Abstract
Objectives. To determine how network-level factors influence individual risk of HIV acquisition, which is key in preventing disease transmission.
Methods. We recruited a cohort of young Black men who have sex with men (n = 618) in Chicago, Illinois, from 2013 to 2016. We identified potential molecular ties via pairwise genetic distance analysis of HIV pol sequences with links inferred between individuals whose sequences were 1.5% or less genetically distant. We defined clusters as 1 or more connections to another individual. We conducted entity resolution between confidant, sexual, referral, and Facebook network data between network types.
Results. Of 266 (43.0%) participants identified as HIV-positive, we obtained 86 (32.3%) genetic sequences. Of these, 35 (40.7%) were linked to 1 or more other sequence; however, none of these were identified in first-, second-, or third-degree confidant and sexual networks. Minimal overlap existed between genetic and Facebook ties.
Conclusions. These results suggest that HIV transmissions may have occurred before elicitation of network data; future studies should expand the data collection timeframe to more accurately determine risk networks. Virtual network data, such as Facebook, may be particularly useful in developing one’s risk environment.
Determining how network-level factors influence individual risk of HIV acquisition is vital to preventing disease transmission. Over the past several years, research has moved beyond the examination of how individual-level risk behaviors are associated with the risk of HIV acquisition to examine the role of network-level risk factors. This is because traditional behavioral factors thought to increase the rate of HIV acquisition—such as substance use1 and condomless sex2—have not sufficiently explained observed differences in risk. Instead, more recent research has begun to focus on network-level analyses and how these characteristics may influence the risk of disease. For example, in one recent study, differences in rate of acquisition of HIV or other sexually transmitted infections were found to have been explained in part by network factors.3 Thus, continued focus on individual risk behaviors is likely to have only limited impact on HIV elimination, suggesting a need to examine the role of network-level risk behaviors in HIV acquisition.
Previous research has leveraged several types of networks to examine factors associated with HIV infection, including social,4,5 sexual,6,7 and molecular networks.8,9 Past work utilizing molecular networks has identified HIV molecular clusters and has examined characteristics associated with both cluster membership and size.8–11 Because of their nature, molecular networks provide no information on HIV-negative individuals and it is unclear the extent to which these networks are relevant to sexual transmission. More recent work has examined how molecular and sexual contact networks overlap and interact,11,12 highlighting the need to include network analyses when formulating prevention policies.13 Sexual contact networks also have their limitations and have been shown to suffer from missing data14 and information bias regarding self-reported sexual history and frequency of risk behaviors.15 Furthermore, past research has demonstrated a reluctance among some participants to provide information regarding sensitive behaviors16,17 with some participants exhibiting emotional distress when asked to report on sexual behaviors.18,19 Perhaps most importantly, sexual networks are highly dynamic in younger populations,7 a finding that challenges the abilities of current approaches to categorize them. Put together, molecular networks and observed sexual networks suffer from several biases and limitations and are highly dynamic, making analysis difficult and any proposed interventions based on these data subject to error.
Compared with sexual networks, social networks provide more complete data and often overlap with sexual networks, particularly among smaller communities such as Black men who have sex with men (MSM).20 Although social network data are still likely to contain missing ties between persons, it is less susceptible to some of the biases observed in sexual contact networks. In addition, nonsexual social networks tend to be more stable over time.7 Examining social context, the influence of social context on behavior, and network formation is vital to the study of sexually transmitted infections, including HIV.13 Past research has suggested that a socio-molecular approach to studying infectious diseases may yield new interventions21; however, little work has been done to characterize the relationship among nonsexual, social, and molecular networks. Understanding how these networks overlap and how each enhances the information provided by the other has the potential to inform prevention strategies and may lead to identification of why disparities in HIV acquisition exist.
In this analysis, we characterized an HIV molecular network among young Black MSM in Chicago, Illinois. We then combined these data with confidant, sexual, and Facebook network data from the same cohort to examine potential overlap between the networks to guide our understanding of how phylogenetic analyses can strengthen and be strengthened by existing network elicitation approaches.
METHODS
In brief, uConnect is a longitudinal population-based cohort study22 that was designed to examine network factors associated with HIV risk and transmission among a sample of young Black MSM. uConnect participants spent most of their time on the South Side of Chicago and the adjacent southern suburbs, which represents the largest contiguous Black community area in the United States.23
Eligibility Criteria
Study respondents were eligible to participate if they
self-identified as African American or Black,
were assigned male at birth,
were aged between 16 and 29 years (inclusive),
reported oral or anal sex with a male partner within the past 24 months,
spent the majority of their time on the South Side of Chicago, and
were willing and able to provide informed consent at the time of the study visit.
Interview
Recruitment utilizing respondent-driven sampling and survey follow-up occurred between August 2013 and January 2016. Surveys were conducted across 3 waves, each separated by 9 months. Interviews were conducted using computer-aided personal interviewing with some portions self-administered. The interview itself involved different types of questions and activities: background and socio-demographic questions, questions related to “hook-up” app use (e.g., Grindr, Jack’d), self-administered scales of substance use, and HIV care continuum measures.
As part of the survey at each wave, participants were asked a series of questions to elicit confidants as well as their past sexual partners, allowing for the separate construction of both a sexual and a confidant network. Participants were asked “Thinking back over the past six months, that is since [month], how many people, including men, women, and transgender women have you had sexual activity with, even if only one time” and “So I can ask some follow-up questions, please list the names of the people with whom you discuss things that are important to you,” to elicit number of sexual partners and confidants, respectively. For sexual partners, participants were asked identifying information and a series of questions regarding their relationship and risk behaviors with each partner. Although participants could claim any number of partners, they were asked to name only their 5 most recent sexual partners in the past 6 months, as well as their main partner, if one existed, for a total of up to 6 recent sexual partners. For confidants, participants were asked identifying information and a series of questions regarding their relationship with up to 5 confidants. We also included network data for all respondent-driven sampling ties (hereafter referred to as “referral network”) as these were each described as either sexual partners or friends or family by the participants included in this study.
In addition to the named confidant and sexual partner questions, an application was developed to extract Facebook friend lists from study respondents.24 Because of changes in Facebook privacy policies, this process was completed only during waves 1 and 2 and was limited to those who consented to this process. In addition, we were only able to obtain 1 degree of information regarding the friends of study participants (i.e., not the friends of their friends).
We performed entity resolution25 for each network type among respondents in the uConnect cohort. At each study wave, respondents were asked detailed information regarding their sexual partners and confidants, including name, age, geographic residence, and other sociodemographics, if known. We then matched these data provided by respondents across all waves to identify unobserved ties that may exist as a result of different respondents naming the same individual. Two separate analysts verified the algorithm used to complete the matching process, with all matches confirmed manually; this has been described in detail elsewhere (Britt Skaathun, oral and written communication, January 15, 2017). For the purposes of this analysis, we only included ties between study respondents as these are the only observable ties that have the potential to overlap between networks.
Molecular network inference.
HIV testing was performed at each study visit via dried blood spots collected from each consenting participant. HIV infection status (including acute infection) was determined by fourth-generation HIV immunoassay (ARCHITECT HIV Ag/Ab Combo assay, Abbott, Wiesbaden, Germany), HIV-1 or HIV-2 antibody differentiation (Multispot HIV-1/-2 Rapid Test, Bio-Rad, Hercules, CA), and viral load testing (ReaLTime HIV-1 assay, Abbott, Wiesbaden, Germany) applied to samples eluted from dry blood spots. Given the limit allowable for elution, HIV pol sequences were obtained from all persons whose viral load was greater than or equal to 2000 copies per milliliter. Specific procedures describing extraction of cell-associated HIV DNA from dried blood spots and HIV pol amplicon sequencing, including number of base pairs analyzed and primers used, have been previously described (Amesika Nyaku, oral communication, February 1, 2017). Those with a viral load less than 2000 copies per milliliter were unable to be sequenced because of the lack of amplifiable virus. For participants whose viruses could not be sequenced, we obtained sequences (if available) collected through routine surveillance by the Chicago Department of Public Health (CDPH). All participants whose data were accessed through CDPH provided a release of information to obtain any HIV-related test results. The first sequence available was obtained from each participant.
All available genetic sequences were aligned to the HXB2 reference sequence using MUSCLE (MUltiple Sequence Comparison by Log-Expectation, European Bioinformatics Institute, Hinxton, Cambridgeshire, UK) multiple sequence alignment in the MEGA version 7.0 software package (Molecular Evolutionary Genetics Analysis, State College, PA). Phylogenetic tree analyses were performed by using the neighbor-joining method, with distance calculated by TN93 analysis. Each individual is represented by a node in the network. Nodes were linked together in the network by a potential transmission event (tie) if the genetic distance between those individuals’ pol sequences was less than or equal to 0.015 nucleotide substitutions per site (≤ 1.5%). A cluster was defined as 2 or more persons linked by 1 or more ties. We performed all cluster visualizations with Visone network visualization software (Karlsruhe Insitute of Technology, Karlsruhe, Germany).
All molecular, confidant, and sexual network analyses were limited to only those named partners who were also study respondents. Molecular network formation was limited to the number of individuals for whom we were able to obtain a viral genetic sequence and we included HIV-negative individuals only if confirmed via laboratory testing.
We combined network data in such a way as to logically arrive at one’s likely risk environment. We began with molecular network data, having identified these individuals as transmission cases. Next, we added the named sexual partner data, or probable cases, to the molecular network. Then the named confidants were added to the network as possible cases, followed by overlapping networks (Figure A, available as a supplement to the online version of this article at http://www.ajph.org).
Sensitivity analysis.
Given the large number of Facebook ties, it is possible that any observed overlap between the social and molecular networks may be attributable to chance alone. To assess this, we ran a series of simulations to test any observed overlap between the networks. Each simulation randomly reassigned the ties in the network. The null hypothesis was that there was no association between the genetic and Facebook networks. We performed 10 000 simulations to obtain an estimate of the distribution of overlap between the networks and evaluated the likelihood of the observed percentage overlap between the networks relative to this distribution, yielding a 1-sided simulation P value. Furthermore, we also considered the complete social and sexual network data (Figure B, available as a supplement to the online version of this article at http://www.ajph.org) to investigate whether molecular ties may connect otherwise disparate network components.
RESULTS
The uConnect cohort included 618 participants, of whom 266 (43.0%) were identified as HIV-positive by the end of the study. Among the 266 HIV-positive individuals, 139 had a detectable viral load and, although challenges occurred with collection of the dried blood spots, we successfully sequenced the pol region of the viral genome for 42 (30.2%) of these individuals. We obtained a further 44 viral sequences from CDPH HIV surveillance data for a total of 86 (61.9%) available sequences. We observed no differences in sociodemographics, risk behaviors, or network position among participants who either did or did not have a sequence available nor based on network type. Characteristics of those with genetic sequence data are presented in Table 1 and are stratified by presence in a molecular cluster.
TABLE 1—
Sample Characteristics Stratified by Presence in an HIV Molecular Cluster Among Young Black Men Who Have Sex With Men in Chicago, IL, With an Available Viral Genetic Sequence: uConnect, 2013–2016
| Characteristic | Total, No. (%) or Mean (Range) | Not in a Cluster,a No. (%) or Mean (Range) | In a Cluster,a No. (%) or Mean (Range) | Pb |
| Total | 86 (100) | 55 (64.0) | 31 (36.0) | . . . |
| Demographics | ||||
| Currently insured | 42 (48.8) | 29 (52.7) | 13 (41.9) | .34 |
| Currently a student | 24 (27.9) | 15 (27.3) | 9 (29.0) | .96 |
| Low incomec | 72 (83.7) | 45 (83.3) | 27 (90.0) | .40 |
| Housing instabilityd | 28 (32.6) | 16 (29.1) | 12 (40.0) | .31 |
| Sexual identity | .66 | |||
| Gay | 66 (77.7) | 42 (77.8) | 24 (77.4) | |
| Bisexual | 14 (16.5) | 8 (14.8) | 6 (19.4) | |
| Straight or other | 5 (5.9) | 4 (7.4) | 1 (3.2) | |
| Years since HIV diagnosis | 2.69 (0.75–7) | 3.11 (0.75–7) | 2.32 (0.75–6) | .22 |
| Mental healthe: depressive symptoms | 11 (12.8) | 9 (16.4) | 2 (6.5) | .19 |
| Risk behaviorsd | ||||
| Condomless sex | 49 (57.0) | 33 (60.0) | 16 (51.6) | .45 |
| Group sex | 22 (25.6) | 16 (29.1) | 6 (20.0) | .36 |
| Grindr or Jack’d profile | 36 (41.9) | 40 (78.4) | 16 (45.7) | .71 |
| Grindr or Jack’d usage | .76 | |||
| Never | 50 (58.1) | 31 (60.8) | 19 (54.3) | |
| Use ≤ 2 times per week | 10 (11.6) | 5 (9.8) | 5 (14.3) | |
| Use > 2 times per week | 26 (30.2) | 15 (29.4) | 11 (31.4) | |
| Drug used | ||||
| Marijuanaf | .96 | |||
| Never | 21 (24.4) | 13 (23.6) | 8 (25.8) | |
| Intermittent | 30 (34.9) | 19 (34.6) | 11 (35.5) | |
| Heavy | 35 (40.7) | 23 (41.8) | 12 (38.7) | |
| Other substance used,g | 23 (26.7) | 16 (29.1) | 7 (22.6) | .51 |
| Network degreeh | ||||
| Molecular network | 1.6 (1–9) | 0 (0) | 1.6 (1–9) | . . . |
| Confidant network | 4.0 (1–9) | 4.2 (1–9) | 3.6 (1–7) | .08 |
| Sexual network | 6.4 (1–15) | 6.6 (1–15) | 6.1 (1–14) | .24 |
| Facebook network | 33.6 (2–98) | 34.7 (2–98) | 32.0 (2–83) | .68 |
Note. The sample size was n = 86.
A cluster was defined as 2 or more persons whose pol sequences were ≤ 1.5% genetically distant.
Using χ2 analysis.
Defined as < $20 000 per year.
In the past 12 months.
Using the Brief Symptom Inventory 18.
Intermittent use is defined as marijuana use less than and including weekly use; heavy use is defined as at least once per day.
Includes the use of ecstasy/molly/E, poppers, cocaine/crack, heroin, psychedelics, methamphetamines, off-label prescription drugs.
Degree is defined as the number of ties each individual had to other study participants in the network (e.g., 1 individual tied to 3 other individuals would have a value of 3).
Most participants had low income (n = 72; 83.7%), self-identified as gay (n = 59; 68.7%), and reported condomless sex in the past 12 months (n = 49; 57.0%). Forty-one percent of individuals had a profile on “hook-up” apps, such as Grindr or Jack’d, of whom 30.2% used the services more than 2 times per week. We also observed an overall mean time since HIV diagnosis of 2.69 years with no significant difference found among those who were and were not in a cluster. Of those with a viral genetic sequence, 35 (40.6%) were present in a molecular cluster, resulting in 55 molecular ties between participants (Figure A, see green lines). Furthermore, on average, 4.8 sexual partners were claimed in the past 6 months (range: 0–35; SD = 6.5) with 13 (15.1%) of those with available sequences claiming more than 6 sexual partners in the past 6 months (mean = 17.9; range: 7–35; SD = 8.0).
The observed pairwise genetic distance ranged from 0.1% to 14.5% (0.001–0.145 nucleotide substitutions per site; Figure C, part A, available as a supplement to the online version of this article at http://www.ajph.org). Table 2 depicts ties that are overlapping, or lack thereof, between each of the network types (i.e., molecular, social, sexual, and referral networks) within the genetic distance threshold. No overlapping ties existed between the molecular network and first-degree social, sexual, or referral networks. We also observed an absence of tie overlap between the molecular network and second- and third-degree confidant network as well as the molecular network and second- and third-degree sexual network. In addition, we observed no overlap between networks when we considered the full social or sexual networks of all 618 participants. Furthermore, when we examined potential overlap beyond the genetic distance threshold (i.e., > 0.015 nucleotide substitutions per site), we observed 12 molecular and social network ties to have overlapped (Figure C, part B, available as a supplement to the online version of this article at http://www.ajph.org) although we found no ties to overlap between the molecular and sexual networks. We did, however, find rather consistent overlap of 45% to 50% among the social, sexual, referral, and Facebook networks. Out of 12 individuals with both molecular and Facebook data, we found that 3 (25.0%) of the ties overlapped between the networks. In the sensitivity analysis, we found that, under the null hypothesis of no association between the networks, the mean number of overlapping ties was 1.16.
TABLE 2—
Total Number of Overlapping Ties Between Each Type of Network Among Young Black Men Who Have Sex With Men in Chicago, IL, With an Available HIV Genetic Sequence: uConnect, 2013–2016
| Network Type,a No. (%) |
|||||
| Molecular (n = 12) | Social (n = 719) | Sexual (n = 403) | Referral (n = 716) | Facebook (n = 8296) | |
| Molecular | . . . | . . . | . . . | . . . | . . . |
| Social | 0 (0.0) | . . . | . . . | . . . | . . . |
| Sexual | 0 (0.0) | 309 (43.0) | . . . | . . . | . . . |
| Referral | 0 (0.0) | 321 (44.6) | 121 (30.0) | . . . | . . . |
| 3 (25.0) | 371 (51.6) | 199 (49.3) | 284 (39.7) | . . . | |
Note. The sample size was n = 86.
Network ties include only those to other study participants.
Table A (available as a supplement to the online version of this article at http://www.ajph.org) presents network metrics for each network type with the molecular and Facebook networks having the highest, and similar, density (0.092 and 0.099, respectively), the sexual network having a high average distance (mean = 11.63; SD = 6.77), and the Facebook network having high transitive triads (n = 73 948).
DISCUSSION
In this study, we present new information regarding the extent to which several types of social networks overlap with HIV molecular networks. First, we found that molecular ties have limited overlap with both named confidant and named sexual partners. Second, we found that slightly less than half of the time molecular ties overlap with Facebook friend ties. These results suggest that a participant’s risk network may not be limited to just the sexual connections within the past 6 months, but may extend beyond these observed ties. Combining social and molecular network data in a socio-molecular approach may more accurately reflect one’s true risk environment and provide information for public health departments to engage individuals not within molecular clusters.
Developing strategies for combining network data in a socio-molecular approach may yield novel strategies for targeting HIV prevention, especially among populations most vulnerable to HIV. Young Black MSM exhibit fewer risk-taking behaviors and use fewer illicit substances than do their White counterparts, yet they have higher rates of HIV transmission,26 making social network interventions particularly relevant as opposed to traditional behavioral interventions. In addition, past research has shown that role shifting between social and sexual ties is common,20 particularly among MSM. In this study, we observed overlap between the social and sexual networks; however, we observed none between the molecular and social or sexual networks. In addition, we observed that, in the complete social and sexual networks of all 618 participants, the molecular ties represented wholly independent ties, connecting otherwise unconnected individuals. Past research has found an overlap between named partners and molecular links; however, this work included older populations, heterosexuals, and people who inject drugs who may have more stable sexual or injecting networks than young MSM.12
The difference observed in our study may be a result of obtaining viral sequences only for a third of those who are HIV-positive in our cohort. In addition, limited overlap may also be attributable to having too short of a framing window (e.g., partners in the past 6 months), especially given that the mean time since diagnosis was more than 2 years ago. Furthermore, in younger populations with a high number of sexual partners, it may be difficult to elicit the transmission chain with name generation. Future research should assess how survey questions can be better tailored to specific populations to develop a more complete picture of one’s network.
We performed several data checks to ensure accuracy of the results. We found a normal distribution of distances (Figure C, part A), similar to those of previous studies.27 In addition, we did find that some named confidants or sexual partners had overlapping ties with our genetic data; however, these were outside our threshold genetic distance. The minimum observed genetic distance for overlapping molecular and social or sexual networks was 6.0% (0.060 nucleotide substitutions per site), similar to past studies,27 confirming that our threshold did not arbitrarily remove any potential overlap (Figure C, part B). Furthermore, the high rate of use of Grindr and Jack’d by this population may suggest that participants are seeking more anonymous sex than previously thought and that the sexual networks of participants expand well beyond what was captured in this study. Past research has shown that, compared with those who do not have a profile on either Grindr or Jack’d, those who do are significantly more likely to be diagnosed with HIV.28
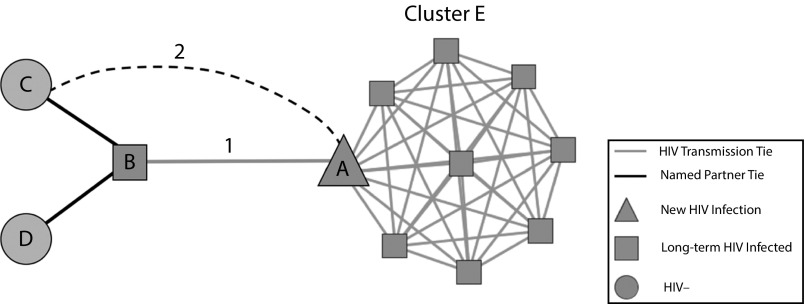
It may also be possible to use a socio-molecular network approach to proactively prevent local HIV molecular clusters from growing by incorporating molecular network data into current partner-tracing efforts (Figure 1). Figure 1 depicts an example of the expansion of current partner services to include molecular network analyses based upon our findings. Currently, when a new HIV infection (node B) is identified by local public health departments, disease intervention specialist officers request that the newly infected individual name their current and past sexual partners. These named partners are then contacted and advised to seek HIV testing and counseling. Here, we are suggesting an additional step to partner-tracing efforts, “network tracing.” In this instance, the newly infected individual’s (node A) viral sequence can be analyzed by using phylogenetic methodologies, yielding a molecular cluster of potentially unnamed or unknown partners (tie 1 and cluster E). Given their close proximity to a molecular cluster, previously named partners (nodes C and D) of the long-term HIV-infected individual (node B) could then be recontacted and advised to again seek HIV testing and counseling, or prioritized to begin pre-exposure prophylaxis use. This method is useful because it is feasible that nodes C and D will come into contact with the molecular clusters given that tie 1 is supported 25.0% of the time by Facebook data and tie 2 is supported 10.0% of the time.
FIGURE 1—
Suggested “Network Tracing” Model for Proactively Incorporating Molecular Network Data Into Current Partner-Tracing Methods in Local Public Health Departments
Note. In this method, molecular ties would be used to identify previously unknown links between HIV-positive individuals. Previously named partners could then be recontacted to advise further HIV testing or to encourage use of pre-exposure prophylaxis or postexposure prophylaxis to prevent transmission clusters from growing.
The data presented in this study suggest that there is potential for the individuals from one’s social network to enter, and grow, HIV molecular clusters. To prevent growth of local molecular clusters, next-generation partner-tracing efforts could be enhanced in local health departments and their delegate agencies to identify individuals for additional prevention services with minimal additional effort and using data that are widely becoming available.29 In addition, while current efforts aimed at getting all at-risk individuals on pre-exposure prophylaxis should be continued, the methods presented here may allow public health departments to determine those who should be most prioritized for pre-exposure prophylaxis engagement. In this manner, HIV-negative individuals who are found to be in close proximity to large molecular clusters can be prioritized. Future research should assess whether the network-tracing method is comparable in scope and feasibility to traditional partner-tracing efforts.
Our data should be viewed in the context of their limitations. First, we were able to obtain viral sequences for only a third of those who were diagnosed with HIV; thus, the molecular network is likely incomplete, yet there were no differences between those who had a sequence and those who did not. Related to this, there is potential for incomplete elicitation of the social and sexual networks using the methods in this analysis; therefore, we may not be observing overlap when it may, in fact, exist. We were also unable to determine whether our molecular ties were indirect or direct, nor were we able to determine the direction of transmission. In addition, we only have survey data on the named confidants and sexual partners who are also study respondents and not on the other partners they named; thus, we are not viewing a complete risk network.
Even with these limitations, we were able to draw meaningful conclusions from our data. We have shown that it is necessary to incorporate many sources of information to better illuminate an individual’s risk environment. We have also shown that social media networks, such as Facebook, may be useful in developing a more complete picture of one’s risk environment. Finally, we presented a case example of how social network data can enhance molecular network data for next-generation partner services approaches—“network services.”
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants R21MH098768, R01DA033875, and R01DA039934. The dried blood spot assay development and validation was supported by NIH grants P30-AI-027757 and UM1-AI-68636 and -06701.
We would like to thank all study participants for the time and effort required to recruit their network members and take part in the interview. We would like to thank Audrey Wong, Jose Ortega, Eleanor Espinosa, Carol Gallardo, Corey Scherrer, and Glenda Daza for laboratory technical support.
Note. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The sponsor had no involvement in the conduct of the research or the preparation of the article.
HUMAN PARTICIPANT PROTECTION
This study was reviewed and approved by University of Chicago institutional review board.
Footnotes
See also Dennis, p. 1443.
REFERENCES
- 1.Millett GA, Flores SA, Peterson JL, Bakeman R. Explaining disparities in HIV infection among Black and White men who have sex with men: a meta-analysis of HIV risk behaviors. AIDS. 2007;21(15):2083–2091. doi: 10.1097/QAD.0b013e3282e9a64b. [DOI] [PubMed] [Google Scholar]
- 2.Eaton LA, Kalichman SC, Cherry C. Sexual partner selection and HIV risk reduction among Black and White men who have sex with men. Am J Public Health. 2010;100(3):503–509. doi: 10.2105/AJPH.2008.155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laumann EO, Youm Y. Racial/ethnic group differences in the prevalence of sexually transmitted diseases in the United States: a network explanation. Sex Transm Dis. 1999;26(5):250–261. doi: 10.1097/00007435-199905000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Schneider J, Michaels S, Bouris A. Family network proportion and HIV risk among Black men who have sex with men. J Acquir Immune Defic Syndr. 2012;61(5):627–635. doi: 10.1097/QAI.0b013e318270d3cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider J, Cornwell B, Jonas A et al. Network dynamics of HIV risk and prevention in a population-based cohort of young Black men who have sex with men. Netw Sci. 2017;5(3):1–29. [Google Scholar]
- 6.Amirkhanian YA. Social networks, sexual networks and HIV risk in men who have sex with men. Curr HIV/AIDS Rep. 2014;11(1):81–92. doi: 10.1007/s11904-013-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider J, Cornwell B, Jonas A et al. Network dynamics and HIV risk and prevention in a population-based cohort of young Black men who have sex with men. Netw Sci. 2017;5(3):1–29. [Google Scholar]
- 8.Lubelchek RJ, Hoehnen SC, Hotton AL, Kincaid SL, Barker DE, French AL. Transmission clustering among newly diagnosed HIV patients in Chicago, 2008 to 2011: using phylogenetics to expand knowledge of regional HIV transmission patterns. J Acquir Immune Defic Syndr. 2015;68(1):46–54. doi: 10.1097/QAI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan E, Oster A, Townsell S, Peace D, Benbow N, Schneider JA. Movement of HIV-1 infection through transmission networks of younger persons in Chicago, Illinois. Public Health Rep. doi: 10.1177/0033354916679988. 2016;2017;132(1):48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oster AM, Pieniazek D, Zhang X et al. Demographic but not geographic insularity in HIV transmission among young Black MSM. AIDS. 2011;25(17):2157–2165. doi: 10.1097/QAD.0b013e32834bfde9. [DOI] [PubMed] [Google Scholar]
- 11.Little SJ, Kosakovsky Pond SL, Anderson CM et al. Using HIV networks to inform real time prevention interventions. PLoS One. 2014;9(6):e98443. doi: 10.1371/journal.pone.0098443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wertheim JO, Kosakovsky Pond SL, Forgione LA et al. Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog. 2017;13(1):e1006000. doi: 10.1371/journal.ppat.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adimora AA, Schoenbach VJ. Social context, sexual networks, and racial disparities in rates of sexually transmitted infections. J Infect Dis. 2005;191(suppl 1):S115–S122. doi: 10.1086/425280. [DOI] [PubMed] [Google Scholar]
- 14.Ghani AC, Donnelly CA, Garnett GP. Sampling biases and missing data in explorations of sexual partner networks for the spread of sexually transmitted diseases. Stat Med. 1998;17(18):2079–2097. doi: 10.1002/(sici)1097-0258(19980930)17:18<2079::aid-sim902>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 15.Catania JA, Gibson DR, Chitwood DD, Coates TJ. Methodological problems in AIDS behavioral research: influences on measurement error and participation bias in studies of sexual behavior. Psychol Bull. 1990;108(3):339–362. doi: 10.1037/0033-2909.108.3.339. [DOI] [PubMed] [Google Scholar]
- 16.Des Jarlais DC, Paone D, Milliken J et al. Audio-computer interviewing to measure risk behaviour for HIV among injecting drug users: a quasi-randomised trial. Lancet. 1999;353(9165):1657–1661. doi: 10.1016/s0140-6736(98)07026-3. [DOI] [PubMed] [Google Scholar]
- 17.Kurth AE, Martin DP, Golden MR et al. A comparison between audio computer-assisted self-interviews and clinician interviews for obtaining the sexual history. Sex Transm Dis. 2004;31(12):719–726. doi: 10.1097/01.olq.0000145855.36181.13. [DOI] [PubMed] [Google Scholar]
- 18.Catania JA. A framework for conceptualizing reporting bias and its antecedents in interviews assessing human sexuality. J Sex Res. 1999;36(1):25–38. [Google Scholar]
- 19.Rossi PH, Wright JD, Anderson AB. Handbook of Survey Research. Vol 14. Bingley, UK: Emerald Publishing Group; 1990. [Google Scholar]
- 20.Tieu H-V, Liu T-Y, Hussen S et al. Sexual networks and HIV risk among Black men who have sex with men in 6 U.S. cities. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0134085. e0134085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasylyeva TI, Friedman SR, Paraskevis D, Magiorkinis G. Integrating molecular epidemiology and social network analysis to study infectious diseases: towards a socio-molecular era for public health. Infect Genet Evol. 2016;46:248–255. doi: 10.1016/j.meegid.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khanna AS, Michaels S, Skaathun B et al. Preexposure prophylaxis awareness and use in a population-based sample of young Black men who have sex with men. JAMA Intern Med. 2016;176(1):136–138. doi: 10.1001/jamainternmed.2015.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. 2005–2009 American Community Survey 5-Year Estimates. Suitland, MD: US Census Bureau; 2011.
- 24.Khanna AS, Schumm P, Schneider JA. Facebook network structure and awareness of preexposure prophylaxis among young men who have sex with men. Ann Epidemiol. 2017;27(3):176–180. doi: 10.1016/j.annepidem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Getoor L, Machanavajjhala A. Entity resolution for big data. Oral presentation at: 19th Association for Computing Machinery’s Special Interest Group on Knowledge Discovery and Data Mining; August 11, 2013; Chicago, IL.
- 26.Tobin KE, Kuramoto SJ, Davey-Rothwell MA, Latkin CA. The STEP into Action study: a peer-based, personal risk network-focused HIV prevention intervention with injection drug users in Baltimore, Maryland. Addiction. 2011;106(2):366–375. doi: 10.1111/j.1360-0443.2010.03146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DM, May SJ, Tweeten S et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23(2):225–232. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goedel WC, Duncan DT. Geosocial-networking app usage patterns of gay, bisexual, and other men who have sex with men: survey among users of Grindr, a mobile dating app. JMIR Public Health Surveill. 2015;1(1):e4. doi: 10.2196/publichealth.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention. Detecting, investigating, and responding to HIV transmission clusters. 2017. Available at: http://test.datamonkey.org/hivtrace. Accessed February 1, 2017.