Abstract
Bone marrow stromal (a.k.a. mesenchymal stem) cells (BMSCs) can differentiate into osteoblasts (OBs), adipocytes, or chondrocytes. As BMSCs undergo OB differentiation, they up-regulate mitochondrial oxidative phosphorylation (OxPhos). Here, we investigated the mechanism(s) connecting mitochondrial OxPhos to OB differentiation. First, we found that treating BMSC-like C3H10T1/2 cells with an OxPhos inhibitor reduces their osteogenic potential. Interestingly, ATP levels were not reduced, as glycolysis compensated for the decreased OxPhos. Thus, mitochondria support OB differentiation not only by supplying ATP, but also by other mechanisms. To uncover these mechanisms, we stimulated OxPhos in C3H10T1/2 cells by replacing media glucose with galactose and observed that this substitution increases both OxPhos and osteogenesis even in the absence of osteoinducers. β-Catenin, an important signaling pathway in osteogenesis, was found to be responsive to OxPhos stimulation. β-Catenin activity is maintained by acetylation, and mitochondria generate the acetyl donor acetyl-CoA, which upon entering the Krebs cycle is converted to citrate capable of exiting mitochondria. Cytosolic citrate is converted back to acetyl-CoA by ATP citrate lyase (ACLY). We found that inhibiting ACLY with SB204990 (SB) reverses the galactose-induced β-catenin activity and OB differentiation. This suggested that acetylation is involved in β-catenin activation after forced OxPhos stimulation, and using immunoprecipitation, we indeed detected SB-sensitive β-catenin acetylation. Both β-catenin acetylation and activity increased during osteoinduction coincident with OxPhos activation. These findings suggest that active mitochondria support OB differentiation by promoting β-catenin acetylation and thus activity.
Keywords: β-catenin, mitochondria, acetylation, mesenchymal stem cells (MSCs), osteoblast, osteogenesis, bone marrow mesenchymal stem cells, acetylation
Introduction
There is emerging evidence that the metabolic state of a stem cell impacts differentiation. Glycolysis is preferred when stem cells are undifferentiated, whereas mitochondrial oxidative phosphorylation (OxPhos) 2 is increased during differentiation. We and others have shown that mitochondrial OxPhos is up-regulated in bone marrow stromal (a.k.a. mesenchymal stem) cells (BMSCs) as they undergo osteogenic differentiation (1–3) and that mitochondrial dysfunction impairs this process (4). Although the importance of the relationship between osteogenic differentiation and mitochondrial function is well-documented, the mechanism that links the two phenomena has yet to be identified. The most obvious hypothesis is that active OxPhos, which is significantly more efficient than glycolysis in ATP synthesis, supports the higher energy demands of osteogenic differentiation by supplying additional ATP. However, mitochondria are also important signaling components of the cell as they regulate cellular second messengers, e.g. calcium, as well as epigenetic signals (5–7). In this study, we aimed to identify the mechanism that links mitochondria to osteogenic differentiation.
A number of signaling pathways regulate the differentiation of BMSCs into bone-forming osteoblasts (OBs) (8, 9). One important signaling pathway is the canonical Wnt/β-catenin pathway. Wnt/β-catenin signaling is activated during the stem cell commitment stage but is inhibited during the maturation stage of OBs (10). Mice lacking Wnt/β-catenin in osteoprogenitors and OBs displayed low bone mass, further implicating the importance of this pathway for osteogenesis (11, 12). β-Catenin activity is regulated by post-translational modifications, such as phosphorylation and acetylation. In the absence of Wnt ligands, β-catenin is phosphorylated, ubiquitinated, and degraded in the cytosol. However, when Wnt ligands bind to their receptors, the resulting signaling cascade causes hypophosphorylation of β-catenin, decreased ubiquitination, and thus stabilization. Stabilized β-catenin accumulates in the cytosol and then translocates to the nucleus where it binds with transcriptional coactivators and regulates expression of the canonical Wnt target genes. Although phosphorylation promotes β-catenin ubiquitination and degradation, acetylation masks the ubiquitination sites, thus preventing β-catenin ubiquitination and degradation and promoting its activity (13). Acetylation has also been shown to stabilize and promote activities of other key osteogenic factors, Runx2 and osterix (14, 15). Acetyl CoA (Ac-CoA) is the substrate for protein acetylation reactions by acetyltransferases. The major source of Ac-CoA is the mitochondrial TCA cycle. Glucose-derived pyruvate is converted to Ac-CoA, which then enters the TCA cycle and is converted to citrate. Citrate can be transported from mitochondria into the cytosol where it is converted back to Ac-CoA by ATP citrate lyase (ACLY) enzyme. Thus, it is likely that mitochondrially derived Ac-CoA is the link between cellular bioenergetic and differentiation pathways.
In this study, we comprehensively explored this prospective link between mitochondrial activity and osteogenic differentiation. Our data demonstrate that mitochondrial activity in BMSC-like cells promotes β-catenin signaling via its acetylation and induces osteogenic differentiation, and blocking the link between mitochondrial activity and β-catenin acetylation reduces osteogenic differentiation.
Results
Active mitochondria are necessary for osteogenesis
Cells can produce energy in the form of ATP either via glycolysis only or via glycolysis followed by mitochondrial OxPhos. The process of osteogenesis is an energy-consuming process, making energy production very important. As BMSCs undergo osteogenesis, they activate mitochondrial OxPhos (1–3, 16), although whether this phenomenon is absolutely required for OB differentiation remains to be proven (17, 18). We aimed to determine whether mitochondrial OxPhos was necessary for osteogenesis of C3H10T1/2 BMSC-like, osteogenic (C3H) cells.
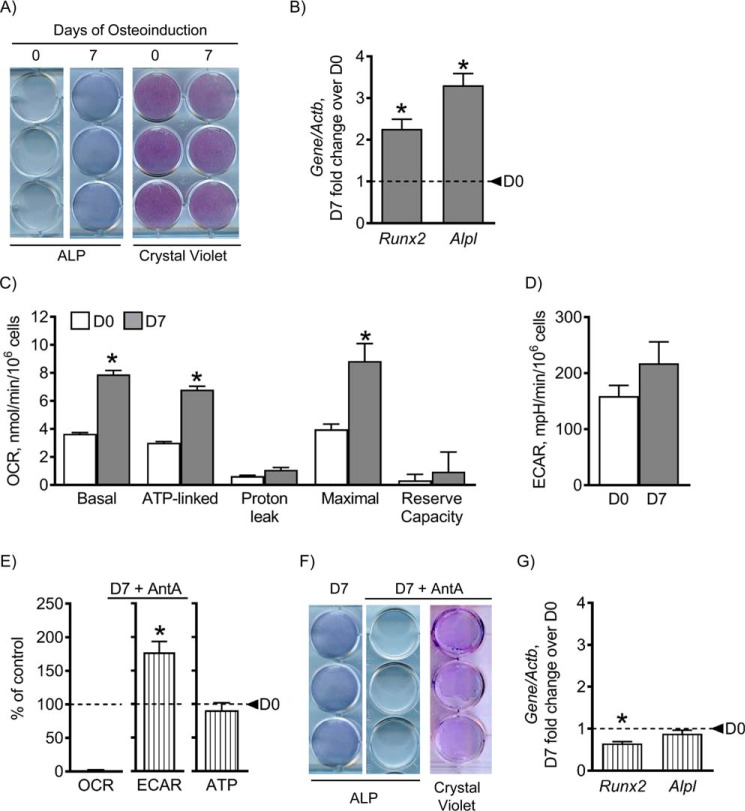
C3H cells were left undifferentiated or osteoinduced for 7 days and then assessed for osteogenic markers by specific staining and real-time reverse transcription (RT)-PCR (Fig. 1, A and B). After 7 days in osteogenic medium, alkaline phosphatase (ALP) staining and osteogenic markers Runx2 and Alpl were significantly increased, indicating osteogenic induction (Fig. 1, A and B).
Figure 1.
Mitochondrial OxPhos is necessary for osteogenesis. C3H cells were osteoinduced for 0 and 7 days using medium containing ascorbate and β-glycerol phosphate. Staining for osteoblast marker ALP and total cell crystal violet stain showed that cells up-regulated ALP at day 7 (D7), indicating osteogenesis (A). Gene expression of osteogenic markers Runx2 and Alpl is significantly increased at day 7, further indicating osteogenesis (B). OCR, a readout of mitochondrial OxPhos, was significantly increased in basal, ATP-linked, and maximal respiration at day 7 (C), whereas there was no change in glycolytic rates as measured by ECAR (D). When treated with AntA, a mitochondrial OxPhos inhibitor, at 0.5 μm, mitochondrial OxPhos is undetectable, and glycolysis is significantly increased, indicating a glycolytic shift with AntA, without noticeable effect on ATP levels (E). Treatment with AntA inhibited osteogenesis as shown by a reduced ALP stain and reduced gene expression of osteogenic markers (F and G). This indicates that mitochondrial OxPhos is required for osteogenesis. Dashed lines indicate day 0 (D0). Data are means. Error bars represent S.E. (n = 3–5). *, p < 0.05 compared with day 0 control.
To determine which metabolic pathway was used throughout osteogenesis, mitochondrial OxPhos or glycolysis only, we performed Seahorse metabolic profiling. Using metabolic inhibitors, we determined the relative contribution of mitochondrial OxPhos and glycolysis. After 7 days of osteoinduction, there was an increase in mitochondrial OxPhos, whereas there was no change in glycolytic lactate as indicated by oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), respectively (Fig. 1, C and D).
To confirm the importance of mitochondrial OxPhos for osteogenesis, we treated cells with antimycin A (AntA), an inhibitor of complex III of the mitochondrial respiratory chain, which shuts down mitochondrial OxPhos. When C3H cells were treated with AntA during osteoinduction, OCR dramatically decreased, whereas ECAR increased, most likely as a compensatory measure. Importantly, ATP content was not significantly affected by AntA treatment. This is not surprising given the fact that although glycolysis is less efficient than OxPhos it is significantly faster (19). This AntA-mediated inhibition of OxPhos and increase in glycolysis led to a reduction in osteogenesis as shown by a decrease in ALP staining and in expression of osteogenic markers Runx2 and Alpl. There was no effect on cell viability as indicated by the unchanged crystal violet staining (Fig. 1, E–G). Overall, these data indicate that activation of mitochondrial OxPhos is required for osteogenesis of C3H cells, and this requirement is not only to satisfy ATP demands.
Inducing mitochondrial OxPhos by replacing media glucose with galactose stimulates osteogenesis
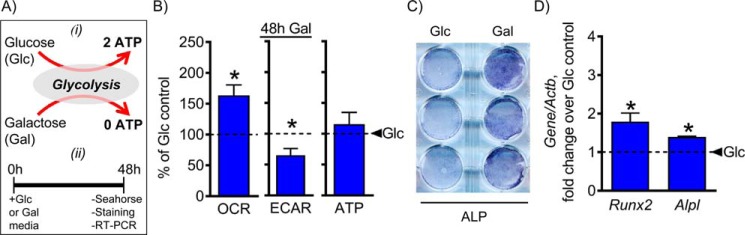
Our data so far indicate that osteogenic cells require active mitochondria during osteoinduction, and this is not just for ATP production. Mitochondria actively participate in various cellular processes, including signaling along with life and death decisions (5–7). We therefore wanted to determine whether forced activation of OxPhos alone had any effect on osteogenic differentiation. In 1925, Otto Warburg (20) was able to demonstrate that osteosarcoma cells utilize 18-fold less glycolysis when galactose is used as a sugar source instead of glucose. It is now understood that galactose leads to activation of mitochondrial OxPhos due to the energetic demands of galactose breakdown through glycolysis. Utilization of galactose through glycolysis results in a net production of zero ATP, leading to a compensatory up-regulation of OxPhos in the presence of exogenous mitochondrial OxPhos substrates, such as pyruvate and glutamine (Fig. 2A). We therefore investigated the effect of inducing mitochondrial OxPhos by replacing media glucose with galactose in C3H cells.
Figure 2.
Inducing mitochondrial OxPhos by replacing media glucose with galactose stimulates osteogenesis. Glucose replacement with galactose in the presence of mitochondrial substrates pyruvate and glutamine is a known strategy to induce mitochondrial OxPhos (A). C3H10T1/2 cells were incubated with galactose for 48 h and then assessed. Treatment with galactose resulted in an increase in OCR accompanied by a decrease in ECAR, indicating a reliance on mitochondrial OxPhos in galactose medium (B). Activation of OxPhos by replacing media glucose with galactose was sufficient to increase ALP staining and expression of osteogenic markers Runx2 and Alpl (C and D). The dashed line indicates glucose control. Data are means. Error bars represent S.E. (n = 3–5). *, p < 0.05 compared with 48-h glucose control. Glc, glucose (control); Gal, galactose.
Cells were incubated in a medium containing either glucose or galactose for 48 h and then assessed for metabolic preference and osteogenicity. Galactose treatment led to increased mitochondrial OxPhos and decreased lactate production when compared with glucose control (Fig. 2B). In addition, the galactose treatment increased osteogenic potential as indicated by an increase in ALP staining and expression of osteogenic markers Runx2 and Alpl (Fig. 2, C and D). Although the induction of Runx2 and Alpl after 48 h in galactose medium (179 ± 22 and 139 ± 2%, respectively) was less pronounced than the induction of Runx2 and Alpl after 7 days in osteogenic medium (226 ± 23 and 331 ± 28% respectively), it was still significant when compared with controls. Of note, incubation of cells for 48 h in regular nonosteogenic glucose-containing medium did not significantly change the expression of the studied genes when compared with day 0 undifferentiated cells. Overall, these data indicate that when glucose in medium is replaced by galactose there is an induction of mitochondrial OxPhos and stimulation of osteogenesis even in the absence of any osteoinducers in the medium.
Up-regulation of mitochondrial OxPhos by replacing media glucose with galactose stimulates β-catenin signaling
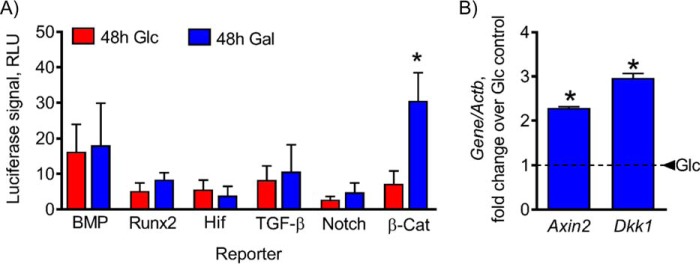
As shown above, activation of OxPhos by replacing media glucose with galactose was sufficient to stimulate osteogenesis in C3H cells. Osteogenesis is a tightly regulated process controlled by signaling mechanisms, such as β-catenin, bone morphogenic protein (BMP), and others (8). To determine which mechanisms were affected by forced activation of OxPhos, we used luciferase reporters to investigate the following pathways involved in regulation of osteogenic signaling: BMP/Smad, Runx2, hypoxia-inducible factor (HIF), transforming growth factor β (TGF-β)/Smad, Notch, and β-catenin. C3H cells transfected with each of these reporters were incubated for 48 h in glucose or galactose medium and then assessed for luciferase signal. Of all the reporters tested, only β-catenin responded to the stimulation of OxPhos by galactose treatment (Fig. 3A). Furthermore, expression of β-catenin target genes Axin2 and Dkk1 was significantly up-regulated (Fig. 3B). It is important to note that although Dkk1 is a negative regulator of β-catenin signaling it is also a transcriptional target and used as a readout of β-catenin activity. These data show that β-catenin is the primary osteogenic signaling pathway induced by mitochondrial OxPhos after galactose treatment.
Figure 3.
Up-regulation of mitochondrial OxPhos by replacing glucose with galactose stimulates β-catenin signaling. C3H cells were transfected with the indicated reporters and assessed for luciferase signal after 48-h incubation in either glucose (Glc) or galactose (Gal) medium. Stimulation of OxPhos by replacing media glucose with galactose resulted in significant up-regulation of the β-catenin (β-cat) reporter (A) and expression of β-catenin target genes Axin2 and Dkk1 (B). The dashed line indicates glucose control. Data are means. Error bars represent S.E. (n = 3–5). *, p < 0.05 compared with 48-h glucose control. RLU, relative luciferase units.
β-Catenin acetylation is stimulated by mitochondrial OxPhos activation
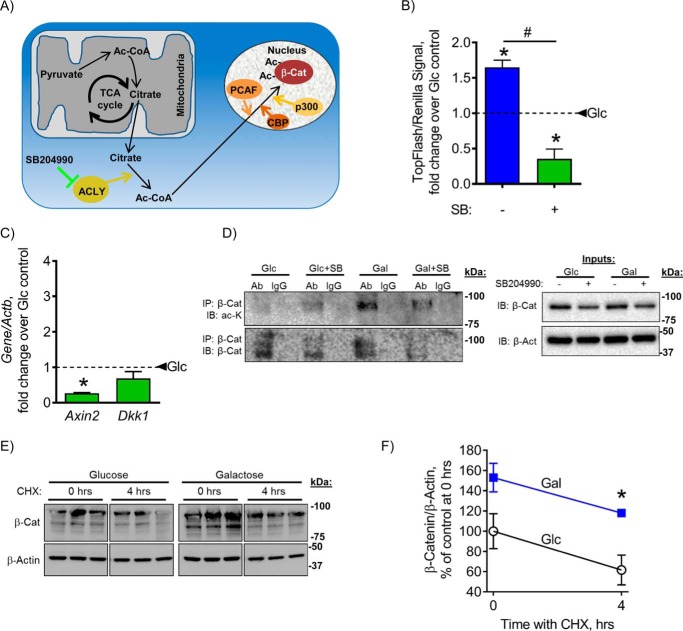
β-Catenin is regulated by post-translational modifications, the most common being phosphorylation and acetylation. Phosphorylation of β-catenin marks the protein for degradation by aiding in the targeting of β-catenin to become ubiquitinated (13). β-Catenin acetylation by p300/CBP-associated factor (PCAF) has the opposite effect in that it activates β-catenin by competing with ubiquination sites on Lys-19 and Lys-49 (13). In addition, β-catenin acetylation by p300 and CREB-binding protein (CBP) on Lys-49 only (21) or on Lys-354 only (22) does not affect its stability but improves its interactions with cofactors and its transcriptional activity. Acetylation depends on cytosolic levels of its substrate, Ac-CoA. Ac-CoA is produced in mitochondria from pyruvate and converted to citrate in the TCA cycle. In highly active mitochondria, excess citrate can translocate to the cytosol where it is converted back to Ac-CoA by ACLY enzyme. This ACLY-dependent cytosolic Ac-CoA is then used by p300, CBP, and PCAF acetyltransferase enzymes to acetylate and thus activate β-catenin (23). SB204990 (SB) is a specific inhibitor of ACLY enzyme (24). SB treatment leads to a decrease in cytosolic Ac-CoA level and is thus expected to decrease acetylated and active β-catenin levels (Fig. 4A). We sought to determine whether the observed activation of β-catenin after stimulation of mitochondrial OxPhos was due to its acetylation.
Figure 4.
β-Catenin acetylation and stabilization are stimulated by mitochondrial OxPhos activation. Citrate, derived from Ac-CoA in active mitochondria, translocates to the cytosol where it is converted back to Ac-CoA by ACLY enzyme. This ACLY-dependent cytosolic Ac-CoA is then used to acetylate and thus activate β-catenin (β-cat). ACLY is inhibited by SB (A). Galactose-induced activation of β-catenin reporter activity (B) and expression of β-catenin target genes (C) in C3H cells were reversed by inhibition of ACLY with SB. Galactose treatment resulted in increased acetylated β-catenin, which was sensitive to SB (D). Cell lysates were immunoprecipitated with an anti-β-catenin antibody and probed with anti-anti-acetylated lysine (ac-K) antibody. Blots are representatives of five. The rate of β-catenin degradation was not affected in galactose medium as evident from the chase experiment using CHX and Western blotting (E) quantitated with densitometry (F). In B and C, dashed lines indicate glucose control. Data are means. Error bars represent S.E. (n = 3–5). *, p < 0.05 compared with 48-h glucose (Glc) control; #, p < 0.05, 48-h galactose (Gal) without SB compared with 48-h galactose with SB. IP, immunoprecipitation; IB, immunoblotting; Ab, antibody.
Inhibition of ACLY with SB for 48 h in C3H cells reversed the stimulatory effect of galactose on β-catenin activity (Fig. 4B) and expression of β-catenin target genes Axin2 and Dkk1 (Fig. 4C). These data confirm that the galactose-induced activation of β-catenin depends on ACLY and thus on mitochondrially derived Ac-CoA. To verify that these effects were not cell line–specific, we repeated some of the above experiments in an osteogenic cell line, MC3T3 E1. MC3T3 E1 cells incubated with galactose for 48 h showed increases in OCR measured in a Seahorse metabolic profiler, in mineralization measured with alizarin red, and in β-catenin activity measured with TopFlash reporter (Fig. S1). These effects were reversed by inhibition of ACLY with SB204990. These data indicate that the effect of galactose-induced activation of OxPhos on β-catenin and osteogenesis is not cell type–specific.
Next, to directly measure β-catenin acetylation, we performed β-catenin immunoprecipitation followed by detection of acetylated β-catenin with anti-acetylated lysine antibody. Galactose treatment resulted in an increase of acetylated β-catenin, which was sensitive to SB (Fig. 4D).
Finally, to assess the effect on β-catenin protein stability, we performed a “chase” experiment using cycloheximide (CHX) to inhibit de novo protein synthesis. Cells were incubated in glucose or galactose medium for 48 h and then treated with CHX for 0 or 4 h. Western blotting for β-catenin was then performed, and β-catenin levels were measured with densitometry (Fig. 4, E and F). The assay showed that incubating cells in galactose medium did not affect the β-catenin degradation rate. This suggests that in our case it was likely β-catenin interaction with its cofactors and its activity, but not its stability, that were increased by its acetylation. Higher β-catenin levels observed in galactose medium may be explained by the ability of Wnt/β-catenin signaling to self-regulate and induce positive feedback, leading to a buildup of β-catenin protein. Altogether, these data show that stimulation of mitochondrial OxPhos induces β-catenin acetylation and thus activation and that this effect depends on ACLY enzyme activity.
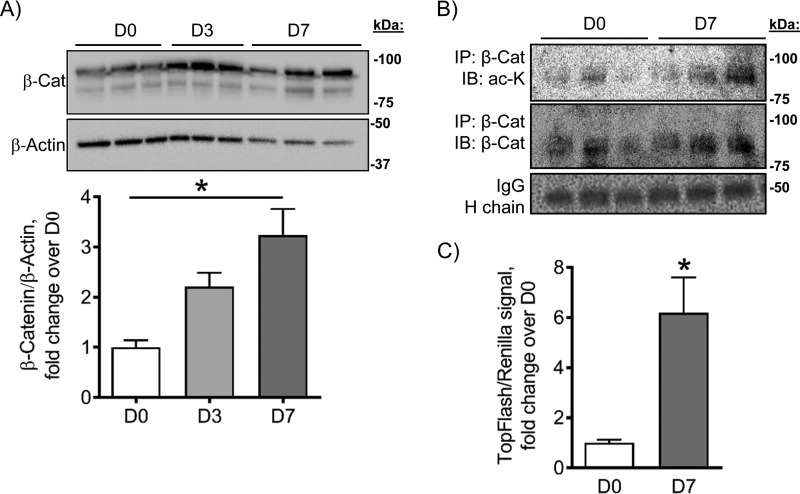
β-Catenin protein acetylation increases during osteogenesis
β-Catenin is necessary for osteogenesis, especially during the early stages of osteoblast differentiation (10). We observed that mitochondrial OxPhos is activated during osteoblast differentiation and that activation of mitochondrial OxPhos alone is sufficient to induce β-catenin acetylation and activation, leading to induction of osteoblast differentiation. We therefore wanted to determine the changes in total β-catenin and acetylated β-catenin levels during the early stages of osteogenic differentiation. C3H cells were incubated in osteogenic medium for 0, 3, and 7 days, and cell protein was collected. As shown in Fig. 1, by day 7 of osteoinduction, these cells express the early osteoblast marker ALP. Fig. 1 also shows that by day 7 of osteoinduction C3H cells activate mitochondrial OxPhos.
We found that β-catenin protein levels in C3H cells progressively increase during the studied time course of early osteogenesis, reaching significance at day 7 when mitochondrial OxPhos is active (Fig. 5A). To determine whether this increase in β-catenin protein levels was accompanied by an increase in acetylation of β-catenin, we performed immunoprecipitation of β-catenin followed by immunoblotting for acetylated lysine. After 7 days of osteoinduction, there was an increase in β-catenin acetylation levels (Fig. 5B). In addition, activity levels of β-catenin increased after 7 days of osteoinduction as measured by a luciferase assay (Fig. 5C). These data indicate that β-catenin protein, acetylation, and activity levels increase during osteogenesis, and this increase is coincident with activation of mitochondrial OxPhos.
Figure 5.
β-Catenin protein, acetylation, and activity levels increase during osteogenesis. β-Catenin protein levels in C3H cells increase during osteogenesis, reaching significance at day 7 (D7) (A). After 7 days of osteoinduction, cells had increased levels of acetylated β-catenin as shown by immunoprecipitation (IP) (B) along with an increase in β-catenin activity as shown by luciferase assay (C). Blots are representative of two blots, and each blot contains three biological replicates. Data are means. Error bars represent S.E. (n = 3–6). *, p < 0.05 compared with day 0 (D0) control. IB, immunoblotting.
Discussion
In this work, we aimed to determine whether activation of mitochondrial OxPhos is required for osteogenic differentiation of progenitor cells (C3H10T1/2) and the mechanism underlying such a requirement. We show evidence that activation of mitochondrial OxPhos is both necessary and sufficient for osteogenesis of these cells. Forcing cells to use OxPhos by replacing media glucose with galactose stimulated osteogenic differentiation even in the absence of osteogenic inducers. In addition, we found that stimulation of mitochondrial OxPhos led to acetylation and activation of a key osteogenic signaling molecule, β-catenin, thus leading to an increase in osteogenesis.
Recently, there has been increased interest in the energetic profile of bone-forming cells and their progenitors. There has also been a growing interest in determining the energy metabolism of both embryonic and somatic stem cells in general as they differentiate (25). In the case of osteoblasts, these stem cells are referred to as BMSCs or skeletal stem cells (26). The exact metabolic profile of BMSCs during osteogenic differentiation continues to be debated. Our laboratory and others have reported a preference for mitochondrial OxPhos as BMSCs differentiate to osteoblasts (1–3, 16). However, there are other reports stating that the metabolic preference for this process is primarily glycolysis (17, 18). As we show in Fig. 1 and in previous work, our data consistently show that mitochondrial OxPhos is activated during osteogenesis, whereas glycolysis is not significantly affected (1). There are two possible explanations for this discrepancy in the field: 1) differences in incubation environment (normoxia versus hypoxia) and/or 2) differences in culture media as we discuss in detail in our previous publication (1). Thus, to avoid any artifacts caused by the in vitro conditions, we utilize physiological levels of substrates (see “Experimental procedures”) and culturing conditions most relevant to BMSCs in vivo.
To confirm that activation of mitochondria alone is sufficient to induce osteogenesis, we utilized galactose medium. Using galactose as a sugar is a known way to force cells to use mitochondrial OxPhos (20). By simply replacing glucose with galactose, we achieved stimulation of osteogenesis, thus supporting the conclusion that active mitochondria are not only necessary but also sufficient for osteogenesis.
Next, we wanted to determine whether there was a known osteogenic signaling pathway specifically responsive to this stimulation of mitochondrial OxPhos by galactose treatment. Many signaling pathways are known to be involved in osteogenic signaling, including BMP, Runx2, HIF-1, TGF-β, Notch, and β-catenin (8, 27, 28). We determined that the pathway stimulated by mitochondrial OxPhos is the β-catenin signaling pathway. We recognize that we did not test an exhaustive list of all osteogenic signaling pathways; however, we chose a panel of signaling pathways that are most noted in osteogenesis.
Osteogenic factors β-catenin, osterix, and Runx2 are known to be acetylated during osteogenesis, leading to their activation and stabilization (13–15). To determine whether mitochondrial OxPhos activated β-catenin by acetylation, we took advantage of the acetylation inhibitor SB (24). We showed that β-catenin acetylation is increased after stimulation of mitochondrial OxPhos due to galactose treatment. Other β-catenin post-translational modifications should be studied in the future, including phosphorylation and ubiquitination. However, based on the competition between ubiquitin and acetyl groups at the same lysine positions, we believe that the results we see with acetylation will be directly inverse to ubiquitination. Phosphorylation of β-catenin precedes ubiquitination, so we also expect to see a similar outcome.
We investigated whether an increase in mitochondrial OxPhos was accompanied by changes in β-catenin acetylation. β-Catenin protein, acetylation, and activity levels increased by day 7 of osteogenesis. It was recently reported that histone acetylation of Wnt genes drives osteogenesis (29). This work also highlighted the importance of β-catenin during osteogenesis by underlining β-catenin inactivity during osteoporotic conditions. Therefore, our data are also supported by the recent studies regarding β-catenin and acetylation during osteogenesis. It should be noted that another recent report by Karner et al. (30) from Long's group demonstrated a decrease in nuclear Ac-CoA levels due to blocking glucose-derived pyruvate from entering the mitochondrial TCA cycle in response to Wnt. Our data complement this report as well. First, both studies show a direct link among Wnt/β-catenin signaling, acetylation, and bioenergetics. Second, we observed a similar acute bioenergetic response of BMSCs to Wnt stimulation, namely activation of glycolysis (31). 3 Wnt acts on stem and progenitor cells where OxPhos is not yet very active, and by stimulating glycolysis it may promote initial activation and proliferation of these cells. At this early stage, decreased acetylation is beneficial as it helps to down-regulate expression of adipogenic and chondrogenic genes (30). Our study targets later stages of OB differentiation where OxPhos is already activated but where Wnt stimulation may not be as robust. Under these circumstances, mitochondria support β-catenin and possibly other osteogenic pathways by providing Ac-CoA and promoting acetylation. In sum, we show evidence that increased β-catenin acetylation is the mechanism of osteogenesis driven by mitochondrial OxPhos. This further underlines the importance of mitochondria in bone while also providing a new therapeutic avenue to explore.
Experimental procedures
Cell culture
C3H10T1/2 and MC3T3 E1 cells were purchased from ATCC and expanded in DMEM/low-glucose medium (Gibco) with glutamine, 10% fetal bovine serum, and 1% penicillin-streptomycin (Gibco), hereafter referred to as LG-DMEM. We used LG-DMEM containing physiological levels of glucose (5 mm) to avoid any artifacts caused by supraphysiological levels of glucose. For all experiments, we used cells from passages 5–10 as recommended by the manufacturer and from previous handling experience. The addition of 50 μg/ml ascorbate and 10 mm β-glycerol phosphate (both from Sigma) to LG-DMEM induced osteogenesis. To confirm osteogenesis, cells were stained with osteoblast-specific ALP stain and alizarin red stain for mineralization as described previously (1). In addition, cells were stained with crystal violet to determine total cell count. In substrate replacement experiments, glucose was replaced with galactose (Sigma) at 5 mm. Cells were treated with galactose or glucose medium for 48 h before collection. To inhibit ACLY and thus β-catenin acetylation, we used SB (R&D Systems) at a concentration of 100 μm. To assess the effect on β-catenin protein stability, we used CHX from Calbiochem at a concentration of 70 μm. To determine mitochondrial contribution to osteogenesis, AntA was added to osteogenic medium at a concentration of 0.5 μm.
Real-time RT-PCR
Total RNA was isolated using the RNeasy kit (Qiagen) and reverse transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad). cDNA was subjected to real-time RT-PCR. The primer pairs used for genes of interest are outlined in Table 1. Real-time RT-PCR was performed in the RotorGene system (Qiagen) using SYBR Green (Quanta). The expression of genes of interest was normalized to expression of Actb (β-actin).
Table 1.
Primers used for real time RT-PCR analysis
All primers are written from 5′ to 3′ (left to right).
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Actb | AGC CAT GTA CGT TGC TAT CC | CGT AGC ACA GCT TCT CCT TAA T |
| Runx2 | TCC GGA ATG CCT CTG TTA TGA | ACT GAG GCG GTC AGA GAA CAA ACT |
| Alpl | TGC AGT ACG AGC TGA ACA GGA ACA | TCC ACC AAA TGT GAA GAC GTG GGA |
| Axin2 | GAT GTC TGG CAG TGG ATG TTA G | GAC TCC AAT GGG TAG CTC TTT C |
| Dkk1 | GAG GGG AAA TTG AGG AAA GC | AGC CTT CTT GTC CTT TGG TG |
Metabolic profiling
OCR and ECAR were measured using Seahorse XF24 (Seahorse Bioscience). Cells were plated on Seahorse 24-well plates 48 h before the experiment at a density of 50,000 cells/well. Immediately before the experiment, medium was replaced with unbuffered DMEM-XF medium containing glutamine at 1 mm and either glucose at 5 mm or galactose at 5 mm and pyruvate at 1 mm (pH 7.4). A baseline measurement of OCR and ECAR was taken, and then an inhibitory analysis was performed using injections of oligomycin (Olig) at 1 μm, FCCP at 0.5 μm and AntA at 1 μm. The following OxPhos and glycolytic indexes were calculated: basal respiration (OCRpre-Olig − OCRpost-AntA), ATP-linked respiration (OCRpre-Olig − OCRpost-Olig), maximal respiration (OCRpost-FCCP − OCRpost-AntA), respiratory capacity (OCRpostFCCP − OCRpre-Olig), proton leak (OCRpost-Olig − OCRpost-AntA), and basic glycolysis (ECARpre-Olig).
Luciferase assay
Cells were grown in 96-well plates and transfected using X-tremeGENE DNA transfection reagent (Roche Applied Science) for 48 h prior to collection with the following promoter reporters at a ratio of 2 μl of X-tremeGENE to 1 μg of DNA: 12xSBE (BMP), 6xOSE (Runx2), HRE (HIF), 6xSBE (TGF-β), RBPjK (Notch), and TopFlash (β-catenin). Reporters were added at 100 ng/well. The Renilla luciferase vector pRL-TK (Promega) was cotransfected at 20 ng/well as a reference. After 48 h, cells were collected in passive lysis buffer (Promega). Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol in a BioTek plate reader in luminometer mode. The firefly luciferase signal was normalized to Renilla luciferase signal and expressed as relative luminescence units.
Immunoprecipitation
200 μg of protein per sample were precleared with EZview Red Protein G Affinity Gel beads (Sigma) and then mixed with either β-catenin antibody (BD Transduction Laboratories) at 250 μg/ml or control nonimmune IgG immobilized on the above beads in “immunoprecipitation buffer” (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.5% Nonidet P-40) supplemented with protease inhibitors. The reactions were nutated overnight at 4 °C. Immunocomplexes were washed in “immunoprecipitation wash buffer” (100 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1% Triton X-100) and resuspended in 2× Laemmli buffer. The inputs were also mixed with 2× Laemmli buffer, and then the immunoprecipitation reactions and inputs were boiled for 10 min and spun down. The supernatants were subjected to immunoblotting as described below.
Western blotting
Cells were lysed with lysis buffer containing protease inhibitors and subjected to 4–12% SDS-PAGE followed by transfer to polyvinylidene difluoride membranes and blocking in 5% dry milk. For β-catenin detection, blots were probed with mouse monoclonal total β-catenin antibody (above) at a concentration of 1:500 and horseradish peroxidase–conjugated goat anti-mouse antibody (Bio-Rad) at a concentration of 1:3000. For acetylated β-catenin detection, blots were probed with mouse monoclonal acetylated lysine antibody (Thermo Scientific) at a concentration of 1:500 and horseradish peroxidase–conjugated goat anti-mouse antibody (Bio-Rad) at a concentration of 1:3000. β-Catenin and acetylated lysine detection was developed with West Femto Substrate (Thermo Scientific). To verify equal loading, blots were reprobed with anti-β-actin antibody (Sigma) at 1:2000. Bands were measured with densitometry using ImageJ software. Signal was normalized to β-actin.
Statistical analysis
Three to six independent experiments were done to derive each panel of the figures as determined by power analysis. Power analysis was carried out using the following formula,
| (Eq. 1) |
where n is sample size, α is type I error, β is type II error, Δ is effect size, σ is standard deviation, and Z is a constant (32). Data were analyzed using Prism 5.01 (GraphPad Software). Mean values and standard errors were calculated, and the statistical significance (p < 0.05) was established using either Student's t test when two variables were compared or one-way analysis of variance when more than two variables were compared based on normal spread of our data.
Author contributions
B. H. S., M. B., N. W., L. S., and R. A. E. formal analysis; B. H. S., M. B., L. S., and R. A. E. validation; B. H. S., M. B., N. W., L. S., and R. A. E. investigation; B. H. S., M. B., N. W., and R. A. E. visualization; B. H. S., M. B., N. W., L. S., and R. A. E. methodology; B. H. S., M. B., L. S., and R. A. E. writing-original draft; B. H. S., M. B., L. S., and R. A. E. writing-review and editing; R. A. E. conceptualization; R. A. E. resources; R. A. E. data curation; R. A. E. software; R. A. E. supervision; R. A. E. funding acquisition; R. A. E. project administration.
Supplementary Material
Acknowledgments
We thank both the Center for Musculoskeletal Research and the Mitochondrial Interest Group for fruitful discussions and Dr. Paul Brookes for help with Seahorse metabolic profiler.
This work was supported by the University of Rochester Department of Orthopedics as well as the National Institutes of Health through NIAMS Grants K01 AR064610 and R01 AR072601 and National Center for Advancing Translational Sciences (NCATS) Grant UL1 TR000042) (to R. A. E.) and NCATS Grant TL1-TR000096 (to L. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
B. H. Shares, M. Busch, N. White, L. Shum, and R. A. Eliseev, unpublished data.
- OxPhos
- oxidative phosphorylation
- ACLY
- ATP citrate lyase
- AntA
- antimycin A
- BMP
- bone morphogenic protein
- BMSC
- bone marrow stromal (a.k.a. mesenchymal stem) cell
- CBP
- CREB-binding protein
- CREB
- cAMP-response element–binding protein
- CHX
- cycloheximide
- ECAR
- extracellular acidification rate
- HIF
- hypoxia-inducible factor
- OB
- osteoblast
- OCR
- oxygen consumption rate
- PCAF
- p300/CBP-associated factor
- Runx2
- runt-related transcription factor 2
- SB
- SB204990
- TGF-β
- transforming growth factor β
- TCA
- tricarboxylic acid
- C3H
- C3H10T1/2 BMSC-like, osteogenic
- ALP
- alkaline phosphatase
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone.
References
- 1. Shum L. C., White N. S., Mills B. N., Bentley K. L., and Eliseev R. A. (2016) Energy metabolism in mesenchymal stem cells during osteogenic differentiation. Stem Cells Dev. 25, 114–122 10.1089/scd.2015.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forni M. F., Peloggia J., Trudeau K., Shirihai O., and Kowaltowski A. J. (2016) Murine mesenchymal stem cell commitment to differentiation is regulated by mitochondrial dynamics. Stem Cells 34, 743–755 10.1002/stem.2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen C.-T., Shih Y.-R. V., Kuo T. K., Lee O. K., and Wei Y.-H. (2008) Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 26, 960–968 10.1634/stemcells.2007-0509 [DOI] [PubMed] [Google Scholar]
- 4. Baker N., Boyette L. B., and Tuan R. S. (2015) Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone 70, 37–47 10.1016/j.bone.2014.10.014 [DOI] [PubMed] [Google Scholar]
- 5. Sandhir R., Halder A., and Sunkaria A. (2017) Mitochondria as a centrally positioned hub in the innate immune response. Biochim. Biophys. Acta 1863, 1090–1097 10.1016/j.bbadis.2016.10.020 [DOI] [PubMed] [Google Scholar]
- 6. Cherry C., Thompson B., Saptarshi N., Wu J., and Hoh J. (2016) 2016: a “mitochondria” odyssey. Trends Mol. Med. 22, 391–403 10.1016/j.molmed.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 7. Vafai S. B., and Mootha V. K. (2012) Mitochondrial disorders as windows into an ancient organelle. Nature. 491, 374–383 10.1038/nature11707 [DOI] [PubMed] [Google Scholar]
- 8. Marie P. J. (2008) Transcription factors controlling osteoblastogenesis. Arch. Biochem. Biophys. 473, 98–105 10.1016/j.abb.2008.02.030 [DOI] [PubMed] [Google Scholar]
- 9. Raouf A., and Seth A. (2000) Ets transcription factors and targets in osteogenesis. Oncogene 19, 6455–6463 10.1038/sj.onc.1204037 [DOI] [PubMed] [Google Scholar]
- 10. Valenti M. T., Dalle Carbonare L., and Mottes M. (2016) Osteogenic differentiation in healthy and pathological conditions. Int. J. Mol. Sci. 18, E41 10.3390/ijms18010041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodda S. J., and McMahon A. P. (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- 12. Day T. F., Guo X., Garrett-Beal L., and Yang Y. (2005) Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 8, 739–750 10.1016/j.devcel.2005.03.016 [DOI] [PubMed] [Google Scholar]
- 13. Gao C., Xiao G., and Hu J. (2014) Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci. 4, 13 10.1186/2045-3701-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeon E.-J., Lee K.-Y., Choi N.-S., Lee M.-H., Kim H.-N., Jin Y.-H., Ryoo H.-M., Choi J.-Y., Yoshida M., Nishino N., Oh B.-C., Lee K.-S., Lee Y. H., and Bae S.-C. (2006) Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 281, 16502–16511 10.1074/jbc.M512494200 [DOI] [PubMed] [Google Scholar]
- 15. Lu J., Qu S., Yao B., Xu Y., Jin Y., Shi K., Shui Y., Pan S., Chen L., and Ma C. (2016) Osterix acetylation at K307 and K312 enhances its transcriptional activity and is required for osteoblast differentiation. Oncotarget 7, 37471–37486 10.18632/oncotarget.9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pietilä M., Lehtonen S., Närhi M., Hassinen I. E., Leskelä H.-V., Aranko K., Nordström K., Vepsäläinen A., and Lehenkari P. (2010) Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells. Tissue Eng. Part C Methods 16, 435–445 10.1089/ten.tec.2009.0247 [DOI] [PubMed] [Google Scholar]
- 17. Lecka-Czernik B., and Rosen C. J. (2015) Energy excess, glucose utilization, and skeletal remodeling: new insights. J. Bone Miner. Res. 30, 1356–1361 10.1002/jbmr.2574 [DOI] [PubMed] [Google Scholar]
- 18. Karner C. M., and Long F. (2018) Glucose metabolism in bone. Bone 115, 2–7 10.1016/j.bone.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng J. (2012) Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (review). Oncol. Lett. 4, 1151–1157 10.3892/ol.2012.928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warburg O. (1925) The metabolism of carcinoma cells. J. Cancer Res. 9, 148–163 10.1158/jcr.1925.148 [DOI] [Google Scholar]
- 21. Wolf D., Rodova M., Miska E. A., Calvet J. P., and Kouzarides T. (2002) Acetylation of β-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277, 25562–25567 10.1074/jbc.M201196200 [DOI] [PubMed] [Google Scholar]
- 22. Chocarro-Calvo A., García-Martínez J. M., Ardila-González S., De la Vieja A., and García-Jiménez C. (2013) Glucose-induced β-catenin acetylation enhances Wnt signaling in cancer. Mol. Cell 49, 474–486 10.1016/j.molcel.2012.11.022 [DOI] [PubMed] [Google Scholar]
- 23. Lévy L., Wei Y., Labalette C., Wu Y., Renard C. A., Buendia M. A., and Neuveut C. (2004) Acetylation of β-catenin by p300 regulates β-catenin-Tcf4 interaction. Mol. Cell. Biol. 24, 3404–3414 10.1128/MCB.24.8.3404-3414.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pearce N. J., Yates J. W., Berkhout T. A., Jackson B., Tew D., Boyd H., Camilleri P., Sweeney P., Gribble A. D., Shaw A., and Groot P. H. (1998) The role of ATP citrate-lyase in the metabolic regulation of plasma lipids. Hypolipidaemic effects of SB-204990, a lactone prodrug of the potent ATP citrate-lyase inhibitor SB-201076. Biochem. J. 334, 113–119 10.1042/bj3340113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loeffler J., Duda G. N., Sass F. A., and Dienelt A. (2018) The metabolic microenvironment steers bone tissue regeneration. Trends Endocrinol. Metab. 29, 99–110 10.1016/j.tem.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 26. Bianco P., and Robey P. G. (2015) Skeletal stem cells. Development 142, 1023–1027 10.1242/dev.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Majidinia M., Sadeghpour A., and Yousefi B. (2018) The roles of signaling pathways in bone repair and regeneration. J. Cell. Physiol. 233, 2937–2948 10.1002/jcp.26042 [DOI] [PubMed] [Google Scholar]
- 28. Wan C., Shao J., Gilbert S. R., Riddle R. C., Long F., Johnson R. S., Schipani E., and Clemens T. L. (2010) Role of HIF-1α in skeletal development. Ann. N.Y. Acad. Sci. 1192, 322–326 10.1111/j.1749-6632.2009.05238.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jing H., Su X., Gao B., Shuai Y., Chen J., Deng Z., Liao L., and Jin Y. (2018) Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis. 10.1038/s41419-017-0231-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karner C. M., Esen E., Chen J., Hsu F.-F., Turk J., and Long F. (2016) Wnt protein signaling reduces nuclear acetyl-CoA levels to suppress gene expression during osteoblast differentiation. J. Biol. Chem. 291, 13028–13039 10.1074/jbc.M115.708578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Esen E., Chen J., Karner C. M., Okunade A. L., Patterson B. W., and Long F. (2013) WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 17, 745–755 10.1016/j.cmet.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kadam P., and Bhalerao S. (2010) Sample size calculation. Int. J. Ayurveda Res. 1, 55–57 10.4103/0974-7788.59946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.