Abstract
Gene duplication is an important driver for the evolution of new genes and protein functions. Duplication of DNA-dependent RNA polymerase (Pol) II subunits within plants led to the emergence of RNA Pol IV and V complexes, each of which possess unique functions necessary for RNA-directed DNA Methylation. Comprehensive identification of Pol V subunit orthologs across the monocot radiation revealed a duplication of the largest two subunits within the grasses (Poaceae), including critical cereal crops. These paralogous Pol subunits display sequence conservation within catalytic domains, but their carboxy terminal domains differ in length and character of the Ago-binding platform, suggesting unique functional interactions. Phylogenetic analysis of the catalytic region indicates positive selection on one paralog following duplication, consistent with retention via neofunctionalization. Positive selection on residue pairs that are predicted to interact between subunits suggests that paralogous subunits have evolved specific assembly partners. Additional Pol subunits as well as Pol-interacting proteins also possess grass-specific paralogs, supporting the hypothesis that a novel Pol complex with distinct function has evolved in the grass family, Poaceae.
Keywords: DNA-dependent RNA polymerase V, RNA-directed DNA methylation, gene duplication, Poaceae
Introduction
Eukaryotic organisms possess three multisubunit DNA-dependent RNA polymerase complexes (Pol I–III), which are each responsible for transcription of a subset of cellular RNA. Plants encode two additional DNA-dependent RNA polymerases (Pol IV and V), which are specialized for RNA-directed DNA methylation (Haag and Pikaard 2011). RNA Pol IV produces 24-nucleotide small RNAs that are bound by ARGONAUTE 4 (AGO4) proteins. These siRNAs then guide AGO4 to sites of Pol V transcription and recruit de novo methylation machinery to the locus (Wierzbicki et al. 2009). The carboxy terminal domain (CTD) of Pol V helps to recruit AGO4 through an AGO-binding platform (El-Shami et al. 2007; Lahmy et al. 2016).
DNA-dependent RNA polymerases are composed of multiple subunits, which are named NRPxn, where x = A–E for Pols I–V, respectively, and n = 1–12 for the largest to smallest subunit, respectively (Zhou and Law 2015). Pol II, IV, and V share many of their 12 subunits, but also possess unique subunits that confer their distinct functions (Huang et al. 2009; Lahmy et al. 2009; Ream et al. 2009; Haag et al. 2014). The largest subunit of Pol IV (NRPD1) and Pol V (NRPE1) arose through duplication of the largest Pol II subunit (NRPB1) (Luo and Hall 2007). The second, fourth, fifth, and seventh subunits have also duplicated and specialized for Pol IV and V at different times during land plant evolution (Tucker et al. 2010; Huang et al. 2015). In addition, the Argonaute-binding platform in the CTD of NRPE1 is evolving more rapidly than other regions of the protein (Trujillo et al. 2016).
Gene duplication is a common occurrence in eukaryotic genomes and is an important source of biological complexity (Conant and Wolfe 2008). Most duplicates (paralogs) are lost because there is no selection for retention of redundant subunits, although in rare cases paralogs can be retained to fix heterozygosity or to increase dosage (Hahn 2009). More commonly, retained paralogs have diverged in function and become nonredundant, either by partitioning multiple functions of the single-copy ancestral gene between the paralogs (subfunctionalization), or by one paralog evolving novel function (neofunctionalization).
Here, we identify retained duplicates of multiple polymerase subunits and polymerase-associated proteins in Poaceae, the family that contains cereal grasses. Phylogenetic analysis of the two largest subunits indicates positive selection for one paralog following the duplication, consistent with retention via neofunctionalization. Analysis of selection at sites of subunit interaction raises the possibility that evolution favored specific polymerase assemblies. The CTDs of paralogous subunits are also characteristically distinct. Together these results suggest that a sixth distinct RNA polymerase complex exists in this critical plant family.
Results
Poaceae Members Encode Paralogous Pol V Subunits
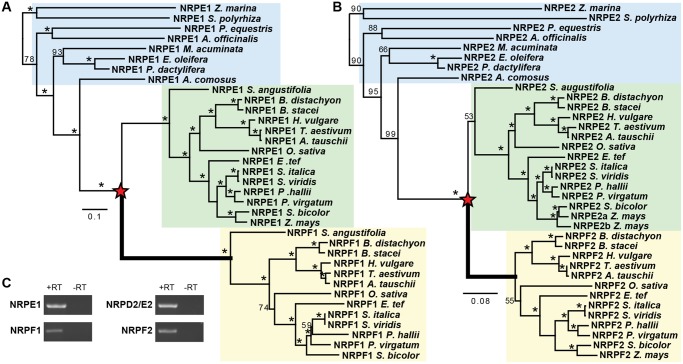
Previous studies investigating Pol V evolution revealed the presence of two NRPE1 paralogs in some monocot genomes (Trujillo et al. 2016). However, this observation was restricted to species within Poaceae as most of the sequenced monocot genomes are members of this agriculturally important family. To understand the timing of this duplication, we identified putative NRPE1 homologs across the monocot lineage. Phylogenetic analysis of these sequences reveals a single NRPE1 ortholog in non-Poaceae monocots and two well-supported clades of NRPE1-like sequences within Poaceae (fig. 1A). Ananus comosus (pineapple) in Bromeliaceae, a sister lineage to Poaceae in the order Poales, contains a single NRPE1 ortholog. Importantly, AcNRPE1 is equally diverged from both Poaceae NRPE1-like clades, indicating that duplication of NRPE1 occurred after the divergence of the two families. Conversely, Streptochaeta angustifolia, an early diverging member of the Poaceae, has two NRPE1-like sequences, indicating the duplication occurred in an early ancestor of extant grasses. We have designated the paralog along the longer branch as NRPF1.
Fig. 1.
Duplications of Pol V subunits are coincident with the emergence of the grass family Poaceae. The evolutionary relationships of NRPE1 (A) and NRPD/E2 (B) within the monocot lineage demonstrate that monocots outside of the Poaceae family have a single gene copy (blue box), while most members of Poaceae have paralogous genes (green and yellow boxes). Phylogenetic trees were inferred by maximum likelihood analysis of mRNA sequence in the catalytic domain (regions B to H). Bootstrap support values <100 are shown and red stars mark the inferred duplications. Thick branches indicate positive selection (P < 0.05). Full species names and gene accession numbers are listed in supplementary table 1, Supplementary Material online. (C) RT-PCR demonstrates that all Oryza sativa paralogs are expressed.
NRPE1 and NRPF1 copies were recovered from every Poaceae genome we assessed, with the exception of Z. mays, which lacks a full-length NRPF1 (fig. 1A). A NRPF1 homolog is present in the genome, but it appears to be a pseudogene in all Z. mays varieties, we analyzed (B73, PH207, CML247, and B104), possibly due to insertion of a transposon within the coding sequence (supplementary fig. 1, Supplementary Material online). Analysis of teosinte (Zea mays ssp parviglumis), the wild progenitor of cultivated Z. mays, revealed that this pseudogenization occurred prior to domestication. It is not clear why Z. mays has lost NRPF1, since retention of this gene in every other grass genome, we assessed indicates that these paralogs are not redundant.
In addition to NRPE1, Pol V is distinguished from Pol II by the smaller subunits NRPD/E2, NRPD/E4, NRPE5, and NRPD/E7. We therefore determined if these subunits are also duplicated within Poaceae. We found no evidence of duplication for NRPD/E4, NRPE5, or NRPD/E7, however the second largest subunit, NRPD/E2, which is shared by RNA Pol IV and V, has also duplicated within monocots (fig. 1B).
A single copy of NRPD/E2 is present in A. comosus, and this sequence is sister to two well-supported clades of Poaceae specific orthologs. As with the largest subunit, the clade with the longer branch has been designated as NRPF2 since this long branch indicates it is more dissimilar to the ancestral NRPE1 than its paralog. It is clear that the NRPD/E2 duplication occurred early in Poaceae diversification, however whether this duplication was simultaneous or subsequent to the NRPE1 duplication is not clear. One NRPD/E2 paralog was identified in S. angustifolia, and this is sister to all other grass NRPD/E2 sequences, suggesting that NRPF2 might have been lost in S. angustifolia. However, it is also possible that S. angustifolia diverged from other grasses prior to duplication giving rise to the NRPD/E2 and NRPF2 paralogs since support for the placement of the single S. angustifolia NRPD/E2 homolog is weak.
Triplication of NRPD/E2 was reported in Z. mays (Sidorenko et al. 2009; Stonaker et al. 2009; Haag et al. 2014). With the addition of other Poaceae homologs, we show that these three copies arose through a subsequent duplication of NRPD/E2 in the maize (or possibly maize + sorghum) lineage, yielding NRPD/E2a, NRPD/E2b, and NRPF2 (previously called NRPD/E2c).
We confirmed expression of NRPE1, NRPF1, NRPD/E2, and NRPF2 paralogs in Oryza sativa floral tissue (fig. 1C). Publicly available expression data also support the expression of both paralogs of each subunit (supplementary fig. 2, Supplementary Material online), confirming that none of the paralogs are pseudogenes. Public expression data also indicate that the paralogs differ in expression level and pattern, supporting our hypothesis that grasses contain novel Pol subunits with nonredundant functions.
NRPF1 Has Distinct CTD Characteristics
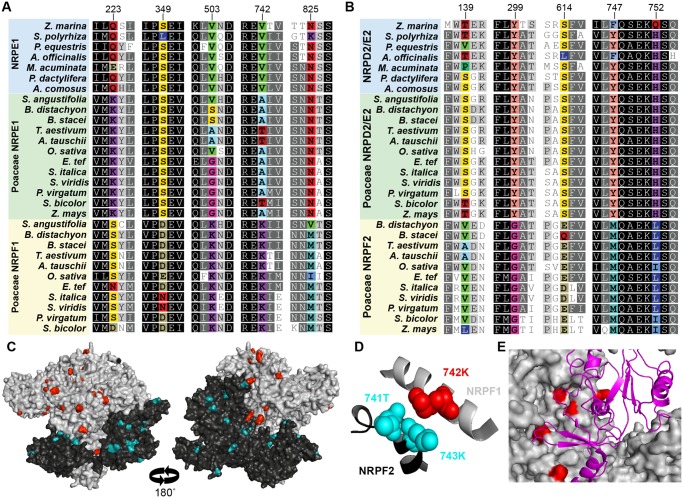
The largest subunits of Pols II, IV, and V have unique C-terminal domains (CTD). NRPB1 has a large region composed exclusively of heptad repeats; NRPD1 and NRPE1 both contain a domain of unknown function with similarity to Defective Chloroplast and Leaves (DeCL) genes at the extreme C-terminus, but only NRPE1 contains an Ago-binding platform between the catalytic and DeCL domains (Pontier et al. 2005; El-Shami et al. 2007; Huang et al. 2015). The Ago-binding platform is repetitive and disordered, and contains numerous Ago-hook motifs for association with AGO4 (Trujillo et al. 2016). Because the NRPE1 Ago-binding platform is evolutionarily labile and only closely related sequences can be aligned, CTDs were not included in the phylogenetic analysis that identified NRPE1 and NRPF1 clades. However, the same two clades are identified when only characteristics of the CTDs are considered.
NRPE1 CTDs possess an Ago-binding platform similar to that found in NRPE1 from non-Poaceae species, namely a long region with numerous Ago hook motifs. In NRPF1 proteins this region is reduced in length and in number of Ago hook motifs (fig. 2 and supplementary table 3, Supplementary Material online). However, NRPF1 CTDs are not as short as NRPD1 CTDs, and all but one NRPF1 ortholog contain at least one Ago hook in its CTD, as well as two Ago hook motifs in the catalytic region. This change in CTD character maps to the branch leading to the NRPF1 clade on the NRPE1/NRPF1 gene tree, suggesting it occurred immediately following duplication. The Ago-binding platform is required for accumulation of Pol V transcripts at many genomic loci (Wendte et al. 2017), and the change in this domain following duplication further indicates that NRPF1 is a nonredundant paralog of NRPE1.
Fig. 2.
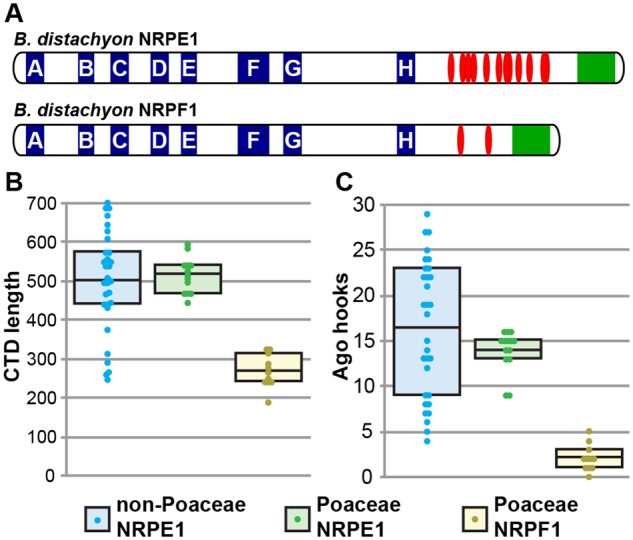
Structural divergence of the CTDs between NRPE1 and NRPF1 paralogs. (A) Diagram of NRPE1 and NRPF1 from B. distachyon. BdNRPE1 retains a canonical Ago-binding platform between the catalytic A–H domains (blue square) and the DeCL domain (green square). The Ago-binding platform contains many Ago hooks (red ovals). In contrast, BdNRPF1 has a shorter CTD that lacks Ago hook motifs. (B and C) Poaceae NRPE1 and non-Poaceae NRPE1 CTDs are similar in length and number of Ago hooks, while NRPF1 CTDs are shorter and contain fewer Ago hooks. Data points for 31 non-Poaceae, 14 Poaceae NRPE1, and 11 Poaceae NRPF1 are shown as colored circles; boxes represent the interquartile range and the mean is shown by a black bar.
Pol VI Paralogs Experienced Positive Selection following Duplication
Gene duplication allows evolutionary changes that can result in a paralog with a novel function (neofunctionalization) or paralogs that partition the original function (subfunctionalization), hypotheses that can be distinguished based on the pattern of selection following duplication. During subfunctionalization, both branches experience relaxed selection as a low number of nonsynonymous substitutions accumulate. During neofunctionalization, one branch maintains purifying selection and has few nonsynonymous substitutions, while the neofunctionalizing paralog experiences positive selection and accumulates many nonsynonymous substitutions (Hahn 2009).
We observed that the branch leading to the NRPF1 clade is longer than that of the Poaceae NRPE1 clade, suggesting that NRPF1 and NPRE1 experienced different selective pressure following duplication. A longer branch length could result from relaxed selection permitting the accumulation of substitutions; alternatively, positive selection on specific substitutions might have driven the progression toward a novel function. We distinguished between these possibilities with a branch-sites model (Zhang et al. 2005), which indicated that positive selection occurred along the branch leading to the NRPF1 and NRPF2 clades while NRPE1 and NRPD/E2 subunits retained purifying selection.
The branch-sites test identified 12 codons with a high likelihood of being under positive selection on the branch leading to NRPF1 and numerous additional sites when all branches of the clade are evaluated. (fig. 3A and supplementary table 4, Supplementary Material online). Similarly, the NRPF2 branch has 9 sites predicted to be under positive selection immediately following the duplication, and 59 sites across the whole clade (fig. 3B and supplementary table 5, Supplementary Material online). Indels also exist between NRPE1 and NRPF1 orthologs, suggesting that structural differences were also selected following duplication (supplementary fig. 3, Supplementary Material online). Only two sites were identified as under positive selection for NRPE1 and no sites for NRPD/E2 (supplementary tables 4 and 5, Supplementary Material online). Positive selection along the NRPF1 and NRPF2 branches and predominantly purifying selection for NRPE1 and NRPD/E2 supports the hypothesis of paralog retention due to neofunctionalization, and suggests that in Poaceae, NRPE1 and NRPD/E2 maintain the ancestral functions, while NRPF1 and NRPF2 may have evolved novel functions.
Fig. 3.
Sites under positive selection cluster on the surface of Pol VI subunits. Alignment of monocot largest (A) and second largest (B) subunits illustrates a subset of the residues that are under positive selection following gene duplication (colored). Remaining residues are colored based on sequence conservation. Numbers at top are the amino acid position in the Oryza sativa NRPF1 and NRPF2 and correspond with supplementary tables 4 and 5, Supplementary Material online. (C) Residues under positive selection (colored) are found on the surface of homology-based structures of NRPF1 (gray) and NRPF2 (black). (D) Residues under positive selection also occur at the interface between subunits, as demonstrated by 742K in NRPF1, which is directly opposite 741T and 743K in NRPF2. (E) Several residues under positive selection are found where the largest subunit (gray) interacts with the fifth subunit (magenta ribbon).
Sites under Positive Selection Are Exposed and Predict Additional Subunit Duplications
We mapped the predicted sites under positive selection on a homology-based model of O. sativa paralogs to evaluate the structural significance of particular substitutions (fig. 3C). NRPE1 and NRPF1 were aligned and modeled to the largest subunit of S. pombe RNA pol II holoenzyme (PDB: 3H0G chain A) with 100% confidence and sequence identity of 23%. The second largest subunit paralogs were analyzed in the same manner with O. sativa NRPD/E2 and NRPF2 mapping with 100% confidence and 36–37% sequence identity to a bovine RNA Pol II structure (PDB: 5FLM chain B).
Most sites predicted to be under positive selection were located on the surface of subunits or at interaction faces with other polymerase subunits, suggesting that the substitutions do not impact the overall structure of the subunits, but might impact assembly of the holoenzyme (supplementary tables 4 and 5, Supplementary Material online). In one case, residues under selection in NRPF1 and NRPF2 are predicted to interact, suggesting that they might be compensatory, and selection may have acted to restrict the number of possible holoenzyme assemblies (fig. 3D). Specific assembly of Pol subunits would indicate that not only are the paralogous subunits functionally nonredundant, they assemble into a unique polymerase complex, a Pol VI.
Based on the position of selected sites on the structure model, we hypothesized additional duplications of smaller subunits. Six selected sites are in the region that interacts with the fifth subunit (fig. 3E) and four sites near the interaction region for the ninth subunit (supplementary tables 4 and 5, Supplementary Material online). Although there is no evidence for duplication of the Pol V-specific NRPE5, we identified multiple copies of NRPB/D5 in Poaceae (supplementary fig. 4, Supplementary Material online). There is also evidence for a duplication of NRPB/D/E9 within the monocots (supplementary fig. 5, Supplementary Material online), although this duplication is difficult to resolve given the limited sequence information in this small subunit. Duplication of NRPB/D5 suggests that a Poaceae-specific Pol VI might have assembled in a modular fashion, using paralogous modules/subunits from both Pol IV and Pol V.
Pol V-Associated Proteins Are Also Duplicated in Poaceae
Canonical RdDM involves RNA Pol V interacting with numerous other proteins to accomplish DNA methylation. If Pol VI has a novel function that is diverged from Pol V, we might expect duplication and neofunctionalization of Pol V interacting proteins. We therefore investigated the evolution of known Pol V-interacting proteins, including the small RNA-binding protein AGO4 (El-Shami et al. 2007), the transcriptional elongation factor SPT5-like (Huang et al. 2009), and the SWI/SNF-related helicase DRD1 (Law et al. 2010; Zhong et al. 2012).
AGO4 associates with the NRPE1 CTD, enabling base-pairing of AGO4-bound small RNAs with Pol V transcripts (Wierzbicki et al. 2009), or with the Pol V transcription bubble (Lahmy et al. 2016). In A. thaliana, AGO4 is a part of a group of Argonautes including the deeply conserved AGO6, and Brassicaceae-specific AGO8 and AGO9 (Zhang et al. 2015; Rodríguez-Leal et al. 2016). In addition to an AGO6 group, we detected three well-supported clades of AGO4 orthologs within grasses arising from nested duplications at the base of Poaceae (fig. 4). AGO4a and AGO15 are sister groups that share high sequence similarity and are located within a few kilobases of one another, suggesting that a tandem duplication of one paralog occurred following whole genome duplication. Most predicted AGO15 sequences consist of partial, fragmented coding sequences, and there is no evidence for AGO15 protein accumulation in rice (Wu et al. 2010), suggesting that OsAGO15 might be a pseudogene. Correspondingly, we were able to detect expression of OsAGO4a and OsAGO4b, but not OsAGO15 (fig. 4B). Public expression data indicate that rice and maize AGO4 orthologs are broadly expressed (supplementary fig. 6, Supplementary Material online), where they bind to different groups of small RNAs (Wu et al. 2010). Genetic data in maize also indicates that despite the fact that ZmAGO4a (ZmAGO119) and ZmAGO4b (ZmAGO104) have broad and overlapping expression patterns, these paralogs are not redundant (Singh et al. 2011).
Fig. 4.
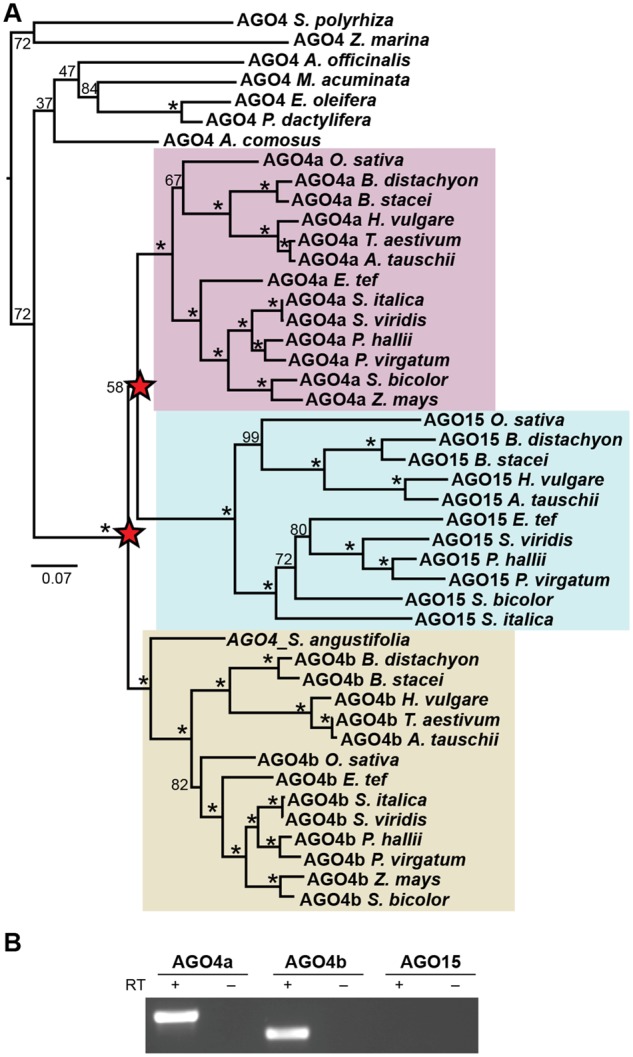
Nested duplications of the AGO4 locus result in three paralogs in most grasses. (A) Comparison of AGO4-orthologous sequences in monocots demonstrates that grasses contain multiple AGO4 paralogs and that these duplications were coincident with the emergence of the Poaceae family. Maximum likelihood phylogenetic tree of AGO4 related nucleotide sequences in monocots. Support values for branches with <100 bootstrap support are marked. Red stars mark the inferred duplication events. (B) RT-PCR demonstrates expression of AGO4a and AGO4b, but not AGO15, in rice.
SPT5L, a duplicate of the Pol II transcription elongation factor SPT5, interacts with Pol V and contains its own Ago-binding platform in its carboxy terminus (Bies-Etheve et al. 2009; Lahmy et al. 2016). Although SPT5L is the paralog that interacts with Pol V, we do not detect a duplication of this gene. Rather, SPT5, which interacts with Pol II and Pol IV, has undergone a duplication at the base of Poaceae (supplementary fig. 7, Supplementary Material online). This observation is additional evidence that not all genes associated with Pol V activity were duplicated in grasses, and further supports the hypothesis that a sixth polymerase complex formed through duplication of both Pol IV and Pol V machinery.
RNA Pol IV and V transcription is assisted through interaction with helicase proteins of the DRD1-like family (Kanno et al. 2004; Smith et al. 2007; Law et al. 2011). We discovered Poaceae-specific duplications within the DRD1 and CLSY3/4 clades, which give rise to paralogs we have named DRD1-like (DRD1L) and CLSY5, respectively (supplementary fig. 8, Supplementary Material online). We discovered DRD1L and CLSY5 copies only in Poaceae species, though DRD1 and CLSY trees suggest the duplication predates the evolution of the grasses. We take the current placement as a preliminary assessment in need of more data from additional species to more fully resolve these gene tree topologies. Whether one or both helicases are required for transcription by a grass-specific Pol VI remains to be determined.
DRD1 interacts with RDM1 and DMS3 to form the DDR complex, which is required for RdDM (Law et al. 2010). We identified only a single copy of RDM1 and DMS3 in Poaceae, further demonstrating that many components of Pol V machinery remain in single copy, while specific components of Pol VI and Pol V duplicated in grasses. The duplication of Pol IV and Pol V interacting protein further supports our hypothesis that grasses contain a distinct sixth polymerase with unique activity.
Discussion
Our evolutionary analysis of DNA-dependent RNA polymerases within the monocot lineage of land plants identified duplications of multiple subunits and polymerase-associated proteins. Non-Poaceae sequences are equally diverged from paralogous Poaceae sequences, supporting our conclusion that these duplications are coincident with the rho whole genome duplication at the base of grasses (McKain et al. 2016). Most genes return to single copy following whole genome duplication, therefore retention of duplicated genes is evidence for the formation of nonredundant protein function (Hahn 2009). Verified expression of NRPE1, NRPF1, NRPD/E2, and NRPF2 (fig. 1C); unique CTD sequences for NRPE1 and NRPF1 (fig. 2); and phylogenetic evidence of positive selection on NRPF1 and NRPF2 (fig. 3A and B) indicate that these paralogous subunits are not merely redundant, but rather are likely to encode unique functions.
Many of the sites with evidence for positive selection are at interfaces where Pol subunits interact (fig. 3D and E), suggesting that in addition to selection for unique activity, there might have been selection for faithful assembly of subunits into unique complexes (Beilstein et al. 2015). However, we cannot exclude the possibility that NRPF2 interacts with multiple largest subunits. This idea is supported by biochemical evidence from Z. mays, in which NRPD/E2 and NRPF2 display differential association with NRPD1 (Haag et al. 2014). However, pseudogenization of NRPF1 in Z. mays makes this species a poor representative for other grasses and further validation of Pol subunit associations is required in a different grass species. The signature of selection at predicted interacting sites, as well as the coordinated duplication of multiple subunits, leads us to hypothesize that not only do NRPF1 and NRPF2 encode novel functions but that they assemble into a unique polymerase complex, Pol VI.
We identified duplications of many Pol V subunits and interacting proteins, but we also found duplications of proteins that are not specific to Pol V. For example, we detected duplication of NRPB/D5, but not NRPE5 (supplementary fig. 3, Supplementary Material online). Similarly, the Pol V-specific transcription elongation factor SPT5L is not duplicated, but the paralogous SPT5, which interacts with Pol II and Pol IV, occurs in multiple copies (supplementary fig. 6, Supplementary Material online). Duplication of Pol II/IV proteins suggest that Pol VI formed through neofunctionalization of both Pol IV and Pol V subunits (fig. 5), although we cannot exclude the hypothesis that duplication of Pol II/IV subunits reflects subfunctionalization or neofunctionalization of Pol IV activity. Pol IV and Pol V both generate noncoding transcripts from otherwise silent DNA, but they differ in their speed, accuracy, and processivity (Wierzbicki et al. 2008; Zhai, Bischof, et al. 2015; Marasco et al. 2017). Conservation of key enzymatic residues, including the metal binding sites, indicates that Pol VI is capable of transcription, although such activity and how it differs from Pol IV and Pol V remain to be studied.
Fig. 5.
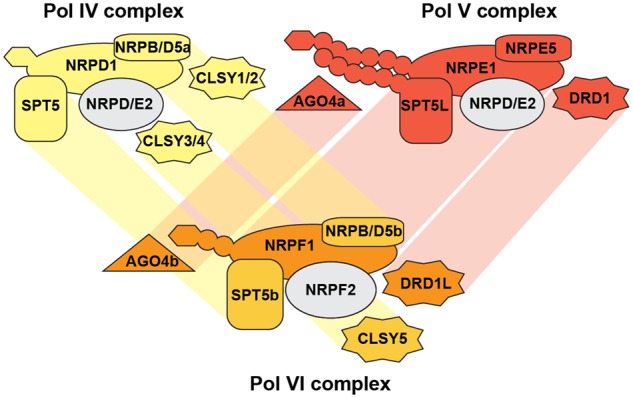
Proposed Pol assembly in grasses. We propose that NRPF1 and NRPF2 subunits assemble into Pol VI with a paralog of NRPB/D5. The Pol VI complex might function with specific paralogs of SPT5, DRD1, CLSY3/4, and AGO4, highlighting the use of paralogous proteins from both Pol V and Pol IV complexes. Paralogous subunits are connected by shaded boxes.
One key difference between Pol IV and Pol V is association with AGO4. The NRPE1 CTD contains numerous Ago-hooks for interaction with AGO4. Likewise, its binding partner SPT5L also contains numerous Ago hooks and associates with AGO4 (Bies-Etheve et al. 2009; Trujillo et al. 2016). In contrast, neither NRPD1 nor SPT5 contain Ago-hook motifs for AGO4 interaction. NRPF1 orthologs possess only a few Ago hooks, suggesting that Pol VI might interact with AGO proteins, but in a manner distinct from the Pol V-AGO4 interaction. Duplication of SPT5 rather than SPT5L also hints that numerous Ago hooks are not necessary for Pol VI function.
The biological role of Pol VI remains to be determined, but its similarity to Pol IV and Pol V along with the presence of Ago hooks suggests a role in small RNA biology while expression data indicate that Pol VI might accumulate during reproductive development (supplementary fig. 2, Supplementary Material online). Pol VI might be required for the biosynthesis or function of a number of novel small RNA classes, including highly expressed endosperm-specific siRNA loci in rice (Rodrigues et al. 2013), 24-nt phased siRNAs required for microspore development (Zhai, Zhang, et al. 2015; Fei et al. 2016), or rice “long” miRNAs (Wu et al. 2010). The potential role of Pol VI in reproductive development could make it an important target of agricultural and biotechnology manipulation.
Grasses are one of the most successful radiations of land plants, covering vast areas of natural habitat and agricultural land and forming the bulk of the human diet. We are acutely dependent on grasses, both for food and environmental stability, and it is therefore critical to understand the unique gene regulatory mechanisms of this family. Our discovery of a novel sixth polymerase in Poaceae uncovers a previously unknown aspect of grasses and offers an opportunity to learn more about this important plant family.
Materials and Methods
Ortholog Identification
Published NRPE1 ortholog sequences (Trujillo et al. 2016) were retrieved from Phytozome versions 11 and 12 (Goodstein et al. 2012). Additional sequences, including homologs of NRPE1, NRPD1, NRPF1, NRPB2, NRPD/E2, NRPB/D5, NRPE5, NRPB9, AGO4, SPT5/SPT5L, and DRD1/CLSY1-like, were obtained through BLAST or TBLASTX queries against whole genome sequences in Phytozome, CoGE (Lyons and Freeling 2008), and Ensembl Plants (Kersey et al. 2018) using O. sativa nucleotide sequences. Streptochaeta angustifolia and Zea mays ssp. parviglumis are available at http://gif-server.biotech.iastate.edu/arnstrm/mhufford/streptochaeta.html and http://gif-server.biotech.iastate.edu/arnstrm/mhufford/parviglumis.html, last accessed July 31, 2018.
In unannotated genomes, or when gene model predictions were incomplete, coding sequences were predicted using FGENESH+ (Softberry Inc. New York, NY) with O. sativa protein sequence as the homology template, followed by manual curation. Orthology was confirmed by reciprocal BLAST searches against the O. sativa genome and with phylogenetic analysis. Where species-specific duplications were detected (often due to polyploidization), only one full-length coding sequence was used for downstream analysis. All genes included in this study are listed in supplementary table 1, Supplementary Material online.
Phylogenetic Analysis
Nucleotide sequences were aligned by translation using MUSCLE in Geneious version 6.1.8 (Kearse et al. 2012). Where necessary, manual curation was performed to correct alignments. Maximum likelihood phylogenetic trees were inferred with RAxML version 7.2.8 (Stamatakis et al. 2008) using full-length coding sequences for most genes. For the largest subunits (NRPE1 and NRPF1), only the catalytic region from domains B–H were aligned. A General Time Reversible (GTR) model with gamma distributed rate of heterogeneity was implemented, and support values were based on 100 bootstrap replicates.
The branch-sites test for positive selection was performed using PAML version 4.9c codeml (Yang 2007). Branches under positive selection were determined by likelihood ratio test (χ2). Parameter space was explored with various starting ω values (0.2, 0.4, 0.6, 0.8, and 1.0) to determine effect on likelihood calculation under M2 (branch-sites) model. Robustness of likelihood values was evaluated by three replications of each analysis under each parameter set.
Expression Analysis
Total nucleic acids were isolated from O. sativa inflorescence as previously described (Grover et al. 2017), followed by Turbo DNase-free treatment (Ambion). First-strand cDNA was synthesized using SuperScript III (Invitrogen) with either polyT or random hexamer primers. Ortholog expression was determined through PCR using primers in supplementary table 2, Supplementary Material online.
Structural Modeling
Protein homology models of O. sativa NRPE1, NRPF1, NRPD/E2, and NRPF2 were generated by Phyre2 intensive modeling (http://www.sbg.bio.ic.ac.uk/phyre2, last accessed July 31, 2018) (Kelley et al. 2015) to the S. pombe (PDB: 3H0G) or Bos taurus (PDB: 5FLM) Poll II holoenzyme structures for the largest and second largest subunits, respectively. Modeled subunits were then aligned based on interaction of the cognate S. pombe subunits and interaction between the subunits was analyzed. Sites under positive selection were visualized in PyMol Molecular Graphics System (Schrödinger, LLC). Residues of O. sativa NRPE1 and NRPF1 were compared with homologous residues of NRPB1 in S. pombe and NRPE1 in A. thaliana with known functional importance. Experimentally derived information regarding subunit interaction regions and specific interacting residues was retrieved through UniProt database (http://www.uniprot.org/, last accessed July 31, 2018) (Bateman et al. 2017).
Supplementary Material
Acknowledgments
The authors thank Dr May Khanna and Dr Samantha Perez-Miller for assistance with visualization of homology models. We also thank Dr Elizabeth Kellogg for support in the assembly of S. angustifolia and Z. mays ssp parviglumis genomes and Dr Lynn Clark for providing S. angustifolia tissue. This work was supported by the National Science Foundation (IOS-1546825 to R.A.M. and M.A.B.) and the National Institutes of Health (T32-GM008659 to J.T.T.).
References
- Bateman A, Martin MJ, O’Donovan C, Magrane M, Alpi E, Antunes R, Bely B, Bingley M, Bonilla C, Britto R.. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45(D1):D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein MA, Renfrew KB, Song X, Shakirov EV, Zanis MJ, Shippen DE.. 2015. Evolution of the telomere-associated protein POT1a in arabidopsis thaliana is characterized by positive selection to reinforce protein-protein interaction. Mol Biol Evol. 325:1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T.. 2009. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 106:649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 912:938–950. [DOI] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi M-A, Jacobsen SE, Cooke R, Lagrange T.. 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2120:2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Yang L, Liang W, Zhang D, Meyers BC.. 2016. Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J Exp Bot. 6721:6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(D1):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover JW, Kendall T, Baten A, Burgess D, Freeling M, King GJ, Mosher RA. (2018). Maternal components of RNA–directed DNA methylation are required for seed development in Brassica rapa. Plant J. 944: 575–582. [DOI] [PubMed] [Google Scholar]
- Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, et al. 2014. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep. 91:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS.. 2011. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 128:483–492. [DOI] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 1005:605–617. [DOI] [PubMed] [Google Scholar]
- Huang L, Jones AME, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC.. 2009. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 161:91–93. [DOI] [PubMed] [Google Scholar]
- Huang Y, Kendall T, Forsythe ES, Dorantes-Acosta A, Li S, Caballero-Pérez J, Chen X, Arteaga-Vázquez M, Beilstein MA, Mosher RA.. 2015. Ancient origin and recent innovations of RNA polymerase IV and V. Mol Biol Evol. 327:1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ.. 2004. Involvement of Putative SNF2 Chromatin Remodeling Protein DRD1 in RNA-Directed DNA Methylation. Curr Biol. 149:801–805. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2812:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE.. 2015. The Phyre2 web portal for protein modelling, prediction, and analysis. Nat Protoc. 106:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, et al. 2018. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 46(D1):D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Bies-Etheve N, Laudié M, Feng S, Jobet E, Hale CJ, Cooke R, Hakimi M, Angelov D, et al. 2016. Evidence for ARGONAUTE4-DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 3023:2565–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T.. 2009. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 1063:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE.. 2010. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol. 2010:951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE.. 2011. SHH1, a Homeodomain protein required for DNA Methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA Polymerase IV. PLoS Genet. 77:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Hall BD.. 2007. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 641:101–112. [DOI] [PubMed] [Google Scholar]
- Lyons E, Freeling M.. 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 534:661–673. [DOI] [PubMed] [Google Scholar]
- Marasco M, Li W, Lynch M, Pikaard CS.. 2017. Catalytic properties of RNA polymerases IV and V : accuracy, nucleotide incorporation and rNTP/dNTP discrimination. Nucleic Acids Res. 4519:11315–11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain MR, Tang H, McNeal JR, Ayyampalayam S, Davis JI, dePamphilis CW, Givnish TJ, Pires JC, Stevenson DW, Leebens-Mack JH.. 2016. A phylogenomic assessment of ancient polyploidy and genome evolution across the Poales. Genome Biol Evol. 84:1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi M-A, Lerbs-Mache S, Colot V, Lagrange T.. 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 1917:2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu J-K, Hagen G, Guilfoyle TJ, Paša-Tolić L, Pikaard CS.. 2009. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 332:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D.. 2013. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci U S A. 11019:7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Castillo-Cobián A, Rodríguez-Arévalo I, Vielle-Calzada J-P.. 2016. A primary sequence analysis of the ARGONAUTE protein family in plants. Front Plant Sci. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, Kermicle J, Jurcin D, Brzeski J, Cai Y, Chandler VL.. 2009. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 511:e1000725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D.. 2011. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 232:443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC.. 2007. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 195:1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 575:758–771. [DOI] [PubMed] [Google Scholar]
- Stonaker JL, Lim JP, Erhard KF, Hollick JB.. 2009. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 511:e1000706.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo JT, Beilstein MA, Mosher RA.. 2016. The Argonaute-binding platform of NRPE1 evolves through modulation of intrinsically disordered repeats. New Phytol. 2124:1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SL, Reece J, Ream TS, Pikaard CS.. 2010. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol. 750:285–297. [DOI] [PubMed] [Google Scholar]
- Wendte JM, Haag JR, Singh J, McKinlay A, Pontes OM, Pikaard CS.. 2017. Functional dissection of the Pol V largest subunit CTD in RNA-directed DNA methylation. Cell Rep. 1913:2796–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS.. 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 1354:635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS.. 2009. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 41:630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y.. 2010. DNA methylation mediated by a MicroRNA pathway. Mol Cell 383:465–475. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee T, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. 2015. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 1632:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan G-L, Walbot V, Meyers BC.. 2015. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci U S A. 11210:3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xia R, Meyers BC, Walbot V.. 2015. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol. 27:84–90. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2212:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE.. 2012. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 199:870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Law JA.. 2015. RNA Pol IV and V in gene silencing: rebel polymerases evolving away from Pol II’s rules. Curr Opin Plant Biol. 27:154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.