Abstract
Interleukin (IL)-1β plays a critical role in IL-6β– and transforming growth factor β (TGFβ)–initiated Th17 differentiation and induction of Th17-mediated autoimmunity. However, the means by which IL-1 regulates various aspects of Th17 development remain poorly understood. We recently reported that IL-1β enhances STAT3 phosphorylation via NF-κB–mediated repression of SOCS3 to facilitate Il17 transcription and Th17 differentiation, identifying an effect of IL-1 signaling on proximal events of STAT3 signaling. Here, we show that IL-1β promotes STAT3 binding to key cis-elements that control IL-17 expression. Additionally, we demonstrate that the IL-1–induced NF-κB factor RelA directly regulates the Il17a/f loci in cooperation with STAT3. Our findings reveal that IL-1 impacts both proximal signaling events and downstream interactions between transcription factors and cis-regulatory elements to promote Il17a/f transcription and Th17 differentiation.
Keywords: chromatin immunoprecipitation (ChiP), DNA-protein interaction, NF-kappa B (NF-KB), STAT3, interleukin 17A (IL-17 or IL-17A), interleukin 1 (IL-1)
Introduction
Differentiation of Th17 cells requires activation of TGFβ 2 and IL-6 signaling (1–3). Although multiple transcription factors positively regulate Th17 development, IL-6–induced STAT3 and RORγt are the principal determinants of Th17 lineage commitment (2, 4). The cytokine IL-1β impacts very early events in Th17 development by inducing genes that program Th17 differentiation (Irf4, Rorc, Il1r1, and Il23r) and counteracting the inhibitory influence of IL-2 (5–8). Deficiency of the IL-1 receptor 1 (IL-1R1) (Il1r1) results in diminished Th17 responses in vivo and resistance to experimental autoimmune encephalitis development (5–7), highlighting the importance of IL-1β to the Th17 developmental pathway.
In addition to their roles in promoting Th17 lineage development and TCR-driven IL-17 secretion, the STAT3-inducing cytokines IL-6, IL-21, and IL-23 permit TCR-independent IL-17 production (9–11). Whereas neither IL-6, IL-21, nor IL-23 alone can elicit IL-17 production from Th17 effector cells (12, 13), in combination with IL-1β these cytokines are powerful inducers of IL-17. Analogous activation pathways exist for Th1 and Th2 cells, which respond to signals from a STAT-activating factor plus an IL-1 family cytokine to induce transcription of effector cytokines. T helper 1 (TH1) cells exhibit robust IFN-γ production in response to stimulation with IL-12 plus IL-18 (14, 15). Similarly, Th2 cells produce IL-13 upon exposure to IL-2, IL-7, or TSLP in combination with IL-33 (9). Several studies have evaluated the nature of the synergism between IL-12 and IL-18 in TCR-independent IFN-γ production (10, 16, 17). IL-18 cannot itself induce Ifng; its role is in augmenting the effects of STAT4 on Th1 development (15). IL-12 activates the transcription factors STAT4 and AP-1, and as an IL-1 family member, IL-18 has been demonstrated to activate the NF-κB pathway (18). These signals converge on sequences in and around the gene encoding IFN-γ to induce its expression. Balasubramani et al. (17) described differential utilization of cis-elements in the regulation of the Ifng contingent on the method of stimulus. Demonstration that IL-12–induced STAT4 is required to recruit the NF-κB factor RelA to key cis-regulatory elements for enhancement of Ifng transcription reinforces the notion that one aspect of IL-12 and IL-18 synergy is at the level of DNA binding.
In this study, we report that multiple STAT-dependent enhancer elements govern Il17a/f expression, and we demonstrate that IL-1β not only heightens IL-23–induced STAT3 binding to these elements but also enables recruitment of STAT3 to additional regulatory sites contained within the extended Il17 locus and induces NF-κB factors that directly regulate Il17a/f transcription in collaboration with STAT3. Our data build on previous studies demonstrating that the IL-1 induced NF-κB pathway is operative in the reinforcement of STAT3 activation through effects on SOCS3 and Jak2 and determine that in addition to its involvement in early Th17 differentiation events, IL-1β serves to promote acute transcription of Il17a/f by exerting direct effects on these gene loci.
Results
IL-1 promotes STAT3 recruitment to distal cis-regulatory elements that regulate Il17a and Il17f transcription
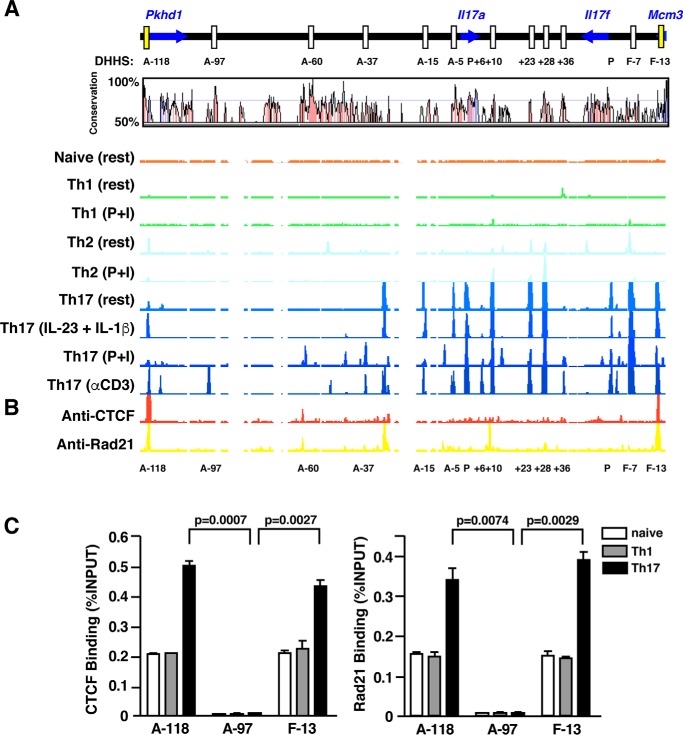
We previously observed that treatment of Th17 effector cells with IL-1 (α or β) can induce IL-17 production independently of TCR stimulation in a manner dependent on STAT3, and we noted that IL-17 production was maximal when IL-1 and IL-23 were used in combination (11). In an effort to understand how IL-1 signaling could impact STAT3-dependent transcription of Il17a and Il17f, we first performed comparative long-range DNase I hypersensitivity (DHS) mapping of the murine Il17a—Il17f gene locus in naïve and in vitro-polarized Th1, Th2, and Th17 cells to identify potential cis-regulatory elements (19). Prominent DNase I HS peaks localized to an ∼200-kb region surrounding the Il17a/f genes (Fig. 1A). Naïve CD4+ cells as well as Th1 and Th2 cells were largely devoid of DNase I HS sites, but Th17 cells exhibited 13 discernable peak clusters that corresponded well with evolutionarily conserved noncoding sequences (CNS). Most of the hypersensitivity peaks were present in resting polarized Th17 cells, indicating that the majority of cis-regulatory elements in the Il17a/f locus are remodeled during Th17 differentiation. TCR stimulation of polarized Th17 cells, whether by anti-CD3 or the TCR signaling surrogate phorbol ester (PMA) plus ionomycin, induced additional hypersensitive sites at CNS elements A−97, A−37, and A+6, suggesting a role for these regions as acutely activated, stimulus-dependent Il17a/f enhancers. Although TCR stimulation induced alterations in chromatin structure at the aforementioned CNS, accessibility of these stimulus-dependent hypersensitivity sites was not induced by cytokine co-signaling (IL-23 + IL-1β); the accessibility profile of the extended Il17a/f locus in cytokine-stimulated cells closely mirrored that of resting Th17 cells, suggesting that signals downstream of the IL-23 and IL-1 receptors likely act on enhancer elements accessible in resting Th17 cells, and they are unlikely to act at CNSs A−97, A−37, and A+6.
Figure 1.
DNase-hypersensitivity profiles and CTCF/Rad21 binding at the murine Il17a-Il17f loci of naïve, Th1, Th2, and Th17 cells. A, long range DNase hypersensitivity mapping was performed on naïve, Th1, Th2, and Th17 cells. CD4+ T cells isolated from C57/BL6 mice were cultured under Th1 polarizing conditions for 5 days or in Th2 differentiation media for 2 weeks. CD4+ cells isolated from Il17f Thy1.1/Thy1.1 reporter mice were used for Th17 polarization. After 6 days of culture, Thy1.1+ Th17 cells were isolated and rested overnight prior to restimulation. Harvested cells were either left unstimulated (rest), restimulated with PMA + ionomycin (P+I), subject to anti-CD3 treatment (αCD3), or stimulated with rIL-23 + rIL-1β (IL-23 + IL-1β) before DNase I digestion, labeling, and hybridization to custom arrays for DNase-chip analysis. DNase I hypersensitivity profiles are displayed using the IGB browser (Affymetrix) and are aligned with a corresponding VISTA plot demonstrating percentage similarity between mouse and human sequences. CNSs are defined as noncoding regions of at least 100 bp in length exhibiting greater than 70% sequence homology between species. Positions of CNSs are relative to the transcription start of the mouse Il17a or Il17f gene. B, ChIP-chip analysis of CTCF and Rad21/cohesin occupancy across the extended Il17a–Il17f loci. Thy1.1+ cells generated from Il17f Thy1.1/Thy1.1 reporter mice underwent chromatin immunoprecipitation (ChIP) with an antibody directed against murine CTCF or Rad21. ChIP samples and input material were subject to whole genome amplification and hybridized to custom-tiled arrays. Regions of CTCF and Rad21 binding are visualized using the IGB browser (Affymetrix). C, CTCF and Rad21ChIP analysis in naïve, Th1, and Thy1.1+ Th17 cells. Results are the mean ± S.E. of duplicate determinations and are representative of two independent experiments; results were quantified using real-time PCR, and data are expressed as percentage of input DNA recovered.
To verify that we had likely identified all cis-elements involved in the regulation of Il17a/f transcription, we probed the region for putative insulator elements by assessing the binding of the CCCTC-binding factor (CTCF) and a subunit of its associated cohesin, Rad21. CTCF has been shown to be required for barrier and enhancer-blocking activities of many insulator elements (20), and cohesin is hypothesized to organize chromatin structure in a manner that facilitates insulator function (21). We mapped CTCF and Rad21 sites by ChIP-chip (Fig. 1B) and found that strong signals for CTCF and Rad21 co-localized to only two sites in the extended Il17a/f locus: DNase I-hypersensitive sites (DHS) A−118 and F−13. DHS A−118 represents an evolutionarily-conserved sequence (CNS), but DHS F−13 lies in a region of noncoding DNA that lacks sequence conservation across species. The locations of these sites, flanking the Il17a/f loci beyond the most distal Th17-specific DNase-hypersensitive regions and situated in or adjacent to the neighboring genes Pkhd1 and Mcm3, respectively, are consistent with a role for A−118 and F−13 as boundary elements. Notably, CTCF and Rad21 binding at DHSs A−118 and F−13 were greatest in Th17 cells but were also evident in naïve CD4 and Th1 cells (Fig. 1C). Although all CTCF-binding sites exhibited Rad21 binding, we detected additional prominent Rad21-binding sites near regions of DNase hypersensitivity (A−34 and A+8) that did not correspond to areas of sequence conservation. It is possible that these regions represent foci of cohesin binding that attribute to cohesin's role in sister chromatid interactions and/or serve a distinct function in gene regulation. As a regulator of higher-order chromatin structure, cohesin contributes to the formation of chromatin loops (21, 22) and therefore could be involved in a number of different interactions between cis-regulatory elements that facilitate gene regulation. In that regard, DNase I hypersensitivity at CNS A−118 was only induced in Th17 cells following activation by TCR or cytokine signaling, suggesting its possible participation in activation-dependent chromatin looping and/or cis-regulatory activity.
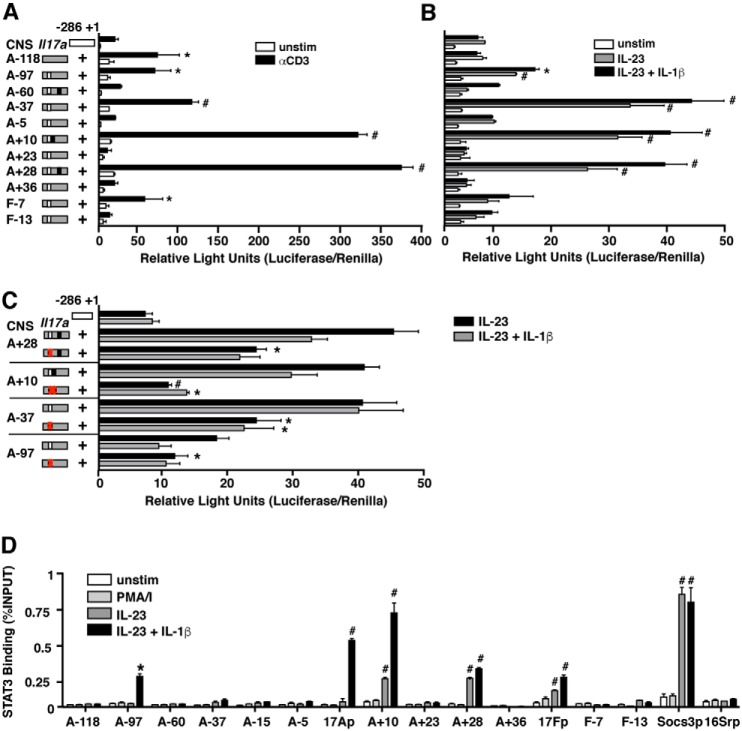
To assess the function of the cis-elements identified in the Il17a/f locus, we utilized a promoter–reporter assay in primary murine Th17 cells. The indicated CNS elements were subcloned upstream of a minimal Il17a or Il17f promoter, and firefly luciferase expression from transfected cells was used as a surrogate for transcriptional activity. CNSs A−118, A−97, A−37, A+10, A+28, and F−7 exhibited significant enhancement of promoter activity upon restimulation with anti-CD3 (Fig. 2A), indicating an ability of these cis-elements to augment TCR-driven Il17a gene transcription. Enhancer activity was retained for CNSs A−97, −37, +10, and +28 under conditions of cytokine restimulation (Fig. 2B), with a significant increase in luciferase activity by addition of IL-1β over IL-23 alone. Similar data were obtained for all CNSs relative to a 295-bp fragment of the Il17f promoter (data not shown).
Figure 2.
Distal cis-elements regulate Il17a transcription in a STAT3-dependent manner. A fragment of the Il17a promoter was linked to a firefly luciferase reporter in the presence or absence of an additional sequence corresponding to a CNS element in the extended Il17a-Il17f loci. Plasmids were transfected into 5-day polarized primary murine Th17 cells along with a Renilla luciferase vector to control for transfection efficiency. Transfected cells were rested 14–18 h and then either left unstimulated or restimulated with anti-CD3 for 4 h (A) or 10 μg/ml cytokine (B and C) for 1 h. Luciferase readings were taken, and data are expressed as relative light units, normalized to Renilla luciferase activity. White boxes indicate predicted STAT-binding sites, and black boxes indicate predicted NF-κB-binding elements. *, p < 0.05, and #, p < 0.01 versus promoter alone under same manner of stimulation. C, luciferase activity of Th17 cells transfected with reporter constructs with (red X in box) or without (open box) mutated STAT-binding sites. Data (mean ± S.E. of duplicate samples) are representative of at least three independent experiments. *, p < 0.05, and #, p < 0.01 versus intact CNS construct. D, CD4+ T cells were isolated from Il17fThy1.1/Thy1.1 reporter mice and either assayed directly or grown under Th17 conditions for 6 days. Following Thy1 isolation and overnight rest, ChIP was performed with antibody directed against STAT3 or IgG in cells that were either left unstimulated, stimulated with IL-23 (3 ng/ml), or IL-23 plus IL-1β (10 μg/ml) for 1 h or PMA + ionomycin (PMA/I) for 4 h. Real-time PCR was performed on immunoprecipitated DNA using primer sets designed to detect the indicated CNS elements and promoter regions. The Socs3 and 16S ribosomal promoters were used as positive and negative controls, respectively. qPCR values were normalized to input DNA, and values representing relative STAT3 binding are expressed as n-fold increase versus naïve CD4. Results are the mean ± S.E. of two to five experiments. *, p < 0.05, and #, p < 0.01 versus STAT3 recruitment to the 16S ribosomal promoter (16Srp).
We examined the enhancer CNS sequences for predicted transcription factor-binding sites and found that a composite STAT/Bcl6-binding site was unique to the four cis-elements that enhanced cytokine-driven Il17 transcription with CNS A+10 also possessing overlapping nuclear factor of activated T cells and NF-κB sites (Fig. S1). Based on this finding, as well as the knowledge that STAT3 is integral to Il17a/f transcription (23–25), we mutated the putative STAT sites contained within CNSs A−97, A−37, A+10, and A+28. These mutations significantly diminished cytokine-driven enhancer activity for all of the CNSs tested (Fig. 2C), consistent with the capacity of these cis-elements to bind STAT3 and function in acute transcriptional regulation following cytokine stimulus.
Previous studies have demonstrated STAT3 binding to cis-elements spanning 90 kb of the extended Il17a/f locus (26–28). To confirm these results and assess for recruitment of STAT3 to genomic elements more distal, we performed chromatin immunoprecipitation (ChIP) analysis for STAT3 binding across a region encompassing ∼180 kb of sequence surrounding Il17a and Il17f. Consistent with previous reports, we detected STAT3 binding at CNSs A+10, A+28, and the Il17f promoter following stimulation with IL-23. (Fig. 2D). Notably, however, we found that addition of IL-1β had a marked effect on STAT3 binding across the locus; compared with stimulation with IL-23 alone, IL-23 plus IL-1β induced both significant increases in STAT3 recruitment to CNSs A+10 and A+28 and substantial STAT3 binding at A−97 and the Il17a promoter that was not detected in the absence of IL-1 signaling. Although enhancer activity associated with accessible sites at CNSs A−118, A−37, and F−7 did not exhibit STAT3 binding, it is likely that these elements exert effects on Il17a/f transcription through interactions with other transcription factors not yet identified. Collectively, these results indicate that enhancers at CNSs A−97, A+10, A+28, and the Il17a and Il17f promoters bind STAT3 to regulate Il17a/f transcription following activation, and factors induced by IL-1R signaling enhance STAT3 binding to key cis-regulatory elements participating in the Il17a/f transcription.
NF-κB factors RelA and c-Rel induced by IL-1 signaling cooperate with STAT3 to regulate Il17a and Il17f transcription
Given the observation that IL-1 treatment enhanced STAT3 binding to cis-elements regulating Il17a/f expression, we wished to further examine the nature of the synergism between IL-1R and Jak/STAT signals. IL-1R signaling is initiated when IL-1α or IL-1β binds to the IL-1R1 subunit, enabling recruitment of an accessory receptor subunit, IL-1RAP. Ligand-induced juxtaposition of TIR domains on the cytoplasmic tails of IL-1R subunits recruits myeloid differentiation primary response protein 88 (MYD88), and subsequently IL-1R–associated kinase 4 (IRAK4), tumor necrosis factor receptor-associated factor 6 (TRAF6), as well as additional downstream signaling molecules (29). Activation of IL-1R signaling typically results in activation of the NF-κB and mitogen-activated protein kinase pathways (30); given that previous studies have reported functional interactions between NF-κB and STAT proteins (9, 17), we hypothesized that IL-1–induced NF-κB factors regulate Il17a/f transcription in cooperation with STAT3. Although previous studies support a role for TCR-induced NF-κB signaling in regulation of Rorc expression and Th17 differentiation (31, 32), neither effects on Il17a/f transcription nor a role for IL-1–mediated NF-κB signaling in Th17 differentiation have been directly assessed.
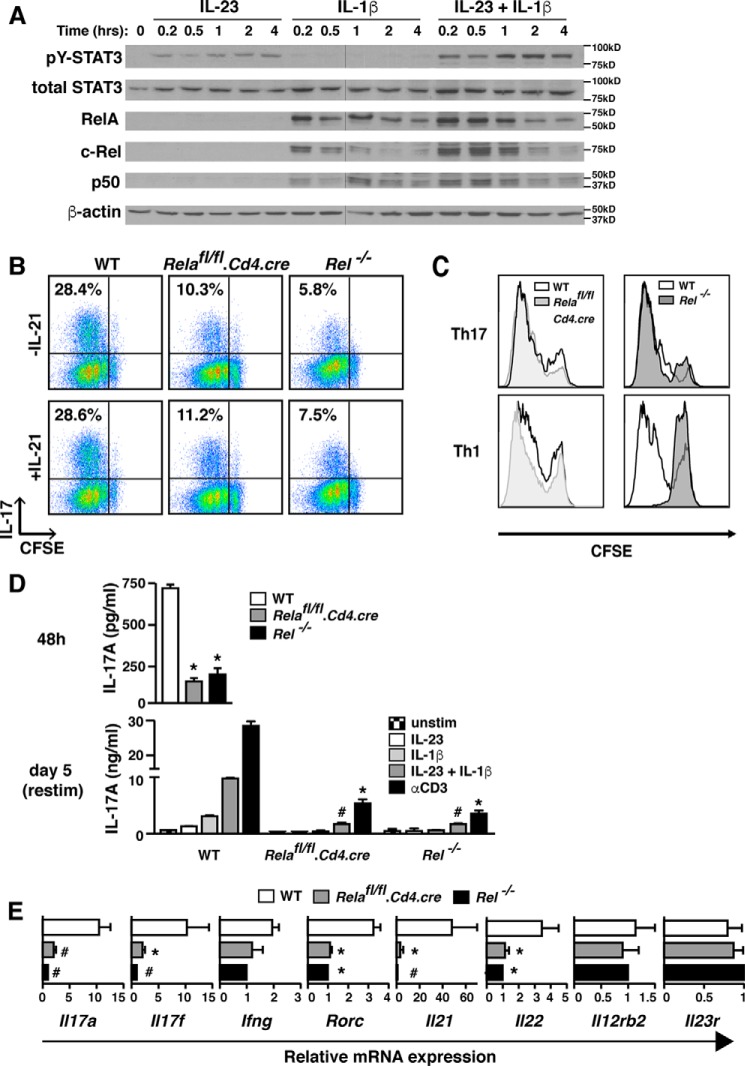
We first assessed the NF-κB factors activated by IL-1 signaling in Th17 cells and compared the kinetics of NF-κB nuclear localization with that of STAT3 phosphorylation induced by concomitant IL-23 signaling (Fig. 3A and Fig. S2). We limited our study to RelA, c-Rel, and p50, as these NF-κB factors are expressed in T cells (33–35). IL-1β triggered NF-κB nuclear import of NF-κB proteins RelA, c-Rel, and p50 and was not significantly altered by the addition of IL-23. Nuclear translocation of the NF-κB proteins took place within 10 min of cytokine exposure, and STAT3 tyrosine phosphorylation occurred concordantly (8). Peak expression of NF-κB proteins in the nucleus of Th17 cells paralleled that of STAT3 activation, and expression of both NF-κB and STAT3 began to decline between 2 and 4 h following cytokine stimulus. Thus, the kinetics of nuclear localization of trans-factors activated by the IL-1 and IL-23 receptors are similar in Th17 cells.
Figure 3.
IL-1β induces NF-κB factors that regulate Il17a/f transcription and Th17 differentiation. A, C57BL/6 CD4+ T cells were Th17-polarized for 5 days before live cell isolation was performed using a Ficoll gradient, and cells were rested overnight. On day 6, Th17 cells were restimulated for the indicated time frames with IL-23 (5 ng/ml) and/or IL-1β (20 ng/ml), and nuclear extracts were prepared. Nuclear translocation of STAT3 and the NF-κB factors RelA, c-Rel, and p50 were evaluated by immunoblotting. Blots were stripped and reprobed with β-actin as a loading control. The splice site at which two separate blots were merged is indicated with a black vertical line. B, FACS-sorted naïve CD4+ T cells from Relafl/fl.Cd4.cre+, Rel−/−, or WT B6 littermate mice were labeled with CFSE and cultured under Th17-polarizing conditions in the absence or presence of IL-21 (10 ng/ml) for 3 days. Cells were stimulated with PMA plus ionomycin stimulation for 5 h in the presence of monensin before intracellular cytokine staining for IL-17A. Proliferation was assessed by quantifying CFSE uptake. Flow cytometry plots are gated on live CD4+ cells, and numbers represent the percentage of cells present in the designated quadrant. Data are representative of at least three independent experiments. C, CD4+ T cells isolated from WT, Relafl/fl.Cd4.cre+, or Rel−/− mice were differentiated under Th1- or Th17-polarizing conditions (± addition of CFSE). On day 3 of polarization, proliferation was assessed by CFSE analysis. Plots are gated on live CD4+ T cells. D, naïve CD4+ T cells from the indicated mice were cultured under Th17 conditions as in Fig. 1 (for 3 days as in A and assayed directly (top panel) or for 5 days and assayed following restimulation (48 h) as indicated (bottom panel). The amount of IL-17 in culture supernatants was quantified by ELISA. Results represent mean ± S.E. of three independent experiments. *, p < 0.05. and #, p < 0.01 versus WT IL-17A production. E, FACS-sorted naïve CD4+ from Relafl/fl.Cd4.cre+, Rel−/−, or WT B6 mice were cultured under Th17 conditions for 72 h and then processed for mRNA quantification by real-time PCR for the indicated genes. Data were normalized to β2m and are expressed as relative difference (n-fold) compared with WT; values for Rel−/− are set at 1. Results represent mean ± S.E. of three independent experiments. *, p < 0.05, and #, p < 0.01 versus WT mRNA expression.
We next examined Th17 cells derived from T cells deficient in either RelA (17, 36) or c-Rel (34) to evaluate the impact of these factors on IL-17 production induced by IL-1 signaling. Consistent with a previous report (37), deficiency of both RelA and c-Rel results in decreased IL-17 production from Th17-polarized cells (Fig. 3, B, D, and E). Although c-Rel is critical for Il21 regulation (35), addition of exogenous IL-21 did not correct the deficit in IL-17 production observed in Relafl/fl.CD4-cre+ or Rel−/− mice (Fig. 3B). Notably, although c-Rel–deficient Th1 cells had severely impaired proliferative capacity (34, 35), Th17 proliferation was unaffected by deficiency of either RelA or c-Rel (Fig. 3C). Deficiency of either RelA or c-Rel resulted in similar impairment of IL-17A protein production during Th17 differentiation (Fig. 3D, upper panel) and upon restimulation of Th17 effector cells, whether by TCR or cytokine pathways (Fig. 3D, lower panel). Real-time PCR analysis revealed that mRNA expression of several additional Th17-associated genes was reduced in RelA and c-Rel–deficient Th17 cells, including Il17f, Rorgt, Il21, and Il22 but not others (Il23r) (Fig. 3E). This indicates that RelA and c-Rel impact early events in Th17 differentiation as well as acute transcription of Il17 in polarized effector cells.
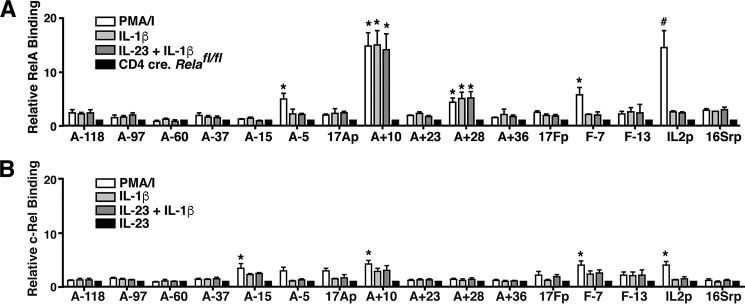
To determine whether RelA or c-Rel binds to the cis-elements in the Il17a and Il17f genes, we performed ChIP analysis to assess binding of these factors across the extended Il17a/f locus (Fig. 4, A and B). We found that like STAT3, RelA binds to the CNS A+10 enhancer element following cytokine and TCR stimulation to affect Il17a/f transcription. RelA also bound to CNS A+28, another potential focus of interaction with STAT3. RelA binding was detected at the proximal CNS elements A−5 and F−7 under TCR stimulation only. c-Rel weakly and/or variably bound to CNSs A+10, A−15, F−13, and the Il17a/f promoters. Taken together, these results point to a role for the NF-κB factor RelA, but not c-Rel, in STAT3-mediated regulation of Il17a/f transcription.
Figure 4.
IL-1β-induced NF-κB factors bind CNS to directly regulate Il17a/f transcription. A and B, CD4+ T cells were isolated from C57Bl/6 mice and either assayed directly or grown under Th17-polarizing conditions for 6 days. Following Ficoll separation and overnight rest, ChIP was performed with antibody directed against NF-κB p65 (RelA) (A) or IgG in cells that were either left unstimulated or stimulated with 10 μg/ml cytokine (IL-1β or IL-23 plus IL-1β) or PMA + ionomycin (PMA/I) for 2 h. The ChIP assay was performed on RelA-deficient Th17 cells generated from Relafl/fl.CD4.cre+ mice for comparison. Real-time PCR was performed on immunoprecipitated DNA using primer sets designed to detect the indicated CNS elements, promoter regions, and the Il2 and 16S ribosomal promoters as in Fig. 3. qPCR values were normalized to input DNA, and values representing relative RelA binding are expressed as n-fold increase over unstimulated Th17. Results are the mean ± S.E. of at least three independent experiments. *, p < 0.05, and #, p < 0.01 relative to values obtained for RelA-deficient Th17 cells. B, c-Rel ChIP analysis on Th17 cells either left unstimulated or restimulated with PMA + ionomycin, IL-1β, or IL-23 + IL-1β for 2 h. Data were normalized to input DNA and are expressed relative to resting Th17 cells. The Il2 and 16S ribosomal promoters were used as positive and negative controls. Results represent mean ± S.E. of four independent experiments. *, p < 0.05, and #, p < 0.01 versus values obtained for IL-23 stimulation.
Although physical interaction between RelA and STAT3 has been reported (38), we did not detect direct interaction between RelA and STAT3 or RelA and p300 following IL-23 + IL-1β stimulation of Th17 cells in co-immunoprecipitation studies (data not shown).
Discussion
In this study, we have performed a functional survey of CNSs spanning the murine Il17a—Il17f locus. In so doing, we have identified six Il17a/f enhancer elements, with a clear dichotomy of utilization in response to TCR and cytokine stimuli. CNSs A−97, −37, +10, and +28 exhibited STAT-dependent enhancer activity that underscores their importance in cytokine-induced transcriptional activation. CNSs A−97, +10, +28, and the Il17a and -f promoters were found to directly interact with STAT3 to acutely regulate Il17a/f transcription. Although CNS A−37 did not bind STAT3 in response to IL-23/IL-1β stimulation of Th17 effector cells, its ability to function as an enhancer in response to cytokine in vitro and the presence of a well-conserved putative STAT-binding site suggests that perhaps this element has interactions with STAT3 early in Th17 development.
Co-occupancy of the NF-κB factor RelA with STAT3, BATF, IRF4, and p300 (28) at CNSs A+10 and A+28 implies that these sites may be regulatory nodes that facilitate interaction between key transcription factors regulating Il17 expression. This supposition is bolstered by our data illustrating Rad21/cohesin binding in close proximity to CNS A+10. CTCF and Rad21 have important roles in directing chromatin interactions by looping DNA, and the presence of a CTCF- or Rad21-binding site near an enhancer element likely denotes a site of physical association between regulatory DNA sequences and trans-factors bound to them. We have defined the limits of the Il17 locus by demonstrating CTCF and Rad21 occupancy of two cis-elements flanking the genes adjacent to Il17a and -f. Furthermore, the capacity of CNS A−118 to bind the insulator-associated factors CTCF/Rad21 and to enhance Il17 transcription in response to TCR stimuli in conjunction with data indicating that absence of this region correlates with lack of expression of a Thy1.1 reporter in a BAC-transgenic mouse model 3 suggest this element may function as a locus control region.
We have shown that IL-1β treatment augmented binding of IL-23–induced STAT3 to genomic regulatory elements and allowed for additional foci of STAT3–DNA interaction to emerge. These observations likely reflect the enhanced duration and amplitude of STAT3 phosphorylation that we reported previously (8) and indicate that STAT3 signal amplification downstream of IL-1R ligation positions the Il17a/f locus for greater accessibility.
The rapid kinetics of the effect implies JAK/STAT signal modification is the major mechanism by which IL-1 heightens IL-23–induced STAT3 occupancy and chromatin remodeling. Our data suggest that potentiation of pSTAT3 action via IL-1 and NF-κB–mediated repression of SOCS3 creates a feed-forward signaling loop that enhances and sustains transcription factor occupancy at Il17a/f. Ciofani et al. (28) found that STAT3-deficient Th17 cells exhibit reduced p300 occupancy across the Il17a/f locus in response to TGFβ + IL-6 stimulation and STAT3 co-localizes with p300 to multiple Il17 enhancers, suggesting that STAT3 associates with p300 and/or other histone acetyltransferases to recruit transcriptional co-activators and induce Il17 transcription.
The fact that deficiency of RelA and c-Rel attenuated expression of multiple Th17-associated genes and greatly diminished IL-17 production early in the course of Th17 development indicates that IL-1 signaling influences Th17 programming as well as acute transcription of Il17. The transcription factors IRF4 and BATF bind multiple Il17 cis-regulatory elements in uncommitted T helper cells (28) suggesting that they mediate initial chromatin assembly. Previous reports indicate that IL-1β and NF-κB induce IRF4 and BATF expression (39), respectively, and thus it is possible that early requirement of IL-1 to achieve optimal Th17 differentiation is related to recruitment of these pioneer factors to Il17a/f. The NF-κB factors c-Rel and RelA have also been reported to bind to and activate the promoters that control RORγ and RORγt expression (32), thereby influencing Th17 specification. Through induction of RORγt, IRF4, and BATF, NF-κB factors indirectly support activation of STAT3, which is crucial for Il17 transcription and Th17 development. Mice and humans deficient in STAT3 have severely impaired Th17 differentiation (6, 23–25). This is because, in addition to directly binding to regulatory sites within the Il17 locus, STAT3 controls expression of multiple genes involved in Th17 lineage specification, among them Rorc, Irf4, Batf, Il21, Il21r, Il23, and Il23r (26).
Our discoveries about the Il17a/f loci parallel existing data on Ifng transcriptional regulation. Multiple cis-elements regulating Ifng exhibit STAT4-dependent binding of RelA (17). This is in striking contrast to the observed phenomenon at the Il17a/f locus, where IL-1 signaling enhances STAT3 recruitment to CNSs. However, a similar dichotomy exists in that there is modular utilization of cis-elements in TCR-induced versus cytokine-induced transcription of the Ifng and Il17a/f cytokine genes. Akin to the Ifng locus, the Il17a/f locus is regulated by the NF-κB factor RelA and to a lesser extent by c-Rel. The intergenic Il17a/f CNSs A+10 and A+28 are targets of RelA, STAT3, and several other Th17 lineages specifying transcription factors (26–28), highlighting their importance as regulatory nodes that function to regulate Il17 transcription. Our previous studies revealing IL-1–induced augmentation of STAT3 activation together with this study's observation that NF-κB factors bind to critical cis-elements to directly regulate Il17 transcription demonstrate that IL-1β can exert significant influence over both differentiation and transcriptional events to impact T helper cell development.
Taken together, our findings significantly extend knowledge about regulation of the Il17a and Il17f cytokine genes. We demonstrate that STAT3 and NF-κB factors directly regulate these genes downstream of IL-23 and IL-1β signaling, and IL-1β enhances the influence of STAT3 on the Il17a/f locus through both NF-κB–mediated STAT3 signal amplification and increasing local chromatin accessibility. These events are of relevance to host defense, as rapid cytokine-induced mobilization of T helper cells facilitates clearance of pathogens. These data also add to mounting evidence that interference with IL-1 and/or NF-κB signaling is a tenable strategy for treatment of Th17-associated autoimmune inflammatory diseases.
Experimental procedures
Mice
C57BL/6, B6.OT-II TCR transgenic (OT-II), B6.129S1-Il12btm/Jm/J (IL-12p40−/−), B6.Il10−/−, and B6.FVB-Tg (EIIa-cre)C5379Lmgd/J mice were purchased from The Jackson Laboratory and/or bred at the University of Alabama at Birmingham.
The generation of Il17f Thy1.1/Thy1.1 reporter mice was described previously (40), as was the creation of Relafl/fl.Cd4-cre+ mice (17). B6.Rel−/− mice used in some studies were a kind gift from Ranjan Sen (NIA, National Institutes of Health). All animal breeding and experimentations were carried out in accordance with institutional regulations.
CD4+ T cell isolation and culture
Spleens and lymph nodes were isolated from the indicated strains of mice, and CD4+ T cells were purified by positive selection with magnetic beads (Invitrogen) before culture with OVA peptide (5 μg/ml) or anti-CD3 (2.5 μg/ml) and irradiated CD4-depleted splenocytes at a ratio of 1:7 (Th17) or 1:5 (Th1 and Th2) CD4 cells/APCs. T cells were cultured in Iscove's media containing 10% fetal bovine serum, 100 IU/ml penicillin, 100 mg/ml streptomycin, 1 mm sodium pyruvate, 1× nonessential amino acids, 2.5 mm β-mercaptoethanol, 2 mm l-glutamine. Th17 polarizations were carried out using neutralizing antibodies to IFN-γ and IL-4 (10 μg/ml), with addition of 20 ng/ml IL-6 (R&D Systems) and 2–5 ng/ml rhTGF-β1 (R&D Systems). Th1 cultures were supplemented with 10 ng/ml IL-12 and 10 μg/ml anti-IL-4, and Th2 cultures were generated using 1000 units/ml IL-4 and 10 μg/ml anti-IFN-γ. In some experiments cells were stimulated with plate-bound anti-CD3 (clone 145-2C11) plus soluble anti-CD28 (clone 27.51, eBioscience) at the indicated concentrations, or underwent polarization with anti-CD3/CD28-coated beads (Invitrogen) according to the manufacturer's instructions.
Plasmids, CD4 cell transfection, and promoter–reporter assay
Il17a and -f promoter fragments were cloned into the pGL3 basic luciferase vector (Promega) using XhoI and HindIII restriction sites with the following primer sequences: Il17a promoter 286-bp fragment Fwd GCTACTCGAGGCAAAGCATCTCTGTTCAGC and Il17a promoter 286-bp Rev CGTAAAGCTTGCGTCCTGATCAGCTGGTGC; Il17f promoter 295-bp fragment Fwd GCATCTCGAGAAAGGTAATGGGAGTGGAAG and Il17f promoter 295-bp fragment Rev GCATAAGCTTGGTTTCTCCAATGGCTGCTTC. Similarly, MluI and NheI restriction sites were used to cassette various CNS elements upstream of the indicated promoter fragment using the following primer sequences: CNS A−118 Fwd CTTGCCATCTTTCCTTCTTG and CNS A−118 Rev CTGTCTTGCCTTCAGTGC; CNS F−13 Fwd GAGACACAGGAAAGGAGAGG and CNS F−13 Rev GGAGCAGAGATTACTCAATGACAG; CNS A−97 Fwd GTTTCTTGTGCCTTCTCTTG and CNS A−97 Rev CAAGGTTGGGCATTGAGC; CNS F−7 Fwd GCAAGACTGGAAAGGAGAAACATC and CNS F−7 Rev GCACAGCCTCTTCGTTTG; CNS A−60 Fwd GCCTAACTGTCAGAAAGTCACC and CNS A−60 Rev GCTGAGTTCTTCTCCCCTTAC; CNS A−37 Fwd ATGGAGCATTTCAGCAGGC and CNS A−37 Fwd Rev ATGCTTCCTGCCTTGATG; CNS A−5 Fwd ATCCTTCATCATAGCAGCC and CNS A−5 Rev TGAATACTTGCGTGGCAG; CNS A+10 Fwd ACTTGCTGCTCTCACGGAAG and CNS A+10 Rev CCTGAACAGAACACCAATGG; CNS A+28 Fwd GCTATCTCTCCAGCCCTAAG and CNS A+28 Rev CAGGCTAATCTTGGGAATG; CNS A+23 Fwd CGTAACGCGTCAGAACAAGTCACCTGCTG and CNS A+23 Rev CGTAGCTAGCCCTGTGATTTCCTCATTGG; CNS A+36 Fwd CGTAACGCGTTCCTACTGTGATGACCAGGC and CNS A+36 Rev CGTAGCTAGCAGTCCATCCTCAATGTGGC. Primers used for mutation of predicted STAT-binding sites include: CNS A+10 STAT mut Fwd GTGCAGTGACTAAAAGGAGAGTCCTCGAGGATAAAGTAACCTACC and CNS A+10 STAT mut Rev GGTAGGTTACTTTATCCTCGAGGACTCTCCTTTTAGTCACTGCAC; CNS A+28 STAT mut Fwd CCTGGCTGAGGAGAACGGAGAATCCCTTTGTGATCTTTCAGTCC and CNS A+28 STAT mut Rev GGACTGAAAGATCACAAAGGGATTCTCCGTTCTCCTCAGCCAGG; CNS A−97 STAT mut Fwd CATCATACACTAATTGTGAGTGAACTTTGTAGCCTTTTGTAGATC and CNS A−97 STAT mut Rev GATCTACAAAAGGCTACAAAGTTCACTCACAATTAGTGTATGATG; CNS A−37 STAT mut Fwd CAGAGGCCCTAGCCGCAAGCTGTCTGGACTCAGCTGGTCAAG and CNS A−37 STAT mut Rev CCTGACCAGCTGAGTCCAGACAGCTTGCGGCTAGGGCCTCTG. Clones for each construct were screened by restriction digest and sequenced to verify their authenticity. Th17 cells were generated as described, and 5–6 days following polarization between 1 and 3 million CD4 cells were transfected using mouse nucleofactor kit (Lonza) and an AMAXA electroporator. 20 μg of promoter–reporter construct DNA was administered to cells along with 1 μg of pRL-TK to allow for normalization. Following transfection, cells were either rested for 2–3 h and restimulated with plate-bound anti-CD3 (1 μg/ml), rested overnight and restimulated with between 3 and 10 ng/ml IL-23 ± 10 μg/ml IL-1β, or were left unstimulated. The dual-luciferase kit (Promega) was used to perform the luciferase assay according to manufacturer's instructions. Cells were lysed in passive lysis buffer, and relative light units were assessed for both firefly and Renilla using a Turner Systems TD 20/20 luminometer. Each transfection was performed in triplicate, and data represent a minimum of three independent experiments.
Flow cytometric analysis
CD4 T cells were collected and, where indicated, stimulated with PMA (50 ng/ml; Sigma) and ionomycin (750 ng/ml; Calbiochem) for 5 h in the presence of Golgi Plug (BD Biosciences). Intracellular staining was performed as described previously (1, 8, 11, 17, 40, 41). LIVE/DEAD Fixable Green Dead Cell Stain (Invitrogen) was used to exclude dead cells in flow cytometric analyses extracellularly. Phycoerythrin (PE)-conjugated anti-CD90.1 (OX-7) and anti-IL-17A (TC11-18H10) were purchased from BD Biosciences; APC-conjugated anti-IFN-γ (XMG1.2) and PE-Cy7–conjugated anti-CD4 (GK1.5) were purchased from eBioscience. Samples were acquired on an LSRII instrument (BD Biosciences), and data were analyzed using CellQuest Pro (BD Biosciences) or FlowJo software (Tree Star Inc.).
RNA isolation, cDNA synthesis, and real-time PCR
mRNA was extracted from T cells using TRIzol (Invitrogen) and treated with DNA-free (Ambion). cDNA synthesis was performed using Superscript III first-strand synthesis system (Invitrogen). Real-time PCR was performed on a Bio-Rad iCycler with TaqMan primer pairs and probes specific for cDNAs of Il23r, Il12rβ2, Ifng, Il17a, Il17f, Il22, Rorc, Il21, and β2-microglobulin (β2m). Primer sequences used were as described (1, 8, 11, 17, 40, 41), with the following additional primers/probes: Socs3 forward primer AGTGCAGAGTAGTGACTAAACATTACAAGA, Socs3 reverse primer AGCAGGCGAGTGTAGAGTCAGAGT, and Socs3 probe CGGCCTCCGAGGCGGCTCT. Reactions were run in triplicate and normalized to β2m expression.
DNase hypersensitivity mapping
DNase–ChIP samples were prepared as described previously (19, 41). In brief, nuclei were isolated form 5 × 10 (7) cells and subjected to digestion with the enzyme DNase I (Roche Applied Science) over a range of concentrations (0–12 units). EDTA was added to halt DNA digestion, and the nuclei were embedded in 1% InCert-agarose (Lonza) at a 1:1 ratio of volumes preceding overnight incubation at 37 °C in LIDS buffer (1% lithium dodecyl sulfate, 10 mm Tris-HCl, pH 7.5, 100 mm EDTA). Agarose plugs were washed extensively in 50 mm EDTA before digestion with T4 DNA polymerase (New England Biolabs). Blunted DNA fragments were then extracted, labeled with biotinylated linkers, captured, and amplified by ligation-mediated PCR to prepare for hybridization to microarrays. Data were visualized using IGB browser (17).
Chromatin immunoprecipitation
All ChIP experiments were performed using a ChIP assay kit (Millipore), as described previously (8, 17, 41). Relative recruitment was determined using RT-PCR, and ΔΔCt values were expressed as fold change over indicated reference cell type or displayed as a percentage of input DNA. Inputs were appropriately diluted to assist in normalization, and primers used were previously described (8, 17, 41), with the addition of the following: CNS A−118 forward CCAAACCTAAACACAAGGAGAAATC, CNS A−118 reverse TCTTGTGGCCAATATTTGCATT, and CNS A−118 probe CAGGCTGCGAAGACAACGCAGG; CNS F−13 forward AGGGAAAATCCCCCAAGAGA, CNS F−13 reverse TGTGGGCTTAGCTTCTGCATT, and CNS F−13 probe CCTGCTGCCACCTTGTGATGACTTGA; Socs3 promoter forward AGTGCAGAGTAGTGACTAAACATTACAAGA, Socs3 promoter reverse AGCAGGCGAGTGTAGAGTCAGAGT, and Socs3 promoter probe CGGCCGGGCAGTTCCAGGA. Antibodies used in ChIP include the following: STAT3 (Cell Signaling catalog no. 9132); RelA (Millipore catalog no. 06-418); c-Rel (Millipore catalog no. 09-040); CTCF (Millipore catalog no. 07-729); and Rad21 (Abcam catalog no. ab992). Samples were prepared for ChIP-chip analysis as described previously (17, 41).
ELISA and luminex assays
Th17 cells were incubated with indicated cytokines such as IL-1β (10 ng/ml), IL-6 (20 ng/ml), IL-18 (50 ng/ml), IL-23 (10 ng/ml), TGF-β (5 ng/ml), and IL-12 (10 ng/ml), all purchased from R&D Systems, in the presence or the absence of coated anti-CD3 (10 μg/ml) (clone 145-11). Supernatants were collected after 48 h, and the production of IL-17A or IFN-γ was detected by color development (TM-Blue; Sigma) of HRP-avidin substrate (Vector Laboratories) followed by incubation of antibodies directed against mouse IL-17A (BD Biosciences) or IFN-γ (BD Biosciences) and biotinylated anti-mouse IL-17A (BD Biosciences) or biotinylated anti-mouse IFN-γ (BD Biosciences). The amounts of cytokine were determined from standard curves established with serial dilutions of recombinant murine IL-17A or IFN-γ (R&D Systems). Luminex analysis for serum cytokines was carried out using Milliplex mouse cytokine immunoassay kit (Millipore catalog no. MPXMCYTO70KPMX32). Luminex analysis for phosphotyrosine-705–STAT3, phosphoserine-727–STAT3, and total STAT3 was carried out using Milliplex MAPmates (Millipore catalog nos. 46-623, 46-624, and 46-625) according to the manufacturer's protocol. Plates were read using a Luminex 100TM instrument.
Immunoblotting
Th17 cells were prepared as described previously. After 4 (Th1) or 5 days (Th17 and Th2), viable CD4 T cells were purified on a Ficoll gradient and activated with IL-12 (0.1–0.5 ng/ml) or IL-23 (4–5 ng/ml) for the indicated time. Cell lysates were prepared in lysis buffer (RIPA buffer: 50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing a protease and phosphatase inhibitor mixture (Pierce). Protein was quantified by the Bradford assay before equivalent amounts were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Primary antibody directed against RelA (Santa Cruz Biotechnology, sc-372x), c-Rel (Santa Cruz Biotechnology, sc-71x), NF-κB p50 (Santa Cruz Biotechnology, sc-1190x), or β-actin (Abcam catalog no. 6276) were utilized. HRP-conjugated donkey anti-rabbit or HRP-conjugated anti-mouse antibody (Affinity Bioreagents) were used to detect target protein by ECL detection kit (GE Healthcare or Pierce SuperSignal Dura).
Statistical analyses
Statistical significance was evaluated using the two-tailed unpaired t test. Unless indicated, all p values <0.05 were considered significant.
Author contributions
S. K. W., A. B., R. S., Y. S., G. E. C., R. D. H., and C. T. W. conceptualization; S. K. W., C. L. Z., R. S., Y. S., G. E. C., R. D. H., and C. T. W. resources; S. K. W., A. B., R. S., Y. S., G. E. C., N. M. W., R. D. H., and C. T. W. data curation; S. K. W., A. B., R. S., Y. S., G. E. C., R. D. H., and C. T. W. formal analysis; S. K. W., A. B., R. S., Y. S., G. E. C., R. D. H., and C. T. W. supervision; S. K. W., Y. S., G. E. C., R. D. H., and C. T. W. funding acquisition; S. K. W., A. B., C. L. Z., R. S., Y. S., G. E. C., N. M. W., R. D. H., and C. T. W. methodology; S. K. W., G. E. C., R. D. H., and C. T. W. writing-original draft; S. K. W., Y. S., G. E. C., R. D. H., and C. T. W. writing-review and editing; C. L. Z., G. E. C., R. D. H., and C. T. W. software; C. L. Z., G. E. C., R. D. H., and C. T. W. visualization.
Supplementary Material
Acknowledgments
We thank Drs. E. Benveniste, H. Qin, R. Lorenz, D. Chaplin, R. Cron, L. Justement, T. Ryan, J. Deshane, and members of the Weaver laboratory for their critical comments and suggestions. We thank J. Oliver, M. Blake, B. Parsons, D. O'Quinn, and S. Sinclair for their expert technical assistance.
This work was supported by National Institutes of Health Grants R01AI035783 and R01DK103744 (to C. T. W.) and R01DK115172 (to C. T. W. and R. D. H.), and the UAB Medical Scientist Training Program (to S. K. W.), supported by National Institutes of Health Grant T32 GM008361. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S2.
S. K. Whitley, A. Balasubramani, C. Zindl, R. Sen, Y. Shibata, G. E. Crawford, N. M. Weathington, R. D. Hatton, and C. T. Weaver, unpublished data.
- TGFβ
- transforming growth factor
- IL
- interleukin
- TCR
- T cell receptor
- Fwd
- forward
- Rev
- reverse
- qPCR
- quantitative PCR
- PE
- phycoerythrin
- CNS
- conserved noncoding sequence
- PMA
- phorbol 12-myristate 13-acetate
- CTCF
- CCCTC-binding factor
- HS
- hypersensitivity
- DHS
- DNase I-hypersensitive site
- APC
- allophycocyanin
- β2m
- β2-microglobulin.
References
- 1. Mangan P. R., Harrington L. E., O'Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., Hatton R. D., Wahl S. M., Schoeb T. R., and Weaver C. T. (2006) Transforming growth factor-β induces development of the T(H)17 lineage. Nature 441, 231–234 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- 2. Zhou L., Ivanov I. I., Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., and Littman D. R. (2007) IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- 3. Ghoreschi K., Laurence A., Yang X. P., Tato C. M., McGeachy M. J., Konkel J. E., Ramos H. L., Wei L., Davidson T. S., Bouladoux N., Grainger J. R., Chen Q., Kanno Y., Watford W. T., Sun H. W., et al. (2010) Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature 467, 967–971 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., and Littman D. R. (2006) The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T cell helper cells. Cell 126, 1121–1133 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- 5. Chung Y., Chang S. H., Martinez G. J., Yang X. O., Nurieva R., Kang H. S., Ma L., Watowich S. S., Jetten A. M., Tian Q., and Dong C. (2009) Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30, 576–587 10.1016/j.immuni.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laurence A., Tato C. M., Davidson T. S., Kanno Y., Chen Z., Yao Z., Blank R. B., Meylan F., Siegel R., Hennighausen L., Shevach E. M., and O'Shea J. J. (2007). Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- 7. Sutton C., Brereton C., Keogh B., Mills K. H., and Lavelle E. C. (2006) A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203, 1685–1691 10.1084/jem.20060285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basu R., Whitley S. K., Bhaumik S., Zindl C. L., Schoeb T. R., Benveniste E. N., Pear W. S., Hatton R. D., and Weaver C. T. (2015) IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the Th17 cell-iTreg cell balance. Nat. Immunol. 16, 286–295 10.1038/ni.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo L., Wei G., Zhu J., Liao W., Leonard W. J., Zhao K., and Paul W. (2009) IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. U.S.A. 106, 13463–13468 10.1073/pnas.0906988106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Zhu H., Murphy T. L., Ouyang W., and Murphy K. M. (2001) IL-18-stimulated GADD45 β required in cytokine-induced, but not TCR-induced, IFN-γ production. Nat. Immunol. 2, 157–164 10.1038/84264 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y. K., Landuyt A. E., Lobionda S., Sittipo P., Zhao Q., and Maynard C. L. (2017) TCR-independent functions of Th17 cells mediated by the synergistic actions of cytokines of the IL-12 and IL-1 families. PLoS One 12, e0186351 10.1371/journal.pone.0186351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., and Schaper F. (2003) Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 374, 1–20 10.1042/bj20030407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nurieva R., Yang X. O., Martinez G., Zhang Y., Panopoulos A. D., Ma L., Schluns K., Tian Q., Watowich S. S., Jetten A. M., and Dong C. (2007) Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 10.1038/nature05969 [DOI] [PubMed] [Google Scholar]
- 14. Murphy K. M., Ouyang W., Farrar J. D., Yang J., Ranganath S., Asnagli H., Afkarian M., and Murphy T. L. (2000) Signaling and transcription in T helper development. Annu. Rev. Immunol. 18, 451–494 10.1146/annurev.immunol.18.1.451 [DOI] [PubMed] [Google Scholar]
- 15. Robinson D., Shibuya K., Mui A., Zonin F., Murphy E., Sana T., Hartley S. B., Menon S., Kastelein R., Bazan F., and O'Garra A. (1997) IGIF does not drive Th1 development but synergizes with IL-12 for interferon-γ production and activates IRAK and NFκB. Immunity 7, 571–581 10.1016/S1074-7613(00)80378-7 [DOI] [PubMed] [Google Scholar]
- 16. Yang J., Murphy T. L., Ouyang W., and Murphy K. M. (1999) Induction of interferon-γ production in Th1 CD4+ T cells: evidence for two distinct pathways for promoter activation. Eur. J. Immunol. 29, 548–555 10.1002/(SICI)1521-4141(199902)29:02%3C548::AID-IMMU548%3E3.0.CO%3B2-Z [DOI] [PubMed] [Google Scholar]
- 17. Balasubramani A., Shibata Y., Crawford G. E., Baldwin A. S., Hatton R. D., and Weaver C. T. (2010) Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity 33, 35–47 10.1016/j.immuni.2010.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dinarello C. A. (1999) IL-18: A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J. Allergy Clin. Immunol. 103, 11–24 10.1016/S0091-6749(99)70518-X [DOI] [PubMed] [Google Scholar]
- 19. Crawford G. E., Davis S., Scacheri P. C., Renaud G., Halawi M. J., Erdos M. R., Green R., Meltzer P. S., Wolfsberg T. G., and Collins F. S. (2006) DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods 3, 503–509 10.1038/nmeth888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dorsett D. (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 21, 199–206 10.1016/j.gde.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wendt K. S., and Peters J. M. (2009) How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 17, 201–214 10.1007/s10577-008-9017-7 [DOI] [PubMed] [Google Scholar]
- 22. Lee G. R., Kim S. T., Spilianakis C. G., Fields P. E., and Flavell R. A. (2006) T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24, 369–379 10.1016/j.immuni.2006.03.007 [DOI] [PubMed] [Google Scholar]
- 23. Holland S. M., DeLeo F. R., Elloumi H. Z., Hsu A. P., Uzel G., Brodsky N., Freeman A. F., Demidowich A., Davis J., Turner M. L., Anderson V. L., Darnell D. N., Welch P. A., Kuhns D. B., Frucht D. M., et al. (2007) STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357, 1608–1619 10.1056/NEJMoa073687 [DOI] [PubMed] [Google Scholar]
- 24. Milner J. D., Brenchley J. M., Laurence A., Freeman A. F., Hill B. J., Elias K. M., Kanno Y., Spalding C., Elloumi H. Z., Paulson M. L., Davis J., Hsu A., Asher A. I., O'Shea J., Holland S. M., et al. (2008) Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776 10.1038/nature06764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang X. O., Panopoulos A. D., Nurieva R., Chang S. H., Wang D., Watowich S. S., and Dong C. (2007) STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282, 9358–9363 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- 26. Durant L., Watford W. T., Ramos H. L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H. W., Kanno Y., Powrie F., and O'Shea J. J. (2010) Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang X. P., Ghoreschi K., Steward-Tharp S. M., Rodriguez-Canales J., Zhu J., Grainger J. R., Hirahara K., Sun H. W., Wei L., Vahedi G., Kanno Y., O'Shea J. J., and Laurence A. (2011) Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C. N., Muratet M., Newberry K. M., Meadows S., Greenfield A., Yang Y., Jain P., et al. (2012) A validated regulatory network for Th17 cell specification. Cell 151, 289–303 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sims J. E., and Smith D. E. (2010) The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10, 89–102 10.1038/nri2691 [DOI] [PubMed] [Google Scholar]
- 30. Suzuki N., Suzuki S., and Yeh W. C. (2002) IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol. 23, 503–506 10.1016/S1471-4906(02)02298-6 [DOI] [PubMed] [Google Scholar]
- 31. Molinero L. L., Cubre A., Mora-Solano C., Wang Y., and Alegre M. L. (2012) T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 18529–18534 10.1073/pnas.1204557109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruan Q., Kameswaran V., Zhang Y., Zheng S., Sun J., Wang J., DeVirgiliis J., Liou H. C., Beg A. A., and Chen Y. H. (2011) The Th17 immune response is controlled by the Rel-RORγ-RORγT transcriptional axis. J. Exp. Med. 208, 2321–2333 10.1084/jem.20110462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Köntgen F., Grumont R. J., Strasser A., Metcalf D., Li R., Tarlinton D., and Gerondakis S. (1995) Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9, 1965–1977 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- 34. Liou H. C., Jin Z., Tumang J., Andjelic S., Smith K. A., and Liou M. L. (1999) c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int. Immunol. 11, 361–371 10.1093/intimm/11.3.361 [DOI] [PubMed] [Google Scholar]
- 35. Chen G., Hardy K., Bunting K., Daley S., Ma L., and Shannon M. F. (2010) Regulation of the IL-21 gene by the NF-κB transcription factor c-Rel. J. Immunol. 185, 2350–2359 10.4049/jimmunol.1000317 [DOI] [PubMed] [Google Scholar]
- 36. Steinbrecher K. A., Harmel-Laws E., Sitcheran R., and Baldwin A. S. (2008) Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 180, 2588–2599 10.4049/jimmunol.180.4.2588 [DOI] [PubMed] [Google Scholar]
- 37. Visekruna A., Huber M., Hellhund A., Bothur E., Reinhard K., Bollig N., Schmidt N., Joeris T., Lohoff M., and Steinhoff U. (2010) c-Rel is crucial for the induction of Foxp3+ regulatory CD4+ T cells but not Th17 cells. Eur. J. Immunol. 40, 671–676 10.1002/eji.200940260 [DOI] [PubMed] [Google Scholar]
- 38. Lee H., Herrmann A., Deng J. H., Kujawski M., Niu G., Li Z., Forman S., Jove R., Pardoll D. M., and Yu H. (2009) Persistently-activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell 15, 283–293 10.1016/j.ccr.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hwang E. S. (2010) Transcriptional regulation of T Helper 17 cell differentiation. Yonsei Med. J. 51, 484–491 10.3349/ymj.2010.51.4.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee Y. K., Turner H., Maynard C. L., Oliver J. R., Chen D., Elson C. O., and Weaver C. T. (2009) Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 10.1016/j.immuni.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mukasa R., Balasubramani A., Lee Y. K., Whitley S. K., Weaver B. T., Shibata Y., Crawford G. E., Hatton R. D., and Weaver C. T. (2010) Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32, 616–627 10.1016/j.immuni.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.