Abstract
Objective
To demonstrate and test the validity of a novel deep-learning-based system for the automated detection of pulmonary nodules.
Materials and Methods
The proposed system uses 2 3D deep learning models, 1 for each of the essential tasks of computer-aided nodule detection: candidate generation and false positive reduction. A total of 888 scans from the LIDC-IDRI dataset were used for training and evaluation.
Results
Results for candidate generation on the test data indicated a detection rate of 94.77% with 30.39 false positives per scan, while the test results for false positive reduction exhibited a sensitivity of 94.21% with 1.789 false positives per scan. The overall system detection rate on the test data was 89.29% with 1.789 false positives per scan.
Discussion
An extensive and rigorous validation was conducted to assess the performance of the proposed system. The system demonstrated a novel combination of 3D deep neural network architectures and demonstrates the use of deep learning for both candidate generation and false positive reduction to be evaluated with a substantial test dataset. The results strongly support the ability of deep learning pulmonary nodule detection systems to generalize to unseen data. The source code and trained model weights have been made available.
Conclusion
A novel deep-neural-network-based pulmonary nodule detection system is demonstrated and validated. The results provide comparison of the proposed deep-learning-based system over other similar systems based on performance.
Keywords: deep learning, pulmunary nodule detection, image recognition, computer-aided detection
BACKGROUND
Lung cancer is the leading cause of cancer mortality throughout the world.1 Regular screening of high-risk individuals using low-dose computed tomography (CT) has been shown to reduce mortality in lung cancer patients.2 Errors in cancer diagnosis are the most costly and detrimental type of diagnosis errors3 with type II errors being particularly common in lung cancer diagnosis.4 Subject to errors in both reading and interpretation, lung cancer diagnosis is highly error prone; however, many errors from diagnosis based on CT imagery can be reduced with a second reader.5 Due to the large number of individuals at high risk of lung cancer, regular screening with or without the assistance from a second reader could impose significant workflow and workload challenges for radiologists and clinical staff. Instead, computer-aided detection (CAD) systems have the potential to aid radiologists in lung cancer screening by reducing reading times or acting as a second reader.
CAD systems for pulmonary nodule detection consist of 2 essential tasks: candidate generation (CG) and false positive reduction (FPR).6 Various methods have been proposed for CG, including thresholding-based methods,7–9 shape-based methods,10,11 rule-based methods,12 and filtering-based methods.13 The best results for this task have been demonstrated using a 2D multiscale enhancement filter.14–16 Methods for FPR are also varied, and include methods similar to those used for CG.17–19 Neural networks were first used for FPR of nodule candidates over 20 years ago20,21 and have been accepted as a leading method for FPR in radiographs for over a decade.22,23
In recent years, deep neural networks (DNNs) have been responsible for significant improvements in a variety of complex tasks. The potential of DNNs for such improvements was first demonstrated at the 2012 ImageNet general image classification competition by Krizhevsky et al.24 Krizhevsky’s team used the first DNN in the competition’s history, winning by such a large margin that each of the top 10 teams in the following year’s competition were also using DNNs. In the years since, deep learning has been applied to a variety of medical imaging tasks,25,26 many of which involve segmentation.27–30 For some medical imaging tasks, deep learning methods have been demonstrated performing at or above the level of licensed experts.31,32 DNNs have also been applied for FPR,33–37 and just recently for pulmonary nodule segmentation.38,39
Since the 2012 ImageNet competition, substantial progress has been made developing increasingly complex DNNs that have surpassed human-level performance in general image classification tasks.40 In 2014, Szegedy et al41 demonstrated an accuracy of 93.33% using an extensive DNN comprised of 22 layers. A key feature of this network, dubbed GoogLeNet, was the inclusion of new components called inception modules that enabled the use of multiple convolutions in each layer of the network. In 2015, He et al42 demonstrated an architecture for deep residual learning using a DNN comprised of 152 layers to achieve an accuracy of 96.43%. This architecture, dubbed a residual network, employed a new feature for learning residual functions with reference to layer inputs.
Recent studies on nodule detection that utilize machine learning approaches, such as neural networks6,34,39 and support vector machines (SVMs),12,15,43,44 have several deficiencies. For example, several prior studies37,39,43,44 exclude high resolution scans, several studies37,39 use 20-year-old deep learning architectures,45 and several34,37,39 exclude small nodules and non-nodules from system evaluation. To address these deficiencies, we present a complete CAD system that uses novel, 3D adaptations of recent deep learning research,46–48 achieves performance comparable to the best existing systems, and exceeds the performance of all previous deep learning systems.
MATERIALS AND METHODS
Dataset
The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI) compiled a repository of CT scans specifically for the development of CAD systems for lung cancer.49,50 These data are made available by the National Cancer Institute’s Cancer Imaging Archive under the Creative Commons Attribution 3.0 Unported License.50 The LIDC-IDRI repository is comprised of 1018 openly available CT scans collected from 5 participating institutions with each of the scans including annotations by 4 radiologists. Lesions identified by radiologists were labeled as large nodule (≥ 3mm), small nodule (< 3mm), or non-nodule. A total of 2669 lesions were marked as a large nodule by at least 1 radiologist, with 928 lesions being marked as large nodule by all 4 radiologists.
Although a large number of CAD systems have been demonstrated,12,34,43,44,51–53 comparing their relative performance has been challenging due to the lack of an objective evaluation framework. The LUNA16 dataset6 was created in part to address this issue. It is a collection of 888 thin-slice CT scans (ie, slice thickness ≤ 3mm) of consistent slice spacing from the LIDC-IDRI dataset.54 These scans were randomly divided into 10 bins for cross-validation purposes, and these bins were used in this study to enable reproducibility and aid in objective comparative evaluation. The small and non-nodule objects, typically ignored by other systems, are included as part of the testing set for this system, resulting in the inclusion of 30 513 items for evaluation, and the detection of these objects is considered a false positive.
Method
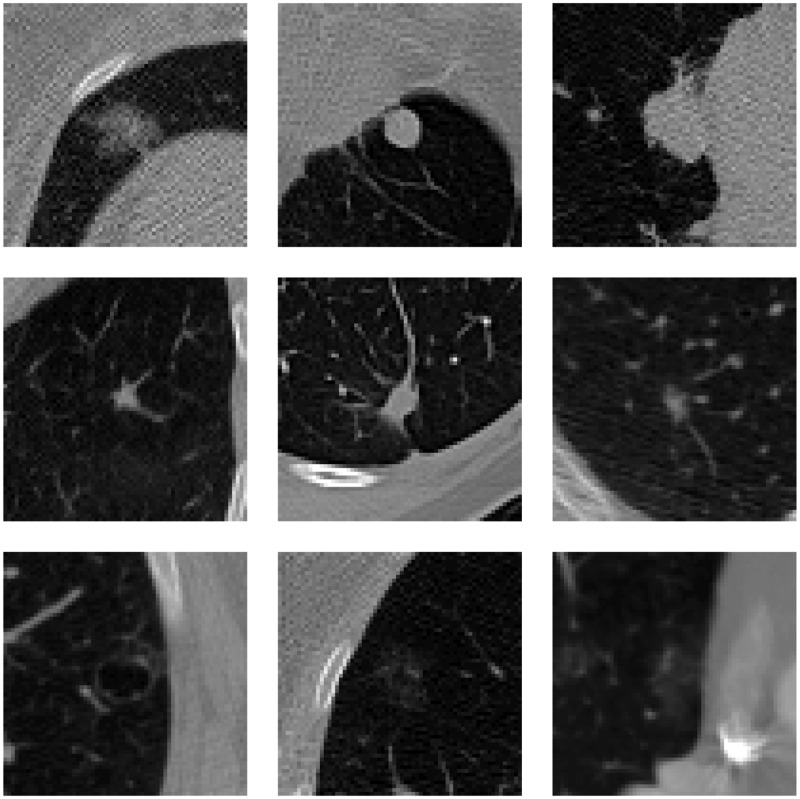
As depicted in Figure 1, pulmonary nodules can vary greatly in density, shape, size, etc. A typical pulmonary nodule exists as an isolated lesion1 in the parenchyma of the lung. Pulmonary nodules are frequently found attached to the pleural wall or vascular structures. Such juxtapleural and juxtavascular nodules, depicted in the first and second rows of Figure 1, respectively, can complicate automated nodule detection. Other nodules that can be challenging for CAD systems to detect are low-density nodules, as seen in the third row of Figure 1. Due to the variety and unique challenges, CAD systems have had difficulty achieving high performance for all varieties
Figure 1.
Different types of pulmonary nodules: juxtapleural (top row), juxtavascular (middle row), and low-density (bottom row).
Deep learning offers unique advantages over traditional methods for nodule detection. Rather than relying on mathematical techniques or hand-crafted features, deep learning enables learning internal feature representations directly from the input data.55,56 The multiple processing layers in DNNs allow for learning these internal representations of the input data with multiple levels of abstraction.57 These characteristics enable DNNs to learn feature representations for a large variety of features hierarchically, ie, at different scales, and combine them for learning more complex features (eg, nodules). This capability is well suited for the unique challenges posed by nodule variety in pulmonary nodule detection.
We use DNNs for each of the 2 essential tasks of computer-aided pulmonary nodule detection. The first DNN is used for volume-to-volume prediction of nodules, ie, segmentation, to identify potential pulmonary nodules within the input CT scan. These potential nodules that are generated are considered candidate nodules. This segmentation generates a large number of candidate nodules, many of which are false positives. The second DNN is used for reducing false positives among these nodule candidates through a binary classification of nodules and non-nodules. Figure 2 describes the system including the 2 primary tasks and the associated subtasks for each DNN.
Figure 2.
a) Process diagram of the proposed detection system. b) Segmented volume of the lungs.
The CT scans from the LUNA16 dataset range from 0.461 mm to 0.977 mm in pixel size and from 0.6 mm to 3.0 mm in slice thickness. Thus, the features depicted in the raw CT scans represent a scaled version of the actual features corresponding to the specifications of the imaging equipment used. To ensure uniformity of the data processed by the system, we begin with an isotropic resampling of the images, using third-order spline interpolation to resize all voxels to a uniform size of 1 mm3. As lung cancer screening is concerned solely with the lungs, a large portion of thoracic CT scans is irrelevant to lung cancer detection. To reduce computational expense, we next segment the lung region using a threshold-based segmentation technique2 that isolates the lung and allows us to ignore the extraneous regions of the original scan. Figure 2b depicts the lungs following segmentation. A bounding box is created circumscribing the lung, and this region of interest is input into the CG DNN.
To date, only 1 study has demonstrated the use of DNNs for the segmentation of pulmonary nodules,38 and DNNs have not been previously used in CG. The U-Net architecture58 is a DNN architecture used for generating high-quality segmentations of medical imagery from small training datasets using heavy augmentation. The U-Net architecture has also been extended to 3D imaging applications successfully.46 In this study, we adapt and apply this 3D U-Net architecture specifically for the segmentation of relatively small volumes in CT scans such as pulmonary nodules. The architecture, depicted in Figure 3a, comprises 135.6 million trainable parameters.
Figure 3.
4 . a) Modified 3D U-Net architecture used for the CG segmentation. b) 3D residual network architecture used for FPR.
The U-Net-inspired DNN architecture that we use for CG acts as a volume-to-volume prediction (ie, segmentation) network that takes an input volume of 64x64x64 and outputs a volume of 32x32x32. CT scans and the regions of interest generated from the lung segmentation are much larger than 64x64x64. As the output is 32x32x32, a 16-unit buffer was created on all sides of the region of interest in order to generate an output volume corresponding with every voxel in the region of interest. A sliding window technique was used with a step size of 32 for each axis to iteratively progress through the region of interest, extracting 64x64x64 volumes for input to the DNN. This required the application of the DNN nearly a thousand times in generating candidate nodules for a single scan. The output volume was an array of probabilities. A threshold was applied setting all values less than 0.13 to zero. The remaining probabilities were converted to a binary array, and labels were generated for the adjacent voxels, yielding an array of numbered volumes. Centroids were computed from these labeled volumes for evaluation and for generating output candidate nodules. During this process, a variety of features were also extracted from nodules, including the volume in voxels of each nodule candidate.
The features extracted during the centroid computation for each of the candidate nodules were used as a threshold as an initial FPR technique. The only feature selected for use in this step was nodule volume due to its strength in discriminating among positive and false positive nodules. The optimal threshold value was determined empirically to be 8 mm3. Nodule candidates smaller in volume than 8 mm3 were deemed false positives. Examples are shown in Figure 4. Following the features-based FPR step, remaining candidate nodules were processed with the DNN FPR module. The DNN model used here involved a complex DNN architecture for the simple task of binary classification. The architecture was based on the work by Szegedy et al59 in combining the inception network architecture41 with residual learning elements.42 The architecture used in our proposed model is adapted from the Inception-ResNet architectures proposed by Szegedy et al, albeit adapted to 3D images. Rather than using leaky rectified linear units as activation functions, scaled exponential linear units, recently demonstrated as better than standard rectified linear units for very deep networks,47 were used, as initial testing indicated performance improvements. Consequently, Gaussian initialization and alpha dropout were used to improve convergence and performance.
Figure 4.
Top: actual nodules. Bottom: corresponding segmented volumes of generated candidates for diameter sizes 6 mm, 12 mm, and 20 mm.
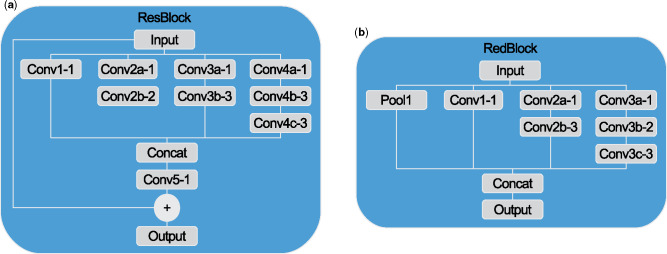
The reduction and residual blocks (Figures 5a and b) provide further details on the architecture of the network (Figure 3b). The reduction block in Figure 5a was adapted from the wider “Reduction-B” block from Inception-ResNet-v1, 1 of 3 Inception-ResNet architectures proposed by Szegedy et al.48 The residual block was adapted from the “Inception-ResNet-A” block of Inception-ResNet-v1 with an extra convolution layer similar to the residual inception blocks of Inception-ResNet-v4. Unlike Inception-ResNet-v1, the entire architecture of our proposed model was built from only 2 of these reduction blocks and 4 of these residual blocks. Each of the convolution layers depicted in the overall architecture applies 1x1x1 convolutions. These are included between concurrent residual blocks to maintain a constant number of features. A smaller input size of 32x32x32 was used to speed up training, as it did not affect the results. The entire architecture was comprised of 1.28 million trainable parameters.
Figure 5.
Architectures for a) 3D reduction blocks and b) 3D residual blocks. Convolutions were uniform in 3 dimensions of size denoted by the hyphenated number in the diagrams.
Training5,6,7
We used 10-fold cross-validation consistent with the cross-validation experimental design used by Setio et al.37 For each of the 10 folds of the cross-validation, 9-fold cross-validation was conducted, and the 10th bin was held out as test data. The best performing model from the 9-fold cross-validation was evaluated on the test data. This enabled the use of all 888 scans as test data.
Training for the CG network was conducted with all available positive nodules8 for each of the 9-fold cross-validations conducted. The nodule candidates generated for each of the validation bins during each cross-validation were used to train the FPR network. Over 1000 positive examples were used for each of the 10 9-fold cross-validations.
A 3D array of training labels was required, as the CG network was a volume-to-volume prediction network. Approximate binary labels for each nodule were generated as spheres based on the diameter of each nodule. Heavy data augmentation was employed during training, involving random translation up to 14 units along each axis, random rotation about each axis up to 90°, and random flipping about each axis. Implementation of this augmentation required the application of the same random transformations to both the input image and its associated labeled array.
The Sorensen-Dice coefficient,60,61 or simply the Dice coefficient (DC), was used to measure the spatial overlap between the predicted volume and the ground truth for the purposes of optimization. It is defined as:
where P is the set of prediction results, and G is the set of ground truth. It ranges in value from 0 to 1, with 1 indicating perfect prediction accuracy. In order to optimize the network architecture, the objective function, J(θ), was defined as:
The Adam optimizer,62 with an initial learning rate of 1e−5, was used for minimizing J(θ). In order to minimize overfitting, the dropout technique63 and batch normalization64 were used for regularization. As noted, the CG architecture included over 100 million trainable parameters. This effectively restricted the batch size to 2, significantly slowing training.9 Due to this, each epoch required training 400 minibatches. Models were trained for 210 epochs. Upon completion of the training, the models exhibiting lowest values were evaluated systematically in order to identify the best models at a minimal computational expense. Results were recorded, and the process was repeated until performance gains plateaued.
The FPR network was trained as a binary classifier with the candidates generated from the best performing models of each validation bin during cross-validation. These candidate nodules were labeled as either nodule or non-nodule.10 Data augmentation was used involving random translation along each axis up to 2 units, random rotation of up to 15° about each axis, and random flipping about each axis. Data were centered by subtracting the image mean and normalized by the image standard deviation.
Categorical cross-entropy was used as an objective function and was minimized using the AdaDelta optimizer.65 Again, to minimize overfitting, dropout63 and batch normalization64 were used. With the architecture including over a million trainable parameters, a batch size of 64 was trained per epoch. During training, only positive examples were included in the validation dataset in order to prioritize sensitivity. Consequently, a large number of well performing models were saved, and specificity was assessed among these.
RESULTS
By conducting 10 9-fold cross-validations, each scan and nodule in the dataset could be evaluated as test data. To evaluate test data, scans were first evaluated with the CG module. The resulting candidate nodules were determined to be a direct hit, a near hit, or a miss. A candidate nodule was classified a “direct hit” when the center of the true nodule was within the segmented volume of the candidate nodule. A nodule was treated as “near hit” when the center of the candidate nodule was within 0.75D of the true nodule.15 Direct hits and near hits were taken as positive candidates. All candidate nodules generated were then evaluated with the FPR module.
The results from the cross-validation and the test data are reported in Table 1. This table reports only mean values of sensitivity, false positives per scan (FPPS), and receiver operating characteristic area under the curve (ROC AUC). The full test results for each of the 10 cross-validations are reported in Supplementary Appendix A in Table 3. The cross-validation results are depicted for comparison only and, unless otherwise noted, all references to results in the paper refer to the test results. The full cross-validation results for the 8 values reported in Table 1 are presented in Tables 4 through 9 in Supplementary Appendix A, which also reports the cross-validation results for each of the 90 cross-validation systems.
Table 1.
Results
| Task | Dataset | Sensitivity | FPPS | ROC AUC |
|---|---|---|---|---|
| CG | Cross-validation | 0.9516 | 30.68 | – |
| Test | 0.9477 | 30.39 | – | |
| FPR | Cross-validation | 0.9594 | 1.815 | 0.9849 |
| Test | 0.9421 | 1.789 | 0.9835 | |
| Complete system | Cross-validation | 0.9129 | 1.815 | 0.9372 |
| Test | 0.8929 | 1.789 | 0.9324 |
The free-response ROC curve (FROC) is commonly used in lieu of the traditional ROC curve for cases such as pulmonary nodule detection in which 1 or more abnormalities may be present for each evaluated case.66,67Figure 6 depicts an FROC curve for the system plotted on a logarithmic scale. Recent work on pulmonary nodule CAD systems6,33,37 has featured a metric referred to as the “competition performance metric” (CPM), which is based on the FROC curve for evaluating CAD systems.68 This metric is computed by taking the mean sensitivity value for each of the 7 labeled positions on the x axis in Figure 6. The CPM for the system was 0.8135.
Figure 6.
FROC curve for the complete system and depicted on a logarithmic scale11 of base 2 from 0.125 to 8.0.
DISCUSSION
A comparison of the features and performance of pulmonary nodule CAD systems is reported in Table 2. In this table, a range of values for the proposed system’s sensitivity is reported in order to enable comparisons with the various systems. The middle value is the raw result of the FPR module. The top and bottom values are sensitivities taken from the FROC curve for 1.0 and 4.0 FPPS, respectively. This table includes no studies that exclusively use the LUNA16 Challenge evaluation framework due to its limitations. Detailed explanation for this is provided in Supplementary Appendix B. Results reported by Setio et al37 and van Ginneken et al34 for CAD systems employing deep learning for FPR presented sensitivities based on false positive rates from FROC curves. Table 2 demonstrates the improvements in the performance of the proposed systems over these 2 systems. Setio et al37 reported a CPM of 0.722 and was the only one of the studies listed in Table 2 to report this metric. As reported earlier, the CPM in this study is 0.8135. Shaukat et al15 demonstrated excellent performance for their complete system, which was heavily influenced by the strong performance of their FPR module, and is discussed further in Supplementary Appendix C. These results suggest that future work could explore the combination of deep learning systems for feature extraction and SVMs for classification. As reported in Table 2, among the studies included for comparison, only Setio et al37 and Jacobs et al52 used the complete number of scans for test data. While Setio et al37 used existing CG systems, Jacobs et al52 simply evaluated existing systems. Shaukat et al15 did, however, use 30% of their total dataset for test, which is substantial. Of the other studies involving a large number of scans, Hamidian et al39 used 25 scans for test while Firmino et al12 and van Ginneken et al34 strictly reported cross-validation results. We are not aware of another novel system in the literature to have been evaluated as rigorously. A complete comparison and discussion of results for each of the tasks can be found in Supplementary Appendix C.
Table 2.
Comparison of pulmonary nodule CAD systems
| # Scans | Sensitivity | FPPS | CG | FPR | Thickness | Small/non-nodules | |
|---|---|---|---|---|---|---|---|
| Proposed system | 888 | 0.8446 | 1.00 | 3D DNN | 3D DNN | <=3.0mm | Yes |
| 0.8929 | 1.79 | ||||||
| 0.9273 | 4.00 | ||||||
| Shaukat et al15 | 850 | 0.9269 | 1.91 | Filter-based segmentation | Specified features w/ SVM classifier | 1.0-3.0 mm | Yes |
| Hamidian et al39 | 509 | 0.8000 | 15.28 | 3D CNN | 3D CNN | 1.5-3.0 mm | Yes |
| Firminio et al12 | 420 | 0.9440 | 7.04 | Segmentation and rule-based | HOG features and SVM classifier | <=3.0mm | No |
| Jacobs et a.52 | 888 | 0.8200 | 3.10 | Comparison of 3 systems (commercial and academic) | <=3.0mm | Yes | |
| Setio et al37 | 888 | 0.7820 | 1.00 | Ensemble of existing systems | Multi-view CNN | <=3.0mm | Yes |
| 0.8790 | 4.00 | ||||||
| Van Ginneken et al34 | 865 | 0.7300 | 1.00 | Existing system | Comparison of 3 existing DNNs | <=2.5mm | No |
| 0.7600 | 4.00 | ||||||
| Choi and Choi44 | 84 | 0.9545 | 6.76 | Dot-enhancement filter | AHSN features and SVM classifier | 1.0-3.0 mm | Yes |
| Choi and Choi43 | 58 | 0.9276 | 2.27 | Block segmentation | Specified features w/ SVM classifier | 1.0-3.0 mm | Yes |
Strengths and limitations
Strengths of the study include the robust experimental design, the large test dataset, and the methodology. Particularly, retaining 30 513 false positive items to evaluate the system strongly distinguishes this study from a large number of current deep learning studies for nodule detection. A further strength of the study is the open availability of the source code and the trained weights for the 180 different models for which results are reported in Supplementary Appendix A.
Despite the use of a substantial amount of test data, the lack of evaluation on an external dataset remains a significant limitation. Further limitations of the dataset are described by Armato et al49 and include the lack of clinical information and the fact that scans were not all scored by the same radiologists. Other limitations concern limited analysis of segmentation quality due to the focus on the CAD system and limited computational capacity due to the high computational costs.
CONCLUSION
We present a novel, fully automated system for the detection of pulmonary nodules that was extensively validated using each scan in the dataset for both training and test. As described earlier, the CG and FPR modules each independently demonstrated results that outperform other reported or known systems for their respective tasks. Overall, the results indicate that the system is comparable with the best existing systems.
The proposed CAD system is designed for detecting pulmonary nodules. Extending it to quantify malignancy and offer diagnostic predictions is an important research direction for future work. Such a system could be evaluated rigorously with a large external dataset—a significant limitation of this study—using data from the National Lung Screening Trial.2,69
FUNDING
Support for this work was provided by Auburn University. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
CONTRIBUTORS
RG developed the model and did coding for the model under supervision of AG. RG and AG developed the manuscript and performed revisions. DP edited the manuscript and provided feedback.
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgments
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI database used in this study. The following institutions were responsible for the LIDC/IDRI: Weill Cornell Medical College, the University of California, Los Angeles, the University of Chicago, the University of Iowa, and the University of Michigan.
Footnotes
Lesions in the lungs less than 3 cm are nodules.
Detailed explanation of the lung segmentation technique can be found in Appendix B.
We observed a large number of voxels with predicted values of magnitude lower than the maximum probability of 1.0, and to reduce noise, we ignored all values that were more than an order of magnitude below 1.0.
The notation for each layer in the architectures presented in Figure 3 indicates the type of layer and its shape. Each layer is represented by a 4D tensor for the 4 dimensions: the width of the network layer, and the x, y, and z dimensions of the input volume.
The source code used for both training and evaluation is available at https://github.com/rossgritz/cad17.
DNN training was conducted using Keras with a Tensorflow backend.
Further details regarding training are presented in Appendix B.
No negative examples were used because the Dice coefficient could be computed only from positive examples.
More information regarding training can be found in Appendix B.
There were substantially more negative examples generated during CG. To handle this, the system was designed such that approximately equal numbers of positive and negative examples were used for each training epoch. The examples used for each epoch were sampled randomly from a single directory containing all negative examples and n copies of all positive examples prior. n was selected such that the total number of positive and negative examples in the directory were roughly equal.
The same data are depicted for log base 10 in Figure 7 of Appendix A.
REFERENCES
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 1365: E359.. [DOI] [PubMed] [Google Scholar]
- 2. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011; 3655: 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singh H, Sethi S, Raber M, Petersen LA.. Errors in cancer diagnosis: current understanding and future directions. J Clin Oncol 2007; 2531: 5009–18. [DOI] [PubMed] [Google Scholar]
- 4. Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 2012; 30722: 2418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peldschus K, Herzog P, Wood SA, et al. Computer-aided diagnosis as a second reader: spectrum of findings in CT studies of the chest interpreted as normal. Chest 2005; 1283: 1517–23. [DOI] [PubMed] [Google Scholar]
- 6. Setio AAA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 2017; 42: 1–13. [DOI] [PubMed] [Google Scholar]
- 7. Messay T, Hardie RC, Rogers SK.. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 2010; 143: 390–406. [DOI] [PubMed] [Google Scholar]
- 8. Choi W-J, Choi T-S.. Genetic programming-based feature transform and classification for the automatic detection of pulmonary nodules on computed tomography images. Inform Sci 2012; 212: 57–78. [Google Scholar]
- 9. Armato SG, Giger ML, Moran CJ, et al. Computerized detection of pulmonary nodules on CT scans. Radiographics 1999; 195: 1303–11. [DOI] [PubMed] [Google Scholar]
- 10. Dehmeshki J, Ye X, Lin X, et al. Automated detection of lung nodules in CT images using shape-based genetic algorithm. Comput Med Imaging Graph 2007; 316: 408–17. [DOI] [PubMed] [Google Scholar]
- 11. Ye X, Lin X, Dehmeshki J, et al. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Trans Biomed Eng 2009; 567: 1810–20. [DOI] [PubMed] [Google Scholar]
- 12. Firmino M, Angelo G, Morais H, Dantas MR, Valentim R.. Computer-aided detection (CADe) and diagnosis (CADx) system for lung cancer with likelihood of malignancy. Biomed Eng Online 2016; 151: 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suárez-Cuenca JJ, Tahoces PG, Souto M, et al. Application of the iris filter for automatic detection of pulmonary nodules on computed tomography images. Comput Biol Med 2009; 3910: 921–33. [DOI] [PubMed] [Google Scholar]
- 14. Li Q, Sone S, Doi K.. Selective enhancement filters for nodules, vessels, and airway walls in two‐and three‐dimensional CT scans. Med Phys 2003; 308: 2040–51. [DOI] [PubMed] [Google Scholar]
- 15. Shaukat F, Raja G, Gooya A, Frangi AF.. Fully automatic and accurate detection of lung nodules in CT images using a hybrid feature set. Med Phys 2017; 447: 3615–29. [DOI] [PubMed] [Google Scholar]
- 16. Tan M, Deklerck R, Jansen B, Bister M, Cornelis J.. A novel computer‐aided lung nodule detection system for CT images. Med Phys 2011; 3810: 5630–45. [DOI] [PubMed] [Google Scholar]
- 17. Ge Z, Sahiner B, Chan H-P, et al. Computer‐aided detection of lung nodules: false positive reduction using a 3D gradient field method and 3D ellipsoid fitting. Med Phys 2005; 328: 2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boroczky L, Zhao L, Lee KP.. Feature subset selection for improving the performance of false positive reduction in lung nodule CAD. IEEE Trans Inf Technol Biomed 2006; 103: 504–11. [DOI] [PubMed] [Google Scholar]
- 19. Gurcan MN, Sahiner B, Petrick N, et al. Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer‐aided diagnosis system. Med Phys 2002; 2911: 2552–8. [DOI] [PubMed] [Google Scholar]
- 20. Wu YC, Doi K, Giger ML, Metz CE, Zhang W.. Reduction of false positives in computerized detection of lung nodules in chest radiographs using artificial neural networks, discriminant analysis, and a rule-based scheme. J Digit Imaging 1994; 74: 196–207. [DOI] [PubMed] [Google Scholar]
- 21. Lo S-C, Lou S-L, Lin J-S, et al. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging 1995; 144: 711–8. [DOI] [PubMed] [Google Scholar]
- 22. Suzuki K, Li F, Sone S, Doi K.. Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose CT by use of massive training artificial neural network. IEEE Trans Med Imaging 2005; 249: 1138–50. [DOI] [PubMed] [Google Scholar]
- 23. Suzuki K, Armato SG, Li F, Sone S, Doi K.. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low‐dose computed tomography. Med Phys 2003; 307: 1602–17. [DOI] [PubMed] [Google Scholar]
- 24. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Bartlett P, Pereira F., Burges C.J.C, et al. eds. Advances in Neural Information Processing Systems: 26th Annual Conference on Neural Information Processing Systems 2012; 2012: 1097–105. Lake Tahoe, CA, USA.
- 25. Cireşan DC, Giusti A, Gambardella LM.. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv 2013; 16(Pt 2): 411–8. [DOI] [PubMed] [Google Scholar]
- 26. Roth HR, Lu L, Seff A, et al. A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. Med Image Comput Comput Assist Interv 2014; 17(Pt 1): 520–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen H, Dou Q, Yu L, et al. VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018; 170: 446–55. [DOI] [PubMed] [Google Scholar]
- 28. Prasoon A, Petersen K, Igel C, et al. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 2013; 16(Pt 2): 246–53. [DOI] [PubMed] [Google Scholar]
- 29. Ciresan D, Giusti A, Gambardella LM, et al. Deep neural networks segment neuronal membranes in electron microscopy images. In: Bartlett P, Pereira F., Burges C.J.C, et al. eds. Advances in Neural Information Processing Systems: 26th Annual Conference on Neural Information Processing Systems 2012; 2012: 2843–51.
- 30. Yu L, Yang X, Chen H, et al. Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. AAAI 2017: 66–72. [Google Scholar]
- 31. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 31622: 2402–10. [DOI] [PubMed] [Google Scholar]
- 32. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 5427639: 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dou Q, Chen H, Yu L, et al. Multi-level contextual 3D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans Biomed Eng 2017; 647: 1558–67. [DOI] [PubMed] [Google Scholar]
- 34. van Ginneken B, Setio AA, Jacobs C, Ciompi F.. Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. In: Biomedical Imaging (ISBI), IEEE 12th International Symposium on Biomedical Imaging 2015: 286–9. Brooklyn, NY, USA. [Google Scholar]
- 35. Gruetzemacher R, Gupta A.. Using Deep Learning for Pulmonary Nodule Detection & Diagnosis. Americas Conference on Information Systems; 2016. San Diego, CA, USA. [Google Scholar]
- 36. Shen W, Zhou M, Yang F, et al. Multi-scale convolutional neural networks for lung nodule classification. Inf Process Med Imaging 2015; 24: 588–99. [DOI] [PubMed] [Google Scholar]
- 37. Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 2016; 355: 1160–9. [DOI] [PubMed] [Google Scholar]
- 38. Wang S, Zhou M, Liu Z, et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 2017; 40: 172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamidian S, Sahinerb B, Petrickb N, et al. Spring M. 3D convolutional neural network for automatic detection of lung nodules in chest CT. Proc SPIE Int Soc Opt Eng 2017; 10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He K, Zhang X, Ren S, et al. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In: proceedings from the IEEE International Conference on Computer Vision, 2015: 1026–34. Santiago, Chile.
- 41. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: proceedings from the IEEE Conference on Computer Vision and Pattern Recognition, 2015: 1–9. Boston, MA, USA.
- 42. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: proceedings from the IEEE Conference on Computer Vision and Pattern Recognition, 2016: 770–8.
- 43. Choi W-J, Choi T-S.. Automated pulmonary nodule detection system in computed tomography images: a hierarchical block classification approach. Entropy 2013; 152: 507–23. [Google Scholar]
- 44. Choi W-J, Choi T-S.. Automated pulmonary nodule detection based on three-dimensional shape-based feature descriptor. Comput Methods Programs Biomed 2014; 1131: 37–54. [DOI] [PubMed] [Google Scholar]
- 45. LeCun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE 1998; 8611: 2278–324. [Google Scholar]
- 46. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer, 2016: 424–32.
- 47. Klambauer G, Unterthiner T, Mayr A, et al. Self-normalizing neural networks. Advances in Neural Information Processing Systems 30 (NIPS 2017), 1-10, Long Beach, CA, USA.
- 48. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA.. Inception-v4, inception-ResNet and the impact of residual connections on learning. AAAI 2017: 4278–84. [Google Scholar]
- 49. Armato SG, McLennan G, Bidaut L, et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011; 382: 915–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013; 266: 1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bergtholdt M, Wiemker R, Klinder T.. Pulmonary nodule detection using a cascaded SVM classifier. SPIE Med Imaging 2016; 9785: 978513. [Google Scholar]
- 52. Jacobs C, Rikxoort EM, Murphy K, et al. Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database. Eur Radiol 2016; 267: 2139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tan M, Deklerck R, Cornelis J, et al. Phased searching with NEAT in a time-scaled framework: experiments on a computer-aided detection system for lung nodules. Artif Intell Med 2013; 593: 157–67. [DOI] [PubMed] [Google Scholar]
- 54. Samuel G, Armato III GM, Bidaut L, et al. Data from LIDC-IDRI: The cancer imaging archive. 2015; 10.7937/K9/TCIA.2015.LO9QL9SX, Accessed January 2016. [DOI]
- 55. Rumelhart DE, Hinton GE, Williams RJ.. Learning representations by back-propagating errors. Cogn Model 1988; 53: 1. [Google Scholar]
- 56. Rumelhart DE, Hinton GE, Williams RJ. Learning internal representations by error propagation. Technical Report, Defense Technical Information Center document, 1985. http://www.dtic.mil/dtic/tr/fulltext/u2/a164453.pdf
- 57. LeCun Y, Bengio Y, Hinton G.. Deep learning. Nature 2015; 5217553: 436–44. [DOI] [PubMed] [Google Scholar]
- 58. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: proceedings from the International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer, 2015: 234–41.
- 59. Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning. AAAI 2017: 4278–4284. San Francisco, CA, USA.
- 60. Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol Skr 1948; 5: 1–34. [Google Scholar]
- 61. Dice LR. Measures of the amount of ecologic association between species. Ecology 1945; 263: 297–302. [Google Scholar]
- 62. Kingma D, Ba J. Adam: A method for stochastic optimization. Proceedings of the 3rd International Conference on Learning Representations 2014: 1-15. San Diego, CA, USA.
- 63. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014; 151: 1929–58. [Google Scholar]
- 64. Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Proceedings of the 32 nd International Conference on Machine Learning 2015; JMLR: W&CP volume 37, 1-9 , Lille, France.
- 65. Zeiler MD. ADADELTA: an adaptive learning rate method. CoRR abs/1212.5701 2012.
- 66. DeLuca PM, Wambersie A, Whitmore GF.. Extensions to conventional ROC methodology: LROC, FROC, and AFROC. J ICRU 2008; 81: 31–5. [DOI] [PubMed] [Google Scholar]
- 67. Bandos AI, Rockette HE, Song T, et al. Area under the free‐response ROC curve (FROC) and a related summary index. Biometrics 2009; 651: 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Niemeijer M, Loog M, Abramoff MD, et al. On combining computer-aided detection systems. IEEE Trans Med Imaging 2011; 302: 215–23. [DOI] [PubMed] [Google Scholar]
- 69. National Lung Screening Trial Research Team. The national lung screening trial: overview and study design. Radiology 2011; 2581: 243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.