Abstract
Posttranscriptional repression of UDP-glucuronosyltransferase (UGT) 2B7 and 2B15 expression by microRNAs (miRNAs) may be an important mechanism underlying inter-individual variability in drug glucuronidation. Furthermore, the UGT2B15 3’-UTR contains a common SNP (rs3100) that could influence miRNA binding. The aim of this study was to identify the complete complement of miRNAs that could regulate UGT2B7 and UGT2B15 expression through binding to the reference and/or variant 3’-UTRs. Luciferase reporter plasmids containing either the reference or variant 3’-UTRs were screened against a 2,048 human miRNA library to identify those miRNAs that decrease luciferase activity by at least 30% when co-transfected into HEK293 cells. Six novel miRNAs (miR-1293, miR-3664–3p, miR-4317, miR 513c-3p, miR-4483, and miR-142–3p) were identified that repressed the reference UGT2B7 3’-UTR, while twelve novel miRNAs (miR-770–5p, miR-103b, miR-3924, miR-376b-3p, miR-455–5p, miR-605, miR-624–3p, miR-4712–5p, miR-3675–3p, miR-6500–5p, miR-548as-3p, and miR-4292) repressed both the reference and rs3100 variant UGT2B15 3’-UTR. Deletion and mutagenesis studies confirmed the binding site location of each miRNA. Although the UGT2B15 rs3100 SNP was located within the miR-376c-3p response element, there was no effect on miRNA binding. miR 142–3p, miR-3664–3p, miR-4317, miR-455–5p, miR-376c-3p, miR-770–5p, miR-3675–3p, miR-331–5p, miR-605, and miR-376b-3p transcript levels were measured by quantitative PCR and correlated with UGT2B7 and UGT2B15 enzyme activities in 27 human liver samples. A significant negative correlation (Rs = -0.53; p = 0.005) was demonstrated between hepatic miR-455–5p transcript levels and UGT2B15-mediated S-oxazepam glucuronidation activities. Thus, the UGT2B7 and UGT2B15 3’-UTRs contain miRNA response elements for multiple miRNAs that may contribute to variable drug glucuronidation.
Keywords: UDP-glucuronosyltransferase, UGT2B7, UGT2B15, glucuronidation, microRNA, functional genomics
1. Introduction
The UDP-glucuronosyltransferases (UGTs) comprise a large family of conjugation enzymes capable of detoxifying a wide variety of both endogenous and exogenous substrates. The UGTs consist of 19 functional enzymes in humans that are subdivided by genetic similarity into three subfamilies, UGT1A, UGT2A, and UGT2B.The UGT2B subfamily includes 7 functional enzymes in humans (UGT2B4, 2B7, 2B10, 2B11, 2B15, 2B17, and 2B28), located on chromosome 4q13 which are expressed as individual genes each with a unique 3′-untranslated region (3’-UTR) [1]. The UGTs are critical for the efficient elimination of the majority of the top 200 prescribed drugs in the United States, ranked second only to the cytochrome P450 enzymes [2]. UGT2B7 is the most commonly listed UGT enzyme known to contribute tothe glucuronidation of the top 200 prescribed drugs [2]. Examples of clinically important drugs that are eliminated by UGT2B7-mediated glucuronidation include morphine and zidovudine (also called 3′-azido-3′-deoxythymidine or AZT) [3].Furthermore, UGT2B7 plays a major role in the glucuronidation of endogenous compounds such as steroids and retinoids [4–6]. UGT2B15 contributes to the glucuronidation of androgenic steroids and various drugs e.g. oxazepam and lorazepam [7, 8]. Interestingly the major elimination pathway of oxazepam in humans isthrough stereoselective hepatic glucuronidation mediated primarily by the UGT2B15 and UGT2B7 enzymes [8]. Specifically in human liver S-oxazepam is mainly glucuronidated by UGT2B15 while R-oxazepam is glucuronidated by both UGT2B7 and UGT1A9 [8, 9]. Furthermore, the glucuronidation of this drug by human subjects is known to be polymorphic although the molecular mechanisms responsible for this variability are only partly understood [9].
We demonstrated previously that the single nucleotide polymorphism (SNP) c.735A>G (UGT2B7*1c; rs28365062) in the UGT2B7 exon 2 explained only part of the relatively high interindividual variability in the clearance of a UGT2B7-specific probe substrate namely zidovudine, observed in HIV patients [10]. Furthermore, results from different groups showed that the SNPs located in the UGT2B7 promoter and coding region cannot explain completely the observed variability in the UGT2B7 mediated glucuronidation of various drugs [11, 12]. Previous work from our lab identified SNP D85Y (253G>T, rs1902023) in exon 1 of the UGT2B15 gene [13]. Moreover we demonstrated that the high frequency variant allele D85Y (allele frequency 47%) was a major determinant of the observed inter-individual variability ofoxazepam glucuronidation in vivo [14]. However, SNP D85Y could explain only 34% of the observed interindividual variability of hepatic oxazepam glucuronidation in vivo [14]. Consequently, other trans-acting factors could regulate UGT2B7 and UGT2B15 expression and contribute to this variability.
In recent years, microRNAs (miRNAs) have emerged as a crucial suppressive regulator of gene expression. miRNAs are a class of small non-coding RNAs that have been shown to control gene expression via imperfect base pairing to the 3’ UTR of target genes [15]. Accumulating data demonstrate that miRNAs modulate the expression of drug disposition genes, including cytochrome P450 enzymes, UGTs, and transporters [16]. Recently we showed using an unbiased genome-wide screen that five novel miRNA response elements (MREs) are located in the common UGT1A 3’-UTR [17]. In addition two recent studies identified and validated the functional MREs of miR-376c and miR-331–5p to the 3’-UTRs of UGT2B15 and UGT2B17 in prostate cancer cells [18, 19]. However there is still a large gap in our knowledge regarding the role of miRNAs in the regulation of two important members of the UGT2B subfamily, namely UGT2B15 and UGT2B7. Essentially we lack knowledge of the full complement of miRNAs other than miR-376c and miR-331–5p which are involved in the direct regulation of UGT2B7 and UGT2B15. Moreover, the studies that are based on a bioinformatics scan of the target gene 3’-UTR to identify novel miRNA candidates tend to bias the results to those miRNAs that match best using available bioinformatics algorithms, which have a significant false negative discovery rate because of the imperfect binding of miRNAs to their targets.Consequently, alternate unbiased genome-wide approaches are likely needed to reveal the full complement of miRNAs regulating the UGT2B7 and UGT2B15 enzyme.
The objective of the current work was to identify the complete complement of miRNAs that could regulate UGT2B7 and UGT2B15 expression through binding to the respective 3’-UTRs. For this purpose, we employed the luciferase reporter system to perform a genome-wide screen of the UGT2B7 and UGT2B15 3’-UTRs against a miRNA mimic library representing all of the available human miRNAs sequenced so far. This was followed by identification and functional validation of the miRNA response elements in the 3’-UTRs by a combination of in silico analysis and luciferase reporter construct assays. Furthermore we determined whether a common SNPin the UGT2B15 3’-UTR (rs3100) that was previously associated with UGT2B15 mRNA allelic imbalance [14] can disrupt miRNA binding. Finally, we measured the transcript amounts of the candidate miRNAs in human liver samples to determine whether they are correlated with hepatic UGT2B7 or UGT2B15 activities.
2. Materials and methods
2.1. Chemicals and reagents.
The miRNA mimic library (miRIDIAN microRNA Mimic Library, v.19), the miRNA mimic negative controls (Negative controls 1 and 2), the short interfering RNA directed against the UGT2B7 and UGT2B15 3’-UTR [positive controls: (siUGT2B7), 5’- AGA AGT CCA CTG ACA GTA T-3’ – (siUGT2B15) 5’- GAC CAA ATA GGA ACA GCT CCA -3’], and the transfection reagent DharmaFECT Duo were purchased from GE Dharmacon (Lafayette, CO).The pMIR-REPORT plasmid was from Life Technologies (Grand Island, NY) and the Renilla luciferase vector pRL-CMV was purchased from Promega (Madison, WI). All synthesized DNA oligos used in this study were obtained from Integrated DNA Technologies, Inc. (Coralville, IA).
2.2. Cell lines and culture conditions.
HEK293 cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Life Technologies). Cells were grown in a humidified chamber at 37oC and 5% CO2.
2.3. Construction of reporter plasmids.
The pMIR-UGT2B7 and pMIR-UGT2B15 luciferase plasmids containing the entire UGT2B7 3’-UTR (hg38, chr4: 69,112,737–69,113,408) and UGT2B15 3’-UTR (hg38, chr4: 68,646,196–68,647,103; contains the rs3100 c.1761 C allele) were de novo synthesized by Blue Heron (Bothell, WA). For the generation of all deletion constructs and the pMIR-UGT2B15 rs3100 c.1761 T allele variant vector we used the KAPA HiFi Polymerase (Kapa Biosystems, Inc., Wilmington, MA) together with the pMIR-UGT2B15 luciferase plasmid and the respective set of primers (Table 1). All the triplicated constructs were generated using synthesized 5’-end phosphorylated oligonucleotides that were annealed with their complementary sequence (Table 1) and cloned into the pMIRREPORT vector at the HindIII sites directly downstream of the firefly luciferase gene.All plasmids used in this study were verified by Sanger sequencing performed at the Washington State University Genomics Core.
Table 1.
List of primers used in this study. Underlined are shown the HindIII restriction sites used for cloning in the pMIR-REPORT vector. Bold are shown the respective MREs for each miRNA. All oligonucleotides used for the generation of the triplicate and deletion constructs were 5’-end phosphorylated.
| Primer | Sequence (5’ – 3’) | Comment |
|---|---|---|
| ΔMRE miR-142–3p F | GAAAATAAAAAATAAGATAAAGCCTTATGAGCTCG | Used to generate the pMIR-UGT2B7 Δ142 construct. |
| ΔMRE miR-142–3p R | GCCACAAAATATTTCTAACCATGAACTGGG | |
| miR-142–3p MRE 3× F |
AGCTTAATGAAGAAAACACTACG TATGTCAATGAAGAAAACACTACG TATGTCAATGAAGAAAACACTACGA |
Used to generate the 142 3× F/R constructs. |
| miR-142–3p MRE 3× R |
AGCTTCGTAGTGTTTTCTTCATTGAC
ATACGTAGTGTTTTCTTCATTGACATACG TAGTGTTTTCTTCATTA |
|
| ΔMRE miR-513c-3p F | TGAACTTATATTGAAATTTGTTGTTCTAATTCAC | Used to generate the pMIR-UGT2B7 Δ513 construct. |
| ΔMRE miR-513c-3p R | TCATAAGGCTTTATCTTATTTTTTATTTTCCG | |
| miR-513c-3p MRE 3× F |
AGCTTGCTCGTATTGAAATTTGTGAGCTA
GCTCGTATTGAAATTTGTGAGCTA GCTCGTATTGAAATTTGTA |
Used to generate the 513c 3× F/R constructs. |
| miR-513c-3p MRE 3× R |
AGCTTACAAATTTCAATACGAGCTAGCTC
ACAAATTTCAATACGAGCTAGCTC ACAAATTTCAATACGAGCA |
|
| ΔMRE miR-1293/4483 F | GTTCATGGTTAGAAATATTTTGTGGCAATG | Used to generate the pMIR-UGT2B7 Δ1293/4483 construct. |
| ΔMRE miR-1293/4483 R | AAATTTTTATTTGACAAAGTAAATTTTGAAAAG | |
| miR-1293/4483 MRE 3× F |
AGCTTGTTTTTCAGAGATTTACCACCCA
TATGTCGTTTTTCAGAGATTTACCACCCA TATGTCGTTTTTCAGAGATTTACCACCCAA |
Used to generate the 1293/4483 3× F/R constructs. |
| miR-1293/4483 MRE 3× R |
AGCTTTGGGTGGTAAATCTCTGAAAAAC
GACATATGGGTGGTAAATCTCTGAAAAAC GACATATGGGTGGTAAATCTCTGAAAAACA |
|
| ΔMRE miR-3664–3p F | CAAAAAAAAAAAAAGAAAAAAAAATCTTTTCAAAATTTAC | Used to generate the pMIR- UGT2B7 Δ3664 construct. |
| ΔMRE miR-3664–3p R | ATCACAATCTTTCTTGCTGGAATAAACTG | |
| miR-3664–3p MRE 3× F |
AGCTTGCAAGATTTCTTCTTCCTGAGA
TATGTCGCAAGATTTCTTCTTCCTGAGA TATGTCGCAAGATTTCTTCTTCCTGAGAA |
Used to generate the 3664 3× F/R constructs. |
| miR-3664–3p MRE 3× R |
AGCTTTCTCAGGAAGAAGAAATCTTGC
GACATATCTCAGGAAGAAGAAATCTTGC GACATATCTCAGGAAGAAGAAATCTTGCA |
|
| ΔMRE miR-376c-3p F | AATGATTTTTAAGCTATGAAAAATACAATGGG | Used to generate the pMIR-UGT2B15 Δ376c construct. |
| ΔMRE miR-376c-3p R | AGCTAAAGTACGTATTAAATCCCTGG | |
| miR-376c-3p MRE 3× F |
AGCTTGAATTATTCTATGTCCCGGAA
GAATTATTCTATGTCCCGGAA GAATTATTCTATGTCA |
Used to generate the 376c 3× F/R constructs. |
| miR-376c-3p MRE 3× R |
AGCTTGACATAGAATAATTCTTCCGG
GACATAGAATAATTCTTCCGGGACATAGAAT AATTCA |
|
| ΔMRE miR-3924 F | TAGCTAACCTGCACGTTGTGCACATGTACCC | Used to generate the pMIR-UGT2B15 Δ3924 construct. |
| ΔMRE miR-3924 R | TGTTGGCGTGCTGCATCCAGTAACTCGTC | |
| miR-3924 MRE 3× F |
AGCTTTGGCACATGTATACATATGGATCCT
TGGCACATGTATACATATGGATCCT TGGCACATGTATACATATGA |
Used to generate the 3924 3× F/R constructs. |
| miR-3924 MRE 3× R |
AGCTTCATATGTATACATGTGCCAAGGATC
CATATGTATACATGTGCCAAGGATCCATATGTAT ACATGTGCCAA |
|
| ΔMRE miR-548as-3p F | ATGGAGGATTTCCTTTTTCCTGTGACAAAAC | Used to generate the pMIR-UGT2B15 Δ548 construct. |
| ΔMRE miR-548as-3p R | GAAATAAAGGAGGAGTCCCATCTTTCAGTC | |
| miR-548as-3p MRE 3× F |
AGCTTAGCATGGAGGGTTTTAAGACATA
AGCATGGAGGGTTTTAAGACATA AGCATGGAGGGTTTTAAA |
Used to generate the 548as 3× F/R constructs. |
| miR-548as-3p MRE 3× R |
AGCTTTTAAAACCCTCCATGCTTATGTC
TTAAAACCCTCCATGCTTATGTC TTAAAACCCTCCATGCTA |
|
| ΔMRE miR-455–5p F | ATACATATGTAGCTAACCTGCACGTTGTGC | Used to generate the pMIR-UGT2B15 Δ455 construct. |
| ΔMRE miR-455–5p R | AGTAACTCGTCATTTAACATTAGGTATATCTCCAAATGC | |
| miR-455–5p MRE 3× F |
AGCTTGGATGCAGCACGCCAACATGGCACATGTGACATA
GGATGCAGCACGCCAACATGGCACATGTGACATA GGATGCAGCACGCCAACATGGCACATGTA |
Used to generate the pMIR-UGT2B15 Δ455 construct. |
| miR-455–5p MRE 3× R |
AGCTTACATGTGCCATGTTGGCGTGCTGCATCC
TATGTCACATGTGCCATGTTGGCGTGCTGCATCC TATGTCACATGTGCCATGTTGGCGTGCTGCATCCA |
|
| ΔMRE miR-624–3p F | AAGACAAAATTTATTTTCCAGGGATTTAATACGTAC | Used to generate the pMIR-UGT2B15 Δ624 construct. |
| ΔMRE miR-624–3p R | GTGAAAAGATGTTTTGTCACAGGAAAAAGGAAATCC | |
| miR-624–3p MRE 3× F |
AGCTTAACTTACCTTGTTGACAAACTTACCTTGTT GACAAACTTACCTTGTTA |
Used to generate the 624 3× F/R constructs. |
| miR-624–3p MRE 3× R |
AGCTTAACAAGGTAAGTTTGTCAACAAGGTAAGTTT GTCAACAAGGTAAGTTA |
|
| ΔMRE miR-6500–5p F | TTTGGAGATATACCTAATGTTAAATGACGAGTTACTGG | Used to generate the pMIR-UGT2B15 Δ6500 construct. |
| ΔMRE miR-6500–5p R | TTTCATAGCTTAAAAATCATTGACATAGAATAATTCAGC | |
| miR-6500–5p MRE 3× F |
AGCTTAATACAATGGGGGGAAGGATAGCAGACATA
AATACAATGGGGGGAAGGATAGCAGACATA AATACAATGGGGGGAAGGATAGCAA |
Used to generate the 6500 3× F/R constructs. |
| miR-6500–5p MRE 3× R |
AGCTTTGCTATCCTTCCCCCCATTGTATTTATGTC
TGCTATCCTTCCCCCCATTGTATTTATGTC TGCTATCCTTCCCCCCATTGTATTA |
|
| ΔMRE miR-4712–5p F | GCAGCACGCCAACATGGCACATGTATAC | Used to generate the pMIR-UGT2B15 Δ4712 construct. |
| ΔMRE miR-4712–5p R | AACATTAGGTATATCTCCAAATGCTATCCTTCCC | |
| miR-4712–5p MRE 3× F |
AGCTTAAATGACGAGTTACTGGATGACATA
AAATGACGAGTTACTGGATGACATA AAATGACGAGTTACTGGATA |
Used to generate the 4712 3× F/R constructs. |
| miR-4712–5p MRE 3× R |
AGCTTATCCAGTAACTCGTCATTTTATGTC
ATCCAGTAACTCGTCATTTTATGTC ATCCAGTAACTCGTCATTTA |
|
| ΔMRE miR-3675–3p F | TACCTAATGTTAAATGACGAGTTACTGGAdTGC | Used to generate the pMIR-UGT2B15 Δ3675 construct. |
| ΔMRE miR-3675–3p R | TTGTATTTTTCATAGCTTAAAAATCATTGACATAGAATAATTC | |
| miR-3675–3p MRE 3× F |
AGCTTTGGGGGGAAGGATAGCATTTGGAGATACATAGC
TGGGGGGAAGGATAGCATTTGGAGATACATAGC TGGGGGGAAGGATAGCATTTGGAGATAA |
Used to generate the 3675 3× F/R constructs. |
| miR-3675–3p MRE 3× R |
AGCTTTATCTCCAAATGCTATCCTTCCCCCCAGCTATG
TATCTCCAAATGCTATCCTTCCCCCCAGCTATG TATCTCCAAATGCTATCCTTCCCCCCAA |
|
| ΔMRE miR-605–5p F | TACGTACTTTAGCTGAATTATTCTATGTCAATG | Used to generate the pMIR-UGT2B15 Δ605 construct. |
| ΔMRE miR-605–5p R | AGGTAAGTTGTGAAAAGATGTTTTGTCACAGG | |
| miR-605–5p MRE 3× F |
AGCTTTGTTAAGACAAAATTTATTTTCCAGGGATTTAA
GCTAGCTGTTAAGACAAAATTTATTTTCCAGGGATTTAA GCTAGCTGTTAAGACAAAATTTATTTTCCAGGGATTTAAA |
Used to generate the 605 3× F/R constructs. |
| miR-605–5p MRE 3× R |
GCTTTTAAATCCCTGGAAAATAAATTTTGTCTTAACA
GCTAGCTTAAATCCCTGGAAAATAAATTTTGTCTTAACA GCTAGCTTAAATCCCTGGAAAATAAATTTTGTCTTAACAsA |
|
| ΔMRE miR-376b-3p F | AATACAATGGGGGGAAGGATAGCATTTG | Used to generate the pMIR-UGT2B15 Δ376b construct. |
| ΔMRE miR-376b-3p R | TGACATAGAATAATTCAGCTAAAGTACG | |
| miR-376b-3p MRE 3× F |
AGCTTATGATTTTTAAGCTATGAAAGAGTGC
ATGATTTTTAAGCTATGAAAGAGTGC ATGATTTTTAAGCTATGAAAA |
Used to generate the 376b 3× F/R constructs. |
| miR-376b-3p MRE 3× R |
AGCTTTTTCATAGCTTAAAAATCATGCACTC
TTTCATAGCTTAAAAATCATGCACTC TTTCATAGCTTAAAAATCATA |
|
| ΔMRE miR-770–5p F | TGCAGCACGCCAACATGGCACATGTATACATATG | Used to generate the pMIR-UGT2B15 Δ770 construct. |
| ΔMRE miR-770–5p R | ATTAGGTATATCTCCAAATGCTATCCTTCCCC | |
| miR-770–5p MRE 3× F |
AGCTTGTTAAATGACGAGTTACTGGAGATACT
GTTAAATGACGAGTTACTGGAGATACT GTTAAATGACGAGTTACTGGAA |
Used to generate the 770 3× F/R constructs. |
| miR-770–5p MRE 3× R |
AGCTTTCCAGTAACTCGTCATTTAACAGTATC
TCCAGTAACTCGTCATTTAACAGTATC TCCAGTAACTCGTCATTTAACA |
|
| ΔMRE miR-4292 F | TTTAATACGTACTTTAGCTGAATTATTCTATGTCAATG | Used to generate the pMIR-UGT2B15 Δ4292 construct. |
| ΔMRE miR-4292 R | AATAAATTTTGTCTTAACAAGGTAAGTTGTGAAAAG | |
| miR-4292 MRE 3× F |
AGCTTTTCCAGGGATAGCTTCCAGGGA TAGCTTCCAGGGATAGCA |
Used to generate the 4292 3× F/R constructs. |
| miR-4292 MRE 3× R |
AGCTTGCTATCCCTGGAAGC
TATCCCTGGAAGCTATCCCTGGAAA |
|
| ΔMRE miR-103b F | AGCATTTGGAGATATACCTAATGTTAAATGACG | Used to generate the pMIR-UGT2B15 Δ103b construct. |
| ΔMRE miR-103b R | AAAAATCATTGACATAGAATAATTCAGCTAAAGTACG | |
| miR-103b MRE 3× F |
AGCTTTTTTTAAGCTATGAAAAATACAATGGGGGG
AAGGATTGTACCTTTTTAAGCTATGAAAAATACAATGGGGGG AAGGATTGTACCTTTTTAAGCTATGAAAAATACAATGGGGGG AAGGATA |
Used to generate the 103b 3× F/R constructs. |
| miR-103b MRE 3× R |
AGCTTATCCTTCCCCCCATTGTATTTTTC
ATAGCTTAAAAAGGTACAATCCTTCCCCCCATTGTATTTTTC ATAGCTTAAAAAGGTACAATCCTTCCCCCCATTGTATTTTTC ATAGCTTAAAAAA |
|
| ΔMRE miR-331–5p F | ATGTTAAATGACGAGTTACTGGATGC | Used to generate the pMIR-UGT2B15 Δ331 construct. |
| ΔMRE miR-331–5p R | TTCCCCCCATTGTATTTTTCATAGC | |
| miR-331–5p MRE 3× F |
AGCTTGGATAGCATTTGGAGATATACCTAACGCCTT
GGATAGCATTTGGAGATATACCTAACGCCTT GGATAGCATTTGGAGATATACCTAAA |
Used to generate the 331 3× F/R constructs. |
| miR-331–5p MRE 3× R |
AGCTTTTAGGTATATCTCCAAATGCTATCCAAGGCG
TTAGGTATATCTCCAAATGCTATCCAAGGCG TTAGGTATATCTCCAAATGCTATCCA |
|
| rs3100 F | GGGATTTAATACGTACTTTAGTTGAATTATTCTATGTCAATG | Used to generate the pMIR-UGT2B15 rs3100 construct. |
| rs3100 R | CATTGACATAGAATAATTCAACTAAAGTACGTATTAAATCCC | |
| rs7660719 F | GAACTCCACTCATAGTATGAACATTGTTTTGCAAATAC | Used to generate the pMIR-UGT2B15 rs7660719 construct. |
| rs7660719 R | GTATTTGCAAAACAATGTTCATACTATGAGTGGAGTTC | |
| rs13145346 F | GGTGCAGCACACTGTGTGTGAGCTGAAGCAGGGC | Used to generate the pMIR-UGT2B15 rs13145346 construct. |
| rs13145346 R | GCCCTGCTTCAGCTCACACACAGTGTGCTGCACC |
2.4. Luciferase reporter assays.
For all luciferase assays HEK293 cells were co-transfected with a firefly luciferase construct, the pRL-CMV Renilla luciferase control vector and the miRNA mimic or one of the mimic controls (negative or positive) as described previously [17].
2.5. Quantitative Real-time PCR (qPCR).
Total RNA was isolated from human liver samples using Trizol (Life Technologies). Mature miRNAs were isolated from individual liver samples of our human liver bank (n=27) using the mirVana™ miRNA Isolation Kit, with phenol (Life Technologies) according to the manufacturers’ instructions. The source of the liver bank tissues have been published recently [20, 21]. These de-identified liver samples (Table 2) were obtained from publicly available sources, including the National Disease Research Interchange (Philadelphia, PA) and the Liver Tissue Procurement and Distribution Service (Minneapolis, MN). The use of these tissues for this study was approved by the Washington State University Institutional Review Board. mRNA and mature miRNA concentrations were determined by qPCR using reverse transcribed total RNA. For mRNA, cDNAs were first synthesized from total RNA using TaqMan Reverse Transcription Reagents (Life Technologies). qPCR was performed using TaqMan Gene Expression Master Mix and Taqman primer-probe mixes from Life Technologies, including UGT2B15 (Hs00870076_s1) and GAPDH (Hs02758991_g1). Mature miRNAs were also quantified from total RNA after reverse transcription with miRNA-specific primers and MultiScribe Reverse Transcriptase (Life Technologies). qPCR was performed using TaqMan Universal Master Mix II, no UNG and Taqman miRNA assays from Life Technologies (Table 3). All assays were performed in triplicate. Gene expression was compared to an endogenous internal control (GAPDH for mRNA or the geometric mean of miR-1521 3p/miR-23b for miRNA as described previously [22]. There was not commercially available a set of primers and probes of the Applied Biosystems™ TaqMan™ microRNA assays for miR-4292 and miR-3924. Thus we used the GTEx database (http://www.gtexportal.org/) to determine the hepatic transcript amounts of these miRNAs. The ΔCt values for each miRNA were calculated this way for all of the samples in our human liver bank (n=27). Subsequently these values were normalized to the liver sample with the highest amount for each miRNA. These normalized values were then used to determine the correlation between the miRNA amounts in human liver samples and the hepatic activities of UGT2B7 and UGT2B15.
Table 2.
Demographics of liver donors (n=27).
| Donor identity | Age | Gender | Ethnicitya | Cause of death | Smoking | Alcoholb | Other drug exposurec |
|---|---|---|---|---|---|---|---|
| LV2 | 8 | F | A | Head trauma | No | No | |
| LV3 | 5 | M | C | Meningitis | No | No | Dopamine, penicillin |
| LV4 | 24 | F | C | Auto accident | Dopamine | ||
| LV5 | 20 | M | C | Head trauma | Yes | No | Nafcillin, mannitol, furosemide, erythromycin, nystatin, neomycin |
| LV6 | 35 | F | C | Head trauma | No | No | None |
| LV7 | 21 | M | A | Head trauma | Yes | Yes | DDAVP |
| LV16 | 45 | F | H | Stroke | No | No | Dopamine |
| LV17 | 6 | M | C | Auto accident | No | No | Dopamine |
| LV23 | 74 | M | C | Colon cancer | |||
| LV25 | 49 | F | C | Stroke | Yes | No | Diazepam, furosemide |
| LV30 | 40 | F | C | Head trauma | Yes | Yes | Mannitol, furosemide. phentolamine |
| LV32 | 45 | F | C | Stroke | Yes | Yes | Prednisone, synthroid, phenytoin, naproxen, diazepam |
| LV36 | 43 | M | C | Stroke | No | No | Memiddlerolol, dexamethasone, hydrocortisol |
| LV39 | 36 | M | C | Head trauma | Yes | Yes | Cefazolin, norcuronium, phytonadione, enalapril, metoclopramide |
| LV40 | 34 | M | C | Gun shot wound | Yes | No | Crack cocaine, dopamine, norcuronium, fentanyl, midazolam |
| LV41 | 43 | M | C | Head trauma | No | Yes | Dopamine, furosemide, neosynephrine, acetaminophen |
| LV42 | 35 | M | C | Encephalopathy | No | No | Dopamine, norepinephrine, naloxone |
| LV43 | 24 | M | C | Blunt trauma | No | No | |
| LV45 | 75 | F | C | Stroke | No | No | Dopamine, vasopressin, furosemide, mannitol, vecuronium |
| LV46 | 49 | M | C | Stroke | Yes | Yes | Occasional crack cocaine and marijuana |
| LV48 | 68 | F | C | Stroke | No | No | Synthroid, celecoxib, fluoxetine, atenolol, omeprazole |
| LV49 | 21 | M | C | Head trauma | Yes | No | Crack cocaine, marijuana |
| LV50 | 37 | M | C | Head trauma | Yes | Yes | Marijuana |
| LV51 | 62 | M | C | Stroke | No | No | Dopamine, norepinephrine, insulin, DDAVP, cefasolin |
| LV53 | 54 | M | C | Brain abscess | No | No | Simvastatin, acetaminophen, alprazolam, flurazepam, venlafaxine |
| LV54 | 63 | M | C | Stroke | No | Yes | Dopamine, norepinephrine, nitroprusside |
| LV55 | 46 | M | C | Stroke | No | No |
C, Caucasian; A, African-American; H, Hispanic.
History of consumption of more than 14 drinks per week.
DDAVP, desmopressin acetate.
Table 3.
List of Taqman miRNA assays (Life Technologies) used to measure the hepatic concentrations of miRNAs.
| miRNA | Assay ID |
|---|---|
| miR-4483 | 461903_mat |
| miR-142-3p | 000464 |
| miR-4317 | 243568_mat |
| miR-3664-3p | 464880_mat |
| miR-1293 | 002298 |
| miR-513c-3p | 472280_mat |
| miR-152-3p | 000475 |
| miR-23b | 002126 |
| miR-331-5p | 002233 |
| miR-770-5p | 002002 |
| miR-103b | 121115_mat |
| miR-376b-3p | 001102 |
| miR-376c-3p | 002122 |
| miR-455-5p | 001280 |
| miR-605 | 466447_mat |
| miR-624-3p | 002430 |
| miR-4712-5p | 463735_mat |
| miR-3675-3p | 461795_mat |
| miR-6500-5p | 474759_mat |
| miR-548as-3p | 471738_mat |
2.6. Statistical analysis.
Statistical analysis was performed with a two-tailed Student’s t test using SigmaPlot Version 12 (Systat Software, Inc., San Jose, CA). Correlation analysis was performed by a Spearman’s rank method. A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. A genome-wide approach identifies novel miRNAs that decrease the UGT2B7 and UGT2B15 3’-UTR-dependent luciferase reporter activity.
To identify candidate miRNAs involved in the regulation of UGT2B7 and UGT2B15 expression via direct binding to the respective 3’-UTRs, we used a commercially available miRNA mimic library (miRIDIAN microRNA Mimic Library, v.19) that contained all human miRNAs sequenced at that time (2,048 miRNAs as listed in miRBase version 19). We transiently co-transfected pMIR-UGT2B7 or the pMIR-UGT2B15 luciferase reporter plasmids and the pRL-CMV vector (Renilla transfection control) together with the miRNA mimics or the respective positive (siUGT2B7, siUGT2B15) and negative controls into HEK293 cells. This cell line is ideal for the heterologous expression of UGT2B7 and UGT2B15 since the mRNA transcript amounts of these enzymes in HEK293 cells are very low [23].
Subsequently we determined the effect of each miRNA mimic on the UGT2B7 and UGT2B15 3’-UTR-dependent luciferase activities and classified them according to the degree of decrease of the relative UGT 3’-UTR-mediated luciferase activity. We chose to study further the miRNAs that showed at least 30% decrease in the relative UGT 3’-UTR-dependent luciferase activity in the initial library screen [19 miRNAs for UGT2B7 (Table 4 and 20 miRNAs for UGT2B15 (Table 5)]. Although miR-331–5p and miR-376c-3p caused a 38% and 54% decrease respectively in UGT2B15 3’-UTR-dependent luciferase activity we did not study these miRNAs further since recent studies demonstrated that they can regulate UGT2B15 expression [18, 19]. After counter-screening with the empty pMIR-REPORT vector to exclude nonselective effects, only six miRNAs for UGT2B7 (miR-1293, miR-3664–3p, miR-4317, miR-513c-3p, miR-4483, and miR-142–3p) and twelve for UGT2B15 (miR-770–5p, miR-103b, miR-3924, miR-376b-3p, miR-455–5p, miR-605, miR-624–3p, miR-47125p, miR-3675–3p, miR-6500–5p, miR-548as-3p, and miR-4292) were found to significantly reduce the luciferase activity of cells transfected with pMIR-UGT2B7 and pMIR-UGT2B15 luciferase reporter plasmids (Fig. 1A, 1B).
Table 4.
miRNAs identified to decrease the UGT2B7-3’-UTR-mediated luciferase reporter activity by at least 30% through library screening. Decrease in luciferase activity is represented relative to the average luciferase activities of non-transfected HEK293 cells and cells transfected with the miRNA mimic negative controls. In bold are the 6 novel candidate miRNAs identified in this study. Other listed miRNAs were not evaluated further since they showed a similar amount of inhibition of luciferase activity in a counterscreen of cells transfected with luciferase reporter lacking the UGT1A-3’UTR and so were considered false positives.
| miRNA | Decrease in luciferase activity relative to the miRNA negative control (%) |
|---|---|
| miR-18b-3p | 65.4 |
| miR-3687 | 65.1 |
| miR-34b-5p | 54.6 |
| miR-30c-2–3p | 54.5 |
| miR-30c-1–3p | 50.0 |
| miR-1293 | 44.2 |
| miR-550a-3–5p | 43.9 |
| miR-3670 | 40.8 |
| miR-3664–3p | 40.1 |
| miR-4317 | 39.5 |
| miR-129–5p | 38.4 |
| miR-4531 | 36.5 |
| miR-513c-3p | 36 |
| miR-4483 | 31.6 |
| miR-1294 | 33.3 |
| miR-4259 | 31.5 |
| miR-28–3p | 30.6 |
| miR-4419a | 30.3 |
| miR-142–3p | 30 |
Table 5.
miRNAs identified to decrease the UGT2B15’-UTR-mediated luciferase reporter activity by at least 30% through library screening. Decrease in luciferase activity is represented relative to the average luciferase activities of non-transfected HEK293 cells and cells transfected with the miRNA mimic negative controls. In bold are the 12 novel candidate miRNAs identified in this study. Underlined are miR-376–3p and miR-331–5p which had been identified in a previous study by an in silico approach. Other listed miRNAs were not evaluated further since they showed a similar amount of inhibition of luciferase activity in a counter-screen of cells transfected with luciferase reporter lacking the UGT1A-3’UTR and so were considered false positives.
| miRNA | Decrease in luciferase activity relative to the miRNA negative control (%) |
|---|---|
| miR-1262 | 62.3 |
| miR-331–5p | 62.1 |
| miR-770–5p | 54.4 |
| miR-103b | 53.9 |
| miR-3942–5p | 53.9 |
| miR-3924 | 52.9 |
| miR-30c-2–3p | 50.1 |
| miR-376b-3p | 47.8 |
| miR-376c-3p | 46.4 |
| miR-30c-1–3p | 45.1 |
| miR-455–5p | 42.6 |
| miR-605 | 42.6 |
| miR-624–3p | 41.6 |
| miR-4712–5p | 40.6 |
| miR-3675–3p | 40.6 |
| miR-6500–5p | 39.7 |
| miR-548as-3p | 38.7 |
| miR-4292 | 37.9 |
| miR-744–5p | 35.5 |
| miR-3678–5p | 31.1 |
Figure 1.
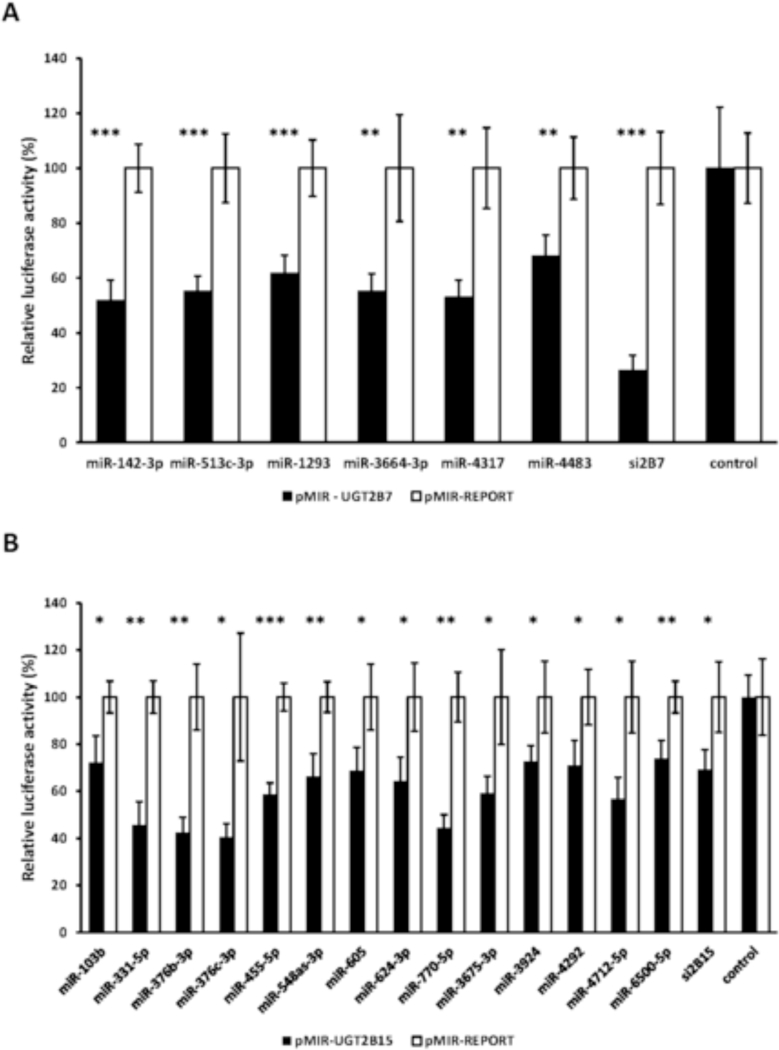
miRNAs identified through a functional genomics library screen that reduce UGT2B7 (1A) and UGT2B15 3’-UTR (1B) luciferase reporter (pMIR-UGT2B7and pMIR-UGT2B15) activities by at least 30% without affecting activity of the empty vector (pMIR). Shown are relative luciferase activities (firefly/Renilla ratio normalized to the pMIR control ratio) of HEK293 cells co-transfected with the pMIR-UGTB7/ pMIR-UGT2B15 or pMIR and with each miRNA mimic, a positive control (siUGT2B7, siUGT2B15: short interfering RNAs directed against the UGT2B7 and UGT2B15 3’ UTRs respectively) or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
3.2. The UGT2B7 and UGT2B15 3’-UTRs contain functional microRNA response elements (MREs) for the novel candidate miRNAs.
miRanda and RNAhybrid are well-verified online bioinformatics programs that predict the location of MREs in the 3’-UTRs of target mRNAs [24–26]. Using these programs, we identified putative MREs in the UGT2B7 and UGT2B15 3’-UTRs for each of the candidate miRNAs (Fig. 2). To confirm the functionality of the candidate MREs in the 3’-UTRs we generated constructs that carried the full length UGT2B7 and UGT2B15 3’-UTRs with a deletion (Δ) of the predicted MREs cloned directly downstream of the firefly luciferase gene. We also cloned each predicted MRE sequence as a triplicate in the forward or reverse (negative control) orientation in the pMIR-REPORT firefly luciferase vector. We then transiently co-transfected the full length, deletion, triplicate constructs or the empty pMIR-REPORT vector with miRNA mimics) or the miRNA mimic negative controls into HEK293 cells and quantified the luciferase activity of each construct. As shown in Figure 3 all miRNA mimics tested significantly reduced the luciferase activity of constructs carrying the full length UGT2B7 and UGT2B15 3’-UTRs (left set of columns in each panel A-T). However, these reductions were abrogated when the respective MRE was deleted (Figure 3; second set of columns from left in each panel A-T). Moreover, cells transfected with the miRNA mimics and pMIR constructs carrying the MREs in the forward orientation showed a significant decrease in the luciferase activity compared with the control, while those with MREs in the reverse orientation showed no miRNA mimic effect (Figure 3; third and fourth set of columns from left in each panel A-T). As expected, none of the miRNA mimics affected luciferase activity of the empty pMIR-REPORT vector (Figure 3; right set of columns in each panel A-T).
Figure 2.
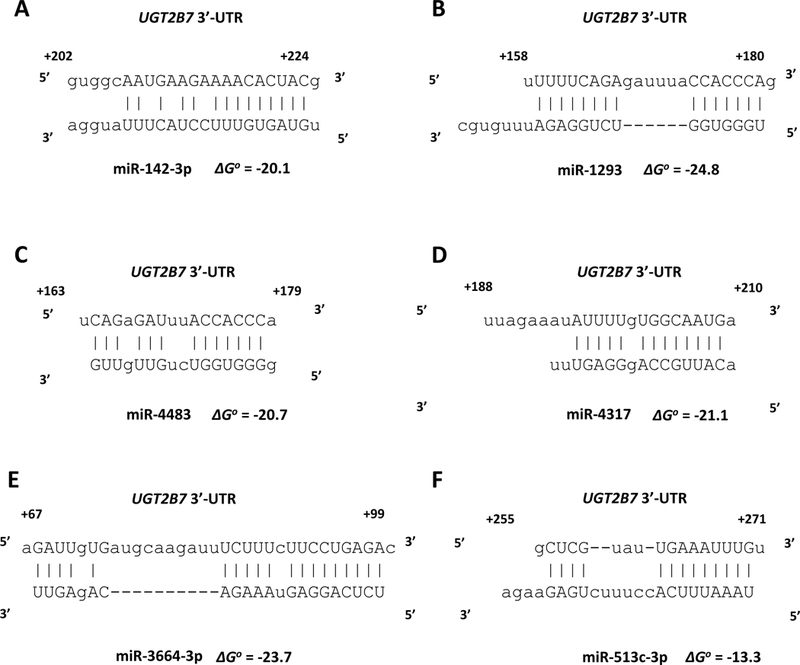
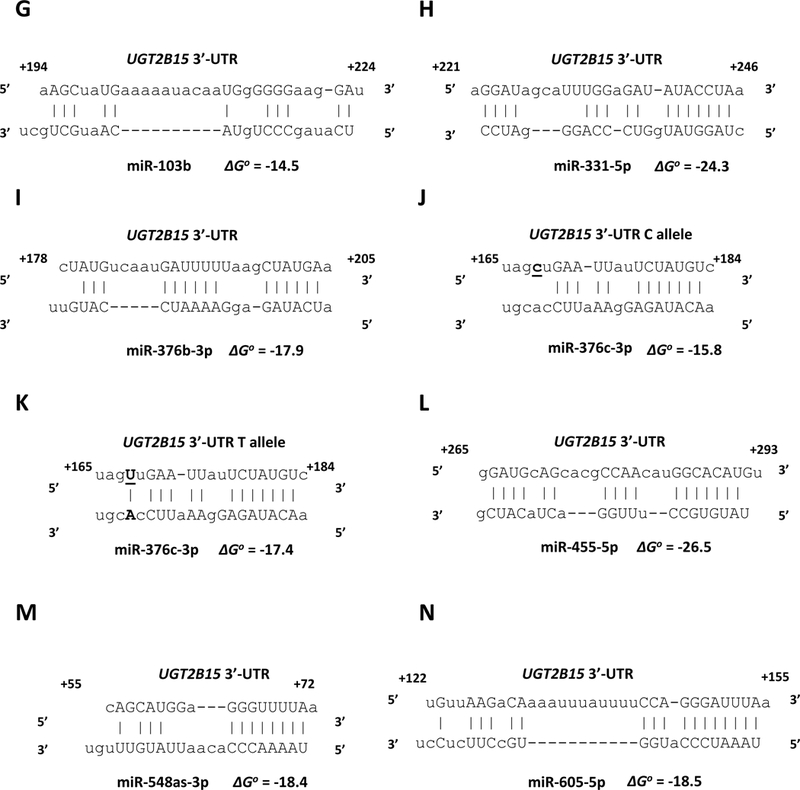
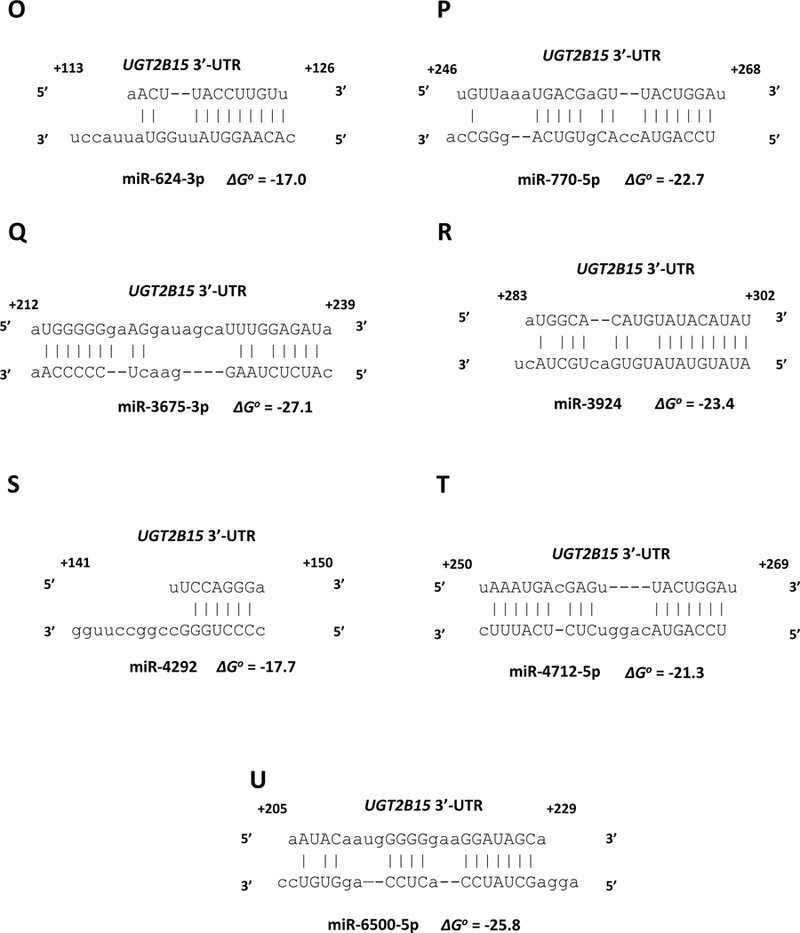
microRNA response elements identified in the UGT2B7 3’-UTR (A-F) and UGT2B15 3’-UTR (G-U) by in silico analysis for the miRNAs shown to reducethe UGT2B7 and UGT2B15-dependent 3’-UTR luciferase reporter activity by at least 30% without affecting an empty luciferase reporter. Nucleotide positions in the mRNA are indicated relative to the last nucleotide of the stop codon. The binding free energy values (ΔGo) between the miRNAs and the UGT2B7 and UGT2B15 3’-UTRs are expressed in kcal/mol and were calculated using RNAhybrid.
Figure 3.
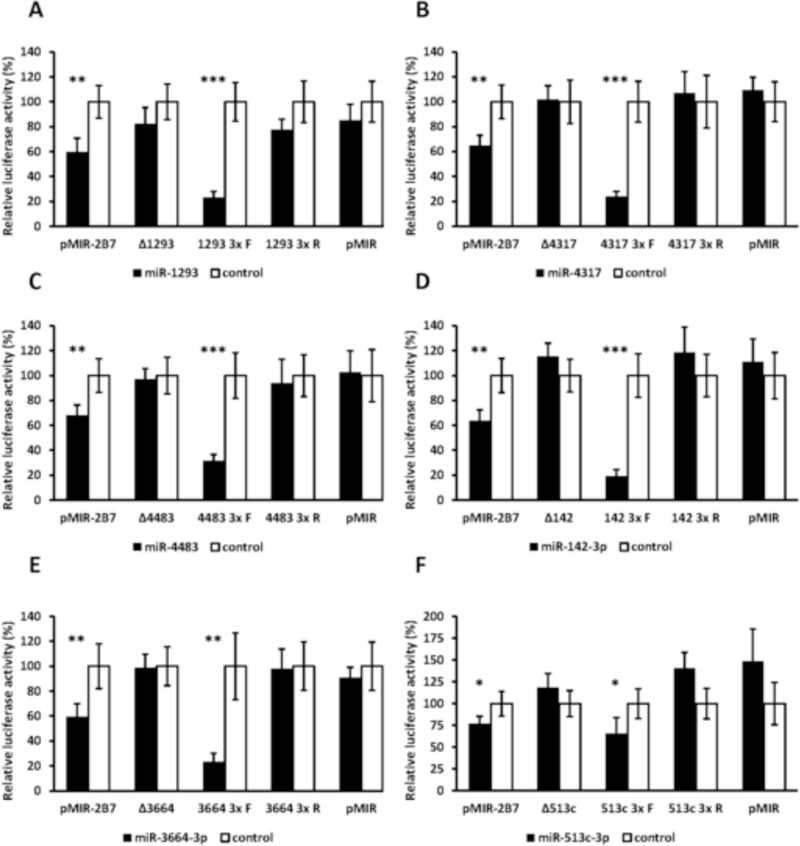
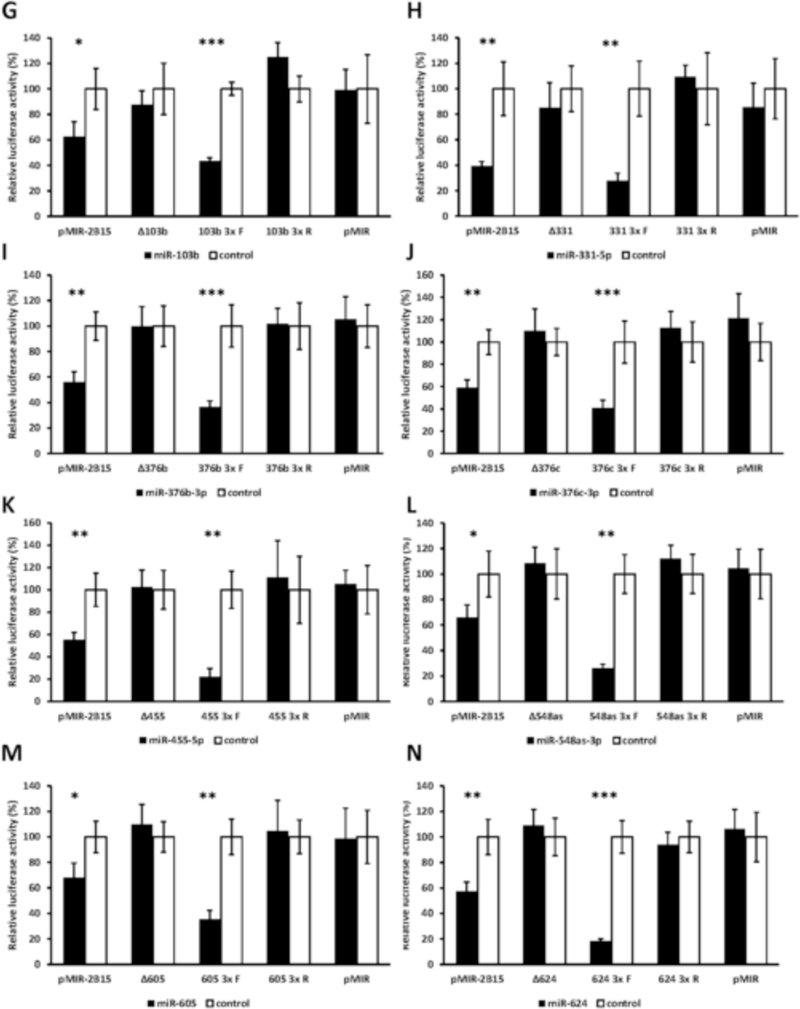
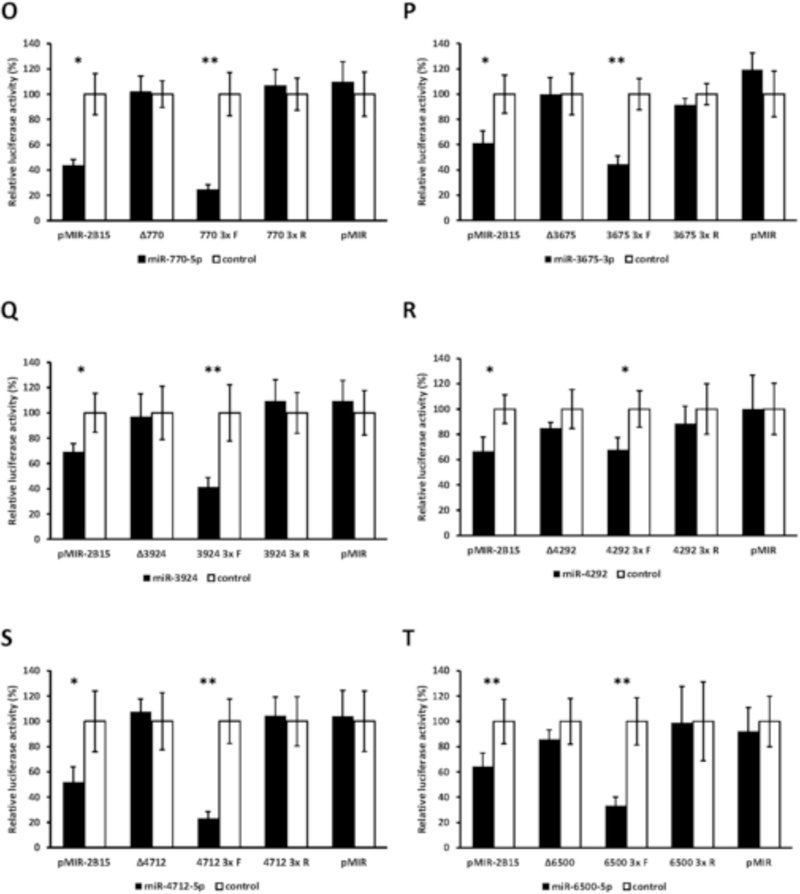
Functional validation of novel miRNA response elements (MREs; shown in Figure 2) in the UGT2B7 and UGT2B15 3’-UTRs. For each miRNA evaluated luciferase reporter constructs were generated containing either the full length UGT2B7 (panels A-E) or UGT2B15 3’-UTR (panels F-T), a deletion (Δ) of the predicted MRE, or the triplicated MRE in the forward or reverse (negative control) orientation. Shown are relative luciferase activities (firefly/Renilla ratio normalized to the miRNA control) of HEK293 cells co-transfected with the luciferase reporter and either the miRNA mimic or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
3.3. Effect of the rs3100 single nucleotide polymorphism (SNP) in the UGT2B15 3’-UTR on miRNA binding.
We next sought to determine whether the common SNP rs3100 c.1761 C/T [27] present in the UGT2B15 3’-UTR had an effect on binding of any of the candidate UGT2B15-regulating miRNAs. Interestingly, in silico analysis using the bioinformatics program miRanda determined that the rs3100 SNP was located within the predicted miR-376c-3p MRE (Fig. 2, panels J, K). Furthermore, the variant c.1761T allele (Fig. 2, panel K) appeared to create an additional base-pairing site between the UGT2B15 3’-UTR and miR-376c-3p when compared with the reference c.1761C allele (Fig. 2, panel J). Consequently, lower luciferase activities would have been expected for the pMIR-UGT2B15-variant construct compared with the reference pMIR-UGT2B15. However, as shown in Figure 4 luciferase activities were reduced by all the candidate UGT2B15-regulating miRNAs (including miR-376c-3p) in both the pMIR-UGT2B15 reference and variant constructs to the same extent indicating this SNP does not affect regulation by any of these miRNAs.
Figure 4.
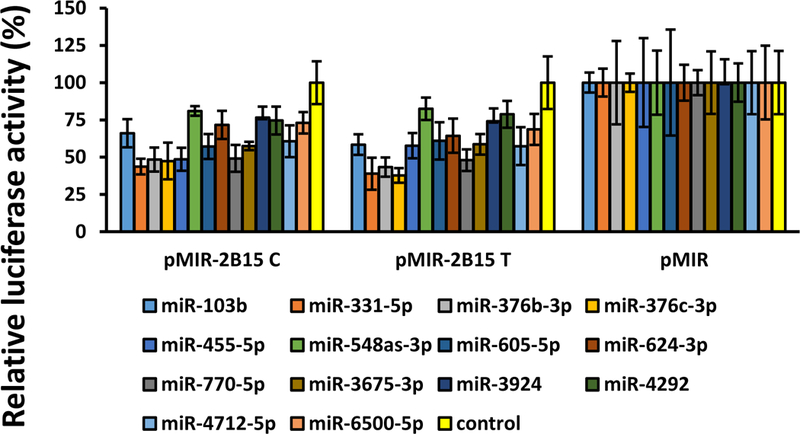
Effect of the common UGT2B15 3’-UTR rs3100 SNP on the binding of miRNAs to the reference 3’-UTR.
3.4. Expression of candidate miRNAs in human liver bank samples and correlation with hepatic UGT2B7 and UGT2B15 activity.
qPCR was then used to determine which of the candidate miRNAs are normally expressed in human liver. Mean (± SD) threshold cycle (Ct) values for each miRNA were determined using RNA extracted from 27 of our human liver bank samples (Table 2). For the UGT2B7 candidate miRNAs relatively high transcript amounts (lowest Ct values) were observed only for miR-142–3p (25.5 ± 1.1). Relatively low transcript amounts were observed for miR-3664–3p (36.1 ± 1.5) and miR-4317 (36.4 ± 1.7), while miR-513c-3p, miR-1293, and miR-4483 showed no measurable expression in any of the liver samples tested (Ct value over 40).
For the UGT2B15 candidate miRNAs relatively high transcript amounts (low Ct values) were observed for miR-455–5p (27.8 ± 0.6), miR-376c-3p (29.9 ± 1.4), miR-770–5p (29.7 ± 0.3), and miR-3675–3p (29.9 ± 0.4). Relatively low transcript amounts (high Ct values) were observed for miR-331–5p (32.3 ± 0.8), miR-605 (35.7 ± 1), and miR-376b-3p (37.6 ± 1.4). We did not measure miR-3924 and miR-4292 transcripts since publicly available RNA-seq data (http://www.gtexportal.org/) indicated that both these microRNAs are not expressed in human liver. Moreover recent results from a different group demonstrated that miR-4292 is not expressed in human liver [28].
Finally, we sought to determine whether expression levels of any of the candidate miRNAs were negatively associated with glucuronidation activities selective for UGT2B7 (zidovudine glucuronidation, morphine-3-glucuronidation, and morphine-6glucuronidation) and UGT2B15 (S-oxazepam glucuronidation) when measured in the same human liver bank samples. These glucuronidation activities had been previously reported in [3, 13]). As shown in Table 6 although hepatic UGT2B7 activities were not negatively correlated with any of the miRNAs assayed, UGT2B15mediated S-oxazepam glucuronidation activity showed a statistically significant negative correlation (Rs = −0.53; p = 0.005) with miR-455–5p levels (Table 7, Fig. 5).
Table 6.
Correlation between UGT2B7 activities and hepatic miRNA transcript levels in samples from a human liver bank (n=27). UGT2B7 activities including zidovudine glucuronidation, morphine-3-glucuronidation, and morphine-6-glucuronidation (results previously reported in [3]). Shown are the Spearman correlation coefficients (Rs) and the associated p values.
| UGT2B7 activity | ||||||
|---|---|---|---|---|---|---|
| miRNA | Zidovudine glucuronidation | Morphine-3-glucuronidation | Morphine-6-glucuronidation | |||
| Rs | p | Rs | p | Rs | p | |
| miR-142–3p | −0.2 | 0.31 | −0.02 | 0.92 | 0.04 | 0.84 |
| miR-3664–3p | 0.28 | 0.16 | 0.1 | 0.62 | 0.12 | 0.55 |
| miR-4317 | 0.41 | 0.03 | −0.06 | 0.76 | −0.04 | 0.84 |
Table 7.
Correlation between hepatic miRNA transcript levels and UGT2B15mediated S-oxazepam glucuronidation activities in samples from a human liver bank (n=27). S-oxazepam glucuronidation activities were previously reported in [13]. Shown are the Spearman correlation coefficients (Rs) and the associated p values.
| S-oxazepam glucuronidation | ||
|---|---|---|
| Rs | p | |
| miR-331–5p | −0.2 | 0.31 |
| miR-376c-3p | −0.05 | 0.8 |
| miR-455–5p | −0.53 | 0.005 |
| miR-605 | 0.05 | 0.78 |
| miR-770–5p | −0.05 | 0.8 |
| miR-3675–3p | −0.08 | 0.7 |
| miR-103b | 0.03 | 0.9 |
Figure 5.
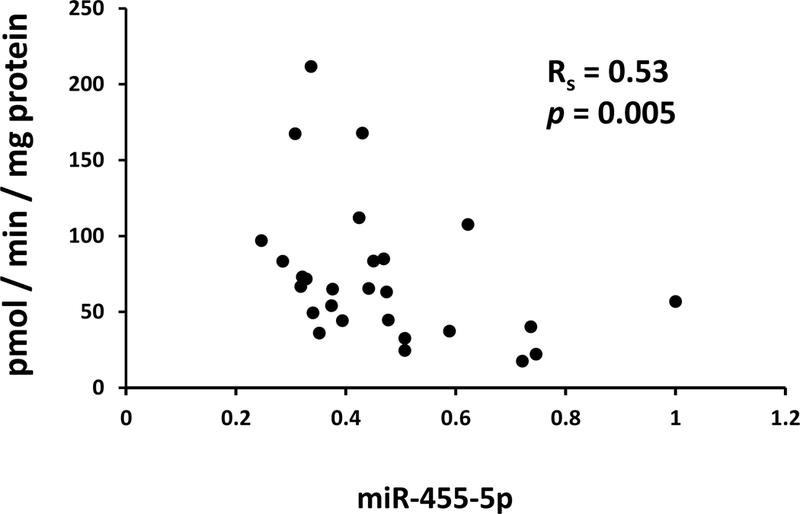
Scatter plot showing a negative correlation between transcript levels of miR-455–5p measured by quantitative PCR in total RNA extracted from 27 human livers and UGT2B15-mediated S-oxazepam glucuronidation activities measured in microsomes prepared from the same livers. Also shown is the Spearman correlation coefficient (Rs) and associated p-value.
4. Discussion
The most important novel findings of our study were the discovery and validation of 6 novel MREs for miR-1293, miR-3664–3p, miR-4317, miR-513c-3p, miR-4483, and miR-142–3p in the UGT2B7 3’-UTR, and 12 novel MREs for miR-770–5p, miR-103b, miR-3924, miR-376b-3p, miR-455–5p, miR-605, miR-624–3p, miR-4712–5p, miR-3675–3p, miR-6500–5p, miR-548as-3p, and miR-4292 in the UGT2B15 3’-UTR The location of each MRE in the UGT2B7 and UGT2B15 3’-UTRs are shown in Fig. 6 as well as the MREs for miR-376c and miR-331–5p, which have been previously characterized [18, 19].
Figure 6.
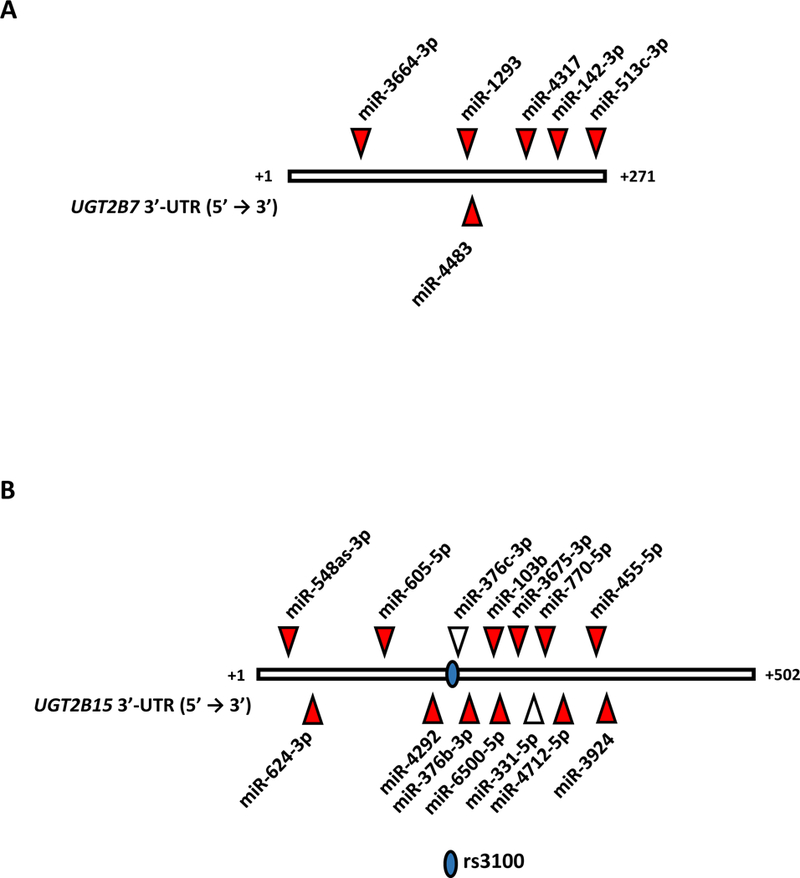
Locations of known miRNA response elements (MREs) in the reference UGT2B7 3’-UTR (A) and UGT2B15 3’-UTR (B). Indicated are the positions of the novel MREs validated in this study (red triangles) and the MREs for miR-376c-3p and miR-331–5p, which had been reported previously [18, 19]. Also indicated is theposition of the UGT2B15 3’-UTR rs3100 SNP.
We showed that half of the miRNAs we identified were expressed in human liver samples. More specifically 3 out of 6 candidate miRNAs for UGT2B7 and 7 out of 14 candidate miRNAs for UGT2B15 had detectable transcript amounts in human liver samples. Correlation analysis demonstrated that UGT2B7 activity did not show statistically significant correlation with any of the miRNAs while UGT2B15 activity showed a significant negative correlation with miR-455–5p hepatic levels (Fig. 5). While this suggests that miR-455–5p may contribute to inter-individual variability in hepatic UGT2B15 activity, the number of liver samples used for this analysis was limited (n=27). Thus, it will be necessary to study more human liver samples to clarify the contribution of miRNAs to the variable hepatic UGT2B7 and UGT2B15 activities in vivo.
Previous results from our laboratory demonstrated that miR-103b and miR-376b-3p have functional MREs in the common UGT1A 3’-UTR [17]. In the current work we showed that miR-103b and miR-376b-3p were also among the miRNAs with functional MREs in the UGT2B15 3’-UTR. Thus, it is plausible that a network of miRNAs contributes to the regulation of the expression of various UGT isoforms. A genome-wide approach is required to validate this hypothesis and evaluate the role of specific miRNAs in the observed interindividual variability in the expression of UGT enzymes.
UGT2B7 activity, protein, and mRNA levels were previously demonstrated to be significantly lower in livers and kidneys obtained from diabetic patients when compared matched tissues from non-diabetic patients [29]. A meta-analysis of miRNA expression profiling studies identified miR-142–3p among the top miRNAs that were significantly and consistently upregulated in blood samples of patients with type 2 diabetes [30]. Taken together these results imply that the observed decrease of hepatic and renal UGT2B7 activity and expression in diabetic patients could be due to increased miR-142–3p expression. However, a detailed analysis of the hepatic and renal UGT2B7 activity and expression and the blood and/or tissue concentrations of miR-142–3p in a large number of samples would be required to prove the correlation between UGT2B7 altered expression and the increased levels of this miRNA in diabetes.
In recent work we used the same in vitro system to perform a genome-wide screen of the miRNA mimic library with the pMIR-REPORT luciferase vector carrying the reference UGT1A 3’-UTR or the variant 3’-UTR [17]. This way we demonstrated that SNP rs10929303 C>T located in the common UGT1A 3’-UTR created a novel MRE for miR-21–3p. Reporter gene assays by another group showed that the UGT2B15 3’-UTR rs3100 c.1761C (reference) allele resulted in lower UGT2B15-dependent luciferase reporter activity compared with the (variant) c.1761T allele in liver, breast, colon, and prostate cancer cell lines [27]. Those authors suggested that this SNP might be located within a miRNA binding site that could influence UGT2B15 expression. Consequently we had hypothesized that the rs3100 c.1761T allele destroys the MRE of a miRNA that normally binds to the UGT2B15 3’-UTR rs3100 c.1761C allele. After screening for miRNAs that bind to the rs3100 c.1761C or c.1761T alleles, only a single miRNA (miR-376c-3p) was identified with a predicted MRE that included the rs3100 SNP. However, in silico analysis showed that the c.1761T allele (Fig. 2, panel K) enhances rather that reduces base-pairing to miR-376c-3p when compared with the c.1761C allele (Fig. 2, panel J). Despite this in silico prediction we were not able to experimentally demonstrate a significant difference in UGT2B15-dependent luciferase activity between the rs3100 c.1761C and c.1761T constructs when coexpressed with miR-376c-3p, or with any other miRNA demonstrated to bind to the UGT2B15 3’-UTR (Fig. 4). This lack of effect of rs3100 on miR-376c-3p binding may be because it is not located within the critical miRNA seed sequence (nucleotides 2–7). Regardless, it is still possible that rs3100 affects UGT2B15 expression through influencing binding of another (yet to be identified) miRNA, or perhaps indirectly through another mechanism.
There are several limitations to our study. We used a 30% cutoff for luciferase reporter inhibition in the initial library screen to focus our efforts on those miRNAs most likely to influence UGT2B7 and UGT2B15 expression. This threshold was sensitive enough to identify miR-376c-3p and miR-331–5p, which were the only other miRNAs with a functionally validated MRE [18, 19]. This cutoff also allowed us to minimize the significant amount of counter-screening that was needed since approximately 80% of candidate miRNAs were subsequently found to inhibit reporters lacking the UGT 3’-UTRs (i.e. false positives). However, it is possible that some of the miRNAs evaluated in our initial screen that showed less than 30% inhibition of reporter activity could also be important regulators of UGT2B7 and UGT2B15 expression. Furthermore, overexpression and knock-down experiments of these miRNAs in the appropriate cell line and in primary human hepatocytes are required to demonstrate their functional role in the regulation of UGT2B7 and UGT2B15 expression and enzyme activity.
In summary we used a genome-wide approach to identify functional MREs in the UGT2B7 and UGT2B15 3’-UTRs. The genome-wide approach we employed could be further used to identify candidate miRNAs that bind directly in the 3’-UTR of other hepatic genes of pharmacological importance.
5. Acknowledgements
This work was supported by the National Institutes of Health through National Institute of General Medical Sciences Grant NIH-R01-GM102130 (to M.H.C.) and the National Institute of Diabetes and Digestive and Kidney Diseases Grant N01-DK-7–0004 (to the Liver Tissue Cell Distribution System, University of Minnesota and University of Pittsburgh). Dr. Court was also supported by the William R. Jones endowment to Washington State University College of Veterinary Medicine.
Abbreviations:
- UGT,
UDP-glucuronosyltransferase
- 3’-UTR
3’-untranslated region
- SNP
single nucleotide polymorphism
- miRNA
microRNA
- qPCR
quantitative real-time PCR
- MRE
miRNA response element
6. References
- [1].Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW, Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily, Pharmacogenet Genomics. 15 (2005) 677–685. [DOI] [PubMed] [Google Scholar]
- [2].Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE, Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios, Drug Metab Dispos. 32 (2004) 1201 1208. [DOI] [PubMed] [Google Scholar]
- [3].Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ, Evaluation of 3'-azido-3'-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism, Drug Metab Dispos. 31 (2003) 1125–1133. [DOI] [PubMed] [Google Scholar]
- [4].Barbier O, Belanger A, Inactivation of androgens by UDP-glucuronosyltransferases in the human prostate, Best Pract Res Clin Endocrinol Metab. 22 (2008) 259–270. [DOI] [PubMed] [Google Scholar]
- [5].Jin C, Miners JO, Lillywhite KJ, Mackenzie PI, Complementary deoxyribonucleic acid cloning and expression of a human liver uridine diphosphate-glucuronosyltransferase glucuronidating carboxylic acid-containing drugs, J Pharmacol Exp Ther. 264 (1993) 475–479. [PubMed] [Google Scholar]
- [6].Radominska-Pandya A, Little JM, Czernik PJ, Human UDP-glucuronosyltransferase 2B7, Curr Drug Metab. 2 (2001) 283–298. [DOI] [PubMed] [Google Scholar]
- [7].Chen F, Ritter JK, Wang MG, McBride OW, Lubet RA, Owens IS, Characterization of a cloned human dihydrotestosterone/androstanediol UDPglucuronosyltransferase and its comparison to other steroid isoforms, Biochemistry. 32 (1993) 10648–10657. [DOI] [PubMed] [Google Scholar]
- [8].Court MH, Duan SX, Guillemette C, Journault K, Krishnaswamy S, Von Moltke LL, Greenblatt DJ, Stereoselective conjugation of oxazepam by human UDP glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9, Drug Metab Dispos. 30 (2002) 1257–1265. [DOI] [PubMed] [Google Scholar]
- [9].Patel M, Tang BK, Grant DM, Kalow W, Interindividual variability in the glucuronidation of (S) oxazepam contrasted with that of (R) oxazepam, Pharmacogenetics. 5 (1995) 287–297. [DOI] [PubMed] [Google Scholar]
- [10].Kwara A, Lartey M, Boamah I, Rezk NL, Oliver-Commey J, Kenu E, Kashuba AD, Court MH, Interindividual variability in pharmacokinetics of generic nucleoside reverse transcriptase inhibitors in TB/HIV-coinfected Ghanaian patients: UGT2B7*1c is associated with faster zidovudine clearance and glucuronidation, J Clin Pharmacol. 49 (2009) 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bhasker CR, McKinnon W, Stone A, Lo AC, Kubota T, Ishizaki T, Miners JO, Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7) at amino acid 268: ethnic diversity of alleles and potential clinical significance, Pharmacogenetics. 10 (2000) 679–685. [DOI] [PubMed] [Google Scholar]
- [12].Holthe M, Klepstad P, Zahlsen K, Borchgrevink PC, Hagen L, Dale O, Kaasa S, Krokan HE, Skorpen F, Morphine glucuronide-to-morphine plasma ratios are unaffected by the UGT2B7 H268Y and UGT1A1*28 polymorphisms in cancer patientson chronic morphine therapy, Eur J Clin Pharmacol. 58 (2002) 353–356. [DOI] [PubMed] [Google Scholar]
- [13].Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, Greenblatt DJ, UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver, J Pharmacol Exp Ther. 310 (2004) 656- [DOI] [PubMed] [Google Scholar]
- [14].He X, Hesse LM, Hazarika S, Masse G, Harmatz JS, Greenblatt DJ, Court MH, Evidence for oxazepam as an in vivo probe of UGT2B15: oxazepam clearance is reduced by UGT2B15 D85Y polymorphism but unaffected by UGT2B17 deletion, Br J Clin Pharmacol. 68 (2009) 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chekulaeva M, Filipowicz W, Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells, Curr Opin Cell Biol. 21 (2009) 452–460. [DOI] [PubMed] [Google Scholar]
- [16].Yu AM, Tian Y, Tu MJ, Ho PY, Jilek JL, MicroRNA Pharmacoepigenetics: Posttranscriptional Regulation Mechanisms behind Variable Drug Disposition and Strategy to Develop More Effective Therapy, Drug Metab Dispos. 44 (2016) 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Papageorgiou I, Court MH, Identification and validation of microRNAs directly regulating the UDP-glucuronosyltransferase 1A subfamily enzymes by a functional genomics approach, Biochem Pharmacol. 137 (2017) 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Margaillan G, Levesque E, Guillemette C, Epigenetic regulation of steroid inactivating UDP-glucuronosyltransferases by microRNAs in prostate cancer, J Steroid Biochem Mol Biol. 155 (2016) 85–93. [DOI] [PubMed] [Google Scholar]
- [19].Wijayakumara DD, Hu DG, Meech R, McKinnon RA, Mackenzie PI, Regulation of Human UGT2B15 and UGT2B17 by miR-376c in Prostate Cancer Cell Lines, J Pharmacol Exp Ther. 354 (2015) 417–425. [DOI] [PubMed] [Google Scholar]
- [20].Court MH, Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system, Drug Metab Rev. 42 (2010) 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, Duan SX, Greenblatt DJ, Lee WM, The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure, J Pharmacol Exp Ther. 345 (2013) 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lamba V, Ghodke-Puranik Y, Guan W, Lamba JK, Identification of suitable reference genes for hepatic microRNA quantitation, BMC Res Notes. 7 (2014) 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T, Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines, Drug Metab Dispos. 36 (2008) 1461–1464. [DOI] [PubMed] [Google Scholar]
- [24].Betel D, Wilson M, Gabow A, Marks DS, Sander C, The microRNA.org 35 36 resource: targets and expression, Nucleic Acids Res. 36 (2008) D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS, MicroRNA targets in Drosophila, Genome Biol. 5 (2003) R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R, Fast and effective prediction of microRNA/target duplexes, RNA. 10 (2004) 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Swart M, Dandara C, Genetic variation in the 3'-UTR of CYP1A2, CYP2B6, CYP2D6, CYP3A4, NR1I2, and UGT2B7: potential effects on regulation by 52 53 microRNA and pharmacogenomics relevance, Front Genet. 5 (2014) 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sun C, Southard C, Olopade OI, Di Rienzo A, Differential allelic expression of c.1568C > A at UGT2B15 is due to variation in a novel cis-regulatory element in the 3'UTR, Gene. 481 (2011) 24–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, Keller A, Distribution of miRNA expression across human tissues, Nucleic Acids Res. 44 (2016) 3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dostalek M, Court MH, Hazarika S, Akhlaghi F, Diabetes mellitus reduces activity of human UDP-glucuronosyltransferase 2B7 in liver and kidney leading to decreased formation of mycophenolic acid acyl-glucuronide metabolite, Drug Metab Dispos. 39 (2011) 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhu H, Leung SW, Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies, Diabetologia. 58 (2015) 900–911. [DOI] [PubMed] [Google Scholar]


