Abstract
Background
To evaluate the prognostic impact of HE4 expression in patients with cancer.
Materials and methods
We searched the PubMed, Web of Science, Chinese National Knowledge Infrastructure and WangFang databases for publications concerning HE4 expression in patients with cancer. The correlation of HE4 expression level with overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS) was analyzed.
Results
In this meta-analysis, 29 studies, with a total of 4,235 patients, were included. Our results showed that HE4 expression was significantly associated with poorer OS (hazard ratio [HR] =2.15, 95% confidence interval [CI] =1.77–2.62, P<0.001). Further subgroup analysis found that this correlation was not affected by race (White: HR =1.92, 95% CI =1.53–2.39, P<0.001; Asian: HR =2.62, 95% CI =2.06–3.35, P<0.001) or tumor types (endometrial cancer: HR =2.91, 95% CI =1.86–4.53, P<0.001; ovarian cancer: HR =1.82, 95% CI =1.50–2.22, P<0.001; lung cancer: HR =2.31, 95% CI =1.54–3.47, P<0.001). Our meta-analysis showed that HE4 overexpression was significantly associated with DFS (HR =2.50, 95% CI =1.86–3.37, P<0.001) and PFS (HR =1.27, 95% CI =1.11–1.45, P=0.001).
Conclusion
These results suggest that expression of HE4 was associated with a worse prognosis in patients with cancer. HE4 is a potential novel prognostic factor in patients with cancer.
Keywords: HE4, cancer, prognosis, meta-analysis
Introduction
Cancer is a global health problem associated with increasing mortality rates, in spite of advances in diagnostic and therapeutic approaches.1 Several pathological parameters and specific blood tumor markers have been proposed as predictive prognostic factors in cancer.2,3 However, the high incidence of cancer-related deaths indicates a need for reliable and efficient biomarkers for patient stratification and treatment selection.
Human epididymis protein 4 (HE4), also known as whey-acidic-protein four-disulfide core protein-2 (WFDC2), is a member of the protease inhibitor family with immune protective effects and is a promising novel cancer biomarker.4,5 HE4 has been approved by the US Food and Drug Administration as a new tumor marker for the diagnosis of early stage ovarian cancer.6 Several cancer types7–9 are associated with HE4 overexpression in the serum and tissues. HE4 overexpression is also associated with cancer progression, and its prognostic value has been investigated in several published studies. However, the results remain controversial, and therefore this meta-analysis was performed to accurately assess the prognostic value of HE4 expression in cancer patients.
Materials and methods
Search strategy
The research databases PubMed, Web of Science, Chinese National Knowledge Infrastructure and WangFang databases were searched independently by two authors (Cong Dai and Yi Zheng) to obtain all relevant papers published as of August 2017. The following search terms were used: “Human Epididymis Protein 4 or HE4” and “neoplasms or cancer or tumor” and “prognosis.” No language restrictions were applied. Furthermore, references within the retrieved relevant articles were screened to identify potentially eligible studies. Disagreements were resolved by iteration, discussion, and consensus between the two authors.
Inclusion and exclusion criteria
Studies were considered eligible for inclusion if they met the following criteria: 1) studies evaluated the relationship of HE4 expression in patients with cancer with detailed information about overall survival (OS), disease-free survival (DFS), or progression-free survival (PFS); 2) selected cancer cases were pathologically confirmed, and 3) the study provided a hazard ratio (HR) with the corresponding confidence interval (CI), or sufficient data to calculate it. The exclusion criteria were as follows: 1) duplicate publications; 2) animal studies; 3) articles without usable data; 4) reviews, case reports, letters, and conference abstracts without original data.
Data extraction
Two independent reviewers (Cong Dai and Yi Zheng) extracted the details of included studies using a standardized form, and any disagreements were resolved through discussion with a third reviewer (Zhijun Dai). The following information was recorded: first author’s surname, year of publication, number of patients, patient source, tumor types, HE4 assessment method, sample, prognostic outcomes, analytical method, and HR with the corresponding 95% CI.
Methodological quality of the studies
Quality assessment of included studies was conducted independently by two authors (Cong Dai and Tian Tian) following the Newcastle–Ottawa Scale (NOS) criteria,10 and any disagreements were resolved by discussion with a third reviewer (Zhijun Dai). The NOS criteria were scored on the basis of three aspects: 1) subject selection, 2) comparability of subject, and 3) clinical outcome. NOS scores may range from 0 to 9, and a score ≥6 indicates high quality.
Statistical methods
Included studies were divided into three groups on the basis of the parameter that was reported: OS, DFS, and PFS. According to the cutoff values provided by the authors of each study, HE4 expression was designated as “high” or “low.” HR and 95% CI were used to assess the association between HE4 expression and OS, DFS, and PFS. HRs obtained from studies were used directly in further analyses. For studies where HR values were not included explicitly, Kaplan–Meier survival curves or other methods were used to derive HRs from available data.11 Data from the Kaplan–Meier survival curves were read using the Engauge Digitizer version 4.1 software. Heterogeneity among studies was determined by the χ2 test and Q test. If there was no significant heterogeneity (I2≤50% or P≥0.05), a fixed-effect model was used; if significant heterogeneity was found to exist (I2>50% or P<0.05), a random-effects model was used. We further conducted subgroup analyses by race, tumor type, sample, method, and HR estimate. Sensitivity analysis was performed by omitting individual studies to examine the reliability of the results. Probable publication bias was assessed by Egger’s and Begg’s test.12,13 All P-values were two-sided, and P<0.05 was considered statistically significant. Statistical calculations were performed using STATA 14.0 (StataCorp LLC, College Station, TX, USA).
Results
Search results and study characteristics
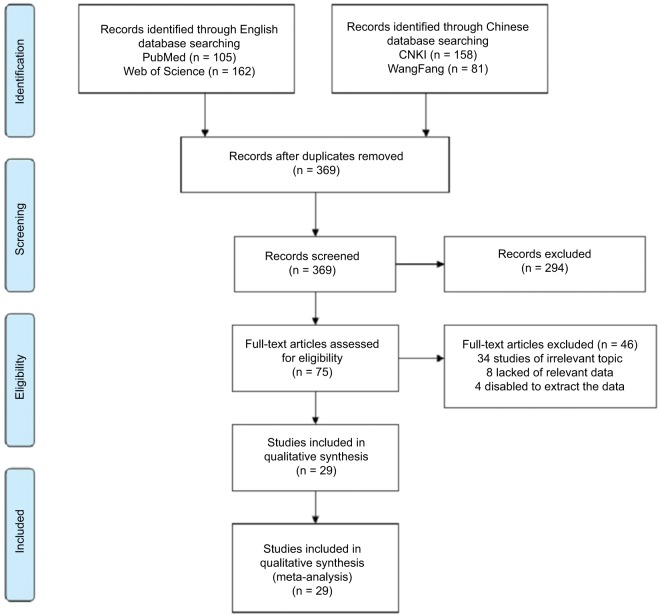
A total of 369 articles from the primary literature were searched in PubMed, Web of Science, CNKI, and WangFang databases. References within the retrieved relevant articles had been screened, but there were no more potentially eligible studies. As shown in Figure 1, 340 studies were excluded because they were irrelevant to the analysis or because the primary outcome was insufficient. Finally, 29 available studies were included in this meta-analysis.7–9,14–39
Figure 1.
Flowchart of the selection of the studies in the meta-analysis.
The characteristics of the 29 studies are summarized in Table 1. Of the 29 publications, 23 assessed the relationship between HE4 expression and OS, eight studies evaluated the association between HE4 expression and DFS, and eleven evaluated PFS. A total of 4,235 patients from People’s Republic of China, the Netherlands, Italy, Denmark, United States of America, France, Australia, Germany, Japan, Canada, Korea, and Sweden were enrolled with sample numbers ranging from 23 to 373. HE4 status in tumors was assessed by various methods: immunohistochemistry (IHC) (6 studies), electrochemiluminescence immunoassay assay (ECLIA) (2 studies), enzyme immu noassay assay (EIA) (15 studies), and Chemiluminesent Microparticle Immunoassay (CMIA) (5 studies). Study quality assessment, as per the Newcastle–Ottawa quality assessment scale, yielded scores ranging from 6 to 9, with a mean score of 7.6.
Table 1.
Main characteristics of all studies included in the meta-analysis
| Study | Year | Patient source | Number of patients | Tumor types | Method | Sample | Cutoff | Outcome | M/U | HR (95% CI) | Quality |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Stiekema et al7 | 2017 | Netherlands | 88 | EC | ECLIA | Blood | 60 pmol/L for women <40 years, 75 pmol/L for patients between 40 and 60 years of age, and 90 pmol/L for patients >60 years of age | OS | M | 7.37 (2.16–25.1) | 9 |
| Orsaria et al14 | 2016 | Italy | 105 | OC | IHC | Tissue | H-score value >1 | OS | M | 1.82 (0.81–4) | 8 |
| Aarenstrup Karlsen et al8 | 2016 | Denmark | 198 | OC | IHC | Tissue | – | OS | U | 1.44 (1.01–2.0) | 8 |
| PFS | 1.49 (1.06–2.11) | ||||||||||
| Lan et al15 | 2015 | People’s Republic of China | 218 | LC | EIA | Blood | 20.5 ng/mL | OS | M | 3.78 (2.23–7.34) | 8 |
| Deng et al16 | 2015 | People’s Republic of China | 65 | EC | IHC | Tissue | – | OS | U | 2.51 (0.66–9.53) | 7 |
| Li et al17 | 2015 | People’s Republic of China | 102 | EC | IHC | Tissue | H-score value >2 | OS | U | 1.85 (0.37–9.28) | 6 |
| Lee et al18 | 2015 | United States of America | 53 | OC | – | Tissue | – | OS | U | 1.72 (0.87–3.46) | 6 |
| Lamy et al9 | 2015 | France | 346 | LC | EIA | Blood | 140 pm | OS | U | 1.96 (1.53–2.53) | 8 |
| Guo et al19 | 2015 | United States of America | 243 | GC | IHC | Tissue | H-score value >1 | OS | U | 1.62 (1.00–2.62) | 7 |
| Lou et al20 | 2014 | United States of America | 153 | LC | EIA | Blood | 65–83 pm | OS | U | 1.08 (0.87–1.36) | 6 |
| PFS | 0.95 (0.78–1.17) | ||||||||||
| Jiang et al21 | 2014 | People’s Republic of China | 100 | LC | EIA | Blood | 7.26 ng/mL | OS | M | 3.65 (2.75–11.98) | 7 |
| Brennan et al22 | 2014 | Australia | 373 | EC | CMIA | Blood | H-score >75% | DFS | M | 2.40 (1.19–4.38) | 9 |
| Zhang et al23 | 2014 | People’s Republic of China | 191 | LC | EIA | Blood | 91.63 pmol/L | OS | M | 2.15 (1.49–3.12) | 7 |
| Braicu et al24 | 2014 | Germany | 73 | OC | ECLIA | Blood | 250 pm | OS | U | 3.33 (1.03–10.7) | 6 |
| Zheng et al25 | 2013 | People’s Republic of China | 112 | OC | EIA | Blood | 415.5 pmol/L | OS | U | 2.17 (1.11–4.23) | 8 |
| Liu et al26 | 2013 | People’s Republic of China | 169 | LC | EIA | Blood | 83.90 pm | OS | M | 2.2 (0.8–5.9) | 7 |
| Zanotti et al27 | 2012 | Italy | 190 | EC | CMIA | Blood | 51 pm | OS | M | 2.78 (1.16–6.63) | 9 |
| DFS | 2.49 (1.13–5.49) | ||||||||||
| PFS | 2.66 (1.10–6.45) | ||||||||||
| Yamashita et al28 | 2012 | Japan | 74 | LC | EIA | Blood | 50.3 pm | DFS | U | 3.28 (0.57–18.84) | 7 |
| Trudel et al29 | 2012 | Canada | 136 | OC | CMIA | Blood | 394 pmol/L | OS | M | 1.67 (1.08–2.59) | 9 |
| PFS | 1.32 (0.87–1.99) | ||||||||||
| Mutz-Dehbalaie et al30 | 2012 | Austria | 183 | EC | CMIA | Blood | 81 pmol/L | OS | M | 2.41 (1.17–4.97) | 8 |
| DFS | 1.59 (0.82–3.08) | ||||||||||
| Kong et al31 | 2012 | Korea | 80 | OC | EIA | Blood | 98.7 pm | PFS | M | 1.47 (1.02–2.1) | 8 |
| Kalapotharakos et al32 | 2012 | Sweden | 312 | OC | EIA | Blood | 405 pm | OS | M | 2.02 (1.1–3.8) | 9 |
| Hu et al33 | 2012 | People’s Republic of China | 76 | OC | EIA | Blood | 208 pmol/L | PFS | U | 1.72 (0.92–3.21) | 6 |
| Steffensen et al34 | 2011 | Denmark | 139 | OC | EIA | Blood | 140 pm | OS | M | 3.17 (1.41–7.10) | 9 |
| PFS | 1.77 (1.03–3.04) | ||||||||||
| Yamashita et al35 | 2011 | Japan | 137 | LC | IHC | Tissue | H-score >75% | OS | M | 5.5 (1.8–17.2) | 8 |
| DFS | 3.7 (1.7–8.4) | ||||||||||
| Paek et al36 | 2011 | Korea | 45 | OC | EIA | Blood | 70 pm | PFS | M | 2.24 (1.14–6.84) | 8 |
| Han et al37 | 2011 | United States of America | 23 | OC | EIA | Blood | 74 pm | PFS | U | 1.97 (0.61–6.39) | 7 |
| Bignotti et al38 | 2011 | Italy | 153 | EC | EIA | Blood | – | OS | U | 3.74 (0.43–32.45) | 8 |
| PFS | 1.78 (0.30–10.44) | ||||||||||
| DFS | 2.43 (0.87–6.77) | ||||||||||
| Bandiera et al39 | 2011 | Italy | 98 | OC | CMIA | Blood | 43.8 pm | OS | M | 3.98 (1.35–11.75) | 8 |
| DFS | 2.46 (1.09–5.56) | ||||||||||
| PFS | 2.77 (1.12–6.85) | ||||||||||
Abbreviations: M, multivariate analysis; U, univariate analysis; HR, hazard ratio; EC, endometrial cancer; OC, ovarian cancer; LC, lung cancer; GC, gastric cancer; ECLIA, electrochemiluminescence immunoassay assay; IHC, immunohistochemistry; EIA, enzyme immunoassay assay; CMIA, chemiluminescent microparticle immunoassay; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival.
Quantitative synthesis
HE4 expression and OS in cancers
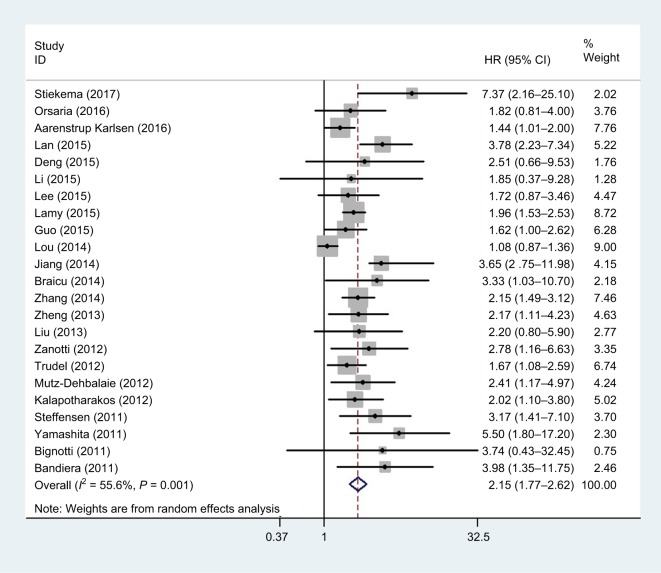
Overall, 23 studies, including 3,564 patients, reported a relationship between OS and HE4 expression level. As heterogeneity among the studies was statistically significant (P=0.001, I2=55.6%), a random-effects model was used. The pooled HR for OS showed that the overexpression of HE4 was significantly associated with reduced OS in cancers (HR =2.15, 95% CI =1.77–2.62, P<0.001, Table 2 and Figure 2).
Table 2.
Main meta-analysis results for OS
| Analysis | Number of studies | Number of patients | Model | HR (95% CI) | P-value | Heterogeneity
|
|
|---|---|---|---|---|---|---|---|
| I2 (%) | P-value | ||||||
|
| |||||||
| OS | 23 | 3,564 | Random | 2.15 (1.77–2.62) | 0.000 | 55.6 | 0.001 |
| Race | |||||||
| White | 15 | 2,470 | Random | 1.92 (1.53–2.39) | 0.000 | 55.9 | 0.004 |
| Asian | 8 | 1,094 | Fixed | 2.62 (2.06–3.35) | 0.000 | 0.0 | 0.587 |
| Tumor types | |||||||
| EC | 6 | 781 | Fixed | 2.91 (1.86–4.53) | 0.000 | 0.0 | 0.719 |
| OC | 9 | 1,226 | Fixed | 1.82 (1.50–2.22) | 0.000 | 0.0 | 0.515 |
| LC | 7 | 1,314 | Random | 2.31 (1.54–3.47) | 0.000 | 82.4 | 0.000 |
| GC | 1 | 243 | – | 1.62 (1.00–2.62) | – | – | – |
| Sample | |||||||
| Blood | 16 | 2,661 | Random | 2.34 (1.82–3.02) | 0.000 | 65.7 | 0.000 |
| Tissue | 7 | 903 | Fixed | 1.67 (1.32–2.11) | 0.000 | 0.0 | 0.488 |
| Method | |||||||
| ECLIA | 2 | 161 | Fixed | 4.86 (2.09–11.34) | 0.000 | 0.0 | 0.358 |
| IHC | 6 | 850 | Fixed | 1.66 (1.30–2.13) | 0.000 | 8.1 | 0.365 |
| EIA | 10 | 1,893 | Random | 2.20 (1.61–3.01) | 0.000 | 73.1 | 0.000 |
| CMIA | 4 | 607 | Fixed | 2.10 (1.51–2.91) | 0.000 | 0.0 | 0.402 |
| HR estimate | |||||||
| Multivariate analysis | 13 | 2,066 | Fixed | 2.47 (2.05–2.97) | 0.000 | 13.4 | 0.310 |
| Univariate analysis | 10 | 1,498 | Fixed | 1.50 (1.31–1.71) | 0.000 | 47.8 | 0.045 |
Abbreviations: HR, hazard ratio; OS, overall survival; EC, endometrial cancer; OC, ovarian cancer; LC, lung cancer; GC, gastric cancer; ECLIA, electrochemiluminescence immunoassay assay; IHC, immunohistochemistry; EIA, enzyme immunoassay assay; CMIA, chemiluminescent microparticle immunoassay.
Figure 2.
Forest plot of hazard ratio for the association of HE4 expression and overall survival.
We also performed subgroup analysis as per race, tumor type, sample, analysis method, and HR estimate (Table 2). Subgroup analysis showed that the correlation between OS and HE4 expression did not differ by race (White: HR =1.92, 95% CI =1.53–2.39, P<0.001; Asian: HR =2.62, 95% CI =2.06–3.35, P<0.001) and tumor type (endometrial cancer: HR =2.91, 95% CI =1.86–4.53, P<0.001; ovarian cancer: HR =1.82, 95% CI =1.50–2.22, P<0.001; lung cancer: HR =2.31, 95% CI =1.54–3.47, P<0.001). Subgroup analysis, based on pooled data pertaining to sample, method, and HR estimate, also demonstrated that there was a significant association between OS and HE4 expression. Subgroup analysis of HR estimates found no significant heterogeneity (multivariate analysis: I2=13.4, univariate analysis: I2=47.8).
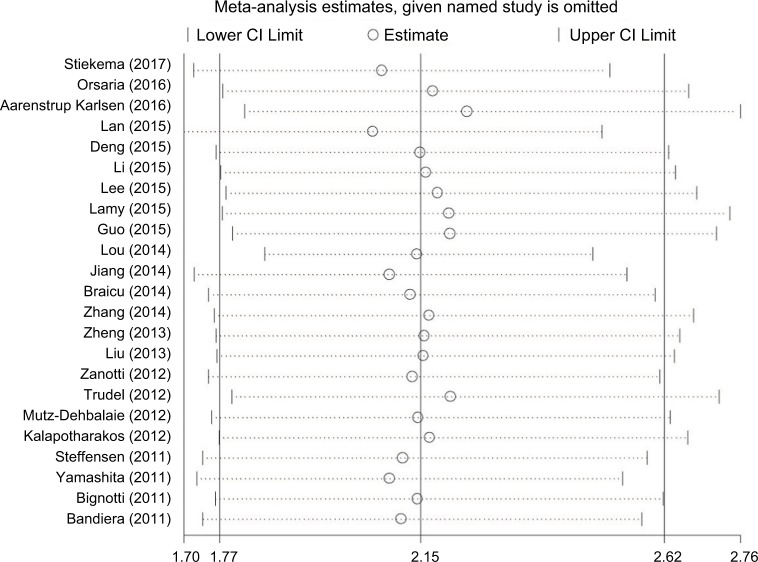
Included studies were sequentially removed to investigate whether any single study could have an influence on the pooled results. The results of the sensitivity analyses showed (Figure 3) that there was no influence of any single study on stable pooled HR.
Figure 3.
Sensitivity analysis of pooled HRs on the association between HE4 expression and overall survival.
HE4 expression and DFS in cancers
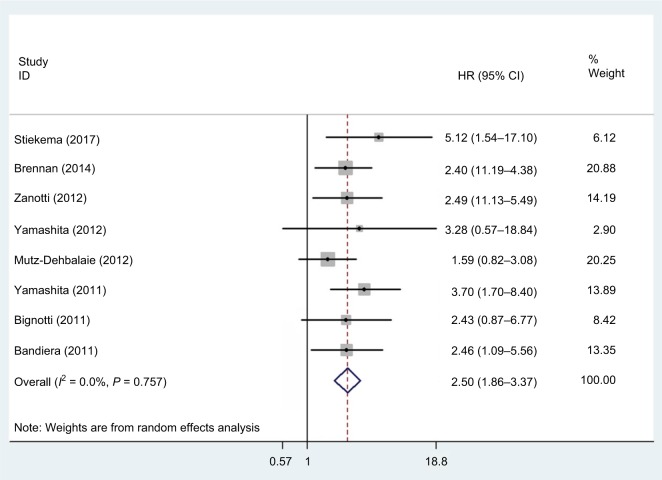
Eight studies, with a total of 1,296 patients, provided results pertaining to DFS. There was no significant heterogeneity (P=0.757, I2=0.0%) among the studies, so a fixed-effect model was used to calculate the pooled HR and 95% CI. Our results showed that increased HE4 expression was significantly associated with poorer DFS (HR =2.50, 95% CI =1.86–3.37, P<0.001) (Figure 4).
Figure 4.
Forest plot of hazard ratio for the association of HE4 expression and disease-free survival.
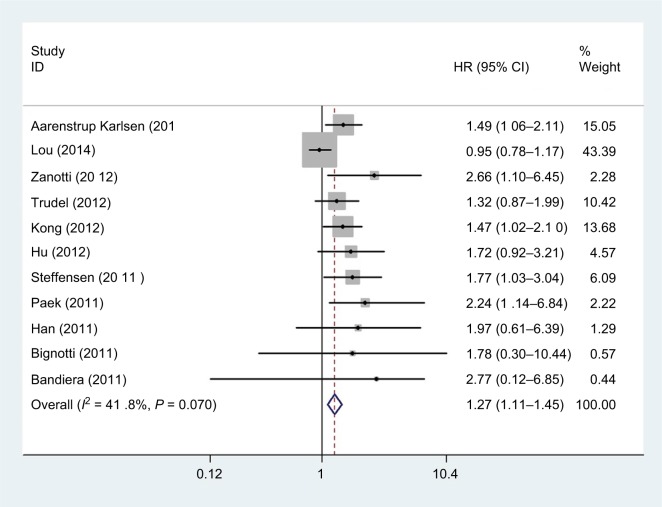
HE4 expression and PFS in cancers
As shown in Figure 5, there were eleven studies, comprising a total of 1,291 patients, that provided results regarding PFS. The pooled data demonstrated that there was a significant association between HE4 expression and PFS (HR =1.27, 95% CI =1.11–1.45, P=0.001). There was no significant heterogeneity (P=0.070, I2=41.8%) among the studies, so a fixed-effect model was used.
Figure 5.
Forest plot of hazard ratio for the association of HE4 expression and progression-free survival.
Publication bias
In this meta-analysis, both the Begg’s and the Egger’s tests were performed to assess if any publication bias existed in the included studies. Publication bias was observed in studies reporting OS (P=0.051, 0.000) and PFS (P=0.755, 0.003) but not in those reporting DFS (P=0.174, 0.149). The Begg’s plots for the effect of HE4 expression level on OS are shown in Figure 6.
Figure 6.
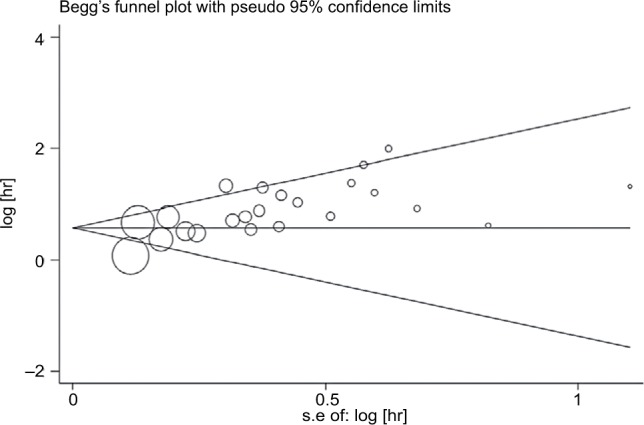
Funnel plots of publication bias for all of the included studies reported with overall survival.
Discussion
HE4 is a new tumor biomarker, which has been a subject of intense research in recent years. HE4, originally discovered by Kirchhoff in the human distal epididymal epithelial cells,40 is located on chromosome 20 at 20q12-13 and contains five exons and four introns.41 It contains a gene encoding protein domains that have homology with whey acidic protein, by which the product encoded is mainly protease inhibitor.42 As a member of the protease inhibitor family, it has an inhibitory effect on cell proliferation. Previous studies have reported that HE4 overexpression significantly promotes tumor cell apoptosis and adhesion and inhibits cell proliferation, migration, and invasiveness.43 Further, Kong et al44 found in vitro that this antitumor effect may be achieved by regulating the mitogen-activated protein kinase and phosphoinositide 3-kinase/AKT signal transduction pathways. Recently, further studies have been carried out to investigate the association between HE4 overexpression and prognosis in several tumors.45,46 However, the studies were inconclusive because of small sample sizes and inconsistencies in results. Therefore, to evaluate the relationship between HE4 expression and prognosis of cancer patients, we conducted this meta-analysis to mitigate sample size problems of individual studies and enhance the statistical power.
In the present meta-analysis, we analyzed 29 studies, including 3,564 patients, with OS data from 23 studies, DFS data from eight studies, and PFS data from eleven studies. The results indicated that there was no significant difference in the OS, DFS, and PFS outcomes based on HE4 expression status. There was significant heterogeneity in OS across the included studies. In order to ascertain the reason for the heterogeneity, we performed sensitivity analyses, and the results showed that the stable pooled HR was not significantly affected by any individual study. However, subgroup analyses revealed that the heterogeneity may have been due to the HR estimate methods used. Differences in the baseline characteristics of patients and in the HE4 expression cutoff values may have also contributed to the observed heterogeneity. However, for want of relevant data, it was not possible to determine the contribution of each of the above factors to the heterogeneity. In addition, we also found that the correlation between HE4 expression and OS of cancer patients was not affected by race, tumor type, sample source, detection method, or HR estimation method, and we therefore believe that HE4 may serve as a reliable and novel parameter for prognostication and a promising target for anticancer therapy in cancers.
Several previous meta-analyses have been conducted to research the association between HE4 expression and diagnosis and prognosis of cancer patients. For example, Zhong et al47 identified eight studies that involved 1,412 lung cancer patients and showed that high serum HE4 level was a marker of poor prognosis in lung cancer patients, particularly in patients of Asian origin. Compared with previous studies, our meta-analysis has several limitations as well as advantages. Our study is the first meta-analysis to review the role of HE4 in the OS, DFS, and PFS in several cancer types. In addition, to ensure the reliability of results, we have increased the number of studies included in the analysis.
Although we made every effort to conduct a comprehensive analysis, our study has several limitations. First, we tried to analyze the association between HE4 expression and prognosis in all cancer types, but the majority of included studies focused on endometrial cancer, ovarian cancer, or lung cancer. Hence, suitably designed larger future studies are needed to confirm our results. Second, when we evaluated the relationship between OS and HE4 expression, there was obvious publication bias, possibly because positive results are more likely to be published than are negative results. Last, the cutoff value for HE4 expression differed between studies, which may have led to heterogeneity. A standardized baseline value to designate positive/high HE4 expression status is thus needed. Meanwhile, the collection time and survival times were not standard, and this may be one of the sources of heterogeneity.
Conclusion
We found that increased expression of HE4 indicated poor survival outcomes in patients with cancer. Therefore, HE4 is a potential novel prognostic factor in cancer patients.
Acknowledgments
This work was supported by the National Social Science Foundation of China (16BGL183), the Natural Science Foundation of Shaanxi Province (2015JM8415), and the Fundamental Research Funds for the Central Universities of China (2011jdhz55).
Footnotes
Author contributions
ZD, CD, and JL designed the research. CD, YL, and TT contributed to the literature search. PX, MW, and YD carried out the data extraction. YW, ZZ, and QH contributed to statistical analysis. CD and YZ wrote the manuscript. ZD and CD contributed to revision of the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Abe S, Kawai K, Ishihara S, et al. Prognostic impact of carcinoembryonic antigen and carbohydrate antigen 19-9 in stage IV colorectal cancer patients after R0 resection. J Surg Res. 2016;205(2):384–392. doi: 10.1016/j.jss.2016.06.078. [DOI] [PubMed] [Google Scholar]
- 3.Jiang H, Tang E, Xu D, et al. Development and validation of nomograms for predicting survival in patients with non-metastatic colorectal cancer. Oncotarget. 2017;8(18):29857–29864. doi: 10.18632/oncotarget.16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speeckaert MM, Speeckaert R, Delanghe JR. Human epididymis protein 4 in cancer diagnostics: a promising and reliable tumor marker. Adv Clin Chem. 2013;59:1–21. doi: 10.1016/b978-0-12-405211-6.00001-2. [DOI] [PubMed] [Google Scholar]
- 5.Karlsen NS, Karlsen MA, Høgdall CK, Høgdall EV. HE4 tissue expression and serum HE4 levels in healthy individuals and patients with benign or malignant tumors: a systematic review. Cancer Epidemiol Biomarkers Prev. 2014;23(11):2285–2295. doi: 10.1158/1055-9965.EPI-14-0447. [DOI] [PubMed] [Google Scholar]
- 6.Chang X, Ye X, Dong L, et al. Human epididymis protein 4 (HE4) as a serum tumor biomarker in patients with ovarian carcinoma. Int J Gynecol Cancer. 2011;21(5):852–858. doi: 10.1097/IGC.0b013e31821a3726. [DOI] [PubMed] [Google Scholar]
- 7.Stiekema A, Lok C, Korse CM, et al. Serum HE4 is correlated to prognostic factors and survival in patients with endometrial cancer. Virchows Arch. 2017;470(6):655–664. doi: 10.1007/s00428-017-2115-1. [DOI] [PubMed] [Google Scholar]
- 8.Aarenstrup Karlsen M, Høgdall C, Nedergaard L, et al. HE4 as a predictor of adjuvant chemotherapy resistance and survival in patients with epithelial ovarian cancer. APMIS. 2016;124(12):1038–1045. doi: 10.1111/apm.12625. [DOI] [PubMed] [Google Scholar]
- 9.Lamy PJ, Plassot C, Pujol JL. Serum HE4: An independent prognostic factor in non-small cell lung cancer. PLoS One. 2015;10(6):e0128836. doi: 10.1371/journal.pone.0128836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 11.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 14.Orsaria M, Londero AP, Marzinotto S, di Loreto C, Marchesoni D, Mariuzzi L. Placental type alkaline phosphatase tissue expression in ovarian serous carcinoma. Cancer Biomark. 2016;17(4):479–486. doi: 10.3233/CBM-160665. [DOI] [PubMed] [Google Scholar]
- 15.Lan WG, Hao YZ, Xu DH, Wang P, Zhou YL, Ma LB. Serum human epididymis protein 4 is associated with the treatment response of concurrent chemoradiotherapy and prognosis in patients with locally advanced non-small cell lung cancer. Clin Transl Oncol. 2016;18(4):375–380. doi: 10.1007/s12094-015-1375-y. [DOI] [PubMed] [Google Scholar]
- 16.Deng L, Gao Y, Li X, et al. Expression and clinical significance of annexin A2 and human epididymis protein 4 in endometrial carcinoma. J Exp Clin Cancer Res. 2015;34:96. doi: 10.1186/s13046-015-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Gao Y, Tan M, et al. Expression of HE4 in Endometrial cancer and its clinical significance. Biomed Res Int. 2015;2015:437468–8. doi: 10.1155/2015/437468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YC, Huang CC, Lin DY, Chang WC, Lee KH. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ. 2015;3:e1386. doi: 10.7717/peerj.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo YD, Wang JH, Lu H, et al. The human epididymis protein 4 acts as a prognostic factor and promotes progression of gastric cancer. Tumour Biol. 2015;36(4):2457–2464. doi: 10.1007/s13277-014-2858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou E, Johnson M, Sima C, et al. Serum biomarkers for assessing histology and outcomes in patients with metastatic lung cancer. Cancer Biomark. 2014;14(4):207–214. doi: 10.3233/CBM-140399. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y, Wang C, Lv B, Ma G, Wang L. Expression level of serum human epididymis 4 and its prognostic significance in human non-small cell lung cancer. Int J Clin Exp Med. 2014;7(12):5568–5572. [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan DJ, Hackethal A, Metcalf AM, et al. ANECS Group Serum HE4 as a prognostic marker in endometrial cancer--a population based study. Gynecol Oncol. 2014;132(1):159–165. doi: 10.1016/j.ygyno.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zheng Y, Jiang L, Zhao Z, Liu W. The prognostic value of serum HE4 in lung cancer. Chin J Radiol Med Prot. 2014;34(6):423–426. [Google Scholar]
- 24.Braicu EI, Chekerov R, Richter R, et al. HE4 expression in plasma correlates with surgical outcome and overall survival in patients with first ovarian cancer relapse. Ann Surg Oncol. 2014;21(3):955–962. doi: 10.1245/s10434-013-3347-1. [DOI] [PubMed] [Google Scholar]
- 25.Zheng H, Gao YN, Gao WJ, Gao M, Yan X. Expression of serum human epididymis protein 4 in epithelial ovarian cancer and its correlation with prognosis. Zhonghua Zhong Liu Za Zhi. 2013;35(6):445–449. [PubMed] [Google Scholar]
- 26.Liu W, Yang J, Chi PD, et al. Evaluating the clinical significance of serum HE4 levels in lung cancer and pulmonary tuberculosis. Int J Tuberc Lung Dis. 2013;17(10):1346–1353. doi: 10.5588/ijtld.13.0058. [DOI] [PubMed] [Google Scholar]
- 27.Zanotti L, Bignotti E, Calza S, et al. Human epididymis protein 4 as a serum marker for diagnosis of endometrial carcinoma and prediction of clinical outcome. Clin Chem Lab Med. 2012;50(12):2189–2198. doi: 10.1515/cclm-2011-0757. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita S, Tokuishi K, Moroga T, et al. Serum level of HE4 is closely associated with pulmonary adenocarcinoma progression. Tumour Biol. 2012;33(6):2365–2370. doi: 10.1007/s13277-012-0499-8. [DOI] [PubMed] [Google Scholar]
- 29.Trudel D, Têtu B, Grégoire J, et al. Human epididymis protein 4 (HE4) and ovarian cancer prognosis. Gynecol Oncol. 2012;127(3):511–515. doi: 10.1016/j.ygyno.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Mutz-Dehbalaie I, Egle D, Fessler S, et al. HE4 is an independent prognostic marker in endometrial cancer patients. Gynecol Oncol. 2012;126(2):186–191. doi: 10.1016/j.ygyno.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Kong SY, Han MH, Yoo HJ, et al. Serum HE4 level is an independent prognostic factor in epithelial ovarian cancer. Ann Surg Oncol. 2012;19(5):1707–1712. doi: 10.1245/s10434-011-1943-5. [DOI] [PubMed] [Google Scholar]
- 32.Kalapotharakos G, Asciutto C, Henic E, Casslén B, Borgfeldt C. High preoperative blood levels of HE4 predicts poor prognosis in patients with ovarian cancer. J Ovarian Res. 2012;5(1):20. doi: 10.1186/1757-2215-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu S, Wang M, Gao J, Qi J. The clinical value of serum HE4 for prognosis evaluation in patients with ovarian cancer. China Cancer. 2012;21(8):638–640. [Google Scholar]
- 34.Steffensen KD, Waldstrøm M, Brandslund I, Jakobsen A. Prognostic impact of prechemotherapy serum levels of HER2, CA125, and HE4 in ovarian cancer patients. Int J Gynecol Cancer. 2011;21(6):1040–1047. doi: 10.1097/IGC.0b013e31821e052e. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita S, Tokuishi K, Hashimoto T, et al. Prognostic significance of HE4 expression in pulmonary adenocarcinoma. Tumour Biol. 2011;32(2):265–271. doi: 10.1007/s13277-010-0118-5. [DOI] [PubMed] [Google Scholar]
- 36.Paek J, Lee SH, Yim GW, et al. Prognostic significance of human epididymis protein 4 in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol. 2011;158(2):338–342. doi: 10.1016/j.ejogrb.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Han JJ, Yu M, Houston N, Steinberg SM, Kohn EC. Progranulin is a potential prognostic biomarker in advanced epithelial ovarian cancers. Gynecol Oncol. 2011;120(1):5–10. doi: 10.1016/j.ygyno.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bignotti E, Ragnoli M, Zanotti L, et al. Diagnostic and prognostic impact of serum HE4 detection in endometrial carcinoma patients. Br J Cancer. 2011;104(9):1418–1425. doi: 10.1038/bjc.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandiera E, Romani C, Specchia C, et al. Serum human epididymis protein 4 and risk for ovarian malignancy algorithm as new diagnostic and prognostic tools for epithelial ovarian cancer management. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2496–2506. doi: 10.1158/1055-9965.EPI-11-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirchhoff C, Habben I, Ivell R, Krull N. A major human epididymis-specific cDNA encodes a protein with sequence homology to extracellular proteinase inhibitors. Biol Reprod. 1991;45(2):350–357. doi: 10.1095/biolreprod45.2.350. [DOI] [PubMed] [Google Scholar]
- 41.Bingle L, Singleton V, Bingle CD. The putative ovarian tumour marker gene HE4 (WFDC2), is expressed in normal tissues and undergoes complex alternative splicing to yield multiple protein isoforms. Oncogene. 2002;21(17):2768–2773. doi: 10.1038/sj.onc.1205363. [DOI] [PubMed] [Google Scholar]
- 42.Clauss A, Lilja H, Lundwall A. A locus on human chromosome 20 contains several genes expressing protease inhibitor domains with homology to whey acidic protein. Biochem J. 2002;368(Pt 1):233–242. doi: 10.1042/BJ20020869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao L, Cheng HY, Dong L, et al. The role of HE4 in ovarian cancer: inhibiting tumour cell proliferation and metastasis. J Int Med Res. 2011;39(5):1645–1660. doi: 10.1177/147323001103900507. [DOI] [PubMed] [Google Scholar]
- 44.Kong X, Chang X, Cheng H, Ma R, Ye X, Cui H. Human epididymis protein 4 inhibits proliferation of human ovarian cancer cells via the mitogen-activated protein kinase and phosphoinositide 3-kinase/AKT pathways. Int J Gynecol Cancer. 2014;24(3):427–436. doi: 10.1097/IGC.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 45.Chudecka-Głaz A, Rzepka-Górska I, Wojciechowska I. Human epididymal protein 4 (HE4) is a novel biomarker and a promising prognostic factor in ovarian cancer patients. Eur J Gynaecol Oncol. 2012;33(4):382–390. [PubMed] [Google Scholar]
- 46.Lee S, Choi S, Lee Y, Chung D, Hong S, Park N. Role of human epididymis protein 4 in chemoresistance and prognosis of epithelial ovarian cancer. J Obstet Gynaecol Res. 2017;43(1):220–227. doi: 10.1111/jog.13181. [DOI] [PubMed] [Google Scholar]
- 47.Zhong H, Qian Y, Fang S, Yang L, Li L, Gu W. HE4 expression in lung cancer, a meta-analysis. Clinica Chimica Acta. 2017;470:109–114. doi: 10.1016/j.cca.2017.05.007. [DOI] [PubMed] [Google Scholar]