Abstract
Overlapping genes in viruses maximize the coding capacity of their genomes and allow the generation of new genes without major increases in genome size. Despite their importance, the evolution and function of overlapping genes are often not well understood, in part due to difficulties in their detection. In addition, most bioinformatic approaches for the detection of overlapping genes require the comparison of multiple genome sequences that may not be available in metagenomic surveys of virus biodiversity. We introduce a simple new method for identifying candidate functional overlapping genes using single virus genome sequences. Our method uses randomization tests to estimate the expected length of open reading frames and then identifies overlapping open reading frames that significantly exceed this length and are thus predicted to be functional. We applied this method to 2548 reference RNA virus genomes and find that it has both high sensitivity and low false discovery for genes that overlap by at least 50 nucleotides. Notably, this analysis provided evidence for 29 previously undiscovered functional overlapping genes, some of which are coded in the antisense direction suggesting there are limitations in our current understanding of RNA virus replication.
Keywords: virus, overlapping reading frame, overprinting, overlapping gene
Introduction
Gene overlap occurs when two or more genes share the same region of a nucleotide sequence in a genome. This occurs frequently in viruses, especially those with RNA genomes, but has also been observed in bacteria and in eukaryotes including humans (Smith et al. 1977; Keese and Gibbs 1992; Veeramachaneni et al. 2004; Nakayama et al. 2007). The high prevalence of gene overlap in viruses has been attributed to two complementary theories: gene “compression” and gene “novelty.” Compression theory argues that the size of viral genomes is constrained by factors such as high mutation rates and the small capsid structure housing the genetic material. This constrained genome size subsequently exerts selection pressure on genes to overlap to maximize genetic potential (Belshaw et al. 2007; Chirico et al. 2010). Gene novelty theory asserts that the constrained nature of viral genomes, combined with their limited noncoding regions, makes the generation of new genes difficult without major changes in genomic structure or input from the host genome. Mutations in current genes that generate a new open reading frame (ORF) then allow the generation of new genes within an established older gene in a process called “overprinting” (Keese and Gibbs 1992; Sabath et al. 2012; Brandes and Linial 2016). These theories are not mutually exclusive and both processes may be operating in virus genomes. Overlapping genes may also function as a mechanism for regulating gene expression and reduce the probability of mutation fixation in overlapping areas as the resident genes may have competing selection pressures (Krakauer 2000; Dreher and Miller 2006). Due to these evolutionary constraints, overlapping genes frequently encode proteins with accessory functions that play important roles in pathogenicity or spread (Rancurel et al. 2009).
Overlapping genes were first detected following the discovery that the cumulative length of protein sequences in bacteriophage ϕ174 exceeded the length of the genome (Barrell et al. 1976). Today, the detection of overlapping genes still largely relies on laboratory methods that isolate, sequence, and align individual proteins to reference genomes (Fellner et al. 2015). These and other potential laboratory methods such as ribosome profiling (Irigoyen et al. 2016) are costly and time intensive, making large scale screening and identification of overlapping genes expensive. Necessarily, these factors have led to the development of bioinformatics and theoretical methods for the analysis of overlapping genes that rely on genome sequence analyses alone. For example, synonymous sites that exhibit a reduced nucleotide substitution rate are indicative of functional overlapping proteins; because these substitutions affect two proteins they are usually expected to be deleterious and hence are observed at a reduced rate (Firth and Brown 2005, 2006; Jagger et al. 2012). A number of other properties of overlapping genes have been used as effective bioinformatics makers, such as synonymous codon dissimilarity between newly generated overlapping genes and the remainder of the genome (Pavesi et al. 1997, 2013), and the restriction that particular codon sequence orders place on alternative reading frames (Lebre and Gascuel 2017). For example, the reverse complementary nucleotide sequence for two adjacent tyrosines (TAT/C and TAT/C) will be A/GTA and A/GTA, which always creates a stop codon (either a TAA or TAG after a reading frame shift of 1 nucleotide). Although these properties help in the development of bioinformatics techniques to discover unknown overlapping genes, they are restricted by their requirement for multiple genomic sequences or by their poor sensitivity. With the rapid rise of metagenomics to discover new viruses (Bekal et al. 2011; Ballinger et al. 2014; Shi et al. 2016, 2018), efficient and sensitive approaches of identifying overlapping genes that require genome sequence information alone will be essential.
Herein, we present a new statistical method for detecting overlapping genes in different reading frames that relies on only a single nucleotide sequence of a gene or genome. We apply this method to a large scale computational screening of all available (linear) RNA virus genomes. The method estimates the theoretical expected length of ORFs before a stop codon is reached in all reading frames within an established gene. If an ORF exists of much greater length than predicted by this expected length, then we surmise that there has been selection against the accumulation of stop codons that shorten the putative ORF. We conclude that this constitutes evidence that the ORF in question provides functional benefit to the virus. Despite its simplicity, we show that this method is a powerful way to detect functional overlapping genes that can be readily applied to large scale computational screening of all known viruses and to viruses newly discovered through metagenomics.
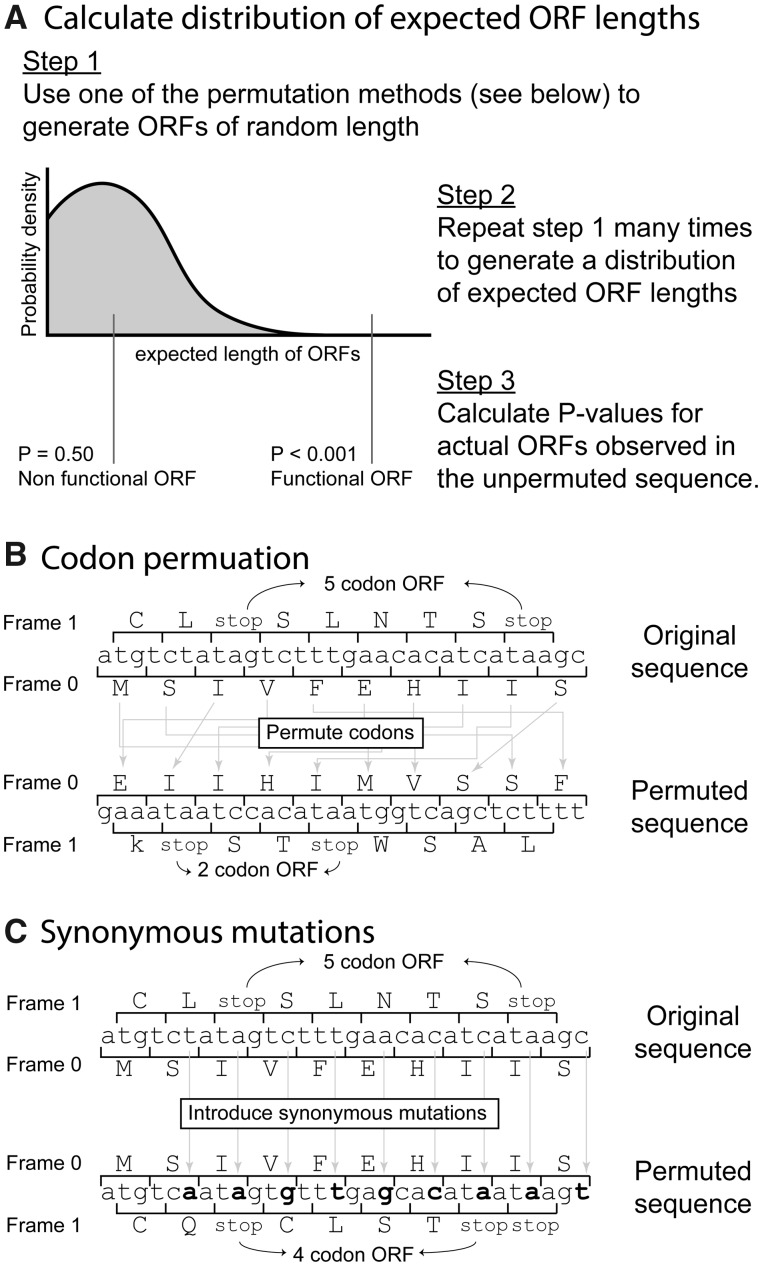
New Approaches
Our rationale is that overlapping ORFs with functional benefit will result in negative selection against nucleotide substitutions that introduce stop codons within that gene. Accordingly, the length of ORFs (as measured by the distance between bookend stop codons) is likely to be larger when they are functional than what would be expected by random chance alone where stop codons could be introduced without penalty. Hence, the defining characteristic of our method for the detection of overlapping genes is identifying ORFs larger than expected by chance alone (fig. 1A). We developed three tests for estimating the distribution of expected ORF lengths. Briefly, in the first test, we estimate the expected length of ORFs by permuting codon positions in the original reading frame and then measuring ORF lengths in other reading frames. This process is repeated to generate an expected distribution of ORF lengths (codon permutation test, fig. 1B). In the second test, instead of permuting codon positions, the codon order is unchanged and nucleotide substitutions that would introduce synonymous mutations in the original reading frame are randomly generated (synonymous mutation test, fig. 1C), before measuring ORF lengths in the other reading frames. In the third test, referred to as the combined test, the P values for both the codon permutation test and the synonymous mutation test must fall below some cut-off value.
Fig. 1.
Method to detect frameshifted open reading frames (ORFs) in viruses. (A) The expected ORF length based on codon composition is calculated. ORFs longer than expected by random chance are identified. The expected ORF length is estimated by one of three tests. For the codon permutation test (B) the codon sequence on the original frame is permuted and ORF lengths on alternative reading frames measured for each permutation. For the synonymous mutation test (C), codons that preserve the original amino acid sequence are randomly generated and the length of ORFs on alternative reading frames subsequently measured (note that codon replacement is not restricted to the example mutations shown in the figure, all of which occur in the third nucleotide positions, and that codon replacement with the original codon is also possible). The third test requires both the codon permutation test and the synonymous mutation test P values to be below some cut-off value.
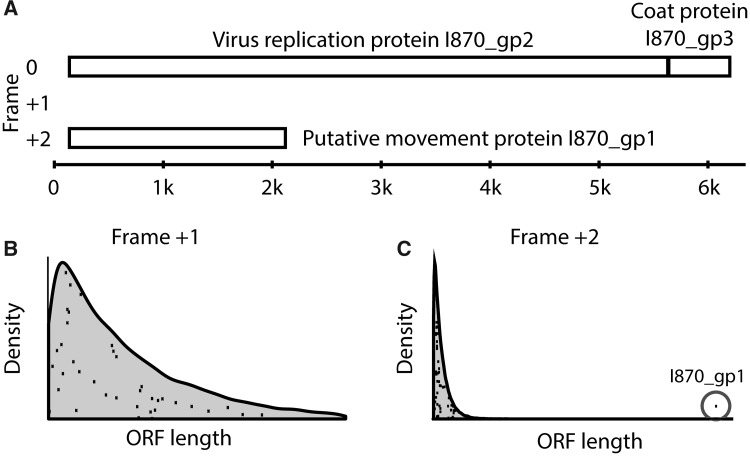
To demonstrate the applicability of this method, we first considered Andean potato latent virus that contains a known overlapping gene. Andean potato latent virus is a positive-sense single-stranded RNA virus (family Tymoviridae) in which the I870_gp1 gene, that encodes the putative movement protein, is overlapping. I870_gp1 is 665 codons long, is located on frame +2, and is largely contained within the larger (1832 codon) I870_gp2 gene that encodes the enzymes necessary for virus replication (methyltransferase, endopeptidase, helicase, and polymerase) (fig. 2A). We calculated the distribution of expected ORF lengths on frames +1 and +2 using the codon permutation test. The distributions of these lengths are shown with the shading in figure 2B and C. The actual ORF lengths on frames +1 and +2 on the unpermuted I870_gp2 gene are represented by black dots on top of the theoretical distribution. For frame +1, we observe 37 ORFs, all of which have lengths within expected ranges (P = 0.92). A different picture is observed in frame 2+. Although there are 62 ORFs, 61 of which have lengths within the expected range, there is a single ORF whose length far exceeds the expected distribution of lengths (P < 0.0001); this is correctly identified as I870_gp2. The synonymous mutation test produces similar results in this example.
Fig. 2.
Proof of concept for ORF detection using Potato Latent virus as an example. (A) Schematic of genomic structure for the Potato Latent virus. This virus contains a known overlapping gene I870_gp1 in Frame 2+. (B) The expected distribution of ORF lengths in frame 1+ (shaded area) calculated by the permutation test, and the actual open reading frame (ORF) lengths in frame 1+ (black dots). (C) The expected distribution of ORF lengths in frame 2+ (shaded area) calculated by permutation test, and the actual ORF lengths on frame 1+ (black dots). The known frameshifted gene, I870_gp1, was clearly identified using the permutation test as its length was much larger than that expected by chance alone (P < 0.0001).
Results
Sensitivity and False Discovery Rate
To explore the possibility of using this method to screen for candidate overlapping ORFs, we calculated both the sensitivity and false discovery rate of the codon permutation test, the synonymous mutation test, and a combined test that requires an ORF to be larger than expected by both the codon permutation and synonymous mutation test. As there are too few coding regions within a single genome to estimate the sensitivity and false discovery rate with sufficient precision, we estimated the population sensitivity and false positive rate across a subset of viruses (linear RNA viruses) known to contain many overlapping genes (see Materials and Methods section). Accordingly, whole genome sequences were downloaded from 2548 reference linear RNA viruses available on GenBank; this produced a total of 6408 coding regions that were used to estimate the sensitivity and false discovery rate of each test.
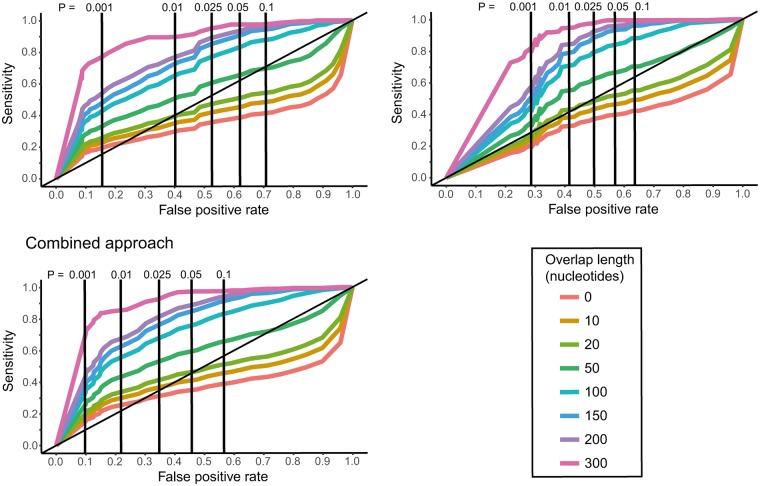
The codon permutation, synonymous mutation and combined test all rely on detecting overlapping ORFs that are larger than expected by random chance. Consequently, the sensitivity of these tests will depend on how much of a gene is overlapping (denoted as overlap length). The sensitivity and false discovery rate will also be dependent on the P value cut-offs used to determine if an ORF is larger than expected by random chance, with higher P values providing higher sensitivity at a cost of greater false discovery. To understand these dependencies, receiver operator characteristic (ROC) curves were generated across a range of P values, and across a range of overlapping gene lengths for all three tests (fig. 3). The number of true overlapping ORF’s used in this sensitivity set ranged from 958 for overlapping ORF lengths greater than 0 nucleotides, to 199 for overlapping ORF lengths greater than 300 nucleotides.
Fig. 3.
Receiver operator characteristic curves showing the sensitivity and false discovery rate of each test, for different P value cut-off values, and different minimum overlapping lengths in nucleotides. Table 1 shows the precise values for this for minimum overlapping lengths of 50, 100, 200, and 300 nucleotides, and P value cutoffs of 0.01 and 0.001.
We find that the three test (codon permutation, synonymous mutation, and combined) have similar sensitivities for P value cut-offs between 0.001 and 0.10, with the synonymous mutation generally having the highest sensitivity, followed by the codon permutation test and then the combined test (fig. 3, Table 1). However, for all three tests we also find that sensitivities are generally insufficient when the overlapping length is below 50 nucleotides in length (<17 codons), but improve considerably as the overlapping length increases above 50 nucleotides. Importantly, the three tests show considerable differences in false discovery rates, with the synonymous mutation tests showing the highest (worst) rates and the combined test with the lowest (best) rates. As the highest and lowest sensitivity tests (synonymous mutation and combined, respectively) are also the tests with the corresponding highest and lowest false discovery rates, we used the standard measure of a diagnostic tool—the area under the curve—to compare which test gave the best sensitivity and false discovery rate combinations. The area under the curve here will lie between 0 and 1, with a value of 0.5 indicating a screening tool of no benefit, and a value of 1 indicating a perfect screening tool with no error. Accordingly, we find that the combined test consistently has the best sensitivity and false discovery rate combinations across all minimum overlap lengths, with an area under the curve increasing from 0.59 to 0.89 as the minimum overlap length increases from 50 to 300 nucleotides (table 1). This demonstrates that the combined test is a successful screening tool with both high sensitivity and relatively low false discovery.
Table 1.
Sensitivity, false discovery, and area under the curve for each test across a range of P value cut-offs and overlapping lengths.
| Test | Minimum overlap length (nucleotides) | Sensitivity |
False discovery rate |
Area under the curve | ||
|---|---|---|---|---|---|---|
| P = 0.001 | P = 0.01 | P = 0.001 | P = 0.01 | |||
| Codon perm. | 50 | 0.33 | 0.51 | 0.16 | 0.40 | 0.56 |
| Codon perm. | 100 | 0.43 | 0.65 | 0.69 | ||
| Codon perm. | 200 | 0.54 | 0.77 | 0.78 | ||
| Codon perm. | 300 | 0.77 | 0.90 | 0.87 | ||
| Synonymous mut. | 50 | 0.35 | 0.55 | 0.29 | 0.41 | 0.55 |
| Synonymous mut. | 100 | 0.45 | 0.71 | 0.67 | ||
| Synonymous mut. | 200 | 0.57 | 0.84 | 0.74 | ||
| Synonymous mut. | 300 | 0.82 | 0.95 | 0.83 | ||
| Combined | 50 | 0.25 | 0.43 | 0.097 | 0.22 | 0.59 |
| Combined | 100 | 0.33 | 0.56 | 0.72 | ||
| Combined | 200 | 0.43 | 0.68 | 0.81 | ||
| Combined | 300 | 0.69 | 0.86 | 0.89 | ||
Comparison to Synplot2
Synplot2 is a commonly used bioinformatic approach to identify overlapping genes by detecting reduced variability at synonymous sites (Firth 2014). Although powerful, this method is necessarily constrained by the requirement for multiple sequences of sufficient diversity to robustly detect overlapping genes. In contrast, our method requires only a single sequence, and can therefore be applied in many situations where Synplot2 would be inviable. Quantitative comparisons of sensitivity and false discovery between the methods are difficult, as the factors associated with sensitivity in Synplot2 (sequence diversity, recombination, and window size) are not present in our method. Therefore, to make this comparison informative, we apply our method to the Synplot2 validation data set (table 1 from Firth 2014) and report the results in table 2. This validation consists of 21 gene overlaps with a minimum overlap length of 108 nucleotides. We find that using a P value cut of value of 0.01, the codon permutation method, synonymous mutation method, and combined approach detects 12, 12, and 10 of the gene overlaps, respectively. These results are in agreement with our previous sensitivity estimates. For example, Figure 3 shows that the combined approach will have ∼50% sensitivity for overlaps of at least 100 nucleotides when a P value cut-off of 0.01 is used.
Table 2.
Comparison of the codon permutation, synonymous mutation and combined methods to Synplot2 for the Synplot2 validation data set (table 1 from Firth 2014).
| Taxon | RefSeq | Gene overlap | Genomic location (nt) | ORF length (nuc) | Codon perm. P value | Synonymous mut. P value |
|---|---|---|---|---|---|---|
| Picornaviridae, Cardiovirus, Theilovirus | NC_001366.1 | L/L* | 1081–1551 | 470 | 0.0005 | 0.002 |
| Arteriviridae, | NC_001961.1 | GP2/GP3 | 12696–12843 | 147 | 0.87 | 0.52 |
| Arterivirus, PRRSV | GP3/GP4 | 13241–13460 | 219 | 0.06 | 0.03 | |
| Bromoviridae, Cucumovirus, Cucumber mosaic virus | NC_002035.1 | ORF2a/2b | 2419–2660 | 241 | 0.002 | 0.0007 |
| Hepeviridae, Hepevirus, HEV | NC_001434.1 | CP/ORF3 | 5123–5453 | 330 | 0.15 | 0.02 |
| Betaflexiviridae, Capillovirus, Apple stem grooving virus | NC_001749.2 | replicase-CP/MP | 4787–5749 | 962 | < 0.0001 | <0.0001 |
| Betaflexiviridae, Trichovirus, Apple chlorotic leaf spot virus | NC_001409.1 | MP/CP | 6784–7100 | 316 | 0.004 | <0.0001 |
| Alphaflexiviridae, Potexvirus, Pepino mosaic virus | NC_004067.1 | TGB2/TGB3 | 5340–5488 | 148 | 0.19 | 0.23 |
| Sobemovirus, Rice yellow mottle virus | NC_001575.2 | replicase/CP | 3447–3607 | 160 | 0.57 | 0.56 |
| Nodaviridae, Betanodavirus, Striped jack nervous necrosis virus | NC_003448.1 | replicase/B2 | 2756–2983 | 227 | 0.15 | 0.007 |
| Tombusviridae, Tombusvirus, Tomato bushy stunt virus | NC_001554.1 | MP/p19 | 3888–4406 | 518 | <0.0001 | <0.0001 |
| Birnaviridae, Aquabirnavirus, Infectious pancreatic necrosis virus | NC_001915.1 | VP5/VP2 | 120–514 | 394 | 0.002 | 0.0005 |
| Birnaviridae, Avibirnavirus, Infectious bursal disease virus | NC_004178.1 | VP5/VP2 | 130–533 | 403 | 0.002 | 0.03 |
| Reoviridae, Orthoreovirus, Mammalian orthoreovirus 3 | NC_004277.1 | σ1/σ1s | 71–433 | 362 | 0.002 | 0.001 |
| Totiviridae, Totivirus, Saccharomyces cerevisiae virus L-A | NC_003745.1 | gag/pol | 1964–2072 | 108 | 0.88 | 0.97 |
| Bunyaviridae, Orthobunyavirus, La Crosse virus | NC_004110.1 | N/NSs | 101–379 | 278 | 0.01 | 0.03 |
| Paramyxoviridae, Morbillivirus, Measles virus | NC_001498.1 | P/C | 1829–2389 | 560 | 0.0001 | <0.0001 |
| P/V | 2499–2705 | 206 | 0.18 | 0.007 | ||
| Paramyxoviridae, Respirovirus, Human parainfluenza virus 3 | NC_001796.2 | P/C | 1794–2393 | 599 | <0.0001 | <0.0001 |
| P/V | 2505–2903 | 398 | 0.0001 | 0.0003 | ||
| Paramyxoviridae, Rubulavirus, Mumps virus | NC_002200.1 | P/V | 2442–2653 | 211 | 0.02 | 0.02 |
| Picornaviridae, Cardiovirus, Theilovirus | NC_001366.1 | L/L* | 1081–1551 | 470 | 0.0005 | 0.002 |
| Arteriviridae, Arterivirus, PRRSV | NC_001961.1 | GP2/GP3 | 12696–12843 | 147 | 0.87 | 0.52 |
| GP3/GP4 | 13241–13460 | 219 | 0.06 | 0.03 | ||
| Bromoviridae, Cucumovirus, Cucumber mosaic virus | NC_002035.1 | ORF2a/2b | 2419–2660 | 241 | 0.002 | 0.0007 |
| Hepeviridae, Hepevirus, HEV | NC_001434.1 | CP/ORF3 | 5123–5453 | 330 | 0.15 | 0.02 |
Newly Discovered Overlapping ORFs
We next screened for previously undiscovered overlapping genes by using the combined test and a P value cut-off of 0.001. This cut-off was chosen as only 9.7% of any discoveries are estimated to be a false positive (table 1). We find evidence for 40 undocumented functional overlapping ORFs within all reference genomes of linear RNA viruses. Of these 40 ORFs, two had been previously described in Synplot2’s RNA screening in 2014 (Firth 2014). Investigating these overlapping ORFs further reveals that although some of these novel ORFs were not annotated within GenBank, they were not necessarily undiscovered, as some existed within the NCBI protein databases. To remove these already discovered or hypothesized overlapping ORFs, we performed a protein BLAST search of the 38 undocumented overlapping ORFs and found that nine had previously been discovered but were not annotated within the reference genome, thereby leaving 29 newly discovered functional overlapping ORFs from our method (table 3, supplementary materials S2 and S5, Supplementary Material online). Of these newly discovered ORFs, we would expect approximately three to be false discoveries. To test if we can detect homologs of the 29 newly discovered overlaps in other species, we aligned their protein sequence against the NCBI nt database using tblastn (supplementary material S4, Supplementary Material online). We filtered the results to only include alignments with a similarity of at least 90% and where the alignment was at least 90% the length of the ORF (Material and Methods section). While all the ORFs aligned against the species of origin as expected, two ORFs also aligned to different species. In the first of these ORFs, from Sosuga virus, we detected hits in two artificial expression vectors. In the second ORF, from Bovine parainfluenza virus 3, we identified an additional alignment in the closely related Swine parainfluenza virus 3, most likely in a homologous position (supplementary material S4, Supplementary Material online).
Table 3.
New putative ORF discoveries made here.
| Family, virus name | RefSeq | Coding region | Coding product | ORF position | Reading frame | ORF length (nuc) |
|---|---|---|---|---|---|---|
| Reoviridae, Aedes pseudoscutellaris reovirus | NC_007673 | 17..1054 | VP8 | 510–890 | +1 | 126 |
| Betaflexiviridae, Ligustrum necrotic ringspot virus | NC_010305 | 6604..6924 | Triple gene block protein | 6605–6919 | +1 | 105 |
| Unassigned, Circulifer tenellus virus 1 | NC_014360 | 643..4044 | Proline-alanine-rich protein | 1652–2647 | +1 | 331 |
| Rhabdoviridae, Infectious hematopoietic necrosis virus | NC_001652 | 1466..2158 | Polymerase-associated protein | 1690–2085 | +2 | 131 |
| Paramyxoviridae, Bovine respirovirus 3 | NC_002161 | 1784..3574 | Phosphoprotein P | 2500–3021 | +2 | 173 |
| Pneumoviridae, Avian metapneumovirus | NC_007652 | 6111..7868 | Attachment glycoprotein | 6560–7675 | +2 | 371 |
| Unassigned, Cassava virus C | NC_013112 | 186..1055 | Putative movement protein | 209–646 | +2 | 145 |
| Unassigned, Circulifer tenellus virus 1 | NC_014360 | 643..4044 | Proline-alanine-rich protein | 645–1757 | +2 | 370 |
| Unassigned, Halastavi arva RNA virus | NC_016418 | 828..6278 | Replicase protein | 1610–2155 | +2 | 181 |
| Reoviridae, Spissistilus festinus reovirus | NC_016874 | 9..3740 | RNA directed RNA polymerase | 380–1267 | +2 | 295 |
| Paramyxoviridae, Bat Paramyxovirus Eid_hel/GH-M74a/GHA/2009 | NC_025256 | 2053..4665 | Phosphoprotein | 2958–3479 | +2 | 173 |
| Arenaviridae, Okahandja mammarenavirus | NC_027137 | 58..339 | Z protein | 60–332 | +2 | 91 |
| Potyviridae, Sweet potato virus 2 | NC_017970 | 118..10518 | Polyprotein | 118–1119 | −c0 | 333 |
| Filoviridae, Marburg marburgvirus | NC_024781 | 5941..7986 | Glycoprotein | 6046–6753 | −c0 | 235 |
| Rhabdoviridae, Oak-Vale virus | NC_025399 | 3393..4988 | Putative glycoprotein | 3837–4721 | −c0 | 294 |
| Virgaviridae, Macrophomina phaseolina tobamo-like virus | NC_025674 | 208..6594 | RNA-dependent RNA polymerase | 406–1422 | −c0 | 338 |
| Rhabdoviridae, Northern cereal mosaic virus | NC_002251 | 142..1437 | Nucleocapsid protein | 810–1436 | −c1 | 208 |
| Flaviviridae, Nhumirim virus | NC_024017 | 103..10440 | Polyprotein | 2328–4454 | −c1 | 708 |
| Rhabdoviridae, Infectious hematopoietic necrosis virus | NC_001652 | 2999..4525 | Glycoprotein | 3555–4037 | −c2 | 160 |
| Tombusviridae, Hibiscus chlorotic ringspot virus | NC_003608 | 2603..3277 | Hypothetical protein | 2745–3248 | −c2 | 167 |
| Paramyxoviridae, Tioman virus | NC_004074 | 2033..2667 | W protein | 2147–2665 | −c2 | 172 |
| Paramyxoviridae, Tioman virus | NC_004074 | 2033..3188 | Phosphoprotein | 2038–2595 | −c2 | 186 |
| Totiviridae, Magnaporthe oryzae virus 1 | NC_006367 | 575..2815 | Putative coat protein | 1314–1931 | −c2 | 205 |
| Alphaflexiviridae, Hydrangea ringspot virus | NC_006943 | 5549..6022 | Virally coded protein | 5553–6020 | −c2 | 156 |
| Tymoviridae, Scrophularia mottle virus | NC_011537 | 127..1980 | Putative movement protein | 776–1438 | −c2 | 220 |
| Peribunyaviridae, Simbu orthobunyavirus | NC_018477 | 50..325 | Nonstructural protein | 60–323 | −c2 | 87 |
| Reoviridae, Umatilla virus | NC_024503 | 13..3912 | RNA-dependent RNA polymerase | 1826–2515 | −c2 | 229 |
| Paramyxoviridae, Sosuga virus | NC_025343 | 1908..3105 | Phosphoprotein | 1913–2542 | −c2 | 210 |
| Paramyxoviridae, Salmon aquaparamyxovirus | NC_025360 | 2535..3667 | V protein | 2538–3164 | −c2 | 209 |
The 29 discovered ORFs ranged from 87 to 708 codons in length, with a median and interquartile range of 195.5 (157–279.2) codons; 13 were transcribed in the same direction (sense, frames +1 and +2) as the original gene with 17 coded in the opposite direction on complementary nucleotides (antisense frames −c0, −c1, and −c2, supplementary material S1, Supplementary Material online). In addition, 18 of the ORFs were located completely within their reference coding region, eight lay on the boundary and four encompassed the entire coding region, suggesting that the reference coding region may lie completely within the larger discovered ORF. Of these discovered ORFs, a number are of particular interest and discussed in more detail below.
Nhumirim Virus
The largest detected ORF was 708 codons long and located within Nhumirim virus, a positive-sense flavivirus recently isolated from mosquitoes in Brazil (Pauvolid-Correa et al. 2015). Unexpectedly, this ORF is coded on a reverse complementary reading frame (−c1), which means that unlike the other proteins in this virus, transcription must occur from a negative-sense RNA template. This finding invites further investigation of the potential mechanisms by which transcription of reverse complementary reading frames might occur in positive-sense RNA viruses. In addition, the 26th codon in this ORF is a methionine (a common start codon) suggesting that a large component of the 708 codons may be transcribed.
Bovine Respirovirus Virus 3
A 173 codon long ORF was detected within the phosphoprotein P coding gene of Bovine respirovirus virus 3 (single-stranded negative-sense RNA virus, family Paramyxoviridae). This +2 reading frame ORF was particularly interesting because although its protein alignment didn’t match any Bovine respirovirus virus proteins, it did align with V proteins and RNA editing derivatives within the phosphoprotein P gene of other paramyxoviruses (Galinski et al. 1992; Wells and Malur 2008). These derivatives may play an important role in virus replication (Durbin et al. 1999), virulence (Huang et al. 2003), and/or the disruption of interferon expression (Roth et al. 2013), and the discovery is in agreement with that claims all three reading frames in the P gene of Bovine respirovirus are expressed (Pelet et al. 1991).
Tioman Virus
The method also detected two new ORFS of length 160 and 131 codons in phosphoprotein in another paramyxovirus, Tioman virus. Although one of these ORFs was in the same sense (+2 reading frame) the other was in the reverse complementary frame (−c2).
Smallest ORFs Detected
The three smallest ORFs detected were in Ligustrum necrotic ringspot virus, a positive-sense virus from the Betaflexiviridae, Okahandja mammarenavirus, a negative-sense virus from the Arenaviridae, and Simbu orthobunyavirus, a vector-borne negative-sense virus from the Peribunyaviridae. These ORFs had lengths 105, 91, and 87 codons, and were in frames +1, +2, and −c2, respectively. Interestingly, these were three of the four detected ORFs that completely encompass their relatively short reference genes, suggesting that the reference gene may be entirely located within the ORFs discovered by this method.
Discussion
We present a simple new method that uses a single genome sequence to detect candidates for overlapping genes. The method assumes that functional ORFs are longer than expected by random chance as they experience selective pressure against mutations that introduce stop codons. We quantify this by using three ways to estimate the null distribution for ORFs lengths within each reading frame of a gene, and use the null to identify those ORFs significantly longer than predicted by random chance. This approach has a number of advantages over current bioinformatics methods to detect overlapping genes. In addition to being simple and quick, it only requires a single genome sequence. This is in contrast to other bioinformatic methods that require multiple sequences to estimate and compare nucleotide or codon diversity. This feature allows the method to be applied much more broadly in both metagenomics projects where genomes of new viruses are frequently only present in a single copy (Bekal et al. 2011; Ballinger et al. 2014; Shi et al. 2016), and also in screening scenarios such as demonstrated herein. The method is best suited to refine regions of the genome that contain candidates for functional overlapping genes, after which these regions can be further tested for functionality with more resource intensive laboratory methods such as protein isolation, ribosomal profiling (Michel et al. 2012; Ingolia 2016), and studying the effects of introduced knock out mutations (Chung et al. 2008).
A second important feature of our method is the relatively high sensitivity to detect overlapping genes, whilst maintaining acceptable false discovery rates. This is best achieved by using the combined test where newly detected ORFs must be larger than expected by both the codon permutation and synonymous mutation tests. The combined test is advantageous as true positives are readily detected by both tests, so the constraint of requiring both tests to detect the ORF does not impact the sensitivity. However, the combined test does substantially reduce the false positives rate, as false positives detected by one test are frequently excluded by the other. There is also scope to further reduce false discovery by modifying our method, or by imposing post analysis constraints, for example by calculating ORF lengths from start codon to stop codon rather than between two stop codons. This was not considered for the screening results here due to variation in alternative start codons among viruses, but would be an important optimization in more targeted screening. One caveat to this method (and other bioinformatics approaches) is that sensitivity depends on the size of overlap, with smaller regions of overlap being more difficult to detect. Unlike other methods, however, we explicitly calculated the sensitivity for many lengths of overlap and find that a length of at least 50 nucleotides (17 codons) is required before the method becomes effective. As this length increases to 300 nucleotides (100 codons), the method becomes a very powerful diagnostic tool as measured by an area under the curve equal to 0.89. The estimate of this method’s sensitivity and false discovery rates for an overlapping gene detection method is a strength, as although sensitivity can be calculated for other methods, false discovery estimation is often neglected and rarely reported due to a lack of negative controls. When it is reported, it is usually based on estimates of type 1 error rates of P values, rather than comparison to a negative control as we have done in here.
To demonstrate the utility of our method’s effectiveness for overlapping gene screening, we individually analyzed all reference linear RNA genomes available on GenBank. This provided evidence for 29 undocumented overlapping ORFs of which we expect only 3 to be false positives, although all should be verified experimentally. One notable ORF identified here is the exceptionally long (708 codons) antisense ORF in Nhumirim virus, a single-stranded positive-sense RNA virus from the family Flaviviridae, which suggests that this virus may employ a novel method of transcription and clearly merits further investigation. We also identified several undiscovered ORFs in the phosphoprotein P within the Paramyxoviridae family, a region known to frequently contain overlapping genes in other reading frames due to RNA editing. Within Bovine respirovirus virus 3, the ORF codon sequence discovered here aligned with many V proteins of other members of the Paramyxoviridae. As the V protein in these viruses also overlaps with the phosphoprotein P protein, this suggests that the V protein also exists in Bovine respirovirus virus 3. In other Paramyxoviridae, notably Tioman virus, we also identified antisense ORFs in the phosphoprotein.
The detection of 17 antisense ORFs is notable. Antisense overlaps have been shown to exist in a number viruses that use DNA as a replication intermediate including those in the Herpesviridae (Ward et al. 1996), REP/ORF3 in Porcine circovirus 2 (He et al. 2012) and HBZ/p12 and HBZ/p30 in Human T lymphotropic virus 1 (Arnold et al. 2006). They are also suspected to occur in many more viruses with DNA intermediaries, including a long suspected antisense protein (asp) in HIV-1 (Torresilla et al. 2015; Cassan et al. 2016). In addition, they have been infrequently suggested to occur in RNA viruses that do not use DNA intermediates, such as a more than 100 amino acid (a) overlapping antisense hypothetical protein in Rice black streaked dwarf virus (dsRNA) (Zhang et al. 2001), a 96 aa overlapping antisense hypothetical protein in Lymphocytic Choriomeningitis Mammarenavirus (-ssRNA) (Salvato et al. 1989), and a possible 167 aa overlapping antisense ORF called “NEG8” in human influenza A virus (Clifford et al. 2009; Sabath et al. 2011). Our method can be used to investigate these further. For example, in the case of NEG8 we find that a 167 codon ORF on in the NEG8 reading frame (−c2) is highly statistically unlikely by both the codon permutation and synonymous mutation methods, providing further evidence for a functional benefit of this ORF. Interestingly, however, a further frameshift of 1 nucleotide (frame −c1) would make ORFs of such lengths much more likely (P = 0.02 and 0.03 for codon permutation and synonymous mutation methods respectively), demonstrating the importance of the expected ORF lengths on every individual reading frame, rather than just the sense direction. Furthermore, when applying our method to HIV-1, we find that the possible antisense ORF (asp) is not substantially longer than expected by chance alone (P = 0.06 and P = 0.04 for codon permutation and synonymous mutation methods, respectively) in that reading frame.
Our results do indicate that antisense ORFs are present at levels higher than currently expected. This does not necessarily mean that a transcribed protein is functional, although its presence may be indicative of some functional benefit, such as regulating expression by diverting ribosomes (Pelechano and Steinmetz 2013; Beltran and Garcia de Herreros 2016). Importantly, our method’s high detection of antisense ORFs are in contrast to other bioinformatic screening methods which have been shown to have poor sensitivity to antisense ORFs by computer simulations. This is because synonymous mutations in frame +0 impact the reverse complementary frame (specifically frame −c2) much less than other reading frames (Mir and Schober 2014). Although this feature would impact the sensitivity of our synonymous mutation test, as it would for all current methods, the codon permutation test will not impacted by this, and could be used in isolation when specifically screening for antisense ORFs.
Overlapping genes play an important role in viral evolution (Simon-Loriere et al. 2013), and are particularly prevalent in RNA viruses with small genomes. However, the study of overlapping genes is limited by detection methods that either have high laboratory costs, or require enough sequences to make reliable substitution rate comparisons. Our simple, but powerful, permutation and synonymous mutation method requires only a single genome sequence and is computationally quick to run. These properties make it an ideal choice for identifying candidate ORFs in screening situations such as metagenomics viral discovery projects, or applied to large genome databases such as we have done here.
Materials and Methods
Data Collection
Whole genome sequences were downloaded for all viruses available from the NCBI FTP site ftp://ftp.ncbi.nlm.nih.gov/genomes/Viruses. Of the 5757 viral genomes, 2548 were RNA viruses with linear genomes, and these were selected for analysis. All 6408 coding regions (annotated with a CDS in Genbank) were analyzed, excluding 291 regions annotated with a “join” indicating some form of midsequence frameshift in the established gene such as a ribosomal slippage (leaving 6117 coding regions analyzed).
Reading Frame Definitions
The following notation is used to identify the different reading frames (supplementary material S1, Supplementary Material online): +0 is the original reading frame; +1 or +2 is a frameshift of 1 or 2 nucleotides, respectively (5′ to 3′ transcription); −0, −1, or −2 a frameshift of 0, 1 or 2 nucleotides, respectively, after the coding sequence has been reversed (i.e. 3′ to 5′ transcription); +c0, +c1 or +c2 is a frameshift of 0, 1 or 2 nucleotides, respectively, on the complement of the coding sequence (3′ to 5′ transcription); and −c0, −c1 or –c2 is a frameshift of 0, 1 or 2 nucleotides, respectively, on the complement and reversed coding sequence (5′ to 3′ transcription). +0, +1, +2, +c0, +c1, +c2 are considered the only viable reading frames as transcription on these frames occur in the 5′ to 3′ direction.
Tests to Identify Frameshifted Genes
For each coding region and for each viable alternative reading frame (+1,+2,−c0, −c1, −c2) we performed the following analysis (summarized in fig. 1A). First, the length of ORFs between the stop codons “TGA,” “TAG,” and “TAA” on that specific alternative reading frame was calculated. Then, 20,000 new coding sequences were created by either randomly permuting the codons in frame +0 (codon permutation test; fig. 1B), or for each amino acid in reading frame +0, randomly choosing a replacement codon (for which the original codon is a possible candidate) that encodes the same amino acid (synonymous mutation test; fig. 1C). For each of these 20,000 new coding sequences, the length of ORFs between stop codons in that alternative reading frame was calculated again. The lengths of ORFs over all 20,000 randomly generated coding sequences were pooled to calculate a theoretical distribution of the length of ORFs on that specific alternative reading frame. For each ORF length L in the original unpermuted coding sequence, the probability of observing a length as large or larger by random chance alone is calculated using this theoretical distribution as follows:
where C is the empirical cumulative distribution function of the theoretical distribution of lengths calculated by permuting codons in the original coding sequence: that is, C(L) is the probability of sampling an ORF length less than L on that specific reading frame. The value 1 − C(L) has an interpretation similar to that of a P value testing whether or not the length L is sampled from the theoretical distribution of lengths calculated earlier. To correct this “P value” for the total number of alternative reading frame ORFs, s, in the original coding sequence the following equation is used:
This adjustment is analogous to a Bonferroni adjustment of P values, here correcting for the number of ORFs within a reading frame. Small P values for an ORF are interpreted as evidence that the ORF in question is larger than expected by random chance alone and therefore provides evidence that there has been negative selection against mutations that introduce stop codons in this ORF. From this, we can also infer that the ORF is of functional benefit to the virus. The third test, denoted as the “combined test,” requires that the P value for both the codon permutation test and the synonymous mutation test be below some cut-off value.
The method was only applied to ORFs on alternative reading frames that exist totally or partially within the parent ORF. When ORFs on alternative reading frames extended beyond the parent ORF boundaries, its length was truncated to the length contained with the parent ORF.
This analysis was performed using R (version 3.3.2; R_Core_Team 2016) and required the packages seqinr (Delphine Charif et al. 2017) and ggplot2 (Hadley Wickham and RSudio 2016). An R file of the functions used to calculate the P values for both tests is provided in supplementary material S3, Supplementary Material online and is available on Github at (https://github.com/TimSchlub/Frameshift).
Sensitivity and False Discovery Rate
The sensitivity (true positive rate) is measured as the proportion of known overlapping genes within the downloaded reference genomes that are detected using our method. An ORF identified with our method was considered a true positive if it was located on the same reading frame and overlapped with a gene already annotated in the reference genome. As the sensitivity of our method will be dependent on the extent of overlap, we calculated the sensitivity for detecting previously annotated overlapping genes where the minimum nucleotide length of overlap is 1, 10, 20 50, 100, 150, 200, and 300 nucleotides. The false discovery rate calculation is more complex as the absence of an annotated overlapping gene does not exclude its biological presence so that distinguishing between false positives and new discoveries is not possible without extensive laboratory work. To overcome this, we conservatively estimated the false discovery rate of our tests by using the nonviable 3′–5′ reading frames (−0, −1, −2, +c0, +c1, and +c2, supplementary material S1, Supplementary Material online) as a negative control. That is, as detected ORFs on these frames cannot be transcribed into proteins and are therefore false positives, they serve as an estimate to what proportion of detected ORFs on viable reading frames are similarly not functional.
Identifying Homologous Sequences
A search for sequences homologous to the 29 newly discovered overlapping ORFs was performed by aligning their protein sequences to the NCBI nt database using tblastn (TBLASTN 2.6.0+). The e-value threshold was set to 0.001 while all other settings were set to their default values. The results were stored in a SQLite3 database (3.24.0). For homologous sequence detection the alignments were filtered to include only alignments with similarity ≥90% and length ≥90% of the corresponding ORF sequence. From each filtered alignment, the NCBI accession for the query (ORF) and subject were extracted and the corresponding taxid and lineage obtained using NCBI Entrez. A python tool taxmax.py was developed (https://gitlab.com/janpb/taxmax.git) to compare the NCBI lineage from each ORF and its aligned sequence. The similarity between two lineages is described as a score between 0 and 1. A score of 0 indicates no similarity between lineages while a score of 1 indicates both sequences have the same NCBI lineage. For each alignment The alignments positions were compared to check for orthologous positions.
Supplementary Material
Acknowledgments
This work was supported by an ARC Australian Laureate Fellowship awarded to ECH (FL170100022). The authors acknowledge the University of Sydney HPC service at The University of Sydney for providing high performance computing resources that have contributed to the research results reported within this article. We thank an anonymous reviewer for suggesting the synonymous mutation method.
References
- Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, Green PL.. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 10710:3976–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger MJ, Bruenn JA, Hay J, Czechowski D, Taylor DJ.. 2014. Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes. J Virol. 8816:8783–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell BG, Air GM, Hutchison CA 3rd. 1976. Overlapping genes in bacteriophage phiX174. Nature 2645581:34–41. [DOI] [PubMed] [Google Scholar]
- Bekal S, Domier LL, Niblack TL, Lambert KN.. 2011. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol. 92(Pt 8):1870–1879. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Pybus OG, Rambaut A.. 2007. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 1710:1496–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Garcia de Herreros A.. 2016. Antisense non-coding RNAs and regulation of gene transcription. Transcription 72:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Linial M.. 2016. Gene overlapping and size constraints in the viral world. Biol Direct 11:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan E, Arigon-Chifolleau AM, Mesnard JM, Gross A, Gascuel O.. 2016. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc Natl Acad Sci U S A. 11341:11537–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N, Vianelli A, Belshaw R.. 2010. Why genes overlap in viruses. Proc Biol Sci. 2771701:3809–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Miller WA, Atkins JF, Firth AE.. 2008. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A. 10515:5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford M, Twigg J, Upton C.. 2009. Evidence for a novel gene associated with human influenza A viruses. Virol J. 6:198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Miller WA.. 2006. Translational control in positive strand RNA plant viruses. Virology 3441:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, McAuliffe JM, Collins PL, Murphy BR.. 1999. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology 2612:319–330. [DOI] [PubMed] [Google Scholar]
- Fellner L, Simon S, Scherling C, Witting M, Schober S, Polte C, Schmitt-Kopplin P, Keim DA, Scherer S, Neuhaus K.. 2015. Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evol Biol. 15:283.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE. 2014. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 4220:12425–12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Brown CM.. 2005. Detecting overlapping coding sequences with pairwise alignments. Bioinformatics 213:282–292. [DOI] [PubMed] [Google Scholar]
- Firth AE, Brown CM.. 2006. Detecting overlapping coding sequences in virus genomes. BMC Bioinformatics 7:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MS, Troy RM, Banerjee AK.. 1992. RNA editing in the phosphoprotein gene of the human parainfluenza virus type 3. Virology 1862:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JL, Dai D, Zhou N, Zhou JY.. 2012. Analysis of putative ORF3 gene within porcine circovirus type 2. Hybridoma (Larchmt) 313:180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Krishnamurthy S, Panda A, Samal SK.. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J Virol. 7716:8676–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 1651:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N, Firth AE, Jones JD, Chung BY, Siddell SG, Brierley I.. 2016. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 122:e1005473.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL.. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 3376091:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese PK, Gibbs A.. 1992. Origins of genes: “big bang” or continuous creation? Proc Natl Acad Sci U S A. 8920: 9489–9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer DC. 2000. Stability and evolution of overlapping genes. Evolution 543:731–739. [DOI] [PubMed] [Google Scholar]
- Lebre S, Gascuel O.. 2017. The combinatorics of overlapping genes. J Theor Biol. 415:90–101. [DOI] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV.. 2012. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2211:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir K, Schober S.. 2014. Selection pressure in alternative reading frames. PLoS One 910:e108768.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Asai S, Takahashi Y, Maekawa O, Kasama Y.. 2007. Overlapping of genes in the human genome. Int J Biomed Sci. 31:14–19. [PMC free article] [PubMed] [Google Scholar]
- Pauvolid-Correa A, Solberg O, Couto-Lima D, Kenney J, Serra-Freire N, Brault A, Nogueira R, Langevin S, Komar N.. 2015. Nhumirim virus, a novel flavivirus isolated from mosquitoes from the Pantanal, Brazil. Arch Virol. 1601:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, De Iaco B, Granero MI, Porati A.. 1997. On the informational content of overlapping genes in prokaryotic and eukaryotic viruses. J Mol Evol. 446:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A, Magiorkinis G, Karlin DG.. 2013. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of Deltaretroviruses. PLoS Comput Biol. 98:e1003162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM.. 2013. Gene regulation by antisense transcription. Nat Rev Genet. 1412:880–893. [DOI] [PubMed] [Google Scholar]
- Pelet T, Curran J, Kolakofsky D.. 1991. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. Embo J. 102:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Core_Team. 2016. R: a language and environment for statistical computing. Version 3.3.2. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D.. 2009. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. j Virol. 8320:10719–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JP, Li JK, Morrey JD, Barnard DL, Vollmer AH.. 2013. Deletion of the D domain of the human parainfluenza virus type 3 (HPIV3) PD protein results in decreased viral RNA synthesis and beta interferon (IFN-beta) expression. Virus Genes 471:10–19. [DOI] [PubMed] [Google Scholar]
- Sabath N, Morris JS, Graur D.. 2011. Is there a twelfth protein-coding gene in the genome of influenza A? A selection-based approach to the detection of overlapping genes in closely related sequences. J Mol Evol. 73(5–6):305–315. [DOI] [PubMed] [Google Scholar]
- Sabath N, Wagner A, Karlin D.. 2012. Evolution of viral proteins originated de novo by overprinting. Mol Biol Evol. 2912:3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato M, Shimomaye E, Oldstone MB.. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 1692:377–384. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, et al. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. [DOI] [PubMed] [Google Scholar]
- Shi M, Zhang YZ, Holmes EC.. 2018. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 243:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E, Holmes EC, Pagan I.. 2013. The effect of gene overlapping on the rate of RNA virus evolution. Mol Biol Evol. 308:1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Brown NL, Air GM, Barrell BG, Coulson AR, Hutchison CA 3rd, Sanger F.. 1977. DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174. Nature 2655596:702–705. [DOI] [PubMed] [Google Scholar]
- Torresilla C, Mesnard JM, Barbeau B.. 2015. Reviving an old HIV-1 gene: the HIV-1 antisense protein. Curr HIV Res. 132:117–124. [DOI] [PubMed] [Google Scholar]
- Veeramachaneni V, Makałowski W, Galdzicki M, Sood R, Makałowska I.. 2004. Mammalian overlapping genes: the comparative perspective. Genome Res. 142:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PL, Barker DE, Roizman B.. 1996. A novel herpes simplex virus 1 gene, UL43.5, maps antisense to the UL43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J Virol. 705:2684–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G, Malur A.. 2008. Expression of human parainfluenza virus type 3 PD protein and intracellular localization in virus infected cells. Virus Genes 373:358–367. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. Package ‘ggplot2’ for R. Elegant Graphics for Data Analysis. Springer-Verlag New York. Version 2.2.1.
- Zhang HM, Chen JP, Adams MJ.. 2001. Molecular characterisation of segments 1 to 6 of Rice black-streaked dwarf virus from China provides the complete genome. Arch Virol. 14612:2331–2339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.