Abstract
The altitudinal effects on the distributions of phyllosphere fungal assemblages in conspecific plants remain poorly elucidated. To address this, phyllosphere fungal communities associated with Mussaenda shikokiana were investigated at four sites across a 350 m elevation gradient in a subtropical forest by employing Illumina metabarcoding of the fungal internal transcribed spacer 2 (ITS2) region. Our results demonstrated that phyllosphere fungal assemblages with a single host possessed high taxonomic diversity and multiple trophic guilds. OTU richness was significantly influenced by elevation. The elevation gradient also entailed distinct shifts in the community composition of phyllosphere fungi, which was significantly related to geographical distance and mean annual temperature (MAT). Additionally, comparison of phyllosphere fungal networks showed reduced connectivity with increasing elevation. Our data provide insights on the distribution and interactions of the phyllosphere fungal community associated with a single host along a short elevation gradient.
Keywords: Phyllosphere fungi, Biodiversity, Elevation gradient, Mussaenda shikokiana, High-throughput sequencing
Introduction
The fungal kingdom represents one of the largest and most diverse eukaryotic lineages with estimates of around 5.1 million extant species (Blackwell, 2011; Truong et al., 2017). Fungi function as important ecosystem components that govern global carbon cycling, plant pathology, development and reproduction (Tedersoo et al., 2014). Although fungi directly impact ecosystem functions, only a fraction of the total fungal species have been described (Truong et al., 2017). Therefore, it is essential to obtain adequate taxonomic and ecological knowledge of members of the fungal lineage to advance basic ecology and to recognize and predict the future consequences of environmental changes (Bryant et al., 2008; Geml et al., 2014).
The distribution pattern of species communities along altitudinal gradients is a fundamental theme of ecology and biogeography, mainly because altitudinal gradients are often characterized by different climatic conditions and complex changes in abiotic and/or biotic factors over short geographic distances (Bryant et al., 2008; Ren et al., 2018; Yao et al., 2017). Although there is a long history of studies that have been conducted on the elevational patterns of vascular plants and animals (Bryant et al., 2008), similar studies have only recently been made possible for microbial communities with the advent of culture-independent molecular techniques (Datlof et al., 2017; Lanzen et al., 2016; Wainwright et al., 2017; Zhang et al., 2018c). Several studies of the elevation patterns of fungal communities showed no consistent conclusions. In a tropical montane cloud forest, Looby, Maltz & Treseder (2016) documented increasing alpha-diversity of soil fungi and a significant shift in the community composition with increasing elevation. A similar elevational pattern was observed in soil fungal communities in the Italian Alps (Siles & Margesin, 2016). However, opposite patterns of species richness were found for ectomycorrhizal fungi in the Hyrcanian forests of northern Iran (Bahram et al., 2012) and for arbuscular mycorrhizal fungi in a Tibetan alpine grassland (Liu et al., 2015). In addition, a unimodal pattern, with a mid-elevation peak in diversity, was detected in ectomycorrhizal fungal communities on the northwest slopes of the Fuji mountains (Miyamoto et al., 2014). In a study conducted in the Cairn Gorm mountains in Scotland, Jarvis, Woodward & Taylor (2015) found that elevation did not influence ectomycorrhizal fungal richness of Scots pine, but strongly influenced community composition. Along gradients in the Changbai mountains in China, neither the species richness nor community composition of soil fungi were significantly correlated with elevation (Shen et al., 2014). These different distribution patterns may partly be explained by difference in environmental variables, including edaphic qualities (Geml et al., 2014; Jarvis, Woodward & Taylor, 2015; Shen et al., 2014; Siles & Margesin, 2016), mean annual temperature and precipitation (Bahram et al., 2012). However, most studies focused only on soil or root associated fungal assemblages, and few studies have investigated the altitudinal effects on the distribution of phyllosphere fungi (Cordier et al., 2012).
In natural environments, microorganisms usually live within complex ecological networks rather than existing in isolation (Faust & Raes, 2012). Co-occurrence patterns, which describe how species within a community coexist, are omnipresent and are crucial for understanding the structure of microbial communities and for revealing the potential interactions between microbial taxa (Kay et al., 2018; Ma et al., 2016). Network analysis, which can identify and describe taxon co-occurrence patterns in large datasets, has proven to be a powerful method that has been recently applied for characterization of bacterial communities across a wide variety of habitats (Baldassano & Bassett, 2016); including soil (Barberan et al., 2012; Jiao et al., 2016), ocean (Milici et al., 2016), gut (Liggenstoffer et al., 2010; Zhang et al., 2018b; Zhang et al., 2014), activated sludge (Ju et al., 2014), and mammalian milk (Li et al., 2017). These studies have uncovered previously unknown co-occurrence patterns, thereby offering new insight into the distribution of species associations. Several processes may be responsible for inter-taxa associations including niche differentiation, shared environmental requirements and phylogenetic relatedness (Kay et al., 2018; Zhang et al., 2018a). Relative to bacterial communities, there have been far fewer studies conducted using high throughput sequencing data to detect the co-occurrence patterns of fungal communities. By analyzing community composition of arbuscular mycorrhizal fungi across Europe, Bouffaud et al. (2016) suggested that closely-related taxa are more likely to co-occur than distantly-related species. Moreover, He et al. (2017) found greater linkage in soil fungal networks when the seasonal precipitation was manipulated, suggesting that inter-species interactions may be intensified to adapt to differences in water availability. In addition, a study conducted on the networks of fungal communities in soils where there has been a prolonged potato monoculture demonstrated scale-free, small world and modular properties (Lu et al., 2013). Scale-free network usually share an important property: a few fungi have a tremendous number of connections to other fungi, whereas most fungi have just a handful. Small world network usually showed small average path length and high clustering coefficient, which indicated that the fungus in this community has close interaction. Modularity is one measure of network structure. Networks with high modularity have dense connections between the nodes within modules but sparse connections between nodes in different modules.
Phyllosphere fungi, which occupy leaves superficially or intracellularly without causing visible symptoms, are important factors affecting plant health and ecosystem function (Drake, White Jr & Belanger, 2018; Gomes et al., 2018; Jumpponen & Jones, 2009). For example, Phyllosphere fungi can protect host plants from pathogen damage (Arnold et al., 2003; Mejia et al., 2008), enhance tolerances against herbivores (Albrectsen et al., 2010; Estrada, Wcislo & Van Bael, 2013; Hartley & Gange, 2009), and play an essential role in the initial decomposition of leaves following senescence (Guerreiro et al., 2018; Kembel & Mueller, 2014; Voriskova & Baldrian, 2013). Although the elevational distribution patterns of phyllosphere fungal communities have been previously described in the Gave Valley of the French Pyrenees (Cordier et al., 2012), such an examination of the elevational co-occurrence patterns of phyllosphere fungi in a subtropical climate has not yet been performed.
Mussaenda shikokiana Makino is a mostly autogamous plant species that is widely distributed throughout the warmer parts of China and Japan (Chen, Luo & Zhang, 2013). The floral diversity and pollination biology of M. shikokiana has been described in a previous study (Chen, Luo & Zhang, 2013), but little is known about the community structure of its phyllosphere fungal assemblages. To assess this, we used Illumina MiSeq sequencing data of fungal ribosomal internal transcribed spacer 2 (ITS2) to analyze the changes in the composition and inter-taxa interactions of the phyllosphere fungal communities inhabiting M. shikokiana along an altitudinal gradient in a subtropical forest. Specifically, we tried to address the following questions: (1) How does the richness and community composition of the phyllosphere fungal assemblage change in response to the elevation gradient? (2) Does the network topology of the fungal community shift significantly with elevation?
Materials and Methods
Study site and sampling
This study was conducted in the Laoshan Nature Reserve of Jingxiu, which is located in Guangxi province, China. This region possesses a subtropical moist climate and abundant vegetation. The annual mean rainfall is 1,824 mm, and the mean annual temperature is 17.0 °C with the lowest and highest temperatures in January (13.8 °C) and July (21.7 °C), respectively. Here, M. shikokiana, an erect shrub that possesses yellow hermaphrodite flowers and thinly papery leaves, occurs at the southern margin of its range at a mean altitude of 1000 m above sea level (Chen, Luo & Zhang, 2013; Chen, Luo & Zhang, 2014). The leaves of M. shikokiana contains several biologically active compounds that significantly stimulate immune activity and has been used as a Chinese folk medicine (Takeda, Nishimura & Inouye, 1977). Sampling was conducted at four different altitudinal stands (838 m, 934 m, 1,078 m and 1,185 m), each of which contained a high proportion of this species. Mean annual temperature (MAT) and mean annual precipitation (MAP) for each site were obtained from the WorldClim global climate data set with a resolution of 30 s (http://worldclim.org/version2; Table S1). At each elevation, we selected five individuals (>10 m apart from each other) that were in the fruiting stage (September; 2014). Nine mature and asymptomatic leaves from the middle of three current-year shoots were randomly collected from each individual plant, and immediately placed in sterile plastic bags containing sufficient silica gel for quick drying. The samples were then brought back to the laboratory and stored at −80 °C until DNA extraction.
Illumina MiSeq sequencing preparation
Homogenization and mechanical cell disruption of the freeze-dried samples were carried out by grinding with liquid nitrogen using sterilized mortars and pestles. The resulting powder was then transferred to tubes with CTAB (2% CTAB, 100 mM Tris–HCl, 1.4 M NaCl, 20 mM EDTA, pH 8.0) extraction buffer preheated to 68 °C. DNA extraction was performed following the protocol modified by Cota-Sanchez, ReMarchuk & Ubayasena (2006). Briefly, an equal volume of chloroform–isoamylol mixture was added to each tube and mixed thoroughly to form an emulsion. The mixture was spun at 13,000 g for 12 min and the aqueous phase was removed into a fresh centrifuge tube. The aqueous phase containing DNA was re-extracted using chloroform:isoamyl alcohol (24:1) until no interface was visible. DNA was then precipitated with an equal volume of isopropanol. The resulting genomic DNA was precipitated at 10,000 g for 3 min and the DNA pellet was washed with 70% ethanol three times. Finally, the DNA pellet was resuspended in 100 µL TE buffer. A NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to quantify the extracted DNA.
To amplify the fungal ITS2 region of the nuclear ribosomal repeat unit, a semi-nested PCR approach was employed. Initial amplification of the entire ITS region was accomplished with conventional primers ITS1F (CTTGGTCATTTAGAGGAAGTAA) (Gardes & Bruns, 1993) and ITS4 (TCCTCCGCTTATTGATATGC) (White et al., 1990). The first round of amplification was carried out in 25 µL reaction volumes containing final concentrations of 0.2 mM dNTP, 0.4 µM of each primer, 2.5 mM MgSO4, 5 ng template DNA and 1 unit KOD-Plus-Neo DNA polymerase (TOYOBO, Osaka, Japan). Thermal cycling conditions for this amplification were as follows: 30 cycles of 94 °C (1 min), 56 °C (50 s), and 68 °C (1 min) after an initial denaturation at 94 °C (4 min), and followed by a final extension at 68 °C for 10 min. The PCR products were diluted 50 times and 1 µL of each dilution was used as the template for the second amplification of the fungal ITS2 region. The primer fITS7 (GTGARTCATCGAATCTTTG) (Ihrmark et al., 2012) and reverse primer ITS4 extended with a unique sample identification barcode were used for the second stage of PCR reaction, which was performed under the same amplification conditions. Negative PCR controls (in which the template DNA was replaced with sterile water) were used to evaluate whether there was contamination during the entire extraction and PCR process. Each sample was amplified in triplicate to minimize PCR biases and the three replicate products were mixed in equal volumes into a general sample that was purified by using an Ultra-Clean PCR cleanup kit (Mo Bio, Carlsbad, CA, USA). The samples were pooled with an equal molar amount (100 ng) from each sample and adjusted to 10 ng µl−1. The library was applied to an Illumina MiSeq PE250 platform for sequencing using the paired end ( 2 × 250 bp) option.
Bioinformatics workflow
The trimming, sorting and quality filtering of reads were processed using the QIIME Pipeline (Caporaso et al., 2010). All sequences were demultiplexed into samples based on unique identification barcodes. FLASH-1.2.8 software (Magoc & Salzberg, 2011) was used to combine overlapping paired-end reads. The ITS2 region from the remaining sequences was identified and extracted using the fungal ITSx software (Bengtsson-Palme et al., 2013). Chimeras were detected and eliminated using the UCHIME program (Edgar et al., 2011) referenced with the UNITE database ( https://unite.ut.ee/; (Koljalg et al., 2013). Non-chimera sequences that passed the filtering processes were binned into operational taxonomic units (OTUs) at a 97% similarity cutoff using the UPARSE pipeline (Edgar et al., 2011). All singletons OTU were excluded from the dataset. Subsequently, the taxonomic identity of each OTU was determined based on BLAST search against the UNITE reference database, and a BLAST e-value <1e−was a criterion for matching. Further, OTUs occurring in only one sample and OTUs with <10 reads were removed because those rare sequences may have been derived from contaminations or PCR/sequencing errors (Toju, Tanabe & Ishii, 2016). For downstream analyses, each sample was rarefied to the minimum sequencing depth by randomly selecting subsets of reads implemented in the MOTHUR program (Schloss et al., 2009).
Community analyses
To evaluate the comprehensiveness of the sampling strategy, the taxa accumulation curves for each site were calculated by employing the “specaccum” function in the vegan R package (Harrison et al., 2016; Oksanen et al., 2016). Vectors representing horizontal spatial structure were obtained by an analysis of principal coordinates of neighbor matrices (PCNM) (Borcard & Legendre, 2002) based on the geographical coordinates using the “PCNM” package (Legendre et al., 2010). Linear regression models were used to test the relationship between fungal OTU richness and elevation.
To identify the effects of elevation, PCNM vectors, and climate factors on phyllosphere fungal OTU richness, we first constructed generalized linear mixed models (GLMM) included elevation site as a random effect. However, we found that random factors explaining no variation in the data, so we switched to construct generalized linear models (GLM). These models were then examined by analysis of variance (ANOVA) tests. Moreover, the effects of those factors on fungal community structure was assessed by permutational multivariate analysis of variance (PERMANOVA) with site as the random effect using the vegan R package (9999 random permutations). Non-metric multidimensional scaling (NMDS) was performed with the Bray–Curtis distance based on a Hellinger-transformed matrix. Subsequently, the “envfit” function of the vegan R package was used to fit the elevation variable into the NMDS plot. Altitudinal distances, geographical distance and Bray-Curtis dissimilarity matrices were analyzed for correlations to evaluate the distance-decay rates, and Mantel tests were employed to assess the significance of the distance-decay regressions from zero (9999 random permutations) (Oono, Rasmussen & Lefevre, 2017). The appropriate trophic categories (symbiotroph, pathotroph and saprotroph) of each OTU were inferred by searching against the FUNGuild data base (Nguyen et al., 2016b).
To further investigate the phyllosphere fungal taxonomic distribution, significant taxonomic differences between the four elevation sites were identified using Linear Discriminant Analysis (LDA) effect size (LEfSe) (Segata et al., 2011), which introduces the non-parametric factorial Kruskal–Wallis (KW) sum-rank test (α = 0.05) to identify taxa having significant differential abundances between four elevation stands (employing one-against-all comparisons) (Bokulich et al., 2014). A logarithmic LDA score of 3.0 was set as the threshold for discriminative features.
To identify the indicator OTUs for each elevation site, LEfSe analysis and Legendre’s INDVAL analysis were combined as per Hubbard et al. (2018). Legendre’s INDVAL procedure was performed in R using the “labdsv” package (Sapkota et al., 2015). The fungal OTUs with an indicator value >0.25 and a significant P values (P < 0.05) were considered indicator species (Dufrene & Legendre, 1997; Sapkota et al., 2015). However, OTUs with low abundance hold little indicator information (Rime et al., 2015). To avoid random effects caused by rare OTUs, only those with >500 reads were defined as true key players (Eusemann et al., 2016).
Network analyses
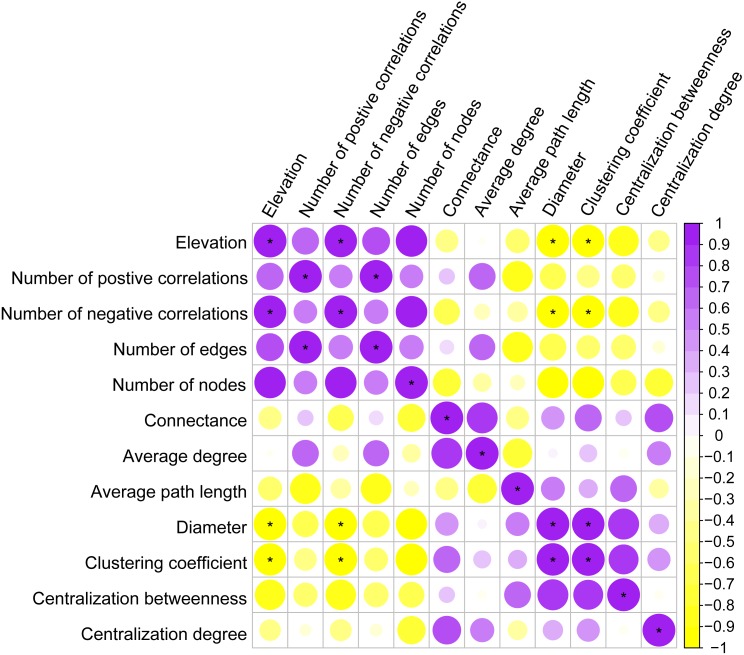
To remove poorly represented OTUs and reduce network complexity, only abundant OTUs with a proportion of the total sequences over 0.02% were kept in the OTU table. The co-occurrence network was inferred based on the Spearman’s Rho between the pairwise OTUs matrix constructed by the psych R package (Revelle, 2017). The P-values for multiple testing were calculated by using the false discovery rate (FDR) controlling procedure as per Benjamini & Hochberg (1995). A valid co-occurrence event was considered to be robust if the correlation coefficient r >—0.6— and if it was statistically significant at P < 0.05 (Widder et al., 2014). Network images for each elevation site were generated with Gephi (Bastian, Heymann & Jacomy, 2009). To describe the topology properties of the co-occurrence networks, the feature set (number of positive correlations, number of negative correlations, number of vertices, average degree, average path length, centralization betweenness, diameter, centralization degree and clustering coefficient) was calculated with the igraph R package (Csardi, 2006). Network diameter is a measure of how integrated the network is (Proulx, Promislow & Phillips, 2005). A network that has multiple fractured components would have a smaller average diameter (Lawes et al., 2017). Spearman correlations and significance tests (level at p < 0.05) between those properties were calculated and plotted using the corrplot R package (Taiyun & Viliam, 2017).
Results
Data characteristics
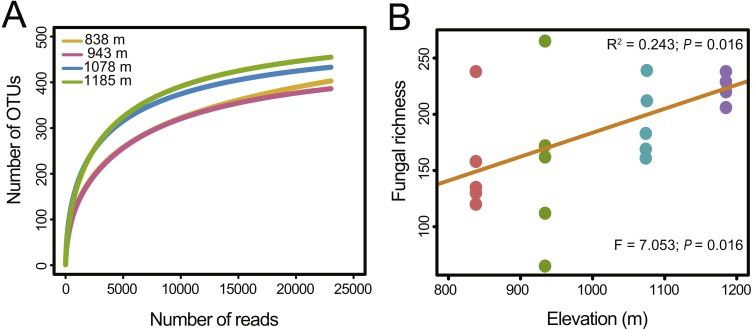
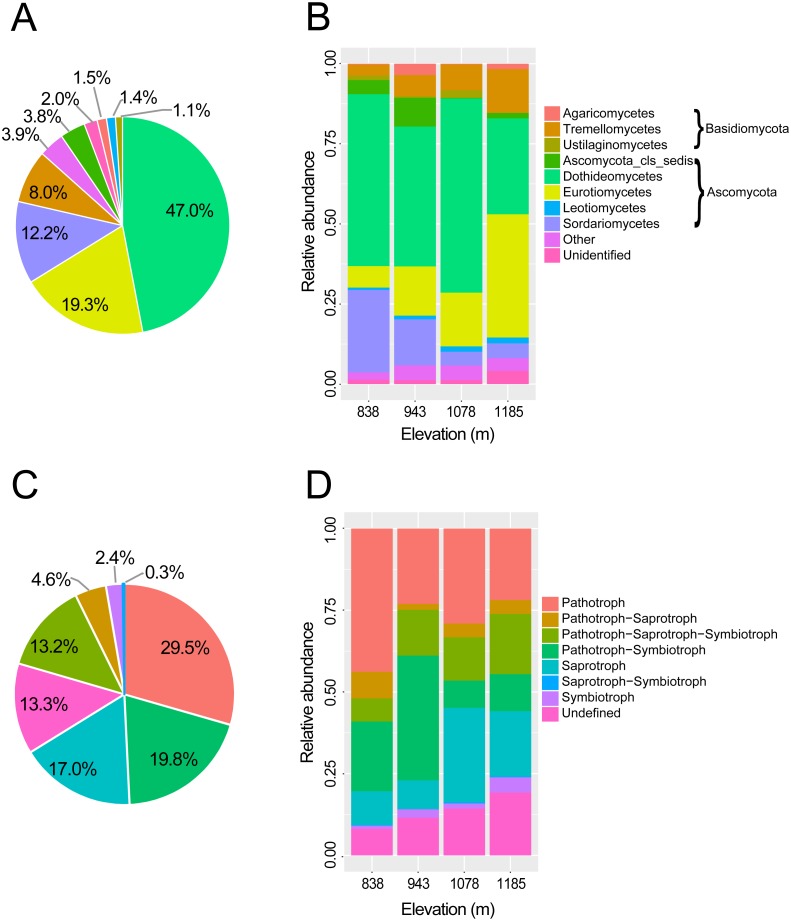
In total, 451,726 raw reads were obtained using Illumina MiSeq sequencing of the ITS2 amplicon libraries. After a series of filterings, 256,258 high-quality reads were clustered into 792 operational taxonomic units (OTUs) at a 97% sequence similarity cutoff. Finally, the sequence number of each sample was rarefied to 4606, resulting in a normalized dataset that contained 708 fungal OTUs. The abundance distribution of the OTUs was highly skewed, with plentiful rare and only a small number of very abundant OTUs. The 50 most abundant OTUs accounted for 72.4% of all sequences. The rarefaction and species accumulation curves reached a saturation plateau, indicating that our sampling and sequencing depths were sufficient to capture the majority of the phyllosphere fungal OTUs at each elevation site (Fig. 1A). Fungal communities were strongly dominated by members of the phylum Ascomycota (84.1% of all reads), followed by Basidiomycota (13.9%). The relative abundance of Basidiomycota increased slightly with elevation. At the class level, the most abundant OTUs were assigned to Dothideomycetes (47.0%), Eurotiomycetes (19.3%), Sordariomycetes (12.3%) and Tremellomycetes (8.0%) (Fig. 2A). Different community compositions between elevations were observed at this taxonomic level (Fig. 2B). For example, the abundance of the Eurotiomycetes and Tremellomycetes was higher at the highest elevation than at other sites, while Ustilaginomycetes species were hardly found at 1,185 m. Among the 52 identified orders, the three most abundant were Capnodiales (27.3%), Chaetothyriales (19.3%) and Pleosporales (11.9%). A total of 210 genera were classified, and Cladosporium (15.4%) was the most abundant genus in the M. shikokiana phyllosphere fungal community. The taxonomic assignment of each OTU is provided in Table S2.
Figure 1. Fungal richness along the elevation gradient.
(A) OTU accumulation curves of the phyllosphere fungal communities at four elevation sites. (B) OTU richness of phyllosphere fungal communities at each site. The results from the linear regressions and the one-way analysis of variance (ANOVA) are indicated in the upper and lower part of the figure, respectively.
Figure 2. Taxonomic composition and trophic guilds of the fungal communities.
Pie charts showing the distribution of sequences of (A) fungal classes and (C) fungal trophic guilds. Bar charts showing the taxonomic composition of the phyllosphere fungal communities in four elevation sites at (B) taxonomic class level and (D) tropic guilds. The fungal classes with a relative abundance of <0.1% were classified to “other”.
At the genus level, 71.9% of OTUs (509 of 708) were assigned to functional groups in FUNGuild database (Nguyen et al., 2016b). Dominant guilds were classified in seven groups of trophic modes (Fig. 2B). Pathotrophic fungi were the highest in abundance (29.5% of all sequences), followed by the pathotroph-symbiotroph group (19.8%), and the saprotroph group (17.0%; Fig. 2C). Compositions of trophic guilds were also different between elevations (Fig. 2D). The relative abundance of the pathotrophs was highest at the lowest elevation, while the symbiotrophs were more abundant at the highest elevation than at other sites.
Variation in fungal richness and community composition
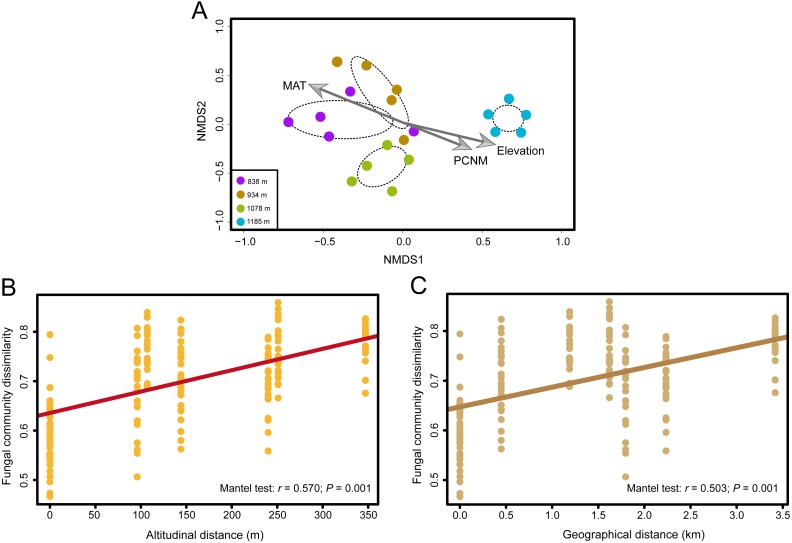
Fungal OTU richness increased significantly with increasing elevation (R2 = 0.243, P = 0.016; Fig. 1B). The generalized linear model (GLM) also supported a significant effect of elevation on M. shikokiana phyllosphere fungal richness (F = 6.484, P = 0.021; Table S3). The nonmetric multidimensional scaling (NMDS) plot revealed a general clustering according to elevation site, and the members of the 1,185 m stand were clearly separated from the other clusters (Fig. 3A). The fit of the variable selected by the “envfit” function showed that patterns of community similarity were strongly structured by elevation (R2 = 0.736, P = 0.001) followed by PCNM vector (R2 = 0.534, P = 0.003) and MAT (R2 = 0.739, P = 0.001). The significant effects of those factors on the community structure were further reflected in the results by the permutational multivariate analysis of variance (PERMANOVA), as ∼16.0%, 12.2% and 10.9% of community variation could be explained by elevation, PCNM and MAT, respectively (Table 1). Furthermore, Mantel tests indicated that fungal community dissimilarities were significantly positively correlated with altitudinal distance (r = 0.570, P = 0.001; Fig. 3B) and geographical distance (r = 0.503, P = 0.001; Fig. 3C), respectively.
Figure 3. The effects of elevation gradient, geographical distance and climate factor on the fungal community composition.
(A) Nonmetric multidimensional scaling (NMDS) ordination of variation in community structure (Bray–Curtis dissimilarity) of the phyllosphere fungal assemblages. The arrow represents significant (P < 0.01) vectors chosen by the “envfit” function to fit the fungal communities. (B) Correlations between altitudinal distance and Bray–Curtis dissimilarities of the phyllosphere fungal community. (C) Correlations between geographical distance and Bray-Curtis dissimilarities of the phyllosphere fungal community. Mantel test results are shown on the figures.
Table 1. Relative importance of elevation and geographical distance on the phyllosphere fungal community composition as revealed by permutational multivariate analysis of variance (PERMANOVA).
| Variable | DF | SS | F-statistics | R2 | P value |
|---|---|---|---|---|---|
| Elevation | 1 | 0.765 | 4.218 | 0.160 | *** |
| PCNM | 1 | 0.584 | 3.219 | 0.122 | *** |
| MAT | 1 | 0.519 | 2.861 | 0.109 | *** |
| Residuals | 16 | 2.900 | 0.608 | ||
| Total | 19 | 4.767 | 1.000 |
Notes.
- DF
- degrees of freedom
- SS
- sum of squares
- PCNM
- principal coordinates of neighbor matrices
- MAT
- mean annual temperature
P < 0.001.
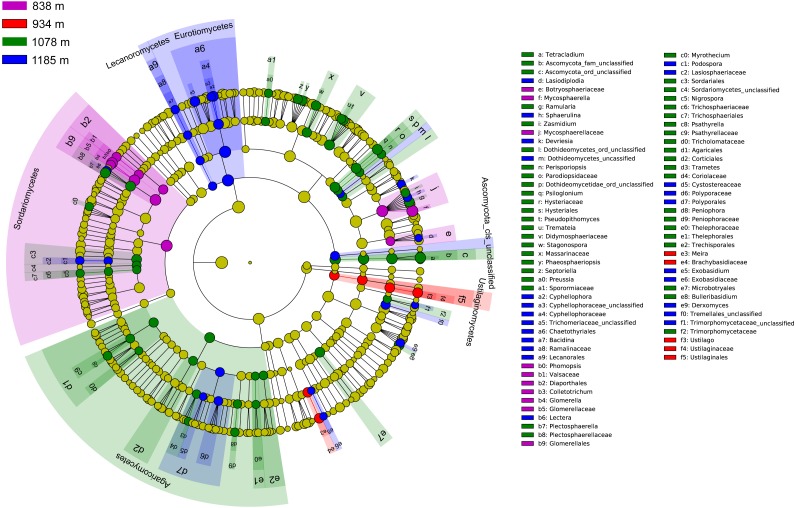
The community compositions of phyllosphere fungi between the four elevation sites were significantly different, as is shown by the linear discriminant analysis effect size (LEfSe) cladogram (Fig. 4). These results indicate that many groups at different taxonomic levels can be significantly distinguished along the elevation gradient. For example, Sordariomycetes, Ustilaginomycetes, and Agaricomycetes appeared as main discriminant clades at 838 m, 934 m, and 1,078 m, respectively. Eurotiomycetes and Lecanoromycetes are the most differentially abundant fungal classes at 1,185 m. At the taxonomic order level, Glomerellales, Diaporthales and Chaetothyriales were significantly enriched at 838 m; Ustilaginales was found to predominate at 934 m; Hysteriales, Sordariales, Microbotryales, Agaricales, Corticiales and Thelephorales were more abundant at 1,078 m; and Chaetothyriales, Polyporales and Lecanorales presented differential abundances at 1,185 m.
Figure 4. Cladogram showing significant discriminative taxa of the phyllosphere fungi associated with Mussaenda shikokiana along the elevation gradient.
The filled circles from inside to out indicate the taxonomic levels with phylum, class, order, family and genus. Circles or nodes shown in color corresponding to that of different elevation sites represented a significantly more abundant group.
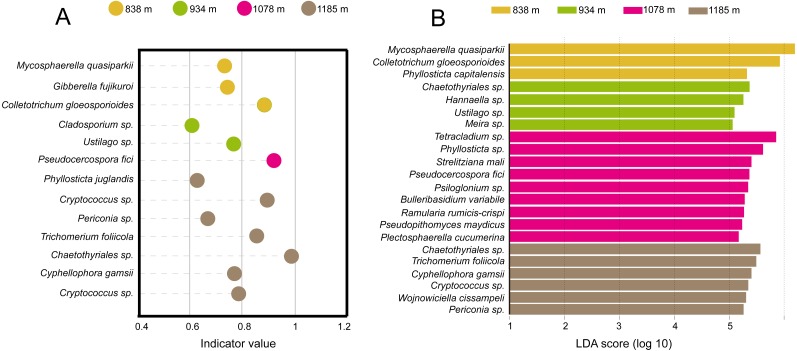
Taken together, the LEfSe and INDVAL analyses detected a total of four indicator OTUs (Mycosphaerella quasiparkii, Gibberella fujikuroi, Colletotrichum gloeosporioides and Phyllosticta capitalensis) that were significantly associated with the phyllosphere at 838 m; five indicator OTUs (Ustilago sp., Cladosporium sp., Meira sp. Hannaella sp. and Chaetothyriales sp.) at 934 m; nine indicator OTUs (Plectosphaerella cucumerina, Pseudopithomyces maydicus, Ramularia rumicis-crispi, Bulleribasidium variabile, Psiloglonium sp., Pseudocercospora fici, Strelitziana mali, Phyllosticta sp. and Tetracladium sp.) at 1,078 m; and eight indicator OTUs (Cryptococcus sp., Cyphellophora gamsii, Chaetothyriales sp., Trichomerium foliicola, Periconia sp., Cryptococcus sp., Phyllosticta juglandis and Wojnowiciella cissampeli) at 1,185 m (Fig. 5).
Figure 5. Identification of indicator species based on (A) INDVAL analysis and (B) LEfse analyses.
Indicator fungi with INDVAL indicator value >0.25 or LDA scores >3 at four elevation sites.
Key topological properties of the co-occurrence networks
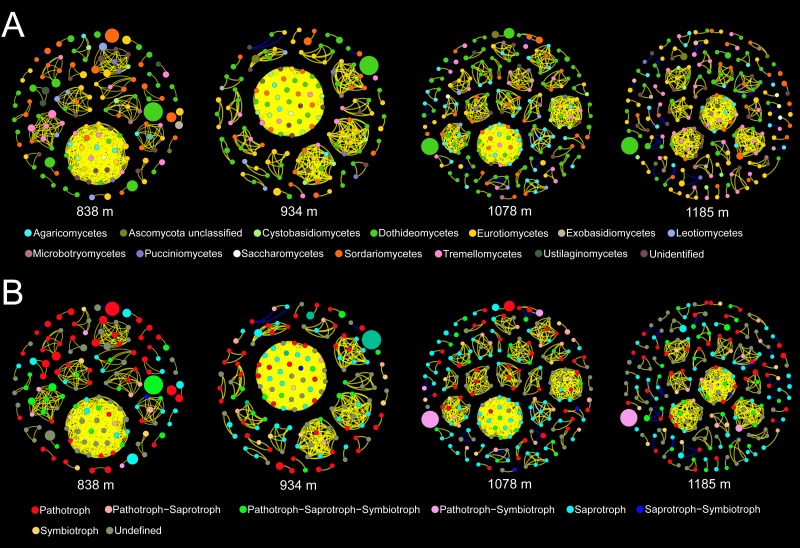
Co-occurrence patterns of the phyllosphere fungal communities were constructed for each elevation site (Fig. 6). Statistically significant and strong correlations were visualized as fungal association networks. Topological properties delineated the interaction pattern nodes, and were used to distinguish differences between the network structures of the four elevation groups. The clustering coefficient, which reflects how well nodes are connected with neighboring ones, decreased significantly with increasing elevation (Fig. 7). This indicated that the network became more discrete and sparser with increasing elevation. The network diameters of the M. shikokiana phyllosphere fungal communities showed a significantly decrease with elevation (Fig. 7). In total, over 98% of all correlations were positive. However, the negative links were significantly positive related to elevation (Fig. 7), indicating an increasing mutual exclusion between the fungal species.
Figure 6. Co-occurrence networks for the phyllosphere fungal communities at each elevation site.
The node color indicates the corresponding taxonomic assignment at class level (A) and trophic mode (B). The node sizes are proportional to the OTU abundances and the color of each line reflects positive (yellow) or negative (blue) associations.
Figure 7. Spearman correlations between topological properties of the co-occurrence networks and elevation gradient.
∗ indicates P < 0.05.
Discussion
Overall taxonomic diversity and community richness
A taxonomically diverse assemblage of phyllosphere fungi (708 OTUs) was detected in M. shikokiana, with taxa from numerous genera (210) and families (134). The OTU richness was higher than that of the needle mycobiome of Picea glauca at the northern treeline in Alaska (Sapkota et al., 2015), but lower than that of the phyllosphere fungal community of Fagus sylvatica in the French Pyrenees (Cordier et al., 2012). The phyllosphere fungal communities were largely composed of Ascomycota and dominated by members of Dothideomycetes, as previously reported in foliar fungal studies. For example, Yang et al. (2016a) found that the phyllosphere fungi of Betula ermanii in the Changbai mountains were dominated by Ascomycota, and in particular by members of Dothidiomycetes. Harrison et al. (2016) also revealed that the majority of fungal OTUs associated with the leaves of Sequoia sempervirens in the western United States were assigned to the Dothideomycetes. Similar observations have been documented in the foliar fungal communities of Eucalyptus grandis (Kemler et al., 2013), Picea glauca (Eusemann et al., 2016), Picea abies (Nguyen et al., 2016a), Metrosideros ploymorpha (Zimmerman & Vitousek, 2012) and Hopea ferrea (Izuno et al., 2016). However, different dominant taxa were detected in Fagus sylvatica (Cordier et al., 2012; Siddique & Unterseher, 2016; Unterseher et al., 2016), and several bryophytes species (Davey et al., 2013), indicating different environmental filtration and host selection across distinct geographic origins and plant species (Yang et al., 2016a; Peršoh, 2015; Rodriguez et al., 2009)).
Most fungal taxa identified here could be assigned to different functional guilds. The seven main ecological guilds (pathotroph, symbiotroph, saprotroph, pathotroph-saprotroph, pathotroph-symbiotroph, saprotroph-symbiotroph and saprotroph-pathotroph-saprotroph) were all found in our study. Many fungal species belong to more than one of the ecological categories, which is very common in fungal kingdom. For example, Fusarium culmorum could be a plant pathogen causing crown rot in cereals (Hollaway et al., 2013), but also could be a symbiotic fungal endophyte conferring salt tolerance to Leymus mollis (Rodriguez et al., 2008). Our result is consistent with a recent study on the fungal endophytes of Fagus sylvatica, and suggest undeniable advantages of high-throughput sequencing over traditional cultivation in terms of uncovering a good representation of the major functional guilds as well as rare fungal taxa (Siddique, Khokon & Unterseher, 2017). In addition to saprotrophic and symbiotrophic taxa, numerous pathotrophic fungi (29.5%) were detected on M. shikokiana leaves. Several indicator species, including Gibberella fujikuroi, Ramularia rumicis-crispi, Mycosphaerella quasiparkii, Plectosphaerella cucumerina and Strelitziana mali also have been recorded as fungal phytopathogens (De Vos et al., 2014; Petriacq, Stassen & Ton, 2016; Wardle & Lindahl, 2014). In accordance with this, Sapkota et al. (2015) found that host-specific pathogens lived in a “sea” of nonspecific fungi in the phyllosphere of six Poaceae species. Notably, several endophytic fungi may be latent pathogens (Vacher et al., 2016). Colletotrichum gloeosporioides, a significant indicator at 838 m, has previously been reported as an endophytic and symbiotic fungus that serves as a biocontrol agent for Theobroma cacao pathogens (Mejia et al., 2008), and is itself also an important plant pathogen causing anthracnose (Phoulivong et al., 2010). Alvarez-Loayza et al. (2011) found that high light environments can trigger the production of H2O2 by Diplodia mutila, a common asymptomatic endophyte in Iriartea deltoidea, resulting in pathogenicity due to constraining niche-space filling of the plant species.
Effects of elevation on fungal richness and community composition
Our results show that phyllosphere fungal richness and the community structure of fungi associated with M. shikokiana are affected significantly by elevation. These findings agree with the patterns of fungal endophytes of B. ermanii in the Changbai mountains (Yang et al., 2016b) and of F. sylvatica phyllosphere fungi in the French Pyrenees (Cordier et al., 2012). Shifts in community composition along the elevational gradient were significantly related to changes in temperature. Many studies showed that altitudinal distribution patterns of fungal assemblages often associate with climatic variables and environmental factors that co-vary with elevation. The variations in incidence and the abundance of F. sylvatica phyllosphere fungi were significantly correlated with variation in average temperature (Cordier et al., 2012). The endophytic community composition of M. ploymorpha can be driven by rainfall along elevation gradient (Zimmerman & Vitousek, 2012). The ectomycorrhizal fungal richness on Mount Kinabalu in Malaysia is highest in the elevation zone with the most available water (Geml et al., 2017). Jarvis, Woodward & Taylor (2015) also suggested that significant climatic niche partitioning between ectomycorrhizal fungi is associated with a single host along a short (300 m) elevation gradient. Therefore, variation in some climatic parameters that we did not record in this study—such as humidity, wind exposure or UV radiation along the elevation gradient—may contribute to the distribution patterns of phyllosphere fungi in M. shikokiana. The indicator species at the elevation of 1,185 m include the yeasts Cryptococcus spp. and the melanized black yeast Cyphellophora gamsii, all of which possess a strong ability to withstand UV radiation (Sapkota et al., 2015), which corresponds to the fact that solar radiation is higher at higher elevations. In addition to the importance of elevation site parameters in shaping fungal communities, abundant research has suggested a significant interplay between phyllosphere fungi and their host plants (Eusemann et al., 2016). Whipps et al. (2008) summarized that the development of phyllosphere microbiota must become involved in the phenotypic traits (leaf chemistry, physiology or morphology) exhibited by host plants. For instance, elevated leaf carbon availability could significantly enhance the phyllosphere fungal richness of B. ermanii along the elevation gradient due to broadening of niche space associated with the phyllosphere environment (Yang et al., 2016b). Furthermore, some phyllosphere fungi are dormant saprotrophs involved in the decomposition of senescent leaves (Vacher et al., 2016). The significant indicator species at the highest elevation in our study, Cyphellophora gamsii, is a saprotrophic fungus usually found in leaf litter (Crous et al., 2016; Madrid et al., 2016). Leaf litter at higher elevations is usually more recalcitrant because of increased nutrient limitation and leaf thickness; these features may facilitate the presence of specialized fungi capable of breaking down recalcitrant organic matter (Looby, Maltz & Treseder, 2016). This pattern further supports a greater richness of fungi released to the soil and air through decomposition, which may lead to increases in phyllosphere fungal richness by horizontal transmission (Osono & Mori, 2003). In addition to altitudinal distance, geographical distance also plays an important role in shaping phyllosphere fungal community composition, indicating dispersal limitation or environment isolation over a local scale (Hanson et al., 2012).
Co-occurrence networks of phyllosphere fungi
By providing a vivid and simplified version of the complex ecological interactions that shape microbial communities, network analyses of the co-occurrence patterns of significant taxa may help to deepen our understanding of the altitudinal distribution patterns of phyllosphere fungal assemblages along elevation gradients (Eiler, Heinrich & Bertilsson, 2011). Our results showed that the phyllosphere fungal network of M. shikokiana shifted between different elevation sites. Most of the correlations between fungal OTUs (838 m: 99%, 934 m: 99%, 1,078 m: 98%, and 1,185 m: 97%) were identified as co-occurrences (positive correlations). Dominant positive associations suggest that most fungal taxa may synergistically act or share similar ecological niches in the phyllosphere environment (Zhang et al., 2018a). A prevalence of positive correlations has also been found in other microbial networks (Aschenbrenner et al., 2017; Chow et al., 2014), although a much lower proportion was found in soil fungal communities. It is possible that nutrient use efficiency and mechanisms underlying fungal community assembly in soil are markedly different from those in the phyllosphere (Peršoh et al., 2018).
Although negative correlations, which suggest co-exclusion between two taxa, were much rarer than positive ones, the number of negative links increased significantly with elevation. This trend may indicate a more competitive relationship between phyllosphere fungi at higher elevations. In phyllosphere microbial communities, competition for limited space and nutrient resources, as well as the production of metabolic compounds and interference with hormone signaling pathways, are the major mechanisms by which microbial inhabitants inhibit each other (Vorholt, 2012). This result agrees with the finding that networks of endophytic communities are almost unconnected in more northern, temperate regions (Guerreiro et al., 2018). Interestingly, many negative interactions occurred between pathogenic members and other taxa; a similar finding has been reported in endophytic fungal communities (Peršoh, 2013). This may help to maintain community stability and plant health. Fokkema (1993) suggested that competition between pathogens and saprotrophs acts as a natural occurring biocontrol process in the phyllosphere. This complex microbial interaction might possess barrier effects enhancing the plant’s resistance to pathogenic infections in the phyllosphere (Vorholt, 2012). Negative correlations can finally result in a less stable network structure (Lawes et al., 2017). This increasing tendency of negative correlations also could be partly responsible for less coherent and compact network structure at higher elevation, as shown by clustering coefficients and diameters (Figs. 6 and 7). Lower elevations may provide more favorable conditions for fungal interaction and niche sharing. Higher elevations are often associated with harsher environmental conditions for plant growth, such as stronger ultraviolet exposure and lower temperatures (Pauchard et al., 2009). The decrease in connectivity and integrity of the co-occurrence networks may indicate a change towards stochastic assembly processes, prevention of biotic communication, or shifts in biogeochemical pathways due to environmental stress (Lawes et al., 2017).
Conclusions
In summary, this study reports high-throughput sequencing data that suggests that an elevation gradient of only 350 m can entail significant shifts in OTU richness, community composition, and co-occurrence patterns of phyllosphere fungi in a single host species. The phyllosphere fungal assemblages inhabiting M. shikokiana show high diversity in taxonomic classification and trophic stages. Fungal richness showed a pattern of monotonal increase along an elevation gradient. In addition, elevation is a critical predictor of the fungal community structure. Shifts in community composition along an elevation gradient were clearly related to MAT, which is suggestive of significant temperature niche partitioning among phyllosphere fungal species. A distance-decay pattern was also found in the fungal assemblages. Co-occurrence networks became less centralized and more fractured with increasing elevation. However, limited information on abiotic and biotic factors that covary with elevation demands further investigation to determine the exact drivers and mechanisms of this altitudinal partitioning.
Supplemental Information
DF, degrees of freedom, SS, sum of squares, PCNM, principal coordinates of neighbor matrices, and MAT: mean annual temperature. ∗ indicates P < 0.05.
Acknowledgments
We are grateful to Prof. Liangdong Guo for field and lab assistance. We thank Zhengkai He for assistance in sampling.
Funding Statement
This work was supported by the Chinese Academy of Sciences (XDA13020504), Ministry of Science and Technology of China (Grant No. 2013FY111200), and National Natural Science Foundation of China (31570314 and U1603231). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Xin Qian conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper.
Liang Chen, Xiaoming Guo, Dan He and Miaomiao Shi contributed reagents/materials/analysis tools.
Dianxiang Zhang conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
Representative sequences of the fungal OTUs are available in European Nucleotide Archive (ENA) under accession number: PRJEB25945.
Data Availability
The following information was supplied regarding data availability:
The raw data are provided in the Supplemental Files.
References
- Albrectsen et al. (2010).Albrectsen BR, Björkén L, Varad A, Hagner Å, Wedin M, Karlsson J, Jansson S. Endophytic fungi in European aspen (Populus tremula) leaves—diversity, detection, and a suggested correlation with herbivory resistance. Fungal Diversity. 2010;41:17–28. doi: 10.1007/s13225-009-0011-y. [DOI] [Google Scholar]
- Alvarez-Loayza et al. (2011).Alvarez-Loayza P, White Jr JF, Torres MS, Balslev H, Kristiansen T, Svenning JC, Gil N. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLOS ONE. 2011;6:e16386. doi: 10.1371/journal.pone.0016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold et al. (2003).Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15649–15654. doi: 10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner et al. (2017).Aschenbrenner IA, Cernava T, Erlacher A, Berg G, Grube M. Differential sharing and distinct co-occurrence networks among spatially close bacterial microbiota of bark, mosses and lichens. Molecular Ecology. 2017;26:2826–2838. doi: 10.1111/mec.14070. [DOI] [PubMed] [Google Scholar]
- Bahram et al. (2012).Bahram M, Polme S, Koljalg U, Zarre S, Tedersoo L. Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytologist. 2012;193:465–473. doi: 10.1111/j.1469-8137.2011.03927.x. [DOI] [PubMed] [Google Scholar]
- Baldassano & Bassett (2016).Baldassano SN, Bassett DS. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Scientific Reports. 2016;6:26087. doi: 10.1038/srep26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan et al. (2012).Barberan A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. The Isme Journal. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian, Heymann & Jacomy (2009).Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. International conference on weblogs and social media.2009. [Google Scholar]
- Bengtsson-Palme et al. (2013).Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sanchez-Garcia M, Ebersberger I, De Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods in Ecology and Evolution. 2013;4:914–919. doi: 10.1111/2041-210x.12073. [DOI] [Google Scholar]
- Benjamini & Hochberg (1995).Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Blackwell (2011).Blackwell M. The fungi: 1, 2, 3... 5.1 million species? American Journal of Botany. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Bokulich et al. (2014).Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcard & Legendre (2002).Borcard D, Legendre P. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling. 2002;153:51–68. doi: 10.1016/S0304-3800(01)00501-4. [DOI] [Google Scholar]
- Bouffaud et al. (2016).Bouffaud ML, Creamer RE, Stone D, Plassart P, Van Tuinen D, Lemanceau P, Wipf D, Redecker D. Indicator species and co-occurrence in communities of arbuscular mycorrhizal fungi at the European scale. Soil Biology & Biochemistry. 2016;103:464–470. doi: 10.1016/j.soilbio.2016.09.022. [DOI] [Google Scholar]
- Bryant et al. (2008).Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Colloquium paper: microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(Suppl 1):11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso et al. (2010).Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, YatSunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Luo & Zhang (2013).Chen S, Luo ZL, Zhang DX. Self-pollination in buds and homostyly inMussaenda shikokiana(Rubiaceae), a monomorphic species in a distylous clade. Journal of Systematics and Evolution. 2013;51:731–742. doi: 10.1111/jse.12031. [DOI] [Google Scholar]
- Chen, Luo & Zhang (2014).Chen S, Luo Z, Zhang D. Pre- and post-zygotic reproductive isolation between co-occurring Mussaenda pubescens var. alba and M. shikokiana (Rubiaceae) Journal of Integrative Plant Biology. 2014;56:411–419. doi: 10.1111/jipb.12140. [DOI] [PubMed] [Google Scholar]
- Chow et al. (2014).Chow CE, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. The Isme Journal. 2014;8:816–829. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier et al. (2012).Cordier T, Robin C, Capdevielle X, Fabreguettes O, Desprez-Loustau ML, Vacher C. The composition of phyllosphere fungal assemblages of European beech (Fagus sylvatica) varies significantly along an elevation gradient. New Phytologist. 2012;196:510–519. doi: 10.1111/j.1469-8137.2012.04284.x. [DOI] [PubMed] [Google Scholar]
- Cota-Sanchez, ReMarchuk & Ubayasena (2006).Cota-Sanchez JH, ReMarchuk K, Ubayasena K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Molecular Biology Reporter. 2006;24:161–167. doi: 10.1007/Bf02914055. [DOI] [Google Scholar]
- Crous et al. (2016).Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PW, Heykoop M, Martin MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gene J, Giraldo A, Guarnaccia V, Guarro J, Hernandez-Restrepo M, Kolarik M, Manjon JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JH, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Cmokova A, Dimitrov RA, Dyakov MY, Duenas M, Dutta AK, Esteve-Raventos F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GE, Held BW, Jurjevic Z, Kaewgrajang T, Latha KP, Lombard L, Luangsa-Ard JJ, Lyskova P, Mallatova N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordonez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sanchez A, Sarria GA, Shin HD, Silva BD, Silva GA, Smith MT, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ. Fungal planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi (2006).Csardi G. The igraph software package for complex network research. Interjournal Complex Systems. 2006:1695. [Google Scholar]
- Datlof et al. (2017).Datlof EM, Amend AS, Earl K, Hayward J, Morden CW, Wade R, Zahn G, Hynson NA. Uncovering unseen fungal diversity from plant DNA banks. PeerJ. 2017;5:e3730. doi: 10.7717/peerj.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey et al. (2013).Davey ML, Heegaard E, Halvorsen R, Kauserud H, Ohlson M. Amplicon-pyrosequencing-based detection of compositional shifts in bryophyte-associated fungal communities along an elevation gradient. Molecular Ecology. 2013;22:368–383. doi: 10.1111/mec.12122. [DOI] [PubMed] [Google Scholar]
- De Vos et al. (2014).De Vos L, Steenkamp ET, Martin SH, Santana QC, Fourie G, Van der Merwe NA, Wingfield MJ, Wingfield BD. Genome-wide macrosynteny among Fusarium species in the Gibberella fujikuroi complex revealed by amplified fragment length polymorphisms. PLOS ONE. 2014;9:e114682. doi: 10.1371/journal.pone.0114682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, White Jr & Belanger (2018).Drake I, White Jr JF, Belanger FC. Identification of the fungal endophyte of Ammophila breviligulata (American beachgrass) as Epichloë amarillans. PeerJ. 2018;6:e4300. doi: 10.7717/peerj.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrene & Legendre (1997).Dufrene M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecological Monographs. 1997;67:345–366. doi: 10.1890/0012-9615(1997)067[0345:Saaist]2.0.Co;2. [DOI] [Google Scholar]
- Edgar et al. (2011).Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler, Heinrich & Bertilsson (2011).Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. The Isme Journal. 2011;6:330. doi: 10.1038/ismej.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada, Wcislo & Van Bael (2013).Estrada C, Wcislo WT, Van Bael SA. Symbiotic fungi alter plant chemistry that discourages leaf-cutting ants. New Phytologist. 2013;198:241–251. doi: 10.1111/nph.12140. [DOI] [PubMed] [Google Scholar]
- Eusemann et al. (2016).Eusemann P, Schnittler M, Nilsson RH, Jumpponen A, Dahl MB, Wurth DG, Buras A, Wilmking M, Unterseher M. Habitat conditions and phenological tree traits overrule the influence of tree genotype in the needle mycobiome-Picea glauca system at an arctic treeline ecotone. New Phytologist. 2016;211:1221–1231. doi: 10.1111/nph.13988. [DOI] [PubMed] [Google Scholar]
- Faust & Raes (2012).Faust K, Raes J. Microbial interactions: from networks to models. Nature Reviews: Microbiology. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Fokkema (1993).Fokkema NJ. Opportunities and problems of control of foliar pathogens with micro-organisms. Pesticide Science. 1993;37:411–416. doi: 10.1002/ps.2780370416. [DOI] [Google Scholar]
- Gardes & Bruns (1993).Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Geml et al. (2017).Geml J, Morgado LN, Semenova-Nelsen TA, Schilthuizen M. Changes in richness and community composition of ectomycorrhizal fungi among altitudinal vegetation types on Mount Kinabalu in Borneo. New Phytologist. 2017;215:454–468. doi: 10.1111/nph.14566. [DOI] [PubMed] [Google Scholar]
- Geml et al. (2014).Geml J, Pastor N, Fernandez L, Pacheco S, Semenova TA, Becerra AG, Wicaksono CY, Nouhra ER. Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Molecular Ecology. 2014;23:2452–2472. doi: 10.1111/mec.12765. [DOI] [PubMed] [Google Scholar]
- Gomes et al. (2018).Gomes T, Pereira JA, Benhadi J, Lino-Neto T, Baptista P. Endophytic and Epiphytic phyllosphere fungal communities are shaped by different environmental factors in a Mediterranean ecosystem. Microbial Ecology. 2018;76(3):668–679. doi: 10.1007/s00248-018-1161-9. [DOI] [PubMed] [Google Scholar]
- Guerreiro et al. (2018).Guerreiro MA, Brachmann A, Begerow D, Peršoh D. Transient leaf endophytes are the most active fungi in 1-year-old beech leaf litter. Fungal Diversity. 2018;89:237–251. doi: 10.1007/s13225-017-0390-4. [DOI] [Google Scholar]
- Hanson et al. (2012).Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. Beyond biogeographic patterns: processes shaping the microbial landscape. Nature Reviews: Microbiology. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Harrison et al. (2016).Harrison JG, Forister ML, Parchman TL, Koch GW. Vertical stratification of the foliar fungal community in the world’s tallest trees. American Journal of Botany. 2016;103:2087–2095. doi: 10.3732/ajb.1600277. [DOI] [PubMed] [Google Scholar]
- Hartley & Gange (2009).Hartley SE, Gange AC. Impacts of plant symbiotic fungi on insect herbivores: mutualism in a multitrophic context. Annual Review of Entomology. 2009;54:323–342. doi: 10.1146/annurev.ento.54.110807.090614. [DOI] [PubMed] [Google Scholar]
- He et al. (2017).He D, Shen WJ, Eberwein J, Zhao Q, Ren LJ, Wu QLL. Diversity and co-occurrence network of soil fungi are more responsive than those of bacteria to shifts in precipitation seasonality in a subtropical forest. Soil Biology and Biochemistry. 2017;115:499–510. doi: 10.1016/j.soilbio.2017.09.023. [DOI] [Google Scholar]
- Hollaway et al. (2013).Hollaway GJ, Evans ML, Wallwork H, Dyson CB, McKay AC. Yield loss in cereals, caused by Fusarium culmorum and F. pseudograminearum, is related to fungal DNA in soil prior to planting, rainfall, and cereal type. Plant Disease. 2013;97:977–982. doi: 10.1094/PDIS-09-12-0867-RE. [DOI] [PubMed] [Google Scholar]
- Hubbard et al. (2018).Hubbard CJ, Brock MT, Van Diepen LT, Maignien L, Ewers BE, Weinig C. The plant circadian clock influences rhizosphere community structure and function. The Isme Journal. 2018;12:400–410. doi: 10.1038/ismej.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrmark et al. (2012).Ihrmark K, Bodeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandstrom-Durling M, Clemmensen KE, Lindahl BD. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiology Ecology. 2012;82:666–677. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- Izuno et al. (2016).Izuno A, Kanzaki M, Artchawakom T, Wachrinrat C, Isagi Y. Vertical structure of phyllosphere fungal communities in a tropical forest in Thailand uncovered by high-throughput sequencing. PLOS ONE. 2016;11:e0166669. doi: 10.1371/journal.pone.0166669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, Woodward & Taylor (2015).Jarvis SG, Woodward S, Taylor AF. Strong altitudinal partitioning in the distributions of ectomycorrhizal fungi along a short (300 m) elevation gradient. New Phytologist. 2015;206:1145–1155. doi: 10.1111/nph.13315. [DOI] [PubMed] [Google Scholar]
- Jiao et al. (2016).Jiao S, Liu ZS, Lin YB, Yang J, Chen WM, Wei GH. Bacterial communities in oil contaminated soils: biogeography and co-occurrence patterns. Soil Biology and Biochemistry. 2016;98:64–73. doi: 10.1016/j.soilbio.2016.04.005. [DOI] [Google Scholar]
- Ju et al. (2014).Ju F, Xia Y, Guo F, Wang Z, Zhang T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environmental Microbiology. 2014;16:2421–2432. doi: 10.1111/1462-2920.12355. [DOI] [PubMed] [Google Scholar]
- Jumpponen & Jones (2009).Jumpponen A, Jones KL. Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytologist. 2009;184:438–448. doi: 10.1111/j.1469-8137.2009.02990.x. [DOI] [PubMed] [Google Scholar]
- Kay et al. (2018).Kay GM, Tulloch A, Barton PS, Cunningham SA, Driscoll DA, Lindenmayer DB. Species co-occurrence networks show reptile community reorganization under agricultural transformation. Ecography. 2018;41:113–125. doi: 10.1111/ecog.03079. [DOI] [Google Scholar]
- Kembel & Mueller (2014).Kembel SW, Mueller RC. Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany. 2014;92:303–311. doi: 10.1139/cjb-2013-0194. [DOI] [Google Scholar]
- Kemler et al. (2013).Kemler M, Garnas J, Wingfield MJ, Gryzenhout M, Pillay KA, Slippers B. Ion torrent PGM as tool for fungal community analysis: a case study of Endophytes in Eucalyptus grandis reveals high taxonomic diversity. PLOS ONE. 2013;8:e81718. doi: 10.1371/journal.pone.0081718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg et al. (2013).Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Duenas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lucking R, Martin MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Poldmaa K, Saag L, Saar I, Schussler A, Scott JA, Senes C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Lanzen et al. (2016).Lanzen A, Epelde L, Blanco F, Martin I, Artetxe U, Garbisu C. Multi-targeted metagenetic analysis of the influence of climate and environmental parameters on soil microbial communities along an elevational gradient. Scientific Reports. 2016;6:28257. doi: 10.1038/srep28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawes et al. (2017).Lawes JC, Dafforn KA, Clark GF, Brown MV, Johnston EL. Multiple stressors in sediments impact adjacent hard substrate habitats and across biological domains. Science of the Total Environment. 2017;592:295–305. doi: 10.1016/j.scitotenv.2017.03.083. [DOI] [PubMed] [Google Scholar]
- Legendre et al. (2010).Legendre P, Borcard D, Blanchet G, Dray S. PCNM: PCNM spatial eigenfunction and principal coordinate analyses. R package version 2.1/r822010
- Li et al. (2017).Li Z, Wright AG, Yang Y, Si H, Li G. Unique bacteria community composition and co-occurrence in the milk of different ruminants. Scientific Reports. 2017;7:40950. doi: 10.1038/srep40950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggenstoffer et al. (2010).Liggenstoffer AS, Youssef NH, Couger MB, Elshahed MS. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. The Isme Journal. 2010;4:1225. doi: 10.1038/ismej.2010.49. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2015).Liu L, Hart MM, Zhang J, Cai X, Gai J, Christie P, Li X, Klironomos JN. Altitudinal distribution patterns of AM fungal assemblages in a Tibetan alpine grassland. FEMS Microbiology Ecology. 2015;91:fiv078–fiv078. doi: 10.1093/femsec/fiv078. [DOI] [PubMed] [Google Scholar]
- Looby, Maltz & Treseder (2016).Looby CI, Maltz MR, Treseder KK. Belowground responses to elevation in a changing cloud forest. Ecology and Evolution. 2016;6:1996–2009. doi: 10.1002/ece3.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2013).Lu LH, Yin SX, Liu X, Zhang WM, Gu TY, Shen QR, Qiu HZ. Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biology & Biochemistry. 2013;65:186–194. doi: 10.1016/j.soilbio.2013.05.025. [DOI] [Google Scholar]
- Ma et al. (2016).Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, Brookes PC, Xu J, Gilbert JA. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. The Isme Journal. 2016;10:1891–1901. doi: 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid et al. (2016).Madrid H, Hernandez-Restrepo M, Gene J, Cano J, Guarro J, Silva V. New and interesting chaetothyrialean fungi from Spain. Mycological Progress. 2016;15:1179–1201. doi: 10.1007/s11557-016-1239-z. [DOI] [Google Scholar]
- Magoc & Salzberg (2011).Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia et al. (2008).Mejia LC, Rojas EI, Maynard Z, Van Bael S, Arnold AE, Hebbar P, Samuels GJ, Robbins N, Herre EA. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biological Control. 2008;46:4–14. doi: 10.1016/j.biocontrol.2008.01.012. [DOI] [Google Scholar]
- Milici et al. (2016).Milici M, Deng ZL, Tomasch J, Decelle J, Wos-Oxley ML, Wang H, Jauregui R, Plumeier I, Giebel HA, Badewien TH, Wurst M, Pieper DH, Simon M, Wagner-Dobler I. Co-occurrence analysis of microbial taxa in the atlantic ocean reveals high connectivity in the free-living bacterioplankton. Frontiers in Microbiology. 2016;7:649. doi: 10.3389/fmicb.2016.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto et al. (2014).Miyamoto Y, Nakano T, Hattori M, Nara K. The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. The Isme Journal. 2014;8:1739–1746. doi: 10.1038/ismej.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen et al. (2016a).Nguyen D, Boberg J, Ihrmark K, Stenström E, Stenlid J. Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecology. 2016a;23:97–108. doi: 10.1016/j.Funeco.2016.07.003. [DOI] [Google Scholar]
- Nguyen et al. (2016b).Nguyen NH, Song ZW, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology. 2016b;20:241–248. doi: 10.1016/j.Funeco.2015.06.006. [DOI] [Google Scholar]
- Oksanen et al. (2016).Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package. R package version 2.4-1https://CRANR-project.org/package=vegan 2016
- Oono, Rasmussen & Lefevre (2017).Oono R, Rasmussen A, Lefevre E. Distance decay relationships in foliar fungal endophytes are driven by rare taxa. Environmental Microbiology. 2017;19:2794–2805. doi: 10.1111/1462-2920.13799. [DOI] [PubMed] [Google Scholar]
- Osono & Mori (2003).Osono T, Mori A. Colonization of Japanese beech leaves by phyllosphere fungi. Mycoscience. 2003;44:437–441. doi: 10.1007/S10267-003-0135-Y. [DOI] [Google Scholar]
- Pauchard et al. (2009).Pauchard A, Kueffer C, Dietz H, Daehler CC, Alexander J, Edwards PJ, Arevalo JR, Cavieres LA, Guisan A, Haider S, Jakobs G, McDougall K, Millar CI, Naylor BJ, Parks CG, Rew LJ, Seipel T. Ain’t no mountain high enough: plant invasions reaching new elevations. Frontiers in Ecology and the Environment. 2009;7:479–486. doi: 10.1890/080072. [DOI] [Google Scholar]
- Peršoh (2013).Peršoh D. Factors shaping community structure of endophytic fungi—evidence from the Pinus-Viscum-system. Fungal Diversity. 2013;60:55–69. doi: 10.1007/s13225-013-0225-x. [DOI] [Google Scholar]
- Peršoh (2015).Peršoh D. Plant-associated fungal communities in the light of meta’omics. Fungal Diversity. 2015;75:1–25. doi: 10.1007/s13225-015-0334-9. [DOI] [Google Scholar]
- Peršoh et al. (2018).Peršoh D, Stolle N, Brachmann A, Begerow D, Rambold G. Fungal guilds are evenly distributed along a vertical spruce forest soil profile while individual fungi show pronounced niche partitioning. Mycological Progress. 2018;17:925–939. doi: 10.1007/s11557-018-1405-6. [DOI] [Google Scholar]
- Petriacq, Stassen & Ton (2016).Petriacq P, Stassen JH, Ton J. Spore density determines infection strategy by the plant pathogenic fungus Plectosphaerella cucumerina. Plant Physiology. 2016;170:2325–2339. doi: 10.1104/pp.15.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoulivong et al. (2010).Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, Chukeatirote E, Hyde KD. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Diversity. 2010;44:33–43. doi: 10.1007/s13225-010-0046-0. [DOI] [Google Scholar]
- Proulx, Promislow & Phillips (2005).Proulx SR, Promislow DE, Phillips PC. Network thinking in ecology and evolution. Trends in Ecology & Evolution. 2005;20:345–353. doi: 10.1016/j.tree.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ren et al. (2018).Ren C, Zhang W, Zhong Z, Han X, Yang G, Feng Y, Ren G. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Science of the Total Environment. 2018;610–611:750–758. doi: 10.1016/j.scitotenv.2017.08.110. [DOI] [PubMed] [Google Scholar]
- Revelle (2017).Revelle W. Northwestern University; Evanston: 2017. [Google Scholar]
- Rime et al. (2015).Rime T, Hartmann M, Brunner I, Widmer F, Zeyer J, Frey B. Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Molecular Ecology. 2015;24:1091–1108. doi: 10.1111/mec.13051. [DOI] [PubMed] [Google Scholar]
- Rodriguez et al. (2008).Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y-O, Redman RS. Stress tolerance in plants via habitat-adapted symbiosis. The Isme Journal. 2008;2:404. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- Rodriguez et al. (2009).Rodriguez R, White Jr J, Arnold A, Redman R. Fungal endophytes: diversity and functional roles. New Phytologist. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- Sapkota et al. (2015).Sapkota R, Knorr K, Jorgensen LN, O’Hanlon KA, Nicolaisen M. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist. 2015;207:1134–1144. doi: 10.1111/nph.13418. [DOI] [PubMed] [Google Scholar]
- Schloss et al. (2009).Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata et al. (2011).Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2014).Shen CC, Liang WJ, Shi Y, Lin XG, Zhang HY, Wu X, Xie G, Chain P, Grogan P, Chu HY. Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology. 2014;95:3190–3202. doi: 10.1890/14-0310.1. [DOI] [Google Scholar]
- Siddique, Khokon & Unterseher (2017).Siddique AB, Khokon AM, Unterseher M. What do we learn from cultures in the omics age? High-throughput sequencing and cultivation of leaf-inhabiting endophytes from beech (Fagus sylvatica L.) revealed complementary community composition but similar correlations with local habitat conditions. MycoKeys. 2017;20:1–16. doi: 10.3897/mycokeys.20.11265. [DOI] [Google Scholar]
- Siddique & Unterseher (2016).Siddique AB, Unterseher M. A cost-effective and efficient strategy for Illumina sequencing of fungal communities: a case study of beech endophytes identified elevation as main explanatory factor for diversity and community composition. Fungal Ecology. 2016;20:175–185. doi: 10.1016/j.Funeco.2015.12.009. [DOI] [Google Scholar]
- Siles & Margesin (2016).Siles JA, Margesin R. Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: what are the driving factors? Microbial Ecology. 2016;72:207–220. doi: 10.1007/s00248-016-0748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiyun & Viliam (2017).Taiyun W, Viliam S. R package corrplot: visualization of a correlation matrix (Version 0.84) https://github.com/taiyun/corrplot 2017
- Takeda, Nishimura & Inouye (1977).Takeda Y, Nishimura H, Inouye H. Two new iridoid glucosides from Mussaenda parviflora and Mussaenda shikokiana. Phytochemistry. 1977;16:1401–1404. doi: 10.1016/S0031-9422(00)88791-9. [DOI] [Google Scholar]
- Tedersoo et al. (2014).Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K. Fungal biogeography. Global diversity and geography of soil fungi. Science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- Toju, Tanabe & Ishii (2016).Toju H, Tanabe AS, Ishii HS. Ericaceous plant-fungus network in a harsh alpine-subalpine environment. Molecular Ecology. 2016;25:3242–3257. doi: 10.1111/mec.13680. [DOI] [PubMed] [Google Scholar]
- Truong et al. (2017).Truong C, Mujic AB, Healy R, Kuhar F, Furci G, Torres D, Niskanen T, Sandoval-Leiva PA, Fernandez N, Escobar JM, Moretto A, Palfner G, Pfister D, Nouhra E, Swenie R, Sanchez-Garcia M, Matheny PB, Smith ME. How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytologist. 2017;214:913–919. doi: 10.1111/nph.14509. [DOI] [PubMed] [Google Scholar]
- Unterseher et al. (2016).Unterseher M, Siddique AB, Brachmann A, Persoh D. Diversity and composition of the leaf mycobiome of beech (Fagus sylvatica) are affected by local habitat conditions and leaf biochemistry. PLOS ONE. 2016;11:e0152878. doi: 10.1371/journal.pone.0152878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher et al. (2016).Vacher C, Hampe A, Porte AJ, Sauer U, Compant S, Morris CE. The Phyllosphere: microbial Jungle at the Plant-Climate Interface. Annual Review of Ecology, Evolution, and Systematics, Vol 47. 2016;47:1–24. doi: 10.1146/annurev-ecolsys-121415-032238. [DOI] [Google Scholar]
- Vorholt (2012).Vorholt JA. Microbial life in the phyllosphere. Nature Reviews: Microbiology. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- Voriskova & Baldrian (2013).Voriskova J, Baldrian P. Fungal community on decomposing leaf litter undergoes rapid successional changes. The Isme Journal. 2013;7:477–486. doi: 10.1038/ismej.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright et al. (2017).Wainwright BJ, Zahn GL, Spalding HL, Sherwood AR, Smith CM, Amend AS. Fungi associated with mesophotic macroalgae from the ‘Au‘au Channel, west Maui are differentiated by host and overlap terrestrial communities. PeerJ. 2017;5:e3532. doi: 10.7717/peerj.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle & Lindahl (2014).Wardle DA, Lindahl BD. Ecology. Disentangling global soil fungal diversity. Science. 2014;346:1052–1053. doi: 10.1126/science.aaa1185. [DOI] [PubMed] [Google Scholar]
- Whipps et al. (2008).Whipps JM, Hand P, Pink D, Bending GD. Phyllosphere microbiology with special reference to diversity and plant genotype. Journal of Applied Microbiology. 2008;105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- White et al. (1990).White TJ, Bruns T, Lee S, Taylor J. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Widder et al. (2014).Widder S, Besemer K, Singer GA, Ceola S, Bertuzzo E, Quince C, Sloan WT, Rinaldo A, Battin TJ. Fluvial network organization imprints on microbial co-occurrence networks. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12799–12804. doi: 10.1073/pnas.1411723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016a).Yang T, Sun H, Shen C, Chu H. Fungal assemblages in different habitats in an Erman’s Birch forest. Frontiers in Microbiology. 2016a;7:1368. doi: 10.3389/fmicb.2016.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016b).Yang T, Weisenhorn P, Gilbert JA, Ni Y, Sun R, Shi Y, Chu H. Carbon constrains fungal endophyte assemblages along the timberline. Environmental Microbiology. 2016b;18:2455–2469. doi: 10.1111/1462-2920.13153. [DOI] [PubMed] [Google Scholar]
- Yao et al. (2017).Yao F, Yang S, Wang Z, Wang X, Ye J, Wang X, DeBruyn JM, Feng X, Jiang Y, Li H. Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Frontiers in Microbiology. 2017;8:2071. doi: 10.3389/fmicb.2017.02071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018a).Zhang B, Zhang J, Liu Y, Shi P, Wei G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biology and Biochemistry. 2018a;118:178–186. doi: 10.1016/j.soilbio.2017.12.011. [DOI] [Google Scholar]
- Zhang et al. (2018b).Zhang L, Wu W, Lee YK, Xie J, Zhang H. Spatial heterogeneity and co-occurrence of mucosal and luminal microbiome across swine intestinal tract. Frontiers in Microbiology. 2018b;9:48. doi: 10.3389/fmicb.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014).Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, Ma ZS, Shi P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. The Isme Journal. 2014;8:881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018c).Zhang Z, Luo L, Tan X, Kong X, Yang J, Wang D, Zhang D, Jin D, Liu Y. Pumpkin powdery mildew disease severity influences the fungal diversity of the phyllosphere. PeerJ. 2018c;6:e4559. doi: 10.7717/peerj.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman & Vitousek (2012).Zimmerman NB, Vitousek PM. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13022–13027. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DF, degrees of freedom, SS, sum of squares, PCNM, principal coordinates of neighbor matrices, and MAT: mean annual temperature. ∗ indicates P < 0.05.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are provided in the Supplemental Files.