Abstract
Functional magnetic resonance imaging is capable of estimating functional activation and connectivity in the human brain, and lately there has been increased interest in the use of these functional modalities combined with machine learning for identification of psychiatric traits. While these methods bear great potential for early diagnosis and better understanding of disease processes, there are wide ranges of processing choices and pitfalls that may severely hamper interpretation and generalization performance unless carefully considered. In this perspective article, we aim to motivate the use of machine learning schizotypy research. To this end, we describe common data processing steps while commenting on best practices and procedures. First, we introduce the important role of schizotypy to motivate the importance of reliable classification, and summarize existing machine learning literature on schizotypy. Then, we describe procedures for extraction of features based on fMRI data, including statistical parametric mapping, parcellation, complex network analysis, and decomposition methods, as well as classification with a special focus on support vector classification and deep learning. We provide more detailed descriptions and software as supplementary material. Finally, we present current challenges in machine learning for classification of schizotypy and comment on future trends and perspectives.
Keywords: functional magnetic resonance imaging, feature extraction, neuroimaging, schizotypy, schizophrenia spectrum disorder
Introduction
The study of schizotypy has received substantial interest in the field of psychiatry and psychology and recently developments and increased interest in machine learning for neuroimaging is showing promising applications in computational psychiatry. Theoretically, schizotypy has been conceptualized as an important phenotype for schizophrenia spectrum disorders.1–3 Two competitive theories, the quasi-dimensional and the fully dimensional approach have been proposed to model the construct of schizotypy. The quasi-dimensional approach posits the view that schizotypy is a discontinuity in the general population.2,4 However, recent studies have suggested that this phenotype is distributed along a continuum, ranging from psychological well-being to full-blown psychosis,5–7 supporting the fully dimensional approach that emphasizes the continuity of schizotypy.8 Furthermore, empirical findings demonstrate that individuals with schizotypal traits exhibit similar but attenuated impairments in cognition,9,10 emotion,7,11 and neurological functions10,12 compared with patients with schizophrenia. Likewise, manifestations of these schizotypal phenotypes are found to be robust and stable across time and environment.5,13–15
With implications from the neurodevelopmental model of psychosis in schizophrenia,16,17 Insel18 further delineated 4 stages, ranging from risk to chronic disability. This 4-stage hypothesis highlights the importance of early risk stages for the understanding of the psychopathology to facilitate early detection and intervention strategies for psychosis and mental disorders. Although schizotypy is not explicitly included in Insel’s model, there are important similarities within the cognitive, emotional, and social impairments. This point toward understanding the psychopathology of schizophrenia spectrum disorders through personality traits presented in the general population.
Recently, schizotypy has recently been conceptualized as a construct well beyond the borders of schizophrenia spectrum disorders (e.g., Cohen et al19). These authors argue that the researchers interested in psychosis have mainly followed narrow research avenues, focusing on molecular, neurophysiological, environmental, and cultural correlates of psychotic expression or investigating potential endophenotypes relating to the extreme manifestation of schizotypy to schizophrenia. However, the unique emotional and social manifestations observed in individuals with schizotypy can actually provide insight into the nature of affective and social systems integral to general human functioning. For example, findings from functional neuroimaging have shown that individuals with social anhedonia exhibit significant hypoactivation of the left pulvinar, claustrum, and insula to positive cues in the anticipatory phase of the affective incentive delay task compared with those without social anhedonia.14 Longitudinal studies also suggest that individuals with schizotypal traits have their unique trajectories that may not necessarily develop into full-blown psychosis.20–23 Recently, Wang et al24 identified 4 trajectories of schizotypy; including 2 stable and 2 reactive groups. The “stable low and high schizotypy” groups displayed the best and worst clinical and functional outcomes, respectively. The “high reactive schizotypy” group was characterized by a relatively rapid decline in function, while the “low reactive schizotypy” group was characterized by low scores at baseline of the assessment but with gradual deterioration. These findings suggest that even within the nonclinical sample of schizotypal phenotypes, similar subtypes, and trajectories comparable to the clinical patients with schizophrenia are observed. This highlights the importance of tracking schizotypy longitudinally because of their unique trajectories and outcomes.
Several studies have already applied neuroimaging data to investigate the neurobiological changes related to schizotypy, reporting both structural and functional changes. For example, structural studies have found grey matter volume changes in many of the areas known to be altered in schizophrenia, such as prefrontal, temporal, and cingulate cortex, as well as insula and subcortical regions.25–28 These studies suggest that cortical changes exist on a dimensional continuum across the schizophrenia spectrum, likely to occur pre-onset of psychopathology. Furthermore, studies using functional magnetic resonance imaging (fMRI) to investigate social cognition, have reported similar regional brain activation changes, when comparing participants with different degree of schizotypy, or individuals with high schizotypy compared with controls.29–31 Finally, functional connectivity studies reported similar network changes to that of patients with schizophrenia,32–34 such as altered connectivity between striatum, medial prefrontal cortex (PFC), anterior cingulate (ACC), and insula. Importantly, almost all the above studies, reported different results for the positive and negative dimension of schizotypy, demonstrating the heterogeneity of schizotypy.
The above findings emphasize the important role of schizotypy in psychiatry and psychology. On one hand, schizotypy is considered to be a trait marker for schizophrenia and the study of behavioral and neurobiological bases of schizotypy may help us understand the underlying psychopathology of schizophrenia. This suggests that schizotypy may be an important phenotype for studying schizophrenia spectrum disorders. On the other hand, schizotypy may serve as a unique entity to examine the underlying emotional and social systems in humans. Therefore, a better way to classify this phenotype will be meaningful to schizotypy scholars. However, to our knowledge, there are only few studies which have been identifying schizotypy based on neuroimaging data. Here, machine learning methods can serve to bridge this knowledge gap, and help elucidate the neurobiological abnormalities of at-risk individuals at an early stage of schizophrenia.
Machine Learning in the Field of Schizotypy and Schizophrenia
The overall aim of machine learning is to make computers classify data without being explicitly programmed. Typically, a distinction is made between supervised and unsupervised learning. The former refers to learning using labelled data, with the aim to generalize classification to data with unknown labels. In contrast, unsupervised learning methods explore statistical dependencies in unlabelled data, with the goal of learning structure in the data and possibly cluster data into distinct classes.
Recently, machine learning methods have been used as a neuroimaging-based tool to automatically discriminate individuals in schizophrenia spectrum disorders from healthy people.35–37 Empirical findings suggest that these methods are able to classify schizophrenia patients from healthy controls with an accuracy rate ranging from 75% to 98%.37–40 Furthermore, recent studies have had success with using support vector classification (SVC) to predict the transition of ultra-high risk individuals converting to full-blown psychosis,41–43 and discriminate converters and nonconverters.44,45 However, limited studies have investigated individuals in the stages before onset of the illness.
As for studying schizotypy using machine learning methods, a range of studies have been exploring the neural mechanism related to schizotypy and classified individuals according to different groups. In 2006, Shinkareva et al46 used spatio-temporal dissimilarity maps to classify individuals with high levels of positive schizotypy and controls based on fMRI data from an emotional Stroop task. With the same aim, Modinos et al47 performed SVC on brain activation maps from an emotional task and found the alterations for the emotional circuitry, including amygdala, ACC, and medial PFC, in individuals with high positive schizotypy. For comparison, they also performed statistical parametric mapping (SPM), which did not detect any class differences, indicating the increased sensitivity to subtle changes in risk populations, by using multivariate approaches. From the view of the “full dimensional” model of schizotypy, Wiebels et al48 used partial least square method, to demonstrate the relationship between different facets of schizotypy with gray matter volume changes.
Furthermore, 2 studies have explored schizotypal scores in individuals with subclinical depression and an ultra-high-risk group, respectively. First, Modinos et al49 found significant correlation between the positive dimension of schizotypy and the SVC weights which were obtained when classifying individuals with subclinical depressive symptoms and healthy controls. Secondly, in a longitudinal study, Zarogianni et al45 applied SVC to classify ultra-high-risk individuals into converters and nonconverters. Whereas this study mainly used structural MRI data, it was shown that the classification performance was increased when adding schizotypy scores to the analysis. Finally, other neuroimaging modalities than (f)MRI have been used to investigate schizotypy using machine learning methods. For example, in a study by Jeong et al50 event-related potentials, measured by EEG during an audiovisual emotion perception task, were used for classification of individuals with schizotypy and controls.
To conclude, the research in schizotypy utilizing machine learning shows great promise in terms of improving our understanding of schizotypy, and is of particular relevance for early detection and potential interventions. The main advantage of machine learning methods is that they can offer higher sensitivity than their counterparts based on standard univariate statistics, due to being able to learn the likely complex manifestations of schizotypy in multimodal neuroimaging data. Currently, existing studies are still limited by quite small sample sizes (n = 7–18 in each group45–47,49,50), and there is a risk that the reported classification rates are overfitted to the observed samples. This highlights the importance of having sufficient large sample sizes, and well-balanced groups to enable adequate learning and ensure that the training data is representative. Furthermore, it is important that future studies focus on independent validation of existing results to ensure that findings are generalizable to the population.
Classification and Feature Extraction Methods
In neuroimaging studies, fMRI data are mostly used to measure either activation changes in isolated brain areas, or to estimate functional connectivity (networks coupling) across regions.51 Because fMRI data are recorded in relatively high spatial resolution with a limited number of time points, estimation of activation patterns and in particular connectivity, is in practice quite unstable.52 Therefore, approaches to reduce dimensionality are often considered to improve the stability of the estimated functional activation.53–55 In the current article, we focus on features derived from fMRI, but classification procedures readily generalize to other modalities and multi-modal settings.
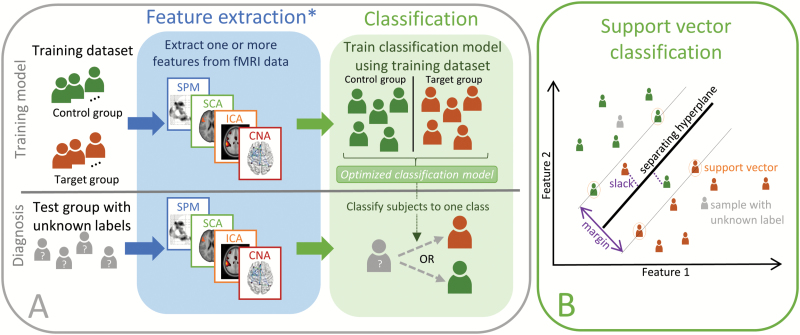
When using supervised learning in the field of neuroimaging, the aim is generally to determine an unknown class label of a subject based solely on the measured imaging data (eg, recorded fMRI data) as illustrated in figure 1, this procedure is also termed classification. In supervised classification, a model discriminating between the known labels in the training data is learned, subsequently enabling application of this model to unlabelled data to predict unknown labels.
Fig. 1.
Classification. The top row in panel A shows how a classification model can be trained on neuroimaging data. First feature extraction methods are used to identify features that can be used to train a classification model on samples with known labels. Once the classification model is trained, it can be applied to features extracted (using the same procedure) from subjects with unknown labels as indicated in the bottom row. *In principle the feature extraction step can be omitted. However, in practice for many imaging modalities (including fMRI), overfitting due to the high dimensionality of the input data will be detrimental to the classification performance. Panel B provides an illustration of the linear soft margin SVC algorithm in a 2-dimensional feature space. The SVC identifies the separating hyperplane that maximizes the margin, this hyperplane is only defined by the support vectors which are samples that are on the margin (marked by a circle). The soft margin SVC allows misclassification to avoid overfitting by introducing slack variables for each misclassified sample (marked with a dotted line). When the SVC is trained the labels of new samples (marked in gray) can be estimated according to the side of the hyperplane on which they reside.
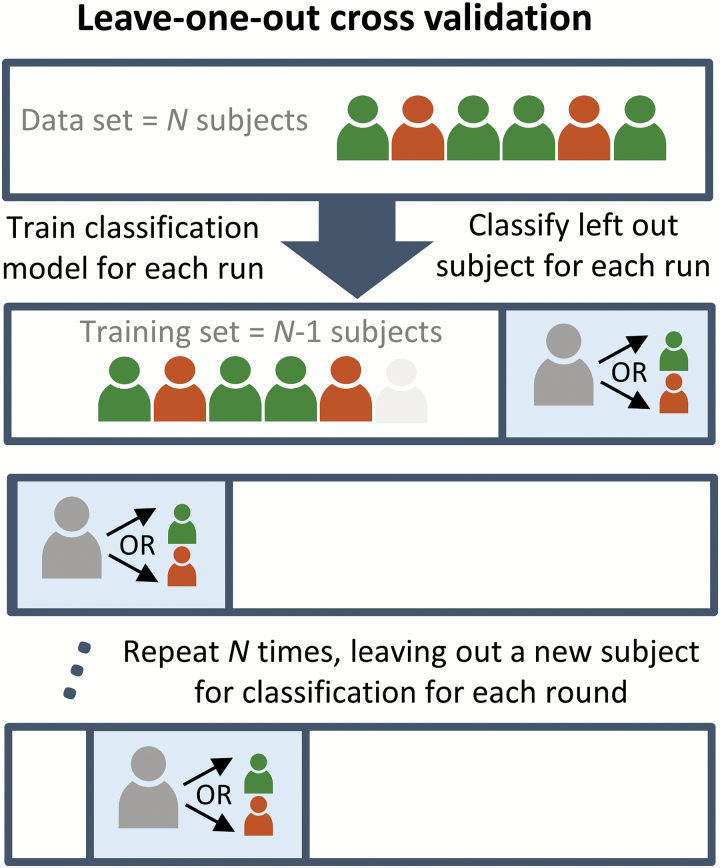
Given a labelled dataset, one can determine the classification performance using cross-validation (CV). The accuracy (rate of correctly identified class labels) is often used as a measure of performance. However, is important to note that this does not provide a full description of the performance, but also sensitivity (also referred to as the true positive rate or recall) and the specificity (the true negative rate) are important quantities. To test if the obtained classification rate is significant, the performance is usually tested against a parametric or empirical null-distribution.56 If the classification step considers several separate classification procedures, corrections for multiple comparisons should be performed when assessing significance. The CV procedure can be considered a simulation of the clinical setting, in which the labels of a set of subjects (test set) are assumed unknown and to be estimated through the training of a classification algorithm on the remaining subjects (training set). A frequently used method is the leave-one-out CV, where only one subject constitutes the test set, and the procedure is repeated for each subject as illustrated in figure 3. The leave-one-out scheme is often preferred because it minimizes the model bias by reserving the maximal amount of data for model training, but is has the disadvantage that there is a higher risk of overfitting to the training data. Therefore, other schemes such as K-fold (dividing the data into K nonoverlapping splits) CV are sometimes preferred. These enable testing of model stability by examining the variability of the identified model across splits. An example is the split half resampling procedure, where the difference between the models in the 2 independent splits can serve as an estimate of the model reproducibility.57
Fig. 3.
Leave-one-out cross-validation. The figure illustrates the leave-one-out LOO CV procedure. For each participant a classification model excluding that particular participant is trained. The model is then used to estimate the class label of the participant. This procedure is repeated for each participant to provide an unbiased estimate of the classification performance. Note that other CV schemes, including more complex nested CV are also possible.
In principle, it is possible to train classification algorithms directly on the raw neuroimaging data. However, due to the high dimensionality of the data compared with the small sample size the input data will appear sparse in the high dimensional space, often referred to as the curse-of-dimensionality. This in turn causes the classification procedure to be too specialized and generalize poorly to the test data, a phenomenon known as overfitting.
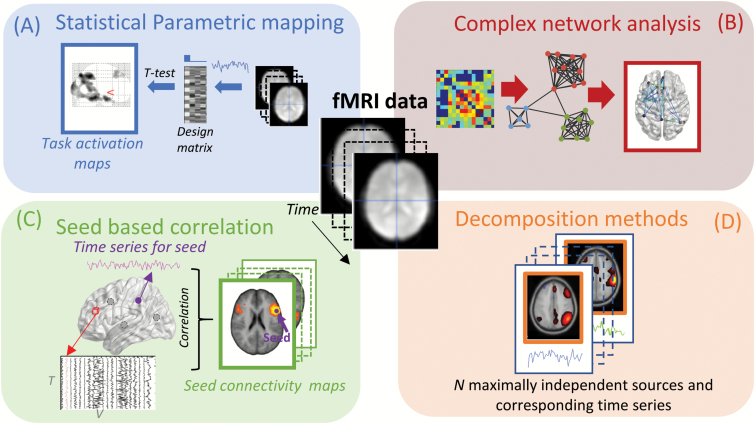
Therefore, classification is typically approached using a 2-step procedure in which features relevant to classification are first identified (see feature extraction steps, as illustrated in figure 2) and subsequently used to train the classification algorithm. The feature extraction step might include feature selection, where features are selected for further training. It is important that feature selection should only use labels from the training dataset, as the evaluation of performance would otherwise be biased and potentially lead to overfitting. Therefore, nested CV schemes, where an additional independent test set is used to estimate optimal features or other free parameters can be advantageous. Overfitting may be mitigated by automated feature selection methods and ensemble learning methods58 such as forward selection, backward elimination, recursive feature elimination,59 decision trees, and random decision forests.60 Also, several toolboxes including scikit-learn,61 Nilearn,62 PRoNTo,63 pyMVPA,64 and the NeuroMiner toolbox used by Koutsouleris et al41 are tailored toward machine learning for neuroimaging and provide tools for automated feature selection.
Fig. 2.
Sketch of 4 feature extraction methods for fMRI. Panel A illustrates statistical parametric mapping, where information about the experimental design is used to test for significant activation in each voxel using a general linear model. Panel B sketches complex network analysis. Here, a network is derived by determining functional connectivity between parcellated brain regions, followed by analysis using graph theoretical measures. In panel C, the seed based correlation approach is illustrated, here the time series from a predetermined brain region is extracted and correlated to the rest of the brain. In panel D, decomposition methods are illustrated where fMRI data are decomposed into spatially independent components with corresponding time series.
Appropriate preprocessing steps are very important before feature extraction, since data which are contaminated by artefacts might not only lead to poor classification performance, but may also cause difficulties in the interpretation of the results. For example, if movement artefacts are more dominant in one of the groups, the classifier might focus on movement artefacts and obtain good classification performance. For further information on common preprocessing steps and software, see supplementary section A.
In the following subsections, we describe a selection of often used feature extraction procedures, and although not covered by this article, additional methods exist, including fALFF,65,66 and methods for estimating regional signal homogeniety.67
Statistical Parametric Mapping
SPM is currently one of the most frequent used methods for analyzing task-based fMRI data. The overall goal of SPM is to localize brain activation that differs significantly between tasks68 as illustrated in figure 2A. The technique is mass univariate, which means that an independent parametric statistical test (t- or f-test) is performed for each voxel separately, typically using the general linear model. The 3 most common software packages for performing parametric mapping are SPM,69 FSL,70 and AFNI.71
When used for classification, the parameter estimates or statistical values (extracted across the entire brain or in regions of interest) are used as classification features, either directly or with an additional feature selection step. An advantage of using SPMs is that the localization of effects is already implied in the features, typically leading to more straightforward interpretation of models. However, because the procedure is essentially univariate it can miss important information shared across a range of variables, and therefore may be less sensitive than feature extraction methods that consider the multivariate structure of the data directly.
Parcellation, Complex Networks, and Seed-Based Analysis
To overcome instability problems due to the low temporal resolution as described above, approaches that parcellate the brain into fewer regions; either defined via atlases72,73 or from data driven clustering methods53,74,75 are often preferred. Functional connectivity features can then be determined between the parcels using a statistical measure such as (partial) correlation or mutual information. The resulting features (typically represented in a symmetric adjacency matrix representing the network coupling between each parcel) can either serve directly as feature for subsequent classification or be used for further extraction of features, eg, in a graph theoretical framework (figure 2B). Often the graphs are binarized by applying a threshold, and global measures such as the node degree distribution (number of connections between parcels/nodes) graph structure via modularity76 or relational modeling77,78 are used to characterize networks. For a more complete description of graph theoretical measures see Bullmore and Sporns.79
A related technique is the simple and intuitive seed-based correlation analysis (SCA),51 which determines the coupling between a number of predefined seeds (based on some a priori hypothesis, from either a localization experiment or the literature). The time series data from each seed is then correlated with all other voxels of the brain, resulting in a whole-brain, voxel-wise functional connectivity map for each seed as shown in figure 2C. For a more detailed description of SCA and how it has been used in resting state fMRI in comparison with data driven methods, please see Cole et al.80 In general, parcellation-based methods are attractive because they generate a more simplistic overview of the data, and often lead to more straightforward interpretation of features. However, the limited flexibility that is implied by fixed parcellation schemes can lead to selection of inappropriate features and result in decreased sensitivity.
Decomposition
Decomposition are unsupervised machine learning methods (sometimes also referred to as data driven methods) that seek to identify latent sources in the data from multiple measurements (ie, fMRI time series). In fMRI, this typically amounts to identifying a relatively low number of underlying spatial sources (typically between 10 and 100) that are associated with time series as illustrated in figure 2D. The procedure can be viewed as a (lossy) compression of the information in the data. The sources are often considered representations of functional networks, because they represent consistent time courses across the brain. A widely accepted method is spatial group independent component analysis (ICA), which results in individual component expressions (sources) across subjects with corresponding time series. Most frequently ICA is performed using one of the open source toolboxes, such as GIFT81 or FSL Melodic.82 Decomposition is advantageous because consistent activation patterns can be captured efficiently and automatically. A potential disadvantage is that interpretation can be challenging because decomposition is prone to also capture prominent nuisance effects in the data including motion and physiological signals such as the cardiac and respiratory cycles. Also, there are typically a wide range of adjustable parameters (such as the number of sources) that are difficult to set manually and can lead to overfitting if considered part of the learning algorithm. More information on decomposition is provided in supplementary section B.
Support Vector Classification
Supervised classification methods seek to identify some function that would enable discrimination between the labels in the training dataset. Importantly, when the input dimensionality is high compared to the number of samples (typically the case in fMRI unless elaborate feature extraction and selection has taken place), it is actually trivial to obtain perfect classification in the training set (overfitting), but the performance may generalize poorly to the test set. Therefore, the real challenge in classification is to ensure that the classification generalizes well to unseen samples.
There are many classification algorithms available. Here, we will focus on the SVC methods,83,84 because they have often been used in previous literature and are readily available in several easy to use software packages.61,85 For further reference and information on other classification methods, see, eg, Schmah et al.86 and Bishop.87
The simplest classification problem is a binary (2 classes) linear classification, where the SVC algorithm attempts to identify a discrimination function expressed as a linear projection across features, where the sign indicates the label. This is most straightforwardly illustrated in the 2-dimensional case, where the so-called separating hyper-plane is a straight line (figure 1B), here it is also evident that there are many lines that would lead to identical classification performance. The SVC chooses the hyperplane that maximizes the margin, ie, the perpendicular distance between the plane the closest data points. The SVC therefore focuses on the points on the margin (samples that are the hardest to classify, also called support vectors), and the classification of new samples only require information about the distances with respect to these so-called support vectors, allowing efficient evaluation. This is often referred to as a solution with sparse support in the training set, where sparsity here refers to samples rather than features. In practice, the soft margin SVC83 is mostly preferred as it allows misclassified samples, to obtain a larger margin, which will increase the stability of the classifier. In this case, the maximization of the margin is traded off against a penalty for misclassified samples which is proportional to the distance to the separating hyperplane. The trade-off is controlled by a parameter (typically referred to as the C-parameter), which has to be selected or determined through an additional nested CV procedure.85 For unbalanced dataset (cases in which the no. samples in each group differs) the class imbalance can be taken into account by weighting the hyperplane such that the imbalance is counteracted (by assigning more weight to the under-represented class). Also, for such datasets the accuracy alone may not be a good performance measure, as even a trivial classifier that always selects the most frequent label would appear to perform well. In these cases, using other metrics such as prediction-recall curves or Matthew’s correlation coefficient are usually more informative.88
Generalization to nonlinear discrimination is typically approached by projecting the data into another space (higher dimensional or even infinitely dimensional) which would enable linear separation. For the SVC, and a range of other classification methods, this can be efficiently implemented using kernels through the so-called kernel-trick. Here, it is sufficient to calculate distances between the samples measured in the projected space (represented in a Gram matrix) which circumvents operating with the potentially high dimensional projection explicitly. Examples of frequently used kernels are the linear kernel (for linear classification), radial basis function kernel and the polynomial kernel. It is important to note that kernels typically introduce at additional parameters that need to be selected or optimized via CV,85 which can exacerbate problems of overfitting.
The classification performance is rarely the only quantity of interest. Often researchers are interested in determining which brain regions are important for classification. For the linear SVC weight maps, or sensitivity maps89 for nonlinear classifiers, are often visualized, as they indicate the importance of each feature for the classification performance. The interpretation of these weight maps is not straightforward as features can actually be important for classification, not because they are directly related to the effect of interest, but rather because they serve to filter out nuisance effects. This issue was highlighted by Haufe et al90 suggesting a procedure for transforming weight maps into more interpretable visualizations for linear classification.
In practice, data labels (eg, schizotypy score) are often determined using questionnaires, which utilizes either continuous or ordinal scales, where it might be difficult to define a clear division between classes. In such cases, it can be attractive to train the algorithms to predict this continuous variable directly. This effectively turns the procedure into a multivariate regression problem. Here, support vector regression91 is analogous with SVC, where the margin is formed by considering how far the predicted value (in the training set) is from the measured value. When considering regression models in place of classification, it should be noted that other performance measures such as the mean absolute error have to be used. Unfortunately, the interpretation of such measures is in general less intuitive than classification rates. Furthermore, evaluation of statistical significance is more involved and researcher most often rely on random permutation tests to form empirical null-distributions.56
To illustrate the classification procedures described above, we have added an illustrative example to the supplementary material, where we used SVC to classify participants into either a low of high social anhedonia group, using features from both SPM and ICA. For more details, please refer to supplementary material section A.
Deep Learning
Deep learning based on neural networks have recently received much attention in the machine learning community, and have also been used to classify neuroimaging data in several general92,93 and clinical settings.94–96 The general philosophy behind deep learning is to train large neural networks with many layers and parameters that take the raw (or in most cases preprocessed) data as input and where the last layer in the network produces an outcome such as classification of subjects. If properly trained the first layers of the network should then represent basic features of the data, that are then refined and specialized in the subsequent layers. As these networks inherently contain many parameters overfitting due to the limited amount of data is a major concern when attempting to train networks. Here, mitigation strategies include regularization, dropout sampling, and weight sharing.97 Another option, is to use transfer learning approaches, which use networks that are pretrained on other datasets (which may even be of a different modality) and only refine weights in the last layers of the network.98 We believe that such strategies, potentially combined with data argumentation99 (where more samples are created using transformations/perturbations of the original data), will be extremely important in the future to ensure the success of deep learning in schizotypy research.
Discussion and Future Perspectives
In the preceding paragraphs, we have motivated the importance for classification of schizotypy, presented previous literature that has used machine learning methods for classification, and described methods for feature extraction and classification. Machine learning approaches have a range of advantages, which make them very attractive for studying early risk stages and subtle differences, as it is the case for schizotypy. A clear example of how these methods can increase the sensitivity to subtle changes, was shown in Modinos et al,47 who found significant alterations in an emotional circuitry in individuals with schizotypy when using SVC, whereas no class difference was detected when using a standard SPM analysis.
However, even though machine learning methods have shown very promising results so far, there are a wide range of pitfalls and challenges that needs to be considered. In the following, we will highlight some of the most important aspects, which should be kept in mind when using machine learning methods for classification of schizotypy or similar early risk groups.
The high dimensionality and typical low sample sizes available in studies represent a challenging problem for machine learning algorithms. Thus, procedures to reduce dimensionality of the input data (feature extraction) and regularization are necessary to ensure good generalization performance. While repeated nested CV procedures are useful for tentatively alleviating data availability issues, initiatives to encourage data sharing across sites are very important to overcome the problems of sparse sample availability.100,101
Appropriate pre-processing can have a profound impact on results and a wide range of choices are available both in terms of methods and parameters.102 This is also true for feature extraction, feature selection and classification steps, and it is important to note that if these choices are considered free parameters of the classification, the problem of overfitting is exacerbated and appropriate procedures to improve generalization such as CV should be considered. The feature extraction method of choice will depend on the research question. If the study is driven by specific hypotheses, it can be an advantage to use feature extraction methods that specifically extract the relevant dimensions of the data. Whereas, if the study is more explorative, decomposition methods may be preferred, as it avoids restriction of the analysis to a set of predefined hypotheses.
In general, the high degree of flexibility in choices of classification pipelines represents a challenge. It is very difficult for researcher to prove that none of the choices biased the reported classification performance (because the pipeline was optimized for classification performance), which might happen even inadvertently. To circumvent these issues, it is highly recommendable that specific hypotheses and detailed analysis procedures are preregistered before studies are commenced. This can be done easily using, eg, the Open Science Framework (https://osf.io/). Note that such preregistration is valuable even for studies with explorative hypotheses. In addition, it is obviously important that studies with negative outcomes are also published, and that specific studies that seek to reproduce previous findings are commenced.
Schizophrenia spectrum disorders are complex and consist of a wide range of symptoms with heterogeneous disease progression across individuals. In practice, this poses challenges in clearly defining disease phenotypes and renders interpretation of potential results difficult. The view of schizotypy as a continuous range of symptoms and traits expressed by individuals, motivate the use of machine learning to predict multiple continuous measures of disease progression. Here, it is natural to consider multivariate regression models, such as support vector regression,91 to directly predict schizotypy traits. Also, to take advantage of the fact that multiple dimensions of schizotypy are often assessed using a variety of rating scales, methods such as partial least squares regression103 can be used to establish compact relations between multivariate neuroimaging data and multiple schizotypy measures.
The use of these tools and more generally applicable methods based on deep learning, represent promising research avenues, which can help us gain a more complete understanding of schizotypy, lead to improved identification of individuals with schizotypy and facilitate appropriate management and intervention for these individuals. Machine learning constitutes a paradigm shift toward quantitative evaluation, where we no longer need to rely on subjective rating and structured interviews. Consequently, the time spend on identification of subtypes of schizophrenia spectrum disorders can be reduced while potentially improving the accuracy in clinical practice.
In summary, classification of schizotypy represents a promising application for the combination of machine learning and neuroimaging, but there are still a range of challenges, in particular, related to how robustness to overfitting, and thereby better generalization performance can be archived. However, if these challenges are appropriately addressed, machine learning can significantly improve our understanding of schizotypy and schizophrenia spectrum disorders. Finally, the emerging field of computational psychiatry had important applications in disease prevention, early diagnosis, identification of drug targets, and individual treatment plans for psychiatric diseases and may revolutionize modern neurology.
Supplementary Material
Supplementary data are available at Schizophrenia Bulletin online.
Funding
R.C.K. was supported by the Beijing Municipal Science & Technology Commission (Z161100000216138), National Key Research and Development Programme (2016YFC0906402), the Beijing Training Project for Leading Talents in S&T (Z151100000315020), and the CAS Key Laboratory of Mental Health, Institute of Psychology.
References
- 1. Meehl PE. Schizotaxia, schizotypy, schizophrenia. Am Psychol. 1962;17:827–838. [Google Scholar]
- 2. Meehl PE. Toward an integrated theory of schizotaxia, schizotypy, and schizophrenia. J Pers Disord. 1990;4:1–99. [Google Scholar]
- 3. Debbané M, Barrantes-Vidal N. Schizotypy from a developmental perspective. Schizophr Bull. 2015;41:S386–S395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Everett KV, Linscott RJ. Dimensionality vs taxonicity of schizotypy: some new data and challenges ahead. Schizophr Bull. 2015;41(suppl 2):S465–S474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kwapil TR, Barrantes-Vidal N, Silvia PJ. The dimensional structure of the Wisconsin Schizotypy Scales: factor identification and construct validity. Schizophr Bull. 2008;34: 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan RCK, Li X, Lai M et al. Exploratory study on the base-rate of paranoid ideation in a non-clinical Chinese sample. Psychiatry Res. 2011;185:254–260. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Lui SSY, Zou LQ et al. Individuals with psychometric schizotypy show similar social but not physical anhedonia to patients with schizophrenia. Psychiatry Res. 2014;216:161–167. [DOI] [PubMed] [Google Scholar]
- 8. Claridge G, Beech T. Fully and quasi-dimensional constructions of schizotypy. In: Schizotypal Personality. New York, NY: Cambridge University Press; 1995:192–216. http://psycnet.apa.org/doi/10.1017/CBO9780511759031.010 [Google Scholar]
- 9. Wang Y, Chan RC, Xin Yu, Shi C, Cui J, Deng Y. Prospective memory deficits in subjects with schizophrenia spectrum disorders: a comparison study with schizophrenic subjects, psychometrically defined schizotypal subjects, and healthy controls. Schizophr Res. 2008;106:70–80. [DOI] [PubMed] [Google Scholar]
- 10. Ettinger U, Mohr C, Gooding DC et al. Cognition and brain function in schizotypy: a selective review. Schizophr Bull. 2015;41(suppl 2):S417–S426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lui SSY, Liu ACY, Chui WWH et al. The nature of anhedonia and avolition in patients with first-episode schizophrenia. Psychol Med. 2016;46:437–447. [DOI] [PubMed] [Google Scholar]
- 12. Chan RCK, Cui H, Chu M et al. Neurological soft signs precede the onset of schizophrenia: a study of individuals with schizotypy, ultra-high-risk individuals, and first-onset schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2017;268:49–56. [DOI] [PubMed] [Google Scholar]
- 13. Chan RCK, Song Shi H, Lei Geng F et al. The Chapman psychosis-proneness scales: consistency across culture and time. Psychiatry Res. 2015;228:143–149. [DOI] [PubMed] [Google Scholar]
- 14. Chan RCK, Li Z, Li K et al. Distinct processing of social and monetary rewards in late adolescents with trait anhedonia. Neuropsychology. 2016;30:274–280. [DOI] [PubMed] [Google Scholar]
- 15. Fonseca-Pedrero E, Debbané M, Ortuño-Sierra J et al. The structure of schizotypal personality traits: a cross-national study. Psychol Med. 2018;48:451–462. [DOI] [PubMed] [Google Scholar]
- 16. Murray RM, Lewis SW. Is schizophrenia a neurodevelopmental disorder?Br Med J (Clin Res Ed). 1987;295:681–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murray RM, Lewis SW. Is schizophrenia a neurodevelopmental disorder?Br Med J (Clin Res Ed). 1988;296:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. [DOI] [PubMed] [Google Scholar]
- 19. Cohen AS, Mohr C, Ettinger U, Chan RC, Park S. Schizotypy as an organizing framework for social and affective sciences. Schizophr Bull. 2015;41(suppl 2):S427–S435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gooding DC, Tallent KA, Matts CW. Clinical status of at-risk individuals 5 years later: further validation of the psychometric high-risk strategy. J Abnorm Psychol. 2005;114:170–175. [DOI] [PubMed] [Google Scholar]
- 21. Gooding DC, Tallent KA, Matts CW. Rates of avoidant, schizotypal, schizoid and paranoid personality disorders in psychometric high-risk groups at 5-year follow-up. Schizophr Res. 2007;94:373–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwapil TR. Social anhedonia as a predictor of the development of schizophrenia-spectrum disorders. J Abnorm Psychol. 1998;107:558–565. [DOI] [PubMed] [Google Scholar]
- 23. Kwapil TR, Gross GM, Silvia PJ, Barrantes-Vidal N. Prediction of psychopathology and functional impairment by positive and negative schizotypy in the Chapmans’ ten-year longitudinal study. J Abnorm Psychol. 2013;122:807–815. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Shi H, Liu W et al. Trajectories of schizotypy and their emotional and social functioning: an 18-month follow-up study. Schizophr Res. 2017. doi:10.1016/j.schres.2017.07.038. [DOI] [PubMed] [Google Scholar]
- 25. Ettinger U, Williams SC, Meisenzahl EM, Möller HJ, Kumari V, Koutsouleris N. Association between brain structure and psychometric schizotypy in healthy individuals. World J Biol Psychiatry. 2012;13:544–549. [DOI] [PubMed] [Google Scholar]
- 26. Wang Y, Yan C, Yin DZ et al. Neurobiological changes of schizotypy: evidence from both volume-based morphometric analysis and resting-state functional connectivity. Schizophr Bull. 2015;41:S444–S454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Modinos G, Mechelli A, Ormel J, Groenewold NA, Aleman A, McGuire PK. Schizotypy and brain structure: a voxel-based morphometry study. Psychol Med. 2010;40:1423–1431. [DOI] [PubMed] [Google Scholar]
- 28. Modenato C, Draganski B. The concept of schizotypy—a computational anatomy perspective. Schizophr Res Cogn. 2015;2:89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Modinos G, Ormel J, Aleman A. Altered activation and functional connectivity of neural systems supporting cognitive control of emotion in psychosis proneness. Schizophr Res. 2010;118:88–97. [DOI] [PubMed] [Google Scholar]
- 30. Wang Y, Liu W, Li Z et al. Dimensional schizotypy and social cognition: an fMRI imaging study. Front Behav Neurosci. 2015;9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanske P, Böckler A, Trautwein FM, Parianen Lesemann FH, Singer T. Are strong empathizers better mentalizers? Evidence for independence and interaction between the routes of social cognition. Soc Cogn Affect Neurosci. 2016;11:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Liu WHH, Li Z et al. Altered corticostriatal functional connectivity in individuals with high social anhedonia. Psychol Med. 2016;46:125–135. [DOI] [PubMed] [Google Scholar]
- 33. Wang Y, Ettinger U, Meindl T, Chan RCK. Association of schizotypy with striatocortical functional connectivity and its asymmetry in healthy adults. Hum Brain Mapp. 2018;39:288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lagioia A, Van De Ville D, Debbané M, Lazeyras F, Eliez S. Adolescent resting state networks and their associations with schizotypal trait expression. Front Syst Neurosci. 2010;4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ardekani BA, Tabesh A, Sevy S, Robinson DG, Bilder RM, Szeszko PR. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum Brain Mapp. 2011;32:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Janousova E, Montana G, Kasparek T, Schwarz D. Supervised, multivariate, whole-brain reduction did not help to achieve high classification performance in schizophrenia research. Front Neurosci. 2016;10:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaufmann T, Skåtun KC, Alnæs D et al. Disintegration of sensorimotor brain networks in schizophrenia. Schizophr Bull. 2015;41:1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arbabshirani MR, Kiehl KA, Pearlson GD, Calhoun VD. Classification of schizophrenia patients based on resting-state functional network connectivity. Front Neurosci. 2013;7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chyzhyk D, Savio A, Graña M. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Netw. 2015;68:23–33. [DOI] [PubMed] [Google Scholar]
- 40. Du W, Calhoun VD, Li H et al. High classification accuracy for schizophrenia with rest and task fMRI data. Front Hum Neurosci. 2012;6:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koutsouleris N, Meisenzahl EM, Davatzikos C et al. Use of neuroanatomical pattern classification to identify subjects in at-risk mental states of psychosis and predict disease transition. Arch Gen Psychiatry. 2009;66:700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koutsouleris N, Borgwardt S, Meisenzahl EM, Bottlender R, Möller HJ, Riecher-Rössler A. Disease prediction in the at-risk mental state for psychosis using neuroanatomical biomarkers: results from the FePsy study. Schizophr Bull. 2012;38:1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nejad AB, Madsen KH, Ebdrup BH et al. Neural markers of negative symptom outcomes in distributed working memory brain activity of antipsychotic-naive schizophrenia patients. Int J Neuropsychopharmacol. 2013;16:1195–1204. [DOI] [PubMed] [Google Scholar]
- 44. Koutsouleris N, Davatzikos C, Bottlender R et al. Early recognition and disease prediction in the at-risk mental states for psychosis using neurocognitive pattern classification. Schizophr Bull. 2012;38:1200–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zarogianni E, Storkey AJ, Johnstone EC, Owens DG, Lawrie SM. Improved individualized prediction of schizophrenia in subjects at familial high risk, based on neuroanatomical data, schizotypal and neurocognitive features. Schizophr Res. 2017;181:6–12. [DOI] [PubMed] [Google Scholar]
- 46. Shinkareva SV, Ombao HC, Sutton BP, Mohanty A, Miller GA. Classification of functional brain images with a spatio-temporal dissimilarity map. Neuroimage. 2006;33:63–71. [DOI] [PubMed] [Google Scholar]
- 47. Modinos G, Pettersson-Yeo W, Allen P, McGuire PK, Aleman A, Mechelli A. Multivariate pattern classification reveals differential brain activation during emotional processing in individuals with psychosis proneness. Neuroimage. 2012;59:3033–3041. [DOI] [PubMed] [Google Scholar]
- 48. Wiebels K, Waldie KE, Roberts RP, Park HR. Identifying grey matter changes in schizotypy using partial least squares correlation. Cortex. 2016;81:137–150. [DOI] [PubMed] [Google Scholar]
- 49. Modinos G, Mechelli A, Pettersson-Yeo W, Allen P, McGuire P, Aleman A. Pattern classification of brain activation during emotional processing in subclinical depression: psychosis proneness as potential confounding factor. PeerJ. 2013;1:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeong JW, Wendimagegn TW, Chang E et al. Classifying schizotypy using an audiovisual emotion perception test and scalp electroencephalography. Front Hum Neurosci. 2017;11:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. [DOI] [PubMed] [Google Scholar]
- 52. Smith SM, Miller KL, Salimi-Khorshidi G et al. Network modelling methods for FMRI. Neuroimage. 2011;54:875–891. [DOI] [PubMed] [Google Scholar]
- 53. Craddock RC, James GA, Holtzheimer PE III, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33:1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Madsen KH, Churchill NW, Mørup M. Quantifying functional connectivity in multi-subject fMRI data using component models. Hum Brain Mapp. 2017;38:882–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith SM, Vidaurre D, Beckmann CF et al. Functional connectomics from resting-state fMRI. Trends Cogn Sci. 2013;17:666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Strother SC, Anderson J, Hansen LK et al. The quantitative evaluation of functional neuroimaging experiments: the NPAIRS data analysis framework. Neuroimage. 2002;15:747–771. [DOI] [PubMed] [Google Scholar]
- 58. Guyon I. Feature Extraction : Foundations and Applications. Berlin, Heidelberg: Springer-Verlag; 2006. [Google Scholar]
- 59. Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 60. Ho TK. Random decision forests. Third International Conference on Document Analysis and Recognition, {ICDAR} 1995, Montreal, Canada, 14–15 August 1995 Vol I 1995:278–282. [Google Scholar]
- 61. Pedregosa F, Varoquaux G, Gramfort A et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 62. Abraham A, Pedregosa F, Eickenberg M et al. Machine learning for neuroimaging with scikit-learn. Front Neuroinform. 2014;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schrouff J, Rosa MJ, Rondina JM et al. PRoNTo: pattern recognition for neuroimaging toolbox. Neuroinformatics. 2013;11:319–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hanke M, Halchenko YO, Sederberg PB, Hanson SJ, Haxby JV, Pollmann S. PyMVPA: a python toolbox for multivariate pattern analysis of fMRI data. Neuroinformatics. 2009;7:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zou QH, Zhu CZ, Yang Y et al. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods. 2008;172:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zang YF, He Y, Zhu CZ et al. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29:83–91. [DOI] [PubMed] [Google Scholar]
- 67. Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage. 2004;22:394–400. [DOI] [PubMed] [Google Scholar]
- 68. Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- 69. PennyTE. Statistical Parametric Mapping: The Analysis of Functional Brain Images. London, United Kingdom: Elsevier; 2007. [Google Scholar]
- 70. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 71. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. [DOI] [PubMed] [Google Scholar]
- 72. Tzourio-Mazoyer N, Landeau B, Papathanassiou D et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. [DOI] [PubMed] [Google Scholar]
- 73. Desikan RS, Ségonne F, Fischl B et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. [DOI] [PubMed] [Google Scholar]
- 74. Goutte C, Toft P, Rostrup E, Nielsen F, Hansen LK. On clustering fMRI time series. Neuroimage. 1999;9:298–310. [DOI] [PubMed] [Google Scholar]
- 75. Churchill NW, Madsen K, Mørup M. The functional segregation and integration model: mixture model representations of consistent and variable group-level connectivity in fMRI. Neural Comput. 2016;28:2250–2290. [DOI] [PubMed] [Google Scholar]
- 76. Newman ME, Girvan M. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69:026113. [DOI] [PubMed] [Google Scholar]
- 77. Nowicki K, Snijders TAB. Estimation and prediction for stochastic blockstructures. J Am Stat Assoc. 2001;96:1077–1087. [Google Scholar]
- 78. Mørup M, Madsen KH, Dogonowski AM, Siebner H, Hansen LK. Infinite relational modeling of functional connectivity in resting state fMRI. In: Lafferty JD, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, eds. Advances in Neural Information Processing Systems, Vol 23; 6–11 December 2010; Vancouver, Canada Curran Associates, Inc., Red Hook, NY; 2010:1750–1758. http://papers.nips.cc/paper/4057-infinite-relational-modeling-of-functional-connectivity-in-resting-state-fmri.pdf [Google Scholar]
- 79. Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. [DOI] [PubMed] [Google Scholar]
- 80. Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front Syst Neurosci. 2010;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. [DOI] [PubMed] [Google Scholar]
- 83. Cortes C, Vapnik V. Support-vector networks. Kluwe Acad Publ. 1995;297:273–297. [Google Scholar]
- 84. Vapnik V, Lerner A. Pattern recognition using generalized portrait method. Autom Remote Control. 1963;24:774–780. [Google Scholar]
- 85. Chang CC, Lin CJ. Libsvm a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 86. Schmah T, Yourganov G, Zemel RS, Hinton GE, Small SL, Strother SC. Comparing classification methods for longitudinal fMRI studies. Neural Comput. 2010;22:2729–2762. [DOI] [PubMed] [Google Scholar]
- 87. Bishop CM. Pattern Recognition and Machine Learning (Information Science and Statistics). Secaucus, NJ: Springer-Verlag New York, Inc; 2006. [Google Scholar]
- 88. Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. [DOI] [PubMed] [Google Scholar]
- 89. Rasmussen PM, Madsen KH, Lund TE, Hansen LK. Visualization of nonlinear kernel models in neuroimaging by sensitivity maps. Neuroimage. 2011;55:1120–1131. [DOI] [PubMed] [Google Scholar]
- 90. Haufe S, Meinecke F, Görgen K et al. On the interpretation of weight vectors of linear models in multivariate neuroimaging. Neuroimage. 2014;87:96–110. [DOI] [PubMed] [Google Scholar]
- 91. Drucker H, Burges CJC, Kaufman L, Smola AJ, Vapnik V. Support vector regression machines. In: Mozer MC, Jordan MI, Petsche T, eds. Advances in Neural Information Processing Systems 9, NIPS; December 2–5, 1996; Denver, CO Cambridge, MA: MIT Press; 1997:155–161. [Google Scholar]
- 92. Plis SM, Hjelm DR, Salakhutdinov R et al. Deep learning for neuroimaging: a validation study. Front Neurosci. 2014;8:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jang H, Plis SM, Calhoun VD, Lee JH. Task-specific feature extraction and classification of fMRI volumes using a deep neural network initialized with a deep belief network: evaluation using sensorimotor tasks. Neuroimage. 2017;145:314–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kim J, Calhoun VD, Shim E, Lee JH. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage. 2016;124:127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Guo X, Dominick KC, Minai AA, Li H, Erickson CA, Lu LJ. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front Neurosci. 2017;11:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vieira S, Pinaya WH, Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev. 2017;74:58–75. [DOI] [PubMed] [Google Scholar]
- 97. Wang B, Klabjan D. Regularization for unsupervised deep neural nets. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17); February 4–9, 2016; San Francisco, CA Ithaca, NY: Cornell University Library; 2017:1–6. http://arxiv.org/abs/1608.04426 [Google Scholar]
- 98. Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, eds. Advances in Neural Information Processing Systems 27 (NIPS ‘14), Vol 27; December 8–13, 2014; Montréal, Canada Curran Associates, Inc.; 2014:3320–3328. http://papers.nips.cc/paper/5347-how-transferable-are-features-in-deep-neural-networks.pdf [Google Scholar]
- 99. Xu Y, Jia R, Mou L et al. Improved relation classification by deep recurrent neural networks with data augmentation. Proceedings of COLING 2016, the 26th International Conference on Computational Linguistics; December 13–16, 2016; Osaka, Japan Ithaca, NY: Cornell University Library; 2016:1461–1470. http://arxiv.org/abs/1601.03651 [Google Scholar]
- 100. Smith SM, Beckmann CF, Andersson J et al. ; WU-Minn HCP Consortium. Resting-state fMRI in the human connectome project. Neuroimage. 2013;80:144–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Biswal BB, Mennes M, Zuo XN et al. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Churchill NW, Oder A, Abdi H et al. Optimizing preprocessing and analysis pipelines for single-subject fMRI. I. Standard temporal motion and physiological noise correction methods. Hum Brain Mapp. 2012;33:609–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. Neuroimage. 1996;3:143–157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.