Abstract
The Nem1–Spo7 protein phosphatase plays a role in lipid synthesis by controlling the membrane localization of Pah1, the diacylglycerol-producing phosphatidate (PA) phosphatase that is crucial for the synthesis of triacylglycerol in the yeast Saccharomyces cerevisiae. By dephosphorylating Pah1, Nem1–Spo7 facilitates its translocation to the nuclear/endoplasmic reticulum membrane for catalytic activity. Like its substrate Pah1, Nem1–Spo7 is phosphorylated in the cell, but the specific protein kinases involved remain to be identified. In this study, we demonstrate that the Nem1–Spo7 complex is phosphorylated by protein kinase A (PKA), which is associated with active cell growth, metabolic activity, and membrane phospholipid synthesis. In vitro phosphorylation of purified Nem1–Spo7 and of their synthetic peptides revealed that both subunits of the phosphatase complex are PKA substrates. Using phosphoamino acid and phosphopeptide-mapping analyses coupled with site-directed mutagenesis, we identified Ser-140 and Ser-210 of Nem1 and Ser-28 of Spo7 as PKA-targeted phosphorylation sites. Immunodetection of the phosphatase complex from the cell with anti-PKA substrate antibody confirmed the in vivo phosphorylations of Nem1 and Spo7 on the serine residues. Lipid-labeling analysis of cells bearing phosphorylation-deficient alleles of NEM1 and SPO7 indicated that the PKA phosphorylation of the phosphatase complex stimulates phospholipid synthesis and attenuates the synthesis of triacylglycerol. This work advances the understanding of how PKA-mediated posttranslational modifications of Nem1 and Spo7 regulate lipid synthesis in yeast.
Keywords: phospholipid, diacylglycerol, triacylglycerol, protein phosphatase, yeast metabolism, Saccharomyces cerevisiae, protein kinase A (PKA), lipid metabolism, Nem1-Spo7 protein phosphatase, PA phosphatase
Introduction
The Nem1–Spo7 protein phosphatase complex, originally identified as a membrane component that participates in nuclear envelope morphogenesis (1), plays an important role in yeast lipid metabolism (2, 3). It principally functions to dephosphorylate Pah1 (4–6), a PA 2 phosphatase enzyme (7) whose molecular function is to catalyze the dephosphorylation of PA to form DAG (8, 9) (Fig. 1A). Through its reaction, the PA phosphatase controls the bifurcation of PA for the synthesis of TAG via DAG or for the synthesis of membrane phospholipids via CDP-DAG (10, 11) (Fig. 1A).
Figure 1.
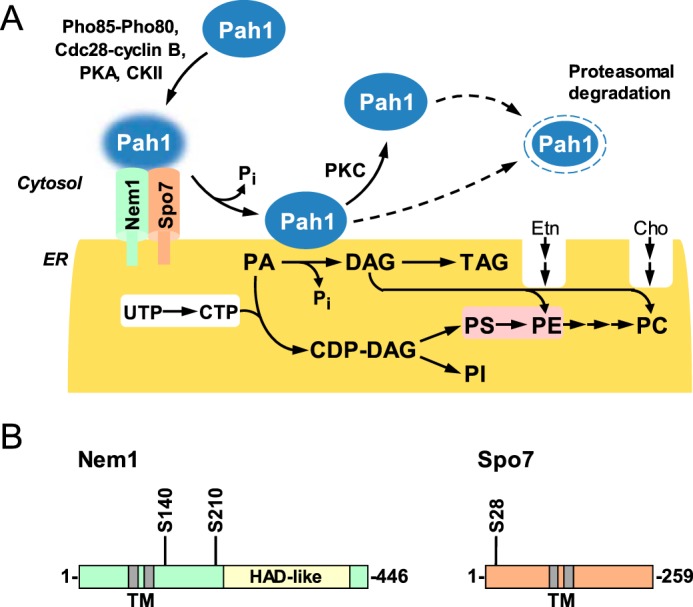
Roles and regulation of Nem1–Spo7/Pah1 phosphatase cascade in lipid synthesis and domains/regions and PKA phosphorylation sites in Nem1 and Spo7. A, Pah1 in the cytosol is phosphorylated by the indicated protein kinases. The phosphorylated Pah1 (indicated by the blue glow) translocates to the ER membrane through its dephosphorylation by the Nem1 (catalytic subunit, green)–Spo7 (regulatory subunit, peach) complex. Dephosphorylated Pah1 that is associated with the ER membrane catalyzes the conversion of PA to DAG, which is then acylated to form TAG. Dephosphorylated Pah1 or PKC-phosphorylated Pah1 that is not phosphorylated at the seven target sites for the Pho85–Pho80 protein kinase is degraded by the proteasome (indicated by the dashed line arrows). PA is also utilized for the synthesis of membrane phospholipids via CDP-DAG. When the CDP-DAG pathway for phospholipid synthesis is blocked, phosphatidylcholine or phosphatidylethanolamine may be synthesized from the DAG derived from the PA phosphatase reaction if cells are supplemented with choline or ethanolamine via the Kennedy pathway. The reactions highlighted in white take place in the cytosol, whereas the reaction highlighted in pink takes place in the mitochondria. CKII, casein kinase II; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol. B, the diagrams show the positions of the two transmembrane (TM)-spanning regions in Nem1 and Spo7, the HAD-like domain that contains the DXDX(T/V) catalytic motif in Nem1, and serine (S) residues identified in this work as the sites phosphorylated by PKA. HAD, haloacid dehalogenase.
Nem1 and Spo7 are integral nuclear/ER membrane proteins; they both possess two transmembrane-spanning domains (1) (Fig. 1B). Nem1 binds to Spo7 through its conserved C-terminal domain, and this association is responsible for the formation of the complex in the membrane bilayer (1) (Fig. 1A). Nem1, which serves as the catalytic subunit, is a member of the haloacid dehalogenase superfamily (12, 13); its phosphatase activity depends on the DXDX(T/V) catalytic motif within its HAD-like domain (1, 4) (Fig. 1B). Spo7, which serves as the regulatory subunit (1), associates with Pah1 to facilitate the formation of the Nem1–Spo7/Pah1 phosphatase complex (14, 15).
Pah1 is a phosphoprotein in the cytosol; it is phosphorylated by multiple protein kinases that include Pho85–Pho80 (16), Cdc28–cyclin B (17), PKA (18), PKC (19), and casein kinase II (20) (Fig. 1A). The phosphorylations by Pho85–Pho80 (16), Cdc28–cyclin B (4, 17), and PKA (18) inactivate Pah1 by causing its retention in the cytosol apart from its substrate PA at the nuclear/ER membrane (Fig. 1A). In addition, the phosphorylations of Pah1 by Pho85–Pho80 (16) and PKA (18) cause the inhibition of its PA phosphatase activity. The phosphorylation by casein kinase II does not have a major effect on the PA phosphatase activity, but it reduces subsequent phosphorylation of Pah1 by PKA (20). The phosphorylation by PKC does not affect the location of Pah1 or its PA phosphatase activity, but instead affects its stability (19). The PKC phosphorylation of Pah1, which is favored when it is not already phosphorylated by Pho85–Pho80, promotes its degradation by the 20S proteasome (19, 21) (Fig. 1A). By removing phosphate groups from Pah1, Nem1–Spo7 facilitates the binding of Pah1 to the ER membrane and to its substrate PA (22) and, at the same time, stimulates PA phosphatase activity (5, 6, 16, 17). The net result of dephosphorylating Pah1 is the increase in TAG synthesis, which occurs at the expense of membrane phospholipid synthesis (5, 6, 16, 17).
Insights into the physiological functions of the Nem1–Spo7/Pah1 phosphatase cascade have been gained through mutant phenotypes. Cells that carry the nem1Δ, spo7Δ, or pah1Δ mutation exhibit the abnormal expansion of the nuclear/ER membrane (1, 4). The expansion of the membrane in cells lacking the Nem1–Spo7 complex or Pah1 has been attributed to an increase in membrane phospholipid synthesis, which happens at the expense of TAG synthesis (7, 23, 24). The increase in phospholipid synthesis correlates with the derepression of phospholipid synthesis gene expression (4, 25), whereas the decrease in TAG synthesis correlates with a decrease in the formation of cytoplasmic lipid droplets (26–28). The loss of Pah1 function gives rise to a multiple assortment of other phenotypes that include the susceptibility of cells to fatty acid–induced toxicity (26), defects in cell wall integrity (29, 30), vacuole fusion and acidification (31, 32), and the inability to grow on nonfermentable carbon sources (7, 33) and at elevated temperatures (4, 7, 33). Cells lacking Pah1 are also hypersensitive to oxidative stress and exhibit an apoptosis phenotype (26) and a decrease in chronological life span (34). Along the lines of the latter phenotypes, mutants lacking Pah1 or the Nem1–Spo7 complex exhibit a defect in autophagy (35, 36). Taken together, these phenotypes emphasize the importance of the Nem1–Spo7/Pah1 phosphatase cascade in the regulation of lipid synthesis and cell physiology. Given the importance of dephosphorylation on Pah1 function, it is evident why cells lacking the Nem1–Spo7 complex exhibit similar phenotypes as cells lacking Pah1.
The physiological function of the Nem1–Spo7 phosphatase is well established, but our understanding of its enzymology and regulation is just emerging. The phosphatase activity depends on Mg2+ ions, a hallmark characteristic of DXDX(T/V) motif-containing enzymes, and has the pH optimum of 5.0 (6, 37). Nem1–Spo7 dephosphorylates all phosphorylated residues on Pah1 (4), and the specificity of the dephosphorylations is in the order of the sites phosphorylated by Pho85–Pho80 > PKA = casein kinase II > Cdc28–cyclin B > PKC (6, 20). That Nem1–Spo7 has the greatest affinity for the sites phosphorylated by Pho85–Pho80 is consistent with the major effects (e.g. increase in membrane association and TAG synthesis and increase in 20S proteasomal degradation) imparted by the alanine mutations of the seven sites phosphorylated by Pho85–Pho80 (5, 16, 17). The acidic pH optimum correlates with the intracellular pH (∼5) of yeast cells as they progress into the stationary phase (6, 37), the phase of growth when Pah1 function and TAG synthesis are maximal and the partitioning of PA toward lipid storage is favored over membrane phospholipid synthesis (24, 37, 38).
Whereas the Nem1–Spo7 phosphatase functions to dephosphorylate Pah1, both subunits of the complex are subject to phosphorylation (39, 40). However, the protein kinases involved have yet to be identified. In the current study, we showed that PKA, the principal protein kinase that transmits signals through the RAS/cAMP pathway in yeast (41, 42), phosphorylates Nem1 and Spo7 in vitro and in vivo. Ser-140 and Ser-210 in Nem1 and Ser-28 in Spo7 were identified as the principal sites of phosphorylation. Lipid compositional analyses of cells bearing phosphorylation-deficient mutations in Nem1 and Spo7 indicated that the PKA phosphorylation of the complex stimulates phospholipid synthesis and attenuates the synthesis of TAG. This work advances the understanding of the posttranslational modifications of Nem1 and Spo7 and the PKA-mediated regulation of lipid synthesis in yeast.
Results
Nem1 and Spo7 are phosphorylated by PKA
The subunits of the Nem1–Spo7 phosphatase complex are known to be phosphorylated in vivo (15, 39, 40), and they possess putative target sites for PKA (43, 44). To address the hypothesis that Nem1 and Spo7 are phosphorylated by PKA, we examined the enzyme–substrate relationship using a purified preparation of the phosphatase complex. Nem1 was expressed as a fusion protein tagged with protein A to facilitate its purification by affinity chromatography with IgG-Sepharose (1). The purified Nem1–Spo7 complex was nearly homogeneous and enzymatically active on its substrate Pah1 phosphorylated by Pho85–Pho80 (6). The phosphorylation of Nem1–Spo7 was catalyzed by bovine heart PKA, which is structurally and functionally like the protein kinase from yeast (45) and has been used to identify the phosphorylation sites of yeast phospholipid metabolism proteins (18, 46–49).
For the in vitro phosphorylation of Nem1–Spo7, the phosphatase complex was incubated with PKA and [γ-32P]ATP, resolved by SDS-PAGE, and transferred to a PVDF membrane. Phosphorimaging analysis of the membrane showed that radioactive phosphate was transferred from [γ-32P]ATP to Nem1 and Spo7 (Fig. 2A, left). Immunoblot analysis with anti-Nem1 and -Spo7 antibodies confirmed the identity of radiolabeled Nem1 and Spo7 (Fig. 2A, right). Phosphoamino acid analysis of 32P-labeled Nem1 and Spo7 showed that they are both phosphorylated on the serine residue (Fig. 2B).
Figure 2.
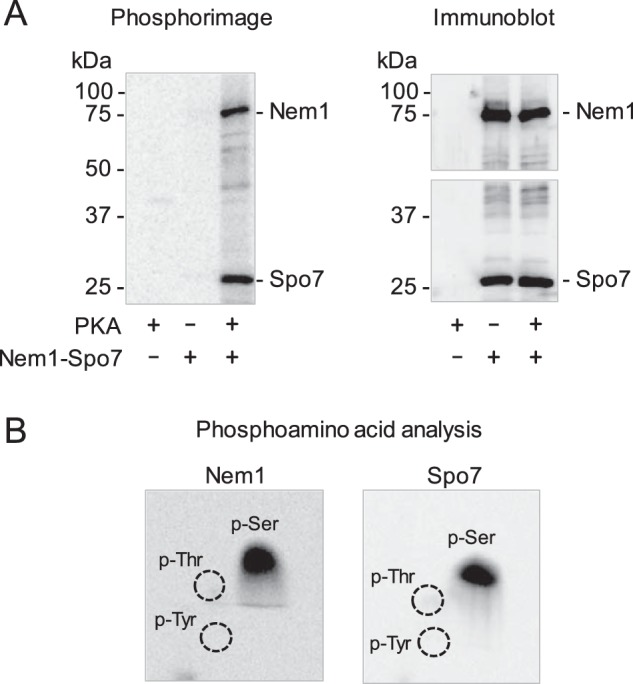
Nem1 and Spo7 are phosphorylated in vitro by PKA on the serine residue. A, Nem1 was co-expressed with Spo7 in yeast cells, and the Nem1–Spo7 complex was purified by IgG-Sepharose affinity chromatography based on the protein A tag on Nem1. The complex (0.5 pmol) was incubated with PKA (10 units) and [γ-32P]ATP (100 μm) at 30 °C for 10 min. Following the reaction, samples were subjected to SDS-PAGE followed by transfer to PVDF membrane. The radioactive phosphorylations of Nem1 and Spo7 were visualized by phosphorimaging (left). The PVDF membrane was cut; the upper and lower portions were subjected to immunoblot analysis using anti-Nem1 and anti-Spo7 antibodies, respectively (right). The positions of Nem1, Spo7, and molecular mass standards are indicated. B, portions of the PVDF membrane containing 32P-labeled Nem1 (left) or Spo7 (right) was incubated with 6 n HCl for 90 min at 110 °C. The acid hydrolysates were separated by two-dimensional electrophoresis on cellulose TLC plates followed by phosphorimaging analysis. The positions of the standard phosphoamino acids phosphoserine (P-Ser), phosphothreonine (P-Thr) (dotted lines), and phosphotyrosine (P-Tyr) (dotted lines) are indicated. The data shown in the figure are representative of three experiments.
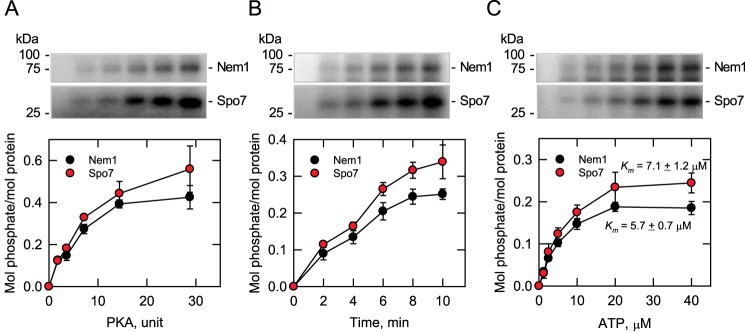
In the in vitro phosphorylation of the Nem1–Spo7 phosphatase complex, PKA activity on the Nem1 and Spo7 substrates depended on the amount of the protein kinase, the time of the reaction, and the concentration of ATP (Fig. 3). Based on the stoichiometry of protein phosphorylation, Nem1 and Spo7 are similarly phosphorylated by PKA (Fig. 3, bottom). Analysis of the data in Fig. 3C according to the Michaelis–Menten equation showed that the apparent Km values for ATP in the phosphorylation of Nem1 and Spo7, respectively, were similar (5.7 ± 0.7 and 7.1 ± 1.2 μm) and within the range of those reported for other PKA substrates of lipid metabolism (10), including Pah1 (18). The kinetic analysis to determine Km values for Nem1 and Spo7 was not carried out due to a limitation in the amount of pure Nem1–Spo7 complex.
Figure 3.
Phosphorylations of Nem1 and Spo7 by PKA are dependent on the amount of PKA, time of reaction, and concentration of ATP. Purified Nem1–Spo7 complex (0.35 pmol) was phosphorylated by PKA using [γ-32P]ATP as described in the legend to Fig. 2. The PKA reaction was conducted by varying the amount of PKA (A), the reaction time (B), and the concentration of ATP (C). A and B, 40 μm ATP; B and C, 2.4 units PKA; A and C, 5 min. The amount of radioactive phosphate incorporated into Nem1 or Spo7 was determined from a standard curve using [γ-32P]ATP. The amounts of Nem1 and Spo7 were determined by comparing their band intensities from a SYPRO Ruby–stained polyacrylamide gel with a standard curve of BSA. The positions of Nem1, Spo7, and molecular mass standards are indicated. The data shown in the plots are averages of three experiments ± S.D. (error bars). The portions of the phosphor images containing the phosphorylated Nem1 or Spo7 that are shown in the figure are representative of three experiments.
Effects of alkaline phosphatase and PKA on Nem1–Spo7 phosphatase activity
We examined the PKA phosphorylation effect of Nem1–Spo7 on its enzyme activity with Pah1 phosphorylated by Pho85–Pho80 (6). The Nem1–Spo7 complex was purified from yeast cells, phosphorylated by PKA, and then assayed for its phosphatase activity on the phosphorylated Pah1. After its phosphorylation, the Nem1–Spo7 complex did not show a major change in phosphatase activity. Considering that Nem1–Spo7 is endogenously phosphorylated, this result may also indicate that the phosphatase complex is sufficiently phosphorylated by PKA in the cell, and its additional in vitro phosphorylation has little effect. To address this possibility, purified Nem1–Spo7 was first treated with alkaline phosphatase to remove its phosphate groups and then phosphorylated by PKA. The dephosphorylation of Nem1–Spo7 resulted in a dose-dependent decrease (33% at its maximum) of its enzyme activity (Fig. 4, left). However, its rephosphorylation by PKA did not restore the enzyme activity (Fig. 4, right). In fact, the rephosphorylation of the Nem1–Spo7 complex by PKA reduced the protein phosphatase activity by 10%. Whereas this modest effect was reproducible, it was not statistically significant. These results also indicated that a site(s) phosphorylated by another protein kinase is responsible for stimulating the phosphatase activity of the complex.
Figure 4.
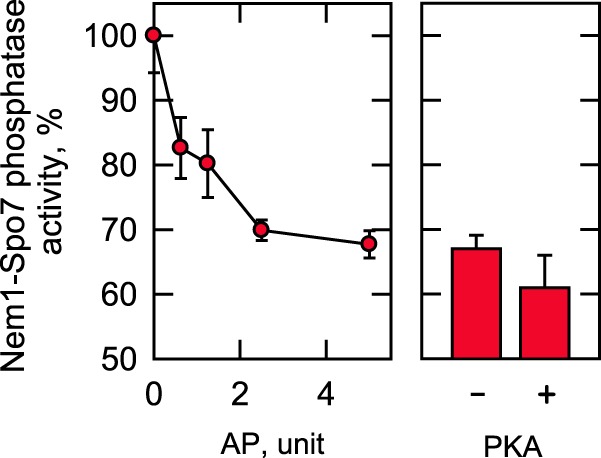
Effects of alkaline phosphatase and PKA on Nem1–Spo7 phosphatase activity. The Nem1 and Spo7 were co-expressed in yeast cells and purified by IgG-Sepharose affinity chromatography based on the protein A tag on Nem1. The Nem1–Spo7 complex was incubated for 10 min with the indicated amounts of alkaline phosphatase (AP) immobilized on agarose. The dephosphorylation of the complex was terminated by removing alkaline phosphatase through filtration. The Nem1–Spo7 phosphatase activity of the dephosphorylated enzyme was then measured by following the release of 32Pi from [32P]Pah1. The Nem1–Spo7 activity was normalized to that of enzyme not incubated with alkaline phosphatase (left). The complex that had been incubated with 5 units of alkaline phosphatase was then incubated with PKA for 15 min, followed by the measurement of the Nem1–Spo7 phosphatase activity. The activity was normalized to that of the alkaline phosphatase–treated enzyme not phosphorylated by PKA (right). The Nem1–Spo7 phosphatase activity that was set at 100% was 12 nmol/min/mg. The data shown are averages of three experiments ± S.D. (error bars).
PKA phosphorylates Nem1 and Spo7 synthetic peptides
We sought to identify which serine residues of Nem1 and Spo7 are phosphorylated by PKA. Analysis of the Nem1 sequence by the web-based servers NetPhosK (43) and NetPhosYeast (44) indicated that several serine residues (e.g. Ser-140, Ser-195, Ser-201, Ser-208, Ser-210, Ser-375, and Ser-376) are target phosphorylation sites. Using synthetic peptides containing the putative serine residues, we examined the in vitro phosphorylation by PKA (Fig. 5A). This analysis showed that the peptide containing Ser-210 was best phosphorylated (3,812 ± 216 nmol/min/mg), followed by the peptide containing Ser-140 (1,876 ± 40 nmol/min/mg). The mutation of Ser-210 or Ser-140 to a nonphosphorylatable alanine residue abolished the peptide phosphorylation, confirming that the serine residues are the target sites. The lack of phosphorylation in the S210A peptide also indicates that Ser-208 in the mutant peptide is not a phosphorylation site. Compared with the peptide containing Ser-210, the peptides containing Ser-195, Ser-375, or Ser-376 were poorly phosphorylated at the level of 5% or lower. The peptide containing the predicted Ser-201 was not phosphorylated by PKA.
Figure 5.
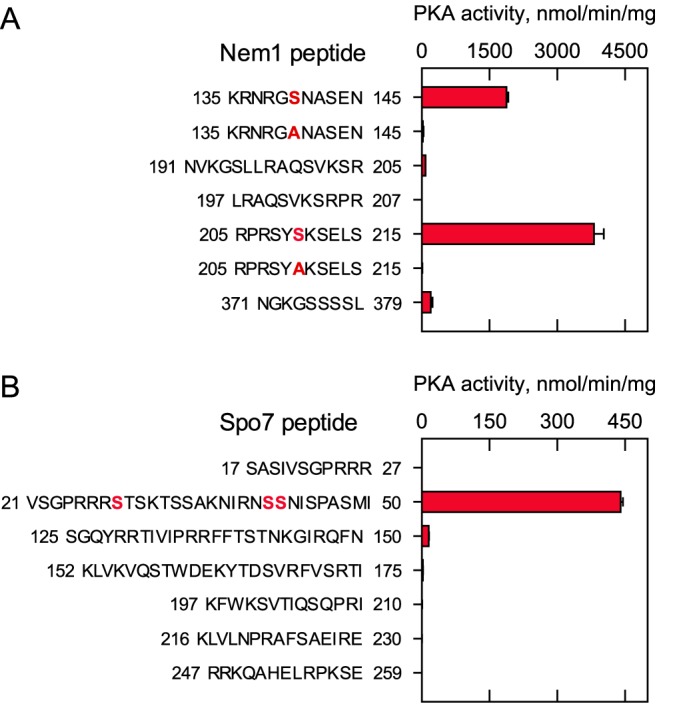
PKA phosphorylates Nem1 and Spo7 synthetic peptides. PKA activity was measured with a 100 μm concentration of the indicated Nem1 (A) or Spo7 (B) peptides using 100 μm [γ-32P]ATP. The enzyme reaction was terminated by spotting the mixture onto P81 phosphocellulose paper, which was then washed with 75 mm phosphoric acid and subjected to scintillation counting. The numbers to the left and right of the Nem1 (A) or Spo7 (B) peptides represent the position of the sequence in the respective proteins. The boldface red residues within the Nem1 peptides designate the serine-to-alanine substitutions in the sequence. The boldface red residues within the Spo7 peptide were identified as phosphorylation sites by MS. The data are the averages of three experiments ± S.D. (error bars).
Using peptide substrates, we characterized the phosphorylation of Ser-140 and Ser-210 by kinetic analysis. In one experiment, the PKA activity on the Ser-140 or Ser-210 peptides was measured at the saturating concentration of ATP by varying the peptide concentration. In another experiment, the enzyme activity was measured at the saturating concentration of the peptides by varying the ATP concentration. In both analyses, the PKA activity followed typical Michaelis–Menten kinetics (data not shown). As shown in Table 1, the apparent Vmax values were 1.4- and 2-fold higher, respectively, with respect to the Ser-210 peptide and ATP when compared with the Ser-140 peptide and ATP. In contrast, the apparent Km values were 32- and 5-fold lower, respectively, for the Ser-210 peptide and ATP than for the Ser-140 peptide and ATP. Accordingly, specificity constants (i.e. Vmax/Km) were 45- and 10-fold higher, respectively, for the Ser-210 peptide and ATP than for the Ser-140 peptide and ATP, indicating that Ser-210 is a much better phosphorylation site when compared with Ser-140.
Table 1.
Kinetic constants for PKA activity using Nem1 synthetic peptides
PKA activity was measured as a function of the concentrations of the peptides Ser-140 (0–2,000 μm) or Ser-210 (0–200 μm) using a set [γ-32P]ATP concentration of 1 mm or as a function of the concentration of [γ-32P]ATP (0–400 μm for Ser-140 or 0–40 μm for Ser-210) using a set concentration of the respective peptide of 1 mm. The enzyme reaction was terminated by spotting the mixture onto P81 phosphocellulose paper, which was then washed with 75 mm phosphoric acid and subjected to scintillation counting. PKA followed typical saturation kinetics; the apparent (app) Vmax and Km values were determined by analysis of the data with the Enzyme Kinetics module of the SigmaPlot software according to the Michaelis–Menten equation. The data are averages of three experiments ± S.D.
| Nem1 peptide |
Vmax(app) |
Km(app) |
Vmax/Km |
|||
|---|---|---|---|---|---|---|
| Peptide | ATP | Peptide | ATP | Peptide | ATP | |
| nmol/min/mg | μm | μm−1 nmol min−1 mg−1 | ||||
| Ser-140a | 3,301 ± 130 | 1,393 ± 50 | 1,508 ± 110 | 77 ± 8 | 2 | 18 |
| Ser-210b | 4,458 ± 240 | 2,836 ± 180 | 47 ± 7 | 15 ± 2 | 95 | 189 |
a 135KRNRGSNASEN145; the numbers to the left and right represent the position of the sequence in the Nem1 protein.
b 205RPRSYSKSELS215; the numbers to the left and right represent the position of the sequence in the Nem1 protein.
The sequence analysis of Spo7 predicts that Ser-28, Ser-41, Ser-42, and Ser-258 are putative phosphorylation sites of PKA. As Spo7 is a relatively small protein consisting of 259 amino acid residues, we synthesized six peptides that cover the entire soluble region of the protein containing serine and threonine residues (Fig. 1B) and analyzed whether they are the substrate of PKA (Fig. 5B). Of the Spo7 peptides, the one corresponding to residues 21–50 served as a PKA substrate. For this peptide containing three putative serine residues, we examined its phosphorylation by LC-MS/MS. This analysis identified Ser-28 as a major site of phosphorylation by PKA and Ser-41 and Ser-42 as minor phosphorylation sites.
Mutagenesis and phosphopeptide-mapping analyses of Nem1 and Spo7 identify their major PKA phosphorylation sites
Based on the peptide phosphorylation data, we examined the full-length complex of Nem1–Spo7 for its phosphorylation by PKA. By site-directed mutagenesis, the serine residues of Nem1 (Ser-140 and Ser-210) and Spo7 (Ser-28, Ser-41, and Ser-42) were changed to the alanine residues. The mutant Nem1 (or Spo7) was coexpressed with WT Spo7 (or Nem1), and the Nem1–Spo7 complex was purified using a protein A tag on the Nem1 subunit by affinity chromatography. The purified Nem1–Spo7 complex was treated with PKA and [γ-32P]ATP, resolved by SDS-PAGE, and analyzed by phosphorimaging (Fig. 6). In the PKA phosphorylation of Nem1, the S210A mutation caused a 60% reduction, whereas the S140A mutation did not show a clear reduction. Compared with the single mutations, the combined mutation of S140A and S210A showed a stronger effect, causing an 80% reduction in the phosphorylation. In the PKA phosphorylation of Spo7, the S28A mutation had the effect of an 85% reduction. However, the S41A, S42A, and S41A/S42A mutations did not affect the phosphorylation of Spo7 (data not shown). Moreover, this analysis showed that the defect of Spo7 phosphorylation increased the phosphorylation of Nem1 by 60%. This indicated that the phosphorylation of Spo7 may prevent the phosphorylation of Nem1.
Figure 6.
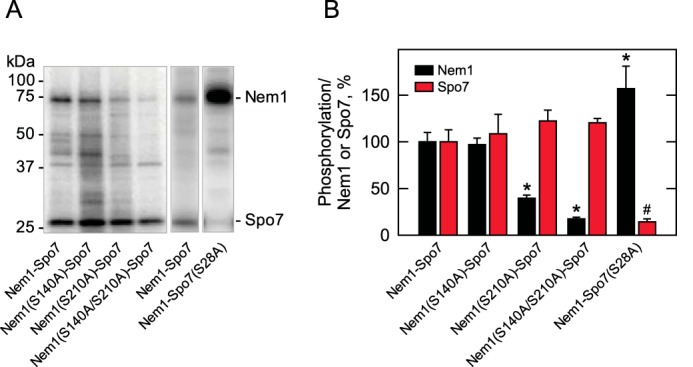
Effects of mutations on the phosphorylations of Nem1 and Spo7 by PKA. The WT and indicated mutant forms of Nem1 or the WT and S28A mutant forms of Spo7 were co-expressed in yeast cells and purified by IgG-Sepharose affinity chromatography based on the protein A tag on Nem1. The Nem1–Spo7 complex was incubated with PKA (20 units) and [γ-32P]ATP (50 μm) at 30 °C for 20 min. Following the reaction, samples were subjected to SDS-PAGE followed by transfer to PVDF membrane. A, the phosphorylations of Nem1 and Spo7 were visualized by phosphorimaging. The positions of Nem1, Spo7, and molecular mass standards are indicated. B, the relative amounts of radioactive phosphate incorporated into Nem1 or Spo7 were quantified by ImageQuant software, whereas the mass amounts of Nem1 and Spo7 were determined by comparing their band intensities from a SYPRO Ruby–stained polyacrylamide gel with a standard curve of BSA. The phosphorylation levels of the mutant proteins were compared with the respective WT proteins that were set at 100%. The phosphor images shown in A are representative of three experiments, whereas the data shown in B are averages of three experiments ± S.D. (error bars). *, p < 0.05 versus WT Nem1; #, p < 0.05 versus WT Spo7.
We further examined the PKA phosphorylation of Nem1–Spo7 by phosphopeptide mapping (Fig. 7). In the analysis of Nem1 (Fig. 7A), the WT protein showed multiple phosphopeptides with a similar extent of phosphorylation, and most of them were lost by the S210A mutation. This result indicates that the multiple phosphopeptides contain the same phosphorylated Ser-210 and suggests that they are produced due to incomplete proteolytic digestion. In contrast, the S140A mutation caused the loss of a single phosphopeptide. The combined mutation of S140A and S210A showed an additive effect on the phosphopeptide map. These results indicate that Ser-210 is a major site of Nem1 phosphorylation by PKA. In the analysis of Spo7 (Fig. 7B), the WT protein showed one major peptide and a few minor peptides. The S28A mutation caused the loss of the major peptide and a minor peptide, indicating that Ser-28 is a major phosphorylation site. In contrast, the S41A, S42A, and S41A/S42A mutations did not cause the loss of the phosphopeptides (data not shown). Taken together, these results indicate that Ser-140 and Ser-210 of Nem1 and Ser-28 of Spo7 are major sites of the Nem1–Spo7 phosphorylation by PKA.
Figure 7.
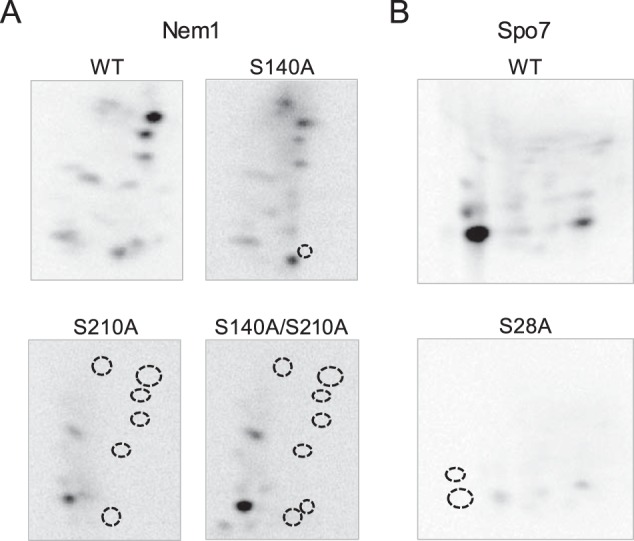
Phosphopeptide-mapping analysis of Nem1 and Spo7 identify their major PKA phosphorylation sites. The WT and indicated mutant forms of Nem1 (A) or the WT and S28A mutant forms of Spo7 (B) were co-expressed in yeast cells and purified by IgG-Sepharose affinity chromatography based on the protein A tag on Nem1. The Nem1–Spo7 complex was incubated with PKA (20 units) and [γ-32P]ATP (100 μm) at 30 °C for 20 min. Following the reaction, samples were subjected to SDS-PAGE followed by transfer to PVDF membrane. The phosphorylated proteins on PVDF membrane were digested with l-1-tosylamido-2-phenylethyl chloromethyl ketone–treated trypsin. The phosphopeptides produced by the proteolytic digestion were separated on cellulose TLC plates by electrophoresis (from left to right) in the first dimension and by chromatography (from bottom to top) in the second dimension. Positions of the phosphopeptides that were absent in the mutant proteins (indicated by the dotted line circles) but present in the WT Nem1 or Spo7 proteins are indicated. The data shown are representative of three experiments.
Nem1 and Spo7 are phosphorylated by PKA in vivo
To determine the in vivo phosphorylation of Nem1–Spo7 by PKA, the phosphatase complex isolated from yeast cells was examined by immunoblotting with an antibody against the phospho-PKA substrate. The immunodetection of Nem1 and Spo7 indicates that both subunits of the phosphatase complex are endogenously phosphorylated by PKA (Fig. 8, left). Because Ser-210 of Nem1 and Ser-28 of Spo7 were identified as major sites of phosphorylation by the protein kinase, we examined the effect of their mutations to nonphosphorylatable alanine residues. Immunoblot analysis of the phosphatase complexes consisting of Nem1(S210A) and Spo7 as well as of Spo7(S28A) and Nem1 showed that the detection of the mutant subunits was greatly reduced by the phospho-PKA substrate antibody (Fig. 8, left), but not by the antibodies against Nem1 and Spo7 (Fig. 8, right). These results indicate that Ser-210 of Nem1 and Ser-28 of Spo7 are major sites of endogenous phosphorylation by PKA.
Figure 8.
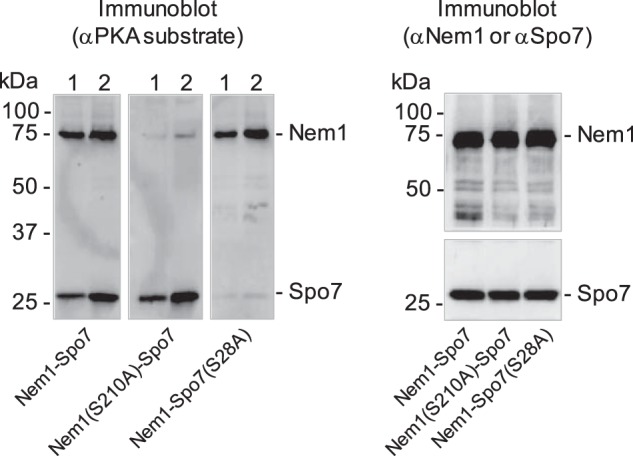
Nem1 and Spo7 are phosphorylated by PKA in vivo. The WT and S210A mutant forms of Nem1 or the WT and S28A mutant forms of Spo7 were co-expressed in yeast cells. Each complex was purified by IgG-Sepharose affinity chromatography based on the protein A tag on Nem1. A, samples (30 ng (lane 1) or 60 ng (lane 2) of the indicated complex) were subjected to SDS-PAGE followed by transfer to PVDF membrane. The membrane was probed with anti-(phosphoserine/phosphothreonine) PKA substrate antibody. B, samples (60 ng of the indicated complex) were subjected to SDS-PAGE followed by transfer to PVDF membrane. The membrane was cut; the upper and lower portions, respectively, were probed with anti-Nem1 and anti-Spo7 antibodies. The positions of Nem1, Spo7, and molecular mass standards are indicated. The data shown in the figure are representative of three experiments.
Effects of phosphorylation-deficient and -mimicking mutations of Nem1 and Spo7 on growth at elevated temperature and on lipid composition
To examine the effect of the PKA phosphorylation site mutations of Nem1 (S140A, S210A, S140D, and S210D) and Spo7 (S28A and S28D) on the physiological function of the Nem1–Spo7 complex, the mutations were introduced singly or in combination into chromosomal NEM1 and SPO7 by gene replacement. For NEM1, its functional allele NEM1-PtA was substituted for consistency with the in vitro analysis of the protein A–tagged Nem1 complexed with Spo7. First, we examined the growth of the cells expressing the mutant alleles of NEM1 and SPO7. As was known previously (1), the nem1Δ or spo7Δ mutations caused a lack of growth at 37 °C, indicating the requirement of both subunits for the function of the Nem1–Spo7 complex (Fig. 9). Unlike the nem1Δ or spo7Δ cells, those harboring the nonphosphorylatable (serine-to-alanine) mutant allele (Fig. 9A) or phosphorylation-mimicking (serine-to-aspartate) allele (Fig. 9B) of NEM1 and SPO7 showed growth at 37 °C that is indistinguishable from that of the control cells. This result indicates that the nonphosphorylatable and phosphorylation-mimicking mutations of Nem1 and Spo7 do not compromise the formation and function of the Nem1–Spo7 complex for cell growth at a higher temperature.
Figure 9.
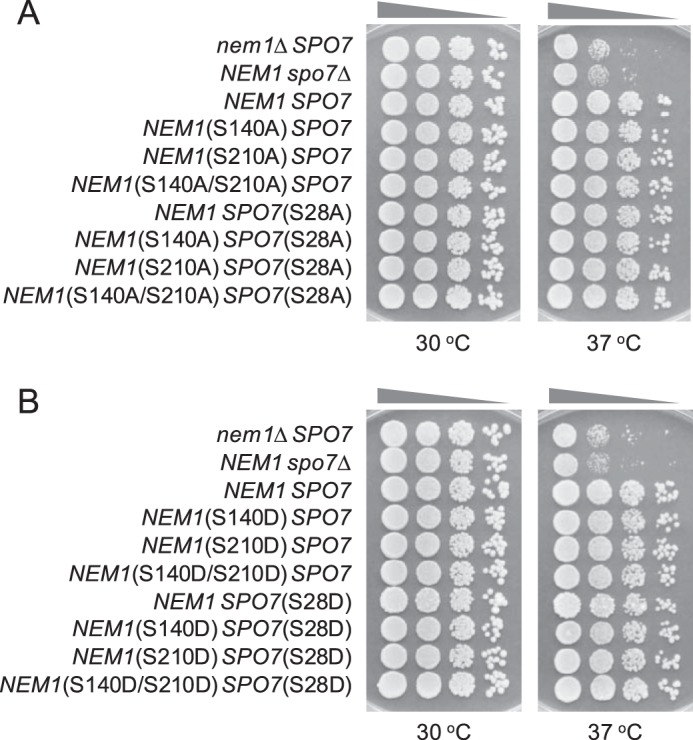
Effect of temperature on the growth of cells harboring the phosphorylation-deficient and -mimicking alleles of NEM1 and SPO7. Yeast strains containing the indicated serine-to-alanine (A) or -aspartate (B) mutations in chromosomal NEM1 and SPO7 were grown at 30 °C to saturation in YEPD medium. The saturated cultures were diluted to a density of 0.67 at A600 nm, followed by 10-fold serial dilutions. The diluted cultures were spotted (2.5 μl) onto YEPD plates and incubated for 3 days at 30 and 37 °C. The data are representative of three independent experiments.
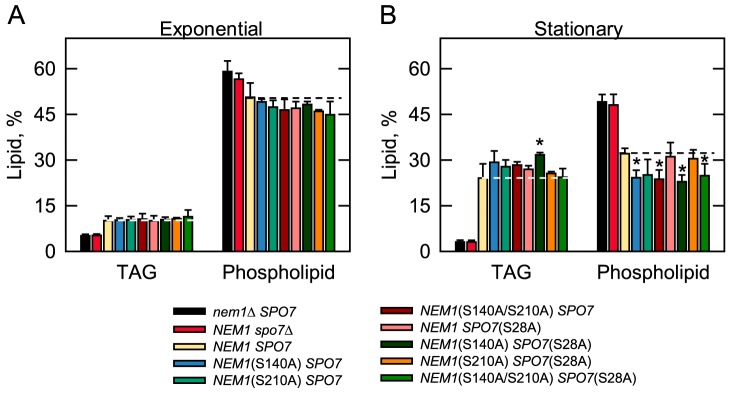
As the major regulator of Pah1 function, the Nem1–Spo7 complex was examined for its function in lipid metabolism. For this analysis, cells bearing the mutant alleles of NEM1 and SPO7 were radiolabeled for lipids with [2-14C]acetate during growth to the exponential and stationary phases. As expected from early studies with WT cells (50, 51), the TAG content of the control cells (NEM1 SPO7, yellow symbols) was 2.4-fold higher in the stationary phase than in the exponential phase (Fig. 10). Compared with the control cells, the nem1Δ (black symbols) and spo7Δ (red symbols) mutants exhibited greatly reduced TAG content in the exponential (50%) and stationary phases (87%) of growth (Fig. 10). These data emphasize the prominent role of the Nem1–Spo7 complex in the regulation of Pah1 function in TAG synthesis. The reduced TAG levels of the nem1Δ and spo7Δ mutants, and particularly in the stationary phase, correlated with the concomitant increase in the levels of membrane phospholipids (Fig. 10B). Together, these results exemplify the importance of the Nem1–Spo7/Pah1 phosphatase cascade in controlling the synthesis of TAG and membrane phospholipids from the common precursor PA (2, 3). In the exponential phase cells, the alanine mutations of the PKA sites in Nem1 and/or Spo7 did not influence the levels of TAG and phospholipids (Fig. 10A). The effects of the phosphorylation site mutations on the amounts of TAG and phospholipids were event in the stationary phase, and particularly for cells harboring the NEM1(S140A) mutation (Fig. 10B). For example, the TAG content of stationary phase cells with the NEM1(S140A) (blue symbols), NEM1(S140A/S210A) (dark red symbols), and NEM1(S140A) SPO7(S28A) (dark green symbols) mutations was 22, 18, and 32% (p < 0.05) greater when compared with that of the control cells (yellow symbols). The phospholipid content of the same cells was 25% (p < 0.05), 26% (p < 0.05), and 29% (p < 0.05) lower, respectively, when compared with the control cells. The phosphorylation-mimicking mutations did not have effects on the lipid composition of exponential or stationary phase cells (data not shown).
Figure 10.
Lipid composition of yeast cells harboring the phosphorylation-deficient alleles of NEM1 and SPO7. Yeast strains containing the indicated serine-to-alanine mutations in chromosomal NEM1 and SPO7 were grown at 30 °C to the exponential (A) and stationary (B) phases of growth in SC medium containing [2-14C]acetate (1 μCi/ml). The lipids were extracted and separated by one-dimensional TLC, and the phosphor images were subjected to ImageQuant analysis. The percentages shown for TAG, and phospholipids were normalized to the total 14C-labeled chloroform-soluble fraction. Each data point represents the average of three experiments ± S.D. (error bars). The data points for cells with nem1Δ SPO7, NEM1 spo7Δ, and NEM1 SPO7 for exponential or stationary phase are the same for comparison. The white and black dashed lines signify the levels of TAG and phospholipid, respectively, in the NEM1 SPO7 control. *, p < 0.05 versus cells with the NEM1 SPO7 control.
Discussion
In this work, we demonstrated that the Nem1–Spo7 phosphatase complex is phosphorylated by PKA, a protein kinase whose activity is associated with active cell growth and increased metabolic activity and an increase in membrane phospholipid synthesis (10, 41, 42). Our in vitro experiments using the purified Nem1–Spo7 complex, as well as peptides derived from their sequences, indicated that both subunits of the complex are substrates for PKA. The immunoblot analysis of the complex derived from cells with the anti-PKA substrate antibody provided evidence that Nem1 and Spo7 are phosphorylated by PKA in vivo. The multifaceted biochemical and molecular genetics approaches used here led to the identifications of Ser-140 and Ser-210 in Nem1 and Ser-28 in Spo7 as sites of phosphorylation by PKA. Phosphoproteomics studies have shown that Ser-210 in Nem1 is a site of phosphorylation, but the protein kinase involved has been unknown (39, 40) until now.
Phosphorylation-deficient mutations in Nem1 and Spo7 were introduced into the chromosome to examine the physiological consequences of the phosphorylations by PKA. The analysis of the cells bearing the mutations indicated that the phosphorylations of the subunits alone, or in combination, are not necessary for their function in vivo. Cells bearing all of the PKA phosphorylation site mutations grew at 37 °C, the restrictive temperature of growth for nem1Δ and spo7Δ mutants that lack the complex (1). Like cells that lack the Nem1–Spo7 complex, cells lacking Pah1 also exhibit a temperature-sensitive phenotype (4, 7, 33). This phenotype is ascribed to the defect in DAG/TAG synthesis, as opposed to the increase in phospholipid synthesis (3). This assertion is based on the observation that temperature sensitivity is not suppressed by the lack of Dgk1, the CTP-dependent DAG kinase that converts DAG back to PA, which is then used to synthesize membrane phospholipids (52, 53). The nuclear/ER membrane expansion phenotype, which is exhibited by cells lacking any component of the Nem1–Spo7/Pah1 phosphatase cascade, is based on elevated phospholipid synthesis; the dgk1Δ mutation suppresses membrane expansion (1, 3, 4, 52). Lipid analysis of cells harboring the phosphorylation-deficient mutations at the PKA sites in Nem1 and/or Spo7 showed that none of the mutations caused a reduction in the synthesis of TAG or a stimulation in phospholipid synthesis. In fact, the mutations caused a small increase and corresponding decrease, respectively, in the relative amounts of TAG and membrane phospholipids. Thus, the PKA site mutations caused a gain-of-function phenotype for the Nem1–Spo7/Pah1 phosphatase cascade, which provides the explanation for why the mutations did not cause loss of growth at the elevated temperature. Also, the mutations would not be expected to cause nuclear/ER membrane expansion or a reduction in lipid droplets, although we did not score the mutants for these phenotypes. The effects of the phosphorylation site mutations on lipid composition were only observed in stationary phase cells, the phase of growth when the Nem1–Spo7/Pah1 phosphatase cascade has its greatest effect on lipid composition (7, 24, 26). Cells harboring the S140A mutation in Nem1 alone, or in combination with the S28A mutation in Spo7, exhibited the greatest effects on lipid composition. Thus, the phosphorylations of Ser-140 in Nem1 and Ser-28 in Spo7 by PKA might be expected to inhibit Nem1–Spo7 phosphatase function and attenuate the synthesis of TAG (Fig. 11). Admittedly, the effects of the mutations on lipid composition are subtle, indicating that the PKA-mediated phosphorylations of Nem1 and Spo7 may be a fine-tuning mechanism to regulate the Nem1–Spo7/Pah1 phosphatase cascade. It is also plausible that the regulation by PKA requires additional phosphorylations by other protein kinases to reveal its full effects on Nem1–Spo7 phosphatase function. For example, the effects of the PKA phosphorylation of Pah1 function are greatly enhanced by the multiple phosphorylations by Pho85–Pho80 (18).
Figure 11.
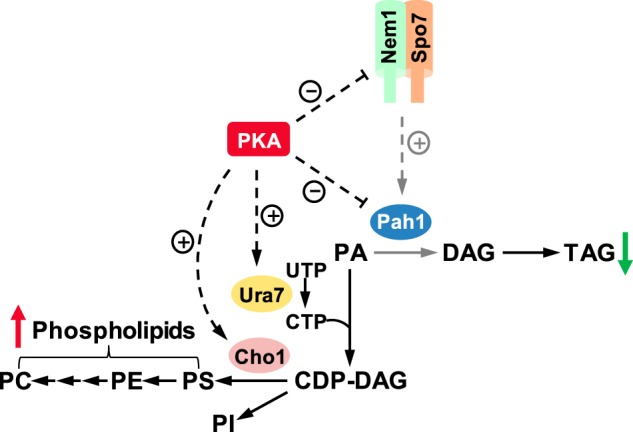
Model for the PKA-mediated regulation of lipid synthesis. The PKA-mediated phosphorylations of the Nem1–Spo7 complex and Pah1 have a negative effect on their functions. The positive effect the Nem1–Spo7 complex has on Pah1 function is attenuated (indicated by the dashed gray arrow) by its phosphorylation. The net effect of the phosphorylations of Nem1–Spo7 and Pah1 by PKA is a decrease in TAG (green arrow) and an increase in phospholipids (red arrow). PKA also has a positive effect on phospholipid synthesis by phosphorylating Ura7 CTP synthetase and Cho1 phosphatidylserine synthase, key enzymes that synthesize and utilize CDP-DAG, respectively.
Ser-195 has been identified as a site in Nem1 that is phosphorylated in cells supplemented with rapamycin (15), the inhibitor of the TORC1 protein kinase complex (54). This phosphorylation, as mediated by an unknown protein kinase, correlates with the dephosphorylation of Pah1 and an increase in TAG content (15), the inference being that the phosphorylation of Ser-195 in Nem1 stimulates its activity to elevate PA phosphatase activity through the dephosphorylation of Pah1 (15). The reduction in Nem1–Spo7 phosphatase activity in response to the alkaline phosphatase treatment might be related to the phosphorylation state of Ser-195, but this possibility has not been addressed. Our data indicated that Ser-195 is not a good target for PKA and that the phosphorylation of Nem1 and/or Spo7 by PKA does not have a stimulatory effect on the protein phosphatase activity of the complex. Instead, the PKA phosphorylation had a small, although not statistically significant, inhibitory effect on Nem1–Spo7 phosphatase activity after its treatment with alkaline phosphatase. The phosphorylation status of Ser-210, the major PKA site in Nem1, is unaffected in cells treated with rapamycin (15). At this point, the connection between the phosphorylations of Nem1 by PKA and the unknown protein kinase that phosphorylates it in response to rapamycin is unclear.
The Nem1–Spo7/Pah1 phosphatase cascade exists in higher eukaryotes, and it is relevant to lipid metabolism (28, 55–58). First, the corresponding PA phosphatase activity in metazoans, and in particular humans, is encoded by three genes named LPIN1, LPIN2, and LPIN3 (59). The molecular function of human lipin 1 as a PA phosphatase enzyme was only discovered after yeast Pah1 was shown to be a PA phosphatase enzyme (7). Subsequently, lipins 1, 2, and 3 from mice were shown to be PA phosphatase enzymes (60). Like Pah1, mouse lipin 1 is a highly phosphorylated enzyme, and the state of its phosphorylation governs its subcellular localization (61–64). Insulin-stimulated phosphorylation of a serine-rich region by an unknown protein kinase facilitates its binding to 14:3:3 proteins for cytoplasmic association (65). TORC1 phosphorylates Ser-106 and Ser-472 (62); the identity of the protein kinases that phosphorylate all of the remaining sites is unknown. Second, an analogous Nem1–Spo7 complex exists in metazoans (28). In humans, CTDNEP1 (C-terminal domain nuclear envelope phosphatase 1) is the catalytic subunit, whereas NEP1-R1 (nuclear envelope phosphatase 1-regulatory subunit 1) is the regulatory subunit (28). Conservation of CTDNEP1–NEP1-R1 function is indicated by its complementing the yeast nem1Δ spo7Δ mutant phenotypes of nuclear/ER membrane expansion and reductions in TAG and lipid droplet formation (28). Moreover, when expressed in human cells, the CTDNEP1–NEP1-R1 dephosphorylates lipin 1 (28). Phosphoproteomics analysis has identified sites in CTDNEP1 that are phosphorylated in vivo (66, 67), and an analysis of mouse and human CTDNEP1 and NEP1-R1 with the NetPhos3.1 server (43) indicates putative sites of phosphorylation by multiple protein kinases including PKA. Whether or not either subunit of the complex is phosphorylated by PKA is unknown, but the conservation of the Nem1–Spo7/Pah1 phosphatase cascade in higher eukaryotes provides impetus for this avenue of investigation.
The work presented here advances our understanding of the posttranslational modification of the yeast Nem1–Spo7 complex by phosphorylation, and it further demonstrates the importance of PKA in modulating lipid metabolism in yeast. The model shown in Fig. 11 illustrates the PKA-mediated phosphorylations of the Nem1–Spo7/Pah1 phosphatase cascade to inhibit TAG synthesis with a concomitant increase in phospholipids. It is also known that PKA has a positive influence on the CDP-DAG–dependent synthesis of phospholipids through the phosphorylations of Ura7 CTP synthetase (46, 68) and Cho1 phosphatidylserine synthase (49), key enzymes that synthesize and utilize CDP-DAG, respectively (10, 11). Thus, by phosphorylating these enzymes, PKA stimulates phospholipid synthesis and, at the same time, inhibits TAG production to ensure cells have enough membrane phospholipids for cell growth.
Experimental procedures
Materials
Difco was the source of growth media. Plasmid DNA purification kits and nickel-nitrilotriacetic acid–agarose resin were from Qiagen. The QuikChange site-directed mutagenesis kit, carrier DNA for yeast transformation, and enzyme reagents for DNA manipulations were obtained from Stratagene, Clontech, and New England Biolabs, respectively. PCR primers were prepared by Genosys Biotechnologies. DNA size ladders, molecular mass protein standards, and reagents for electrophoresis, immunoblotting, and protein determination were purchased from Bio-Rad. Ampicillin, carbenicillin, chloramphenicol, PCR primers, nucleotides, Ponceau S stain, Triton X-100, protease inhibitors (phenylmethylsulfonyl fluoride, benzamidine, aprotinin, leupeptin, and pepstatin), 2-mercaptoethanol, BSA, phosphoamino acid standards, isopropyl-β-d-1-thiogalactoside, l-1-tosylamido-2-phenylethyl chloromethyl ketone–treated trypsin, alkaline phosphatase–agarose, and rabbit anti-protein A antibody (product P3775, lot 025K4777) were from Sigma-Aldrich. SYPRO Ruby protein gel stain was from Invitrogen. IgG-Sepharose, protein A–Sepharose, Q-Sepharose, PVDF membrane, and the enhanced chemifluorescence Western blotting detection kit were from GE Healthcare. Promega was the source of bovine heart PKA catalytic subunit. Nem1 and Spo7 peptides used for PKA phosphorylation assays were synthesized by EZBioLabs. The peptide sequences KESDQNQERKNSVPKKPKA (residues 65–82) and REGARRRKQAHELRPKSE (residues 242–259) at the N-terminal and C-terminal portions of Nem1 and Spo7, respectively, were synthesized and used to raise antibodies in New Zealand White rabbits at BioSynthesis, Inc. Rabbit anti-(phosphoserine/phosphothreonine) PKA substrate antibody (product 9624, lot 18) was from Cell Signaling Technology. Thermo Scientific was the source of alkaline phosphatase–conjugated goat anti-rabbit IgG antibody (product 31340, lot NJ178812). Lipids and TLC plates (cellulose and silica gel 60) were from Avanti Polar Lipids and EMD Millipore, respectively. P81 phosphocellulose paper was from Whatman. Radiochemicals were from PerkinElmer Life Sciences, and scintillation-counting supplies were from National Diagnostics. All other chemicals were reagent grade or better.
Strains and growth conditions
The bacterial and yeast strains used in this study are listed in Table 2. The Escherichia coli strain DH5α was used for the propagation of plasmids. E. coli strains BL21(DE3)pLysS and BL21(DE3) were used to express His6-tagged Pah1 and His6-tagged Pho85 and Pho80, respectively. The bacterial cells were grown at 37 °C in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.0). For the selection of cells carrying plasmids, the growth medium was supplemented with antibiotics (e.g. ampicillin, 100 μg/ml; carbenicillin, 100 μg/ml; chloramphenicol, 34 μg/ml). The expressions of His6-tagged Pah1, His6-tagged Pho85, and Pho80 were induced with 1 mm isopropyl β-d-thiogalactoside. The Saccharomyces cerevisiae 3 strain RS453 or SS1010 was used for the galactose-induced expressions of the WT and mutant forms of the Nem1–Spo7 complex (Nem1 tagged with protein A). The yeast were routinely grown at 30 °C in SC medium containing 2% glucose; appropriate amino acids were omitted from the growth medium to select for cells carrying specific plasmids (69). For the galactose-induced expressions of protein A–tagged Nem1 and Spo7, cells were first grown to the exponential phase in SC medium with 2% raffinose and then grown in the same medium for 8 h with 2% galactose. For the growth spot test, cells were grown on YEPD (1% yeast extract, 2% peptone, and 2% glucose) agar plates (69). Cell numbers in liquid cultures were determined spectrophotometrically at A600 nm. Solid medium plates were prepared by adding agar (1.5% for E. coli and 2% for yeast) into the lipid growth medium.
Table 2.
Strains used in this work
| Strain | Genotype or relevant characteristics | Source/Reference |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacZΔM15Δ (lacZYAargF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Ref. 70 |
| BL21(DE3)pLysS | F− ompT hsdSB (rB−mB−) gal dcm (DE3) pLysS | Novagen |
| BL21(DE3) | F− ompT hsdSB (rB−mB−) gal dcm | Invitrogen |
| S. cerevisiae | ||
| RS453 | MATa ade21 his311,15 leu23,112 trp11 ura352 | Ref. 52 |
| Derivatives | ||
| SS1010 | nem1Δ::HIS3 spo7Δ::HIS3 | Ref. 1 |
| SS1002 | nem1Δ::HIS3 | Ref. 1 |
| WMY161 | nem1Δ::URA3 | This study |
| WMY162 | NEM1-PtA | This study |
| WMY163 | NEM1(S140A)-PtA | This study |
| WMY164 | NEM1(S140D)-PtA | This study |
| WMY165 | NEM1(S210A)-PtA | This study |
| WMY166 | NEM1(S210D)-PtA | This study |
| WMY167 | NEM1(S140A/S210A)-PtA | This study |
| WMY168 | NEM1(S140D/S210D)-PtA | This study |
| GHY67 | spo7Δ::URA3 | This study |
| GHY68 | NEM1-PtA spo7Δ::URA3 | This study |
| GHY69 | NEM1(S140A)-PtA spo7Δ::URA3 | This study |
| GHY70 | NEM1(S140D)-PtA spo7Δ::URA3 | This study |
| GHY71 | NEM1(S210A)-PtA spo7Δ::URA3 | This study |
| GHY72 | NEM1(S210D)-PtA spo7Δ::URA3 | This study |
| GHY73 | NEM1(S140A/S210A)-PtA spo7Δ::URA3 | This study |
| GHY74 | NEM1(S140D/S210D)-PtA spo7Δ::URA3 | This study |
| GHY75 | SPO7(S28A) | This study |
| GHY76 | SPO7(S28D) | This study |
| GHY77 | NEM1-PtA SPO7(S28A) | This study |
| GHY78 | NEM1-PtA SPO7(S28D) | This study |
| GHY79 | NEM1(S140A)-PtA SPO7(S28A) | This study |
| GHY80 | NEM1(S140D)-PtA SPO7(S28D) | This study |
| GHY81 | NEM1(S210A)-PtA SPO7(S28A) | This study |
| GHY82 | NEM1(S210D)-PtA SPO7(S28D) | This study |
| GHY83 | NEM1(S140A/S210A)-PtA SPO7(S28A) | This study |
| GHY84 | NEM1(S140D/S210D)-PtA SPO7(S28D) | This study |
Construction of plasmids and yeast strains
The plasmids used in this study are listed in Table 3. The isolation of chromosomal and plasmid DNA, the digestion and ligation of DNA, and PCR were performed according to standard protocols (70, 71). E. coli (70) and yeast (72) transformations were performed as described previously. pWM210 was constructed from the vector pRS314 at the PstI site by insertion of 1.8-kb GAL1/10-NEM1-PtA, which was released from pRS313-GAL1/10-SPO7. pGH443 was constructed from the vector pRS415 at the XhoI/NotI sites by insertion of the 2.3-kb SPO7 DNA, which was amplified by PCR from strain BY4741. The derivatives of YCplac111-GAL1/10-NEM1-PtA, YCplac111-NEM1-PtA, pWM210, and pGH443 were produced by PCR-mediated site-directed mutagenesis using the plasmid templates.
Table 3.
Plasmids used in this work
| Plasmid | Relevant characteristics | Source/Reference |
|---|---|---|
| YCplac111-GAL1/10-NEM1-PtA | NEM1PtA under control of GAL1/10 promoter in CEN/LEU2 vector | Ref. 4 |
| Derivatives | ||
| pWM201 | NEM1(S140A) | This study |
| pWM202 | NEM1(S210A) | This study |
| pWM203 | NEM1(S140A/S210A) | This study |
| pRS313-GAL1/10-SPO7 | SPO7 under control of GAL1/10 promoter in CEN/HIS3 vector | Ref. 52 |
| pRS314-GAL1/10-SPO7 | SPO7 under control of GAL1/10 promoter in CEN/TRP1 vector | This study |
| Derivatives | ||
| pWM211 | SPO7(S28A) | This study |
| pWM212 | SPO7(S41A) | This study |
| pWM213 | SPO7(S42A) | This study |
| pWM214 | SPO7(S41A/S42A) | This study |
| YCplac111-NEM1-PtA | NEM1PtA inserted into the CEN/LEU2 vector | S. Siniossoglou |
| Derivatives | ||
| pWM204 | NEM1(S140A) | This study |
| pWM205 | NEM1(S140D) | This study |
| pWM206 | NEM1(S210A) | This study |
| pWM207 | NEM1(S210D) | This study |
| pWM208 | NEM1(S140A/S210A) | This study |
| pWM209 | NEM1(S140D/S210D) | This study |
| pRS426 | Multicopy E. coli/yeast shuttle vector with URA3 | Ref. 88 |
| pRS416 | Single-copy E. coli/yeast shuttle vector with URA3 | Ref. 89 |
| pRS415 | Single-copy E. coli/yeast shuttle vector with LEU2 | Ref. 89 |
| pGH443 | SPO7 inserted into pRS415 | This study |
| Derivatives | ||
| pGH444 | SPO7(S28A) | This study |
| pGH445 | SPO7(S28D) | This study |
Yeast strains with chromosomal mutations were generated by one-step gene replacement (73). The nem1Δ::URA3 and spo7Δ::URA3 disruption cassettes, which consist of the URA3 gene flanked with the 50-bp sequences corresponding to the upstream and downstream of the NEM1 or SPO7 coding sequence, were generated by PCR from the template pRS416 and pRS426, respectively. Strain SS1002 (RS453 nem1Δ::HIS3) was transformed with the nem1Δ::URA3 disruption cassette, and the transformants were selected on SC medium lacking uracil. Of the Ura+ transformants, the nem1Δ::URA3 mutant (WMY161) was identified by its lack of growth on SC medium lacking histidine. The nem1Δ::URA3 mutant was then transformed with 2.8-kb NEM1-PtA or its phosphorylation site mutant allele produced from YCplac111-NEM1-PtA and its derivatives, and the resulting transformants were selected on medium containing 5-fluoroorotic acid. The 5-fluoroorotic acid–resistant transformants were analyzed by PCR to confirm gene replacement and by DNA sequencing of the PCR products to confirm the mutation of the NEM1 coding sequence. Strain RS453 or WMY162 (RS453 NEM1-PtA) was transformed with the spo7Δ::URA3 disruption cassette, and the resulting transformants were selected on SC-Ura medium. Of Ura+ transformants, the spo7Δ::URA3 (GHY67) and NEM1-PtA spo7Δ::URA3 (GHY68) mutants were identified by PCR analysis of genomic DNA. The NEM1-PtA spo7Δ::URA3 mutant was then transformed with 2.3-kb SPO7 DNA released from pGH444 or pGH445, and the transformants were selected on 5-fluoroorotic acid medium. The 5-fluoroorotic acid–resistant transformants were analyzed by PCR to confirm gene replacement and by DNA sequencing of the PCR products to confirm mutations in the SPO7 coding sequence. Strains carrying combined mutations of NEM1-PtA and SPO7 were constructed from the NEM1-PtA mutant by disruption of SPO7 with URA3, followed by the marker gene replacement with the SPO7 mutant allele as described above.
Preparation of cell extracts, purification of enzymes, and protein determinations
All steps were performed at 4 °C. Lysates were prepared by disruption of the yeast cells with glass beads (0.5-mm diameter) using a BioSpec Products Mini-BeadBeater-16 (74). The cell disruption buffer contained 50 mm Tris-HCl (pH 7.5), 0.3 m sucrose, 0.15 m NaCl, 10 mm 2-mercaptoethanol, 0.5 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin (7). The WT and mutant forms of the Nem1–Spo7 complex (Nem1 tagged with protein A) were purified from yeast cells by IgG-Sepharose affinity chromatography as described by Siniossoglou et al. (75) with the minor modifications described by Su et al. (6). His6-tagged Pah1 expressed in E. coli was purified by affinity chromatography with nickel-nitrilotriacetic acid–agarose (7) followed by Q-Sepharose chromatography as described by Su et al. (6). The His6-tagged Pho85–Pho80 protein kinase complex expressed in E. coli was purified by nickel-nitrilotriacetic acid–agarose affinity chromatography as described by Jeffery et al. (76). SDS-PAGE analysis indicated that these enzyme preparations were highly purified. The protein content in solution was estimated by the method of Bradford (77) or by ImageQuant analysis of Coomassie Blue–stained or SYPRO Ruby–stained SDS-polyacrylamide gels. BSA was used as the standard in estimating protein concentrations in solution or in polyacrylamide gels.
SDS-PAGE and immunoblotting
SDS-PAGE (78) was routinely performed with a 10 or 12% slab gel, and immunoblotting (79, 80) was performed with a PVDF membrane. The samples for blotting were normalized to total protein loading on SDS-polyacrylamide gels. Ponceau S staining was used to monitor the protein transfer from the gel to the PVDF membrane. The IgG fraction of the rabbit anti-Nem1 and anti-Spo7 antibodies were purified from antisera by protein A–Sepharose chromatography (81) and used for immunoblotting experiments. The antibody preparations were characterized with respect to specificity using the purified preparation of the Nem1–Spo7 complex. The rabbit anti-Nem1 and anti-Spo7 antibodies were used at a protein concentration of 1 μg/ml. The rabbit anti-protein A and anti-(phosphoserine/phosphothreonine) PKA substrate antibodies were used at dilutions 1:1,000 and 1:4,000, respectively. Alkaline phosphatase-conjugated goat anti-rabbit IgG antibody was used at a dilution of 1:5,000. Immune complexes were detected using the enhanced chemifluorescence immunoblotting substrate. Fluorimaging was used to acquire fluorescence signals from immunoblots, and the intensities of the images were analyzed by ImageQuant software. A standard curve was used to confirm that the immunoblot signals were in the linear range of detection.
PKA phosphorylation assays
The phosphorylations of WT and mutant forms of Nem1 and Spo7 complex by PKA were routinely measured in triplicate at 30 °C by following the incorporation of radiolabeled phosphate from [γ-32P]ATP into each protein. The reaction mixture contained 25 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 2 mm DTT, 100 μm [γ-32P]ATP (3,000 cpm/pmol), the indicated amount of Nem1–Spo7 complex, and the indicated amount of PKA in a total volume of 20 μl. The PKA reaction was terminated by the addition of 5× Laemmli sample buffer (78), subjected to SDS-PAGE to separate 32P-labeled Nem1 and Spo7 from [γ-32P]ATP, and transferred to a PVDF membrane. Radioactively labeled Nem1 and Spo7 were visualized by phosphorimaging, and the extents of their phosphorylations were quantified by ImageQuant software. For the phosphorylations of Nem1 and Spo7 peptides, the reactions were terminated by spotting the reaction mixture onto a P81 phosphocellulose paper. The paper was washed three times with 75 mm phosphoric acid and then subjected to scintillation counting. The phosphorylation reactions were linear with time and protein concentration. One unit of PKA activity was defined as 1 nmol/min.
Analysis of phosphoamino acids and phosphopeptides
Pieces of PVDF membrane containing 32P-labeled Nem1 or Spo7 were subjected to hydrolysis with 6 n HCl at 110 °C (for phosphoamino acid analysis) or proteolytic digestion with l-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin (for phosphopeptide-mapping analysis) (82–84). The acid hydrolysates were mixed with standard phosphoamino acids and separated by two-dimensional electrophoresis on cellulose TLC plates. The tryptic digests were separated on the cellulose plates first by electrophoresis and then by TLC (82–84). Radioactive phosphoamino acids and peptides were visualized by phosphorimaging analysis. Nonradioactive phosphoamino acid standards were visualized by ninhydrin staining.
Analysis of phosphorylation sites by mass spectrometry
The PKA-phosphorylated Spo7 peptide was analyzed by MS at the Rutgers Mass Spectrometry Center for Integrative Neuroscience Research. The phosphorylated peptide was analyzed by LC-MS/MS on a Thermo LTQ Orbitrap Velos mass spectrometer. The data were analyzed against the Spo7 sequence using the MASCOT search engine on Proteome Discoverer software.
Nem1–Spo7 phosphatase assay
Nem1–Spo7 phosphatase activity was measured by following the release of 32Pi from [32P]Pah1 (6). The reaction mixture contained 100 mm sodium acetate (pH 5.0), 10 mm MgCl2, 0.25 mm Triton X-100, 1 mm DTT, 0.25 μm phosphorylated Pah1, and the Nem1–Spo7 complex in a total volume of 50 μl. Following 10-min incubation, the reaction was terminated by the addition of 0.5 ml of TCA (20% final concentration) and 0.2 ml of BSA (0.4 mg/ml final concentration); samples were cooled on ice for 15 min to allow precipitation of proteins. The mixture was centrifuged for 20 min at 15,000 × g to separate the reaction product 32Pi from the substrate, and a 0.2-ml aliquot of the supernatant was measured for radioactivity by scintillation counting. The amount of phosphate produced in the reaction was calculated on the basis of the specific activity of the [γ-32P]ATP used to prepare [32P]Pah1. A unit of Nem1–Spo7 phosphatase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of phosphate/min. The Nem1–Spo7 phosphatase reactions, which were conducted in triplicate at 30 °C, were linear with time and protein concentration. To prepare the radioactive substrate, Pah1 was phosphorylated by the Pho85–Pho80 protein kinase complex using 100 μm [γ-32P]ATP (5,000–10,000 cpm/pmol) as described by Choi et al. (16). The Pho85–Pho80 complex was removed from 32P-labeled Pah1 by Q-Sepharose chromatography (6).
Radiolabeling and analysis of lipids
The steady-state labeling of lipids with [2-14C]acetate (85), the extraction of lipids from the radiolabeled cells (86), and their separation one-dimensional TLC (87) were performed as described previously. The resolved lipids were observed by phosphorimaging and quantified by ImageQuant software. The identity of radiolabeled lipids was confirmed by comparison with the migration of authentic standards visualized by staining with iodine vapor.
Analyses of data
SigmaPlot software was used for statistical analyses, where p values < 0.05 were taken as a significant difference. The Enzyme Kinetics module of the SigmaPlot software, which uses the Marquardt–Levenberg algorithm, was used to determine kinetic parameters according to the Michaelis–Menten equation.
Author contributions
W.-M. S., G.-S. H., P. D., and G. M. C. designed the study, analyzed the results, and prepared the manuscript. W.-M. S., G.-S. H., and P. D. performed the experiments.
Acknowledgments
The MS data were obtained from an Orbitrap mass spectrometer funded in part by National Institutes of Health Grant NS046593, for the support of the Rutgers Mass Spectrometry Center for Integrative Neuroscience Research.
This work was supported, in whole or in part, by National Institutes of Health Grant GM050679 from the United States Public Health Service. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
In this paper, “Saccharomyces cerevisiae” is used interchangeably with “yeast.”
- PA
- phosphatidate
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- ER
- endoplasmic reticulum
- PKA
- protein kinase A
- PKC
- protein kinase C
- SC
- synthetic complete
- PVDF
- polyvinylidene difluoride.
References
- 1. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siniossoglou S. (2013) Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim. Biophys. Acta 1831, 575–581 10.1016/j.bbalip.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 3. Pascual F., and Carman G. M. (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831, 514–522 10.1016/j.bbalip.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Hara L., Han G.-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su W.-M., Han G.-S., and Carman G. M. (2014) Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289, 34699–34708 10.1074/jbc.M114.614883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han G.-S., Wu W.-I., and Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin Y.-P., and Carman G. M. (1989) Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 9. Smith S. W., Weiss S. B., and Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 10. Carman G. M., and Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Madera M., Vogel C., Kummerfeld S. K., Chothia C., and Gough J. (2004) The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32, D235–D239 10.1093/nar/gkh117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koonin E. V., and Tatusov R. L. (1994) Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity: application of an iterative approach to database search. J. Mol. Biol. 244, 125–132 10.1006/jmbi.1994.1711 [DOI] [PubMed] [Google Scholar]
- 14. Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G.-S., Reggiori F., Carman G. M., and Siniossoglou S. (2013) Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133 10.1091/mbc.e13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubots E., Cottier S., Péli-Gulli M. P., Jaquenoud M., Bontron S., Schneiter R., and De Virgilio C. (2014) TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex. PLoS One 9, e104194 10.1371/journal.pone.0104194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi H.-S., Su W.-M., Han G.-S., Plote D., Xu Z., and Carman G. M. (2012) Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287, 11290–11301 10.1074/jbc.M112.346023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Su W.-M., Han G.-S., Casciano J., and Carman G. M. (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287, 33364–33376 10.1074/jbc.M112.402339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su W.-M., Han G.-S., and Carman G. M. (2014) Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289, 18818–18830 10.1074/jbc.M114.581462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2016) Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291, 9974–9990 10.1074/jbc.M116.726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2015) Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290, 11467–11478 10.1074/jbc.M115.648659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karanasios E., Han G.-S., Xu Z., Carman G. M., and Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han G.-S., Siniossoglou S., and Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- 24. Pascual F., Soto-Cardalda A., and Carman G. M. (2013) PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288, 35781–35792 10.1074/jbc.M113.525766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han G.-S., and Carman G. M. (2017) Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem. 292, 13230–13242 10.1074/jbc.M117.801720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fakas S., Qiu Y., Dixon J. L., Han G.-S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M. (2011) Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 10.1074/jbc.M111.258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han S., Bahmanyar S., Zhang P., Grishin N., Oegema K., Crooke R., Graham M., Reue K., Dixon J. E., and Goodman J. M. (2012) Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem. 287, 3123–3137 10.1074/jbc.M111.324350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Kapteyn J. C., Roemer T. W., Vo D. H., Bondoc D. C., Hall J., Zhong W. W., et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruiz C., Cid V. J., Lussier M., Molina M., and Nombela C. (1999) A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast 15, 1001–1008 10.1002/(SICI)1097-0061(199907)15:10B%3C1001::AID-YEA400%3E3.0.CO%3B2-T [DOI] [PubMed] [Google Scholar]
- 31. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 10.1074/jbc.M111.317420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sherr G. L., LaMassa N., Li E., Phillips G., and Shen C. H. (2017) Pah1p negatively regulates the expression of V-ATPase genes as well as vacuolar acidification. Biochem. Biophys. Res. Commun. 491, 693–700 10.1016/j.bbrc.2017.07.127 [DOI] [PubMed] [Google Scholar]
- 33. Irie K., Takase M., Araki H., and Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 [DOI] [PubMed] [Google Scholar]
- 34. Park Y., Han G. S., Mileykovskaya E., Garrett T. A., and Carman G. M. (2015) Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290, 25382–25394 10.1074/jbc.M115.680314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rahman M. A., Mostofa M. G., and Ushimaru T. (2018) The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 285, 1840–1860 10.1111/febs.14448 [DOI] [PubMed] [Google Scholar]
- 36. Xu X., and Okamoto K. (2018) The Nem1-Spo7 protein phosphatase complex is required for efficient mitophagy in yeast. Biochem. Biophys. Res. Commun. 496, 51–57 10.1016/j.bbrc.2017.12.163 [DOI] [PubMed] [Google Scholar]
- 37. Barbosa A. D., Sembongi H., Su W.-M., Abreu S., Reggiori F., Carman G. M., and Siniossoglou S. (2015) Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 26, 3641–3657 10.1091/mbc.e15-03-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orij R., Urbanus M. L., Vizeacoumar F. J., Giaever G., Boone C., Nislow C., Brul S., and Smits G. J. (2012) Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae. Genome Biol. 13, R80 10.1186/gb-2012-13-9-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N. J., and Villén J. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Broach J. R., and Deschenes R. J. (1990) The function of RAS genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54, 79–139 10.1016/S0065-230X(08)60809-X [DOI] [PubMed] [Google Scholar]
- 42. Thevelein J. M. (1994) Signal transduction in yeast. Yeast 10, 1753–1790 10.1002/yea.320101308 [DOI] [PubMed] [Google Scholar]
- 43. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., and Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 44. Ingrell C. R., Miller M. L., Jensen O. N., and Blom N. (2007) NetPhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics 23, 895–897 10.1093/bioinformatics/btm020 [DOI] [PubMed] [Google Scholar]
- 45. Toda T., Cameron S., Sass P., Zoller M., and Wigler M. (1987) Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50, 277–287 10.1016/0092-8674(87)90223-6 [DOI] [PubMed] [Google Scholar]
- 46. Yang W.-L., and Carman G. M. (1996) Phosphorylation and regulation of CTP synthetase from Saccharomyces cerevisiae by protein kinase A. J. Biol. Chem. 271, 28777–28783 10.1074/jbc.271.46.28777 [DOI] [PubMed] [Google Scholar]
- 47. Kim K.-H., and Carman G. M. (1999) Phosphorylation and regulation of choline kinase from Saccharomyces cerevisiae by protein kinase A. J. Biol. Chem. 274, 9531–9538 10.1074/jbc.274.14.9531 [DOI] [PubMed] [Google Scholar]
- 48. Sreenivas A., and Carman G. M. (2003) Phosphorylation of the yeast phospholipid synthesis regulatory protein opi1p by protein kinase A. J. Biol. Chem. 278, 20673–20680 10.1074/jbc.M300132200 [DOI] [PubMed] [Google Scholar]
- 49. Choi H.-S., Han G.-S., and Carman G. M. (2010) Phosphorylation of yeast phosphatidylserine synthase by protein kinase A: identification of Ser46 and Ser47 as major sites of phosphorylation. J. Biol. Chem. 285, 11526–11536 10.1074/jbc.M110.100727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taylor F. R., and Parks L. W. (1979) Triacylglycerol metabolism in Saccharomyces cerevisiae relation to phospholipid synthesis. Biochim. Biophys. Acta 575, 204–214 10.1016/0005-2760(79)90022-5 [DOI] [PubMed] [Google Scholar]
- 51. Hosaka K., and Yamashita S. (1984) Regulatory role of phosphatidate phosphatase in triacylglycerol synthesis of Saccharomyces cerevisiae. Biochim. Biophys. Acta 796, 110–117 10.1016/0005-2760(84)90244-3 [DOI] [PubMed] [Google Scholar]
- 52. Han G.-S., O'Hara L., Carman G. M., and Siniossoglou S. (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fakas S., Konstantinou C., and Carman G. M. (2011) DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286, 1464–1474 10.1074/jbc.M110.194308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soulard A., Cohen A., and Hall M. N. (2009) TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21, 825–836 10.1016/j.ceb.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 55. Csaki L. S., Dwyer J. R., Fong L. G., Tontonoz P., Young S. G., and Reue K. (2013) Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 52, 305–316 10.1016/j.plipres.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang P., and Reue K. (2017) Lipin proteins and glycerolipid metabolism: roles at the ER membrane and beyond. Biochim. Biophys. Acta 1859, 1583–1595 10.1016/j.bbamem.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim Y., Gentry M. S., Harris T. E., Wiley S. E., Lawrence J. C. Jr., and Dixon J. E. (2007) A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6596–6601 10.1073/pnas.0702099104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu R., Garland M., Dunaway-Mariano D., and Allen K. N. (2011) Homo sapiens Dullard protein phosphatase shows a preference for the insulin-dependent phosphorylation site of lipin1. Biochemistry 50, 3045–3047 10.1021/bi200336b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Péterfy M., Phan J., Xu P., and Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- 60. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 [DOI] [PubMed] [Google Scholar]
- 61. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., and Lawrence J. C. Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286 10.1074/jbc.M609537200 [DOI] [PubMed] [Google Scholar]
- 62. Chang H. J., Jesch S. A., Gaspar M. L., and Henry S. A. (2004) Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168, 1899–1913 10.1534/genetics.104.032961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boroda S., Takkellapati S., Lawrence R. T., Entwisle S. W., Pearson J. M., Granade M. E., Mullins G. R., Eaton J. M., Villén J., and Harris T. E. (2017) The phosphatidic acid-binding, polybasic domain is responsible for the differences in the phosphoregulation of lipins 1 and 3. J. Biol. Chem. 292, 20481–20493 10.1074/jbc.M117.786574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eaton J. M., Mullins G. R., Brindley D. N., and Harris T. E. (2013) Phosphorylation of lipin 1 and charge on the phosphatidic acid head group control its phosphatidic acid phosphatase activity and membrane association. J. Biol. Chem. 288, 9933–9945 10.1074/jbc.M112.441493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Péterfy M., Harris T. E., Fujita N., and Reue K. (2010) Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J. Biol. Chem. 285, 3857–3864 10.1074/jbc.M109.072488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stuart S. A., Houel S., Lee T., Wang N., Old W. M., and Ahn N. G. (2015) A phosphoproteomic comparison of B-RAFV600E and MKK1/2 inhibitors in melanoma cells. Mol. Cell. Proteomics 14, 1599–1615 10.1074/mcp.M114.047233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., and Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Park T.-S., Ostrander D. B., Pappas A., and Carman G. M. (1999) Identification of Ser424 as the protein kinase A phosphorylation site in CTP synthetase from Saccharomyces cerevisiae. Biochemistry 38, 8839–8848 10.1021/bi990784x [DOI] [PubMed] [Google Scholar]
- 69. Rose M. D., Winston F., and Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 70. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 71. Innis M. A., and Gelfand D. H. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., and White T. J., eds) pp. 3–12, Academic Press, Inc., San Diego [Google Scholar]
- 72. Ito H., Fukuda Y., Murata K., and Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rothstein R. (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194, 281–301 10.1016/0076-6879(91)94022-5 [DOI] [PubMed] [Google Scholar]
- 74. Carman G. M., and Lin Y.-P. (1991) Phosphatidate phosphatase from yeast. Methods Enzymol. 197, 548–553 10.1016/0076-6879(91)97182-X [DOI] [PubMed] [Google Scholar]
- 75. Siniossoglou S., Hurt E. C., and Pelham H. R. (2000) Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX(T/V) motif required for sodium stress response in yeast. J. Biol. Chem. 275, 19352–19360 10.1074/jbc.M001314200 [DOI] [PubMed] [Google Scholar]
- 76. Jeffery D. A., Springer M., King D. S., and O'Shea E. K. (2001) Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80-Pho85 is semi-processive with site preference. J. Mol. Biol. 306, 997–1010 10.1006/jmbi.2000.4417 [DOI] [PubMed] [Google Scholar]
- 77. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 78. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 79. Burnette W. (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 10.1016/0003-2697(81)90281-5 [DOI] [PubMed] [Google Scholar]
- 80. Haid A., and Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 10.1016/S0076-6879(83)96017-2 [DOI] [PubMed] [Google Scholar]
- 81. Harlow E., and Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 82. Boyle W. J., van der Geer P., and Hunter T. (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–149 10.1016/0076-6879(91)01013-R [DOI] [PubMed] [Google Scholar]
- 83. Yang W.-L., and Carman G. M. (1995) Phosphorylation of CTP synthetase from Saccharomyces cerevisiae by protein kinase C. J. Biol. Chem. 270, 14983–14988 10.1074/jbc.270.25.14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. MacDonald J. I. S., and Kent C. (1994) Identification of phosphorylation sites in rat liver CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 269, 10529–10537 [PubMed] [Google Scholar]
- 85. Morlock K. R., Lin Y.-P., and Carman G. M. (1988) Regulation of phosphatidate phosphatase activity by inositol in Saccharomyces cerevisiae. J. Bacteriol. 170, 3561–3566 10.1128/jb.170.8.3561-3566.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- 87. Henderson R. J., and Tocher D. R. (1992) in Lipid Analysis (Hamilton R. J., and Hamilton S., eds) pp. 65–111, IRL Press, New York [Google Scholar]
- 88. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., and Hieter P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- 89. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]