Abstract
Objective
The gold standard for diagnosing sleep disorders is polysomnography, which generates extensive data about biophysical changes occurring during sleep. We developed the National Sleep Research Resource (NSRR), a comprehensive system for sharing sleep data. The NSRR embodies elements of a data commons aimed at accelerating research to address critical questions about the impact of sleep disorders on important health outcomes.
Approach
We used a metadata-guided approach, with a set of common sleep-specific terms enforcing uniform semantic interpretation of data elements across three main components: (1) annotated datasets; (2) user interfaces for accessing data; and (3) computational tools for the analysis of polysomnography recordings. We incorporated the process for managing dataset-specific data use agreements, evidence of Institutional Review Board review, and the corresponding access control in the NSRR web portal. The metadata-guided approach facilitates structural and semantic interoperability, ultimately leading to enhanced data reusability and scientific rigor.
Results
The authors curated and deposited retrospective data from 10 large, NIH-funded sleep cohort studies, including several from the Trans-Omics for Precision Medicine (TOPMed) program, into the NSRR. The NSRR currently contains data on 26 808 subjects and 31 166 signal files in European Data Format. Launched in April 2014, over 3000 registered users have downloaded over 130 terabytes of data.
Conclusions
The NSRR offers a use case and an example for creating a full-fledged data commons. It provides a single point of access to analysis-ready physiological signals from polysomnography obtained from multiple sources, and a wide variety of clinical data to facilitate sleep research.
Keywords: data commons, data integration, data sharing, common data element, data provenance, sleep research, polysomnography, data quality, European Data Format
INTRODUCTION
To advance sleep research,1 the National Institutes of Health (NIH) has funded a number of clinical trials and epidemiological cohort studies, such as the Sleep Heart Health Study,2,3 Childhood Adenotonsillectomy Trial,3–5 Heart Biomarker Evaluation in Apnea Treatment,6 Cleveland Family Study,7,8 Study of Osteoporotic Fractures,9,10 MrOS Sleep Study,3,11,12 Cleveland Children's Sleep and Health Study,13–15 Hispanic Community Health Study/Study of Latinos,16,17 Honolulu-Asia Aging Study of Sleep Apnea,18 and Multi-Ethnic Study of Atherosclerosis,19 and the Jackson Heart Study.20 Polysomnography recordings called polysomnograms (PSGs) from these studies were analyzed by a central Sleep Reading Center. Several of these studies are also part of the Trans-omics in Precision Medicine (TOPMed) initiative,21 a program aimed at generating -omics data on over 100 000 research participants. Collectively, these represent a largely untapped extensive data resource involving human physiology.
In this paper, we describe the design and implementation of the NSRR,22 a system for the structural and semantic harmonization of and web-based access to PSGs and associated clinical data generated from NIH-funded epidemiological cohort studies. The NSRR offers a use case and an example to guide the creation of a full-fledged data commons.23 It provides a single point of access to analysis-ready polysomnography and clinical data to facilitate sleep research. It also serves as an exemplar to promote the FAIR24 principles for advancing data-enabled clinical and translational research.
Sharing clinical research data
Proper sharing and reuse of data sets can help accelerate research. Since early 2000, the NIH mandated policies and regulations requiring the sharing of final research data for larger awards.
However, it is one thing to generate one’s own data and perform one’s own analysis; it is a different matter making data accessible in an analysis-ready form for analysis by an independent researcher. The state of affairs for sharing research data is at best uneven, and at worst underdeveloped, due to multiple reasons and challenges. Efforts needed to meet the challenges have been grossly underestimated. Some investigators may not have sufficient resources or expertise for the proper sharing of their data. Others may not be sharing data in a way that facilitates reuse. There is also a well-known gap in the lack of data standards to ensure interoperability and proper attribution of data collection efforts.
The NIH data commons
The NIH Data Commons25 (or Commons) is an ambitious vision for a shared virtual space to allow digital objects to be stored and computed upon by the scientific community. The Commons would allow investigators to find, manage, share, use, and reuse data, software, metadata, and workflows. It imagines an ecosystem that makes digital objects findable, accessible, interoperable, and reusable (FAIR24). Four components are considered integral parts of the Commons: a computing resource for accessing and processing of digital objects; a “digital object compliance model” that describes the properties of digital objects that enable them to be FAIR; datasets that adhere to the digital object compliance model; and software and services to facilitate access to and use of data.
Creating such a Commons could benefit from many strategies, including a bottom-up and domain-specific approach involving key stakeholders in iterative processes, because the complexities involved in realizing this vision cannot be fully anticipated, and the ultimate product needs to be responsive to the community’s needs. Sample bottom-up efforts include the bioCADDIE26 project aimed at tackling some of the inherent challenges in managing digital object identifiers that could serve the purpose of a Commons. Sample domain-specific efforts include the National Sleep Research Resource, demonstrating a FAIR-oriented digital object environment for the domain of sleep medicine that involves polysomnograms as unique types of digital objects.
The National Sleep Research Resource
Over the last several years, we have developed a system with a single point of access for sharing and reusing large-scale physiological signals for the NIH-funded National Sleep Research Resource (NSRR; R24HL114473). The NSRR offers free and open web access to de-identified data for more than 26 000 subjects, including PSGs and links to risk factor and outcome data for study participants.22
This national repository of sleep data, the first and largest of its kind, is significant because biophysiological data have not been previously made available at such a large and systematic scale. In fact, many sleep studies have been conducted using data from single laboratories, limiting scope, statistical power, and generalizability. The NSRR provides opportunities for investigators to address critical questions about the impact of sleep disorders, using data from scored summary data, annotations, and the actual physiological signal data, on important clinical outcomes, thereby enhancing clinical and translational work in human sleep medicine and physiology.
The NSRR makes data available in two ways. One is through the open access of a standard collection of study metadata (What, Who, When, Funding). The “What” section provides further details in subsection headings About, Data Overview, Protocols and Manuals, Analysis, Equipment, and Publication Links. The second is through the cross-cohort open search interface x-search.net, allowing a user to query and visualize data across multiple datasets. For registered users, a case-control exploration interface allows the user to specify a control cohort and a case cohort in a step-by-step manner to quickly assess the data support for a potential hypothesis.
Overview
This paper describes the contributions of the NSRR along several aspects of the Commons vision: metadata for sleep research digital objects; a collection of annotated sleep data sets; and interfaces and tools for accessing and analyzing such data. More importantly, the NSRR provides the design of a functional architecture for implementing a Sleep Data Commons. The NSRR also reveals complexities and challenges involved in making clinical sleep data conform to the FAIR principles.
APPROACH
We, the NSRR team of informaticians, data managers, computer scientists, and clinical and epidemiological researchers, developed the NSRR using a metadata-guided approach, in the sense that a set of common sleep terms (Sleep Common Data Elements - SCDEs) was created and used for data annotation and mapping, for user interfaces that support browsing and cross-cohort data exploration, and for sleep signal visualization and analysis. The NSRR consists of three main components: (1) annotated datasets; (2) user interfaces for accessing data; and (3) computational tools for the analysis of polysomnography recordings. The NSRR also embeds the management of the approval process for dataset-specific data access and use agreements (DAUAs), evidence of Institutional Review Board (IRB) review for accessing NSRR source data sets (part or whole), and the corresponding access control strategies in a single web portal housed at https://sleepdata.org.
Sleep metadata: common data elements and provenance information
The NSRR uses the metadata-guided approach to achieve uniform semantic interpretation of data elements across the entire spectrum of data integration activities: for annotating source data, for interfaces to query and search data, and for tools that access and assist in analysis.
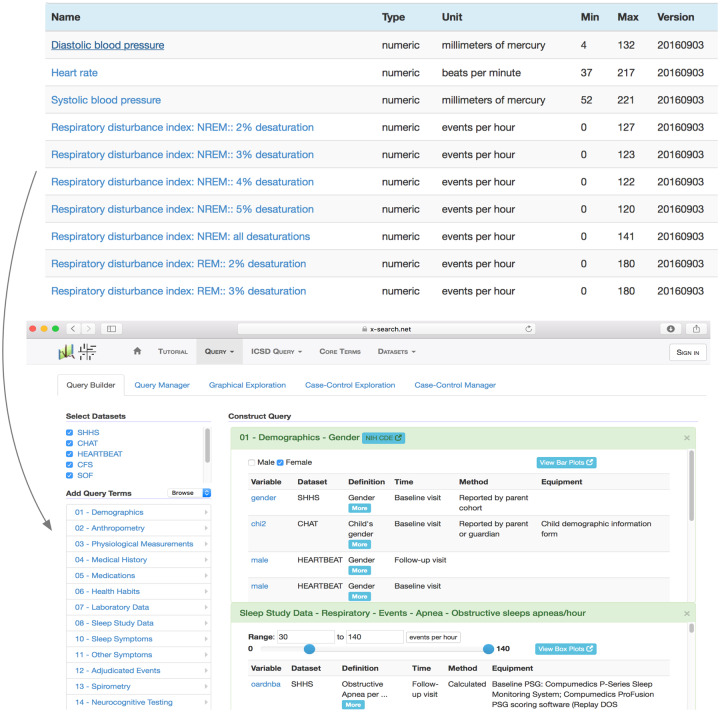
Existing terminological systems do not cover the sleep domain in sufficient detail to meet the goals of the NSRR. For this reason, we developed SCDEs (Figure 1), which consist of more than 900 core sleep terms that capture demographic information, anthropometric parameters, physiologic measurements, medical history elements, sleep study data, sleep symptoms, polysomnogram sleep events, relevant medical history elements, laboratory data, and neurocognitive testing results. The provenance information on the terms includes assessment point (eg baseline visit, follow-up visit), method of data capture (eg direct measurement, calculated, reported by subject), and equipment (eg sensors used for data capture). Whenever possible, the SCDE terms have been linked to coded terms listed in established biomedical terminologies, including the SNOMED CT, the FDA Drug Classification system, the Ontology for Biomedical Investigations, International Classification of Sleep Disorders, and the effort on a Sleep Domain Ontology (SDO27) developed as part of the earlier Physio-MIMI project by the same NSRR team.28–31 Terms appearing as NIH Common Data Elements (CDEs) are also provided for cross-reference.
Figure 1.
Above: Screenshot of NSRR Sleep Common Data Elements with attributes consisting of Name, Type, Unit, Min, Max, and Version. Below: Screenshot of NSRR’s cross-cohort exploration system, guided by the Sleep Common Data Elements (left column). This system is openly accessible at x-search.net.
Functional architecture
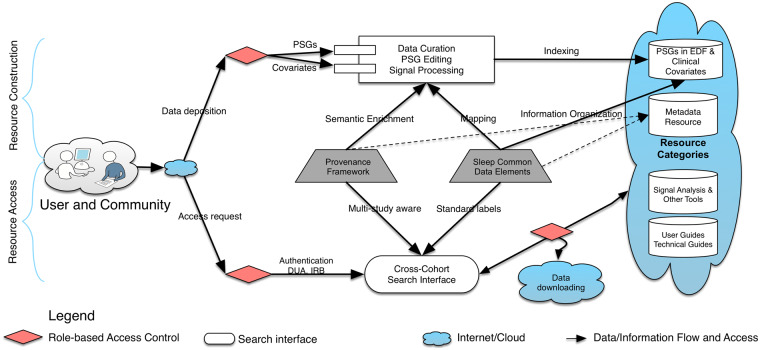
We designed the NSRR functional architecture to flexibly accommodate the deposition of a growing set of new tools and data. To do so, the NSRR functional architecture consists of two main parts: Resource Construction and Resource Access (Figure 2). Resource Construction allows data from individual data sets to be curated, mapped, and integrated into NSRR on a cohort-by-cohort basis over time (retrospectively or prospectively) by the NSRR team. We process two broad categories of data in Resource Construction: study variables as defined by each cohort and transformed and mapped to NSRR’s sleep terms, and PSGs converted to European Data Format (EDF), a standard file format for polysomnography recordings, with precisely annotated sleep events. We also preprocess EDF files to generate derived data with established signal analysis tools such as those for spectral analyses adopted from PhysioNet,32 while keeping the raw data also available. Four categories of resources are made available incrementally: (a) PSGs in EDF and clinical covariates; (b) metadata for sleep research; (c) tools for data analytics and cross-cohort exploration; (d) user guides, technical guides, and study documents originated from individual study (Figure 2).
Figure 2.
Functional architecture representing the connections and interactions among NSRR components. Sleep Common Data Elements play a central role in coordinating and facilitating incremental resource construction (above) and resource access (below).
We developed and used a number of tools for Resource Construction including Spout,33 a data dictionary tool for maintaining and validating information on datasets and their clinical data elements, and Edfize,34 for testing the integrity of the converted EDF files. For Resource Access, the NSRR team provides a gem tool35 that simplifies the downloading of large source files by users with appropriate credentials of data access and use agreements and IRB reviews, which are also tracked and managed through the NSRR web portal.
Workflow for data curation and metadata annotation
We curate study-specific variables from individual cohorts and make them publicly available using a two-stage process. In stage one, our team performs the curation of datasets and their data dictionaries. All variables from each data dictionary are transformed into an NSRR-specific format using an automated script developed by the NSRR team. The script systematically checks the variables after their transformation, against source data for variable-specification quality control, such as conformation to the NSRR naming convention, value set or variable domain matching (value type, range gap, out of range, and outliers), and variable tagging by dataset-specific questionnaire forms where applicable. This systematic quality check against source data is performed iteratively by the NSRR team, resulting in progressively enhanced versions of the variable specifications that are tracked using version control. In stage two, a subset of variables shared across datasets are manually identified and mapped to the SCDE terms to facilitate cross-cohort search and meta-analysis.36 The versions of mappings are also systematically tracked.37
The import of a dataset into the NSRR involves annotation with two types of metadata: study variable metadata and polysomnography recording metadata. Each study variable that has been mapped to an SCDE term is also annotated with provenance metadata abstracted from documents on study processes and methodology. Provenance metadata include the source of the data, the time point at which the data were collected, the method used to collect or abstract the data, the equipment used to collect the data, and any formulas applied to calculate derived values.
Polysomnography recording metadata contain information such as study identifiers, signal channel names, signal units, and signal minimums and maximums that were acquired at the time of signal acquisition based on the configuration of the recording system. Such information is stored in the EDF header and is extracted and mapped using our EDF Editor and Translator38 for analyses. For details of the NSRR metadata extraction pipeline, see Supplementary Appendix A.
Tools for data exploration, visualization, and analysis
The NSRR team developed a number of tools for data exploration and analysis. X-search.net36 enables users to query across multiple datasets using the SCDE terms (Figure 1). The tool Altamira39 is used for web-based rendering of EDF signals, making use of the Edfize library.34 The NSRR also provides a downloading tool called the NSRR gem35 that facilitates the downloading of large datasets. The gem checks the integrity of files to be downloaded, recreates the local folder hierarchy, and manages downloading interruptions.
The data source for x-search.net is a mirrored copy of the NSRR clinical data after data mapping. The NSRR team handles mapping and coding inconsistencies before new datasets (excluding the EDF files) are integrated and become queryable using x-search.net. The data source for Altamira is the same imported copy of the EDF files.
We developed and used the Spout data dictionary management tool33 to generate dataset descriptions for access through the NSRR web portal. Spout generates and updates the specifications and histogram renderings of thousands of variables used in individual NSRR cohorts. An extended list of openly accessible tools appears in Supplementary Appendix A.
Data quality review
The NSRR team reviews new datasets for outlying and implausible values, which are tracked in a Known Issues file in each version-controlled data dictionary repository. Raw EDF files are processed in the EDF Editor and Translator tool to ensure that each file is readable and conforms to EDF specifications. Polysomnograms that cannot be converted successfully to EDF are noted in the dataset documentation.
Additional details about data protection and the hosting environment appear in Supplementary Appendix A.
RESULTS
Annotated and integrated data sets in NSRR
We have integrated data from 10 large, NIH-funded sleep cohort studies (Table 1). In total, the NSRR contains semantically annotated data from 26 808 subjects, and 51 435 linked files of raw or processed signals, including 31 166 EDF files that are available for downloading.
Table 1.
Description of data sets included in NSRR
| Cohort/Study | N, subjects (n, PSGs)* | Objective Sleep Data | Main Study Outcomes | Present in TOPMed |
|---|---|---|---|---|
| Sleep Heart Health Study (SHHS: subsets of ARIC, CHS, FHS, Tucson) | 5600 (8080) | Full PSG | Incident cardiovascular disease | Selected samples (ARIC, FHS) |
| 40+ years | ||||
| Childhood Adenotonsillectomy Trial | 1244 (1639) | Full PSG | Sleep apnea treatment effects on cognition, behavior, and growth | No |
| 5–10 yrs | ||||
| Heart Biomarkers in Apnea Treatment, HeartBEAT | 305 (580) | Oximetry, NP, RIP; ECG | Sleep apnea treatment effects on 24-hour blood pressure and biomarkers | No |
| 45–75 yrs | ||||
| Cleveland Family Study | 1600 (3200) | Oximetry; Thermistry; Chest effort, ECG; Full PSG in n = 700 | Genetics of sleep apnea | Yes |
| 4–96 yrs | ||||
| Study of Osteoporetic Fractures in Older Women (SOF)-Sleep | 460 | Full PSG; actigraphy | Incident dementia, falls, and fractures. | No |
| 75+ yrs | ||||
| Study of Osteoporetic Fractures in Older Men-Sleep (MrOS) | 2991 (4452) | Full PSG; actigraphy | Incident falls, fractures, and cardiovascular disease | No |
| 65+ yrs | ||||
| Cleveland Children’s Sleep and Health Study | 850 (1603) | Oximetry; Thermistry; NP, RIP; ECG in all; Full PSG and actigraphy on n = 504 | Incident obesity and pediatric sleep disorders | No |
| 8–19 yrs | ||||
| Hispanic Community Health Study (HCHS/SOL) | 15 000 | Oximetry, NP, snoring, movement; actigraphy on n = 2000 | Diabetes, cardiovascular disease, neurocognition, hearing loss | Yes |
| 18–74 yrs | ||||
| Honolulu Asian American Asian Sleep Study, HAAS | 700 | Full PSG | Incident cognitive impairment | No |
| 85+ yrs | ||||
| Multiethnic Study of Atherosclerosis (MESA)-Sleep Study | 2200 | Full PSG; actigraphy | Incident cardiovascular disease (including cardiac MRI) | Yes |
| 45–84 yrs |
RIP: inductance plethysmography; NP: nasal pressure.
The existing NSRR datasets were derived from completed prospective research studies, and therefore the clinical data were obtained through direct data collection, questionnaires, or research procedures (eg adjudication of follow-up data and medical records performed during primary data collection at the cohort level). Future extensions of the repository may include data directly obtained from clinical records, which will involve additional efforts to achieve their reusability.
The actigraphy data (usually covering 5 to 7 days) in NSRR are collected from research/medical devices prospectively for a subset of the studies. We have made these data available as summary metrics (the processed output of the devices) and raw data (counts), and are in the process of sharing specific metrics such as the one derived from yielding measurements of diurnal rhythm. The NSRR can also accommodate mobile sensor data as they become available, although the actigraphy data were collected in a controlled setting.
Deployed tools
As of March 2018, the NSRR Tool section40 features 17 tools, 1 R script, and 5 tutorials. While most of these tools have been created by the NSRR team, we recently invited the extended sleep research community to share their signal analysis tools in open source on the NSRR. Tools that have been contributed by external researchers are SpiSOP,41 a standalone tool for sleep EEG analysis that includes slow wave and spindle detection, and a MATLAB algorithm42 for detecting rapid eye movements in REM sleep.
Registered users
A total of 3059 users have registered for the resource, of which 6 are NSRR designated Academic User Group members, and 15 are core team members. Of the remaining 3038 registered users, 901 DAUAs have been submitted, of which 819 (90.9%) have been approved by the NSRR team. On average, the resource approves 17 new DAUAs per month since inception. We noticed a growing trend in the number of approved DAUAs per month in each year: 8 per month for 2014; 14 per month for 2015; 17 per month for 2016; 22 per month for 2017; and 34 per month for 2018.
Evidence of usage
Supplementary Appendix B (Table A) summarizes the numbers of files and data download sizes by registered users for each dataset. Overall, the NSRR has served over 5.8 million files, covering 133 TB of data. On average, over the 47 months since its inception, the NSRR served up approximately 95 000 files per month, with recent data sharing of 1TB per week. This equates to a daily average of 3100 files covering 75 GB of data per day.
Evidence of NSRR usage for scientific research includes research proposals submitted and publications. So far, 13 research proposals using NSRR as a data resource have been submitted or funded. More than 35 publications, identified in the acknowledgements or references section, have appeared in scientific venues, including a recent publication characterizing sleep spindles.43 Additional user feedback information is provided in Supplementary Appendix B.
DISCUSSION
Related efforts on sharing medical time series data
Although NIH has consistently invested in data-sharing resources, such as BioLINCC,44 dbGAP,45 PhysioNet,32 and CVRG,46 there is no single repository for complex time-series physiological data such as those represented by PSGs together with relevant subject-level clinical data. Existing resources also do not contain query and processing tools needed to maximize the usability of the data and the user experience.
PhysioNet32 is an NIBIB/NIGMS supported resource for openly disseminating and exchanging biomedical signals and open-source analysis software. Compared to NSRR, PhysioNet has a limited set of PSGs and lacks comprehensive clinical data.
Measuring degrees of FAIR quality
The NSRR provides a use case on how large-scale domain-specific data sets can be semantically enriched using a metadata-guided approach, a necessary step to make sleep research data FAIR using the following strategies and techniques:
Findable. This FAIR principle requires data objects to be uniquely and persistently identifiable. The NSRR achieves this through the use of Uniform Resource Identifiers (URIs), a string of characters used to identify a resource. For example, “https://sleepdata.org/datasets/shhs/variables/angina” is a string that uniquely identifies NSRR’s Sleep Heart Health Study variable for the number of angina episodes since baseline; “https://sleepdata.org/datasets/shhs/files/polysomnography/edfs/shhs1/shhs1-200008.edf” is a string that uniquely identifies Sleep Heart Health Study subject 200008’s baseline polysomnography recording in EDF format. Effort is underway to incorporate the SCDEs into NIH CDEs, and unique identifiers will be created for terms in the SCDEs.
Accessible: The NSRR provides open access to the entire study metadata, including study provenance information and sleep concepts, events, and variable specifications (see https://github.com/nsrr/cross-dataset-mapping/tree/master/mappings). Access to subject-level data and polysomnogram signal files is facilitated through an online-tracked process involving data use agreement and IRB as a part of the functionalities of the NSRR web portal. Tools for downloading large datasets are also provided by the NSRR team to facilitate access to a large collection of EDF files.
Interoperable: The NSRR uses standard formats for both the polysomnography and clinical data: polysomnography data in EDF, annotation in XML, and clinical data and data dictionaries in Comma Separated Values (CSV). The SCDEs reuse standard terminology from a variety of sources such as SNOMED CT and NIH CDEs. The interoperability between different clinical datasets is achieved by mapping individual datasets to SCDEs, and the mapping files are managed through the publicly available GitHub repository (https://github.com/nsrr/cross-dataset-mapping). The SCDEs also facilitate the exchange of PSG data in EDF: each EDF file derived from the PSGs has an accompanying Extensible Markup Language (XML) file using terms defined in SCDEs, documenting a wide scope of sleep events in the PSGs. This provides standardized interpretation of sleep events across different cohorts. Downstream tools for EDF signal analysis and visualization can all take advantage of the standardized sleep event descriptions given in such XML files.
Reusable: The NSRR data, metadata, and tools are shared through 3 main web resources to promote their reuse: sleepdata.org, github.com/nsrr, and github.com/sleepepi. They are well described and rich enough to support reuse, citation, and automated linkage, as demonstrated through cross-reference with BioLINCC (https://biolincc.nhlbi.nih.gov/studies) and the number of proposals and publications that cite their use of the NSRR resource.
Lessons learned
The NIH Data Commons is a vision laying out a set of coherent higher-level requirements that emphasize FAIR as the required properties for the “digital object compliance model” to fully leverage the computational power afforded by the cloud. However, it leaves open important questions such as how such a vision is to be achieved, who are to implement it, and where the digital object content comes from.
We believe that the Data Commons vision is best achieved by instantiating it through disease- or domain-specific efforts, such as the NCI Genomic Data Commons (GDC47,48). The GDC addresses unique computational challenges and provides researchers uniform analytic pipelines for bioinformatics processes. The NSRR, on the other hand, offers pragmatic experiences in answering questions related to how digital objects in sleep medicine can be developed toward the goal of a sleep data commons, where the digital contents come from, and the type of team composition suited for such a development.
The NSRR functional architecture was designed and implemented to support continuous data integration and sharing. This metadata-guided approach is adaptable to support other similar data integration and sharing needs. For instance, it has been successfully adapted and further enhanced to support prospective data capture, integration, and sharing for an ongoing multi-center project for epilepsy research,49,50 and current efforts are underway to incorporate data from independent research groups (eg Wisconsin Sleep Cohort).
The agile development paradigm, with the hallmark that requirements and solutions evolve through collaborative team efforts, has successfully enabled involvement of the key stakeholders in all phases of software development to ensure the usability of the NSRR web-based application tools. “Version-control everything,” including documentation, metadata, and code repository using GitHub51 turned out to be an effective means for managing the project and facilitating collaboration.
One of the non-technical challenges is obtaining agreements to share data through the NSRR from the “owners” of individual study cohorts. Additional details about this aspect appear in Supplementary Appendix A.
In a way, the NSRR is a data engineering experiment that confirms the notion of a rough 80-20 split data science between data readiness work and data analysis work52,53: higher than expected effort is required to make data accessible and reusable. Two non-technical strategies emerged as indispensable for the success of a project such as the NSRR: one is the team-science, trusted, collaborative, results-driven spirit growing from a long-term partnership among a group of sleep investigators, computer scientists, and informaticians; and the other is the equal partnership, shared leadership roles, and proper intellectual property ownership among the domain-experts from distinct individual disciplines.
Future directions
Shared resources offered by emerging resources such as cloud instances provide promising platforms for the Data Commons. However, simply expanding storage or adding computational power may not allow us to cope with the rapidly expanding volume and increasing complexity of biomedical data. Concurrent efforts must be spent to address digital object organization challenges. To make our approach future proof, we need to continue advancing research in data representation and interfaces for human-data interaction.
A possible next phase of the NSRR is the creation of a universal self-descriptive sequential data format. The idea is to break large, unstructured, sequential data files into minimal, semantically meaningful fragments.54 Such fragments can be indexed, assembled, retrieved, rendered, or repackaged on-the-fly, for multitudes of application scenarios. Data points in such a fragment will be locally embedded with relevant metadata labels, governed by terminology and ontology. Potential benefits of such an approach may include precise levels of data access, increased analysis readiness with on-the-fly data conversion, multi-level data discovery, and support for effective web-based visualization of contents in large sequential files.
CONCLUSIONS
In this paper, we introduced the NSRR, a data-sharing system aimed at fully supporting the FAIR principles, for integrating clinical data and physiological signal data from NIH-funded epidemiological cohort studies in sleep research. We believe that several aspects of the NSRR can help inform progress towards the implementation of a future-proof NIH Data Commons from a domain-specific, usability-informed, bottom-up perspective.
FUNDING
This work is supported by the National Heart, Lung, and Blood Institute (NHLBI R24 HL114473), the National Science Foundation under MRI Grant No.1626364, and the University of Kentucky Center for Clinical and Translational Science (UL1TR001998).
CONTRIBUTORS
GQZ and SR conceptualized and designed this study. RM, LC, ST, SM, and SP implemented the tools. LC, MK, and GQZ developed the sleep metadata framework. MR, RM, DM, SM, and SR processed the data. GQZ wrote the manuscript with contributions from LC, RM, ST, MT, MR, SM, and DM. SR and LC reviewed and contributed to formatting the manuscript.
Conflict of interest
None declared.
Supplementary Material
REFERENCES
- 1. Altevogt BM, Colten HR, ed. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem. Washington, DC: National Academies Press; 2006. [PubMed] [Google Scholar]
- 2. Nieto EJ, O’Connor GT, Rapoport DM, et al. The sleep heart health study: design, rationale, and methods. Sleep 1997; 2012: 1077–85. [PubMed] [Google Scholar]
- 3. Redline S, Sanders MH, Lind BK, et al. Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep 1998; 217: 759–68. [PubMed] [Google Scholar]
- 4. Redline S, Amin R, Beebe D, et al. The Childhood Adenotonsillectomy Trial (CHAT): rationale, design, and challenges of a randomized controlled trial evaluating a standard surgical procedure in a pediatric population. Sleep 2011; 3411: 1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marcus CL, Moore RH, Rosen CL, et al. A randomized trial of adenotonsillectomy for childhood sleep apnea. N Engl J Med 2013; 36825: 2366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gottlieb DJ, Punjabi NM, Mehra R, et al. CPAP versus oxygen in obstructive sleep apnea. N Engl J Med 2014; 37024: 2276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Redline S, Tishler PV, Tosteson TD, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med 1995; 151 (3 Pt 1): 682–7. [DOI] [PubMed] [Google Scholar]
- 8. Redline S, Tishler PV, Schluchter M, et al. Risk factors for sleep-disordered breathing in children: associations with obesity, race, and respiratory problems. Am J Respir Crit Care Med 1999; 159 (5 Pt 1): 1527–32. [DOI] [PubMed] [Google Scholar]
- 9. Cummings SR, Black DM, Nevitt MC, et al. Appendicular bone density and age predict hip fracture in women. JAMA 1990; 2635: 665–8. [PubMed] [Google Scholar]
- 10. Spira AP, Blackwell T, Stone KL, et al. Sleep-disordered breathing and cognition in older women. J Am Geriatr Soc 2008; 561: 45–50. [DOI] [PubMed] [Google Scholar]
- 11. Blank JB, Cawthon PM, Carrion-Petersen ML, et al. Overview of recruitment for the osteoporotic fractures in men study (MrOS). Contemp Clin Trials 2005; 265: 557–68. [DOI] [PubMed] [Google Scholar]
- 12. Orwoll E, Blank JB, Barrett-Connor E, et al. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials 2005; 265: 569–85. [DOI] [PubMed] [Google Scholar]
- 13. Rosen CL, Larkin EK, Kirchner HL, et al. Prevalence and risk factors for sleep-disordered breathing in 8-to 11-year-old children: association with race and prematurity. J Pediatr 2003; 1424: 383–9. [DOI] [PubMed] [Google Scholar]
- 14. Spilsbury JC, Storfer-Isser A, Drotar D, et al. Effects of the home environment on school-aged children's sleep. Sleep 2005; 2811: 1419–27. [DOI] [PubMed] [Google Scholar]
- 15. Hibbs AM, Storfer-Isser A, Rosen C, et al. Advanced sleep phase in adolescents born preterm. Behav Sleep Med 2014; 125: 412–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Redline S, Sotres-Alvarez D, Loredo J, et al. Sleep-disordered breathing in Hispanic/Latino individuals of diverse backgrounds. The Hispanic community health study/study of Latinos. Am J Respir Crit Care Med 2014; 1893: 335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel SR, Weng J, Rueschman M, et al. Reproducibility of a standardized actigraphy scoring algorithm for sleep in a US Hispanic/Latino population. Sleep 2015; 389: 1497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foley DJ, Masaki K, White L, et al. Sleep-disordered breathing and cognitive impairment in elderly Japanese-American men. Sleep 2003; 265: 596–9. [DOI] [PubMed] [Google Scholar]
- 19. Dean DA, Goldberger AL, Mueller R, et al. Scaling up scientific discovery in sleep medicine: the National Sleep Research Resource. Sleep 2016; 395: 1151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sempos CT, Bild DE, Manolio TA.. Overview of the Jackson Heart Study: a study of cardiovascular diseases in African American men and women. Am J Med Sci 1999; 3173: 142–6. [DOI] [PubMed] [Google Scholar]
- 21.Trans-Omics for Precision Medicine (TOPMed) Program. https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed (Accessed July 25, 2017).
- 22. National Sleep Research Resource. https://sleepdata.org (Accessed July 25, 2017).
- 23.What is the (NIH) Commons? https://datascience.nih.gov/TheCommons (Accessed July25, 2017).
- 24. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3: 160018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. NIH Data Commons Pilot Phase. https://commonfund.nih.gov/bd2k (Accessed July25, 2017).
- 26.biomedical and healthCAre Data Discovery Index Ecosystem (bioCADDIE). https://biocaddie.org (Accessed July 25, 2017).
- 27. Sleep Domain Ontology (SDO). https://bioportal.bioontology.org/ontologies/SDO (Accessed July 25, 2017).
- 28.Three New Informatics Pilot Projects to Aid Clinical and Translational Scientists Nationwide. https://www.nih.gov/news-events/news-releases/three-new-informatics-pilot-projects-aid-clinical-translational-scientists-nationwide (Accessed May 29, 2017).
- 29. Arabandi S, Ogbuji C, Redline S, et al. Developing a sleep domain ontology. AMIA Jt Summits Transl Sci Proc 2010: 12–3. [Google Scholar]
- 30. Zhang GQ, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research. AMIA Jt Summits Transl Sci Proc 2010; 2010: 76–80. [PMC free article] [PubMed] [Google Scholar]
- 31. Mueller R, Sahoo S, Dong X, et al. Mapping multi-institution data sources to domain ontology for data federation: the Physio-MIMI approach. AMIA Jt Summits Transl Sci Proc 2011: 38. [Google Scholar]
- 32. Costa M, Moody GB, Henry I, et al. PhysioNet: an NIH research resource for complex signals. J Electrocardiol 2003; 36: 139–44. [DOI] [PubMed] [Google Scholar]
- 33. Spout. https://github.com/sleepepi/spout (Accessed July 25, 2017).
- 34. Edfize. https://github.com/sleepepi/edfize (Accessed July 25, 2017).
- 35. NSRR Ruby Gem. https://github.com/nsrr/nsrr-gem (Accessed July 25, 2017).
- 36. NSRR Cross Dataset Query Interface. https://x-search.net (Accessed July 25, 2017).
- 37.Version controlled repository for NSRR canonical data dictionary and cross-dataset mapping files. https://github.com/nsrr/cross-dataset-mapping/tree/master/mappings (Accessed July 25, 2017).
- 38. Jayapandian CP, Wang W, Morrical MG, et al. RREV: reconfigurable rendering engine for visualization of clinically annotated polysomnograms. IEEE Int Conf Bioinformatics Biomed Proc 2015: 309–16. [Google Scholar]
- 39. Altamira. https://github.com/nsrr/altamira (Accessed July 25, 2017).
- 40. NSRR tools. https://sleepdata.org/tools (Accessed July 25, 2017).
- 41. SpiSOP. http://www.spisop.org/ (Accessed July 25, 2017).
- 42. Rapid Eye Movement Detector. https://osf.io/fd837/ (Accessed July 25, 2017).
- 43. Purcell SM, Manoach DS, Demanuele C, et al. Characterizing sleep spindles in 11,630 individuals from the National Sleep Research Resource. Nat Comms 2017; 8: 15930.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. National Institutes of Health, National Heart, Lung, and Blood Institute. Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC). https://biolincc.nhlbi.nih.gov/home/ (Accessed July 25, 2017).
- 45. Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 2007; 3910: 1181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winslow RL, Saltz J, Foster I, et al. The CardioVascular Research Grid (CVRG) project. AMIA Jt Summits Transl Sci Proc 2011: 77–81. [Google Scholar]
- 47. Grossman RL, Heath AP, Ferretti V, et al. Toward a shared vision for cancer genomic data. N Engl J Med 2016; 37512: 1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jensen MA, Ferretti V, Grossman RL, et al. The NCI Genomic Data Commons as an engine for precision medicine. Blood 2017; 1304: 453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang GQ, Cui L, Lhatoo S, et al. MEDCIS: multi-modality epilepsy data capture and integration system. AMIA Annu Symp Proc 2014; 2014: 1248–57. [PMC free article] [PubMed] [Google Scholar]
- 50. Cui L, Huang Y, Tao S, et al. ODaCCI: Ontology-guided Data Curation for Multisite Clinical Research Data Integration in the NINDS Center for SUDEP Research. AMIA Annu Symp Proc 2016; 2016: 441–50. [PMC free article] [PubMed] [Google Scholar]
- 51. The NSRR team. NSRR GitHub Repository. https://github.com/nsrr (Accessed July 25, 2017).
- 52.Why data preparation is an important part of data science. https://www.dezyre.com/article/why-data-preparation-is-an-important-part-of-data-science/242 (Accessed April 1, 2018).
- 53.Data cleaning is a critical part of the data science process. http://blog.revolutionanalytics.com/2014/08/data-cleaning-is-a-critical-part-of-the-data-science-process.html (Accessed April 1, 2018).
- 54. Li X, Cui L, Tao S, et al. SpindleSphere: a web-based platform for large-scale sleep spindle analysis and visualization. AMIA Annu Symp Proc 2017: 1159–68. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.