Abstract
Multiple sequence alignment is a prerequisite for many evolutionary analyses. Multiple Alignment of Coding Sequences (MACSE) is a multiple sequence alignment program that explicitly accounts for the underlying codon structure of protein-coding nucleotide sequences. Its unique characteristic allows building reliable codon alignments even in the presence of frameshifts. This facilitates downstream analyses such as selection pressure estimation based on the ratio of nonsynonymous to synonymous substitutions. Here, we present MACSE v2, a major update with an improved version of the initial algorithm enriched with a complete toolkit to handle multiple alignments of protein-coding sequences. A graphical interface now provides user-friendly access to the different subprograms.
Keywords: multiple sequence alignment, molecular evolution, phylogenomics, pseudogenes, metabarcoding
Brief Communication
Multiple Alignment of Coding Sequences (MACSE) was the first automatic solution developed to align multiple protein-coding nucleotide sequences based on their amino acid translation while allowing for the occurrence of frameshifts (Ranwez et al. 2011). Its key feature is to align DNA sequences at the nucleotide level, but with the possibility to include gap lengths that are not a multiple of three bases, that is, generating frameshifts, while scoring the resulting nucleotide alignments based on their amino acid translation. This allows one to produce nucleotide alignments that preserve the underlying codon structure while benefiting from the higher similarity of amino acid sequences. Since its first release in 2011, MACSE has been used in multiple contexts including comparative transcriptomic studies (Lan and Pritchard 2016), pseudogene evolution (Delsuc et al. 2015), genome-wide analyses of selection (Assis et al. 2012), metabarcoding analyses (Leray et al. 2013), and phylogenomic pipelines (Bragg et al. 2016).
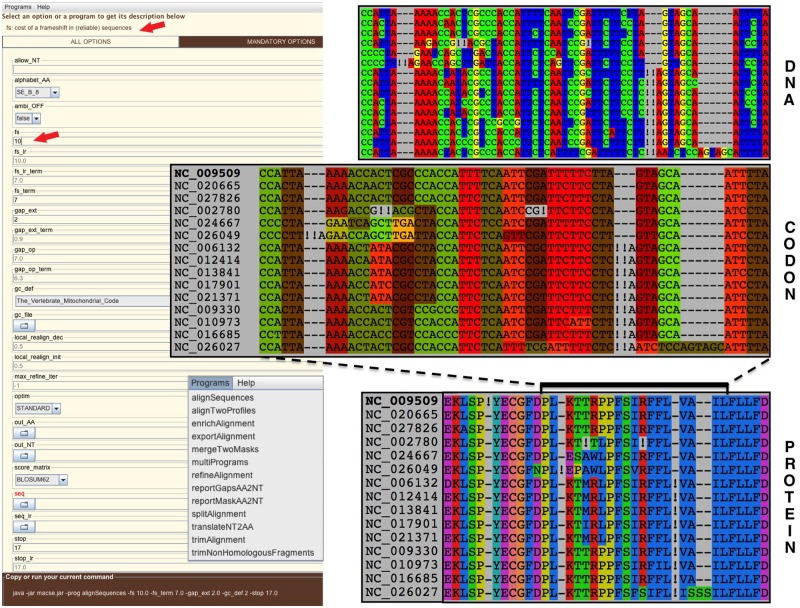
Here we present a major update of MACSE with an improved version enriched by a series of subprograms aimed at facilitating the production and handling of multiple alignments of protein-coding sequences. Altogether, the subprograms implemented in the new MACSE v2 release compose a powerful toolkit now easily accessible through a graphical user interface (fig. 1).
Fig. 1.
The graphical user interface of MACSE v2 (left) allows to select the desired subprogram, to browse the file system for choosing input FASTA files, and to set parameter values. It automatically generates the corresponding command line (bottom left). When the user selects a new subprogram or click on an option field, a brief help related to this program or option is displayed on the top of the interface (red arrows). An exemplar data set of 15 mitochondrial NADH dehydrogenase subunit 3 (nad3) gene sequences of turtles has been aligned by MACSE (parameters shown). The resulting alignment is displayed at the nucleotide (top right), codon (middle), and amino acid (bottom right) levels using SeaView v4.6.4 (Gouy et al.2010). Exclamation marks (!) emphasize the frameshifts detected by MACSE, most of which corresponding to programmed frameshift mutations (Russell and Beckenbach 2008).
The core alignment subprogram (alignSequences) has been improved in performance through a faster estimation of its objective function, namely the SP-score, thanks to recently derived optimal algorithmic solutions (Ranwez 2016). Additional parameters have also been introduced to control the speed/extensiveness ratio of the heuristic search for an alignment optimizing the SP-score. MACSE v2 uses a progressive alignment strategy to obtain an initial draft of the multiple sequence alignment that is subsequently improved using the 2-cut refinement strategy. This widespread strategy, also used for instance by MUSCLE (Edgar 2004), consists of partitioning the current solution into two subalignments that are subsequently realigned. The resulting alignment replaces the previous one if its SP-score is improved and the refinement process stops when no more improvements are found (see Ranwez et al. 2011; Ranwez 2016 for algorithmic details).
A tricky part of multiple sequence alignment is the choice of the elementary cost of each possible event. For instance, the relative costs of gap openings and gap extensions with respect to amino acid substitution strongly impact the final result and no efficient strategy as been found so far to select the ideal costs with respect to the sequences to be aligned (Wheeler and Kececioglu 2007). MACSE requires additional costs for frameshifts and stop codons that are not easier to set than traditional gap-associated costs. We provide default values that have proved to be effective based on our experience. This is further discussed in the MACSE online documentation that provides guidelines for handling specific sequences such as pseudogenes or RNAseq contigs resulting from error prone long read sequencing technologies.
The TrimNonHomologousFragments subprogram was developed to remove long sequence fragments that are unrelated to other sequences. Indeed, positioning long insertions in one or several sequences could drastically slow down and impede the alignment process. Moreover, long insertions may often prove finally useless since they are removed by alignment filtering tools in subsequent analyses. When a compatibility graph of maximum exact match (MEM) is constructed between two genomic sequences, they can be rapidly aligned after identification of the longest weighted path (Hohl et al. 2002). We extended this approach to handle the translation of nucleotide sequences in the three possible coding frames using a compressed amino acid alphabet. This allows identifying and trimming long insertions present in only few sequences, as such regions are rarely part of long MEM paths.
The enrichAlignment subprogram can be used to sequentially add new DNA sequences to an existing alignment. Its input parameters allow defining criteria that the additional sequences should fulfil to be actually incorporated into the final alignment. For instance, sequences can be automatically discarded when, once aligned, they would contain a stop codon, too many gaps, or more than a given number of frameshifts. The original alignment can either be sequentially enriched, or kept unchanged so that all sequences are compared with the same reference alignment. This latter option is especially useful for metabarcoding projects based on markers such as the mitochondrial Cytochrome Oxidase subunit I (cox1) gene. This typically involves enriching a reference alignment containing sequences from databases such as BOLD (Ratnasingham and Hebert 2007) or MIDORI (Machida et al. 2017) with thousands of newly generated sequences.
The reportMaskAA2NT subprogram takes as input a nucleotide alignment and a filtered version of the corresponding amino acid alignment, for example, produced by HMMcleaner (Philippe et al. 2017), and reports this filtering at the codon level. By default, it additionally filters out small sequence fragments mostly surrounded by gaps or filtered nucleotides. Other MACSE v2 subprograms allow performing useful alignment manipulations such as translating sequences using different genetic codes in the same alignment (translateNT2AA); restricting a coding alignment to a subset of sequences and/or sites (splitAlignment, trimAlignment); or refining an existing alignment using the 2-cut strategy to improve its SP-score (refineAlignment).
The command line interface is still key in most analyses that require running MACSE v2 in parallel on hundreds or thousands of data sets using a computing cluster. However, as the number of subprograms and options increased significantly, we now provide a user-friendly graphical interface. This should make it easier for new users to adopt MACSE v2 and hopefully broaden its usage and application scope.
MACSE v2 is Java software freely available under the CECILL license (GPL variant) at https://bioweb.supagro.inra.fr/macse/, last accessed August 22, 2018. MACSE v2 and OMM_MACSE, a pipeline strongly relying on the MACSE v2 toolkit that has been used to align the thousands of orthologous genes in the OrthoMAM database (Douzery et al. 2014), are also available through dedicated web services at http://mbb.univ-montp2.fr/MBB/, last accessed August 22, 2018.
Acknowledgments
We thank François Lechevallier for developing the website hosting MACSE v2, Jimmy Lopez and Khalid Belkhir for deploying MACSE v2 and the OMM_MACSE pipeline as web services on the Montpellier Bioinformatics Platform (MBB), and two anonymous reviewers for comments.
Funding
This work was supported by the Agence Nationale de la Recherche “Investissements d’avenir/Bioinformatique” (ANR-10-BINF-01-02 project Ancestrome), Labex CeMEB “Centre Méditerranéen de l’Environnement et de la Biodiversité” (ANR-10-LABX-0004), and the European Research Council (ERC-2015-CoG-683257 project ConvergeAnt). This is contribution ISEM 2018-151 of the Institut des Sciences de l’Evolution de Montpellier.
References
- Assis R, Zhou Q, Bachtrog D.. 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 411: 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg JG, Potter S, Bi K, Moritz C.. 2016. Exon capture phylogenomics: efficacy across scales of divergence. Mol Ecol Resour. 165: 1059–1068. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Gasse B, Sire J-Y.. 2015. Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta. BMC Evol Biol. 15:148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJP, Scornavacca C, Romiguier J, Belkhir K, Galtier N, Delsuc F, Ranwez V.. 2014. OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol Biol Evol. 317: 1923–1928. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 325: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 272: 221–224. [DOI] [PubMed] [Google Scholar]
- Hohl M, Kurtz S, Ohlebusch E.. 2002. Efficient multiple genome alignment. Bioinformatics 18(Suppl 1): S312–S320. [DOI] [PubMed] [Google Scholar]
- Lan X, Pritchard JK.. 2016. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science 3526288: 1009–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ.. 2013. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool. 101: 34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida RJ, Leray M, Ho SL, Knowlton N.. 2017. Metazoan mitochondrial gene sequence reference datasets for taxonomic assignment of environmental samples. Sci Data 4:170027.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, de Vienne DM, Ranwez V, Roure B, Baurain D, Delsuc F.. 2017. Pitfalls in supermatrix phylogenomics. Eur J Taxon 283:1–25. [Google Scholar]
- Ranwez V. 2016. Two simple and efficient algorithms to compute the SP-score objective function of a multiple sequence alignment. PLoS One 118: e0160043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP.. 2011. MACSE: multiple alignment of coding SEquences Accounting for frameshifts and stop codons. PLoS One 69: e22594.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PD.. 2007. BOLD: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Resour. 73: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RD, Beckenbach AT.. 2008. Recoding of translation in turtle mitochondrial genomes: programmed frameshift mutations and evidence of a modified genetic code. J Mol Evol. 676: 682–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Kececioglu JD.. 2007. Multiple alignment by aligning alignments. Bioinformatics 2313: i559–i568. [DOI] [PubMed] [Google Scholar]