Abstract
TAR DNA-binding protein of 43 kDa (TDP-43) forms pathological aggregates in neurodegenerative diseases, particularly in certain forms of frontotemporal dementia and amyotrophic lateral sclerosis. Pathological modifications of TDP-43 include proteolytic fragmentation, phosphorylation, and ubiquitinylation. A pathognomonic TDP-43 C-terminal fragment (CTF) spanning amino acids 193–414 contains only four lysine residues that could be potentially ubiquitinylated. Here, serial mutagenesis of these four lysines to arginine revealed that not a single residue is responsible for the ubiquitinylation of mCherry-tagged CTF. Removal of all four lysines was necessary to suppress ubiquitinylation. Interestingly, Lys-408 substitution enhanced the pathological phosphorylation of the immediately adjacent serine residues 409/410 in the context of mCherry-CTF. Thus, Lys-408 ubiquitinylation appears to hinder Ser-409/410 phosphorylation in TDP-43 CTF. However, we did not observe the same effect for full-length TDP-43. We extended the mutagenesis study to full-length TDP-43 and performed MS. Ubiquitinylated lysine residues were identified in the nuclear localization sequence (NLS; Lys-84 and Lys-95) and RNA-binding region (mostly Lys-160, Lys-181, and Lys-263). Mutagenesis of Lys-84 confirmed its importance as the major determinant for nuclear import, whereas Lys-95 mutagenesis did not significantly affect TDP-43's nucleo-cytoplasmic distribution, solubility, aggregation, and RNA-processing activities. Nevertheless, the K95A mutant had significantly reduced Ser-409/410 phosphorylation, emphasizing the suspected interplay between TDP-43 ubiquitinylation and phosphorylation. Collectively, our analysis of TDP-43 ubiquitinylation sites indicates that the NLS residues Lys-84 and Lys-95 have more prominent roles in TDP-43 function than the more C-terminal lysines and suggests a link between specific ubiquitinylation events and pathological TDP-43 phosphorylation.
Keywords: proteasome, protein aggregation, protein phosphorylation, ubiquitylation (ubiquitination), mass spectrometry (MS), site-directed mutagenesis, amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), neurodegeneration, frontotemporal dementia, TDP-43
Introduction
The TAR DNA-binding protein of 43 kDa (TDP-43) is the main protein component of pathological inclusions in the brain and spinal cord of almost all cases of amyotrophic lateral sclerosis (ALS) 2 and in ∼45% of frontotemporal lobar degeneration (FTLD-TDP) (1–3). This ubiquitously expressed DNA/RNA-binding protein contains two highly conserved RNA recognition motifs (RRM1 and RRM2), a C-terminal low complexity, glycine-rich domain (GRD) that is important for its protein–protein interactions but also pathological aggregation. TDP-43 has a bipartite nuclear localization signal (NLS) and a nuclear export signal (NES), which allow the protein to shuttle between the nucleus and cytoplasm, although TDP-43 is predominantly located in the nucleus. Under physiological conditions, TDP-43 regulates mRNA processing, stability, trafficking, and miRNA biogenesis, autoregulates its own expression, and translocates into cytoplasmic stress granules under conditions of stress (4, 5).
Many neurodegenerative diseases exhibit insoluble protein inclusions with the characterizing protein often being strongly ubiquitinylated and/or phosphorylated. In ALS and FTLD-TDP, TDP-43 is found ubiquitinylated, hyperphosphorylated, mislocalized to the cytoplasm and cleared from the nucleus, proteolytically cleaved into 25- and 35-kDa C-terminal fragments (CTFs), and aggregated into insoluble inclusions (1, 2, 6). Dual phosphorylation at serine residues 409/410 (Ser-409/410) is a pathological marker for abnormal TDP-43 inclusions (6–8). Moreover, in 1–2% of total ALS cases, more than 50 pathogenic TDP-43 mutations were identified, mainly in the aggregation-prone CTF (9). Also, ubiquitinylated and phosphorylated TDP-43 was found in other neurodegenerative diseases like Alzheimer's disease, corticobasal degeneration, and progressive supranuclear palsy (10–12).
The post-translational modifications that TDP-43 undergoes in disease or disease-mimicking models, i.e. phosphorylation, ubiquitinylation, truncation, SUMOylation, acetylation, oxidation, and deamidation (1, 13–15), have been studied to varying extent. As for phosphorylation, many sites of modification are known (6–8, 13), and the impact on TDP-43 toxicity and aggregation, mainly of Ser-409/410 phosphorylation, was studied to a certain degree (15).
In contrast to the diverse TDP-43 phosphorylation sites, only few specific ubiquitinylation sites have been identified so far (Table 1), and their distinct roles on solubility, localization, and aggregation and also their physiological functions are not understood. Ubiquitinylations may not only influence protein degradation but can also affect protein localization, endocytosis, DNA repair, and protein activity and can mediate protein–protein interactions (16, 17).
Table 1.
Published TDP-43 ubiquitinylation sites
| Ubiquitinylation sites | TDP-43 species | Cell type | Treatment | Method | Refs. |
|---|---|---|---|---|---|
| Lys-102, Lys-114, Lys-145, Lys-181 | Insoluble overexpressed TDP-43 and TDP-S6 | HEK-293 | LC-MS/MS | 30 | |
| Lys-181 | Overexpressed TDP-43-ΔNLS-Lys-145Q | QBI293 | TDP-43 IP followed by nano-LC/nanospray/MS/MS | 67 | |
| Lys-114 | Endogenous | HEK-293 | LC-MS/MS | 35 | |
| Lys-102, Lys-160 | Endogenous | MV4-11 | MG-132 | ||
| Lys-95, Lys-102, Lys-114, Lys-160, Lys-181 | Endogenous | HCT116 | Bortezomib | LC-MS/MS | 34 |
| Lys-102, Lys-114, Lys-121, Lys-140, Lys-145, Lys-160, Lys-176, Lys-181, Lys-263 | Endogenous | Myeloma cell lines | |||
| Lys-79 | Endogenous TDP-43 from Sarko-sylinsoluble human ALS brain | LC-MS/MS | 13 | ||
Specifically for TDP-43, ubiquitinylated species are insoluble and can be induced by inhibition of protein degradation via UPS or autophagy or by other stressors (18–27). TDP-43 is polyubiquitinylated via Lys-48- and Lys-63–linked ubiquitin chains, indicating proteasomal and autophagosomal degradation of the protein (26, 28). We have shown that TDP-43 is ubiquitinylated by the UBE2E family of ubiquitin-conjugating enzymes, and this can be reversed by the ubiquitin isopeptidase Y (29). Parkin was also suggested as a ubiquitin ligase for TDP-43 (28). Four ubiquitinylation sites in or near the RRM1 were identified by Dammer et al. (30), although their inactivation did not alter the amount and solubility of total ubiquitinylated TDP-43, emphasizing that further TDP-43 lysine residues are ubiquitinylated under pathological conditions. Therefore, it is important to identify and characterize these specific ubiquitinylated lysine residues and how their modification affects localization, solubility, and aggregation of TDP-43.
TDP-43 contains 20 lysine residues, most of which are located in the N-terminal section and RRM1, and only few lysine residues reside in TDP-43 domains that are part of truncated CTFs (Fig. 1A). We observed in our previous study that a 35-kDa CTF that includes the RRM2 and GRD (193–414 aa) is strongly ubiquitinylated (29). This CTF contains four lysine residues, narrowing down the possibly ubiquitinylated lysine residues. In this study, we investigated, with the help of single- and multiple-site lysine mutants, which of them are mainly ubiquitinylated and how their modification with ubiquitin affects localization, solubility, and aggregation of the CTF. The mutagenesis study was then extended to full-length TDP-43 (TDP-43 FL) and the hyper-ubiquitinylated K263E mutant TDP-43, including mass spectrometric discovery of novel TDP-43 ubiquitinylation sites. Our extensive systematic study identified no single functionally relevant ubiquitinylation site of TDP-43 but yielded novel detailed findings. In the context of the particularly aggregation-prone mCherry-CTF, removal of the Lys-408 ubiquitinylation site by arginine substitution facilitated phosphorylation of the immediately adjacent Ser-409/410, but that was not detectable in TDP-43 FL. For TDP-43 FL we noticed effects on TDP-43 modifications for the NLS residues Lys-84 and Lys-95 discovered here as novel ubiquitinylation sites.
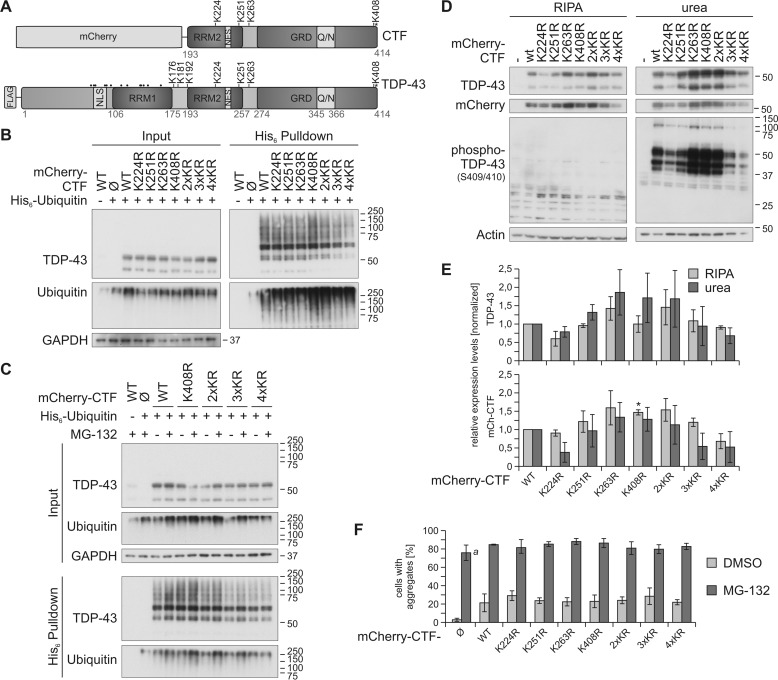
Figure 1.
Site-specific ubiquitinylation of C-terminal lysine residues. A, schematic overview of the mCherry-CTF and FLAG–TDP-43 constructs used in this study. The positions of all 20 lysine residues are depicted; C-terminally located lysine residues are indicated with K, and all other lysine residues are indicated with dots. The phosphorylation site Ser-409/410 is labeled with asterisks. Q/N, glutamate/asparagine-rich prion-like region; and NLS, nuclear localization signal (aa 82–98). B, His6-ubiquitin was co-expressed with mCherry-control (Ø), mCherry-CTFWT, or the indicated mCherry -CTF lysine mutants in HEK293E cells. Cells were lysed harshly with 8 m urea lysis buffer followed by Ni-NTA–affinity purification of His6-ubiquitin–conjugated proteins, and total protein and eluates were analyzed by Western blotting with antibodies detecting TDP-43, ubiquitin, and GAPDH. C, HEK293E cells were transfected with His6-ubiquitin and mCherry-CTFWT, mCherry-CTFK408R, and mCherry-CTFK2/3/4R. After proteasomal inhibition with MG-132 for 2 h, cells were lysed and analyzed as in B. D, sequential extraction of HEK293E cells overexpressing mCherry-CTF lysine mutants and quantification. After 2 days of mCherry-CTF expression, RIPA- and urea-soluble fractions were prepared and analyzed by Western blotting with antibodies against TDP-43, mCherry, and phospho-TDP-43 (Ser-409/410) and actin as loading control. E, quantification of protein levels detected with anti-TDP (upper graph) and anti-mCherry (lower graph), respectively. Data represent the mean ± S.D.; *, p ≤ 0.05 compared with mCherry-CTFWT. F, quantification of the aggregate formation of mCherry-CTF lysine mutants upon proteasomal inhibition (Fig. S1, A and B). 50–250 cells per condition of two independent experiments were analyzed for aggregate formation. The mCherry protein alone formed many tiny aggregates upon proteasomal inhibition, which were included in the quantification (a). Data represent the mean ± S.D.
Results
Site-specific ubiquitinylation of a C-terminal TDP-43 fragment
In our previous study (29), we observed a strong ubiquitinylation of a 35-kDa CTF of TDP-43 amino acids 193–414 with an N-terminal mCherry tag. This CTF contains four of the 20 lysine residues of TDP-43 (Lys-224, Lys-251, Lys-263, and Lys-408; see Fig. 1A). We were interested whether one or several of these potential ubiquitinylation sites are mainly ubiquitinylated and how the removal of these sites affects localization, insolubility, and aggregation of the mCherry-CTF. We performed site-directed mutagenesis to exchange the four lysine residues with arginine residues individually (K224R, K251R, K263R, and K408R) and in combination (2×KR = K224R/K251R, 3×KR = K224R/K251R/K263R, 4×KR = K224R/K251R/K263R/K408R). To isolate and detect ubiquitinylated TDP-43, we have previously established a protocol in which His6-ubiquitin–conjugated proteins are isolated with Ni-NTA affinity purification and analyzed by SDS-PAGE/Western blotting (29, 31). Ubiquitinylated TDP-43 appears as mono- and polyubiquitinylated species in the form of an ∼55-kDa band and a higher molecular weight smear (HMW), respectively. Importantly, with this method we can detect insoluble ubiquitinylated proteins, as this affinity purification allows us to use cell lysates generated in strongly denaturing 8 m urea buffer instead of a mild lysis that is necessary for immunoprecipitation approaches. This is substantial for investigations of TDP-43 ubiquitinylation, because 1) we have shown before that ubiquitinylated TDP-43 is mainly insoluble (29), and 2) the ratio of ubiquitinylated to nonubiquitinylated TDP-43 is quite low and can virtually not be detected in blots of cell lysates (see inputs).
First, we studied whether one or several individual lysine residues are mainly ubiquitinylated in the mCherry-CTF. Therefore, we overexpressed lysine-to-arginine–substituted (ubiquitinylation-deficient) mCherry-CTFs in human embryonic kidney (HEK) 293E cells together with His6-tagged ubiquitin, followed by affinity purification of all His6-ubiquitinylated proteins with Ni-NTA–agarose. Western blot analysis for TDP-43 showed the same steady-state level of all mutants in the total protein fraction (Fig. 1B, input). The removal of single lysine residues did not decrease the overall ubiquitinylation of mCherry-CTF lysine mutants, represented by the HMW TDP-43 species. The ubiquitinylation of the CTFK408R mutant was increased, meaning that ubiquitinylation of one or several of the other three lysine residues should be enhanced in CTFK408R (Fig. 1B, pulldown, HMW). A reduction of CTF ubiquitinylation was detected when at least three lysine residues were mutagenized to arginine (Fig. 1, B and C; 3×KR and 4×KR). However, ubiquitinylation of the lysine-less mCherry-CTF4×KR was not totally lost. Because the mCherry tag has 24 lysine residues itself, we assume that the remaining HMW signal represents mCherry ubiquitinylation. We could not analyze the ubiquitinylation of CTFs untagged or with a short tag because of their extremely low expression levels and rapid degradation rates (our own observations).
Next, we investigated whether the ubiquitinylation of mCherry-CTFK408R, -CTF2×KR, -CTF3×KR, and -CTF4×KR is stabilized by proteasomal inhibition with MG-132 (Fig. 1C). We detected a slightly enhanced ubiquitinylation of CTFWT, CTFK408R, CTF2×KR, and CTF3×KR upon MG-132 treatment, but proteasomal inhibition showed no effect on the HMW signal strength of CTF4×KR (Fig. 1C, His6 pulldown). In summary, there is no single lysine residue that is mainly ubiquitinylated in the CTF, but all can be ubiquitinylated, and at least three ubiquitinylation sites have to be removed to reduce the overall ubiquitinylation of the CTF.
Solubility and aggregation of CTF lysine mutants
In ALS and FTLD, ubiquitinylated TDP-43 and CTFs are found in insoluble, ubiquitinylated, and phosphorylated inclusions (1, 2). Thus, we studied whether elimination of ubiquitinylation sites alters solubility, phosphorylation, and aggregation of mCherry-CTF (Fig. 1, D–F, and Fig. S1). We extracted proteins into RIPA and urea-soluble fractions from HEK293E cells overexpressing the mCherry-CTF lysine mutants (Fig. 1, D and E). Western blot analysis revealed increased insolubility of mCherry-CTFK263R, -CTFK408R, and CTF2×KR, whereas mCherry-CTFK224R and -CTF4×KR were less abundant in the RIPA and urea fractions (Fig. 1, D and E).
Because phosphorylation at Ser-409/410 is a pathological marker for TDP-43 inclusions in ALS and FTLD, we also analyzed the phosphorylation status of the CTF lysine mutants with a specific antibody (6). Ser-409/410 phosphorylation was strongly increased in mCherry-CTFK263R, -CTFK408R, and -CTF2×KR and decreased in mCherry-CTFK224R and -CTF4×KR, reflecting the insolubility pattern of these mutants. Interestingly, the insolubility and phosphorylation of CTF2×KR (CTFK224R/K251R) was increased, although the single mutant CTFK224R showed less insolubility and phosphorylation, and both properties were not altered in CTFK251R. Also, studies of aggregate formation in HEK293E cells by immunofluorescence showed that the enhanced insolubility and phosphorylation of CTFK263R, CTFK408R, and CTF2×KR were not reflected in an increase of cells with TDP-43- and phospho-TDP-43–positive aggregates when compared with CTFWT, neither under control conditions nor upon MG-132 treatment (Fig. 1F and Fig. S1). Moreover, although we detected decreased ubiquitinylation and/or insolubility of CTFK224R and CTF4×KR, the number of cells expressing these mutants with aggregates was not reduced. A possible explanation for this might be that the amount of phosphorylated and aggregated CTF per cell is altered but not the total cell numbers that exhibit aggregated TDP-43. In addition, we would like to point out that we also detected many very tiny inclusions upon MG-132 treatment in cells that expressed the control mCherry protein, which are included in the quantification (Fig. 1F and Fig. S1, B and C). However, these inclusions were phospho-TDP-43 negative (Fig. S1, B and C), and we did not observe similar inclusions in cells expressing mCherry-CTFs.
Although we did not detect an obvious difference in aggregation among the lysine mutants, we found altered solubility and phosphorylation patterns. Importantly, removing more than two ubiquitinylation sites in the 3×KR and 4×KR mutants reduced their insolubility and phosphorylation. Thus, although not a single lysine appears responsible for the shift into the insoluble fraction, the four lysines in CTF cooperatively promote this effect.
Cross-talk between C-terminal ubiquitinylation and phosphorylation of TDP-43 CTF
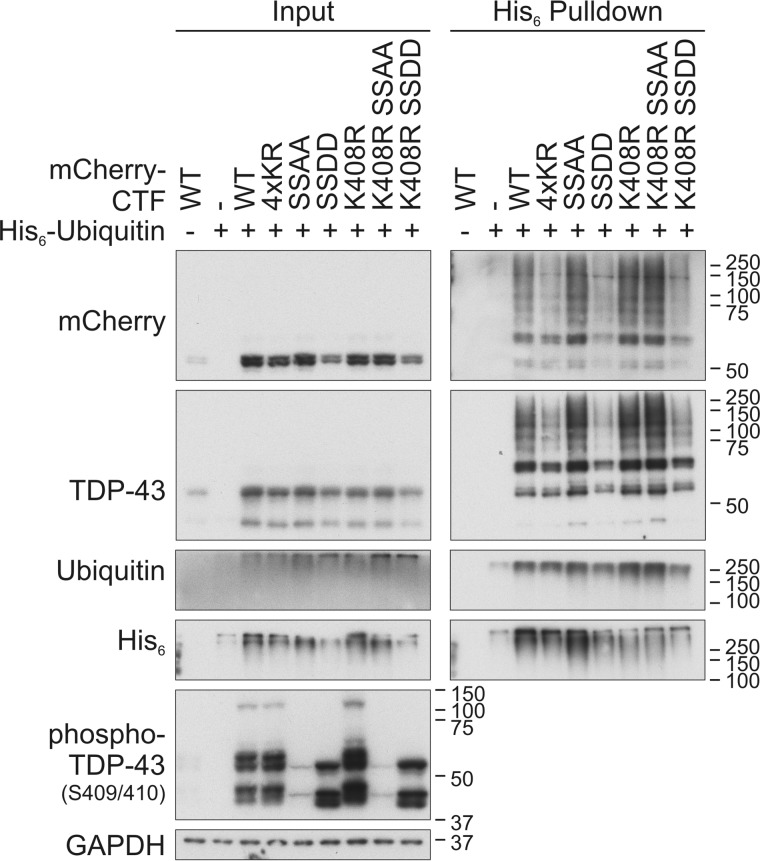
The most C-terminal lysine residue Lys-408 is located in proximity to the phosphorylation site Ser-409/410, and its mutation led to increased phosphorylation of mCherry-CTF (Figs. 1D and 2). Therefore, we asked whether there is a cross-talk between ubiquitinylation at Lys-408 and phosphorylation at Ser-409/410. To study the influence of the Ser-409/410 phosphorylation on the ubiquitinylation of mCherry-CTFK408R, we generated phosphorylation-mimic (SSDD) and phosphorylation-dead (SSAA) mCherry-CTFs in which serine residues 409/410 were mutated to aspartate and alanine residues, respectively. Additionally, the K408R mutation was introduced into the Ser-409/410 mutants. Ni-NTA pulldown of ubiquitinylated CTF mutants showed comparably increased levels of ubiquitinylated CTFK408R as well as of CTFSSAA and CTFK408R/SSAA, but we did not detect a further increase of ubiquitinylation in the double mutant (Fig. 2). The anti-phospho-TDP-43 recognized CTFSSDD on Western blots, indicating the appropriateness of the phosphomimic substitution. Interestingly, CTFSSDD and CTFK408R/SSDD showed strongly decreased ubiquitinylation, but the steady-state levels of these phosphomimic CTF mutants were also reduced (Fig. 2). Thus, Ser-409/410 phosphorylation appears not to be a priming modification enhancing ubiquitinylation of TDP-CTF.
Figure 2.
Cross-talk between C-terminal ubiquitinylation and phosphorylation of CTF. HEK293E cells were double-transfected with CTFWT, CTF4×KR, CTFK408R, and the phospho-mimic- or -dead CTFSSDD or CTFSSAA double mutants (all CTF constructs fused to mCherry) or mCherry-control vector (−). After cell lysis with urea buffer, His6-ubiquitinylated proteins were affinity-purified with Ni-NTA–agarose. Total protein (Input) and Ni-NTA–agarose eluates (His6 Pulldown) were subjected to Western blot analysis with antibodies detecting mCherry, TDP-43, ubiquitin, His6, phospho-TDP-43, and GAPDH as loading control.
The findings of increased ubiquitinylation and phosphorylation of CTFK408R, increased ubiquitinylation of CTFSSAA, and strongly decreased CTFSSDD ubiquitinylation give hints that ubiquitinylation at Lys-408 might impair phosphorylation at serines 409/410, maybe through impeded kinase binding, and that impaired phosphorylation increases insolubility and ubiquitinylation. It is also possible that the amino acid exchange at position 408 to arginine facilitates kinase binding to the C terminus of TDP-43 and thereby increases the phosphorylation at this site. All these mCherry-CTF variants distributed evenly throughout the transfected cells (Fig. S2), confirming that the observed effects are not due to different subcellular localizations.
Impact of C-terminal ubiquitinylation site mutation on ubiquitinylation in full-length TDP-43
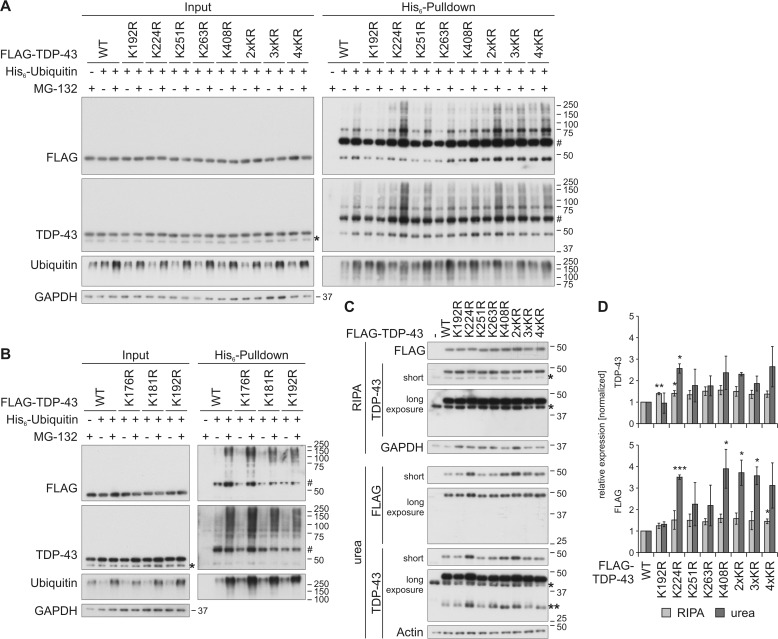
Not a single residue of the four individual lysine residues within the mCherry-CTF turned out to be a main ubiquitinylation site, but at least three lysines had to be removed to reduce ubiquitinylation of the CTF. In ALS, predominantly ubiquitinylated full-length TDP-43 is detected in pathological inclusions in spinal cord motor neurons (6, 32). Thus, we investigated the role of the C-terminal lysine residues on the ubiquitinylation of FLAG–TDP-43 FL (Fig. 3). In addition to the four lysine residues investigated above, we included three further lysine residues (Lys-176, Lys-181, and Lys-192) into our investigations, because the exact cleavage sites of pathological CTFs are not known. Additionally, Lys-181 was identified as ubiquitinylation site by MS in several studies (Table 1). Also, the following multilysine mutants were investigated: 2×KR (K224R/K251R), 3×KR (K224R/K251R/K263R), and 4×KR (K224R/K251R/K263R/K408R).
Figure 3.
Characterization of ubiquitinylation of C-terminal lysine residues in FLAG–TDP-43. A and B, HEK293E cells overexpressing His6-ubiquitin (+) or vector control (−), and FLAG–TDP-43WT or indicated lysine mutants for 48 h were treated with MG-132 (+) or DMSO (−) for 2 h. The urea-soluble lysates were prepared, and His6-ubiquitin–conjugated proteins were pulled down from cell lysates. Total-cell lysates (Input) and Ni-NTA–agarose eluates were analyzed by Western blotting and stained for FLAG, TDP-43, ubiquitin, and GAPDH. Asterisks label endogenous TDP-43, and hash marks indicate monoubiquitinylated TDP-43. C, FLAG–TDP-43WT, C-terminal lysine mutants, or control vector (−) were overexpressed in HEK293E cells for 48 h, and RIPA- and urea-soluble lysates were prepared. The lysates were subjected to Western blot analysis, and the blots were stained with antibodies detecting FLAG and TDP-43 and GAPDH and actin as loading controls. Endogenous TDP-43 is labeled with one asterisk; FLAG–TDP-43 derived 35-kDa CTFs are labeled with two asterisks. D, quantification of at least n = 3 experiments is as in C. Band intensities of TDP-43 (upper graph) and FLAG probings (lower graph) were normalized to WT levels. Data represent the mean ± S.D. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.005.
We analyzed the ubiquitinylation of the lysine-mutated FLAG–TDP-43 proteins by His6-ubiquitin pulldown (Fig. 3, A and B). The proteasome was inhibited with MG-132 administration to stabilize the otherwise very weak ubiquitinylation of FLAG–TDP-43. The steady-state levels of all mutants were not altered. We detected a slightly decreased ubiquitinylation of TDP-43K192R and enhanced ubiquitinylation of TDP-43K176R, TDP-43K224R, and all multilysine mutants, which all contain the K224R mutation. Also, TDP-434×KR ubiquitinylation was not reduced like in CTF4×KR, indicating that in TDP-43 FL the C-terminal lysines are not the only ubiquitinylation sites and that the N-terminal lysines have a higher impact on ubiquitinylation of TDP-43.
We hypothesized that the increased ubiquitinylation of the K224R mutant could lead to or be the result of enhanced insolubility, as we have observed for mCherry-CTFK408R. Therefore, we also investigated the solubility of the mutants, where we observed an increase of insoluble TDP-43K224R, TDP-43K408R, TDP-432×KR, TDP-433×KR, and TDP-434×KR (Fig. 3, C and D), which correlates with the increased ubiquitinylation of these mutants. The formation of 35-kDa CTFs from TDP-43K224R, TDP-43K408R, and TDP-432×KR was also increased in the urea fraction (Fig. 3C, urea, TDP-43 long exposure (**)). We did not observe a decrease of the more insoluble mutants in the soluble fraction, probably due to a high ratio of soluble to insoluble protein. Phosphorylation of the lysine mutants was analyzed, but the level of TDP-43 FL phosphorylation was below the detection limit (data not shown). Finally, all lysine mutants were located in the nucleus like TDP-43WT (Fig. S3).
In conclusion, we identified only one single C-terminal site, Lys-192 in FLAG–TDP-43, by which preventing ubiquitinylation by mutation to arginine led to a detectable decrease of the overall ubiquitinylation of TDP-43. Also, the data from ubiquitinylation studies of mCherry-CTF did not correlate with the data from FLAG–TDP-43, suggesting that different sites are mainly ubiquitinylated and that in TDP-43 FL also other lysines are ubiquitinylated. There may be either no major ubiquitinylation sites among the C-terminal lysine residues investigated here or degradation-targeting ubiquitinylation is not a very site-specific post-translational modification: when one site is not accessible for ubiquitinylation, an alternative site may be chosen.
Mass spectrometric identification of TDP-43 ubiquitinylation sites
Several TDP-43 ubiquitinylation sites were identified in a small number of proteomic approaches (Table 1), and the contribution of their ubiquitinylation to the total ubiquitinylation of TDP-43 was studied only in one case (30). In a next step we sought to confirm our results and, additionally, wanted to identify further TDP-43 ubiquitinylation sites. Thus, we conducted MS analyses of TDP-43 FL. With this approach, we could analyze all possible ubiquitinylation sites of the protein by detecting the diGly motif that is formed at ubiquitinylated lysine residues after trypsin digestion of the purified ubiquitinylated TDP-43 prior to MS analysis. Therefore, we overexpressed N-terminally His6-tagged TDP-43 in HEK293E cells. Because ubiquitinylation often targets proteins for degradation and, thus, mediates fast turnover of the modified proteins, we inhibited the proteasome for 6 h prior to cell lysis to enhance the otherwise very low ratio of ubiquitinylated to nonmodified TDP-43. DMSO-treated cells were used as control. Given that ubiquitinylated TDP-43 is mainly insoluble (29), and to reduce unspecific binding of soluble proteins to the Ni-NTA–agarose, we first extracted the soluble proteins prior to lysis of the insoluble fraction with an 8 m urea lysis buffer. All His6-ubiquitinylated TDP-43 proteins, including any post-translational modifications, were isolated by Ni-NTA–affinity chromatography (Fig. S4, A and B). Western blot analysis showed that TDP-43 was predominantly monoubiquitinylated, and polyubiquitinylation was not as pronounced, as the monoubiquitinylation band at ∼55 kDa was much stronger than the HMW smear (Fig. S4B; TDP-Ub1 versus TDP-Ubn). To distinguish between these two types of TDP-43 ubiquitinylation, we separated the purified TDP-43 by SDS-PAGE, visualized the protein with Coomassie Brilliant Blue staining, and used the gel areas mono (likely monoubiquitinylated) and HMW (likely polyubiquitinylated) (see Fig. S4A) for the detection of the diGly motif by LC-MS/MS analysis.
The coverage of TDP-43 in the four different samples was between 45 and 60% (Table 2 and Fig. S4C). The diGly motif was detected under control conditions (DMSO) at lysine residues Lys-84 and Lys-181 in the TDP-43 mono- and Lys-84, Lys-181, and Lys-263 in the HMW sample (Table 2). Additionally, proteasomal inhibition stabilized the ubiquitinylation at Lys-95, Lys-160, and Lys-263 in the His6-TDP-43 mono samples and Lys-95 and Lys-160 in the His6-TDP-43 HMW samples. In this first analysis, mainly ubiquitinylation sites were identified that are located in the bipartite NLS or RRM1 of TDP-43, with the exception of Lys-181 and Lys-263 for which we could already show that their individual lysine-to-arginine mutation does not reduce the overall TDP-43 ubiquitinylation (see above).
Table 2.
Ubiquitin-modified lysine residues of TDP-43 and ubiquitin, identified by MS/LC analysis in N- and C-terminally His6-tagged TDP-43 purified by Ni-NTA affinity chromatography
Coverages of TDP-43 in analyzed samples are specified as percentage values in parentheses.
| DMSO treatment, analyzed band HMW | DMSO treatment, analyzed band TDP-43 mono | MG-132 treatment, analyzed band HMW | MG-132 treatment, analyzed band TDP-43 mono | ||
|---|---|---|---|---|---|
| TDP-43 | His6-TDP-43 | Lys-84, Lys-181, Lys-263 (45%) | Lys-84, Lys-181 (46%) | Lys-84, Lys-95, Lys-160, Lys-181, Lys-263 (52%) | Lys-84, Lys-95, Lys-160, Lys-181, Lys-263 (60%) |
| TDP-43-His6 | Lys-102, Lys-181 (44%) | Lys-114, Lys-181 (83%) | Lys-84, Lys-95, Lys-102, Lys-160, Lys-181, Lys-263 (62%) | Lys-84, Lys-95, Lys-102, Lys-114, Lys-160, Lys-181, Lys-263 (72%) | |
| Ubiquitin | His6-TDP-43 | Lys-48, Lys-63 | Lys-48 | Lys-48, Lys-63 | Lys-48, Lys-63 |
| TDP-43-His6 | Lys-48, Lys-63 | Lys-11, Lys-48, Lys-63 | Lys-48, Lys-63 | Lys-48, Lys-63 | |
Use of an N-terminal His6 tag in these experiments would not allow pulldown and coverage of putative CTFs derived from the transfected TDP-43. Therefore, we generated C-terminally His6-tagged TDP-43 that also allowed pulldown of a 35-kDa CTF (Fig. S4B, His6 Pulldown, arrow). We performed a second MS analysis with purified TDP-43–His6. The coverage was better for three of the four samples with values between 62 and 83% (Table 2 and Fig. S4C). The Lys-181 ubiquitinylation site was detected again in all four samples, and ubiquitinylation of Lys-84, Lys-95, Lys-160, and Lys-263 was found only upon MG-132 administration, confirming the results from the first analysis (Table 2). In addition to the detected sites of the first analysis, Lys-102 and Lys-114 were identified in the HMW and TDP-43 mono-samples, respectively, independently of the treatment.
Of the four C-terminal ubiquitinylation sites only Lys-263 was identified in the MS analyses, although Lys-224, Lys-251, and Lys-408 were covered. Ubiquitinylation of Lys-192 could not be confirmed, although we found decreased levels of FLAG–TDP-43K192R in our previous experiments (Fig. 3, A and B). However, this lysine residue was not covered in our MS analyses, and therefore, it is still possible that Lys-192 is a real TDP-43 ubiquitinylation site.
It is noteworthy that in both analyses ubiquitin peptides with diGly motifs at Lys-48 and Lys-63 were detected in proteasome-inhibited samples but also in the DMSO control samples (Table 2). These peptides were probably generated from TDP-43 polyubiquitin chains linked via Lys-48 and Lys-63. This finding emphasizes the proteasome and autophagosome targeting function of TDP-43 ubiquitinylation. Neddylation and ISGylation also produce diGly-tagged peptides upon trypsin digestion, but we did not find any peptides matching Nedd8 or ISG15 among the peptides co-enriched with His6-tagged TDP-43. Moreover, ISG15 is only expressed upon interferon-α/β exposure (33), and Neddylation is thought to be only a minor modification (34).
In summary, we identified several ubiquitinylation sites of TDP-43 that are mainly located in the NLS and RRM1 (Fig. 4A), the latter being in accordance with data from four other studies that identified ubiquitinylation sites of TDP-43 (Table 1). Importantly, the ubiquitinylation of the NLS Lys-84 is a novel finding of this study. The modifications of Lys-95 and Lys-160 were detected only upon proteasomal inhibition, which was also found by Kim et al. (34) and Wagner et al. (35), who also applied proteasomal inhibitors. The lack of degradation prevention might be the reason why Lys-95 and Lys-160 ubiquitinylation were not detected in the other two studies (Table 1). However, we could only confirm the ubiquitinylation of one C-terminal lysine residue, Lys-263.
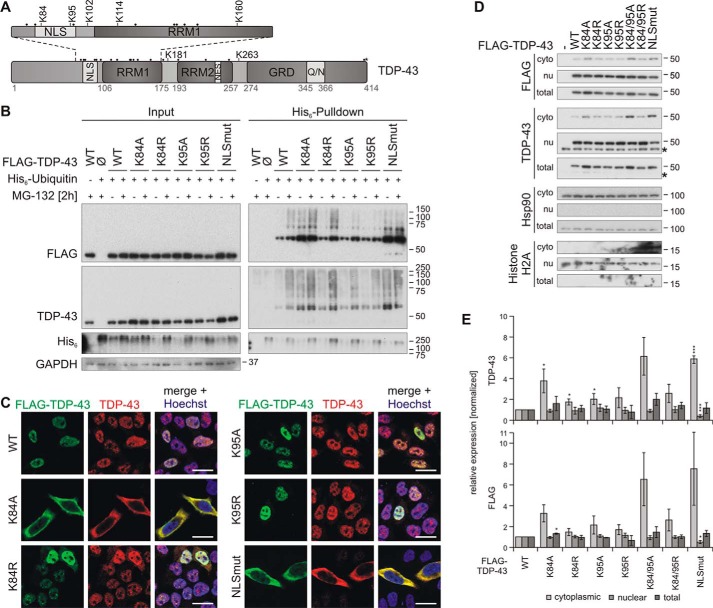
Figure 4.
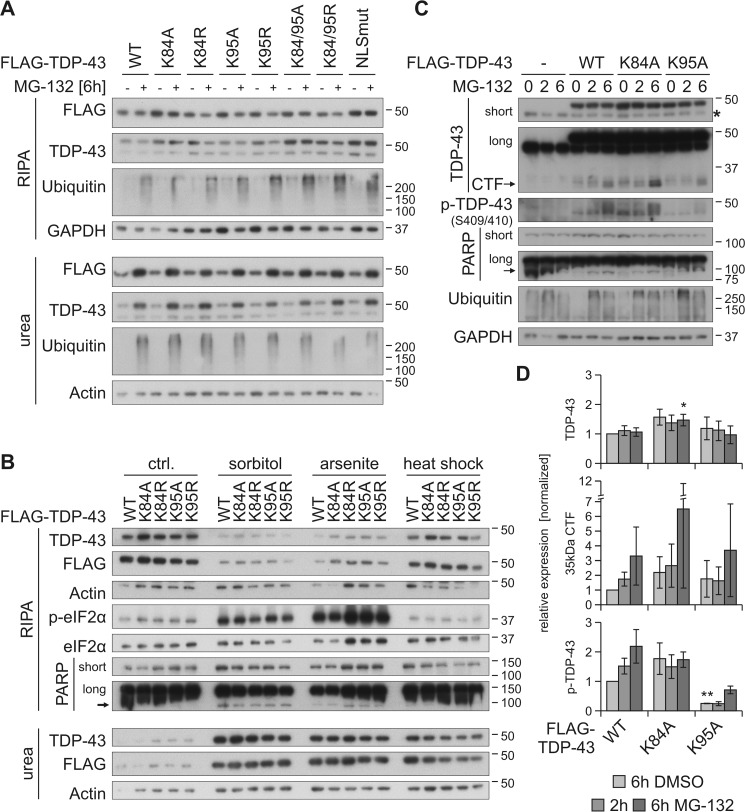
Characterization of TDP-43 NLS lysine mutant ubiquitinylation and subcellular localization. A, schematic overview of TDP-43 ubiquitinylated lysine (K) residues that were identified by MS. All other lysine residues are indicated with dots. B, His6-ubiquitin (+) or vector control (−) and FLAG–TDP-43WT, NLS lysine mutants, or FLAG control vector (Ø) were overexpressed in HEK293E cells. Proteasomal inhibition with MG-132 for 2 h was followed by cell lysis in urea buffer and affinity purification of His6-ubiquitinylated proteins. Total protein (Input) and Ni-NTA–agarose eluates (His6 Pulldown) were subjected to Western blot analysis and stained with antibodies against FLAG, TDP-43, His6, and GAPDH. C, HEK293E cells overexpressing FLAG–TDP-43WT and NLS lysine mutants were immunolabeled with mouse anti-FLAG (green) and rabbit anti-TDP-43 (red), and cell nuclei were counterstained with Hoechst 33342 (blue). Scale bars are 20 μm. D, nuclear–cytoplasmic fractionation was performed on HEK293E cells that overexpressed FLAG–TDP-43WT or the indicated NLS lysine mutants. Cytoplasmic (cyto), nuclear (nu), and total lysates were analyzed by Western blotting with antibodies detecting FLAG, TDP-43, and Hsp90 as cytoplasmic marker, and histone H2A as nuclear marker. Asterisks label endogenous TDP-43. E, quantification of TDP-43 (upper graph) and FLAG (lower graph) signals from at least n = 3 experiments as in D. Band intensities were normalized to the FLAG–TDP-43WT signal of the respective fraction. Data represent the mean ± S.D. *, p ≤ 0.05; ***, p ≤ 0.005.
NLS Lys-95 is ubiquitinylated upon proteasomal inhibition
Next, we studied the ubiquitinylation of the NLS ubiquitinylation sites Lys-84 and Lys-95 in more detail and analyzed the effects of their ubiquitinylation on their localization, solubility, and aggregation. We hypothesized that ubiquitin modification of these sites might have an impact on TDP-43 localization (36). Also, function and solubility of TDP-43 might be altered by possible shifts to the cytoplasm. It was shown before that cytoplasmic relocalization induces aggregation and insolubility of TDP-43 and cell toxicity and also has a negative impact on the function of the protein (37).
First, we investigated whether the identified NLS ubiquitinylation sites Lys-84 and Lys-95 can be detected indirectly with our His6-ubiquitin pulldown approach and how this is altered by proteasomal inhibition. Because TDP-43 contains 20 lysine residues, mutating all but Lys-84 or Lys-95 to see whether these sites are ubiquitinylated would strongly alter the structure of the whole protein. Thus, we used the indirect approach and mutated Lys-84 and Lys-95 either to arginine or alanine. The arginine mutants could theoretically still be recognized and bound by importins due to the preservation of the positive lysine charge, whereas the alanine substitution should not be recognized by them. We overexpressed TDP-43K84R/K84A and TDP-43K95R/K95A mutants, respectively, in HEK293E cells, treated them with MG-132, and isolated the ubiquitinylated proteins from the total urea fraction using Ni-NTA–agarose (Fig. 4B). As a control, the cytoplasmic TDP-43NLSmut (K82A/R83A/K84A) was included. Proteasomal inhibition stabilized the ubiquitinylation of all mutants. The ubiquitinylation of TDP-43K84A was increased, even without MG-132 treatment, and TDP-43K95A and TDP-43K95R ubiquitinylations were reduced upon proteasomal inhibition, confirming that this lysine is a proteasomal degradation-targeting ubiquitinylation site. The effects on the arginine mutant TDP-43K84R were not as pronounced as on the alanine mutant, giving a hint that the pure availability of a certain ubiquitinylation site is not important for its modification but perhaps also the subcellular localization (see below). This is supported by the strong ubiquitinylation of the TDP-43NLSmut mutant, even under basal conditions (Fig. 4B).
Next, we studied whether the subcellular localization of the NLS lysine mutants is indeed altered by performing subcellular fractionations and immunofluorescence stainings of HEK293E cells that overexpress the mutants under nonstressed conditions. Indeed, we found a shift of TDP-43K84A to the cytoplasm in the fractionation and subcellular staining, whereas TDP-43K95A localization was equal to TDP-43WT (Fig. 4, C–E). Also, both arginine mutants TDP-43K84R and TDP-43K95R were mainly located in the nucleus. The localization of a double mutant TDP-43K84A/K95A was also shifted to the cytoplasm, whereas the corresponding arginine mutant TDP-43K84R/K95R was still mainly nuclearly localized (Fig. 4, D and E). Moreover, the total levels of the K84A, K84A/K95A, and NLSmut mutants were increased (Fig. 4D). The cytoplasmic localization of TDP-43K84A also indicates that Lys-84 is the important importin-recognition amino acid in the bipartite NLS of TDP-43, and it also implies that Lys-95 plays no substantial role in the nuclear import of the protein. The cytoplasmic shift of TDP-43K84A alone might alter or impair the solubility, aggregation, and/or function of TDP-43, making it difficult to investigate the impact of ubiquitinylation at this site, even more because we can only investigate the ubiquitinylation indirectly by removing the ubiquitinylation site of interest through mutagenesis. The localization of TDP-43K95A instead was not altered, and even more, ubiquitinylation at this site was only detected upon proteasomal inhibition by MS (Table 2).
In summary, TDP-43 ubiquitinylation at Lys-95 could be confirmed, but modification of this site with ubiquitin did not alter the subcellular localization of the protein.
NLS lysine ubiquitinylation does not alter solubility of TDP-43
So far, we identified decreased ubiquitinylation of TDP-43K95A and TDP-43K84A and a shift into the cytoplasm of TDP-43K84A. Thus, we next investigated whether these alterations affect the solubility, CTF formation, and aggregation of TDP-43, as it was shown before that a shift of TDP-43 to the cytoplasm increases its insolubility (37). First, we overexpressed TDP-43WT and the NLS lysine mutants, additionally inhibited the proteasome for 6 h, and separated the cellular proteins in soluble and insoluble fractions (Fig. 5A). We observed increased levels of TDP-43K84A, TDP-43K84A/K95A, and TDP-43NLSmut in the soluble fraction under control conditions, and proteasomal inhibition shifted all TDP-43 species from the RIPA into the insoluble urea fraction. However, we did not detect altered insolubility of the TDP-43K84A/K84R and TDP-43K95A/K95R mutants compared with TDP-43WT. We also tested whether osmotic, oxidative, or thermal stress might induce differences in the solubility of the NLS lysine mutants (Fig. 5B). Like MG-132, all three conditions, sorbitol, arsenite, and heat shock, shifted more TDP-43 into the insoluble urea fraction, but no differences were seen for the Lys-84 and Lys-95 mutants. Thus, a shift to the cytoplasm or removal of ubiquitinylation sites does not induce increased TDP-43 insolubility in HEK293E cells, neither under control nor stress conditions.
Figure 5.
Prevention of ubiquitinylation at Lys-95 reduces MG-132–stabilized phosphorylation of TDP-43. HEK293E cells were transfected with FLAG–TDP-43WT or NLS lysine mutants and treated with MG-132 (+) or DMSO (−) for 6 h (A) as well as with 0.4 m sorbitol or 200 μm arsenite for 1 h, or subjected to heat shock at 43 °C for 30 min (B). Sequential extraction of the cells was performed, and RIPA and urea lysates were analyzed by Western blotting with antibodies against FLAG, TDP-43, ubiquitin, GAPDH, and actin as loading controls (A). Additional probings of phospho-eIF2α and total eIF2α confirmed stress granule conditions for sorbitol and arsenite treatments and PARP cleavage as a marker of apoptosis (B). C, FLAG–TDP-43WT-, FLAG–TDP-43K84A-, and FLAG–TDP–43K95A-overexpressing HEK293E cells were treated with MG-132 for 2–6 h or DMSO as control (0), followed by lysis with urea buffer. Lysates were subjected to Western blot analysis and stained with antibodies against TDP-43, phospho-TDP-43, PARP, ubiquitin, and GAPDH. D, quantification of FLAG–TDP-43, 35-kDa CTF, and phospho-TDP-43 from at least three experiments as in C. Band intensities were normalized to FLAG–TDP-43WT (6h DMSO), and data represent the mean ± S.D. *, p ≤ 0.05; **, p ≤ 0.01.
Furthermore, we also asked whether the NLS lysine to alanine mutants exhibit altered formation of 35-kDa CTFs, which are frequently observed in HEK293 and other cell lines. The CTFs are degraded rapidly by the proteasome after their formation (24, 38) and become insoluble if they are stabilized by proteasomal inhibition (19, 26, 29, 39). We blocked the proteasome with MG-132 for 2 or 6 h prior to lysis with an 8 m urea buffer, followed by Western blot analysis (Fig. 5C). We detected increased 35-kDa CTF level upon 6-h proteasome inhibition from TDP-43WT and TDP-43K95A, and CTF formation from TDP-43K84A increased even stronger, possibly due to the already higher expression levels of total FLAG–TDP-43 for this mutant (Fig. 5, C and D). Interestingly, although levels of TDP-43WT and TDP-43K95A species were comparable as well as their derived CTFs, the amount of phosphorylated TDP-43K95A was significantly decreased under control and proteasome-inhibited conditions (Fig. 5, C and D).
Because it is thought that these CTFs are generated by caspase-3 cleavage and MG-132 was shown to mediate this activation (40, 41), we also checked the level of activated caspase-3, but we did not observe any cleaved caspase-3 (data not shown). However, proteasomal inhibition induced PARP cleavage (Fig. 5C), a marker for apoptosis, which was shown to be conducted by several caspases, among them caspase-3 (42). We also investigated the aggregate formation upon 6-h proteasomal inhibition of the NLS lysine mutants with immunofluorescence. We found mainly nuclear phospho-TDP-43–positive inclusions in 10–15% of MG-132–treated cells, with no significant differences between the mutants and TDP-43WT (data not shown). Likewise, no difference in cystic fibrosis transmembrane conductance regulator (CFTR) and ribosomal S6 kinase 1 Aly/REF-like target (SKAR) splicing and HDAC6 mRNA stability was observed (data not shown).
In summary, we describe reduced phosphorylation of TDP-43K95A, and increased expression level of TDP-43K84A, probably due to its cytoplasmic localization.
Ubiquitinylation of disease-associated TDP-43 mutants
In principle, ubiquitinylation could be a mechanism conferring pathogenicity of TDP-43 and its clinical mutants. We examined two TDP-43 mutations that introduce additional lysines, namely Q331K and N345K. These two TDP-43 mutants were expressed at the same level as TDP-43WT in transiently transfected HEK293 cells (Fig. S5). No enhanced ubiquitinylation could be detected in His6-ubiquitin pulldown experiments, neither under basal conditions nor after treatment with MG-132 (Fig. S5). Thus, at least for these two disease-associated TDP-43 mutants, the introduction of extra lysines did not lead to excessive ubiquitinylation of TDP-43.
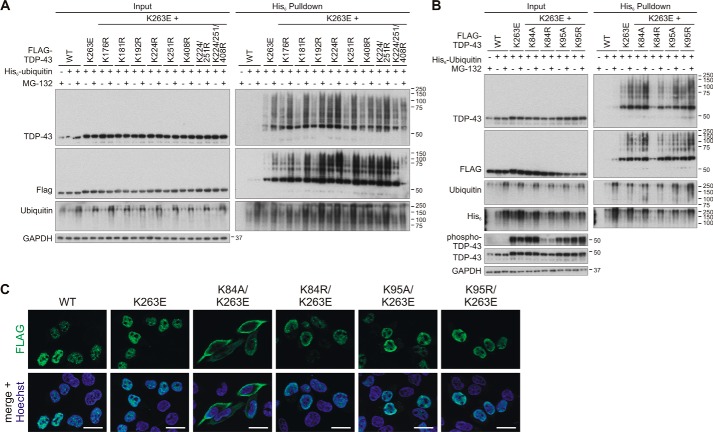
We had previously reported a hyper-ubiquitinylated TDP-43 mutant, K263E (29). Thus, we sought to identify lysine residues that could account for the enhanced ubiquitinylation of TDP-43K253E. We generated a novel set of lysine mutants combined with K263E and analyzed these TDP-43 proteins as described above for TDP-43WT. Combining K263E with a collection of C-terminal lysine single or multiple arginine substitutions, we did not identify any specific residue that could account for the enhanced ubiquitinylation of TDP-43K263E (Fig. 6A). We did notice some enhancement of ubiquitinylation in the K224R/K263E double mutant, similar to the enhanced ubiquitinylation of TDP-43K224R alone (see Fig. 3). Thus, enhanced ubiquitinylation of K224R and K263E might occur in an additive manner. It required the combination of K263E with K224R/K251R/K408R to start seeing reductions of ubiquitinylation, but such multiple mutagenesis to quadruple sites becomes increasingly artificial. We conclude that similar to TDP-43WT, the hyper-ubiquitinylated TDP-43K263E also can be ubiquitinylated at multiple redundant sites.
Figure 6.
Investigation of ubiquitinylation sites for the hyper-ubiquitinylated mutant TDP-43K263E. HEK293E cells were transfected with FLAG–TDP-43WT and FLAG–TDP-43K263E alone or combined with the indicated lysine substitutions localized in the C-terminal part (A) or the NLS sequence (B). His6-ubiquitin (+) or vector control (−) was cotransfected, as indicated. Cells were then treated with MG-132 (+) or DMSO (−) for 2 h. The urea-soluble lysates were prepared, and His6-ubiquitin–conjugated proteins were pulled down from cell lysates. Total cell lysates (Input) and Ni-NTA–agarose eluates were analyzed with Western blotting and stained for FLAG, TDP-43, ubiquitin, and GAPDH, and in B additionally with anti-His6. Lysates samples in B were re-run and subjected to Western blot analysis of phospho-TDP-43. C, HEK293E cells overexpressing FLAG–TDP-43WT and FLAG–TDP-43K263E alone or combined with the indicated Lys-84 and Lys-94 substitutions were immunostained with rabbit anti-FLAG (green). Nuclei were counterstained with Hoechst 33342 (blue). Scale bars correspond to 20 μm.
Combining K263E with substitutions of Lys-84 and Lys-95 showed no reduction of ubiquitinylation for the Lys-95 mutants (Fig. 6B). Interestingly, a remarkable reduction of TDP-43K263E ubiquitinylation was observed in the K263E/K84R double mutant. Likewise, the enhanced Ser-409/410 phosphorylation of TDP-43K263E was reduced when combined with the K84R mutation (Fig. 6B). The slight reduction of TDP-43K84R/K263E expression levels does not seem likely to account for the strong reductions of ubiquitinylation and phosphorylation. Curiously, these effects on TDP-43K263E modifications were not observed when combined with the nonconservative K84A substitution (Fig. 6B). In contrast to the other mutants, introduction of K84A leads to impairment of nuclear import (Fig. 6C). We assume that inside the nucleus, Lys-84 plays a somewhat more prominent role in TDP-43K263E modifications. When mislocalized to cytosolic compartments, the TDP-43 modifications appear to become more promiscuous and less dependent on Lys-84.
Discussion
Here, we investigated C-terminal and NLS-located potential ubiquitinylation sites of TDP-43 and their role in ubiquitinylation, localization, solubility, and aggregation of the protein. We found the following: 1) there is no main C-terminal ubiquitinylation site, but all four lysine residues contribute to the heavy ubiquitinylation of CTF; 2) C-terminal lysine residues are not the main ubiquitinylation sites of TDP-43 FL; 3) ubiquitinylation at Lys-408 might impede the phosphorylation at Ser-409/410 at least in the context of CTF; 4) substitutions of Lys-84 impair nuclear import, whereas Lys-95 ubiquitinylation does not appear to alter cellular distribution and solubility, yet prevention of Lys-95 ubiquitinylation reduces the pathological Ser-409/410 phosphorylation; and 5) Lys-84 might become an important ubiquitinylation residue in the nucleus for TDP-43 mutants such as the hyper-ubiquitinylated TDP-43K263E. An overview of the results is displayed in Table S1.
The identification of ubiquitinylation sites and of the linkage type is important for learning more about the fate of the ubiquitinylated substrate. The localization of ubiquitin post-translational modifications in a certain domain or the type of linkage can give information about the function of ubiquitin attachment besides targeting the protein for degradation, which is the best studied purpose of ubiquitinylation (43). All 20 lysine residues of TDP-43 are conserved among species down to zebrafish and most also in Drosophila melanogaster. However, there are no distinct ubiquitinylation motifs known (34), and sequences around ubiquitinylated lysine residues are not conserved across different species (44). Also, the broad range of E2 and E3 enzymes disfavors the existence of distinct ubiquitinylation motifs. This absence of a clear motif makes it difficult to predict ubiquitinylation sites in silico. Therefore, ubiquitinylation sites are usually identified experimentally by MS. However, in this study we decided to start with a more unconventional approach using site-directed mutagenesis of TDP-43 C-terminal lysine residues, as CTFs are heavily ubiquitinylated (29) but harbor only a restricted number of possible ubiquitinylation sites. Moreover, among the published ubiquitinylation sites (Table 1), only Lys-263 is located in our CTF(193–414), and Lys-181 and Lys-176 are located in the extended C-terminal region of TDP-43 (Fig. 1A). We did not find a single main ubiquitinylation site in our CTF with the site-directed mutagenesis approach. Thus, it is possible that all sites contribute equally to the CTF ubiquitinylation, which indicates that the CTF is not site-specifically ubiquitinylated. Interestingly, MG-132 treatment stabilized the CTF ubiquitinylation poorly, suggesting that the ratio of ubiquitinylated CTF was already so high that a further increase due to proteasomal inhibition was barely visible or that these ubiquitin modifications might be not proteasome but autophagosome targeting. Also, the increased amount of insoluble CTFK263R and CTFK408R might point toward a decreased proteasomal degradation of those mutants due to the lack of UPS-targeting ubiquitinylation sites. Ubiquitinylation of pathological TDP-43 FL and of CTFs in ALS and FTLD should target them for degradation, but the ubiquitinylated TDP-43 species cannot be disposed likely due to decreased efficiency of the protein degradation systems in neurodegenerative diseases and aging (45, 46).
K192R was the only C-terminal lysine mutant that exhibited decreased ubiquitinylation of TDP-43 FL. This site could not be confirmed with MS because the corresponding peptide was not covered (Fig. S4C). Interestingly, Cohen et al. (47) identified Lys-192 as a major acetylation site and suggested that this modification might interfere with nucleic acid binding in the proximal RRM2. Therefore, it is conceivable that ubiquitinylation at Lys-192 indirectly interferes with RNA binding by competing with acetylation at this site. We also identified Lys-181 and Lys-263 as C-terminal ubiquitinylation sites of TDP-43 FL by MS, but not Lys-224, Lys-251, and Lys-408, although these sites were covered. It is possible that the same lysine residues are differentially ubiquitinylated depending on whether they are part of TDP-43 FL or CTF. For instance, TDP-43K224R was strongly ubiquitinylated, and TDP-43K408R ubiquitinylation was not altered, but mCherry-CTFK408R ubiquitinylation was clearly increased, whereas CTFK224R ubiquitinylation was not enhanced. In another study, two TDP-43 variants, TDP-43 FL and a C-terminally truncated TDP-S6(1–276), were depleted of four lysine residues located in or near RRM1 (Lys-102, Lys-114, Lys-145, and Lys-181), and they exhibited different solubility and total ubiquitinylation properties (30). Thus, it is possible that MS of TDP-43 FL could not detect ubiquitinylation sites of the CTF.
Ubiquitinylation at Lys-160 was also detected only upon MG-132 treatment by Wagner et al. (35), supporting a proteasome-targeting function. Interestingly, the Lys-181 ubiquitinylation site was identified in all studies (including ours) using cell line-derived endo- and exogenous TDP-43 (Tables 1 and 2), although we could not confirm a clear decrease of TDP-43K181R ubiquitinylation in our pulldown assay. This suggests that either other lysine residues are ubiquitinylated in compensation, that the ubiquitinylation at Lys-181 only contributes negligibly to the total amount of ubiquitinylated TDP-43, or that the Lys-181–containing peptide is very suitable for detection by MS.
Several of the identified ubiquitinylations sites of TDP-43 are probably degradation targeting as they were detected by inhibiting the proteasome to stabilize ubiquitinylated TDP-43. This was also confirmed by the detection of Lys-48- but also Lys-63–linked polyubiquitins among the isolated insoluble TDP-43 in this study (Table 2) and in previous studies (14, 26, 28). Lys-48 is the typical proteasome targeting polyubiquitin linkage, and Lys-63–linked ubiquitin labels proteins for lysosomal degradation but is also a sign for stress response, DNA damage, or endocytosis (43). As there are other types of polyubiquitin linkage as well as monoubiquitinylation that target the substrate for other functional destinations (17), it is possible that the ubiquitinylation sites detected under control conditions have other functions. Because mono- and different polyubiquitin modifications leave a diGly motif after tryptic digestion prior to MS, this method cannot distinguish between these types of ubiquitinylation.
A challenge of this study was the confirmation of potential ubiquitinylation sites as ubiquitinylated proteins are usually turned over rapidly. Also, in contrast to other post-translational modifications, like phosphorylation or acetylation, ubiquitinylation cannot be mimicked. Thus, we had to employ the indirect approach where a lysine is mutated to arginine (under charge preservation) or alanine, followed by the detection of decreased total ubiquitinylation if the site is indeed a ubiquitinylated lysine residue. This decrease can be small if the protein contains many ubiquitinylation sites or if a neighboring lysine is alternatively ubiquitinylated. Also, little is known about the site-specificity of ubiquitinylations, but a proteomic study showed that ubiquitinylations were considerably increased in about two-thirds of the identified ubiquitinylation sites upon proteasomal inhibition, suggesting a main proteasome-targeting function for these sites (35). This indicates that proteasome-targeting ubiquitinylations might be selective to some extent.
A novel and interesting finding of this study is the increased phosphorylation at Ser-409/410 of CTFK408R, which also exhibits increased ubiquitinylation. This suggests a cross-talk between C-terminal ubiquitinylation and phosphorylation. Ubiquitinylation at Lys-408 could sterically impair kinase binding and prevent phosphorylation, or the lysine to arginine mutation might facilitate kinase binding and thus increase Ser-409/410 phosphorylation. A further possibility is that the stronger ubiquitinylation of the other CTF lysine residues decreased CTF solubility and thus also caused increased phosphorylation. Likewise, the increased phosphorylation might also enhance the CTF insolubility. Providing support for this assumption, several studies demonstrated that phosphorylation at Ser-409/410 decreases TDP-43 degradation, enhances its insolubility and aggregation, and results in neurodegeneration (7, 23, 48, 49). Therefore, the cause and the consequence of the increased phosphorylation of CTFK408R need to be further investigated.
Several studies gave evidence for a direct cross-talk between ubiquitinylation and phosphorylation of the same protein molecule. Noteworthy, ubiquitinylation sites of nuclear proteins in yeast are often in proximity of phosphorylation sites (50). In polyQ diseases, phosphorylation of ataxin-1 at Ser-776 modulates its ubiquitinylation by the C terminus of the hsp70-interacting protein (CHIP) and its aggregation (51). Quite straightforward, phosphorylation is often a prerequisite for degradation-targeting ubiquitinylation of cell cycle-associated or regulatory proteins (cyclins, period circadian protein homologs) by providing a recognition site for E3 ligases (52–54), as the secondary structure is important for E3 recognition of the ubiquitinylation site (55, 56). Thus, there is certain evidence for phosphorylation influencing degradation of proteins, including TDP-43.
In this study, we identified two ubiquitinylation sites that are located in the NLS of TDP-43. The Lys-84 ubiquitinylation site is a novel finding of this study, and the validity of the ubiquitinylation of Lys-95 targeting TDP-43 for proteasomal degradation is supported by another study that found this modification under proteasomal inhibition (34). The localization of these sites in the NLS of TDP-43 suggests that their ubiquitinylation might affect nuclear import either directly by impairing importin-α binding, as is possible for Lys-84, or indirectly by targeting TDP-43 for degradation before it can be shuttled back into the nucleus. In fact, it was shown for several proteins that ubiquitinylation regulates their localization. Mono-ubiquitinylation of choline-phosphate cytidylyltransferase A at Lys-57 in proximity to its NLS results in the disruption of importin-α binding, which facilitates its lysosomal degradation (57), and ubiquitinylation of the NLS and NES is required for nuclear-cytoplasmic trafficking of Paramyxovirinae matrix proteins (58). Also, p53 import is restrained by ubiquitinylation of its NLS lysines 319–321 by the E3 ligase MDM2, which blocks importin-α3 binding (59). Additionally, other post-translational modifications were shown to regulate protein import and export. Arginine methylation of an RGG motif next to the C-terminal PY-NLS of FUS, another RNA-binding protein involved in pathogenesis of a subgroup of ALS and FTLD, impairs transportin binding, and thus its nuclear import (60).
Actually, nucleo-cytoplasmic transport defects recently moved into focus as disease-modifying factor in ALS and FTLD. The formation of pathological TDP-43 inclusions is accompanied by nuclear loss of the protein, which suggests a loss-of-function in disease (1). Also, decreased importin-β levels were detected in nuclei of motor neurons from ALS cases (61), and exportin-2 and importin-α1 are reduced in FTLD-TDP (36). Furthermore, the nuclear import/export machinery is impaired in C9ORF72-related ALS and FTLD cases, and several pathogenic FUS mutations also impede nuclear import (62). Recently, proteins of the import/export machinery and of the nuclear pore complex were detected in insoluble TDP-43 aggregates in several cellular models (63). Thus, post-translational modifications at or adjacent to the NLS and/or NES of TDP-43 might influence its nuclear-cytoplasmic shuttling and thereby could regulate TDP-43 mis-localization and disease progression.
We did not observe a difference in solubility comparing TDP-43WT and TDP-43K95A under basal conditions, although ubiquitinylated TDP-43 exhibits increased insolubility (29). An explanation for this might be that the proportion of ubiquitinylated TDP-43 compared with total TDP-43 is very low as the HMW depicting ubiquitinylated TDP-43 is hardly detectable in Western blots of total protein lysates, and thus, a small decrease of ubiquitinylation is not sufficient to cause a visible shift in solubility of the total protein.
Likewise, the insolubility shifts after cellular stress (proteasomal inhibition or application of osmotic, oxidative, or thermal stress) were not altered by the NLS lysine substitutions. Perhaps forced ubiquitinylation and insolubility occurs at sites beyond Lys-95, as also the enhanced ubiquitinylation of TDP-43K263E was not as much reduced as for TDP-43WT. For TDP-43K263E, surprisingly, we found a strong reduction of ubiquitinylation and phosphorylation in the K84R double mutant, but not for the nuclear import–impaired TDP-43K84A/K263E. Perhaps Lys-84 is a more prominent ubiquitinylation anchor for particular TDP-43 species inside the nucleus, whereas post-translational modifications in cytosolic mislocalizations become even more complex and promiscuous.
Surprisingly, we found reduced phosphorylation of TDP-43K95A at Ser-409/410. We have no straightforward explanation for this observation, as we detected no alterations of subcellular localization and solubility. Perhaps ubiquitinylation at Lys-95 affects distribution of TDP-43 in a subtle manner below the detection limits of our assays and/or regulates accessibility of Ser-409/410 kinases. The other NLS residue Lys-84 appears to be essential for nuclear import, as its substitution to alanine impeded the nuclear import of TDP-43K84A, probably caused by impaired importin binding. On the one hand, this means that Lys-84 ubiquitinylation likely prevents importin-α binding and thus shuttling into the nucleus. On the other hand, this makes it difficult to distinguish between direct and indirect effects on solubility and aggregation due to Lys-84 ubiquitinylation or impaired importin-α binding, respectively. Impaired nuclear import due to NLS ubiquitinylation might contribute to disease pathogenesis, but unfortunately this hypothesis is difficult to prove because ubiquitinylation cannot be mimicked by site-directed mutagenesis.
Taken together, our study identifies ubiquitinylation sites in TDP-43 and highlights an interplay between specific ubiquitinylation events and the pathological phosphorylation of TDP-43.
Experimental procedures
Constructs
pcDNA3.1(−)-FLAG-tagged full-length wild-type (WT) and NLS-mutated TDP-43, pcDNA3.1(−)–mCherry-CTF (aa 193–414), and pCMV–His6-ubiquitin expression constructs were described before (29, 31, 64). The TDP-43 FL and CTF lysine mutants were generated by two-step site-directed mutagenesis of TDP-43WT or CTFWT and cloned into pcDNA3.1(−) with a 5′-FLAG tag or 5′-mCherry tag via BamHI/HindIII. The primer sequences are available on request.
Antibodies
The following antibodies were used in this study for Western blotting (WB) or immunofluorescence stainings (IF): mouse anti-β-actin (WB 1:50.000; catalog no. A5441, Sigma, clone AC-15); mouse anti-GAPDH (WB 1:35.000; catalog no. H86504M, Biodesign International, clone 6C5); mouse anti-FLAG (IF 1:1000; Sigma, clone M2 affinity-purified); rabbit anti-FLAG (IF 1:500; catalog no. 2368, Cell Signaling); peroxidase-conjugated anti-FLAG (WB 1:2000–1:60,000; catalog no. A8592, Sigma, clone M2); mouse anti-His6 (WB 1:3000; catalog no. 27-4710-01, Amersham Biosciences); mouse anti-His6 (WB 1:5.000–10.000; catalog no. MA1-21315, Invitrogen, clone HIS.H8); rabbit polyclonal against histone H2A (WB 1:1000; catalog no. 2578, Cell Signaling); rabbit anti-Hsp90 (WB 1:10,000; catalog no. SPA-846, Stressgen); rabbit anti-eIF2α (WB 1:2000; catalog no. 9722, Cell Signaling); rabbit anti-phospho-eIF2α (WB 1:2000; catalog no. ab4837, Abcam, clone S51); rabbit anti-PARP (WB 1:4000; catalog no. 9542, Cell Signaling); rabbit anti-dsRed “mCherry” (WB 1:5000; catalog no. 632496, Clontech); rabbit anti-TDP-43 (WB 1:10,000, IF 1:1000; catalog no. 10782-2-AP, ProteinTech Group); rat anti-phospho-TDP-43 (Ser-409/410) (WB 1:10, IF 1:50; M. Neumann, clone 1D3); mouse anti-ubiquitin (mono- and poly-) (WB 1:4000; catalog no. MAB1510, Millipore, clone Ubi-1). The secondary HRP-conjugated antibodies for Western blot analysis were purchased from Jackson ImmunoResearch (1:5000–1:30,000), and secondary AlexaFluor-488- or -568-conjugated antibodies for immunofluorescence were from Invitrogen (1:1000–1:2000).
Cell culture, transfection, proteasomal inhibition, and cell stress induction
HEK293E cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS at 37 °C and 5% CO2. Depending on the experiment, 0.1–3 × 106 cells were seeded in 6-well, 6-, 10-, or 15-cm cell culture dishes. After 24 h, the cells were transiently transfected with DNA with FuGENE 6 (Promega) following the manufacturer's instructions. For immunofluorescence studies, the ratio of DNA/FuGENE 6 was 1:4.5 diluted in OptiMEM, and for all other studies, the ratio was 1:3. Further analysis of the cells was performed 24–72 h after the transfection.
For proteasomal inhibition, the medium was withdrawn by suction from the cells, replaced with fresh DMEM, 10% FCS, with 10 μm MG-132 (Sigma), and incubated for the indicated time points. As control, cells were treated with 1:2000 diluted DMSO in DMEM.
To induce osmotic or oxidative stress, cells were cultivated for 1 h in fresh DMEM, 10% FCS with 0.4 m sorbitol or 200 μm sodium arsenite (all Sigma). Heat shock was induced by cultivation for 30 min at 43 °C.
Preparation of cell lysates for Western blotting
HEK293E cells (0.1 × 106 cells/well) grown in a 6×-well were lysed either in RIPA lysis buffer (50 mm Tris/HCl, pH 8.0; 150 mm NaCl; 1% Nonidet P-40; 0.5% deoxycholate; 0.1% SDS; 10 mm NaPPi) containing 1× Complete protease inhibitor mixture (Roche Applied Science) for 30 min on ice or in 8 m urea buffer (10 mm Tris, pH 8.0; 100 mm NaH2PO4; 8 m urea), followed by DNA shearing with a 23-gauge needle. Cell debris was pelleted at 14,000 × g for 15 min at 4 °C, and total protein concentration was determined either with the bicinchoninic acid (BCA) protein assay kit (Pierce) for the RIPA lysates or with the Bradford assay (Bio-Rad) for the urea lysates. 5–10 μg of protein was boiled in 1× Laemmli buffer at 95 °C for 5 min (RIPA lysate) or incubated at room temperature for 10 min (urea lysate) and subjected to Western blot analysis.
Sequential extraction of HEK293E proteins
HEK293E cells (0.35 × 106 cells/dish) cultivated in a 6-cm cell culture dish were lysed in RIPA lysis buffer (50 mm Tris/HCl, pH 8.0; 150 mm NaCl; 1% Nonidet P-40; 0.5% deoxycholate; 0.1% SDS; 10 mm NaPPi) containing 1× Complete protease inhibitor and PhosphoStop (both from Roche Applied Science) and pelleted at 14,000 × g for 15 min at 4 °C. RIPA-insoluble pellets were washed twice with RIPA buffer and lysed in 8 m urea buffer (10 mm Tris, pH 8.0; 100 mm NaH2PO4; 8 m urea). Total protein concentration of RIPA lysates was determined with bicinchoninic acid (BCA) protein assay kit (Pierce). Total protein concentration of urea lysates was determined with the Bradford assay (Bio-Rad). 5 μg of protein of the RIPA fraction was boiled in 1× Laemmli buffer at 95 °C for 5 min, and 2.5–5 μg of the urea fraction was incubated with 1× Laemmli buffer at room temperature for 10 min, and both were analyzed by Western blotting.
Pulldown of total ubiquitinylated proteins with nickel-NTA–affinity chromatography
HEK293E cells (1 × 106 cells/dish) grown in a 10-cm cell culture dish were co-transfected with the indicated constructs and His6-ubiquitin for 48 h. Cells were washed with PBS and lysed with 8 m urea buffer (10 mm Tris, pH 8.0; 100 mm NaH2PO4; 8 m urea) containing 10 mm imidazole. The DNA was sheared by passing the lysate 20 times through a 23-gauge needle, and cell debris was pelleted at 14,000 × g for 15 min at 4 °C. The protein concentration was determined by the Bradford assay (Bio-Rad protein assay), and total protein (500–1000 μg) was incubated with Ni-NTA–agarose beads (Qiagen) for 3 h at room temperature. Beads were washed three times with washing buffer (10 mm Tris, pH 6.3; 100 mm NaH2PO4; 8 m urea) containing 20 mm imidazole, and proteins were eluted in 3× Laemmli buffer at 95 °C for 10 min. Total protein and one-tenth of eluates were subjected to Western blot analysis.
Pulldown of sequentially extracted His6-tagged TDP-43
HEK293E cells (3 × 106 cells/dish) that were cultivated in two 15-cm cell culture dishes were transfected with N- or C-terminally His6-tagged TDP-43 for 48 h. Ubiquitinylation was stabilized by a 6-h MG-132 treatment. The cells were lysed under native conditions in 1000 μl of Nonidet P-40 lysis buffer (50 mm NaH2PO4, pH 8.0; 300 mm NaCl; 1% Nonidet P-40) containing 1× Complete protease inhibitors and 10 mm imidazole for 30 min on ice and pelleted at 14,000 × g for 15 min at 4 °C. The BCA protein assay kit (Pierce) was used to measure the total protein concentration of the Nonidet P-40–soluble fraction. Nonidet P-40–insoluble pellets were washed two times with Nonidet P-40 buffer and lysed in 500 μl of 8 m urea buffer. The DNA was sheared by passing the lysates 20 times through a 23-gauge needle. The protein concentration of the urea-soluble fraction was determined with the Bradford assay (Bio-Rad). The pulldown with 50 μl of Ni-NTA–agarose beads (Qiagen) was performed with 1000 μg of Nonidet P-40 and ∼1500 μg of urea lysates for 4 h at 4 °C (Nonidet P-40 lysates) or room temperature (urea lysates). The beads were washed three times with either Nonidet P-40 buffer or urea wash buffer containing 20 mm imidazole, and proteins were eluted with 50 μl of 3× Laemmli buffer at 95 °C for 10 min. 5 μg of total proteins or one-tenth of eluates were subjected to WB. The remaining eluates were subjected to LC-MS/MS analysis.
LC-MS/MS
Pulldown eluates were subjected to SDS-PAGE. A protein band corresponding to the expected molecular weight of monoubiquitinylated TDP-43 and visible after Coomassie Brilliant Blue stain was excised for MS analysis. In addition, a higher molecular weight region in the gel above the theoretical molecular weight of TDP-43 showing a Coomassie-stained smear, potentially containing polyubiquitinylated isoforms of TDP-43, was excised. Sample preparation and mass spectrometric analysis were performed as described previously (65). To avoid artifacts in the analysis of the diGly-specific mass shifts, cysteine residues were reduced by DTT following carbamylation by chloroacetamide instead of iodoacetamide prior to in-gel proteolysis by trypsin (Promega) (66). Extracted peptides were analyzed by LC-MS/MS using a nanoflow HPLC system (Ultimate 3000 RSLC; ThermoFisher Scientific) coupled to an Orbitrap Fusion (ThermoFisher Scientific) tandem mass spectrometer. Peptides were separated by reversed C-18 chromatography and 120-min gradients. MS1 spectra were acquired in the Orbitrap at 120,000 resolution. Precursors were selected with the top speed method and 3-s cycle time. HCD-MS2 spectra were acquired with linear ion trap detection. For database search, tandem mass spectra were extracted by MSConvert (ProteoWizard version 3.0.6938). Charge state deconvolution and de-isotoping were not performed. All MS/MS samples were analyzed using Mascot (Matrix Science; version 2.5.1). Mascot was set up to search the SwissProt database (selected for Homo sapiens, 2015_03, 20203 entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 ppm. Carbamidomethyl of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine, acetylation of lysine, and di-Gly modification of lysine, observed after tryptic proteolysis of lysine-conjugated ubiquitin, were specified in Mascot as variable modifications.
Nuclear-cytoplasmic fractionation
HEK293E cells grown in a 10-cm cell culture dish (1 × 106 cells/dish) were transfected with the indicated proteins and were harvested 48 h later in ice-cold PBS. A small amount of cells was lysed in RIPA buffer and kept as total protein fraction. The remaining cells were incubated with 1 ml of ice-cold hypotonic buffer (10 mm HEPES/KOH, pH 7.6; 1.5 mm MgCl2; 10 mm KCl; 0.1 mm DTT) + 1× Complete protease inhibitor (Roche Applied Science) for 5 min on ice. Nuclei were released by breaking up the cells with a Dounce tissue homogenizer with 20 strokes with a tight pestle and separated from the soluble cytoplasmic proteins by centrifugation at 228 × g for 5 min at 4 °C. To minimize cross-contamination, half of the supernatant containing the cytoplasmic proteins was transferred into a new reaction tube, and the remaining supernatant was discarded. The cytoplasmic proteins were lysed by adding 5× RIPA buffer (250 mm Tris/HCl, pH 7.4; 750 mm NaCl; 5% Nonidet P-40; 2.5% deoxycholate), followed by clearing of the lysates at 28,000 × g for 10 min at 4 °C. The nuclear pellet was washed two times in hypotonic buffer and further purified by sucrose gradient centrifugation: 400 μl of S3 buffer (0.88 m sucrose, 0.5 mm MgCl2) + 1× Complete protease inhibitor was overlaid with the nuclear pellet resuspended in 400 μl of S1 buffer (0.25 m sucrose, 10 mm MgCl2) + 1× Complete protease inhibitor, and centrifuged at 2800 × g for 10 min. The purified nuclear pellet was lysed in 8 m urea buffer, and DNA was sheared with a 23-gauge needle. The protein concentration of the total and the cytoplasmic fraction was determined with the BCA assay, and the concentration of the nuclear protein fraction was measured with the Bradford assay. 5 μg of each fraction were subjected to Western blot analysis.
Western blot analysis
The denatured protein samples were separated on 7.5, 10, 12.5, or 15% self-cast polyacrylamide gels or on 4–12% BisTris NuPAGE gradient gels (Invitrogen) and then transferred onto Hybond-P polyvinylidene difluoride membranes (Millipore). The membranes were blocked in 5% skim milk/TBST or 5% BSA/TBST and incubated with primary antibody in Western Blocking Reagent (Roche Applied Science) at 4 °C overnight. This was followed by incubation with HRP-conjugated secondary antibodies for 1–2 h at room temperature. The detection of proteins was performed with the Immobilon Western chemiluminescent HRP substrate (Millipore) on Amersham Biosciences HyperfilmTM ECL (GE Healthcare).
Immunofluorescence
HEK293E cells were seeded in 6-well plates (0.1 × 106 cells/well), transfected as indicated on the 2nd day, split 1:5, and seeded on poly-d-lysine (Sigma) and collagen (Cohesion)-coated coverslips on the 3rd day. After 48 h, the cells were fixed with 4% (w/v) PFA/PBS for 25 min, permeabilized with 1% Triton X-100/PBS for 5 min, and blocked for 1 h with 10% normal goat serum/PBS or 5% BSA/PBS at room temperature. The primary antibody incubation was performed in 1% BSA/PBS for 2 h at room temperature, followed by incubation with secondary Alexa-Fluor–conjugated antibodies in the dark for 2 h at room temperature. The nuclei were counterstained with Hoechst 33342 (2 μg/ml/PBS) for 10 min at room temperature, and the coverslips were mounted in fluorescent mounting medium (Dako) onto microscope slides. The cells were analyzed with ApoTome Imaging system and processed with AxioVision software (both Zeiss).
Data analysis, quantifications, and statistics
Quantification analyses of Western blotting signals were done with ImageJ software (version 1.47, National Institute of Health). The statistical significance of the data were calculated with paired, two-sided Student's t test. Probability values below 0.05 (p < 0.05) were considered as statistically significant. Quantitative data are expressed as mean ± S.D. from at least three different experiments, unless otherwise stated.
Author contributions
F. H., C. J. G., and P. J. K. conceptualization; F. H. resources; F. H., C. J. G., and P. J. K. formal analysis; F. H. validation; F. H. investigation; F. H. visualization; F. H., F. v. Z., and C. J. G. methodology; F. H. and P. J. K. writing-original draft; P. J. K. supervision; P. J. K. funding acquisition; M. E. cloned some constructs during internship; F. v. Z. performed mass spectrometry; C. J. G. mass spectrometry.
Supplementary Material
Acknowledgments
We thank the staff of the Core Facility for Medical Bioanalytics at the Institute for Ophthalmic Research (University of Tübingen) for technical assistance in MS, Manuela Neumann for providing phospho-TDP-43 antibody, and Jorge García Morato and Jonas Kosten for critical reading of the manuscript.
This work was supported by the NOMIS Foundation, German Research Foundation (DFG) Grant KA1675/3-2, the German Center for Neurodegenerative Diseases (DZNE) within the Helmholtz Association, and the Hertie Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
- ALS
- amyotrophic lateral sclerosis
- CTF
- C-terminal fragment
- diGly
- diglycine
- eIF2α
- eukaryotic Initiation Factor 2 subunit α
- FL
- full length
- FTLD
- frontotemporal lobar degeneration
- FUS
- Fused in Sarcoma
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- GRD
- glycine-rich domain
- HEK
- human embryonic kidney
- HMW
- higher molecular weight smear
- HRP
- horseradish peroxidase
- NES
- nuclear export signal
- NLS
- nuclear import signal
- PARP
- poly(ADP-ribose) polymerase
- RIPA
- radioimmunoprecipitation assay
- RRM
- RNA recognition motif
- UPS
- ubiquitin proteasome system
- aa
- amino acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- WB
- Western blotting
- Ni-NTA
- nickel-nitrilotriacetic acid
- DMEM
- Dulbecco's modified Eagle's medium
- FCS
- fetal calf serum
- IF
- immunofluorescence.
References
- 1. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 2. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., and Oda T. (2006) TDP-43 is a component of ubiquitin-positive τ-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 10.1016/j.bbrc.2006.10.093 [DOI] [PubMed] [Google Scholar]
- 3. Ling H., Kara E., Bandopadhyay R., Hardy J., Holton J., Xiromerisiou G., Lees A., Houlden H., and Revesz T. (2013) TDP-43 pathology in a patient carrying G2019S LRRK2 mutation and a novel p.Q124E MAPT. Neurobiol. Aging 34, 2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ratti A., and Buratti E. (2016) Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 138, Suppl. 1, 95–111 10.1111/jnc.13625 [DOI] [PubMed] [Google Scholar]
- 5. Aulas A., and Vande Velde C. (2015) Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell. Neurosci. 9, 423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., and Lee V. M. (2009) Phosphorylation of Ser-409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 10.1007/s00401-008-0477-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., and Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 10.1002/ana.21425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F. E., Akiyama H., Hisanaga S., and Hasegawa M. (2008) Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett. 582, 2899–2904 10.1016/j.febslet.2008.07.027 [DOI] [PubMed] [Google Scholar]
- 9. Kapeli K., Martinez F. J., and Yeo G. W. (2017) Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet. 136, 1193–1214 10.1007/s00439-017-1830-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uryu K., Nakashima-Yasuda H., Forman M. S., Kwong L. K., Clark C. M., Grossman M., Miller B. L., Kretzschmar H. A., Lee V. M., Trojanowski J. Q., and Neumann M. (2008) Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol. 67, 555–564 10.1097/NEN.0b013e31817713b5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidson Y. S., Raby S., Foulds P. G., Robinson A., Thompson J. C., Sikkink S., Yusuf I., Amin H., DuPlessis D., Troakes C., Al-Sarraj S., Sloan C., Esiri M. M., Prasher V. P., Allsop D., et al. (2011) TDP-43 pathological changes in early onset familial and sporadic Alzheimer's disease, late onset Alzheimer's disease and Down's syndrome: association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 122, 703–713 10.1007/s00401-011-0879-y [DOI] [PubMed] [Google Scholar]
- 12. Yokota O., Davidson Y., Bigio E. H., Ishizu H., Terada S., Arai T., Hasegawa M., Akiyama H., Sikkink S., Pickering-Brown S., and Mann D. M. (2010) Phosphorylated TDP-43 pathology and hippocampal sclerosis in progressive supranuclear palsy. Acta Neuropathol. 120, 55–66 10.1007/s00401-010-0702-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kametani F., Obi T., Shishido T., Akatsu H., Murayama S., Saito Y., Yoshida M., and Hasegawa M. (2016) Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281 10.1038/srep23281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seyfried N. T., Gozal Y. M., Dammer E. B., Xia Q., Duong D. M., Cheng D., Lah J. J., Levey A. I., and Peng J. (2010) Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol. Cell. Proteomics 9, 705–718 10.1074/mcp.M800390-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buratti E. (2018) TDP-43 post-translational modifications in health and disease. Expert Opin. Ther. Targets 22, 279–293 10.1080/14728222.2018.1439923 [DOI] [PubMed] [Google Scholar]
- 16. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 17. Akutsu M., Dikic I., and Bremm A. (2016) Ubiquitin chain diversity at a glance. J. Cell Sci. 129, 875–880 10.1242/jcs.183954 [DOI] [PubMed] [Google Scholar]
- 18. Lee S., Lee T. A., Song S. J., Park T., and Park B. (2015) Hyperproduction of IL-6 caused by aberrant TDP-43 overexpression in high-fat diet-induced obese mice. FEBS Lett. 589, 1825–1831 10.1016/j.febslet.2015.05.040 [DOI] [PubMed] [Google Scholar]
- 19. van Eersel J., Ke Y. D., Gladbach A., Bi M., Götz J., Kril J. J., and Ittner L. M. (2011) Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS ONE 6, e22850 10.1371/journal.pone.0022850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., and Rouleau G. A. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 10.1038/ng.132 [DOI] [PubMed] [Google Scholar]
- 21. Caccamo A., Majumder S., Deng J. J., Bai Y., Thornton F. B., and Oddo S. (2009) Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 284, 27416–27424 10.1074/jbc.M109.031278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., and Simonsen A. (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brady O. A., Meng P., Zheng Y., Mao Y., and Hu F. (2011) Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J. Neurochem. 116, 248–259 10.1111/j.1471-4159.2010.07098.x [DOI] [PubMed] [Google Scholar]
- 24. Scotter E. L., Vance C., Nishimura A. L., Lee Y. B., Chen H. J., Urwin H., Sardone V., Mitchell J. C., Rogelj B., Rubinsztein D. C., and Shaw C. E. (2014) Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 127, 1263–1278 10.1242/jcs.140087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker A. K., Tripathy K., Restrepo C. R., Ge G., Xu Y., Kwong L. K., Trojanowski J. Q., and Lee V. M. (2015) An insoluble frontotemporal lobar degeneration-associated TDP-43 C-terminal fragment causes neurodegeneration and hippocampus pathology in transgenic mice. Hum. Mol. Genet. 24, 7241–7254 10.1093/hmg/ddv424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urushitani M., Sato T., Bamba H., Hisa Y., and Tooyama I. (2010) Synergistic effect between proteasome and autophagosome in the clearance of polyubiquitinated TDP-43. J. Neurosci. Res. 88, 784–797 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Fan H., Ying Z., Li B., Wang H., and Wang G. (2010) Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Lett. 469, 112–116 10.1016/j.neulet.2009.11.055 [DOI] [PubMed] [Google Scholar]
- 28. Hebron M., Chen W., Miessau M. J., Lonskaya I., and Moussa C. E. (2014) Parkin reverses TDP-43-induced cell death and failure of amino acid homeostasis. J. Neurochem. 129, 350–361 10.1111/jnc.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hans F., Fiesel F. C., Strong J. C., Jäckel S., Rasse T. M., Geisler S., Springer W., Schulz J. B., Voigt A., and Kahle P. J. (2014) UBE2E ubiquitin-conjugating enzymes and ubiquitin isopeptidase Y regulate TDP-43 protein ubiquitination. J. Biol. Chem. 289, 19164–19179 10.1074/jbc.M114.561704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dammer E. B., Fallini C., Gozal Y. M., Duong D. M., Rossoll W., Xu P., Lah J. J., Levey A. I., Peng J., Bassell G. J., and Seyfried N. T. (2012) Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS ONE 7, e38658 10.1371/journal.pone.0038658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 32. Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., and Lee V. M. (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 173, 182–194 10.2353/ajpath.2008.080003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skaug B., and Chen Z. J. (2010) Emerging role of ISG15 in antiviral immunity. Cell 143, 187–190 10.1016/j.cell.2010.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishimura A. L., Zupunski V., Troakes C., Kathe C., Fratta P., Howell M., Gallo J. M., Hortobágyi T., Shaw C. E., and Rogelj B. (2010) Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771 10.1093/brain/awq111 [DOI] [PubMed] [Google Scholar]
- 37. Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., and Lee V. M. (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309 10.1074/jbc.M800342200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pesiridis G. S., Tripathy K., Tanik S., Trojanowski J. Q., and Lee V. M. (2011) A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J. Biol. Chem. 286, 18845–18855 10.1074/jbc.M111.231118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Araki W., Minegishi S., Motoki K., Kume H., Hohjoh H., Araki Y. M., and Tamaoka A. (2014) Disease-associated mutations of TDP-43 promote turnover of the protein through the proteasomal pathway. Mol. Neurobiol. 50, 1049–1058 10.1007/s12035-014-8644-6 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., and Petrucelli L. (2007) Progranulin mediates caspase-dependent cleavage of TAR DNA-binding protein-43. J. Neurosci. 27, 10530–10534 10.1523/JNEUROSCI.3421-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki H., Lee K., and Matsuoka M. (2011) TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. J. Biol. Chem. 286, 13171–13183 10.1074/jbc.M110.197483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lazebnik Y. A., Kaufmann S. H., Desnoyers S., Poirier G. G., and Earnshaw W. C. (1994) Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371, 346–347 10.1038/371346a0 [DOI] [PubMed] [Google Scholar]
- 43. Clague M. J., and Urbé S. (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 10.1016/j.cell.2010.11.012 [DOI] [PubMed] [Google Scholar]
- 44. Chen Z., Chen Y. Z., Wang X. F., Wang C., Yan R. X., and Zhang Z. (2011) Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS ONE 6, e22930 10.1371/journal.pone.0022930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martinez-Lopez N., Athonvarangkul D., and Singh R. (2015) Autophagy and aging. Adv. Exp. Med. Biol. 847, 73–87 10.1007/978-1-4939-2404-2_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruegsegger C., and Saxena S. (2016) Proteostasis impairment in ALS. Brain Res. 1648, 571–579 10.1016/j.brainres.2016.03.032 [DOI] [PubMed] [Google Scholar]
- 47. Cohen T. J., Hwang A. W., Restrepo C. R., Yuan C. X., Trojanowski J. Q., and Lee V. M. (2015) An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845 10.1038/ncomms6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liachko N. F., Guthrie C. R., and Kraemer B. C. (2010) Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J. Neurosci. 30, 16208–16219 10.1523/JNEUROSCI.2911-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y. J., Gendron T. F., Xu Y. F., Ko L. W., Yen S. H., and Petrucelli L. (2010) Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA-binding protein-43 C-terminal fragments. Mol. Neurodegener. 5, 33 10.1186/1750-1326-5-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Catic A., Collins C., Church G. M., and Ploegh H. L. (2004) Preferred in vivo ubiquitination sites. Bioinformatics 20, 3302–3307 10.1093/bioinformatics/bth407 [DOI] [PubMed] [Google Scholar]
- 51. Choi J. Y., Ryu J. H., Kim H. S., Park S. G., Bae K. H., Kang S., Myung P. K., Cho S., Park B. C., and Lee D. H. (2007) Co-chaperone CHIP promotes aggregation of ataxin-1. Mol. Cell. Neurosci. 34, 69–79 10.1016/j.mcn.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 52. Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., and Diehl J. A. (2006) Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-αB crystallin) complex. Mol. Cell 24, 355–366 10.1016/j.molcel.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., Harper J. W., and Elledge S. J. (2001) Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 10.1126/science.1065203 [DOI] [PubMed] [Google Scholar]
- 54. Kai M., Ueno N., and Kinoshita N. (2015) Phosphorylation-dependent ubiquitination of paraxial protocadherin (PAPC) controls gastrulation cell movements. PLoS ONE 10, e0115111 10.1371/journal.pone.0115111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jadhav T., and Wooten M. W. (2009) Defining an embedded code for protein ubiquitination. J. Proteomics Bioinform. 2, 316 10.4172/jpb.1000091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 57. Chen B. B., and Mallampalli R. K. (2009) Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme cytidylyltransferase. Mol. Cell. Biol. 29, 3062–3075 10.1128/MCB.01824-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pentecost M., Vashisht A. A., Lester T., Voros T., Beaty S. M., Park A., Wang Y. E., Yun T. E., Freiberg A. N., Wohlschlegel J. A., and Lee B. (2015) Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog. 11, e1004739 10.1371/journal.ppat.1004739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marchenko N. D., Hanel W., Li D., Becker K., Reich N., and Moll U. M. (2010) Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-α3 binding. Cell Death Differ. 17, 255–267 10.1038/cdd.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dormann D., Madl T., Valori C. F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I. R., Neumann M., and Haass C. (2012) Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 10.1038/emboj.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kinoshita Y., Ito H., Hirano A., Fujita K., Wate R., Nakamura M., Kaneko S., Nakano S., and Kusaka H. (2009) Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 68, 1184–1192 10.1097/NEN.0b013e3181bc3bec [DOI] [PubMed] [Google Scholar]
- 62. Kim H. J., and Taylor J. P. (2017) Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 96, 285–297 10.1016/j.neuron.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chou C. C., Zhang Y., Umoh M. E., Vaughan S. W., Lorenzini I., Liu F., Sayegh M., Donlin-Asp P. G., Chen Y. H., Duong D. M., Seyfried N. T., Powers M. A., Kukar T., Hales C. M., Gearing M., et al. (2018) TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 10.1038/s41593-017-0047-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., et al. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221 10.1038/emboj.2009.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gloeckner C. J., Boldt K., Schumacher A., and Ueffing M. (2009) Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Methods Mol. Biol. 564, 359–372 10.1007/978-1-60761-157-8_21 [DOI] [PubMed] [Google Scholar]
- 66. Nielsen M. L., Vermeulen M., Bonaldi T., Cox J., Moroder L., and Mann M. (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods 5, 459–460 10.1038/nmeth0608-459 [DOI] [PubMed] [Google Scholar]
- 67. Wang P., Wander C. M., Yuan C. X., Bereman M. S., and Cohen T. J. (2017) Acetylation-induced TDP-43 pathology is suppressed by an HSF1-dependent chaperone program. Nat. Commun. 8, 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.