Abstract
Autophagy, a pathway for bulk protein degradation and removal of damaged organelles, represents one of the major responses of cells to stress, thereby exerting a strict control on their correct functioning. Consequently, this process has been involved in the pathogenesis and therapeutic responses of several human diseases. Mitogen-activated protein (MAP) kinase 15 (MAPK15) is an atypical member of the MAP kinase family that recently emerged as a key modulator of autophagy and, through this, of cell transformation. Still, no information is available about signaling pathways mediating the effect of MAPK15 on this process, nor is it known which phase of autophagosome biogenesis is affected by this MAP kinase. Here, we demonstrate that MAPK15 stimulated 5′-AMP–activated protein kinase–dependent activity of UNC-51-like kinase 1 (ULK1), the only protein kinase among the ATG-related proteins, toward downstream substrates and signaling intermediates. Importantly, MAPK15 directly interacted with the ULK1 complex and mediated ULK1 activation induced by starvation, a classical stimulus for the autophagic process. In turn, ULK1 and its highly homologous protein ULK2 are able to transduce MAPK15 signals stimulating early phases of autophagosomal biogenesis in a multikinase cascade that offers numerous potential targets for future therapeutic intervention in cancer and other autophagy-related human diseases.
Keywords: autophagy, vesicles, trafficking, mitogen-activated protein kinase (MAPK), cell signaling
Introduction
Eukaryotic cells are constantly subjected to changes in their micro-environmental conditions. Therefore, they continuously need to adapt to such changes to optimize their survival strategies through the concerted action of different cell-response pathways, whose role is either to allow cells to adapt to novel external conditions or to commit to suicidal mechanisms, when excessively damaged. A key mechanism involved in adaptation to stress is macro-autophagy (hereafter autophagy), an intracellular pathway for bulk protein degradation and removal of damaged organelles (1). Based on its functions, autophagy allows survival in conditions of cellular stress and exerts a strict control on the correct functioning of proteins or even entire organelles. Indeed, it constantly participates in metabolic functions of the cells by eliminating old and damaged organelles, such as mitochondria, and in very basic decisions such as growing and dividing or dying (2). As one of the major responses to cell stress, it is not a surprise that autophagy has been implicated in several pathological conditions such as infections, neurodegenerative diseases, and cancer (3). Several stress-signaling cascades impinge on the activation of the ULK1 (in mammals; Atg1 in yeast) multiprotein complex that participates in the very early phases of autophagosome formation (4) and coordinates the autophagic response to face such stimuli (5). The human ULK1 complex consists of the ULK1 protein kinase itself, the FIP200 (i.e. focal adhesion kinase family kinase interacting protein of 200 kDa) scaffold, and the autophagy-related proteins 13 (ATG13) and 101 (ATG101) (6). Importantly, mammals have four ULK1 homologs, with ULK2 showing the most extensive conservation and partial functional redundancy to ULK1 in the control of conventional autophagy (4).
MAPK15 is an atypical member of the MAP 4 kinase family (7), whose increased expression has been recently correlated with malignant transformation of human gastric mucosa (8) and male germ cell tumors (9). Furthermore, its activity is modulated by relevant human oncogenes (10, 11) and is required for transformation sustained by human colon cancer (12) and chronic myeloid leukemia (CML) cells (10). Interestingly, MAPK15 is also required for proliferating cell nuclear antigen protein stability (13), for controlling reactive oxygen species generation (9), and for telomerase activity (14), whereas its activity and expression are also regulated by DNA damage (15), thus possibly contributing to the maintenance of genomic integrity.
The best-characterized stimulus of autophagy is nutrient deprivation. We have already shown that MAPK15 (ERK8 and ERK7) is able to control autophagy induced by starvation (16). Importantly, we have demonstrated that MAPK15 ability to control autophagy is key for CML and embryonic carcinoma tumor cells to induce in vivo tumor formation (9, 10).
Results
MAPK15 controls phosphorylation of the ULK1 protein
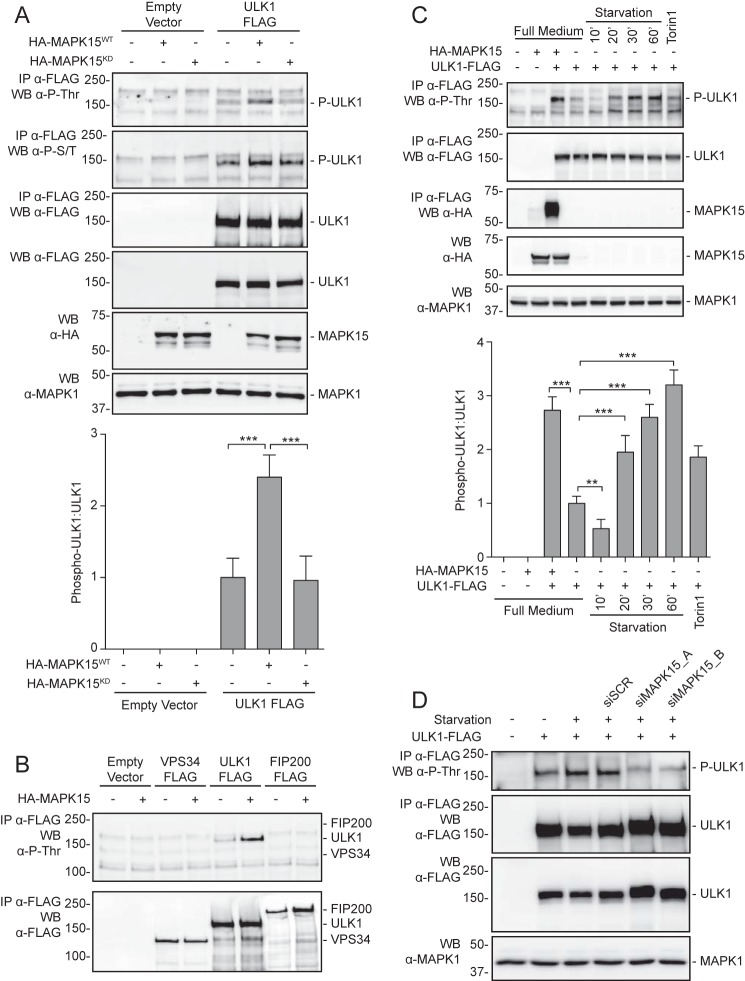
ULK1 is an extensively phosphorylated protein regulated by nutrient cellular status through mTOR and AMPK kinases (6). The ULK1 complex integrates a multitude of pathways converging on its phosphorylation that, in turn, initiate autophagy in response to appropriate stimuli. We therefore asked whether MAPK15, a known regulator of the autophagic process (16), could induce ULK1 activation by impinging on its phosphorylation. As multiple phosphorylation sites have already been described for ULK1 (17, 18), we first decided to use consensus-independent anti-phosphoserine or anti-phosphothreonine antibodies to discriminate general changes in ULK1 phosphorylation induced by MAPK15. We therefore cotransfected, in HEK293T cells, MAPK15 and a FLAG epitope-tagged ULK1 protein and, upon immunoprecipitation with anti-FLAG antibodies, observed a strong increase of ULK1 Ser/Thr phosphorylation, in these experimental conditions (Fig. 1A). Importantly, a MAPK15 kinase-dead (MAPK15KD) mutant was unable to induce ULK1 Ser/Thr phosphorylation while being expressed at comparable levels as the MAPK15 WT (MAPK15WT) protein (Fig. 1A), supporting the specificity of our approach. As a further control, we also cotransfected MAPK15 with FIP200 (a component of the ULK1 complex) and VPS34 (a class III phosphatidylinositol 3-kinase also controlling autophagy) FLAG epitope-tagged proteins and demonstrated that in both cases their phosphorylation was unaffected by MAPK15 under our experimental conditions (Fig. 1B and Fig. S1A).
Figure 1.
Evaluation of MAPK15-dependent ULK1 phosphorylation. A, HEK293T cells were transfected with plasmid encoding for ULK1-FLAG in combination with HA-MAPK15WT, HA-MAPK15KD, or the empty vector. After 24 h, total lysates were subject to anti-FLAG immunoprecipitation (IP) and analyzed by Western blot (WB) analysis (n = 3). ULK1 was analyzed for threonine phosphorylation levels, and scores were plotted on the corresponding graph (lower panels, n = 3). B, HEK293T cells were transfected with different plasmid encoding for VPS34, ULK1, or FIP200 FLAG-tagged proteins in combination with HA-MAPK15 or the empty vector. After 24 h, total lysates were subject to anti-FLAG immunoprecipitation and analyzed by Western blot analysis (n = 3). C, HEK293T cells were transfected with plasmid encoding for ULK1-FLAG in combination with HA-MAPK15 or the empty vector. After 24 h, cells were incubated in full medium, starvation medium (10–60 min), or 250 nm Torin1 (2 h). Total lysates were subject to anti-FLAG immunoprecipitation and analyzed by Western blot analysis (n = 3). ULK1 was analyzed for threonine phosphorylation levels, and scores were plotted on the corresponding graph (lower panel, n = 3). D, HEK293T cells were transfected with MAPK15 siRNA (siMAPK15) or SCR siRNA (siSCR) and, after 48 h, further transfected with plasmid encoding for ULK1 FLAG or the empty vector. After 24 h, cells were incubated in full medium or starvation medium (60 min). Total lysates were subject to anti-FLAG immunoprecipitation and analyzed by Western blot analysis (n = 3).
A number of post-translational modifications, mostly phosphorylations able to modify the activity of the ULK1 complex, have been described in the last few years (4). Most of these modifications act in specific contexts, regulating the kinase activity, the binding partners, and integrating different signaling pathways (5). Nonetheless, we are still far from a clear view of the overall status of phosphorylation of ULK1 and of the synergistic effect of all such modifications (19). To gain insight about the positive or negative role of the observed MAPK15-dependent phosphorylation of ULK1 in the context of the autophagic process, we next compared it with well-established inducers of autophagy, such as starvation and Torin1 (an mTOR inhibitor and inducer of autophagy (20)). Both these treatments strongly increased ULK1 phosphorylation, as detected by our anti-phospho-Ser/Thr antibodies, at levels comparable with MAPK15 overexpression (Fig. 1C), thereby suggesting an overall positive connection with such post-translational modification. Based on these results and on our previous demonstration that starvation requires endogenous MAPK15 expression and activity to induce autophagy (16), we next asked whether endogenous MAPK15 was also required for starvation-dependent increase of ULK1 phosphorylation. Interestingly, two already characterized MAPK15-specific unrelated siRNAs (9, 10, 16) strongly reduced ULK1 phosphorylation induced by starvation (Fig. 1D and Fig. S1B). Overall, our results strongly suggest that MAPK15 may take advantage of an ULK1-dependent pathway to control the autophagic process.
MAPK15 stimulates AMPK-dependent ULK1 activity
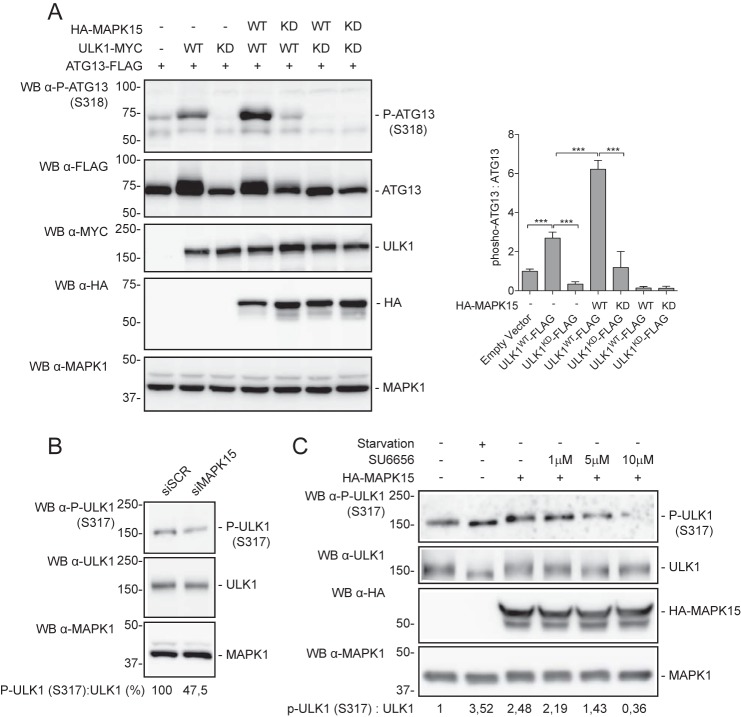
Once activated, ULK1 transduces cellular signals to downstream events by phosphorylating effector proteins involved in autophagy, i.e. BECN1 (21), p62 (22), and ATG101, FIP200, and ATG13 (23). Among them, in a stable isotope labeling by amino acids in cell culture MS experiments, ULK1 was found to phosphorylate human ATG13 at Ser318 in a manner dependent on the Hsp90–Cdc37 chaperone complex (24). Therefore, we took advantage of a commercially available anti-ATG13–pSer318 antibody to demonstrate the ability of MAPK15 for stimulating ULK1 activity by scoring ULK1 phosphorylation of one of its endogenous direct substrates. Indeed, MAPK15 induced an increase in ULK1-dependent ATG13–pSer318 phosphorylation (Fig. 2A). Interestingly, MAPK15KD not only was unable to induce ATG13 phosphorylation, but also reduced basal ULK1-dependent phosphorylation (Fig. 2A), suggesting a dominant-negative effect possibly based on direct protein–protein interaction. Importantly, these data supported a stimulatory role for MAPK15 on ULK1 kinase activity and therefore on its downstream signaling pathways.
Figure 2.
MAPK15 activates ULK1. A, HeLa cells were transfected with plasmid encoding for ATG13-FLAG in combination with HA-MAPK15WT, HA-MAPK15KD, ULK1WT-MYC, ULK1KD-MYC, or the empty vector. After 24 h, total lysates were analyzed by Western blot (WB) analysis (left panel). ATG13 was analyzed for phosphorylation levels (Ser318), and scores were plotted on the graph (right panel, n = 3). B, HeLa cells were transfected with MAPK15 siRNA or SCR siRNA. After 48 h, cells were incubated in starvation medium (60 min), and total lysates were analyzed by Western blot analysis. Endogenous ULK1 was analyzed for phosphorylation (Ser317) and total levels. Inhibition of Ser317 phosphorylation of ULK1 (ratio P-ULK1 (Ser317)/ULK1) induced by MAPK15 down-regulation by specific siRNAs has been expressed as a percentage of the siSCR-transfected sample (control, 100%). C, HeLa cells were transfected with plasmid encoding for HA-MAPK15 or the empty vector (−). After 24 h, cells were treated for 1 h with increasing concentrations of the SU6656 AMPK inhibitor, as indicated. Starved, positive control, cells were incubated in starvation medium for 60 min at 24 h after transfection with an empty vector. Total lysates were analyzed by Western blot analysis (left panel). Endogenous ULK1 was analyzed for phosphorylation (Ser317) and total levels. HA-MAPK15 and MAPK1 levels were also analyzed by Western blotting for normalization purposes.
Under glucose starvation, AMPK promotes autophagy by directly activating ULK1 through phosphorylation of several residues, including Ser317 (4). We therefore hypothesized that MAPK15 could take advantage of this pathway to control autophagy. Therefore, we used a commercially available anti-ULK1–pSer317 antibody as a readout to ascertain whether MAPK15 was able to stimulate AMPK-dependent ULK1 activity, in the context of the autophagic process. Specifically, we first confirmed that endogenous MAPK15 was specifically impinging on AMPK-dependent Ser317 phosphorylation of ULK1 by an siRNA approach (Fig. 2B). Next, we overexpressed MAPK15 and scored endogenous anti-ULK1–pSer317 protein levels also in presence of a specific AMPK inhibitor. Indeed, MAPK15 overexpression was able to induce ULK1-activating phosphorylation at levels comparable with starvation, our positive control, while increasing amounts of the AMPK inhibitor SU6656 (25), and strongly inhibited MAPK15-dependent phosphorylation of Ser317 in ULK1 (Fig. 2C), thus confirming the involvement of AMPK in mediating MAPK15-dependent ULK1 phosphorylation. Importantly, we specifically avoided using the classical AMPK inhibitor, compound C, in this context because of its ability to also inhibiting MAPK15 with greater potency (25).
MAPK15 is part of the ULK1 complex and localized onto autophagosomes
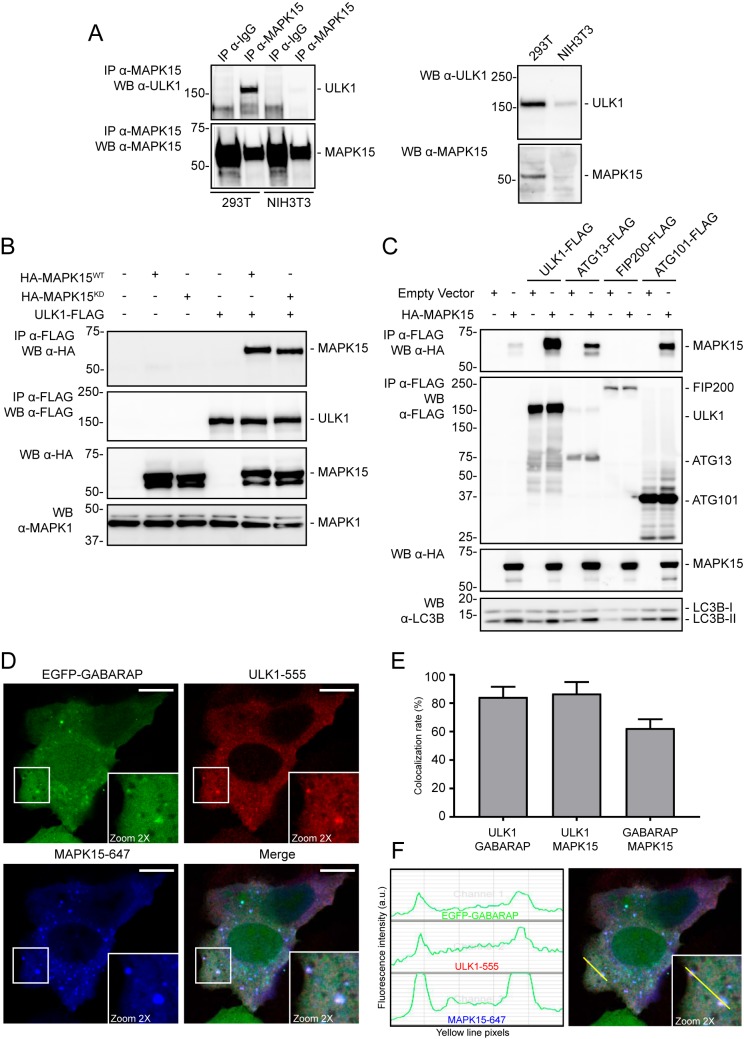
In mammals, ULK1 (and ULK2) may exist in different complexes depending on the metabolic status of the cell and the specific functions, canonical (i.e. autophagy) or noncanonical, with which they participate (26). Based on the role of MAPK15 in autophagy (16) and in regulating ULK1 activity (see above), we sought to investigate the possibility of a direct interaction of this MAP kinase with the ULK1 complex, already involved in controlling the autophagic process (6). To establish an interaction between the two endogenous proteins, we first immunoprecipitated MAPK15 from human 293T cells and next blotted for ULK1 protein, demonstrating coimmunoprecipitation in human cells (Fig. 3A, left panels). As our antibody only recognizes the human protein, as an additional negative control we also performed the same approach in mouse NIH3T3 cells and, as expected, we could detect no relevant coimmunoprecipitation (Fig. 3A, left panels). Importantly, we also showed that the ULK1 protein could be detected in total lysates of both human and mouse cells, whereas MAPK15, as expected, could be detected only in total lysates of human 293T cells (Fig. 3A, right panels). Also, we next demonstrated that such interaction was not affected by MAPK15 kinase activity as an epitope-tagged ULK1 protein could similarly coimmunoprecipitate both MAPK15WT and a MAPK15KD mutant (Fig. 3B), also explaining the ability of the kinase-dead mutant to act as a dominant-negative protein able to inhibit ULK1 basal activity (see Fig. 2A). As ULK1 regulates autophagy as part of a multiprotein complex containing also ATG13, FIP200, and ATG101, we next sought to confirm the interaction of MAPK15 with this specific complex by cotransfecting the MAP kinase with each single FLAG-tagged recognized component, and we proceeded to coimmunoprecipitation by using a FLAG antibody. This approach clearly showed that not only ULK1 but also ATG13 and ATG101 were coimmunoprecipitated by MAPK15 (Fig. 3C), supporting the possibility that this kinase must physically colocalize with the “ULK complex” to stimulate its enzymatic activity upon transduction of upstream stimuli. To further verify this idea, we next examined cellular localization of MAPK15 and ULK1 to autophagosomal compartments. Interestingly, the two kinases colocalized onto GABARAP-positive autophagosomal vesicles (Fig. 3, D–F), confirming our previous hypothesis.
Figure 3.
MAPK15 participates in the ULK1 complex. A, HEK293 and NIH3T3 were harvested and lysed in 1% Nonidet P-40 lysis buffer. The proteins (10 mg per sample) were then subjected to immunoprecipitation (IP) with isotype control IgG (IgG) or MAPK15-specific antibody (custom preparation). Total lysates and immunoprecipitated complexes were subjected to Western blotting (WB) to detect endogenous ULK1 and MAPK15 protein levels. Representative images from three different experiments are shown (n = 3). B, HEK293T cells were transfected with plasmid encoding for ULK1-FLAG in combination with HA-MAPK15WT, HA-MAPK15KD, or the empty vector. After 24 h, total lysates were subject to anti-FLAG immunoprecipitation and analyzed by Western blot analysis (n = 3). C, HEK293T cells were transfected with different plasmids encoding for FLAG-tagged ULK1, ATG13, FIP200, or ATG101 proteins in combination with HA-MAPK15 or the empty vector. After 24 h, total lysates were subject to anti-FLAG immunoprecipitation and analyzed by Western blot analysis (n = 3). D, HeLa cells stably expressing EGFP-GABARAP were transfected with HA-MAPK15 and ULK1-FLAG plasmids and subjected to immunofluorescence analysis after 48 h. Representative images are from three different experiments (n = 3). EGFP-GABARAP is visualized in green, ULK1-FLAG in red, and HA-MAPK15 in blue (upper panels). White squares indicate zoom areas. Scale bars, 10 μm. E, colocalization rate was measured by LAS AF (Leica Microsystem) software, analyzing single cells that express both MAPK15 and ULK1 in addition to EGFP-GABARAP. Thresholds were set at 20% for each channel as suggested from the manufacturer's protocol. Reciprocal colocalization rate between ULK1 and GABARAP, ULK1 and MAPK15, and GABARAP and MAPK15 were evaluated. Measures were obtained by analyzing at least 100 cells/sample from three different experiments. F, colocalization spots were analyzed for fluorescent signal intensity for each channel; the yellow line indicates the measured points on the merged image.
ULK1 and ULK2 mediate MAPK15-dependent stimulation of the autophagic process
MAPK15 activity positively regulates autophagy and is necessary for starvation-dependent induction of this process (16). Based on the key role of ULK1 and of its ULK2 homologous protein in autophagy and on our previously described results, we next investigated the possibility of a direct role for both these proteins in mediating MAPK15 signaling to autophagy. Specifically, we decided to interfere with endogenous ULK1 and ULK2 expression by using a classical siRNA-mediated approach.
Indirect immunofluorescence microscopy, scoring the formation of punctate intracellular vacuoles stained for LC3B, is a key assay to monitor autophagy and to understand its dynamics (27). Therefore, we performed indirect immunofluorescence analysis in HeLa cells overexpressing MAPK15, aimed at scoring an increase in the number of vesicles positive for endogenous LC3B. Importantly, such an approach was performed both in basal conditions and upon treatment with an inhibitor of lysosomal proteases, bafilomycin A1 (BAF), to appreciate autophagic flux in these cells (27) (quantification performed through unbiased analysis using the Quantitation module of the Volocity software). Interestingly, combined transfection of siRNAs recognizing ULK1 and ULK2 strongly reduced MAPK15-dependent autophagic flux (Fig. 4A), whereas, as additional controls, single transfection of siRNA against ULK1 (siULK1; Fig. 4B and Fig. S2) or ULK2 (siULK2, Fig. 4C and Fig. S3) more limitedly impaired MAPK15 induction of autophagy (from 42 and 56% for siULK1 and siULK2, respectively, to 28% for combined siULK1/siULK2 treatment). To confirm these results, we also took advantage of a different LC3B-based assay used to score autophagy, i.e. Western blot analysis of the amount of the lipidated, autophagosome-associated form of LC3B (LC3B-II). Indeed, the combined use of anti-ULK1 and anti-ULK2 siRNA in cells in basal conditions or treated with BAF also strongly reduced MAPK15-dependent autophagic flux (Fig. 4D).
Figure 4.
ULK1/2 mediates MAPK15-dependent increase in LC3-II–positive vesicles. A, HeLa cells were transfected with ULK1 plus ULK2 siRNA (siULK1/ULK2) or SCR siRNA (siSCR) and, after 48 h, further transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated in full medium or with 100 nm BAF (1 h). Cells were fixed and labeled for endogenous LC3B (red) and for HA-positive cells (green) and counterstained with DAPI for nuclei (blue). Scale bars correspond to 7.5 μm except for lower row in which they are 10 μm. LC3B dots per cell were plotted on the graph (right panel, n = 3). B, HeLa cells were transfected with ULK1 siRNA (siULK1) or SCR siRNA (siSCR) and, after 48 h, further transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated in full medium or with 100 nm BAF (1 h). Cells were fixed and labeled for endogenous LC3B (red) and for HA-positive cells (green) and counterstained with DAPI for nuclei (blue). LC3B dots per cell were plotted on the graph (n = 3). Corresponding images are in Fig. S2. C, same as in B, but using ULK2 siRNA (siULK2) instead of siULK1. Corresponding images are in Fig. S3. D, HeLa cells were transfected with ULK1 plus ULK2 siRNA or SCR siRNA and, after 48 h, further transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated in full medium or with 100 nm BAF (1 h). Total lysates were analyzed by Western blot (WB) analysis for endogenous LC3B (n = 3). LC3B-II levels were plotted on the graph (right panel, n = 3).
To further confirm these results and to expand their biological significance also with a different marker of autophagic vesicles, we used SQSTM1/p62, which, in HeLa cells, shows a cytoplasmic distribution in basal conditions, and although degraded at an increased rate, it is re-localized to autophagic vesicles upon activation of autophagy (16). Indeed, MAPK15 also led to an increase in the number of SQSTM1-positive puncta, which, in turn, diminished upon contemporary down-regulation of ULK1 and ULK2 by specific siRNA (Fig. 5).
Figure 5.
ULK1/2 mediates MAPK15-dependent increase in SQSTM1/p62-positive vesicles. HeLa cells were transfected with ULK1 plus ULK2 siRNA or SCR siRNA and, after 48 h, further transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated in full medium or with 100 nm BAF (6 h). Cells were fixed and labeled for endogenous SQSTM1/p62 (red) and for HA-positive cells (green) and counterstained with DAPI for nuclei (blue). Scale bars correspond to 10 μm except for lower row in which they are 25 μm. SQSTM1 puncta per cell were plotted on the graph (right panel, n = 3).
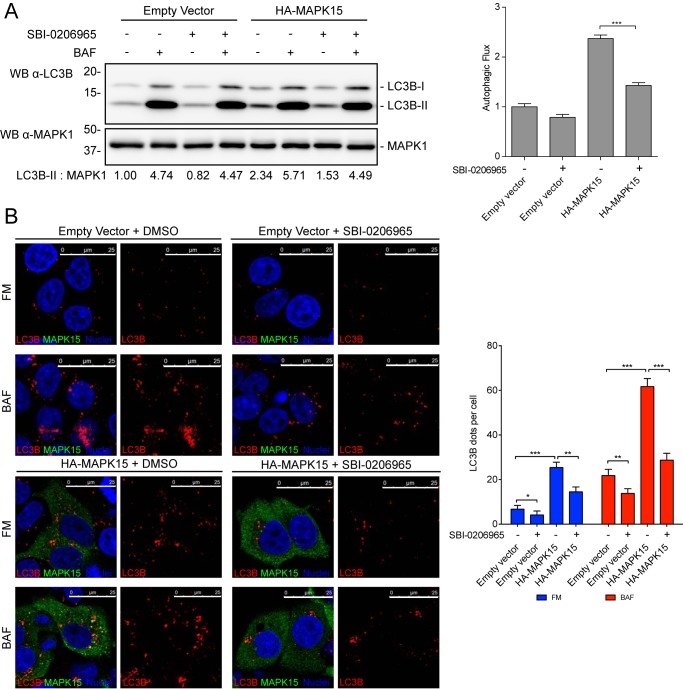
Ultimately, we sought to confirm these data also by using a pharmacological approach. To this aim, we took advantage of an available drug, SBI-0206965, which inhibits both ULK1 and ULK2, with IC50 of 108 and 711 nm, respectively (23), and we used it to score the contribution of these two kinases in the induction of autophagic flux by MAPK15. Indeed, we demonstrated that the ULK1/2 inhibitor efficiently reduced MAPK15-dependent autophagic flux by using both the LC3B-II Western blotting detection assay (Fig. 6A) and the immunofluorescence-based assay detecting the number of autophagosomal vesicles (Fig. 6B). Overall, our data therefore demonstrate that MAPK15 requires endogenous ULK1 and ULK2 proteins to stimulate activation of cellular autophagic machinery.
Figure 6.
Pharmacological approach to evaluate ULK1/2 contribution to autophagy induced by MAPK15. A, HeLa cells were transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated for 6 h in full medium or with 100 nm BAF, in combination with DMSO (carrier) or 10 μm SBI-0206965. Total lysates were analyzed by Western blot (WB) analysis for endogenous LC3B (n = 3). LC3B-II levels were scored below the immunoblots. Autophagic flux evaluation was obtained by the ratio between BAF+ and BAF− of the corresponding sample, and scores were plotted on the graph (right panel, n = 3). B, HeLa cells were transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 48 h, cells were incubated for 6 h in full medium or with 100 nm BAF, in combination with DMSO (carrier) or 10 μm SBI-0206965. Cells were fixed and labeled for endogenous LC3B (red) and for HA-positive cells (green) and counterstained with DAPI for nuclei (blue). Scale bars correspond to 25 μm. LC3B dots per cell were plotted on the graph (right panel, n = 3).
MAPK15 controls early phases of autophagosomal biogenesis through ULK1/2
Although we have clearly demonstrated that MAPK15 controls the rate of autophagic flux in cells in response to starvation (16), as well as in human oncogenes (10), no information is yet available about the specific phase of autophagosomal biogenesis affected by this MAP kinase.
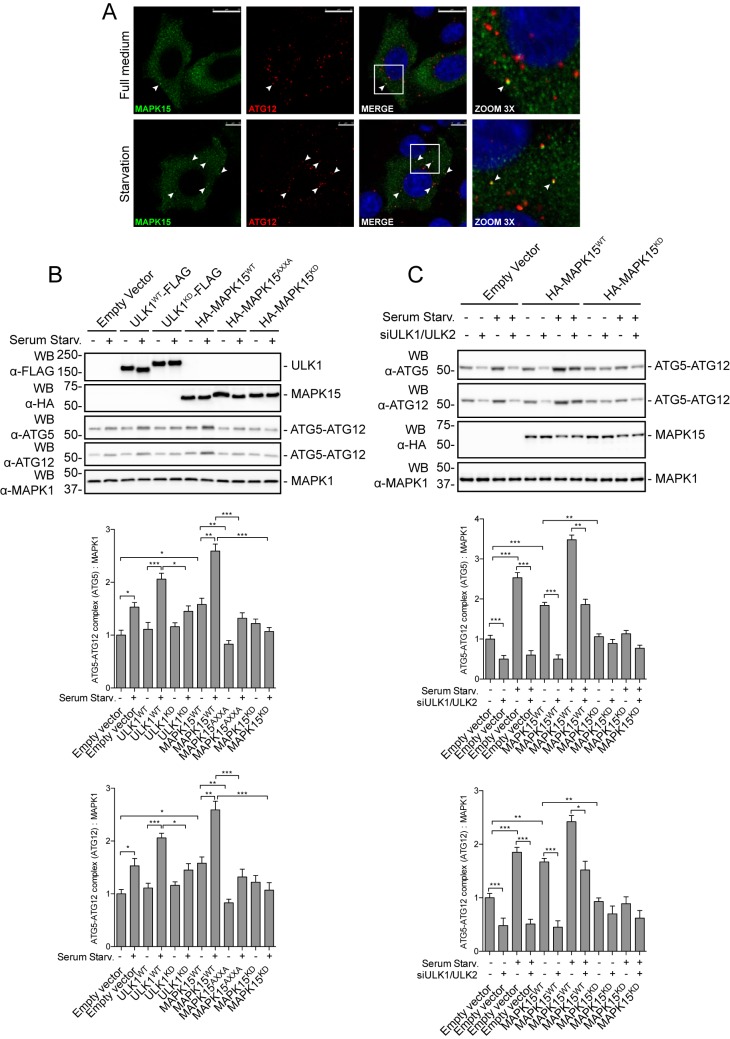
In mammals, the ATG12–ATG5–ATG16L complex predominantly localizes to the outer surface of the isolation membrane, where it is essential for determining the site of LC3 lipidation and for its proper elongation, ultimately dissociating from the membranes at the completion of autophagosome formation (1). Importantly, ATG12 and ATG5 are covalently conjugated to each other thanks to the ubiquitin-like reaction requiring ATP and ATG7 (E1-like) and ATG10 (E2-like) enzymes (1). We therefore decided to investigate whether MAPK15 is localized also onto early autophagosomal vesicles, and whether it may take advantage of ULK1 and ULK2 to stimulate initial phases of their biogenesis. Indeed, MAPK15 colocalized with ATG12 on these vesicles (Fig. 7A), supporting a role for this MAP kinase in the initial phases of autophagy. Next, we also sought to collect functional data supporting the interaction of MAPK15 with the biogenesis of autophagosomes. Specifically, we evaluated the effects of MAPK15 overexpression on the protein levels of endogenous ATG12–ATG5 complexes, which represent a measure of the formation of early-stage autophagosomes, as already demonstrated in our laboratory (28). With this approach, we demonstrated that levels of the ATG12–ATG5 complex increased, in full medium and starvation conditions, upon MAPK15 overexpression, whereas two mutants, MAPK15KD and MAPK15AXXA (specifically affecting the ability of this kinase of inducing autophagy without disturbing its kinase activity) (16), prevented starvation-dependent induction of early autophagosomal biogenesis (Fig. 7B), supporting a role for MAPK15 in this early autophagy step. Also, ULK1 and ULK1KD (kinase-dead mutant) were used as positive and negative controls, respectively, confirming the specificity of our experimental approach.
Figure 7.
MAPK15 controls early phases of autophagosomal biogenesis through ULK1/2. A, HeLa cells were transfected with HA-MAPK15WT plasmid. After 48 h, cells were incubated for 4 h in full medium or serum starvation medium. MAPK15 (green) and ATG12 (red) were labeled to monitor their subcellular localization. Nuclei were visualized in blue. Colocalization is shown in yellow and indicated by white arrowheads. Reference bars correspond to 10 μm. Representative images from three different experiments are shown (n = 3). B, HeLa cells were transfected with plasmid encoding for ULK1WT-FLAG, ULK1KD-FLAG, HA-MAPK15WT, HA-MAPK15AXXA, HA-MAPK15KD, or the empty vector. After 24 h, cells were incubated for 4 h in full medium or serum starvation medium. Total lysates were analyzed by Western blot (WB) analysis for endogenous ATG5 and ATG12 (n = 3). ATG12 levels were plotted on the graph (lower panel). ATG5 levels were plotted on the graph (middle panel). C, HeLa cells were transfected with ULK1 plus ULK2 siRNA or SCR siRNA and, after 48 h, further transfected with plasmid encoding for HA-MAPK15WT, HA-MAPK15KD, or the empty vector. After 24 h, cells were incubated for 4 h in full medium or serum starvation medium. Total lysates were analyzed by Western blot analysis for endogenous ATG5 and ATG12 (n = 3). ATG12 levels were plotted on the graph (lower panel). ATG5 levels were plotted on the graph (middle panel).
Next, in line with our results, we showed that down-regulation of ULK1 and ULK2 by specific siRNA consistently reduced MAPK15-dependent ATG12–ATG5 accumulation both in full medium and in starvation conditions (Fig. 7C), supporting the role of these kinases in mediating the control of MAPK15 on early autophagosome biogenesis. As a control, also in this case MAPK15KD prevented starvation-dependent induction of early autophagosomal biogenesis (Fig. 7C).
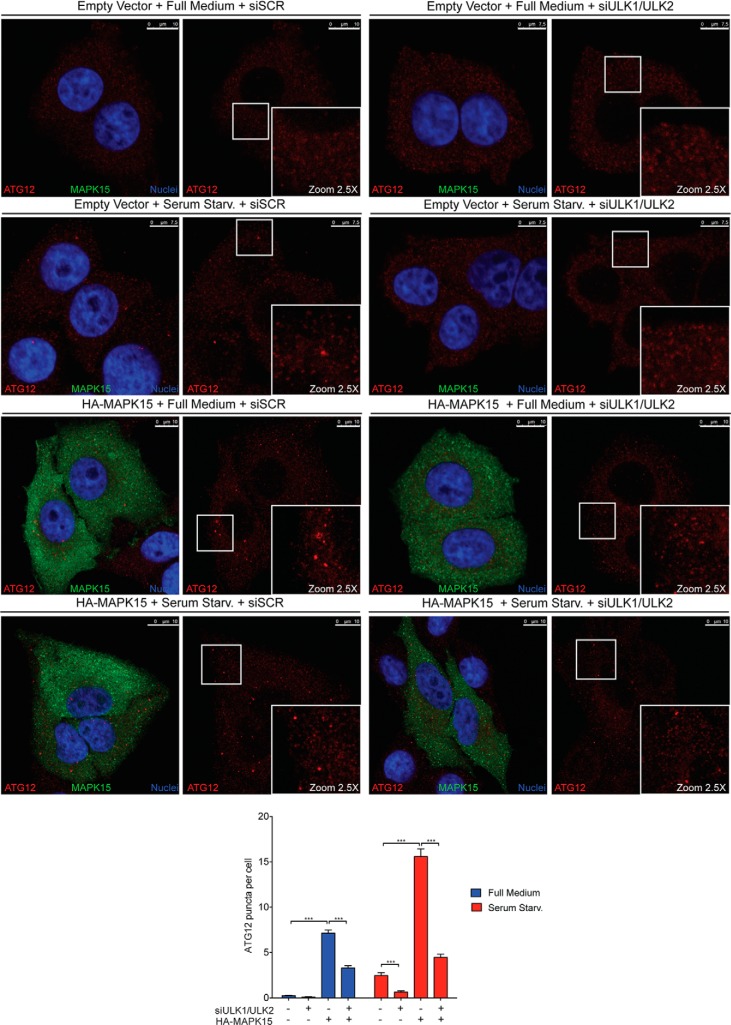
Based on ATG12–ATG5 localization on isolation membranes, puncta formed by these endogenous protein conjugates, detected by immunofluorescence, can be used to monitor the specific formation of early autophagosomal membranes (27). We therefore monitored the formation of ATG12 puncta in full medium and upon serum starvation, a stimulus for early autophagosomal biogenesis (28), and as shown above, we demonstrated that these vesicles strongly increased upon MAPK15 overexpression in both conditions (Fig. 8). Ultimately, this approach also allowed us to confirm the role of ULK1 and ULK2 in mediating MAPK15-dependent induction of early-stage autophagosomes as down-regulation of ULK1/2 by specific siRNA consistently reduced the number of ATG12 puncta induced by MAPK15, both in full medium and upon starvation (Fig. 8).
Figure 8.
MAPK15 controls early phases of autophagosomal biogenesis through ULK1/2. HeLa cells were transfected with ULK1 plus ULK2 siRNA or SCR siRNA and, after 48 h, further transfected with plasmid encoding for HA-MAPK15 or the empty vector. After 24 h, cells were incubated for 4 h in full medium or serum starvation medium. Cells were fixed and labeled for endogenous ATG12 (red) and for HA-positive cells (green) and counterstained with DAPI for nuclei (blue). White squares indicate zoom areas. Scale bars correspond to 10 μm except for Empty Vector + Serum Starvation (Starv.) + siSCR or siULK1/ULK2, and for Empty Vector + Full Medium + siSCR or siULK1/ULK2 in which they are 7.5 μm. ATG12 puncta per cell were plotted on the graph (lower panel, n = 3).
Discussion
ULK1 (and ULK2, where functional redundancy has been demonstrated), in complex with ATG13, FIP200, and ATG101, acts as the enzymatic component of the ULK1 complex involved in autophagy, with different proteins necessary for the formation of the autophagosome already recognized as its substrates (21–24). Still, ULK1 itself is heavily phosphorylated, making this protein a key integration point for stimuli coming from different kinases, whose identity is just starting to be clarified (6, 29, 30), but also increasing the difficulty of identifying a precise fingerprint of phosphorylated residues on ULK1 corresponding to its activation or inhibition. Here, we added MAPK15 to the list of proteins regulating ULK1 activity and, based on the recently established role of this MAP kinase in cell transformation (8–12), better defined a new signaling pathway whose modulation by pharmacological inhibitors hold promises for future innovative therapeutic approaches to cancer. This is particularly important, as autophagy has been repeatedly implicated in cancer development and its response to therapy, giving an advantage to cancer cells to survive drug treatment (31). For this reason, pharmacological approaches able to inhibit autophagy are being actively investigated to enhance the efficacy of different cancer therapies, with hydroxychloroquine (HCQ), a very aspecific inhibitor of autophagy, representing the only clinically approved drug for this purpose to date. Unfortunately, even high doses of HCQ only produce modest inhibition of autophagy in vivo, making urgent the need for alternative drugs targeting this process (32). Indeed, several pre-clinical studies have addressed this problem identifying more specific inhibitors for this process, with potential for cancer therapy. Among them, SBI-0206965 is indeed an ULK1/2 inhibitor and already showed to synergize with rapamycin in inducing tumor cell death (23). Our demonstration that MAPK15 impinges on ULK1/2 activity therefore supports the possibility that inhibitors of this MAP kinase may successfully perform, alone or in combination with other approaches, as specific inhibitors of autophagy in cancer therapy. Indeed, we have already demonstrated that MAPK15 mutants specifically lacking their ability of stimulating autophagy (but maintaining their kinase activity) impair proliferation of embryonic carcinoma and leukemic cells (9, 10), and we are actively pursuing the search for specific MAPK15 inhibitors (33).
Recently, a novel role for ULK1 emerged in the regulation of the endoplasmic reticulum-to-Golgi secretory pathway (34, 35). Specifically, in nutrient-rich conditions (which inhibit autophagy), ULK1 binds SEC16A and phosphorylates it on Ser846, triggering COPII-dependent vesicle traffic (35). Conversely, upon starvation, activation of MAPK15 negatively regulates the same SEC16A-dependent secretory pathway (35) while stimulating autophagy (16) through ULK1 (this paper), suggesting the possibility that MAPK15 may allow ULK1 to differentially regulate autophagy and secretion in normal or stressful conditions. Differential interaction of MAPK15 with the ULK complexes controlling autophagy or secretion may actually represent an intriguing hypothesis to explain the switch between the different ULK1 functions.
Overall, we ultimately expect that inhibition of MAPK15 may actually be extremely beneficial in cancer therapy for its potential to affect both general and selective autophagy, by taking advantage of specific tumor vulnerabilities based on the addiction of cancer cells on these stress-response mechanisms for survival (36).
Experimental procedures
Reagents and antibodies
Bafilomycin A1 (Santa Cruz Biotechnology, sc-201550), Torin1 (LC Laboratories, T-7887), SBI-0206965 (Selleckchem, S7885), and SU6656 (Sigma, S9692) were dissolved in DMSO (Sigma, D8418). For Western blot analysis, the following primary antibodies were used: anti-FLAG (Sigma, F1804); anti-HA (Covance, MMS-101R); anti-MYC (Covance, MMS-150R); anti-LC3B (Nanotools, 0231-1000); anti-MAPK15 (custom preparation (39)); anti-GABARAP (MBL, M135-3); anti-SQSTM1/p62 (BD Biosciences, 610833); anti-phosphothreonine (Cell Signaling, 9386); anti-phosphoserine/threonine (Qiagen, 37430); anti-phospho-ULK1 Ser317 (Cell Signaling, 12753); anti-ULK1 (Santa Cruz Biotechnology, sc-33182); anti-phospho-ATG13 Ser318 (Rockland, 600-401-C49); anti-ATG13 (Rockland, 600-401-C50); anti-ATG12 (Cell Signaling Technology, 2010); anti-ATG5 (Sigma, A0731); and anti-MAPK1/ERK2 (Santa Cruz Biotechnology, sc-154). For confocal immunofluorescence microscopy experiments, the following primary antibodies were used: anti-HA (Roche Diagnostics, 11867423001); anti-MAPK15 (custom preparation) (37); anti-FLAG (Sigma, F1804); anti-LC3B (MBL, M152-3); anti-SQSTM1/p62 (BD Biosciences, 610833); and anti-ATG12 (Cell Signaling Technology, 2010). The following secondary antibodies were used for Western blotting experiments: anti-mouse (Santa Cruz Biotechnology, sc-2004) and anti-rabbit (Santa Cruz Biotechnology, sc-2005) horseradish peroxidase-conjugated IgGs. The following secondary antibodies were used for confocal microscopy experiments: anti-mouse Alexa Fluor 488-conjugated (Life Technologies, Inc., A21202); anti-rabbit Alexa Fluor 488-conjugated (Life Technologies, Inc., A21206); anti-mouse Alexa Fluor 555-conjugated (Life Technologies, Inc., A31570); anti-rabbit Alexa Fluor 555-conjugated (Life Technologies, Inc., A31572); anti-rabbit Alexa Fluor 647-conjugated (Life Technologies, Inc., A31573); anti-rat Alexa Fluor 488-conjugated (Life Technologies, Inc., A11006); and anti-rat Alexa Fluor 594-conjugated (Life Technologies, Inc., A21209).
Cell culture and transfections
HEK293T and HeLa cells were maintained in full medium, composed of DMEM (PAA, E15-009) supplemented with 10% fetal bovine serum (PAA, A15-151), 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin and cultured at 37 °C in an atmosphere of 5% CO2/air. Hanks' medium (ECB4006L), used as starvation medium, was obtained from Euroclone, whereas DMEM (PAA, E15-009) supplemented with 2 mm l-glutamine and 100 units/ml penicillin/streptomycin and without FBS was used as serum starvation medium. For immunofluorescence experiments and Western blot analysis, 2 × 105 cells were seeded in 6-well plates (or 6-cm dishes) and transfected with 1 μg (200 μg in 6-cm dishes) of each expression vector, using Lipofectamine LTX (Life Technologies, Inc., 15338500). All experiments were performed 24–48 h after transfection. For confocal microscopy experiments, 24 h after transfection 2.5 × 104 cells were seeded on coverslips placed in 12-well plates. HeLa cells stably expressing EGFP–GABARAP were generated by transfecting pCEFL EGFP–GABARAP in HeLa cells, selecting with 1.5 mg/ml G418 (Genespin, STS-G418), and screening positive clones by fluorescence microscopy.
Plasmids
HA-MAPK15WT (pCEFL HA-MAPK15WT), HA-MAPK15KD(pCEFL HA-MAPK15KD), HA-MAPK15AXXA (pCEFL HA-MAPK15AXXA), EGFP-GABARAP (pCEFL EGFP-GABARAP), and empty vector (pCEFL) were previously described (16). ULK1-MYC (pRK5 myc ULK1, Addgene 31961) (38), ULK1WT-FLAG (p3×FLAG CMV14 FLAG ULK1, Addgene 24301) (39), ATG13-FLAG (p3×FLAG CMV10 FLAG ATG13, Addgene 22872) (39), VPS34-FLAG (pcDNA4 VPS34 FLAG, Addgene 24398) (40), FIP200-FLAG (p3×FLAG CMV10 FLAG FIP200, Addgene 24300) (41), and ATG101-FLAG (p3×FLAG CMV10 FLAG ATG101, Addgene 22877) (42) were provided by Addgene as the kind gifts from the producing laboratories. ULK1KD-FLAG and ULK1KD-MYC have been created by site-specific mutagenesis of ULK1WT-FLAG and ULK1WT-MYC changing Lys46 in Ile46, generating the kinase-dead ULK1 with K46I, as reported previously (43).
RNAi
MAPK15-specific siRNA (target sequence for MAPK15_A siRNA 5′-GCTTGGAGGCTACTCCC-3′ and for MAPK15_B siRNA 5′-GACAGATGCCCAGAGAACA-3′) and nonsilencing siRNA (scrambled, SCR; target sequence 5′-AATTCTCCGAACGTGTCACGT-3′) were obtained from Qiagen. ULK1-specific siRNAs are ON-TARGET plus SMARTpool (Dharmacon, L-005049-00-0005) and ULK2-specific siRNAs were used as pool (Sigma, SASI_Hs01_00157141, SASI_Hs01_ 00157142, and SASI_Hs01_00157143). HeLa cells were transfected with siRNA at a final concentration of 50 nm using HiPerFect (Qiagen, 301707), according to the manufacturer's instructions. Samples were analyzed, unless specified, 72 h after transfection.
Western blot analysis
Total lysates were obtained and treated as described previously. Briefly, washed cellular pellet fractions were resuspended in MAPK lysis buffer made of 20 mm HEPES (PAA, S11001), pH 7.5, 10 mm EGTA (Sigma, E4378), 40 mm β-glycerophosphate (Sigma, G6501), 1% Nonidet P-40 (Sigma, I3021), 2.5 mm MgCl2 (Sigma, M2670), 2 mm orthovanadate (Sigma, S6508), 2 mm NaF (Carlo Erba, 7681494), 1 mm DTT (IBI, IB21040), and protease inhibitors mixture (Roche Diagnostics, 05056489001). Proteins were quantified by the Bradford assay, and before loading, 5× Laemmli sample buffer was added to the lysates, which were next incubated for 5 min at 95 °C. Alternatively, cells were directly lysed in 1× Laemmli. Lysates were loaded on SDS-polyacrylamide gels, transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore, IPVH00010), probed with the appropriate antibodies, and detected using enhanced chemiluminescence (ECL Prime; GE Healthcare, RPN2232). Images were then acquired with a LAS 4000 imager (GE Healthcare, Italy, Milan). Densitometric analysis of Western blottings was performed with ImageJ 1.43u (National Institutes of Health).
Immunofluorescence
Cells were cultured and treated as described previously. Briefly, they were washed with PBS, then fixed with 4% paraformaldehyde (Sigma, 47608) in 1× PBS for 20 min, and permeabilized with 0.2% Triton X-100 (Sigma, T8787) for 10 min or 100 μg/ml digitonin solution (Life Technologies, Inc., BN2006) for 20 min. Permeabilized cells were incubated with the appropriate primary antibodies for 1 h, washed three times with 1× PBS, incubated for 30 min with appropriate secondary antibodies, and then washed again three times in 1× PBS. Nuclei were stained with a solution of 1.5 μm of 4′,6-diamidino-2-phenylindole (DAPI; Sigma, D9542) in PBS for 5 min. Coverslips were mounted in fluorescence mounting medium (Dako, S3023). Samples were visualized on a TSC SP5 confocal microscope (Leica Microsystems, Mannheim, Germany), installed on an inverted LEICA DMI 6000CS microscope (Leica Microsystems, Mannheim, Germany), and equipped with an oil immersion PlanApo 63 × 1.4 NA objective. Images were acquired using the LAS AF acquisition software (Leica Microsystems).
Immunoprecipitations
Whole-cell lysates (500 μg to 2 mg) were obtained by resuspending washed pellet fractions in the above-mentioned MAPK lysis buffer. Lysates were incubated with appropriate antibodies for 2 h at 4 °C. Then, immunocomplexes were purified by incubating the lysates for 45 min with protein G Mag-Sepharose Xtra (GE Healthcare, 28-9670-70). After five washes, the immunocomplexes were resuspended in 2× Laemmli buffer and subjected to Western blot analysis.
Dot count and statistical analysis
Dot counts for LC3B, SQSTM1, and AGT12 were performed with the Quantitation Module of Volocity software (PerkinElmer Life Sciences) and identifying specific fluorescence intensity range and size range (44). Dot counts were subjected to statistical analysis. Measurements were obtained by analyzing at least 400 cells per sample from three different experiments. Intensitometric analyses of protein levels in immunoblots were performed with ImageJ by evaluating representative images from at least three independent experiments (n = 3). Significance (p value) was assessed by one-way analysis of variance test. Asterisks were attributed for the following significance values: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Author contributions
D. C., F. D., L. S., and M. C. conceptualization; D. C., F. D., S. T., L. S., and M. C. data curation; D. C., F. D., S. T., L. S., and M. C. formal analysis; D. C., L. S., and M. C. supervision; D. C., F. D., S. T., L. S., and M. C. investigation; D. C., F. D., S. T., L. S., and M. C. methodology; D. C., L. S., and M. C. writing-original draft; D. C. and F. D. writing-review and editing; L. S. and M. C. funding acquisition; M. C. project administration.
Supplementary Material
Acknowledgments
We acknowledge the support of Regione Toscana, Istituto Toscano Tumori (ITT). We thank Laura Tinti for technical assistance and instrumental support.
This work was supported by Regione Toscana, Istituto Toscano Tumori (ITT). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- MAP
- mitogen-activated protein
- MAPK
- mitogen-activated protein kinase
- CML
- chronic myeloid leukemia
- KD
- kinase dead
- BAF
- bafilomycin A1
- SCR
- scrambled
- EGFP
- enhanced green fluorescent protein
- DAPI
- 4,6-diamidino-2-phenylindole
- HCQ
- hydroxychloroquine
- DMEM
- Dulbecco's modified Eagle's medium
- mTOR
- mechanistic target of rapamycin
- SCR
- scrambled
- AMPK
- 5′-AMP–activated protein kinase.
References
- 1. Mizushima N., Yoshimori T., and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 2. Boya P., Reggiori F., and Codogno P. (2013) Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravikumar B., Sarkar S., Davies J. E., Futter M., Garcia-Arencibia M., Green-Thompson Z. W., Jimenez-Sanchez M., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Moreau K., Narayanan U., Renna M., et al. (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90, 1383–1435 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- 4. Zachari M., and Ganley I. G. (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61, 585–596 10.1042/EBC20170021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papinski D., and Kraft C. (2016) Regulation of autophagy by signaling through the Atg1/ULK1 complex. J. Mol. Biol. 428, 1725–1741 10.1016/j.jmb.2016.03.030 [DOI] [PubMed] [Google Scholar]
- 6. Lin M. G., and Hurley J. H. (2016) Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68 10.1016/j.ceb.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coulombe P., and Meloche S. (2007) Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773, 1376–1387 10.1016/j.bbamcr.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 8. Jin D. H., Lee J., Kim K. M., Kim S., Kim D. H., and Park J. (2015) Overexpression of MAPK15 in gastric cancer is associated with copy number gain and contributes to the stability of c-Jun. Oncotarget 6, 20190–20203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rossi M., Colecchia D., Ilardi G., Acunzo M., Nigita G., Sasdelli F., Celetti A., Strambi A., Staibano S., Croce C. M., and Chiariello M. (2016) MAPK15 upregulation promotes cell proliferation and prevents DNA damage in male germ cell tumors. Oncotarget 7, 20981–20998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colecchia D., Rossi M., Sasdelli F., Sanzone S., Strambi A., and Chiariello M. (2015) MAPK15 mediates BCR-ABL1-induced autophagy and regulates oncogene-dependent cell proliferation and tumor formation. Autophagy 11, 1790–1802 10.1080/15548627.2015.1084454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iavarone C., Acunzo M., Carlomagno F., Catania A., Melillo R. M., Carlomagno S. M., Santoro M., and Chiariello M. (2006) Activation of the Erk8 mitogen-activated protein (MAP) kinase by RET/PTC3, a constitutively active form of the RET proto-oncogene. J. Biol. Chem. 281, 10567–10576 10.1074/jbc.M513397200 [DOI] [PubMed] [Google Scholar]
- 12. Xu Y. M., Zhu F., Cho Y. Y., Carper A., Peng C., Zheng D., Yao K., Lau A. T., Zykova T. A., Kim H. G., Bode A. M., and Dong Z. (2010) Extracellular signal-regulated kinase 8-mediated c-Jun phosphorylation increases tumorigenesis of human colon cancer. Cancer Res. 70, 3218–3227 10.1158/0008-5472.CAN-09-4306 [DOI] [PubMed] [Google Scholar]
- 13. Groehler A. L., and Lannigan D. A. (2010) A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J. Cell Biol. 190, 575–586 10.1083/jcb.201002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cerone M. A., Burgess D. J., Naceur-Lombardelli C., Lord C. J., and Ashworth A. (2011) High-throughput RNAi screening reveals novel regulators of telomerase. Cancer Res. 71, 3328–3340 10.1158/0008-5472.CAN-10-2734 [DOI] [PubMed] [Google Scholar]
- 15. Klevernic I. V., Martin N. M., and Cohen P. (2009) Regulation of the activity and expression of ERK8 by DNA damage. FEBS Lett. 583, 680–684 10.1016/j.febslet.2009.01.011 [DOI] [PubMed] [Google Scholar]
- 16. Colecchia D., Strambi A., Sanzone S., Iavarone C., Rossi M., Dall'Armi C., Piccioni F., Verrotti di Pianella A., and Chiariello M. (2012) MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 8, 1724–1740 10.4161/auto.21857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alers S., Löffler A. S., Wesselborg S., and Stork B. (2012) Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 32, 2–11 10.1128/MCB.06159-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dorsey F. C., Rose K. L., Coenen S., Prater S. M., Cavett V., Cleveland J. L., and Caldwell-Busby J. (2009) Mapping the phosphorylation sites of Ulk1. J. Proteome Res. 8, 5253–5263 10.1021/pr900583m [DOI] [PubMed] [Google Scholar]
- 19. Zhang L., Ouyang L., Guo Y., Zhang J., and Liu B. (2018) UNC-51-like Kinase 1: from an autophagic initiator to multifunctional drug target. J. Med. Chem. 61, 6491–6500 10.1021/acs.jmedchem.7b01684 [DOI] [PubMed] [Google Scholar]
- 20. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., and Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim J., Lachenmayer M. L., Wu S., Liu W., Kundu M., Wang R., Komatsu M., Oh Y. J., Zhao Y., and Yue Z. (2015) Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 11, e1004987 10.1371/journal.pgen.1004987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egan D. F., Chun M. G., Vamos M., Zou H., Rong J., Miller C. J., Lou H. J., Raveendra-Panickar D., Yang C. C., Sheffler D. J., Teriete P., Asara J. M., Turk B. E., Cosford N. D., and Shaw R. J. (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joo J. H., Dorsey F. C., Joshi A., Hennessy-Walters K. M., Rose K. L., McCastlain K., Zhang J., Iyengar R., Jung C. H., Suen D. F., Steeves M. A., Yang C. Y., Prater S. M., Kim D. H., Thompson C. B., et al. (2011) Hsp90–Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell 43, 572–585 10.1016/j.molcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang B., and Kundu M. (2017) Canonical and noncanonical functions of ULK/Atg1. Curr. Opin. Cell Biol. 45, 47–54 10.1016/j.ceb.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C. M., Adams P. D., Adeli K., Adhihetty P. J., Adler S. G., Agam G., Agarwal R., Aghi M. K., et al. (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd Ed.). Autophagy 12, 1–222 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colecchia D., Stasi M., Leonardi M., Manganelli F., Nolano M., Veneziani B. M., Santoro L., Eskelinen E. L., Chiariello M., and Bucci C. (2018) Alterations of autophagy in the peripheral neuropathy Charcot-Marie-Tooth type 2B. Autophagy 14, 930–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim J., Kundu M., Viollet B., and Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puente C., Hendrickson R. C., and Jiang X. (2016) Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J. Biol. Chem. 291, 6026–6035 10.1074/jbc.M115.689646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amaravadi R. K., Lippincott-Schwartz J., Yin X. M., Weiss W. A., Takebe N., Timmer W., DiPaola R. S., Lotze M. T., and White E. (2011) Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 17, 654–666 10.1158/1078-0432.CCR-10-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chude C. I., and Amaravadi R. K. (2017) Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18, 1279–1289 10.3390/ijms18061279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strambi A., Mori M., Rossi M., Colecchia D., Manetti F., Carlomagno F., Botta M., and Chiariello M. (2013) Structure prediction and validation of the ERK8 kinase domain. PLoS ONE 8, e52011 10.1371/journal.pone.0052011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gan W., Zhang C., Siu K. Y., Satoh A., Tanner J. A., and Yu S. (2017) ULK1 phosphorylates Sec23A and mediates autophagy-induced inhibition of ER-to-Golgi traffic. BMC Cell Biol. 18, 22 10.1186/s12860-017-0138-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joo J. H., Wang B., Frankel E., Ge L., Xu L., Iyengar R., Li-Harms X., Wright C., Shaw T. I., Lindsten T., Green D. R., Peng J., Hendershot L. M., Kilic F., Sze J. Y., et al. (2016) The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol. Cell 62, 491–506 10.1016/j.molcel.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo J., Solimini N. L., and Elledge S. J. (2009) Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 10.1016/j.cell.2009.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossi M., Colecchia D., Iavarone C., Strambi A., Piccioni F., Verrotti di Pianella A., and Chiariello M. (2011) Extracellular signal-regulated kinase 8 (ERK8) controls estrogen-related receptor α (ERRα) cellular localization and inhibits its transcriptional activity. J. Biol. Chem. 286, 8507–8522 10.1074/jbc.M110.179523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., and Kim D. H. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., and Mizushima N. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun Q., Fan W., Chen K., Ding X., Chen S., and Zhong Q. (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., and Mizushima N. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., and Mizushima N. (2009) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- 43. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klionsky D. J., and Eskelinen E. L. (2014) The vacuole versus the lysosome: when size matters. Autophagy 10, 185–187 10.4161/auto.27367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.