Abstract
The 2′–5′-oligoadenylate synthetase (OAS) family of enzymes sense cytosolic dsRNA, a potent signal of viral infection. In response to dsRNA binding, OAS proteins synthesize the second messenger 2′–5′-linked oligoadenylate that activates the latent ribonuclease L (RNase L). RNase L–mediated degradation of viral and cellular RNAs effectively halts viral replication and further stimulates innate immune responses by inducing type I interferon. The OAS/RNase L pathway is therefore central in innate immune recognition and promotion of antiviral host responses. However, the potential for specific RNA sequences or structures to drive OAS1 activation and the molecular mechanisms by which they act are not currently fully understood. Moreover, the cellular regulators of OAS activity are not well defined. Here, we demonstrate that the human cellular noncoding RNA 886 (nc886) activates OAS1 both in vitro and in human A549 cells. We show that a unique structure present only in one of the two structural conformers adopted by nc886 drives potent OAS1 activation. In contrast, the conformer lacking this unique structure activated OAS1 only very weakly. We also found that formation of this OAS1-activating structural motif depends on the nucleotides in the apical-most loop of nc886 and the adjacent helix. These findings identify a cellular RNA capable of activating the OAS/RNase L pathway in human cells and illustrate the importance of structural elements, and their context, in potentiating OAS1 activity.
Keywords: RNA structure, RNA-binding protein, innate immunity, double-stranded RNA (dsRNA), translation regulation, antiviral defense, nc886, oligoadenylate synthetase
Introduction
The innate immune response serves as a critical first line of defense against pathogens, comprising membrane-bound and cytosolic pattern recognition receptors (PRRs) 2 that detect bacterial or viral pathogen-associated molecular patterns (1, 2). Accumulation of double-stranded RNA (dsRNA) in the cell cytoplasm is a potent signal of viral infection and is detected by PRRs, including Toll-like receptors 3, 7, and 8; dsRNA-activated protein kinase (PKR); and the retinoic acid-inducible gene I (RIG-I)-like and 2′–5′-oligoadenylate synthetase (OAS) families of enzymes (3, 4).
OAS enzymes are activated by dsRNA binding and initiate signaling cascades to halt viral replication and establish an antiviral state in the cell. The OAS family includes three catalytically active, 2′-specific nucleotidyltransferases, OAS1/2/3, whose antiviral action is mediated by the latent ribonuclease L (RNase L) (5). Additionally, a catalytically inactive form, OASL, acts in the RIG-I pathway (6). The OAS1/2/3 proteins differ in their number of copies of the OAS domain, although only one copy is catalytically active in each protein (7). In OAS1, binding of short dsRNA (≥17 bp) drives a conformational change that organizes the enzyme-active site, resulting in synthesis of the 2′–5′-linked oligoadenylate (2–5A) second messenger that in turn activates RNase L (8–10). In OAS3, the catalytically inactive N-terminal OAS domain serves as a dsRNA-binding module, whereas its third C-terminal OAS domain carries out 2–5A synthesis. This domain organization makes OAS3 selective for longer dsRNAs (≥50 bp) (11). The immediate consequence of OAS/RNase L pathway activation is the degradation of viral and cellular messenger RNA (mRNA) and ribosomal RNA (rRNA) to halt protein synthesis and thus prevent viral replication (12–16). More recent evidence also suggests that OAS/RNase L–mediated translational arrest arises, in part, via specific cleavage of cellular tRNAs (tRNA) and Y RNAs (17).
Precisely how PRRs distinguish “self” from “nonself” and how basal activity of constitutively expressed PRRs is regulated in the absence of infection are important areas of current investigation. PKR, for example, is well established as a critical sensor of dsRNA during viral infection and as a mediator of other important cellular processes (18–22). Recent evidence suggests that basal PKR activity is suppressed by the ubiquitously expressed 101-nucleotide cellular noncoding RNA 886 (nc886; Fig. 1A) to prevent spurious activation in the uninfected cell while allowing rapid detection of foreign dsRNA when required (23–25).
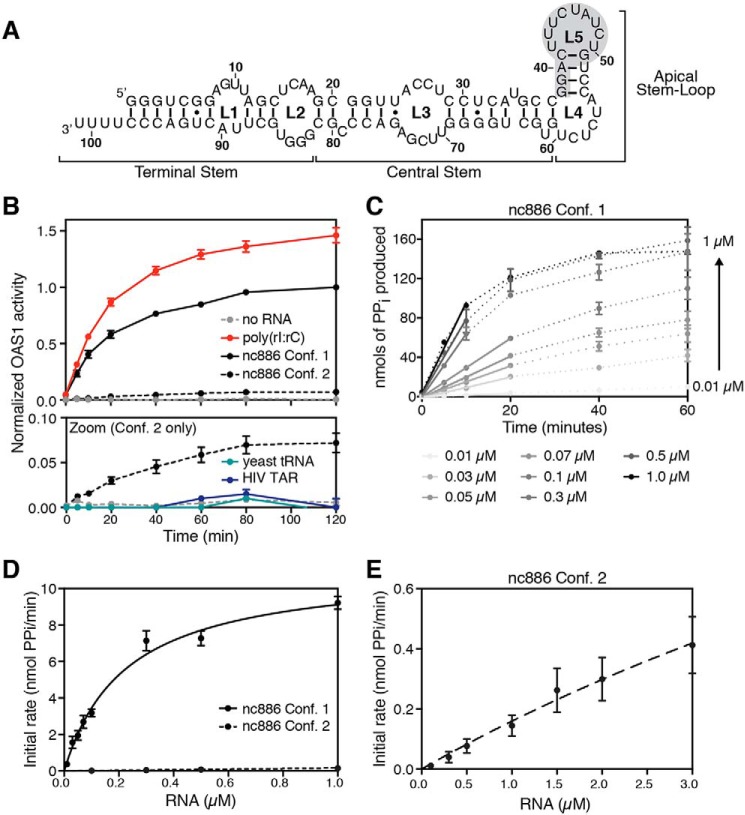
Figure 1.
Two nc886 conformers have starkly different abilities to activate OAS1 in vitro. A, nc886 secondary structure and domain organization. The region of predicted tertiary structure present only in conformer 1 is highlighted by gray shading. B, reaction progress curves from an in vitro chromogenic assay of OAS1 activity performed under previously established conditions (27). nc886 conformer 1 (black line) potently activates OAS1, near the levels of activation for the synthetic dsRNA poly(rI·rC) (red line), whereas conformer 2 (dashed black line) only weakly activates OAS1. The lower panel (Zoom) shows a rescaled plot of the same data to highlight the reaction curves for nc886 conformer 2, the background control lacking RNA (dashed gray line), and two unrelated RNAs as control: yeast tRNA (teal line) and HIV TAR (navy line). Data are normalized to nc886 conformer 1. C, example plot of time-course assays with different RNA concentrations to determine initial rates of PPi production. nc886 conformer 1 time-course assay at each RNA concentration is shown by dotted lines. Data were fit using linear regression analysis on the first 10–20 min of the reaction to obtain the initial rate (slope of the line) used in kinetic analysis. D, kinetic analyses of OAS1 activation over a range of nc886 RNA concentrations. Data were fit using nonlinear regression analysis to obtain the kinetic parameters Vmax and Kapp as described in the text. E, rescaled plot of initial rates of PPi production determined from time-course assays plotted versus RNA concentration shown to highlight the data for the weakly activating nc886 conformer 2.
Our previous work has demonstrated that nc886 RNA adopts two structurally distinct conformers, distinguished by their apical stem-loop structures, that possess opposing activities in the regulation of PKR (26). We also previously reported that the mixture of nc886 conformers appeared to strongly activate OAS1 and that this activity was only modestly reduced by deletion of the 3′-single–stranded pyrimidine (3′-ssPy) motif that more significantly potentiates activity by viral noncoding RNAs (27). Here, we show that the two nc886 conformers exhibit profound differences in their capacity to activate the OAS/RNase L pathway: one nc886 conformer is a potent activator of OAS1 in vitro and in human A549 cells whereas the other conformer activates OAS1 only very weakly. We further show that potent activation depends on the nc886 apical stem-loop structure, highlighting the critical importance of RNA structural features, and their context, in potentiating OAS activity.
Results
OAS1 is potently activated by a single nc886 conformer
nc886 RNA was previously shown to adopt two stable, structurally distinct conformers (26). To dissect the activity of each conformer on OAS1, we purified the individual nc886 conformers by native PAGE. An established in vitro OAS1 activation assay (27, 28) was then used to measure PPi production, the by-product of 2–5A synthesis, in the presence of each nc886 conformer. nc886 conformer 1 strongly activates OAS1 (Fig. 1B), with a level of activity comparable with that measured for poly(rI·rC) under our previously established conditions. Poly(rI·rC) is a synthetic dsRNA that acts as a potent trigger of 2–5A synthesis via activation of OAS/RNase L in vitro and in cells (29). In stark contrast, nc886 conformer 2 has dramatically attenuated capacity to activate OAS1. However, the low level of OAS1 activation by nc886 conformer 2 is nonetheless comparable with that of some structured viral noncoding RNAs, such as adenovirus VA RNAI and Epstein-Barr virus EBER-1 (27), and greater than two unrelated control RNAs, yeast tRNA or HIV TAR RNA, not expected to activate OAS1 (Fig. 1B).
We further probed the ability of each nc886 RNA conformer to promote OAS1 activation using an in vitro OAS1 kinetic analysis of 2–5A synthesis performed over a range of RNA concentrations (Fig. 1, C–E). Initial rates determined from these analyses were used to obtain the kinetic parameters for the apparent RNA dissociation constant (Kapp) and maximum reaction velocity (Vmax) as measures of RNA–OAS1 binding and OAS1 catalytic activity, respectively, for each nc886 RNA conformer. nc886 conformer 1 exhibits a Kapp in the nanomolar range (224 ± 29 nm), whereas that of nc886 conformer 2 was too weak to be determined accurately (> ∼10 μm), resulting in a >50-fold difference between the two nc886 conformers. The OAS1 Vmax measured for nc886 conformer 1 was 11.2 ± 0.6 nmol PPi/min, whereas the data for nc886 conformer 2 could not be reliably fit. Thus, the structural differences in the two conformers dramatically affect the extent of OAS1 activation through effects on both the RNA–OAS1 interaction and OAS1 catalysis, with the most significant impact being on apparent RNA affinity.
nc886 conformer 1 activates the OAS/RNase L pathway in human A549 cells
We next assessed the ability of each nc886 conformer to activate the OAS/RNase L pathway in human A549 cells, which basally express OAS1 and OAS3 (30). A549 cells were transfected with nc886 conformer 1, nc886 conformer 2, or a truncated version of adenoviral VA RNAI (TSΔ21), an RNA of comparable size (99 nts) to nc886 that was previously shown to very weakly activate OAS1 (31–33). Native PAGE analysis was used to confirm the conformational homogeneity of each nc886 RNA immediately prior to transfection (Fig. 2A). At 3 h post-transfection, cells were lysed, and total RNA was analyzed by agarose gel electrophoresis to monitor rRNA cleavage by RNase L (Fig. 2B). Transfection with nc886 conformer 1 resulted in significant rRNA degradation, comparable with poly(rI·rC) dsRNA and consistent with strong activation of the OAS/RNase L pathway. In contrast, nc886 conformer 2 showed no detectable rRNA degradation, with rRNA integrity remaining identical to untransfected and mock-transfected controls. Finally, consistent with its known weak activity in vitro (33), transfection with VA RNAI (TSΔ21) resulted in a small amount of rRNA cleavage. These results thus mirror the relative capacity of each nc886 conformer to activate OAS1 in our in vitro assay (Fig. 1B). Furthermore, nc886-mediated rRNA cleavage activity is completely absent in A549 cells lacking RNase L (RNASEL KO), confirming that the observed rRNA cleavage results exclusively from activation of the OAS/RNase L pathway by nc886 conformer 1 (Fig. 2C).
Figure 2.
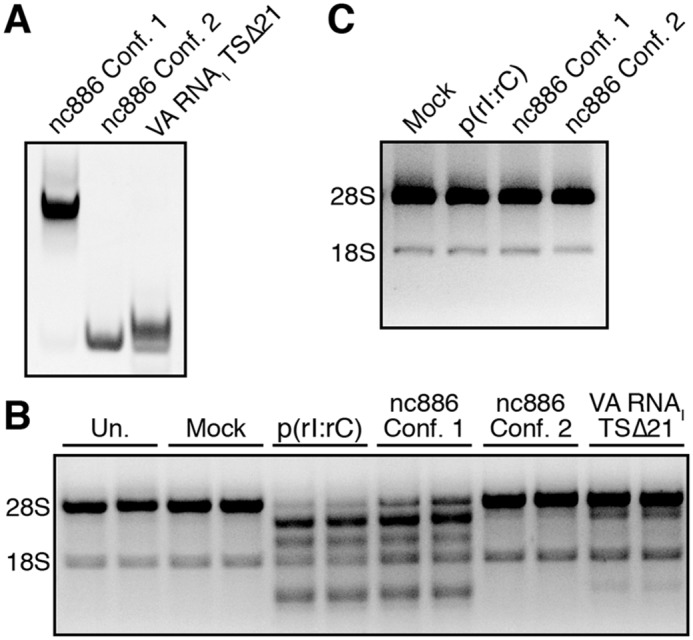
nc886 conformer 1 activates the OAS/RNase L pathway in human A549 cells. A, native PAGE analysis of purified nc886 conformers and the adenoviral VA RNAI (TSΔ21) variant used for transfections. B, agarose gel analysis of total cellular RNA extracted at 3 h post-transfection with the indicated RNA (1 μg/ml) or other treatment: untransfected (Un.) or mock-transfected (Mock). rRNA degradation, based on 28S and 18S rRNA integrity, is only induced by treatment with nc886 conformer 1 and poly(rI·rC). A representative gel is shown for one of three independent replicates, which showed essentially identical results. C, analysis of total cellular RNA extracted from A549 cells lacking RNase L treated as in B, demonstrating that rRNA degradation observed with nc886 conformer 1 and poly(rI·rC) treatment is specifically dependent on activation of the OAS/RNase L pathway.
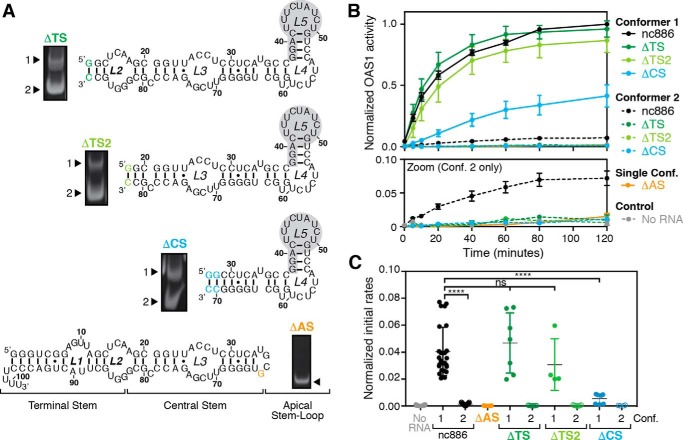
nc886 terminal stem is dispensable for activation of OAS1
To dissect the contributions of different regions of the RNA to OAS1 activation, four nc886 variants were created (Fig. 3A) with truncations of the terminal stem (ΔTS, deletion of nts 1–12 and 88–101; ΔTS2, deletion of nts 1–18 and 82–101), the terminal stem and the central stem (ΔCS, deletion of nts 1–28 and 70–101), or the apical stem-loop (ΔAS, deletion of nts 37–63). As reported previously (26), the ΔAS variant adopts a single conformer, whereas the terminal and central stem variants (ΔTS, ΔTS2, and ΔCS) retain the ability to form two conformers (Fig. 3A). These observations are consistent with conserved terminal and central stem structures between the two WT nc886 conformers and a unique tertiary structure, present in the apical stem-loop of only conformer 1, as the origin of the structural differences between the nc886 conformers (26).
Figure 3.
Apical stem-loop of nc886, but not the terminal stem, is critical for OAS1 activation. A, nc886 secondary structure and domain organization with truncations of the TS, CS, or AS highlighted. Additional sites of altered sequence with respect to WT nc886 (to facilitate in vitro transcription; compare with Fig. 1A) are denoted in the color of the corresponding truncation variant. Gel images adjacent to each RNA secondary structure show native PAGE analysis of the RNA following in vitro transcription. B, reaction progress curves from an in vitro chromogenic assay of OAS1 activity demonstrating that the terminal stem of nc886 is largely dispensable for OAS1 activation by nc886. The lower panel (Zoom) shows a rescaled plot of the same data to highlight the reaction curves for nc886ΔAS and conformer 2 of nc886ΔTS, nc886ΔTS2, and nc886ΔCS, which all completely fail to activate OAS1. The data for each WT nc886 conformer are the same as shown in Fig. 1B. C, plots of initial rates of reaction (first 10–20 min) are shown for the same data as in B. Comparison of initial rate of reaction assessed by one-way ANOVA: p ≤ 0.0001 (****) and not significant (ns; p ≥ 0.05). All data in B and C are normalized to the activity in the presence of nc886 conformer 1.
We tested the ability of each nc886 variant, and each individual RNA conformer where applicable, to activate OAS1 in vitro at a single fixed RNA concentration. These experiments were performed under the same conditions and protein/RNA concentrations as used for WT nc886 (Fig. 1B) and are shown as both reaction progress curves and initial rates of reaction (Fig. 3, B and C, respectively). nc886ΔAS fails to activate OAS1, with the level of 2–5A synthesis comparable with the control reaction with no RNA. This result indicates that the structure formed by the apical stem-loop of nc886 is essential for activation of OAS1. In contrast, conformer 1 of both the ΔTS and ΔTS2 variants retains near WT activity suggesting that in the context of conformer 1, the terminal stem is largely dispensable for OAS1 activation. However, the larger truncation of ΔCS results in a conformer 1 variant with dramatically reduced ability to activate OAS1 (Fig. 3, B and C). Finally, similar to the ΔAS variant, and in contrast to the weakly activating WT nc886 conformer 2, conformer 2 of each terminal or central stem deletion variant completely fails to activate OAS1 (Fig. 3, B and C). Collectively, these data reveal that deletion of the nc886 terminal stem fully ablates OAS1 activation in the context of conformer 2 but has only a very minor impact on the activity of conformer 1 RNAs. Critically, only the largest deletion (ΔCS) significantly impacts OAS1 activation in the context of conformer 1 suggesting that this region forms a critical part of the OAS1-binding site required for potent OAS1 activation.
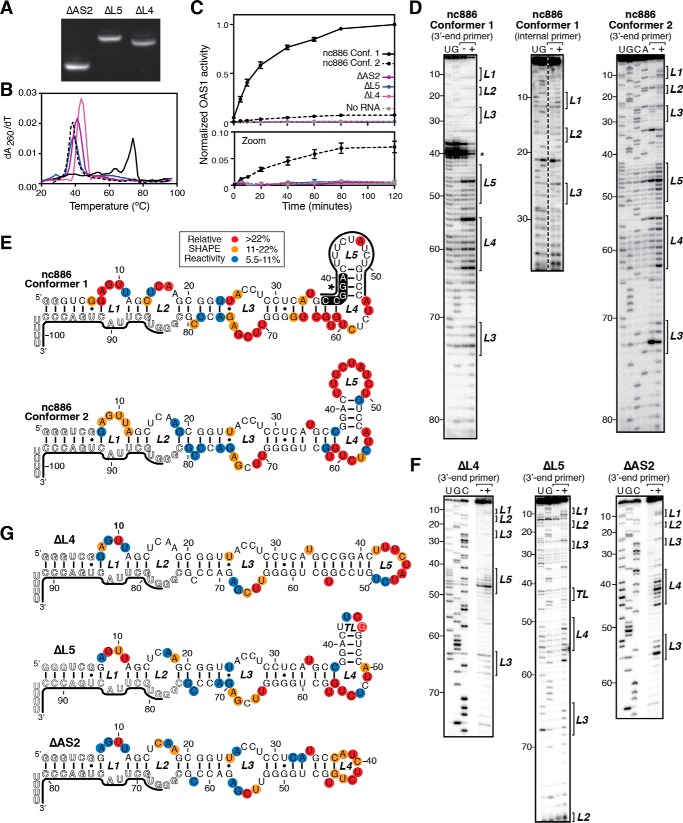
OAS1 activation depends on the nc886 apical stem-loop
To further examine the role of the nc886 apical stem-loop sequence and structure on OAS1 activation, we created three additional variants with smaller truncations of this region. Using RNA secondary structures of the WT nc886 RNA conformers previously experimentally defined using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (26), we created variants lacking loop 4 (ΔL4), loop 5 (ΔL5), or loop 5 and the adjacent helix (ΔAS2). Each of these new variants adopted a single conformation as determined by native PAGE analysis (Fig. 4A) suggesting loss of the capacity to form the apical stem-loop structure of conformer 1. RNA variants were also assessed by UV thermal melting analysis, and each was found to more closely resemble WT nc886 conformer 2 in RNA stability (Table 1 and Fig. 4B). We then tested each RNA variant for its ability to activate OAS1 in vitro and found that all apical stem-loop variants failed to activate OAS1 above the level of the control lacking RNA (Fig. 4C). SHAPE analyses were carried out for each RNA variant and compared with previously published SHAPE analyses of the WT nc886 conformers (Fig. 4, D–G). All the apical stem-loop variants closely resemble each other and full-length nc886 conformer 2, aside from the specifically deleted nucleotides. The results for each of these RNAs are thus distinct from full-length nc886 conformer 1, which exhibits loss of loop 5 reactivity and NMIA-independent stops on the 5′-side of the helix adjacent to loop 5, indicative of a stable structure and potential long-range tertiary interactions. Together, these data suggest that the specific apical stem-loop sequence and structure play a critical role in OAS1 activation by either conformer, being essential for both potent activation by conformer 1 and for the weak activation by conformer 2.
Figure 4.
Activation of OAS1 depends on an intact nc886 RNA apical stem. A, native PAGE analysis of purified variant RNAs: truncation of loop 5 (ΔL5), loop 4 (ΔL4), or the entire apical stem-loop (ΔAS2). B, UV melting profiles of each isolated nc886 RNA variant: nc886 conformer 1 (solid black line); nc886 conformer 2 (dashed black line); nc886ΔAS2 (solid purple line); nc886ΔL5 (solid blue line); and nc886ΔL4 (solid magenta line). Melting temperatures (Tm) for the major apparent unfolding transitions are listed in Table 1. C, reaction progress curves from an in vitro chromogenic assay of OAS1 activity demonstrating that all variants with altered apical stem-loop structures fail to activate OAS1. The lower panel (Zoom) shows a rescaled plot of the same data to highlight the reaction curves relative to WT nc886 conformer 2 and the background control lacking RNA (dashed gray line). The data for each WT nc886 conformer are the same as shown in Fig. 1B. D, example autoradiograms of sequencing gel analysis of SHAPE probing of WT nc886. Lanes are as follows: −, mock-treated (no NMIA); +, NMIA-treated; and U/G/C/A, sequencing lanes containing the complementary dideoxynucleotide triphosphate. Brackets on the right of the gel images denote the loop regions (L1–L5) in the predicted secondary structures. E, secondary structure map of WT nc886. Experimentally determined SHAPE reactivity was mapped onto the predicted apical stem-loop regions of each structure. F, example autoradiograms of sequencing gel analysis of SHAPE probing of apical stem-loop variants as described in D. G, secondary structure map of apical stem-loop variants nc886ΔL4, nc886ΔL5, and nc886ΔAS2. SHAPE reactivities for full-length nc886 conformer 1, nc886 conformer 2, and nc886ΔL5, presented here to simplify comparisons with the apical-stem variant RNAs generated in this study, were previously reported (26).
Table 1.
Apparent melting temperatures (Tm) for wildtype and variant nc886 unfolding
| RNA | Apparent Tm |
|
|---|---|---|
| 0.5× TBE | HEPES/NaCl | |
| °C | ||
| nc886 conformer 1 | 73.8 | 92.2 |
| nc886 conformer 2 | 38.2 | 58.4 |
| nc886ΔAS2 | 40.9 | 61.2 |
| nc886ΔL4 | 43.6 | 63.9 |
| nc886ΔL5 | 39.0 | 59.4 |
nc886 apical stem-loop structure activates the cellular OAS/RNase L pathway through OAS1
To further probe the role of the unique apical-stem loop structure of nc886 conformer 1 in activation of the cellular OAS/RNase L pathway, we transfected human A549 cells with nc886ΔTS2 conformer 1, the smallest RNA variant that retains the unique structural motif and near WT levels of OAS1 activation, or nc886ΔL5, the largest RNA variant that exhibits complete loss of both the unique apical stem-loop structural motif and OAS1 activation in vitro. Consistent with our in vitro data, nc886ΔTS2 conformer 1 triggers rRNA cleavage, whereas nc886ΔL5 fails to activate the OAS/RNase L pathway in human A549 cells (Fig. 5). The ability of ΔTS2 conformer 1 to activate the OAS/RNase L pathway in human A549 cells also suggests that nc886 can act, at least in part, through OAS1 as OAS3 requires longer (>50 bp) dsRNA (11).
Figure 5.
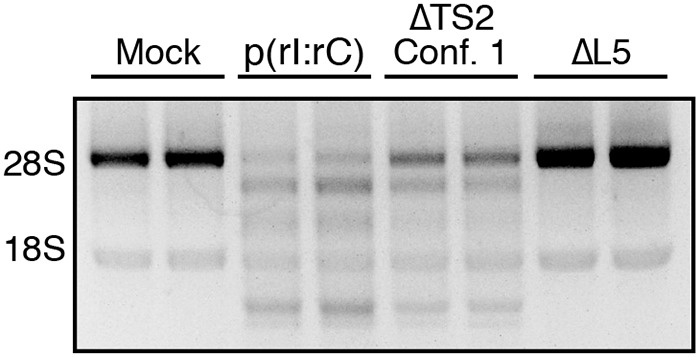
nc886 conformer 1 apical stem structure is sufficient to activate the OAS/RNase L pathway in human A549 cells. Agarose gel analysis was performed of total cellular RNA extracted at 3 h post-transfection with the indicated RNA or following mock transfection (Mock). A representative gel is shown for one of three independent replicates that showed essentially identical results. rRNA degradation, based on 28S and 18S rRNA integrity, is only induced by treatment with nc886ΔTS2 conformer 1 and poly(rI·rC).
Discussion
Nucleic acid sensing in innate immunity plays a critical role in the detection of viral pathogens. The precise regulation of nucleic acid sensors, such as the OAS family of enzymes, is required for both proper cell function in the absence of infection and survival of viral infection. This work demonstrates the ability of a cellular noncoding RNA, nc886, to regulate OAS through potent activation in vitro and in human A549 cells. However, the two RNA conformers adopted by nc886 induce 2–5A synthesis by OAS1 in vitro to markedly different levels. Although nc886 conformer 2 activates OAS1 only weakly, nc886 conformer 1 potently induces 2–5A synthesis in vitro and RNase L–mediated rRNA cleavage in A549 cells. This is, to our knowledge, the first identification of a cellular RNA activator of OAS and raises questions about the biological roles OAS1 may play outside of innate immunity.
The conformational identity of nc886 RNA has profound effects on both RNA–OAS1-binding affinity and OAS1 catalysis. Our kinetic analyses demonstrated a significant impact on apparent affinity with a more than 50-fold difference in Kapp between the two conformers. Moreover, the level of OAS1 catalysis depends on the presence or absence of the predicted tertiary structural motif in the apical stem-loop of nc886 (Fig. 6). OAS1 catalysis in the presence of nc886 conformer 1 appears similar to that previously observed in vitro for defined stem-loop structures derived from the 5′-end of the West Nile Virus genome (34) further supporting the idea that specific RNA structural features, in addition to dsRNA, can activate OAS1. In contrast, nc886 conformer 2 has dramatically attenuated capacity to activate OAS1, and as such, kinetic analyses could not accurately determine Vmax. This weak catalysis, however, mirrors that observed for viral noncoding RNAs produced by Epstein-Barr virus and adenovirus (27, 35).
Figure 6.
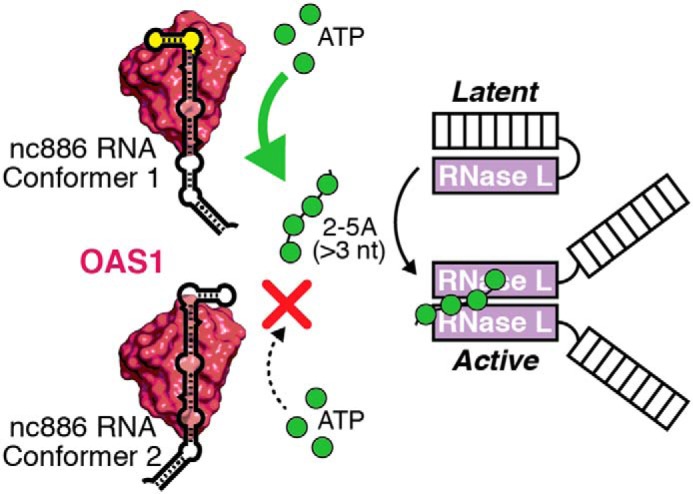
Model for regulation of the OAS/RNase L pathway by nc886. OAS1 can bind to either nc886 conformer, but only conformer 1 induces 2–5A synthesis, which depends on the presence of the unique apical stem structure (highlighted in yellow). 2–5A synthesis leads to the subsequent activation of RNase L through 2–5A-mediated RNase L dimerization.
The RNA features that promote OAS1 activation are currently poorly understood. Although OAS1 has been shown to require at least 18 bp of dsRNA for activation (8), additional molecular features that define a specific RNA as a more potent OAS1 activator have been limited to the identification of a consensus sequence (36) and the 3′-single-stranded pyrimidine (3′-ssPy) motif (27). The effect of structural features, and their context, on RNA-mediated activation of OAS1 has not yet been fully explored. In this study, we show that in the context of nc886, specific RNA structure has major functional consequences for OAS1 activation. Although the two conformers adopted by nc886 share the vast majority of their terminal and central stem structure, potent activation depends on a unique structural motif present in the apical stem-loop of only one conformer adopted by the RNA. Our SHAPE analysis of the WT nc886 conformers revealed that although loop 5 of nc886 conformer 2 adopts the predicted secondary structure, the same nucleotides are unreactive in nc886 conformer 1 suggesting that they take part in base-pairing or long-range interactions. SHAPE reactivity for the 5′-side of the adjacent helix is absent in conformer 1 as this region produces NMIA-independent stops suggesting a highly stable structure that is resistant to unfolding during the RT reaction. Because of these experimental limitations, we were unable to fully map the nucleotide flexibility in the nc886 conformer 1 apical stem-loop to define the folding of this potent activator of OAS1. Our results do, however, clearly show that formation of this structural motif requires nucleotides in loops 4 and 5 because truncations in these regions result in the loss of ability to form two conformers. Future high-resolution structural studies will be essential to shed light on the nature of this unique structural feature and how it is able to drive such potent activation of OAS1.
The varying effects of different truncations on each nc886 RNA conformer suggest that multiple, nonequivalent OAS1-binding sites might exist in nc886. Although deletion of the terminal stem (ΔTS and ΔTS2) has a minimal impact on OAS1 activation by nc886 conformer 1, the same deletions abrogate the ability of nc886 conformer 2 to weakly activate OAS1. The same complete loss of OAS1 activation is also observed for ΔCS conformer 2. However, in contrast to the terminal stem deletion RNAs, this larger truncation also strongly impacts OAS1 activation by conformer 1. This latter observation suggests that the nc886 central stem is critical for OAS1 interaction, and loss of this binding site in the context of nc886 conformer 1 prevents the correct positioning of its unique structural motif to potently drive activation of OAS1. We note that from the site of truncation at G18 to form ΔTS2, the shortest variant with near WT level OAS1 activation, the central stem forms an ∼18-bp duplex with the mismatched loop 3 region at its center. Binding of OAS1 to this region of nc886 could therefore reflect a similar mechanism of action to the potentiation in activity conferred to simple duplex RNAs or viral noncoding RNAs by the 3′-ssPy motif (27). Interestingly, although the presence of the 3′-ssPy motif and the conformer 1 apical stem-loop structure both enhance OAS1 activation by VA RNAI (27) and nc886, respectively, these RNA features have opposite impacts on RNA Kapp. Thus, for structured RNAs, the relationship between apparent OAS1–RNA affinity and the RNA's ability to activate OAS1 appears to be complex. The loss of activity observed for all apical stem-loop mutants suggests that OAS1 requires the nc886 apical stem-loop sequence and/or structure in the context of both conformers. Although this result was expected for conformer 1, where the RNA tertiary structure in this region is responsible for OAS1 activation, it is less clear how the apical stem-loop influences the weak OAS1 activation in the context of conformer 2. The complete loss of activity in ΔL5, compared with WT nc886 conformer 2, further points to a specific interaction of OAS1 with loop 5 of the RNA. One possibility that warrants further exploration is that alterations of the apical stem-loop sequence may generate (or unmask) OAS1 interaction sites in nc886 that bind but fail to activate the enzyme.
nc886 is a ubiquitously expressed noncoding RNA that resides in the cytoplasm where both OAS and RNase L are found (25). It therefore remains to be addressed how nc886 is prevented from spuriously activating the OAS/RNase L pathway and whether tuning the levels of each RNA conformer in the cell could impact OAS1 activity basally and during infection (12, 14, 15). Recently, RNase L was shown to regulate cellular proliferation and migration revealing potential functions of the OAS/RNase L pathway outside of innate immunity (12, 14). Whether nc886 also has a role in these cellular functions requires further study. This work has revealed the requirement for the apical stem-loop of nc886 for OAS1 activation and thus functional overlap in the RNA–protein interactions for nc886-mediated regulation of both OAS and PKR (26). These overlapping but distinct requirements for PKR and OAS1 suggest the potential for competition between the two proteins for nc886 binding or communication between these two arms of the innate immune response via nc886.
There is a growing appreciation for the diverse functions noncoding RNAs play in regulating gene expression, cellular proliferation, apoptosis, and their impacts on human diseases (37, 38). The ability of nc886 to regulate the innate immune response through OAS1 activation hints at a potential new mechanism for cellular noncoding RNA-mediated regulation of nucleic acid sensors in response to viral infection and other cell stresses. In regard to innate immunity, several cellular RNAs have been identified as regulators of nucleic acid sensors, including but not limited to interferon-γ mRNA, cytoskeletal mRNAs, tumor necrosis factor α mRNA, and small nucleolar RNAs (19, 39–41). Furthermore, noncoding RNAs produced by adenovirus and Epstein-Barr virus have been shown to inhibit the function of nucleic acid sensors in innate immunity (42–44). nc886 may represent one of many unknown cellular noncoding RNAs with the potential to impact the immune response to viral infection or other biological processes controlled by nucleic acid sensors.
Conclusions
The work presented here provides new evidence for a novel, cellular noncoding RNA activator of the cytosolic dsRNA sensor OAS1. This noncoding RNA, nc886, adopts two structurally distinct conformers that are not functionally equivalent, and we identified a putative tertiary structure, present in only one conformer of nc886, as being responsible for strongly potentiating the activity of OAS1. Collectively, our results demonstrate that OAS1 activation can be affected by specific RNA structural elements, in addition to dsRNA length or the presence of other signatures such as the 3′-ssPy motif (27) or an OAS1 activation consensus sequence (36, 45). The findings reported here open the way for new studies to address the roles the nc886–OAS1 interaction may play during and outside of viral infection.
Experimental procedures
RNA in vitro transcription and purification
RNAs were transcribed in vitro from linearized plasmid DNA templates using T7 RNA polymerase as described previously (46). Completed transcription reactions were dialyzed against 1× TE buffer (10 mm Tris-HCl, pH 8, and 1 mm EDTA) and RNA purified by native PAGE on 0.5× TBE (44.5 mm Tris-HCl, pH 8.3, 44.5 mm boric acid, and 1 mm EDTA) gels. RNA bands were identified by UV shadowing, excised from the gel, eluted by crushing and soaking in 0.3 m sodium acetate, pH 5.2, and recovered by ethanol precipitation. All RNAs were analyzed by native PAGE after purification and prior to use in assays.
OAS1 expression and purification
Human OAS1 (p41/E16 isoform) was expressed in E. coli BL21 (DE3) as an N-terminal hexa-histidine–tagged SUMO–OAS1 fusion protein (27). Cells were grown in Lysogeny Broth at 37 °C, and expression was induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside at mid-log phase growth (A600 ∼0.5). Growth was continued overnight at 20 °C. Cells were lysed in 50 mm Tris-HCl buffer, pH 8, containing 150 mm NaCl, 10% (v/v) glycerol, 10 mm imidazole, and 1 mm DTT. SUMO–OAS1 fusion protein was purified by sequential Ni2+-affinity and heparin-affinity chromatographies on an ÄKTApurifier 10 system (GE Healthcare). The fusion protein was dialyzed against 50 mm Tris-HCl buffer, pH 8, containing 150 mm NaCl, 10% (v/v) glycerol, and 2 mm DTT and cleaved with SUMO Protease 1 (LifeSensors) leaving a native OAS1 N terminus.
In vitro chromogenic assay of OAS1 activity
2–5A synthesis was monitored by detection of the reaction by-product PPi in an established chromogenic assay of OAS1 activity (27). OAS1 (300 nm) was incubated at 37 °C with 20 μg/ml poly(rI·rC) or 300 nm WT or variant nc886 RNA in 20 mm Tris-HCl buffer, pH 7.4, containing 7 mm MgCl2, 1 mm DTT, and 1.5 mm ATP. Aliquots (10 μl) were removed at time points between 0 and 120 min and quenched with 2.5 μl of 250 mm EDTA pre-dispensed into the wells of a 96-well plate. At completion of the time course, 10 μl of 2.5% ammonium molybdate in 2.5 m H2SO4, 10 μl of 0.5 m β-mercaptoethanol, and water to 100 μl total volume were added to each well. Absorbance at 580 nm was measured using a Synergy4 plate reader (BioTek), and readings were converted to PPi produced by comparison with PPi standards after background subtraction from a blank reaction. For comparisons of OAS1 initial rate of reaction driven by nc886 variants truncated at the terminal stem (ΔTS and ΔTS2), central stem (ΔCS), or the apical stem-loop structure (ΔAS), statistical analysis was by one-way ANOVA with significance assessed by Dunnett's multiple comparisons test in GraphPad Prism 6.
Complete kinetic analyses were performed similarly but using RNA concentrations in the range of 0.01–1 μm (nc886 conformer 1) or 0.1–3 μm (nc886 conformer 2) and measuring PPi production over the initial 10–20 min of the reaction. Linear regression analysis with GraphPad Prism 6 was used to obtain the nanomoles of PPi produced per min at each RNA concentration and the resulting data fit using nonlinear regression analysis with the Michaelis-Menten model in GraphPad Prism 6 to obtain OAS1 Vmax and RNA Kapp values. At least two independent experiments with different preparations of protein and RNA were used for each RNA variant.
OAS/RNase L activation in A549 cells
Human WT and RNase L knockout A549 cells, constructed using CRISPR-Cas9 gene editing technology, as reported previously (47), were cultured in F-12K medium (Gibco/Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Both cell lines tested negative for mycoplasma. For analysis of OAS/RNase L pathway activation, cells (0.3 × 106) were seeded into six-well plates in media lacking antibiotics and after 24 h were transfected with 1 μg/ml nc886 conformer 1, nc886 conformer 2, or poly(rI·rC), or equimolar concentrations of the VA RNAI TSΔ21, nc886 ΔTS2 conformer 1, or nc886 ΔL5 variants, using Lipofectamine 2000 reagent (Invitrogen) for 3 h. Cells were harvested in 350 μl of RLT lysis buffer (Qiagen), and the total RNA was extracted using an RNeasy kit (Qiagen). Total RNA was resolved on 1.5% agarose gels and stained with ethidium bromide to determine integrity of 28S and 18S rRNA. Three independent experiments were performed with essentially identical results.
RNA UV thermal melting analysis
RNA UV melting curves were collected at 260 and 280 nm on a Cary400 UV-visible spectrophotometer (Varian). Samples contained 20–25 μg of RNA in either 0.5× TBE buffer or 20 mm HEPES buffer, pH 7.5, containing 100 mm NaCl. After normalization, the first derivative of each UV absorbance curve (“melting profile”) was calculated for each RNA in GraphPad Prism 6 software to simplify comparisons between RNA variants. The melting temperature (Tm) values (Table 1) correspond to the peaks of the melting profile and were reproducible ±0.5 °C in independent experiments.
SHAPE
SHAPE RNA probing with NMIA was carried out as described previously (26, 48) with the following modifications: reactions were initiated with 1 μl of 130 mm NMIA and run for 45 min at 37 °C. Reverse transcription was carried out with a 5′-end 32P-labeled DNA primer corresponding to the sequence of the 3′-end of full-length nc886 RNA (nucleotides 84–101). For full-length nc886 conformer 1 only, a second primer corresponding to an internal sequence (nucleotides 36–54) was also used for analysis of the 5′-half of the RNA. To determine the position of each SHAPE reactive nucleotide, dideoxynucleotide triphosphate (ddNTP) sequencing was carried out using the radiolabeled primer and untreated RNA. All reactions were resolved on denaturing (urea) sequencing-style polyacrylamide gels, dried, and exposed to a phosphor storage screen. The intensity of bands was analyzed on a Typhoon Trio Imager (GE Healthcare) and quantified using ImageQuant software (GE Healthcare). Following subtraction of background corresponding to reactions without NMIA, reactivity at each nucleotide was normalized, and the values from at least two replicates were averaged and classified as low (5.5–11%), medium (11–22%), and high (>22%) (48). Previously reported SHAPE reactivities for full-length nc886 conformer 1, nc886 conformer 2, and nc886ΔL5 (26) are presented here to simplify comparisons with the apical-stem variant RNAs generated in this study.
Author contributions
B. M. C. investigation; B. M. C. methodology; B. M. C. writing-original draft; B. M. C. and G. L. C. writing-review and editing; G. L. C. conceptualization; G. L. C. funding acquisition.
Acknowledgments
Human A549 and A549 RNASE L K/O cells were generously provided by Drs. Anice C. Lowen and Susan R. Weiss (University of Pennsylvania), respectively. We also thank Dr. Lowen, Dr. John Steel, and Shamika Danzy for assistance with cell culture; Samantha L. Schwartz for providing purified protein for the OAS1 kinetic experiments; Dr. Virginia K. Vachon for sharing unpublished results and many useful discussions throughout the course of this work; and Drs. Anita H. Corbett and Daniel Reines for comments on the manuscript.
This work was supported by an award from the Emory University Research Council (URC) (to G. L. C.), a scholarship from the ARCS Foundation (to B. M. C.), and by BCDB NIGMS Training Grant T32-GM008367 from the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- PRR
- pattern recognition receptor
- 2–5A
- 2′–5′-oligoadenylate
- dsRNA
- double-stranded RNA
- nc886
- noncoding 886 RNA
- OAS
- oligoadenylate synthetase
- PKR
- dsRNA-activated protein kinase
- RNase L
- ribonuclease L
- SHAPE
- selective 2′-hydroxyl acylation analyzed by primer extension
- TS
- terminal stem
- nt
- nucleotide
- SUMO
- small ubiquitin-like modifier
- CS
- central stem
- AS
- apical stem
- ANOVA
- analysis of variance
- NMIA
- N-methylisatoic anhydride
- ssPy
- single-stranded pyrimidine.
References
- 1. Chow J., Franz K. M., and Kagan J. C. (2015) PRRs are watching you: localization of innate sensing and signaling regulators. Virology 479, 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schneider W. M., Chevillotte M. D., and Rice C. M. (2014) Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schlee M., and Hartmann G. (2016) Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsutsui-Takeuchi M., Ushio H., Fukuda M., Yamada T., Niyonsaba F., Okumura K., Ogawa H., and Ikeda S. (2015) Roles of retinoic acid-inducible gene-I-like receptors (RLRs), Toll-like receptor (TLR) 3 and 2′–′ oligoadenylate synthetase as viral recognition receptors on human mast cells in response to viral infection. Immunol. Res. 61, 240–249 10.1007/s12026-014-8617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kristiansen H., Gad H. H., Eskildsen-Larsen S., Despres P., and Hartmann R. (2011) The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 31, 41–47 10.1089/jir.2010.0107 [DOI] [PubMed] [Google Scholar]
- 6. Zhu J., Zhang Y., Ghosh A., Cuevas R. A., Forero A., Dhar J., Ibsen M. S., Schmid-Burgk J. L., Schmidt T., Ganapathiraju M. K., Fujita T., Hartmann R., Barik S., Hornung V., Coyne C. B., and Sarkar S. N. (2014) Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity 40, 936–948 10.1016/j.immuni.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin G., and Keller W. (2007) RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 10.1261/rna.652807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donovan J., Dufner M., and Korennykh A. (2013) Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl. Acad. Sci. U.S.A. 110, 1652–1657 10.1073/pnas.1218528110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang H., Zeqiraj E., Dong B., Jha B. K., Duffy N. M., Orlicky S., Thevakumaran N., Talukdar M., Pillon M. C., Ceccarelli D. F., Wan L. C., Juang Y. C., Mao D. Y., Gaughan C., Brinton M. A., et al. (2014) Dimeric structure of pseudokinase RNase L bound to 2–5A reveals a basis for interferon-induced antiviral activity. Mol. Cell 53, 221–234 10.1016/j.molcel.2013.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y., Whitney G., Donovan J., and Korennykh A. (2012) Innate immune messenger 2–5A tethers human RNase L into active high-order complexes. Cell Rep. 2, 902–913 10.1016/j.celrep.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 11. Donovan J., Whitney G., Rath S., and Korennykh A. (2015) Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc. Natl. Acad. Sci. U.S.A. 112, 3949–3954 10.1073/pnas.1419409112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rath S., Donovan J., Whitney G., Chitrakar A., Wang W., and Korennykh A. (2015) Human RNase L tunes gene expression by selectively destabilizing the microRNA-regulated transcriptome. Proc. Natl. Acad. Sci. U.S.A. 112, 15916–15921 10.1073/pnas.1513034112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brennan-Laun S. E., Ezelle H. J., Li X. L., and Hassel B. A. (2014) RNase-L control of cellular mRNAs: roles in biologic functions and mechanisms of substrate targeting. J. Interferon Cytokine Res. 34, 275–288 10.1089/jir.2013.0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee S., Li G., Li Y., Gaughan C., Baskar D., Parker Y., Lindner D. J., Weiss S. R., and Silverman R. H. (2015) RNase L is a negative regulator of cell migration. Oncotarget 6, 44360–44372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siddiqui M. A., Mukherjee S., Manivannan P., and Malathi K. (2015) RNase L cleavage products promote switch from autophagy to apoptosis by caspase-mediated cleavage of beclin-1. Int. J. Mol. Sci. 16, 17611–17636 10.3390/ijms160817611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Banerjee S., Chakrabarti A., Jha B. K., Weiss S. R., and Silverman R. H. (2014) Cell-type-specific effects of RNase L on viral induction of beta interferon. MBio 5, e00856–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donovan J., Rath S., Kolet-Mandrikov D., and Korennykh A. (2017) Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 10.1261/rna.062000.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yim H. C., Wang D., Yu L., White C. L., Faber P. W., Williams B. R., and Sadler A. J. (2016) The kinase activity of PKR represses inflammasome activity. Cell Res. 26, 367–379 10.1038/cr.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Youssef O. A., Safran S. A., Nakamura T., Nix D. A., Hotamisligil G. S., and Bass B. L. (2015) Potential role for snoRNAs in PKR activation during metabolic stress. Proc. Natl. Acad. Sci. U.S.A. 112, 5023–5028 10.1073/pnas.1424044112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalet A., Gatti E., and Pierre P. (2015) Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Lett. 589, 1539–1545 10.1016/j.febslet.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 21. Kim Y., Lee J. H., Park J. E., Cho J., Yi H., and Kim V. N. (2014) PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev. 28, 1310–1322 10.1101/gad.242644.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchal J. A., Lopez G. J., Peran M., Comino A., Delgado J. R., García-García J. A., Conde V., Aranda F. M., Rivas C., Esteban M., and Garcia M. A. (2014) The impact of PKR activation: from neurodegeneration to cancer. FASEB J. 28, 1965–1974 10.1096/fj.13-248294 [DOI] [PubMed] [Google Scholar]
- 23. Kunkeaw N., Jeon S. H., Lee K., Johnson B. H., Tanasanvimon S., Javle M., Pairojkul C., Chamgramol Y., Wongfieng W., Gong B., Leelayuwat C., and Lee Y. S. (2013) Cell death/proliferation roles for nc886, a non-coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene 32, 3722–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeon S. H., Lee K., Lee K. S., Kunkeaw N., Johnson B. H., Holthauzen L. M., Gong B., Leelayuwat C., and Lee Y. S. (2012) Characterization of the direct physical interaction of nc886, a cellular non-coding RNA, and PKR. FEBS Lett. 586, 3477–3484 10.1016/j.febslet.2012.07.076 [DOI] [PubMed] [Google Scholar]
- 25. Lee K., Kunkeaw N., Jeon S. H., Lee I., Johnson B. H., Kang G. Y., Bang J. Y., Park H. S., Leelayuwat C., and Lee Y. S. (2011) Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 17, 1076–1089 10.1261/rna.2701111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calderon B. M., and Conn G. L. (2017) Human noncoding RNA 886 (nc886) adopts two structurally distinct conformers that are functionally opposing regulators of PKR. RNA 23, 557–566 10.1261/rna.060269.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vachon V. K., Calderon B. M., and Conn G. L. (2015) A novel RNA molecular signature for activation of 2′–5′ oligoadenylate synthetase-1. Nucleic Acids Res. 43, 544–552 10.1093/nar/gku1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Justesen J., and Kjeldgaard N. O. (1992) Spectrophotometric pyrophosphate assay of 2′,5′-oligoadenylate synthetase. Anal. Biochem. 207, 90–93 10.1016/0003-2697(92)90506-3 [DOI] [PubMed] [Google Scholar]
- 29. Li Y., Banerjee S., Goldstein S. A., Dong B., Gaughan C., Rath S., Donovan J., Korennykh A., Silverman R. H., and Weiss S. R. (2017) Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwon Y. C., Kang J. I., Hwang S. B., and Ahn B. Y. (2013) The ribonuclease L-dependent antiviral roles of human 2′,5′-oligoadenylate synthetase family members against hepatitis C virus. FEBS Lett. 587, 156–164 10.1016/j.febslet.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 31. Wahid A. M., Coventry V. K., and Conn G. L. (2008) Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J. Biol. Chem. 283, 17485–17493 10.1074/jbc.M802300200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilson J. L., Vachon V. K., Sunita S., Schwartz S. L., and Conn G. L. (2014) Dissection of the adenoviral VA RNAI central domain structure reveals minimum requirements for RNA-mediated inhibition of PKR. J. Biol. Chem. 289, 23233–23245 10.1074/jbc.M114.550046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meng H., Deo S., Xiong S., Dzananovic E., Donald L. J., van Dijk C. W., and McKenna S. A. (2012) Regulation of the interferon-inducible 2′–5′-oligoadenylate synthetases by adenovirus VA(I) RNA. J. Mol. Biol. 422, 635–649 10.1016/j.jmb.2012.06.017 [DOI] [PubMed] [Google Scholar]
- 34. Deo S., Patel T. R., Dzananovic E., Booy E. P., Zeid K., McEleney K., Harding S. E., and McKenna S. A. (2014) Activation of 2′–5′-oligoadenylate synthetase by stem loops at the 5′-end of the West Nile Virus genome. PLoS One 9, e92545 10.1371/journal.pone.0092545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vachon V. K., and Conn G. L. (2016) Adenovirus VA RNA: an essential pro-viral non-coding RNA. Virus Res. 212, 39–52 10.1016/j.virusres.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 36. Kodym R., Kodym E., and Story M. D. (2009) 2′–5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem. Biophys. Res. Commun. 388, 317–322 10.1016/j.bbrc.2009.07.167 [DOI] [PubMed] [Google Scholar]
- 37. Harries L. W. (2012) Long non-coding RNAs and human disease. Biochem. Soc. Trans. 40, 902–906 10.1042/BST20120020 [DOI] [PubMed] [Google Scholar]
- 38. Mattick J. S. (2011) The central role of RNA in human development and cognition. FEBS Lett. 585, 1600–1616 10.1016/j.febslet.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 39. Cohen-Chalamish S., Hasson A., Weinberg D., Namer L. S., Banai Y., Osman F., and Kaempfer R. (2009) Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat. Chem. Biol. 5, 896–903 10.1038/nchembio.234 [DOI] [PubMed] [Google Scholar]
- 40. Nussbaum J. M., Gunnery S., and Mathews M. B. (2002) The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 30, 1205–1212 10.1093/nar/30.5.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osman F., Jarrous N., Ben-Asouli Y., and Kaempfer R. (1999) A cis-acting element in the 3′-untranslated region of human TNF-α mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13, 3280–3293 10.1101/gad.13.24.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McKenna S. A., Lindhout D. A., Shimoike T., Aitken C. E., and Puglisi J. D. (2007) Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J. Mol. Biol. 372, 103–113 10.1016/j.jmb.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., and Jacobs B. L. (2006) Inhibition of PKR by RNA and DNA viruses. Virus Res. 119, 100–110 10.1016/j.virusres.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 44. McKenna S. A., Kim I., Liu C. W., and Puglisi J. D. (2006) Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 358, 1270–1285 10.1016/j.jmb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 45. Hartmann R., Norby P. L., Martensen P. M., Jorgensen P., James M. C., Jacobsen C., Moestrup S. K., Clemens M. J., and Justesen J. (1998) Activation of 2′–5′ oligoadenylate synthetase by single-stranded and double-stranded RNA aptamers. J. Biol. Chem. 273, 3236–3246 10.1074/jbc.273.6.3236 [DOI] [PubMed] [Google Scholar]
- 46. Linpinsel J. L., and Conn G. L. (2012) in Recombinant and In Vitro RNA Synthesis (Conn G. L., ed) pp. 43–68, Springer Science + Business Media, Berlin/Heidelberg, Germany [Google Scholar]
- 47. Li Y., Banerjee S., Wang Y., Goldstein S. A., Dong B., Gaughan C., Silverman R. H., and Weiss S. R. (2016) Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 2241–2246 10.1073/pnas.1519657113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilkinson K. A., Merino E. J., and Weeks K. M. (2006) Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]