Abstract
The eMERGE Network is establishing methods for electronic transmittal of patient genetic test results from laboratories to healthcare providers across organizational boundaries. We surveyed the capabilities and needs of different network participants, established a common transfer format, and implemented transfer mechanisms based on this format. The interfaces we created are examples of the connectivity that must be instantiated before electronic genetic and genomic clinical decision support can be effectively built at the point of care. This work serves as a case example for both standards bodies and other organizations working to build the infrastructure required to provide better electronic clinical decision support for clinicians.
BACKGROUND
Genetic and genomic sequencing (GS) has the potential to improve medical decision making and outcomes for patients over the course of their lifetimes. However, GS test results are often difficult to track, interpret, and apply, especially as genomic knowledge evolves over time.1,2 Computer-based clinical decision support (CDS) could help address these issues.3–5 In order for CDS to be effective, GS data must be accessible from the electronic health record (EHR). The challenge of making GS data accessible from the EHR was recognized in Phase II of the Electronic Medical Records and Genomics (eMERGE) Network as dependent on the format and complexity of the data.6 Furthermore, most institutions lack the health information technology (HIT) infrastructure to receive and store even basic forms of these data in a manner that would enable timely triggering of CDS rules integrated into routine clinical care. Recognizing these challenges, in 2014, the National Human Genome Research Institute (NHGRI) issued a request for application (RFA) for Phase III of the eMERGE Network.7
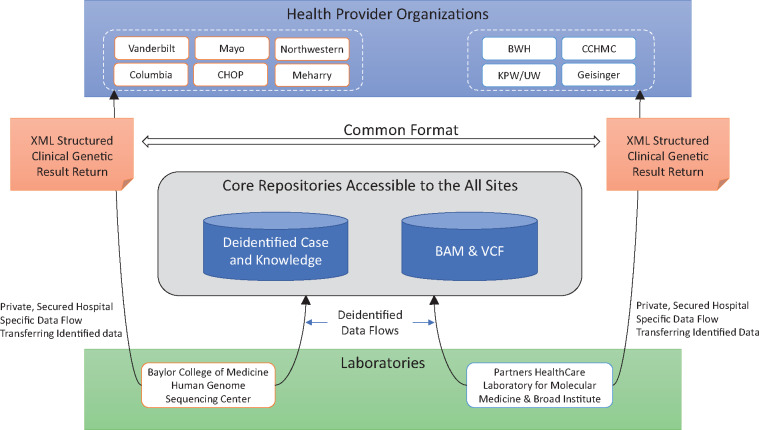
eMERGE Phase III includes 10 health provider organizations (HPO) dispersed throughout the United States. These HPOs are implementing genomic medicine pilot studies centered around the return of results from targeted germline sequencing of 109 genes and approximately 1600 single nucleotide variants (SNVs), from which data derived from up to 68 “actionable” genes and 14 SNVs will be returned to individual participants. The HPOs each order GS tests from 1 of 2 centralized GS testing laboratories, the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) or Partners HealthCare Laboratory for Molecular Medicine (LMM), partnered with the Broad Institute.7 The HPOs have objectives that can be achieved only through the receipt of structured, machine readable GS data from the central laboratories. In addition, research conducted on eMERGE datasets requires secure, centralized access to de-identified test results generated across sites. The network has reached consensus on a network architecture supported by the central laboratories to achieve these objectives (Figure 1).
Figure 1.
eMERGE III Architecture Supporting the Transfer of Genetic and Genomic Results from Central Laboratories to Health Provider Organizations and Core Repositories.
Implementation of a robust architecture for the transfer of results from central laboratories to HPOs is a critical first step to developing broad CDS using GS data that are timely and integrated into routine clinical practice. We aim to provide: (a) a description of our architecture to transfer GS results, and (b) a discussion about the potential use of those GS results.
ARCHITECTURE TO TRANSFER RESULTS FROM LABORATORIES TO EHR ECOSYSTEMS AND SHARED REPOSITORIES
External genetic testing laboratories usually deliver their reports to clinicians via fax or PDF, which are then scanned into their EHRs.8 Genetic information can be highly sensitive because it can identify susceptibility to disease that is not yet manifest, its inherited nature can create privacy issues for families as a whole, and it can be inherently identifying. Further, genetic information can also play a critically important role in care decisions. For these reasons, it is important that clinical IT infrastructure created to facilitate its movement be both secure and robust. Furthermore, it is difficult to programmatically extract structured, machine-readable information from these reports to reliably enable computational benefits. In PDFs, important GS test information is locked away, unavailable for structured display and algorithmic use by the rest of the EHR ecosystem. This issue can be addressed by linking laboratory and healthcare provider computer systems through interfaces that transfer structured data between organizations. However, these types of interfaces are resource intensive to build and maintain.9 eMERGE III aims to help address these challenges by instantiating secure inter-institutional interfaces.
A key challenge to developing a robust architecture for transferring results is accounting for the diversity of processes and technologies used by eMERGE III institutions. For example, the IT infrastructures of the two sequencing laboratories are based on solutions from different vendors. The HPOs also use different IT systems to implement heterogenous processes.
CLINICAL DATA STREAMS TO EHR ECOSYSTEMS
eMERGE III genetic reports are patient specific, contain sensitive data, and are intended for clinical use. Thus, to ensure privacy, they must be transferred only from the laboratory to the ordering HPO and cannot be shared with other HPOs.
We interviewed the HPOs to determine their preferences and capabilities to receive structured GS results. All HPOs required structured data access; no HPOs required real-time data transfers (all could accept 24-hour delayed delivery); all HPOs could download data from sFTP sites; not all HPOs were prepared to accept results through a representational state transfer (REST)-based Fast HealthCare Interoperability Resources (FHIR10) interface. Based on these capabilities, we chose to transfer results from laboratories to HPOs using sFTP (LMM) and an HTTPS-based Simple Storage Service (S3) file transfer provided by DNAnexus11 (BCM-HGSC). We found that coordinating site-to-site access involving multiple security groups took significant time and resources. Different sites have different policies for reviewing infrastructure used to transmit clinical genetic data. We also secured our sFTP sites in a manner that required coordination between the LMM networking team, the LMM infrastructure team, site users, and site network engineers.
Once the method for file transfer was established, we specified a format for the exchange of reported results and associated variant interpretations. We chose to use the GeneInsight format for communication between system components as a starting point for our eMERGE format. The Health Level 7 (HL7) version 2 Genetic Test Results standard Release 2 Implementation Guide was voted as an informative standard in 2013, which extended Release 1, which, in turn, was informed by an earlier version of this message format. This work helped inform the currently active v2 Laboratory Results Interface (LRI) standard, which is presently being targeted as a normative standard for January 2019.
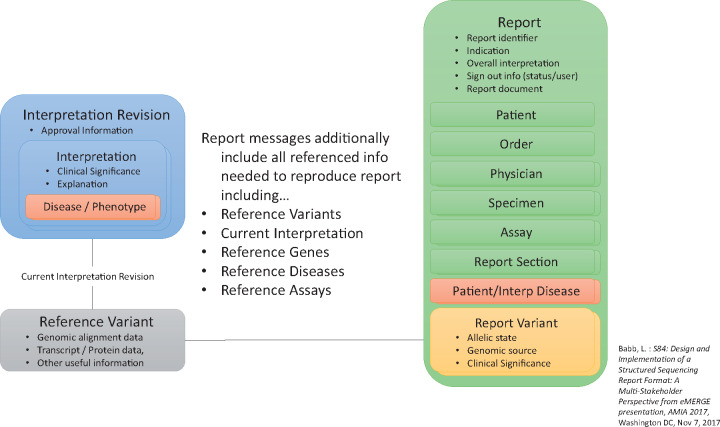
GeneInsight’s parent company, Sunquest®, provided the eMERGE network permission to revise, use, and publicly release this new eMERGE format (schema and examples available at: https://github.com/emerge-ehri/results-schema). As of February 2018, more than 6892 report transfers had occurred using the format, thereby giving us confidence that it is robust. At a high level, the format separates report-specific result concepts from the referenceable concepts on which they depend. This separation of reporting and knowledge concepts is a fundamental principle behind the design of the format. The format is segmented into high-level concepts necessary to reconstitute the results effectively in external systems (Table 1). The Report concept contains the Patient, Order, Physician, Specimen, Reported Variant, and Reported Assay concepts, thus representing the case-specific data. The Reference Variant and Assay (genes and diseases - not shown) concepts are definitional concepts that define the basis for the corresponding reported concepts, but are not fulfilled by any existing standard (Figure 2). The coding systems the eMERGE group chose to use to populate this structure are available here: https://github.com/emerge-ehri/results-schema/blob/master/doc/eMERGE-XML-Schema-Coded-Concepts.pdf. Once agreed upon by the laboratories and sites, each laboratory developed the ability to produce results in this format.
Table 1.
Concepts included in the GeneInsight XML schema for structured genetic and genomic test reports adopted by eMERGE III
| High-Level Concept | Data Fields Contained |
|---|---|
| Report | Case type, edited by, genomic source, indication, patient diseases, interpreted diseases, lab status, overall interp, report document, report identifier, status, type of test |
| Patient | Date of birth, date of death, de-identification flag, affected status, first/middle/last name, patient#/identifier, sex, race-or-ethnicity |
| Order | lab-order#, order date, sender encounter#, sender facility, sender lab control#, sender-order#, sender-patient# |
| Physician | addresses/contact info, first/middle/last/suffix name, Unique Physician Identification Number (UPIN)/National Provider Identifer (NPI) identifiers, primary/referring physician flag |
| Specimen | Label, type, received date, collected date, description, comment, anatomic location, tumor type % tumor, metastatic flag, differentiation (Some fields are included to support somatic variant reporting but eMERGE reports only include germline variation.) |
| Reported Variant (Extends from Reference Variant) | External identifier, allele name, DNA change inHuman Genome Variant Society Nomenclature (HGVS), amino acid change (HGVS), DNA change type, amino acid change type, allele state/zygosity, clinical significance/category, chromosome, compound type, forced incidental flag, gene symbol/identifiers, gene region, genome build name, genomic source, not interpreted flag, nested variants, structural change type, transcript ID |
| Reported Assay (Extends from Reference Assay) | External assay ID/version, test code, test name |
| Reference Variant | Alignments (chromosome, genome build, start/end position, reference/alternate sequence), current lab approved interp revision (approved by/on), interpretations (category, disease, inheritance pattern, text), # of families/times reported by lab, literature references, splicing impact |
| Reference Assay | Background, date introduced, external identifier, methodology, type, test code/name, version# |
Figure 2.
Structured Reports and Variant Interpretations XML Schema.
ESTABLISHING CENTRAL SHARED REPOSITORIES
In addition to establishing result flow to the HPO EHR ecosystems, eMERGE III also needs to support clinical knowledge sharing and research processes across sites. To do so, shared, commonly accessible repositories were established based on the systems already in use within the eMERGE laboratories. One solution for this was enabled by BCM-HGSC, LMM, DNAnexus, and GeneInsight working together to implement and validate interfaces to establish a centralized de-identified case repository (DCR). This repository enables users to browse network-wide de-identified cases and variant knowledge. The DCR is an instance of the GeneInsight Clinical Lab application that allows non-technical users to execute searches, and to generate lists and reports. Users can search for cases based on variants, variant characteristics, genes, chromosomes, reported patient and/or interpreted conditions, eMERGE identifiers, overall interpretations, gender, race or ethnicity, and a variety of other factors. Unlike the direct clinical data transfers, protected health information is stripped from these case records to protect patient privacy. Variant knowledge is also searchable through the DCR. The process of integrating the results from both the BCM-HGSC and LMM into a common repository helped us to improve consistency in formats and coding systems across the laboratories. As of this writing, there are 3595 cases in this repository (See Table 2). This number will expand to include all clinical reports generated through eMERGE III. A centralized repository for raw, de-identified genomic data (ie, variant calling files [VCF] and binary alignment/map [BAM] files) was also established through a data commons infrastructure based in DNANexus. The data commons provides similar underlying functionality to the DCR, but without established query structures and the graphical user interface tailored to the DCR users. The data commons will ultimately grow to include ∼25 000 eMERGE III participants across the 10 HPOs. Clinical reports will not be generated on all negative test results. As a result, the number of cases in the DCR is expected to grow to approximately 16 000 over the course of the project.
Table 2.
Frequency of cases and reports among eMERGE III participating institutions
| Site | Lab | Cases in Commons as of 2/22/2018 | Clinical Reports Electronically Delivered as of 2/22/2018 and Placed in the DCR | Total Cases to Be Sequenced and Placed in Commons | Projected Clinical Reports to Be Electronically Delivered and Placed in the DCR |
|---|---|---|---|---|---|
| Brigham and Women’s Hospital | LMM | 2482 | 36 | 2500 | 91 |
| Children’s Hospital of Philadelphia | BCM-HGSC | 2323 | 267 | 3000 | 3000 |
| Cincinnati Children’s Hospital Medical Center | LMM | 2826 | 56 | 3000 | 160 |
| Columbia University | BCM-HGSC | 1498 | 7 | 2646 | 1500 |
| Geisinger | LMM | 1251 | 184 | 2500 | 414 |
| Kaiser Permanente Washington Health Research Institute | LMM | 1221 | 1221 | 2500 | 1816 |
| Mayo Clinic | BCM-HGSC | 2723 | 655 | 3000 | 3000 |
| Meharry Medical College | BCM-HGSC | 0 | 0 | 500 | 500 |
| Northwestern University | BCM-HGSC | 1785 | 1160 | 3000 | 3000 |
| Vanderbilt University | BCM-HGSC | 2034 | 9 | 2500 | 2500 |
| Total | 18 143 | 3595 | 25 146 | 15 981 |
DISCUSSION
Previous eMERGE efforts examined possible data paths for GS use.6 Distinct from that work, we illustrate an eMERGE Phase III multi-institutional case study for the delivery of consistent, machine-readable GS test results from multiple testing laboratories to multiple HPOs, which rarely occurs in today’s healthcare environment. Transfering results in structured form is a necessary first step to enable robust EHR displays of genetic results12,13 through SMART on FHIR14 and other methods, tethering of the reports to patient portals,13 and use of “computable observations” to trigger CDS rules through mechanisms such as CDS Hooks.15 For example, in the case of a pathogenic variant in a gene associated with Lynch syndrome, initial recommendations can be based on age and previous Lynch syndrome diagnosis. One eMERGE site is working to implement CDS to highlight that prior to diagnosis, patients may need referral for evaluation by a specialized clinic. After the initial diagnosis, then CDS could alert providers if a patient has not received the recommended interventions, for example, colonoscopy at regular intervules, as specified by the Displaying and Integrating Genetic Information into the Electronic Health Record (DIGITizE) Lynch Syndrome Implementation CDS Guilde developed with the Clinical Sequencing Evidence Generating Research (CSER) and eMERGE Consortia.16
One of the key decisions faced in Phase III was determining how best to integrate with the evolving FHIR Genetics Resource Standard. Members of our eMERGE EHR Integration working group are involved in the development of this resource. At the time we made decisions related to our interfaces, the standard was not yet finalized. We chose to use an approach based on a “battle tested” format to enable network connectivity as quickly as possible. In turn, we aimed to provide invaluable feedback to the broader FHIR Genomics Resource Standard effort, which is still evolving. Many aspects of the eMERGE XML format are easily transferable to the HL7 v2 and FHIR formats. However, there are key differences in that our XML format treats variant, gene, assay, genetic disease concepts, and variant interpretation knowledge as separate objects that are referenced from the report object. This allows these objects to be versioned independent of a report. We look forward to working with the FHIR community to evaluate these concepts for incorporation into the FHIR standard.
As with all data-sharing efforts, defining the structure is only part of the challenge. Negotiating and defining the semantic requirements for content that should be placed in fields contained in the data structure is the other half. To increase consistency and avoid managing a separate set of rules, transformations, and translations for each laboratory, a set of core components and fields within the formatted specification was identified and constrained. Ideally, all fields would be standardized and coded precisely and consistently across laboratories. While we have made significant progress towards these objectives, we had to provide laboratories with the ability to make site-specific enhancements and modifications to the format and structure similar to the way FHIR supports the creation of profiles. We anticipate this will remain the case for some time as the field evolves. Iteratively standardizing the content of these types of messages in the face of evolving testing processes will be an important ongoing effort within the fields of clinical genomics and HIT.
The eMERGE III use cases involve conducting broad GS tests largely for screening purposes. The tests are queued and take weeks to months to conduct. Therefore, allowing 24 hours for result transmission does not materially increase test turnaround time. Because all sites could use secure file-transfer-based mechanisms and not all could use REST-based methods, we chose accessibility over speed in this circumstance. However, no site that had the ability to use a REST-based interface preferred a file-transfer-based one. All of these sites either preferred REST or were neutral. There are circumstances in which a REST-based near-real-time interface would have been preferable—for example, in a circumstance in which a clinician orders a pharmacogenomic test to urgently assess the risks of using a particular drug or particular drug dose. As more sites develop the capability to use REST-based interfaces, we believe they will often become a preferred choice because of their faster speed and more robust error handling.
Future efforts will explore whether the transmitted data files described here are sufficient to support CDS implementation requirements and the steps needed to simplify CDS implementations. We will identify any additional issues related to file transmission, coding systems, or the clinical processes themselves that need to be adjusted to achieve the formats for usable GS “computable observations.”
CONCLUSIONS
This initiative serves as a real-world, clinical, multi-institutional example of the process required to establish and validate infrastructure to transfer structured GS reports across a network of HPOs. Because most forms of CDS depend on structured data, the types of network connections we have built will have to be established in other clinical settings before genetic and genomic aware CDS can be provided. The need for this type of network “plumbing” is often an under-appreciated, hidden constraint on the HIT support that can be established in the clinic. Now that we have established these connections, the HPOs in our network can begin to develop genetic aware displays in the EHR ecosystems as well as create new forms of CDS. Furthermore, we believe this working data transmission network facilitating clinical and research process flows across diverse institutions will be invaluable to standards communities. It can serve as an environment for understanding the critical components of these messages and how related standards can evolve and pragmatically drive towards adoption. Our ultimate goal is to lower the barriers to establishing these required network connections in other settings, thereby contributing to the accessibility and distribution of HIT support for precision medicine.
FUNDING
The eMERGE Network was initiated and funded by the NHGRI through the following grants: U01HG8657 (Kaiser Permanente Washington Health Research Institute/University of Washington); U01HG8685 (Brigham and Women’s Hospital); U01HG8672 (Vanderbilt University Medical Center); U01HG8666 (Cincinnati Children’s Hospital Medical Center); U01HG6379 (Mayo Clinic); U01HG8679 (Geisinger Clinic); U01HG8680 (Columbia University Health Sciences); U01HG8684 (Children’s Hospital of Philadelphia); U01HG8673 (Northwestern University); U01HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG8676 (Partners Healthcare/Broad Institute); and U01HG8664 (Baylor College of Medicine). National Library of Medicine training grants T15LM007442 and 5T15LM007450 also supported this work as did grant R138709 from the National Heart, Lung, and Blood Institute.
Conflict of interest
Samuel Aronson and Shawn Murphy: Partners HealthCare receives royalties from sales of GeneInsight software. Lawrence Babb: Employed by GeneInsight, a Sunquest Information Systems company. Sunquest is a commercial laboratory software vendor. Richard Gibbs: Baylor College of Medicine receives payments from Baylor Genetics Laboratories, which provides services for genetic testing. Eric Venner: Cofounder of Codified Genomics, which provides variant interpretation services.
CONTRIBUTORS
Samuel Aronson: Contributed to conceptualization of interface strategy and assessment of fitness for use in both laboratory and provider settings; drafted significant portions of manuscript; will approve final version and be accountable for contents.
Lawrence Babb: Contributed to the description of the implementation strategy and file format used and the assessment of how it would be modified to work for the project; drafted significant portions of the sections surrounding the content of the XML file and statistics related to the historical use of the GeneInsight tool; will approve final version and be accountable for contents.
Darren Ames: Contributed to adaptation of XML schema to a second, distinct pipeline and identified modifications that needed to be made to the schema to support that pipeline; reviewed and will approve final version of the manuscript and be accountable for contents.
Richard A. Gibbs: Contributed to the overall concept of the manuscript and the implementation strategy, as well as drafting of the manuscript; reviewed and will approve final version of the manuscript and be accountable for content.
Eric Venner: Adapted the XML schema to a second, distinct pipeline and identified modifications that needed to be made to the schema to support that pipeline; reviewed the manuscript and will approve and agree to accountability for the final product.
John J. Connelly: Contributed to the overall design; participated in draft revisions; agrees to review and will approve final version and accept accountability for all aspects of the manuscript.
Keith Marsolo: Contributed to the overall dataset and design; participated in drafting/revision; will approve final version and agree to accountability.
Chunhua Weng: Contributed to the conceptualization and discussion of the design of the data flow; actively participated in the preparation of the draft and revisions of the manuscript; agrees to review and approve the final version and accept accountability for all aspects of the manuscript.
Marc S. Williams: Contributed to the conception of the project and participated in the analysis and interpretation of the data; actively participated in the preparation of the draft and revisions of the manuscript; agrees to review and approve the final version and accept accountability for all aspects of the manuscript.
Andrea L. Hartzler: Contributed to the overall dataset and design; participated in drafting and revision; will approve the final version of the work and agree to accountability.
Wayne H. Liang: Contributed to interpretation of data; participated in drafting/revising; agrees to final approval; agrees to accountability.
Ralston, James: Contributed to the interpretation of data; participated in drafting and revision; will approve final version of the work and agree to accountability.
Beth Devine: Contributed to interpretation of data; participated in drafting and revising; will provide final approval and agree to accountability.
Shawn Murphy: Contributed to the overall dataset and design; participated in drafting and revision; will approve final version of the work and agree to accountability.
Christopher Chute: Contributed to the overall dataset and design; participated in drafting and revision; will approve final version of the work and agree to accountability.
Pedro J. Caraballo: Contributed to the overall design of the project including conceptualization of data dependencies for execution of clinical decision support in the EHR and analysis of the results by using the genetic data in the design of CDS in one institution; contributed to the design, editing, and final approval of the manuscript submitted for publication; will approve final version of the work; agrees to be accountable for all aspects of this work.
Iftikhar J. Kullo: Contributed to the overall design of the work; revised the work critically for important intellectual content; will provide final approval; agrees to be accountable for all aspects of the work.
Robert Freimuth: Contributed to the overall concept of the manuscript and the implementation strategy; participated in drafting and revision of the manuscript; reviewed and will approve final version of the manuscript, and will be accountable for content.
Luke Rasmussen: Contributed to the overall dataset and design; participated in drafting and revision; will approve final version of the work and agree to accountability.
Firas Wehbe: Contributed local concept around data flow; contributed to the drafting and revision of the manuscript; will approve final version and agree to be accountable for contents.
Jamie R. Robinson: Contributed to interpretation of data; participated in drafting/revising; agrees to final approval; agrees to accountability.
Josh Peterson: Contributed to the overall design of the project; revised the manuscript for important intellectual content, and will gave final approval and accept accountability for the manuscript.
Ken Wiley: Contributed to conceptualization of interface strategy and assessment of fitness for use in both laboratory and provider settings; will review and approve the final version of the manuscript and accept accountability for the manuscript.
Casey Overby: Contributed to conceptualization and discussion of the design of the data flow; drafted and revised the paper for critically important intellectual content; will review and approve the final version of the manuscript and accept accountability for the manuscript.
Acknowledgments
Brianne Derveloy and Kayla Howell were highly instrumental to the coordination of this work.
REFERENCES
- 1. Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform 2012; 453: 419–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM.. Leveraging the electronic health record to implement genomic medicine. Genet Med 2013; 154: 270–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM.. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin Pharmacol Ther 2017; 1022: 254–64. [DOI] [PubMed] [Google Scholar]
- 4. Tarczy-Hornoch P, Amendola L, Aronson SJ, et al. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic health record. Genet Med 2013; 1510: 824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Welch BM, Eilbeck K, Del Fiol G, Meyer LJ, Kawamoto K.. Technical desiderata for the integration of genomic data with clinical decision support. J Biomed Inform 2014; 51: 3–7. [DOI] [PubMed] [Google Scholar]
- 6. Starren J, Williams MS, Bottinger EP.. Crossing the omic chasm: a time for omic ancillary systems. JAMA 2013; 30912: 1237–8. [published Online First: Epub Date]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Electronic Medical Records and Genomics (eMERGE) Network, Phase III—Study Investigators (U01), RFA-HG-14-025. Secondary The Electronic Medical Records and Genomics (eMERGE) Network, Phase III—Study. https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-14-025.html. Accessed May 22, 2018.
- 8. Shirts BH, Salama JS, Aronson SJ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc 2015; 226: 1231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrides AK, Tanasijevic MJ, Goonan EM, et al. Top ten challenges when interfacing a laboratory information system to an electronic health record: experience at a large academic medical center. Int J Med Inform 2017; 106: 9–16. [DOI] [PubMed] [Google Scholar]
- 10. Health Level 7. FHIR Specification Home Page. Secondary Health Level 7. FHIR Overview. https://www.hl7.org/fhir/overview.html. Accessed May 22, 2018.
- 11. Reid JG, Carroll A, Veeraraghavan N, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics 2014; 151: 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat 2011; 325: 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stuckey H, Williams JL, Fan AL, et al. Enhancing genomic laboratory reports from the patients’ view: a qualitative analysis. Am J Med Genet A 2015; 16710: 2238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mandel JC, Kreda DA, Mandl KD, et al. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016; 235: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDS Hooks—Home. http://cds-hooks.org/. Accessed January 22, 2018.
- 16.DIGITizE Lynch Syndrome CDS Guide. http://www.nationalacademies.org/hmd/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/DIGITizE.aspx Accessed Feburary 21, 2018.