Abstract
The pentose phosphate pathway (PPP) confers protection against oxidative stress by supplying NADPH necessary for the regeneration of glutathione, which detoxifies H2O2 into H2O and O2. RPE functions in the PPP, catalyzing the reversible conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate and is an important enzyme for cellular response against oxidative stress. Here, using structural, biochemical, and functional studies, we show that human d-ribulose 5-phosphate 3-epimerase (hRPE) uses Fe2+ for catalysis. Structures of the binary complexes of hRPE with d-ribulose 5-phosphate and d-xylulose 5-phosphate provide the first detailed molecular insights into the binding mode of physiological ligands and reveal an octahedrally coordinated Fe2+ ion buried deep inside the active site. Human RPE folds into a typical (β/α)8 triosephosphate isomerase (TIM) barrel with a loop regulating access to the active site. Two aspartic acids are well positioned to carry out the proton transfers in an acid-base type of reaction mechanism. Interestingly, mutating Ser-10 to alanine almost abolished the enzymatic activity, while L12A and M72A mutations resulted in an almost 50% decrease in the activity. The binary complexes of hRPE reported here will aid in the design of small molecules for modulating the activity of the enzyme and altering flux through the PPP. —Liang, W., Ouyang, S., Shaw, N., Joachimiak, A., Zhang, R., Liu, Z-J. Conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE.
Keywords: oxidative stress, pentose phospate pathway, metalloenzyme
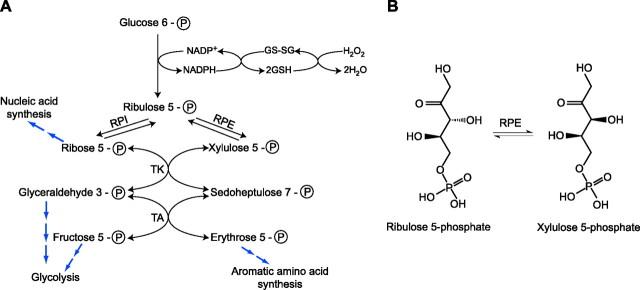
d-ribulose 5-phosphate 3-epimerase (RPE) catalyzes the reversible conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate (Fig. 1) (1). This conversion is important for the assimilation of CO2 by plants, which is carried out via the Calvin cycle (2). In humans, this reaction drives the nonoxidative phase of the pentose phosphate pathway (PPP), which generates precursors such as erythrose 4-phosphate, glyceraldehyde 3-phosphate, and fructose 6-phosphate that are necessary for the synthesis of aromatic amino acids and production of energy (Fig. 1) (3, 4). In addition, a majority of the NADPH used by the human body for biosynthetic purposes is supplied by the PPP (5). Interestingly, RPE has been shown to protect cells from oxidative stress via its participation in PPP for the production of NADPH (6–8). The protection against reactive oxygen species is exerted by NADPH's ability to reduce glutathione, which detoxifies H2O2 into H2O (Fig. 1). Mutants of yeast lacking a functional RPE were shown to be susceptible to oxidative stress (9).
Figure 1.
A) Schematic diagram of the pentose phosphate pathway depicting the role of NADPH in detoxification of H2O2. The pathway produces precursors for the synthesis of nucleic acids, aromatic amino acids, and energy via the glycolytic pathway. B) Reaction catalyzed by RPE. RPE, ribulose 5-phosphate 3-epimerase; RPI, ribose 5-phosphate isomerase; TK, transketolase; TA, transaldolase.
RPE is a metalloenzyme and has been shown to use the divalent Zn2+ ion predominantly for catalysis (10–13). The apo enzyme from Streptococcus pyogenes could be activated by the addition of Zn2+, Mn2+, or Co2+ ions, but not Fe2+ or Mg2+ ions (10). These results suggest that RPE may not be able to use Fe2+ or Mg2+ for catalysis. The 3-dimensional structures of a number of RPE homologues are known (10–14). A comparison of these structures seems to suggest that the overall structure of RPE is conserved. However, none of the structures have been determined in complex with the physiological ligands. The only structure of a RPE homologue solved in complex with a ligand to date, the structure of RPE from S. pyogenes in complex with a substrate analog d-xylitol 5-phosphate (10), confirms results of mutagenesis and isotope exchange studies that implicate a pair of aspartates as the acid/base catalysts (15, 16). A single metal ion is observed octahedrally coordinated in RPEs and has been postulated to stabilize the cis-enediolate reaction intermediate. The human RPE (hRPE) shares 44% sequence identity with the RPE from S. pyogenes. To gain structural insights into the mode of substrate binding and the mechanism of the reversible conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate, we have solved, for the first time, the structures of the binary complexes of hRPE with d-ribulose 5-phosphate and d-xylulose 5-phosphate. Further, we have probed the role of residues surrounding the ligands in catalysis by site-directed mutagenesis and functional assays. Our structural, mutagenesis, and functional studies on hRPE suggest a highly conserved mechanism of catalysis. Interestingly, structural and biochemical evidence indicates that hRPE uses Fe2+ ion for catalysis. These findings have implications for the role of RPE in oxidative stress.
MATERIALS AND METHODS
Protein production
RPE was PCR amplified from human brain cDNA library and cloned into pMD-18T vector (Takara, Beijing, China). After verifying the DNA sequence, full-length RPE (aa 1–228) was subcloned into pMCSG7 vector for expression in Escherichia coli BL 21 (DE3). N-terminal hexa His-tagged RPE was produced by growing the cells in Luria Bertani medium at 37°C for 4 h until OD660nm reached 1.0. Induction was carried out at 16°C for 20 h by adding 0.2 mM IPTG. Cells were harvested by centrifugation and lysed by sonication. Soluble hRPE was purified by Ni-affinity chromatography. After buffer exchange to remove the imidazole, the His tag was removed by treating the protein with TEV protease. Uncut protein and TEV were removed by a second round of Ni-affinity chromatography. The tagless protein was exchanged into a buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM NaCl, using a Superdex G75 size-exclusion column (GE Healthcare, Piscataway, NJ, USA). After concentration using 10-kDa-cutoff centrifugal concentrators, the protein (15–20 mg/ml) was immediately screened for crystallization.
Crystallization and data collection
Crystallization screening was carried out using commercially available sparse matrix screens. Hanging drops (1 μl) containing 0.5 μl protein mixed with 0.5 μl mother liquor were equilibrated over 300 μl reservoir solution and incubated at 16°C. Crystals grown in 25% PEG 3350, 100 mM Bis-Tris (pH 5.5), and 200 mM NaCl diffracted X-rays to 1.70 Å at beamline 19-ID of the Advanced Photon Source (APS; Argonne National Laboratory, Argonne, IL, USA). The binary complexes of hRPE with d-ribulose 5-phosphate and d-xylulose 5-phosphate were obtained by soaking the crystals with ligands in 50% PEG 3350 (Table 1).
Table 1.
Data collection and refinement statistics
| Statistic | RPE | RPE soaked with Ru5P | RPE soaked with Xu5P |
|---|---|---|---|
| Data collection | 19-ID, APS | MicroMax-007IP, IBP | MicroMax-007IP, IBP |
| Wavelength (Å) | 0.9794 | 1.54 | 1.54 |
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 63.77, 46.75, 73.31 | 61.79, 43.54, 72.27 | 62.01, 43.67, 72.66 |
| α, β, γ (deg) | 90.00, 89.98, 90.00 | 90.00, 94.88, 90.00 | 90.0, 94.69, 90.0 |
| Resolution (Å) | 50.00–1.70 (1.76–1.70) | 50.00–2.00 (2.07–2.00) | 50.00–1.95 (2.02–1.95) |
| Rsym (%) | 0.076 (0.279) | 0.054 (0.347) | 0.056 (0.306) |
| I/σI | 36.68 (6.26) | 86.31 (12.25) | 41.52 (7.93) |
| Completeness (%) | 98.5 (96.3) | 99.9 (100.0) | 98.8 (97.8) |
| Redundancy | 7.1 (5.3) | 29.0 (28.0) | 8.5 (8.1) |
| Refinement | |||
| Resolution (Å) | 36.66–1.70 (1.76–1.70) | 25.94–2.00 (2.08–2.00) | 25.22–1.95 (2.02–1.95) |
| Reflections | 46167 (3905) | 26067 (2675) | 27840 (2474) |
| Rwork/Rfree (%) | 14.74/17.42 (14.02/16.73) | 14.72/19.10 (12.55/19.53) | 14.41/19.18 (14.01/21.45) |
| Atoms | |||
| Protein | 3630 | 3621 | 3607 |
| Ligand | 2 Fe | 2 Fe | 2 Fe |
| — | 2 Ru5P | 2 Xu5P | |
| 6 PEG chains | 7 PEG chains | 7 PEG chains | |
| Water | 282 | 154 | 252 |
| RMS deviation | |||
| Bonds | 0.006 | 0.007 | 0.006 |
| Angles | 1.087 | 1.033 | 1.017 |
| Mean B value (Å2) | 17.21 | 32.05 | 23.28 |
| Ramachandran analysis | |||
| Favored region (%) | 368 (96.08) | 346 (95.58) | 379 (95.47) |
| Allowed region (%) | 10 (2.61) | 14 (3.87) | 17 (4.56) |
| Outliers (%) | 3 (0.78) | 2 (0.55) | 2 (0.54) |
Numbers in parentheses represent values for the highest-resolution shell.
Structure determination
Crystals were frozen in liquid nitrogen prior to diffraction testing and data collection. Native diffraction data were collected at a wavelength of 0.979 Å at beamline 19-ID of the APS (Argonne National Laboratory). Data were indexed and scaled to 1.70-Å resolution using HKL2000 (17). The structure was solved by molecular replacement method using Phaser MR (18) with structure of the rice RPE [Protein Data Bank (PDB) code 1H1Y] as a search model. The initial phase was improved with Oasis (19). The asymmetric unit consists of 2 molecules of hRPE. The model was manually improved in Coot (20). Refinement was carried out using Refmac (21) and Phenix (22) alternately. Details of data collection and refinement statistics are listed in Table 1. The quality of the final model was validated with MolProbity (23). Additional data sets were collected near the absorption edge of Fe. Electron density maps calculated from the anomalous differences were used to confirm identity of the metal ion.
Activity assay
The enzymatic activity of hRPE was measured using a commercially available kit from Sigma (St. Louis, MO, USA). Briefly, the production of d-xylulose 5-phosphate was monitored using an enzyme-coupled spectrophotometric assay. The d-xylulose 5-phosphate formed was first converted to glyceraldehyde 3-phosphate and sedoheptulose 7-phosphate using a transketolase. Next, the glyceraldehyde 3-phosphate was converted to dihydroxyacetone phosphate by the action of triosephosphate isomerase (TIM). Finally, the dihydroxyacetone phosphate was converted to glycerol phosphate using a glycerophosphate dehydrogenase. The last step involves the oxidation of NADH to NAD, which can be monitored by reading the absorbance at 340 nm. The reaction mixture consisted of 2 mM of d-ribulose 5-phosphate in 50 mM glycylglycine (pH 7.7), 0.001 mg cocarboxylase/thiamine pyrophosphate, 0.0625 mM NADH, 0.01 U transketolase, 0.01 U of α-glycerophosphate dehydrogenase/TIM, 7.5 mM MgCl2, and appropriate dilution of the enzyme. The change in absorbance recorded at 340 nm represents the rate of conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate. One unit of activity is defined as the amount of enzyme required to convert 1 μmol of d-ribulose 5-phosphate to d-xylulose 5-phosphate under the assay conditions.
Mutagenesis
Point mutations were introduced into hRPE by overlap extension PCR (24) with primers containing intended mutations. The overlap PCR product was ligated into pMCSG7 as described earlier. The mutations were confirmed by nucleotide sequencing.
Metal analysis
X-ray fluorescence scans were performed at the absorption edges for Zn, Ca, Ni, Mg, Co, and Fe at beamline 19-ID of the APS (Argonne National Laboratory). Analysis of the total metal content was carried out using inductively coupled plasma mass spectrometry (ICP-MS; Thermo, San Jose, CA, USA) at the Analysis Center of Tsinghua University (Beijing, China).
Accession codes
The atomic coordinates and structure factor files of the apo-hRPE, hRPE:d-ribulose 5-phosphate complex, and hRPE:d-xylulose 5-phosphate complex have been deposited in the PDB under the accession codes 3OVP, 3OVQ, and 3OVR, respectively.
RESULTS
Overall structure of hRPE
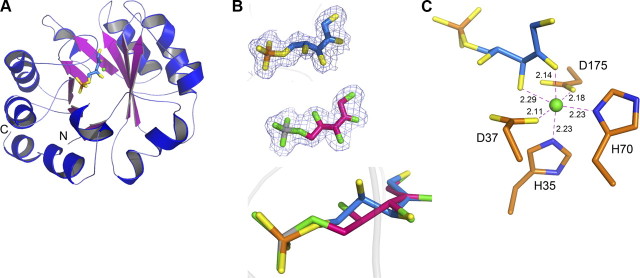
The structure of the apo form of hRPE was solved by molecular replacement using the structure of the rice RpE (Oryza sativa; PDB code 1H1Y) as a search model. The final model, containing residues 4–223 of the enzyme and 282 water molecules, was refined to 1.70-Å resolution. The asymmetric unit consists of a dimer of RPE molecules, which is consistent with the results of the size-exclusion chromatography elution profile of RPE where the protein elutes as a dimer. To confirm the oligomerization state of the protein in solution, we carried out analytical ultracentrifugation analysis of RPE. Results of the sedimentation velocity experiments suggest that hRPE exists as a dimer in solution. The monomers within the dimer are identical. RPE folds into a single domain with a classical TIM-barrel α/β fold (Fig. 2A). A central β sheet made up of 8 parallel strands makes up the core barrel. A helix is inserted between consecutive strands. The βα loops connecting the strands with helices have been known to impart substrate specificities to a wide range of enzymes catalyzing diverse reactions employing the TIM-barrel fold. The overall structure of hRPE closely mirrors the structures of RPE homologues reported previously (10–14). To gain insights into the mode of ligand binding and the nature of the active site residues of hRPE, we soaked the crystals of apo-RPE with the substrate d-ribulose 5-phosphate. The electron density for the substrate was clear and permitted unambiguous placement of the substrate into the active site (Fig. 2B). Further, to unravel the structural basis for the mechanism of catalysis at the molecular level and view the interaction of the product with the enzyme, we solved the structure of hRPE in complex with the product d-xylulose 5-phosphate by soaking the crystals of apo-RPE with the product (Fig. 2B). Except for the position of the loop connecting helice α3 with strand β3, the structures of the apo and binary complexes of RPE with the substrate and product are identical. The loop is seen capping the active site and therefore the binding of the ligand might have caused the movement of the loop. An interesting aspect of the structural studies on hRPE was the nature of the metal ion bound to the enzyme. RPE is a metalloenzyme and requires a divalent metal ion for its activity. Previously, RPE has been shown to carry out catalysis using Co2+, Mn2+, and Zn2+ ions. More important, the enzyme could not use Fe2+ and Mg2+ for catalysis (10). We carried out metal analysis on the hRPE expressed in E. coli. Surprisingly, the results suggested that hRPE binds Fe2+ predominantly. The metal ion seems to have originated from the medium used for the production of RPE. The Fe2+ binds hRPE tightly, and density for the metal was visible even after treatment of the protein with 20 mM EDTA. Structural evidence supports binding of a divalent metal ion into the density observed. An octahedral coordination and the charge of the groups involved in coordination support building of a positively charged divalent ion in the electron density (Fig. 2C). To confirm the nature of the metal ion, we performed an X-ray fluorescence scan of the protein crystal at the absorption edge of Zn2+, Mg2+, Co2+, Ca2+, Ni2+, and Fe2+. The results of the scan suggested that hRPE bound Fe2+ predominantly under the conditions mentioned in Materials and Methods. Finally, in order to confirm the nature and location of Fe atoms in the crystal structure, we collected anomalous data at the Fe peak and low remote wavelengths. Inspection of the anomalous difference electron density map around the Fe binding site confirmed the presence of Fe2+ in the crystal. The statistics of the anomalous data are listed in Supplemental Table S1, and the anomalous difference electron density map is shown in Supplemental Fig. S1.
Figure 2.
Structure of hRPE. A) Cartoon representation of the structure. B) Electron density for d-ribulose 5-phosphate (blue sticks) and d-xylulose 5-phosphate (magenta sticks) contoured at 1 σ. Superimposition of the ligands that were modeled in the density to depict the epimerization at C3 atom. C) Octahedral coordination of the Fe2+ ion. d-Ribulose 5-phosphate is shown as blue sticks, Fe2+ is depicted as a sphere and the amino acids involved in coordination are shown as orange sticks.
Active site of hRPE
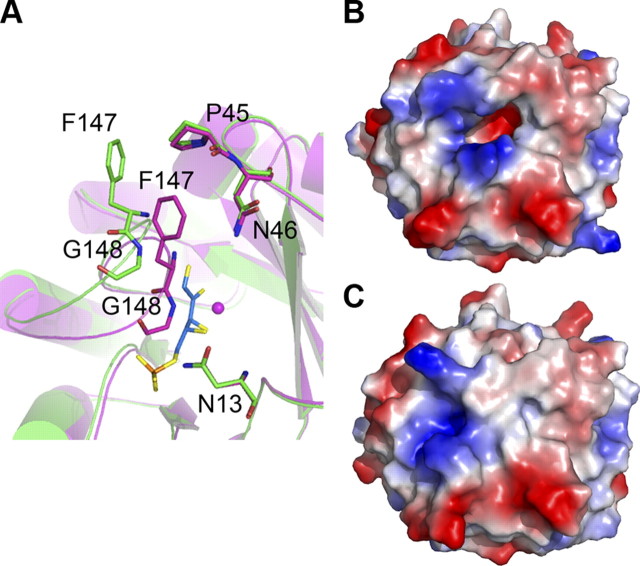
To map the location and gain insights into the architecture of the active site, we solved the structure of binary complexes of hRPE with ribulose 5-phosphate and xylulose 5-phosphate at 1.76- and 1.80-Å resolution, respectively. We compared the structure of the binary complexes with the structure of the apo enzyme. The ligands bind deep inside a narrow tunnel just above the β barrel (Fig. 2). Phe147, Gly148, and Ala149 of the loop region connecting strand β6 with helice α6 are interacting with the ligand in the binary complexes and appear to be capping the active site. The interaction between the pi electron cloud of Phe147 and the γ carbon atom of Pro45 observed in the apo structure is broken as the loop caps the active site in presence of the ligand (Fig. 3A). While the ε1 carbon atom of Phe147 was 3.9 Å from the γ carbon atom of Pro45 in the apo structure, the minimum distance between any of the atoms of Phe147 and Pro45 is now > 4.44 Å in the binary complexes. As a result of this movement of the loop, the ε1 carbon atom of Phe147 is now at a distance of 3.55 Å from the δ2 nitrogen atom of Asn46, and the α carbon of Gly148 is 4.0 Å from the δ1 oxygen atom of Asn13. These new interactions of the aromatic ring of Phe147 with Asn46 and that of Gly148 with Asn13 observed in the binary complexes result in the closure of the active site and isolation of the reactants from the aqueous environment (Fig. 3B, C). The Fe2+ ion occupies an identical position in all three structures. In the structure of the apo enzyme, the Fe2+ ion is tetrahedrally coordinated, with His35, His70, Asp37, and Asp175 participating in the coordination. A similar tetrahedral coordination for a Zn2+ ion has been reported for the apo form of RpE homologs from Plasmodium falciparum, potato, and rice (11, 13, 14). In contrast to the structure of the apo enzyme, the Fe2+ ion is coordinated octahedrally in the binary complexes of the hRPE with the substrate and product. His35, His70, Asp37, Asp175, and oxygens O2 and O3 of the ligand are coordinated to the Fe2+ ion (Fig. 2C).
Figure 3.
Active site of hRPE is capped on ligand binding. A) Structure of the apo-hRPE (green) superimposed on the hRPE–d-ribulose 5-phosphate binary complex (magenta) showing Phe147 and Gly148 closing the active site on binding of d-ribulose 5-phosphate (blue sticks). Fe2+ is shown as a sphere. B, C) Surface electrostatic potential representation of the apo-hRPE (B) and the binary complex (C) showing the open and capped active site, respectively. Blue indicates positive potential; red, negative potential.
The active site pocket can accommodate either d-ribulose 5-phosphate or d-xylulose 5-phosphate without causing any noticeable movement of the side chains of the catalytic residues. A number of amino acids are interacting with the ligands (Fig. 4A). The positions of C1, O1, C2, O2, and the phosphate group of d-ribulose 5-phosphate and d-xylulose 5-phosphate are identical in the binary complexes. The C1 end of both the ligands is localized by a 2.7-Å hydrogen bond between the O1 oxygen atom of the ligands and the backbone amide nitrogen of Gly146. Further, side chains of Met39, Met72, and Met141 constrict the active site around the O1 and C1 atoms, preventing any movement of the ligands around this region. Based on modeling studies, the methionines have been postulated to stabilize the charge on the O2 oxygen during catalysis (14). In the binary complexes of hRPE with d-ribulose 5-phosphate and d-xylulose 5-phosphate, these methionines are at least 3.5 Å from the ligands. Therefore, the methionines are more likely to play a role in imparting substrate specificity by constricting the active site rather than participate directly in catalysis. In addition, they probably assist in docking of the substrate into the active site under optimal orientation for catalysis. The C2 and O2 atoms of d-ribulose 5-phosphate and d-xylulose 5-phosphate are immobilized by bonding with Fe2+ ion, Asp37, His70, and Asp175. In contrast to the C1 and C2 atoms, the positions of the C3 carbon and O3 oxygen are different in the binary complexes simply because the epimerization occurs at this carbon (Fig. 2B). Further, the C4 carbon and the O4 oxygen have risen upwards in the hRPE:d-xylulose 5-phosphate binary complex, as a result of which the O4 is now hydrogen bonded to the hydroxyl group of Ser-10. This hydrogen bond is missing in the hRPE:d-ribulose 5-phosphate binary complex. These subtle differences in the configuration of the C4 atom have implications for the mechanism of catalysis.
Figure 4.
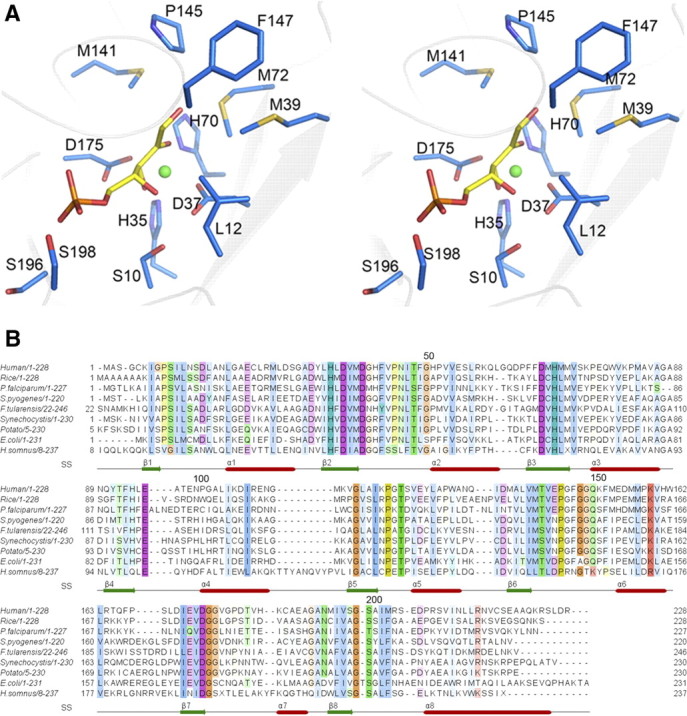
A) Stereo view of the amino acids surrounding d-ribulose 5-phosphate (yellow sticks). Amino acids are shown as blue sticks; Fe2+ is shown as a sphere. B) Alignment of the primary sequence of RPE orthologs deposited in PDB. Conservation has been colored according to Clustal W convention. Secondary structure of hRPE is annotated at bottom.
Catalytically critical residues
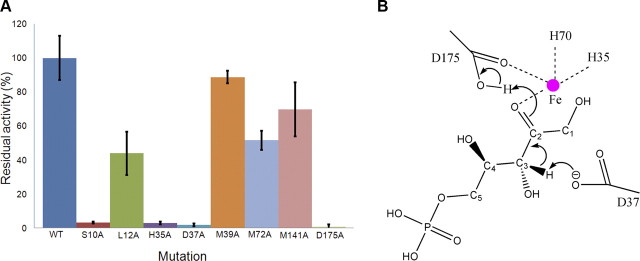
Using the structures of the binary complexes of RPE and primary sequence alignment of RPE orthologs as a guide, we carried out alanine scanning mutagenesis of amino acids surrounding the ligands to determine their role in catalysis (Fig. 4). Accordingly, 9 amino acids were mutated to alanine and expressed under identical conditions in E. coli (Fig. 5A). All the mutants were purified to homogeneity using affinity and size-exclusion chromatography, before being assayed for enzymatic activity under identical conditions. His35, Asp37, His70, and Asp175 are seen coordinating the metal ion in the structures of hRPE (Fig. 4A). Mutating either of them to alanine affected the solubility of the protein. Although the level of expression was similar to that of the wild-type enzyme, > 90% of the mutant enzyme was insoluble, suggesting that the mutations affected the secondary structure of the protein. While some protein could be salvaged to perform activity assays for H35A, D37A, and D175A mutants, H70A mutant was completely insoluble and therefore could not be tested for activity. None of the mutants displayed any significant activity (Fig. 5A). These amino acids are highly conserved among all the orthologs of RPE (Fig. 4B). Mutating similar amino acids in the RPE from S. pyogenes resulted in a loss of catalytic activity (10). Interestingly, mutating Ser-10 to alanine resulted in a dramatic decrease in the activity of the enzyme (Fig. 5A). In the binary complex of xylulose 5-phosphate with RPE, the hydroxyl oxygen of Ser-10 is hydrogen bonded to the C4 oxygen, which is forming a hydrogen bond with Wat30. Further, Wat30 is hydrogen bonded to Wat10, which is linked to the carbonyl oxygen of Ser-10. This hydrogen bonding network of Ser-10 is probably important for the relay of charge. Circular dichroism (CD) analysis of the S10A mutant suggested that the mutation does not affect the secondary structure of the enzyme. Leu12, Asn13, and Met39 together with Pro145-Phe147 are capping the active site. Further, the side chain of Leu12 is interacting with the C4 hydroxyl oxygen of d-xylulose 5-phosphate, backbone amide of Gly148, and the side chains of Met39 and Thr48. L12A mutation probably disrupts these interactions and decreases the hydrophobicity of the region, resulting in a > 55% loss of the enzymatic activity (Fig. 5A). L12A mutation does not affect the secondary structure of the protein as indicated by a CD analysis of the mutant enzyme. Leu12 is highly conserved among the orthologs of RPE. Mutating Met39, Met72, or Met141 to alanine decreased the enzymatic activity (Fig. 5A). These methionines are inside the active site pocket and are within the van der Waal's radii of the substrate. Mutating the methionines to alanine probably perturbs the structure around this region, affecting the optimal docking of the substrate into the active site. While M72A mutation resulted in an almost 50% loss in activity, the loss in activity for M141A was marginal. M39A retained 90% of its activity when compared to that of the wild-type enzyme. CD analysis of all three mutants revealed that the mutation altered the content of the secondary structural elements of the protein. These methionines are highly conserved, with Met72 being absolutely conserved among the orthologs of RPE (Fig. 4B).
Figure 5.
Mechanism of catalysis. A) Residual activity of hRPE mutants. Values plotted are an average of 3 independent experiments performed under identical conditions in duplicates. Error bars = sd. B) Diagrammatic representation of the mechanism of catalysis depicting the proton transfers. For clarity, the coordination of Fe2+ with the hydroxyl group of C3 and the carboxyl oxygen of Asp37 has not been shown.
DISCUSSION
Human RPE uses Fe2+ ions for catalysis
Human RPE consistently bound Fe2+ when produced under conditions mentioned in Materials and Methods. The enzyme was active when assayed for activity using ribulose 5-phosphate as a substrate. Since the Fe2+ ion binds the enzyme tightly, hRPE is probably able to catalyze the reaction using Fe2+ ion. Interestingly, the activity of the S. pyogenes RPE stripped off its metal ion by treatment with EDTA and did not increase on addition of Fe2+ ions. However, addition of Zn2+, Mn2+, and Co2+ under identical conditions resulted in an increase in activity of the enzyme (10). These results suggest that the enzyme may not be able to utilize Fe2+ to catalyze the reaction. Although we are reporting for the first time that hRPE might be Fe2+ dependent or at least be able to bind and use Fe2+ for activity, the nature of the divalent metal ion preferred by RPE under physiological conditions needs to be investigated. In addition, a comparative study of the effect of metal ion on RPE enzymatic activity using enzyme produced in minimal medium supplemented with different divalent metal ions would help understand the specificity of the enzyme for divalent metal ions.
The ability of RPE to confer protection against oxidative stress stems from its role in NADPH/NADP+ homeostasis, which plays a major role in detoxification of the reactive oxygen species (8). The observation that hRPE can utilize Fe2+ ions for catalysis potentially provides another explanation for its role in protection against oxidative stress. A number of essential biological processes result in the formation of hydrogen peroxide during cellular metabolism. Under normal conditions, the hydrogen peroxide is converted to H2O and O2 by peroxidases and catalases (25). Accumulation of hydrogen peroxide during stress has deleterious effects and can lead to cell damage and death. Some of the detrimental effects are a manifestation of the Fenton reaction, where Fe2+ ions react with H2O2, resulting in the formation of highly toxic hydroxyl radicals that can modify amino acids, carbohydrates, lipids, and nucleotides (26). RPE probably helps alleviate this damage by binding free Fe2+ ions, thereby making them unavailable for reaction with H2O2.
Dimer interface is not conserved
RPEs have been reported to exist as dimers or hexamers. RPEs from yeast (1), rice (11), and Plasmodium (13) exist as dimers, while the RPEs from potato (14), Cyanobacterium synechocystis (12), and S. pyogenes (10) assemble into hexamers. Human RPE crystallized as a dimer. Analytical ultracentrifugation analysis confirmed that hRPE exists as a dimer in solution. In the structure of hRPE, amino acids positioned between stands β2 and β3 and between β3 and β4 are seen engaged in intermolecular interactions. Among the 14 amino acids involved in intermolecular interactions within a distance of ≤3.2 Å, only Asp40 and Asn46 are conserved among the orthologs of RPE. Therefore, the dimerization interface observed for hRPE in the crystal structure is not conserved.
Mechanism of catalysis
RPE catalyzes the epimerization of ribulose 5-phosphate to xylulose 5-phosphate via a cis-enediolate intermediate employing an acid-base type of catalytic mechanism. Isotope exchange studies, mutagenesis, and structural studies on RPE homologues reported previously (15, 16) suggest the participation of a pair of aspartic acids, with one acting as a proton donor and the other as a proton acceptor (Fig. 5B). Asp37 is hydrogen bonded to Fe2+ and Ser-10. The deprotonated Asp37 abstracts a proton from the C3 atom of d-ribulose 5-phosphate, resulting in a cis-enediolate intermediate. The excess charge on the O2 atom of the intermediate is probably stabilized by the interactions of the atom with Fe2+ and His70. Asp175 donates a proton to complete the epimerization and formation of d-xylulose 5-phosphate (Fig. 5B). Because of the change in configuration of C3, the positions of C4 and the hydroxyl group at C4 changes. The hydroxyl group is now hydrogen bonded to Ser-10 and Asp37. This assists in the reversal of role for the catalytic aspartates during the conversion of d-xylulose 5-phosphate to d-ribulose 5-phosphate.
In summary, the structures of hRPE reported here provide a clear picture of the architecture of the active site. Structural, mutagenesis, and functional data suggest that RPE uses a highly conserved mechanism for catalysis. Our studies on hRPE uncover an unknown aspect of the enzyme—hRPE can bind and use Fe2+ ions for catalysis. Further functional studies are warranted to elucidate the physiological significance of this finding. This will also help answer the question whether the ability of RPE to bind Fe2+ ions plays a role in protection against oxidative stress.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
Acknowledgments
The authors thank Dr. Keming Tan (Structural Biology Center, Argonne National Laboratory) for the help in collecting anomalous data at the edge of Fe.
This work was funded by the Ministry of Science and Technology of China (grants 2006AA02A316, 2009DFB30310, and 2006CB910901), the National Natural Science Foundation of China (grants 30670427 and 30721003), the Ministry of Health of China (grant 2008ZX10404), a Chinese Academy of Sciences (CAS) research grant (KSCX2-YW-R-127 and INFO-115-D01-2009), and a CAS fellowship for young international scientists (grant 2010Y1SA1). Crystallographic data were collected at beamline 19-ID of APS (Argonne National Laboratory).
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Williamson W. T., Wood W. A. (1966) d-Ribulose 5-phosphate 3-epimerase. In Methods of Enzymology, (, Wood W. A., ed.), Academic Press, New York [Google Scholar]
- 2.Nowitzki U., Wyrich R., Westhoff P., Henze K., Schnarrenberger C., Martin W. (1995) Cloning of the amphibolic Calvin cycle/OPPP enzyme d-ribulose-5-phosphate 3-epimerase (EC 5.1.3.1) from spinach chloroplasts: functional and evolutionary aspects. Plant Mol. Biol. , 1279–1291 [DOI] [PubMed] [Google Scholar]
- 3.Kruger N. J., von Schaewen A. (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr. Op. Plant Biol. , 236–246 [DOI] [PubMed] [Google Scholar]
- 4.Herrmann K. M., Weaver L. M. (2003) The shikimate pathway. Ann. Rev. Plant Physiol. Plant Mol. Biol. , 473–503 [DOI] [PubMed] [Google Scholar]
- 5.Wood T. (1986) Physiological functions of the pentose phosphate pathway. Cell. Biochem. Funct. , 241–247 [DOI] [PubMed] [Google Scholar]
- 6.Thorpe G. W., Fong C. S., Alic N., Higgins V. J., Dawes I. W. (2004) Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U. S. A. , 6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juhnke H., Krems B., Kötter P., Entian K. (1996) Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. MGG , 456–464 [DOI] [PubMed] [Google Scholar]
- 8.Ng C.-H., Tan S.-X., Perrone G. G., Thorpe G. W., Higgins V. J., Dawes I. W. (2008) Adaptation to hydrogen peroxide in Saccharomyces cerevisiae: the role of NADPH-generating systems and the SKN7 transcription factor. Free Radical Biol. Med. , 1131–1145 [DOI] [PubMed] [Google Scholar]
- 9.Tan S.-X., Teo M., Lam Y. T., Dawes I. W., Perrone G. G. (2009) Cu, Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell , 1493–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akana J., Fedorov A. A., Fedorov E., Novak W. R. P., Babbitt P. C., Almo S. C., Gerlt J. A. (2006) d-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ribulose-phosphate binding (β/α)8-barrel superfamily. Biochemistry , 2493–2503 [DOI] [PubMed] [Google Scholar]
- 11.Jelakovic S., Kopriva S., Süss K.-H., Schulz G. E. (2003) structure and catalytic mechanism of the cytosolic d-ribulose-5-phosphate 3-epimerase from rice. J. Mol. Biol. , 127–135 [DOI] [PubMed] [Google Scholar]
- 12.Wise E. L., Akana J., Gerlt J. A., Rayment I. (2004) Structure of d-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 A resolution. Acta Crystallogr. D , 1687–1690 [DOI] [PubMed] [Google Scholar]
- 13.Caruthers J., Bosch J., Buckner F., Voorhis W. V., Myler P., Worthey E., Mehlin C., Boni E., DeTitta G., Luft J., Lauricella A., Kalyuzhniy O., Anderson L., Zucker F., Soltis M., Wim G. J. H. (2006) Structure of a ribulose 5-phosphate 3-epimerase from Plasmodium falciparum. Proteins Struct. Funct. Bioinform. , 338–342 [DOI] [PubMed] [Google Scholar]
- 14.Kopp J., Kopriva S., Süss K.-H., Schulz G. E. (1999) Structure and mechanism of the amphibolic enzyme -ribulose-5-phosphate 3-epimerase from potato chloroplasts. J. Mol. Biol. , 761–771 [DOI] [PubMed] [Google Scholar]
- 15.Chen Y.-R., Larimer F. W., Serpersu E. H., Hartman F. C. (1999) Identification of a catalytic aspartyl residue of d-ribulose 5-phosphate 3-epimerase by site-directed mutagenesis. J. Biol. Chem. , 2132–2136 [DOI] [PubMed] [Google Scholar]
- 16.Davies L., Lee N., Glaser L. (1972) On the mechanism of the pentose phosphate epimerases. J. Biol. Chem. , 5862–5866 [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. , 307–326 [DOI] [PubMed] [Google Scholar]
- 18.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. , 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., Yao D.-Q., Gu Y.-X., Lin Z.-J., Zheng C.-D., Fan H.-F. (2007) OASIS and molecular-replacement model completion. Acta Crystallogr. D , 793–799 [DOI] [PubMed] [Google Scholar]
- 20.Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D , 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D , 240–255 [DOI] [PubMed] [Google Scholar]
- 22.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L.-W., Read R. J., Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D , 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. , W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene , 51–59 [DOI] [PubMed] [Google Scholar]
- 25.Temple M. D., Perrone G. G., Dawes I. W. (2005) Complex cellular responses to reactive oxygen species. Trends Cell. Biol. , 319–326 [DOI] [PubMed] [Google Scholar]
- 26.Valko M., Morris H., Cronin M. T. D. (2005) Metals, toxicity and oxidative stress. Curr. Med. Chem. , 1161–1208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.