altered and generally more active serine (58), cysteine (59, 60), and matrix metallo-proteinases (61–65) landscape in IPF tissue, although much of the attention has focused on extracellular proteolysis. This more pronounced proteolytic environment could contribute to the cytoplasmic shift seen in IPF patients in our study. In vitro, we found that stiffness representative of scarred tissue in IPF (25 kPa) and physiological lung tissue (1 kPa) produced statistically similar abundances of all the LEMD3 fragments we identified (Fig. S6). However, the 46-kDa fragment was nearly twice as abundant on 25-kPa matrices, prompting a need for further investigation. Overall, we have demonstrated that LEMD3 sits at an interesting intersection of mechanical and biochemical cues in pulmonary fibrosis, and LEMD3 expression may be a promising adjunctive avenue for addressing fibrosis-driven cellular sensitivity to TGFβ in combination with pirfenidone.
Our original hypothesis for the stiffness regulation of LEMD3–Smad2/3 complexes was that LEMD3's affinity for Smad2/3 would be negatively regulated by biophysical stress from the actin network transmitted to LEMD3 through the LINC–lamina–LEM set of complexes. However, this hypothesis requires critical re-evaluation in light of our discovery of cytosolic LEMD3 fragments that bind Smad2/3 and the apparent lack of efficacy of cytosol–nuclear and lamin–LEM-disrupting constructs (DN-Kash and DN-LEM) in decreasing LEMD3–Smad2/3 complex formation in the nucleus. We have also observed that LEMD3–Smad2/3 complexes are negatively correlated to actin polymerization. However, it is not obvious how actin polymerization itself would directly modulate LEMD3–Smad2/3 complex formation given that neither LEMD3 nor Smad2/3 has a known direct association with actin. One parsimonious explanation would be that LEMD3 competes for Smad2/3 binding with other actin-regulated proteins, namely Yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) and/or Rho-associated protein kinase (ROCK). YAP/TAZ and ROCK bind Smad2/3 and have been shown to be necessary for nuclear accumulation of Smad2/3 in response to TGFβ stimulus (13, 66–69). ROCK inhibition has been shown to modulate phosphorylation in Smad3's linker region (Ser-203, Ser-207, and Ser-212) (66), whereas TAZ has been shown to bind to the MH1 domain of Smad2 (67). Interestingly, yeast two-hybrid assays indicate that LEMD3 binds to the MH2 domain of both Smad2 and Smad3 (14). Although YAP/TAZ and LEMD3 seem to bind different locations in Smads, YAP/TAZ have been shown to negatively regulate the Smad binding of forkhead box protein H1 (FOXH1), which competes for binding to Smad2/3's MH2 domain with LEMD3 in embryonic stem cell CHIP assays (16, 70). These data indicate that YAP/TAZ, in particular, are promising candidates for modulating LEMD3's ability to bind Smad2/3 in an actin polymerization-dependent fashion.
LEMD3 is also directly implicated in the development of BOS with and without melorheostosis. Our findings that LEMD3 was post-translationally processed proteolytically and has cytosolic forms raises new questions regarding the pathogenesis of BOS. Many of the identified BOS patients possess nonsense or splice-site mutations that eliminate the Smad-binding C-terminal end of LEMD3 (20, 24–31). Interestingly, there are a five missense mutations of unknown clinical significance associated with BOS in NCBI's ClinVar database, which are associated with one of the two deletion mutants (p.Δ294–325 and p.Δ579–647) identified in this study 3 (71). These missense mutations could suggest that alterations to the fragment controlling regions of LEMD3 might play a role in the development of BOS. Additionally, our finding that LEMD3 was processed by a serine protease and the 26S proteosome might suggest additional targets/mechanisms in individuals with BOS who lack mutations in LEMD3 (32).
Finally, we have characterized several N- and C-terminal LEMD3 fragments and demonstrated their varying sensitivities to the following: 1) genetic perturbation through deletion mutants; 2) serine (DCI), cathepsin G, and 26S proteosome (MG-132) inhibitors; 3) lamin integrity/disassembly through Cdk1 inhibition; and 4) substrate stiffness. One particular pair of fragments, the 60-kDa N-terminal FLAG and 46-kDa C-terminal V5 fragments, is particularly interesting because both are sensitive to the same mutation region (p.Δ294–325), the abundance of both fragments is reduced with respect to DCI and MG-132 treatment, and they plausibly sum to the mass of full-length LEMD3. However, these two fragments are differentially regulated by integrity of the lamin network (following inhibition of Cdk1 by roscovitine) and by cathepsin G inhibition, making it less likely that these fragments are generated in a concerted proteolytic reaction. Additionally, we do not have a straight-forward explanation relating the position of our deletion mutations and the fragments' sizes they seemingly control. It is not possible to directly align our Western blotting fragment data and mass spectrometry data because of the unknown distributions of post-translational modifications in LEMD3; however, the fragments' relative masses controlled by either deletion mutant do not logically correspond with the expected size of products from cleavage in those areas.
This is, to our knowledge, the first report of an integral inner nuclear membrane protein processed proteolytically into cytosolic forms. However, our observations fit a broader pattern of a more locationally dispersed role for nuclear envelope integral proteins. Several integral members of the nuclear membrane have alternative splice forms that both include and omit their transmembrane domain(s), including nesprin, lamina-associated polypeptide 2 (LAP2), torsin-1a-interacting protein 1 (LAP1), and nurim (72–75). Moreover, some “localized” elements of the nuclear envelope have been found dispersed throughout the cytosol based on the cellular context. For example, nesprins, structural elements of the outer nuclear membrane that link the cytoskeleton and nucleus, are restricted to the nuclear envelope in myoblasts but not in differentiated myotubes (76, 77), and certain nesprin isoforms are also de-compartmentalized during muscle regeneration in Duchenne muscular dystrophy (77). Additionally, alternatively spliced “KASH-less” nesprins, which lack the nuclear envelope anchoring domain, KASH, have recently been linked to a variety of cytosolic locations, including focal adhesions and actin stress fibers (72), Golgi bodies (78), and RNA-processing bodies (79, 80). Our findings expand on the observed mechanisms that regulate the localization of nuclear envelope proteins in the cell.
Experimental procedures
Primary fibroblast cultures and transfection, human lung biopsies, and other reagents
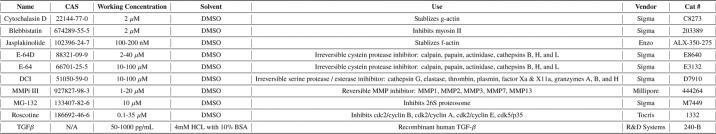
Primary HFFs were procured from the ATCC (ATCC SCRC-1041, ATCC, Manassas, VA) and routinely cultured in DMEM (ThermoFisher Scientific, Waltham, MA) supplemented with 15% FBS and 1% penicillin/streptomycin (ThermoFisher Scientific, Waltham, MA) in 5% CO2 at 37 °C in a humidified incubator to passage 13. CCL210s, a primary human pulmonary fibroblast line, were procured from the ATCC (ATCC CCD-19Lu) and routinely cultured in Eagle's minimal essential media (ThermoFisher Scientific) supplemented with 10% FBS and 1% penicillin/streptomycin (ThermoFisher Scientific). For DNA transfections, 50,000 HFFs were resuspended in 10 μl of Buffer R and electroporated using a 1700-V, 20-ms pulse width, 1-pulse program with 1 μg of DNA in a Neon transfection system (ThermoFisher Scientific). For siRNA transfections, 8000–10,000 cells/cm2 were treated with 25 or 200 nm siRNA with Lipofectamine2000 (ThermoFisher Scientific) according to the manufacturer's recommendations. IPF and non-IPF human lung core biopsies were generously provided by Dr. Eric White and the Lung Tissue Research Consortium (NHLBI: HHSN2682016000021). Selected cores all came from distal sections of lung parenchyma. Human subjects' approval for this tissue was obtained through the University of Michigan Institutional Review Board. This study abides with the Declaration of Helsinki principles. For information on the oligos, antibodies, and uncommon chemicals used in this work, please refer to the Tables 1–3 for supplier and use information.
Table 1.
Table 2.
Primary, secondary, and other antibodies used in this work
Table 3.
Chemicals and recombinant proteins used in this work
Generation of recombinant constructs
LEMD3 constructs were derived from the pSVK3–FLAG–MAN1 construct, kindly provided by Dr. Howard Worman (Addgene plasmid no. 26002) (33). SDM was performed using the Q5 SDM kit (New England Biolabs, Ispwich, MA) according to their protocol, including the use of “NEB BaseChanger” applet to design primers and select PCR conditions. Primers and associated melting points for each mutagenesis are listed in Table 1. Constructs were confirmed with Sanger sequencing (MacrogenUSA, Rockville, MD).
DN LEM construct
DN Kash and DN control plasmids (pCDHEF1–MCS1–puro–mCherry–Nesprin–1αKASH and pCDH–EF1–MCS1–puro–mCherry, respectively) were kindly provided by Dr. Jan Lammerding and are described in detail in Ref. 40. The DN LEM construct was created by Gibson assembly of the N-terminal LEM domain cDNA from LEMD3 (without FLAG) from pSVK3–FLAG–MAN1 into the frame occupied by Nesprin–1αKASH in the pCDHEF1–MCS1–puro–mCherry–Nesprin–1αKASH construct with primers and PCR conditions as described in Table 1.
Western blotting
For nonphosphoprotein Western blottings, cell lysates from tissue culture plastic or from 2- or 25-kPa, 150-cm Petrisoft dishes (Matrigen, Brea, CA) were either harvested directly in 4× protein loading buffer (Licor, Lincoln, NE) supplemented with protease and phosphatase inhibitors (no. A32959, ThermoFisher Scientific) according to the manufacturer's instructions or processed with the NE-PER compartment isolation kit (ThermoFisher Scientific) according to the manufacturer's instructions. Lysates were treated with 0.3 μl of benzonuclease (ThermoFisher Scientific) for 15 min at room temperature with vigorous shaking. Samples were heated to 95 °C for 5 min and run on a 4–12% BisTris gel in 1× MES buffer and run for 70 min at 150 V. The gel was then transferred to a nitrocellulose membrane using the XCell Blot Module (ThermoFisher Scientific) at 25 V for 65 min. Membranes were air-dried for an hour, blocked with 5% nonfat milk in 1× PBS for an hour at room temperature, and then incubated with primary antibodies as described in Table 2 in 1× PBS-T with 1% milk for 16–24 h at 4 °C. Near-IR or HRP-conjugated secondaries as described in Table 2 were then incubated with the blot and imaged subsequently on either an Odyssey CLx system for near-IR dyes (Licor, Lincoln, NE) or a ChemiDoc MP system (Bio-Rad) with SuperSignal West Femto reagents (ThermoFisher Scientific) for HRP secondaries according to the manufacturer's instructions. Band analysis was performed using the associated Licor or Bio-Rad software.
For phosphoprotein Western blots, the procedure above was followed with the following changes. Cells were treated with 100 pg/ml TGFβ 90 min before lysis. Membranes were incubated in and washed in buffers formulated with TBS instead of PBS. Membranes were blocked in 5% BSA in 1× TBS, and all antibody incubations were done in 1% BSA in 1× TBS-T.
For recombinant LEMD3 Western blots, two samples of HFFs were prepared and electroporated as above and plated onto each plastic or hydrogel surface for 12 h (protease inhibitor experiments) or 24 h (LEMD3 fragment abundance and deletion mutant experiments) before harvesting.
qPCR
RNA was isolated from cultured cells using RNeasy Plus Mini kits (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cDNA libraries were constructed using RT2 First Strand kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction. 25-μl qPCRs were prepared using SYBR Green Master Mix (ThermoFisher Scientific) with 250 nm primers, listed in Table 1, and 10 ng of input cDNA. Reactions were carried out on a StepOnePlus (ThermoFisher Scientific) for 40 cycles. Melt curves were visually inspected after each run to confirm a single defined peak in the derivated intensity plot. Data from each run was imported into LinRegPCR, which performed baseline correction and measured PCR efficiency by amplicon group as described previously (81). The relative number of transcripts was calculated by dividing the Cq threshold by the PCR efficiency for the reaction raised to the power of Cq, the cycle number at which the threshold is reached for a given target. For relative quantification between conditions, LEMD3 transcripts were either normalized for each condition to the mean of two reference genes, 18S and ACTB, if the samples were all from a single stiffness condition or normalized to 18S alone if substrate stiffness was an experimental condition.
Smad-driven luciferase assays
HFFs were transfected with Cignal Lenti Reporter virus (Qiagen, Hilden, Germany) with a multiplicity of infection of 40 and subsequently selected using 400 ng/ml puromycin until untransduced HFFs died. Stably transfected HFFs, with or without additional LEMD3-focused genetic treatments as described above, were plated at 10,000 cells/well on an HTS plate (Matrigen, Brea, CA) containing glass surfaces and polyacrylamide gels, which ranged in stiffness from 0.2 to 50 kPa, all functionalized with 10 μg/ml plasma-purified human fibronectin according to the manufacturer's instruction. Cells adhered to the surfaces for 4 h in serum-free DMEM supplemented with 1% BSA and 1% penicillin/streptomycin. Media were then supplemented with the desired amounts of TGFβ and, when applicable, with cytoskeletal agents as described under the “Results.” HFFs were then incubated for 16 h at 37 °C before the luciferase reaction. Wells were supplemented to 2 mm VivoGlo d-luciferin (Promega, Madison, WI) using a plate reader-controlled auto-injector system, and light production was subsequently quantified every 5 min for 30 min on a Synergy H4 plate reader (BioTek, Winooski, VT). The raw luciferase signal was the median intensity measured over this time course minus the intensity from luciferase-transfected cells not treated with TGFβ to account for any TGFβ elaborated and activated by the cells in situ. Cells were then fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and then stained with 1:10,000 Hoechst stain 33342 in 1× PBS for 30 min at room temperature (ThermoFisher Scientific). The raw luciferase signal was then normalized to the nuclear signal, which was inside a linear standard curve from cells plated on glass and on polyacrylamide within each plate. Data for each condition was then fit to a three-parameter logistic equation using a least-squares minimization fit in Prism (GraphPad, La Jolla, CA). For curves that did not converge with this model, individual data points are shown as the mean with the standard error of the mean to illustrate data heterogeneity. Otherwise, the trends across the maximal luciferase signal (“Top” variable from the model) or the mid-transition point observed between “Bottom” and “Top” signals (“EC50” variable from the model, called “transition point” in text), were statistically compared using an ANOVA with “test for trends,” which restricts the hypothesis testing to a particular order of the data sets (e.g. maximum luciferase signal as a function of increasing TGFβ dose or transition point as a function of increased LEMD3 expression). Each data point represents at least three biological replicates.
Proximity ligation assays
Ex vivo PLA
Lung biopsy samples were prepared by OCT embedding flash-frozen tissue (Tissue-Tek, Sakura Finetek, Torrance, CA) and preparing 10-μm cryosections on a CryoStar NX70 (ThermoFisher Scientific). Sections were fixed for 10 min at 4 °C in 2% paraformaldehyde in 1× PBS. Protein–protein PLAs were performed with antibody pairs listed in Table 2 as described previously (42) with the following modifications: all primary antibody incubations were done for 16–24 h at 4 °C; sections were blocked in 0.5% Tween 20, 0.1% Triton X-100, 0.5% gelatin, 5% donkey serum, 2% bovine serum albumin (BSA), and 5 μg/ml poly(dI-dC) DNA in PBS, and sections were actin counter-stained with 1:40 phalloidin-488 (ThermoFisher Scientific) for 30 min at room temperature following the PLA reaction using the IF protocol recommended by Sigma's PLA Resource Center. Slides were analyzed using a Axiovert 200M microscope (Carl Zeiss Microscopy, Jena, Germany) with an UltraVIEW spinning disk (PerkinElmer Life Sciences) and Flash 4.0v2 cMOS camera (Hamamatsu Photonics, Hamamatsu City, Japan). Low resolution images (×20) were collected using a 0.8NA Plan-Apochromat, and high resolution images (×63) were collected using a 1.4NA Plan-Apochromat objective (Carl Zeiss Microscopy, Jena, Germany). Image acquisition and analysis were performed in Volocity (PerkinElmer Life Sciences). To analyze the incidence of LEMD3–Smad2/3 interactions between IPF and non-IPF biopsies, at least six random points were chosen by Volocity in each section and registered to a serial section of the same tissue that was prepared as a no primary antibody PLA control. Tiled images with 20% overlap were acquired at ×20, covering a 628 × 334-μm area with z-slices acquired every 0.6 μm. The PLA signal was processed in Volocity using a consistent intensity threshold to identify PLA puncta, which were then spatially filtered by excluding PLA signal not associated with the actin or DAPI tissue-based signals. The remaining PLA signal was processed using the “subtract” function to remove tissue autofluorescence, measured independently in an unused spectral window (615 nm, width 70 nm). The PLA signal was then normalized to the measured nuclear volume in the image slice. Relative PLA incidence between conditions was calculated by subtracting the normalized PLA signals between experimental and no primary antibody at each co-registered pair of points. High-resolution imaging (×63) of each tissue was performed over at least six randomly chosen areas, each 111 × 106 μm with z-slices acquired every 0.2 μm, to measure the cytosolic and nuclear PLA compartments in tissue. Signal processing was similar to the low-resolution processing described above except that the PLA signal was subdivided using the “exclude non-touching” function in Volocity to measure the nuclear and non-nuclear associated signal.
In vitro PLA
Protein–protein PLA was performed with antibody pairs listed in Table 2 as described previously (42) with the following modifications: cells were plated on fibronectin-coated glass or hydrogels (Matrigen, Brea, CA). All primary antibody incubations were performed for 16–24 h at 4 °C, and cells were counter-stained with 1:40 phalloidin-488 or phalloidin-546 in 1× PBS for 30 min at room temperature and/or further processed for IF imaging using the procedure recommended in Sigma's PLA resource center. For all recombinant LEMD3 and C-terminal fragment experiments, HFFs were electroporated as above and allowed to culture for 24 h on 10 μg/ml fibronectin-coated glass or hydrogels before fixation. For LEMD3–phospho-Smad2/3, LEMD3–Smad2/3 + TGFβ, and Smad2/3–Smad4 PLA assays, cells were treated 90 min before fixation with 50 pg/ml TGFβ for LEMD3- and CTF–Smad2/3 assays, 100 pg/ml TGFβ for LEMD3–phospho-Smad2/3 experiments, and 500 pg/ml TGFβ for Smad2/3–Smad4 experiments. For FLAG post-staining of PLA-processed tissues, cells were blocked with unlabeled AffiniPure donkey anti-rabbit antibodies (Jackson ImmunoResearch, West Grove, PA) at a 1:10 dilution in 1× PBS-T at room temperature for 1 h before further IF processing with 1:1000 rabbit anti-FLAG (no. F7425, Sigma, Darmstadt, Germany) for 1 h in PBS-T at room temperature. Following three washes with PBS-T for 5 min, each sample was stained with 1:500 goat anti-rabbit 546 in PBS-T for 1 h at room temperature, washed again, and then counter-stained as above. For V5 post-staining of PLA-processed samples, cells were incubated with 1:200 647-labeled anti-V5 antibody (R&D Systems, Minneapolis, MN) diluted in PBS-T at room temperature for 1 h before washing three times in PBS-T for 5 min each and then counter-stained as above. Imaging and analysis was performed as described for high-resolution ex vivo PLA signals except that no tissue autofluorescence signal was measured or needed to be subtracted and, when appropriate, other IF signals (e.g. FLAG) were measured in parallel. For cells on Matrigen HTS plates (Fig. 2, C and D), the confluence of cells prevented identifying individual cell boundaries. Instead, the PLA signal was quantified as the PLA volume divided by the nuclear intensity for each image. Each data point represents at least three unique surfaces with at least 12 cells measured per surface. For Smad2/3–Smad4 experiments, only two independent hydrogels were analyzed for the “CTF 500 pg/ml” and “Neon 500 pg/ml” groups due to loss of hydrogels during processing. Each bar in Fig. 5D represents the mean with standard error of the mean for all the cells analyzed from that condition.
Atomic force microscopy and cell morphology
Experiments were conducted on an MFP-3D AFM (Asylum Research, Santa Barbara, CA) on an inverted optical microscope (Nikon, Melville, NY). MCLT O-10 cantilevers (Bruker Nano, Camarillo, CA) were functionalized manually with 4.47-μm polystyrene beads. Probes were calibrated before each experiment using a combination of glass indention and thermal fluctuation measurements. Cellular and hydrogel measurements were acquired using a 2 nanonewton relative force trigger with a tip velocity of 2 μm/s. Hydrogels were measured over a 400 μm2 space without cells, sampled as a 4 × 4 array. Young's modulus was calculated using custom scripts in MATLAB (TheMathWorks, Waltham, MA) that use a linearized Hertz approach for force-indentation measurements 40–60 nm into each surface. Cells and hydrogels were assumed to be incompressible (Poisson's ratio, ν = 0.5). After AFM analysis, cells were fixed in 4% paraformaldehyde for 10 min and stained with 1:40 phalloidin-488 and 1:1000 Hoechst stain 33342 for 30 min. Cell spread area and morphology were analyzed from images taken at ×63 in Volocity (as above). Each data point represents at least three unique surfaces with at least 10 cells per surface.
Mass spectroscopy on LEMD3 fragments
LEMD3 fragments were affinity-purified from RIPA-extracted lysates using FLAG (clone M2, no. A2220, Sigma, Darmstadt, Germany) or V5 (clone V5-10, no. A7345, Sigma, Darmstadt, Germany) affinity agarose according to the instructions of the manufacturer. Purified fragments were separated by SDS-PAGE on a 4–12% BisTris gel in 1× MES buffer and visualized by SimplyBlue staining (ThermoFisher Scientific). In gel digestion, nano-LC-MS/MS, and peptide identification was performed as described previously (82) with the following modifications. Reverse-phase chromatography was performed using an in-house packed column (40 cm long × 75 μm inner diameter × 360 outer diameter, Dr. Maisch GmbH ReproSil-Pur 120 C18-AQ 1.9-μm beads) and a 120-min gradient. The Raw files were searched using the Mascot algorithm (version 2.5.1) against a protein database constructed by combining the FASTA file for LEMD3 with a contaminant database (cRAP, downloaded November 21, 2016 from https://www.thegpm.org/crap/index.html) 4 via Proteome Discoverer 2.1. Only peptide spectral matches with expectation value of less than 0.01 (“high confidence”) were used.
Author contributions
Dwight M. Chambers and T. H. B. conceptualization; Dwight M. Chambers, L. M., P. J. S., and T. H. B. resources; Dwight M. Chambers, L. M., J. J. Z., S. W. C., and Davis M. Chambers data curation; Dwight M. Chambers software; Dwight M. Chambers, L. M., J. J. Z., S. W. C., Davis M. Chambers, and T. H. B. formal analysis; Dwight M. Chambers, P. J. S., and T. H. B. funding acquisition; Dwight M. Chambers, P. J. S., and T. H. B. validation; Dwight M. Chambers, L. M., J. J. Z., S. W. C., and Davis M. Chambers investigation; Dwight M. Chambers, P. J. S., and T. H. B. visualization; Dwight M. Chambers and L. M. methodology; Dwight M. Chambers writing-original draft; Dwight M. Chambers, P. J. S., and T. H. B. project administration; Dwight M. Chambers, L. M., J. J. Z., S. W. C., Davis M. Chambers, P. J. S., and T. H. B. writing-review and editing; P. J. S. and T. H. B. supervision.
Supplementary Material
Acknowledgments
Use of the Bio-Rad ChemiDoc MP system was kindly provided by Dr. Ravi Kane and his laboratory. Use of the StepOnePlus system was kindly provided by Dr. Johnna Temenoff and her laboratory. The siGFP reagent was a kind gift from the lab of Dr. M. G. Finn. We are thankful for helpful conversations regarding PLA with Emmeline Blanchard and for advice regarding protease inhibitors from Chris Rivera, Andrew Shockey, and Akia Parks from Dr. Manu Platt's laboratory. Editing of the manuscript was generously provided by Dr. Chiara Zurla and Dr. Victoria Stefanelli. We acknowledge Dr. Elliot Woods for helpful conversation. The authors thank the Georgia Institute of Technology's Parker H. Petit Institute for Bioengineering and Bioscience, including the Systems Mass Spectrometry Core Facility (Dr. David Smalley), the Cellular Analysis and Cytometry Core (Sommer Durham and Nadia Boguslavsky), and the Biopolymer Characterization Core (Drs. John Robbins and Bettina Bommarius) for instrumentation use and expertise.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL127283 and R01 HL132585 (to T. H. B.) and F30 HL122065 (to Dwight M. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9.
NCBI ClinVar submission accession numbers are SCV000332734.2, SCV000341129.2, SCV000380866.2, SCV000380877.2, SCV000380879.2, and SCV000380878.2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TGFβ
- transforming growth factor-β
- ECM
- extracellular matrix
- oligo
- oligonucleotide
- SDM
- site-directed mutagenesis
- qPCR
- quantitative PCR
- IPF
- idiopathic pulmonary fibrosis
- DCI
- 3,4-dichloroisocoumarin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HFF
- human foreskin fibroblast
- HRP
- horseradish peroxidase
- PLA
- proximity ligation assay
- MMP
- matrix metallo-proteinase
- MMPi
- matrix metallo-proteinase inhibitor
- ANOVA
- analysis of variance
- LINC
- linker of nucleus and cytoskeleton
- BOS
- Buschke-Ollendorf syndrome
- RRM
- RNA recognition motif
- BMP
- bone morphogenic protein
- CTF
- C-terminal fragment
- kPa
- kilopascal
- AFM
- atomic force microscope
- ROCK
- Rho-associated protein kinase
- PSM
- peptide spectral match
- DN
- dominant-negative
- TC
- tissue culture, plastic
- IF
- immunofluorescence.
References
- 1. Zeisberg M., and Kalluri R. (2013) Cellular mechanisms of tissue fibrosis. 1. common and organ-specific mechanisms associated with tissue fibrosis. Am. J. Physiol. Cell Physiol. 304, C216–C225 10.1152/ajpcell.00328.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morikawa M., Derynck R., and Miyazono K. (2016) Tgf-β and the tgf-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8, a021873 10.1101/cshperspect.a021873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walton K. L., Johnson K. E., and Harrison C. A. (2017) Targeting tgf-β mediated smad signaling for the prevention of fibrosis. Front. Pharmacol. 8, 461 10.3389/fphar.2017.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. King T. E. Jr., Bradford W. Z., Castro-Bernardini S., Fagan E. A., Glaspole I., Glassberg M. K., Gorina E., Hopkins P. M., Kardatzke D., Lancaster L., Lederer D. J., Nathan S. D., Pereira C. A., Sahn S. A., Sussman R., et al. (2014) A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N. Engl. J. Med. 370, 2083–2092 10.1056/NEJMoa1402582 [DOI] [PubMed] [Google Scholar]
- 5. Stahnke T., Kowtharapu B. S., Stachs O., Schmitz K. P., Wurm J., Wree A., Guthoff R. F., and Hovakimyan M. (2017) Suppression of tgf-β pathway by pirfenidone decreases extracellular matrix deposition in ocular fibroblasts in vitro. PLoS ONE 12, e0172592 10.1371/journal.pone.0172592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vancheri C., Kreuter M., Richeldi L., Ryerson C. J., Valeyre D., Grutters J. C., Wiebe S., Stansen W., Quaresma M., Stowasser S., Wuyts W. A., and INJOURNEY Trial Investigators. (2018) Nintedanib with add-on pirfenidone in idiopathic pulmonary fibrosis: results of the in journey trial. Am. J. Respir. Crit. Care Med. 197, 356–363 [DOI] [PubMed] [Google Scholar]
- 7. Du J., Paz K., Flynn R., Vulic A., Robinson T. M., Lineburg K. E., Alexander K. A., Meng J., Roy S., Panoskaltsis-Mortari A., Loschi M., Hill G. R., Serody J. S., Maillard I., Miklos D., et al. (2017) Pirfenidone ameliorates murine chronic gvhd through inhibition of macrophage infiltration and tgf-β production. Blood 129, 2570–2580 10.1182/blood-2017-01-758854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wipff P. J., Rifkin D. B., Meister J. J., and Hinz B. (2007) Myofibroblast contraction activates latent tgf-β1 from the extracellular matrix. J. Cell Biol. 179, 1311–1323 10.1083/jcb.200704042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klingberg F., Chow M. L., Koehler A., Boo S., Buscemi L., Quinn T. M., Costell M., Alman B. A., Genot E., and Hinz B. (2014) Prestress in the extracellular matrix sensitizes latent tgf-β1 for activation. J. Cell Biol. 207, 283–297 10.1083/jcb.201402006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olsen A. L., Bloomer S. A., Chan E. P., Gaça M. D., Georges P. C., Sackey B., Uemura M., Janmey P. A., and Wells R. G. (2011) Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G110–G118 10.1152/ajpgi.00412.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park J. S., Chu J. S., Tsou A. D., Diop R., Tang Z., Wang A., and Li S. (2011) The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to tgf-β. Biomaterials 32, 3921–3930 10.1016/j.biomaterials.2011.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi Y., Dong Y., Duan Y., Jiang X., Chen C., and Deng L. (2013) Substrate stiffness influences tgf-β1-induced differentiation of bronchial fibroblasts into myofibroblasts in airway remodeling. Mol. Med. Rep. 7, 419–424 10.3892/mmr.2012.1213 [DOI] [PubMed] [Google Scholar]
- 13. Szeto S. G., Narimatsu M., Lu M., He X., Sidiqi A. M., Tolosa M. F., Chan L., De Freitas K., Bialik J. F., Majumder S., Boo S., Hinz B., Dan Q., Advani A., John R., et al. (2016) Yap/taz are mechanoregulators of tgf-β-smad signaling and renal fibrogenesis. J. Am. Soc. Nephrol. 27, 3117–3128 10.1681/ASN.2015050499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin F., Morrison J. M., Wu W., and Worman H. J. (2005) Man1, an integral protein of the inner nuclear membrane, binds smad2 and smad3 and antagonizes transforming growth factor-β signaling. Hum. Mol. Genet. 14, 437–445 10.1093/hmg/ddi040 [DOI] [PubMed] [Google Scholar]
- 15. Kondé E., Bourgeois B., Tellier-Lebegue C., Wu W., Pérez J., Caputo S., Attanda W., Gasparini S., Charbonnier J. B., Gilquin B., Worman H. J., and Zinn-Justin S. (2010) Structural analysis of the smad2-man1 interaction that regulates transforming growth factor-β signaling at the inner nuclear membrane. Biochemistry 49, 8020–8032 10.1021/bi101153w [DOI] [PubMed] [Google Scholar]
- 16. Bourgeois B., Gilquin B., Tellier-Lebègue C., Östlund C., Wu W., Pérez J., El Hage P., Lallemand F., Worman H. J., and Zinn-Justin S. (2013) Inhibition of tgf-β signaling at the nuclear envelope: characterization of interactions between man1, smad2 and smad3, and ppm1a. Sci. Signal. 6, ra49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bermeo S., Al-Saedi A., Kassem M., Vidal C., and Duque G. (2017) The role of the nuclear envelope protein man1 in mesenchymal stem cell differentiation. J. Cell. Biochem. 118, 4425–4435 10.1002/jcb.26096 [DOI] [PubMed] [Google Scholar]
- 18. Osada S., Ohmori S. Y., and Taira M. (2003) Xman1, an inner nuclear membrane protein, antagonizes bmp signaling by interacting with smad1 in Xenopus embryos. Development 130, 1783–1794 10.1242/dev.00401 [DOI] [PubMed] [Google Scholar]
- 19. Raju G. P., Dimova N., Klein P. S., and Huang H. C. (2003) Sane, a novel lem domain protein, regulates bone morphogenetic protein signaling through interaction with smad1. J. Biol. Chem. 278, 428–437 10.1074/jbc.M210505200 [DOI] [PubMed] [Google Scholar]
- 20. Hellemans J., Preobrazhenska O., Willaert A., Debeer P., Verdonk P. C., Costa T., Janssens K., Menten B., Van Roy N., Vermeulen S. J., Savarirayan R., Van Hul W., Vanhoenacker F., Huylebroeck D., De Paepe A., et al. (2004) Loss-of-function mutations in lemd3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 36, 1213–1218 10.1038/ng1453 [DOI] [PubMed] [Google Scholar]
- 21. Pan D., Estévez-Salmerón L. D., Stroschein S. L., Zhu X., He J., Zhou S., and Luo K. (2005) The integral inner nuclear membrane protein man1 physically interacts with the r-smad proteins to repress signaling by the transforming growth factor-β superfamily of cytokines. J. Biol. Chem. 280, 15992–16001 10.1074/jbc.M411234200 [DOI] [PubMed] [Google Scholar]
- 22. Lin X., Duan X., Liang Y.-Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y.-G., Meng A., and Feng X.-H. (2006) Ppm1a functions as a smad phosphatase to terminate tgfβ signaling. Cell 125, 915–928 10.1016/j.cell.2006.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yadegari M., Whyte M. P., Mumm S., Phelps R. G., Shanske A., Totty W. G., and Cohen S. R. (2010) Buschke-Ollendorff syndrome: absence of lemd3 mutation in an affected family. Arch Dermatol. 146, 63–68 [DOI] [PubMed] [Google Scholar]
- 24. Couto A. R., Bruges-Armas J., Peach C. A., Chapman K., Brown M. A., Wordsworth B. P., and Zhang Y. (2007) A novel lemd3 mutation common to patients with osteopoikilosis with and without melorheostosis. Calcif. Tissue Int. 81, 81–84 10.1007/s00223-007-9043-z [DOI] [PubMed] [Google Scholar]
- 25. Pope V., Dupuis L., Kannu P., Mendoza-Londono R., Sajic D., So J., Yoon G., and Lara-Corrales I. (2016) Buschke-Ollendorff syndrome: a novel case series and systematic review. Br. J. Dermatol. 174, 723–729 10.1111/bjd.14366 [DOI] [PubMed] [Google Scholar]
- 26. Gass J. K., Hellemans J., Mortier G., Griffiths M., and Burrows N. P. (2008) Buschke-Ollendorff syndrome: a manifestation of a heterozygous nonsense mutation in the lemd3 gene. J. Am. Acad. Dermatol. 58, Suppl. 1, S103–S104 10.1016/j.jaad.2007.03.031 [DOI] [PubMed] [Google Scholar]
- 27. Yuste-Chaves M., Cañueto J., Santos-Briz Á., Ciria S., González-Sarmiento R., and Unamuno P. (2011) Buschke-Ollendorff syndrome with striking phenotypic variation resulting from a novel c.2203c>t nonsense mutation in lemd3. Pediatr. Dermatol. 28, 447–450 10.1111/j.1525-1470.2010.01206.x [DOI] [PubMed] [Google Scholar]
- 28. Burger B., Hershkovitz D., Indelman M., Kovac M., Galambos J., Haeusermann P., Sprecher E., and Itin P. H. (2010) Buschke-Ollendorff syndrome in a three-generation family: influence of a novel lemd3 mutation to tropoelastin expression. Eur. J. Dermatol. 20, 693–697 [DOI] [PubMed] [Google Scholar]
- 29. Korekawa A., Nakano H., Toyomaki Y., Takiyoshi N., Rokunohe D., Akasaka E., Nakajima K., and Sawamura D. (2012) Buschke-Ollendorff syndrome associated with hypertrophic scar formation: a possible role for lemd3 mutation. Br. J. Dermatol. 166, 900–903 10.1111/j.1365-2133.2011.10691.x [DOI] [PubMed] [Google Scholar]
- 30. Gutierrez D., Cooper K. D., Mitchell A. L., and Cohn H. I. (2015) Novel somatic mutation in lemd3 splice site results in Buschke-Ollendorff syndrome with polyostotic melorheostosis and osteopoikilosis. Pediatr. Dermatol. 32, e219–e220 10.1111/pde.12634 [DOI] [PubMed] [Google Scholar]
- 31. Kratzsch J., Mitter D., Ziemer M., Kohlhase J., and Voth H. (2016) Identification of a novel point mutation in the lemd3 gene in an infant with Buschke-Ollendorff syndrome. JAMA Dermatol. 152, 844–845 10.1001/jamadermatol.2016.0350 [DOI] [PubMed] [Google Scholar]
- 32. Condorelli A., Musso N., Scuderi L., Condorelli D. F., Barresi V., and De Pasquale R. (2017) Juvenile elastoma without germline mutations in lemd3 gene: a case of Buschke-Ollendorff syndrome?. Pediatr. Dermatol. 34, e345–e346 10.1111/pde.13287 [DOI] [PubMed] [Google Scholar]
- 33. Wu W., Lin F., and Worman H. J. (2002) Intracellular trafficking of man1, an integral protein of the nuclear envelope inner membrane. J. Cell Sci. 115, 1361–1371 [DOI] [PubMed] [Google Scholar]
- 34. Mansharamani M., and Wilson K. L. (2005) Direct binding of nuclear membrane protein man1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J. Biol. Chem. 280, 13863–13870 10.1074/jbc.M413020200 [DOI] [PubMed] [Google Scholar]
- 35. Liu J., Lee K. K., Segura-Totten M., Neufeld E., Wilson K. L., and Gruenbaum Y. (2003) Man1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 100, 4598–4603 10.1073/pnas.0730821100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ostlund C., Sullivan T., Stewart C. L., and Worman H. J. (2006) Dependence of diffusional mobility of integral inner nuclear membrane proteins on a-type lamins. Biochemistry 45, 1374–1382 10.1021/bi052156n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wagner N., Kagermeier B., Loserth S., and Krohne G. (2006) The Drosophila melanogaster lem-domain protein man1. Eur. J. Cell Biol. 85, 91–105 10.1016/j.ejcb.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 38. Starr D. A., and Han M. (2002) Role of anc-1 in tethering nuclei to the actin cytoskeleton. Science 298, 406–409 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- 39. Padmakumar V. C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A. A., Gotzmann J., Foisner R., and Karakesisoglou I. (2005) The inner nuclear membrane protein sun1 mediates the anchorage of nesprin-2 to the nuclear envelope. J. Cell Sci. 118, 3419–3430 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- 40. Lombardi M. L., Jaalouk D. E., Shanahan C. M., Burke B., Roux K. J., and Lammerding J. (2011) The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286, 26743–26753 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caputo S., Couprie J., Duband-Goulet I., Kondé E., Lin F., Braud S., Gondry M., Gilquin B., Worman H. J., and Zinn-Justin S. (2006) The carboxyl-terminal nucleoplasmic region of man1 exhibits a DNA binding winged helix domain. J. Biol. Chem. 281, 18208–18215 10.1074/jbc.M601980200 [DOI] [PubMed] [Google Scholar]
- 42. Zurla C., Jung J., Blanchard E. L., and Santangelo P. J. (2017) in Enhancer RNAs: Methods and Protocols (Ørom U. A., ed) pp. 155–170, Springer, New York [Google Scholar]
- 43. Booth A. J., Hadley R., Cornett A. M., Dreffs A. A., Matthes S. A., Tsui J. L., Weiss K., Horowitz J. C., Fiore V. F., Barker T. H., Moore B. B., Martinez F. J., Niklason L. E., and White E. S. (2012) Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am. J. Respir. Crit. Care Med. 186, 866–876 10.1164/rccm.201204-0754OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harper J. W., Hemmi K., and Powers J. C. (1985) Reaction of serine proteases with substituted isocoumarins: discovery of 3,4-dichloroisocoumarin, a new general mechanism based serine protease inhibitor. Biochemistry 24, 1831–1841 10.1021/bi00329a005 [DOI] [PubMed] [Google Scholar]
- 45. Collins G. A., and Goldberg A. L. (2017) The logic of the 26s proteasome. Cell 169, 792–806 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song J., Tan H., Perry A. J., Akutsu T., Webb G. I., Whisstock J. C., and Pike R. N. (2012) Prosper: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS ONE 7, e50300 10.1371/journal.pone.0050300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peter M., Nakagawa J., Dorée M., Labbé J. C., and Nigg E. A. (1990) In vitro disassembly of the nuclear lamina and m phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61, 591–602 10.1016/0092-8674(90)90471-P [DOI] [PubMed] [Google Scholar]
- 48. Hill C. S. (2009) Nucleocytoplasmic shuttling of smad proteins. Cell Res 19, 36–46 10.1038/cr.2008.325 [DOI] [PubMed] [Google Scholar]
- 49. Massagué J. (2012) Tgfβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jones M. G., Fletcher S., and Richeldi L. (2013) Idiopathic pulmonary fibrosis: recent trials and current drug therapy. Respiration 86, 353–363 10.1159/000356958 [DOI] [PubMed] [Google Scholar]
- 51. Richeldi L., du Bois R. M., Raghu G., Azuma A., Brown K. K., Costabel U., Cottin V., Flaherty K. R., Hansell D. M., Inoue Y., Kim D. S., Kolb M., Nicholson A. G., Noble P. W., Selman M., et al. (2014) Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med. 370, 2071–2082 10.1056/NEJMoa1402584 [DOI] [PubMed] [Google Scholar]
- 52. Liu F., Mih J. D., Shea B. S., Kho A. T., Sharif A. S., Tager A. M., and Tschumperlin D. J. (2010) Feedback amplification of fibrosis through matrix stiffening and cox-2 suppression. J. Cell Biol. 190, 693–706 10.1083/jcb.201004082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu F., Lagares D., Choi K. M., Stopfer L., Marinković A., Vrbanac V., Probst C. K., Hiemer S. E., Sisson T. H., Horowitz J. C., Rosas I. O., Fredenburgh L. E., Feghali-Bostwick C., Varelas X., Tager A. M., and Tschumperlin D. J. (2015) Mechanosignaling through yap and taz drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L344–L357 10.1152/ajplung.00300.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y., Yella J., Chen J., McCormack F. X., Madala S. K., and Jegga A. G. (2017) Unsupervised gene expression analyses identify ipf-severity correlated signatures, associated genes and biomarkers. BMC Pulm. Med. 17, 133 10.1186/s12890-017-0472-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. DePianto D. J., Chandriani S., Abbas A. R., Jia G., N′Diaye E. N., Caplazi P., Kauder S. E., Biswas S., Karnik S. K., Ha C., Modrusan Z., Matthay M. A., Kukreja J., Collard H. R., Egen J. G., et al. (2015) Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax 70, 48–56 10.1136/thoraxjnl-2013-204596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nance T., Smith K. S., Anaya V., Richardson R., Ho L., Pala M., Mostafavi S., Battle A., Feghali-Bostwick C., Rosen G., and Montgomery S. B. (2014) Transcriptome analysis reveals differential splicing events in ipf lung tissue. PLOS ONE 9, e92111 10.1371/journal.pone.0092111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin F., Blake D. L., Callebaut I., Skerjanc I. S., Holmer L., McBurney M. W., Paulin-Levasseur M., and Worman H. J.. Man1, an inner nuclear membrane protein that shares the lem domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275, 4840–4847 [DOI] [PubMed] [Google Scholar]
- 58. Kristensen J. H., Karsdal M. A., Sand J. M., Willumsen N., Diefenbach C., Svensson B., Hägglund P., and Oersnes-Leeming D. J. (2015) Serological assessment of neutrophil elastase activity on elastin during lung ecm remodeling. BMC Pulm. Med 15, 53 10.1186/s12890-015-0048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bühling F., Röcken C., Brasch F., Hartig R., Yasuda Y., Saftig P., Brömme D., and Welte T. (2004) Pivotal role of cathepsin k in lung fibrosis. Am J Pathol 164, 2203–2216 10.1016/S0002-9440(10)63777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Srivastava M., Steinwede K., Kiviranta R., Morko J., Hoymann H. G., Länger F., Buhling F., Welte T., and Maus U. A. (2008) Overexpression of cathepsin k in mice decreases collagen deposition and lung resistance in response to bleomycin-induced pulmonary fibrosis. Respir. Res. 9, 54 10.1186/1465-9921-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fukuda Y., Ishizaki M., Kudoh S., Kitaichi M., and Yamanaka N. (1998) Localization of matrix metalloproteinases-1, -2, and -9 and tissue inhibitor of metalloproteinase-2 in interstitial lung diseases. Lab. Invest. 78, 687–698 [PubMed] [Google Scholar]
- 62. Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., Werb Z., and Bissell M. J. (2005) Rac1b and reactive oxygen species mediate mmp-3-induced emt and genomic instability. Nature 436, 123–127 10.1038/nature03688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yamashita C. M., Dolgonos L., Zemans R. L., Young S. K., Robertson J., Briones N., Suzuki T., Campbell M. N., Gauldie J., Radisky D. C., Riches D. W., Yu G., Kaminski N., McCulloch C. A., and Downey G. P. (2011) Matrix metalloproteinase 3 is a mediator of pulmonary fibrosis. Am. J. Pathol. 179, 1733–1745 10.1016/j.ajpath.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. García-Prieto E., González-López A., Cabrera S., Astudillo A., Gutiérrez-Fernández A., Fanjul-Fernandez M., Batalla-Solís E., Puente X. S., Fueyo A., López-Otín C., and Albaiceta G. M. (2010) Resistance to bleomycin-induced lung fibrosis in mmp-8 deficient mice is mediated by interleukin-10. PLoS ONE 5, e13242 10.1371/journal.pone.0013242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zuo F., Kaminski N., Eugui E., Allard J., Yakhini Z., Ben-Dor A., Lollini L., Morris D., Kim Y., DeLustro B., Sheppard D., Pardo A., Selman M., and Heller R. A. (2002) Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc. Natl. Acad. Sci. U.S.A. 99, 6292–6297 10.1073/pnas.092134099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kamaraju A. K., and Roberts A. B. (2005) Role of rho/rock and p38 map kinase pathways in transforming growth factor-β-mediated smad-dependent growth inhibition of human breast carcinoma cells in vivo. J. Biol. Chem. 280, 1024–1036 10.1074/jbc.M403960200 [DOI] [PubMed] [Google Scholar]
- 67. Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B. M., Dembowy J., Yaffe M. B., Zandstra P. W., and Wrana J. L. (2008) Taz controls smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837–848 10.1038/ncb1748 [DOI] [PubMed] [Google Scholar]
- 68. Samarakoon R., Higgins S. P., Higgins C. E., and Higgins P. J. (2008) Tgf-β1-induced plasminogen activator inhibitor-1 expression in vascular smooth muscle cells requires pp60(c-src)/egfr(y845) and rho/rock signaling. J. Mol. Cell. Cardiol. 44, 527–538 10.1016/j.yjmcc.2007.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grannas K., Arngården L., Lönn P., Mazurkiewicz M., Blokzijl A., Zieba A., and Söderberg O. (2015) Crosstalk between hippo and tgfβ: Subcellular localization of yap/taz/smad complexes. J. Mol. Biol. 427, 3407–3415 10.1016/j.jmb.2015.04.015 [DOI] [PubMed] [Google Scholar]
- 70. Beyer T. A., Weiss A., Khomchuk Y., Huang K., Ogunjimi A. A., Varelas X., and Wrana J. L. (2013) Switch enhancers interpret tgf-β and hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624 10.1016/j.celrep.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 71. Landrum M. J., Lee J. M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., Jang W., Katz K., Ovetsky M., Riley G., Sethi A., et al. (2016) Clinvar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rajgor D., Mellad J. A., Autore F., Zhang Q., and Shanahan C. M. (2012) Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS ONE 7, e40098 10.1371/journal.pone.0040098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Santos M., Domingues S. C., Costa P., Muller T., Galozzi S., Marcus K., da Cruz e Silva E. F., da Cruz e Silva O. A., and Rebelo S. (2014) Identification of a novel human lap1 isoform that is regulated by protein phosphorylation. PLoS ONE 9, e113732 10.1371/journal.pone.0113732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Abascal F., Tress M. L., and Valencia A. (2015) Alternative splicing and co-option of transposable elements: the case of tmpo/lap2α and znf451 in mammals. Bioinformatics 31, 2257–2261 10.1093/bioinformatics/btv132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang W., Bai T., Zhang S., Xu S., Chen H., and Li C. (2017) Isoforms of the nuclear envelope protein nurim are differentially expressed during heart development in mice. Gene 627, 123–128 10.1016/j.gene.2017.06.009 [DOI] [PubMed] [Google Scholar]
- 76. Zhang Q., Skepper J. N., Yang F., Davies J. D., Hegyi L., Roberts R. G., Weissberg P. L., Ellis J. A., and Shanahan C. M. (2001) Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114, 4485–4498 [DOI] [PubMed] [Google Scholar]
- 77. Holt I., Duong N. T., Zhang Q., Lam le T., Sewry C. A., Mamchaoui K., Shanahan C. M., and Morris G. E. (2016) Specific localization of nesprin-1-α2, the short isoform of nesprin-1 with a kash domain, in developing, fetal and regenerating muscle, using a new monoclonal antibody. BMC Cell Biol. 17, 26 10.1186/s12860-016-0105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kobayashi Y., Katanosaka Y., Iwata Y., Matsuoka M., Shigekawa M., and Wakabayashi S. (2006) Identification and characterization of gsrp-56, a novel Golgi-localized spectrin repeat-containing protein. Exp. Cell Res. 312, 3152–3164 10.1016/j.yexcr.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 79. Rajgor D., Mellad J. A., Soong D., Rattner J. B., Fritzler M. J., and Shanahan C. M. (2014) Mammalian microtubule p-body dynamics are mediated by nesprin-1. J. Cell Biol. 205, 457–475 10.1083/jcb.201306076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rajgor D., Hanley J. G., and Shanahan C. M. (2016) Identification of novel nesprin-1 binding partners and cytoplasmic matrin-3 in processing bodies. Mol. Biol. Cell 27, 3894–3902 10.1091/mbc.e16-06-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ruijter J. M., Lorenz P., Tuomi J. M., Hecker M., and van den Hoff M. J. (2014) Fluorescent-increase kinetics of different fluorescent reporters used for qpcr depend on monitoring chemistry, targeted sequence, type of dna input and pcr efficiency. Mikrochim. Acta 181, 1689–1696 10.1007/s00604-013-1155-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rinker T. E., Philbrick B. D., Hettiaratchi M. H., Smalley D. M., McDevitt T. C., and Temenoff J. S. (2018) Microparticle-mediated sequestration of cell-secreted proteins to modulate chondrocytic differentiation. Acta Biomater. 68, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rho H.-W., Lee B.-C., Choi E.-S., Choi I.-J., Lee Y.-S., and Goh S.-H. (2010) Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qpcr. BMC Cancer 10, 240 10.1186/1471-2407-10-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gardinassi L. G., Garcia G. R., Costa C. H. N., Costa Silva V., and de Miranda Santos I. K. F. (2016) Blood transcriptional profiling reveals immunological signatures of distinct states of infection of humans with leishmania infantum. PLOS Negl. Trop. Dis. 10, e0005123 10.1371/journal.pntd.0005123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.