Abstract
Our understanding of kidney disease pathogenesis is limited by an incomplete molecular characterization of the cell types responsible for the organ’s multiple homeostatic functions. To help fill this knowledge gap, we characterized 57,979 cells from healthy mouse kidneys using unbiased single-cell RNA sequencing. Based on gene expression patterns, we infer that inherited kidney diseases that arise from distinct genetic mutations but have similar phenotypic manifestations share the same cell of origin. We also found that the kidney collecting duct in adult mice generates a spectrum of cell types via a newly identified transitional cell. Computational cell trajectory analysis and in vivo lineage tracing revealed that intercalated cells and principal cells undergo transitions mediated by the Notch signaling pathway. In mouse and human kidney disease, these transitions were shifted toward a principal cell fate and were associated with metabolic acidosis.
The kidney is a highly complex organ that performs many diverse functions that are essential for health. It removes nitrogen, water and other waste products from the blood. It controls blood electrolytes and acid-base balance and it secretes hormones that regulate blood composition and blood pressure. The kidney consists of several functionally and anatomically discrete segments. The glomerulus is a specialized group of capillaries that filters the blood and produces the primary filtrate of water and solutes such as sodium, potassium glucose and bicarbonate. The proximal tubules then reabsorb the majority of the water and electrolytes whereas other solutes such as uric acid and other organic anions, potassium and protons are secreted into the filtrate. The Loop of Henle is primarily involved in solute concentration. The distal tubule and the collecting duct are segments where highly regulated solute transport occurs; thus each segment is critical for maintaining electrolyte and water homeostasis.
In the past, kidney cells have been annotated on the basis of their function, anatomical location or by the expression of a small number of marker genes (1), yet these classification systems do not fully overlap. An emerging technology called single-cell transcriptional profiling allows investigators to monitor global gene regulation in thousands of individual cells in a single experiment (2, 3). In principle, this technology could answer central questions in kidney biology and disease pathogenesis because it has the potential to provide four distinct types of information.
First, unbiased single cell clustering can redefine kidney cell types based only on their global transcriptome patterns (4). Such analyses have already been applied to other organs (2, 5–7) and even to whole multicellular organisms (8, 9). These experiments have identified novel cells as well as catalogued marker genes for previously defined cells, indicating that this approach has the power to redefine kidney cell types.
Second, single cell analysis may help dissect the mechanisms underlying common kidney diseases (10, 11). In general, kidney pathologies have been grouped together by their temporal patterns (acute or chronic) or by their target structures (glomerular vs tubular), which has obscured the underlying biology. Previously obtained bulk transcriptome profiles have generated read-outs only for predominant cell populations such as the proximal tubular cells (12). Kidney segment-specific RNAseq analysis of the rat kidney has provided useful resources (13) but single cell analysis can potentially further exploit cell-type specific changes and identify novel cell types during disease modulation, independent of preconceived cellular definitions.
Third, single cell analysis may be able to identify fluctuating states of the same cell type. It is generally believed that terminally differentiated cells have limited plasticity. Most cell plasticity in the adult has been observed in the context of terminal differentiation of progenitor cells, best described in the blood and intestine (14). Yet, such cellular transitions also may be ongoing in collecting duct development (15–17) or even in the adult collecting duct. For example, subtypes of intercalated cells (ICs) can change their functional polarity, and even stem cell-like populations originating from the principal cell types (PCs) may persist in the adult collecting duct and respond to the external stimuli (18, 19), but definitive isolation and definition of these plastic cells is lacking.
Fourth, current models of kidney disease cannot distinguish primary cell autonomous responses from secondary cell non-autonomous responses; in contrast, single cell specific gene expression profiles could potentially identify the readout of disease-associated gene mutations in each cell.
Single cell profiling and unbiased clustering of mouse kidney cells
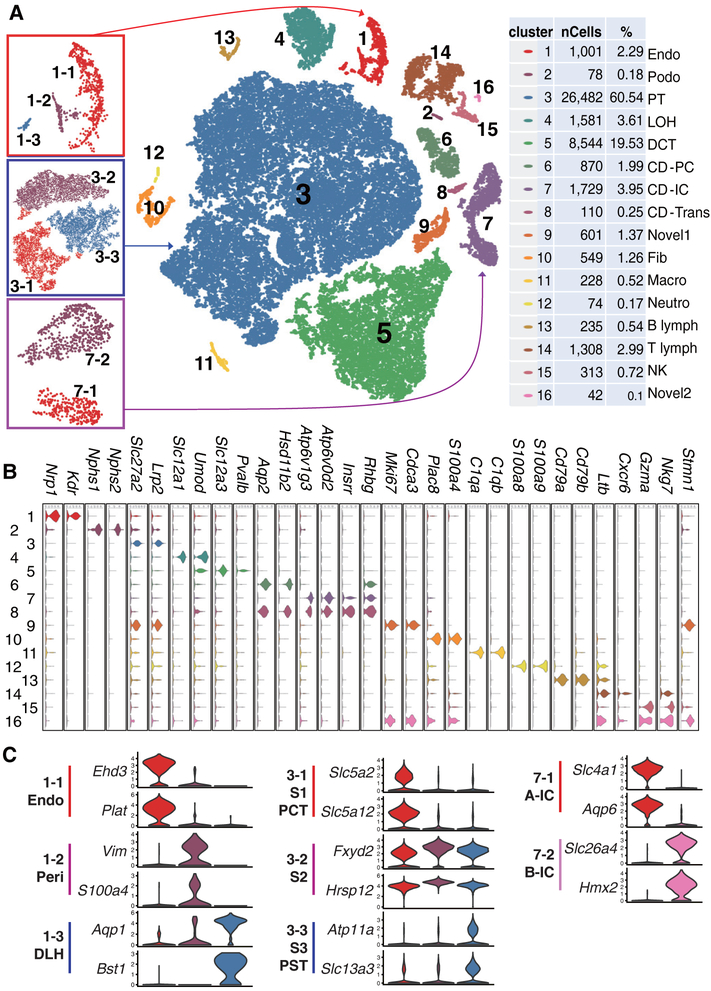
We first catalogued mouse kidney cell types in an unbiased manner using droplet-based single-cell RNA sequencing (20). We isolated and sequenced 57,979 cells from whole kidney cell suspensions of seven healthy male mice. Using stringent quality controls (20), we further analyzed 43,745 cells. Clustering analysis identified 16 distinct cell clusters consisting of as few as 24 cells to as many as 26,482 cells per cluster (the clusters were censored for a minimum of 20 cells) (Fig. 1A).
Fig. 1. Cell diversity in mouse kidney cells delineated by single cell transcriptomic analysis.
(A) Unsupervised clustering demonstrates 16 distinct cell types shown in a tSNE map. Left panels are subclusters of clusters 1, 3, and 7. Percentage of assigned cell types are summarized in the right panel. Endo: containing endothelial, vascular, and descending loop of Henle, Podo: podocyte, PT: proximal tubule, LOH: ascending loop of Henle, DCT: distal convoluted tubule, CD-PC: collecting duct principal cell, CD-IC: CD intercalated cell, CD-Trans: CD transitional cell, Fib: fibroblast, Macro: macrophage, Neutro: neutrophil, NK: natural killer cell. (B and C) Violin plots showing the expression level of representative marker genes across the 16 main clusters. Y-axis is log scale normalized read count. (C) Cluster1 (from panel A left) separates into endothelial cells, pericytes/vascular smooth muscle cells and descending loop of Henle (DLH). Cluster3 (proximal tubules) separates into S1, S2 and S3 segments or PCT (proximal convoluted tubules) and PST (proximal straight tubules). Cluster 7 intercalated cells separates into A and B type intercalated cells.
We next performed several important quality control analyses to validate our map. First, we ensured that cells from seven different batches of mice were distributed evenly in all 16 clusters and each cluster contained cells from more than four experiments (fig. S1). Next, we examined the effect of mitochondrial gene content (fig. S2A). The clustering of cells was not affected by mitochondrial gene content (fig. S2, B-E). Furthermore, genes whose expression positively correlated with mitochondria encoded proteins associated with solute transport (which require abundant energy), rather than cellular stress responses (fig. S3). This indicates that the increased mitochondrial gene count was inherent to specific (proximal and distal tubule) cell types in the kidney. In addition, by testing different clustering methods we found that most methods identified similar cell groups (fig. S4), expressing the same group of marker genes with limited variations in cell separation. Finally, we showed that decreasing the cell number from 40,000 to 10,000 or 3,000 or 1,000 cells (fig. S5A) was associated with increasing uncertainty in cell cluster identification and the loss of rarer cell types (fig. S5, B and C).
Classification of kidney cells based on cell-type specific marker genes
To define the identity of each cell cluster, we generated cluster-specific marker genes by performing differential gene expression analysis (Fig. 1B, fig. S6 and table S1) (20). In many cases, the unbiased cluster identifier was a known cell-type specific marker, such as the vascular endothelial growth factor receptor 2 (Kdr) for endothelial cells, Nephrin (Nphs1) and Podocin (Nphs2) for podocytes, Na/K/2Cl cotransporter (Slc12a1) for ascending loop of Henle and thiazide sensitive sodium-chloride cotransporter (Slc12a3) for distal convoluted tubule (Fig 1B). Immune cells and endothelial cell clusters separated from epithelial cells but the ureteric bud- (cluster 6–8) and metanephric mesenchyme-derived epithelial clusters (cluster 2–5) were more closely aligned (Fig. 1A). While some of the markers were already known, we identified a large number of additional markers, including Cdkn1c and Bcam for podocytes (Fig. 1B and fig. S6B). Further analysis identified eight subclusters within clusters 1, 3 and 7 (Fig. 1C, table S2 and S3). Cluster1 separated into endothelial cells, pericyte/vascular smooth muscle/mesangial-like cells, and descending loop of Henle cells (DLH). Cluster3 (proximal tubules) separated into S1, S2 and S3 segments or proximal convoluted and straight segments (fig. S7). Intercalated cells (cluster 7) separated into A and B type intercalated cells.
To reliably assign a specific cell type to each cell cluster, we first correlated our gene expression results with bulk RNA sequencing data from microdissected rat kidney segments (fig. S8) and microarray data of human immune cell types (fig. S9). To further validate our clustering analysis, we used Nphs2CremT/mG, SclCremT/mG and Cdh16CremT/mG mice as reporter lines to mark podocytes, endothelium and tubule cells with green fluorescent protein (GFP). The GFP expression in these models confirmed the proposed cell identity of our cell clusters (fig. S10). Altogether, our single cell transcriptome atlas provides a molecular definition of 18 previously defined kidney and immune cell types as well as three novel cell types.
Mendelian disease genes show cell-type specificity
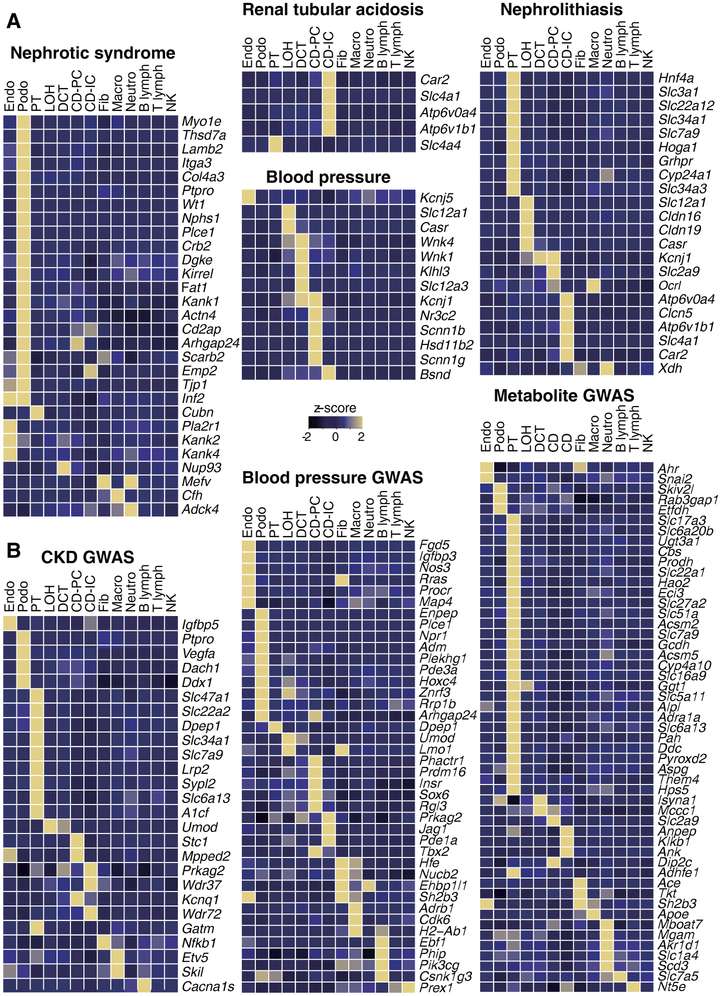
We next tested the hypothesis that hereditary kidney diseases that are characterized by the same phenotypic manifestations originate from the same cell type. We also explored whether the functions of specific cell types in the mouse kidney could be inferred from the expression pattern of human genes, whose loss of function results in kidney disease. We found that the mouse homologs of 21 of 29 genes that have been associated with monogenic inheritance of proteinuria in humans were expressed in only one cell type, namely the podocyte of the glomerulus (Fig. 2A and fig. S11). While earlier studies had implicated defects in endothelial cells and proximal tubules in the development of proteinuria, and indeed functional and structural changes in these cell types can be seen in patients with proteinuria, our results unequivocally show that podocyte dysfunction is the principal reason for proteinuria (21). As another example, we found that the mouse homologs of genes associated with renal tubule acidosis (RTA) in humans were expressed only by intercalated cells (IC) of the collecting duct, confirming the major role of these cells in acid-base homeostasis (Fig. 2A). Furthermore, genes that have been implicated in blood pressure (BP) regulation through analysis of human Mendelian diseases, such as Wnk4, Wnk1, Klh3, Slc12a3, were expressed specifically in the distal convoluted tubule, while Nr3c2, Scnn1b, Scnn1g and Hsd11b2 were specifically expressed by principal cells of the collecting ducts (Fig. 2A, fig. S11 and S13).
Fig. 2. Discrete human disease phenotypes are due to mutations in single specific cell types.
Single cell-type specific average expression of human (A) monogenic disease genes and (B) complex trait genes identified by GWAS. Mean expression values of the genes were calculated in each cluster. The color scheme is based on z-score distribution; the map shows genes with z-score>2. In the heatmap, each row represents one gene and each column is single cell type (defined in Fig. 1). The full list of cell types and genes are shown in fig. S11 and 12.
Following the same logic, we annotated the expression of putative complex trait disease genes that have been associated with blood pressure (BP), chronic kidney disease (CKD) and serum metabolite levels, nephrolithiasis (e.g. Slc34a1) and renal tubular acidosis (e.g. Atp6v1b1) (Fig. 2B, fig. S12 and S13) (22–24). We found that most genes implicated in these traits were expressed only in a single cell type such as collecting duct cells (RTA) and proximal tubule cells (nephrolithiasis). The expression of genes associated with plasma metabolite levels such as Slc17a3 (uric acid), Slc51a (bile acid) and Slc16a9 (carnitine) (22–24) and CKD showed strong enrichment for proximal tubule-specific expression, whereas BP-associated genes were mostly expressed in collecting duct cells. Thus, our single cell transcriptome analysis has highlighted specific cells responsible for specific kidney-related disorders, as well as highlighting the critical functions of these cells.
Identification of a novel cell type in the kidney collecting duct
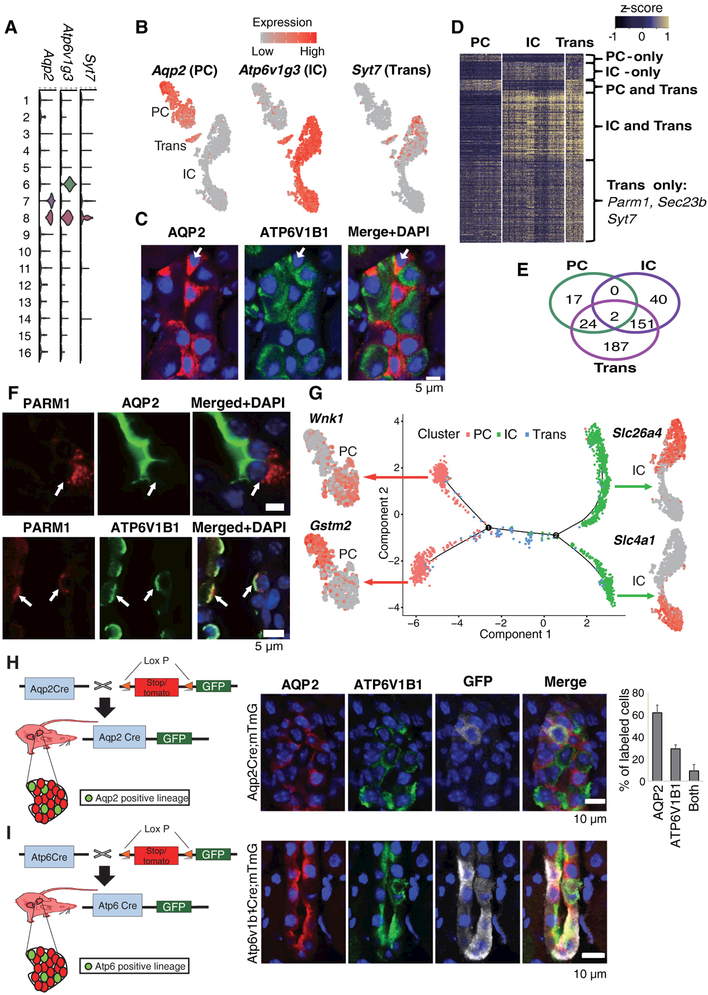
The kidney collecting duct differs from all other kidney epithelia since it originates from the ureteric bud and not from the metanephric mesenchyme. This compartment is comprised of at least three distinct cell types: the principal cells (PC), which are responsible for sodium, water reabsorption and potassium secretion; and the alpha and beta intercalated cells (A-IC and B-IC), which are responsible for acid and alkali secretion, respectively. We identified aquaporin 2 (Aqp2) and H+-ATPase subunit (Atp6v1g3) as the key marker genes for clusters 6 and 7, defining these clusters as PC and IC cells (Fig. 1 and 3A, B).
Fig. 3. Identification of a transitional cell type and a conversion process in the collecting duct.
(A) The expression level of marker genes across the 16 clusters. Y-axis is log scale normalized read count. (B) Gene expression levels in PC (Aquaporin 2, Aqp2), IC (H+ATPase, Atp6v1g3,) and in Transitional cells (Syt7) demonstrated by a tSNE plot. (C) Representative immunofluorescence images of AQP2 (PC marker), ATP6V1B1 (IC marker) and DAPI in the kidney collecting duct. Arrow demonstrates transitory PC/IC cell type expressing AQP2 and ATP6V1B1. (D) Heatmap showing the expression level of differentially expressed genes in collecting duct cell types. Color scheme is based on z-score distribution. (E) Venn diagram showing the overlaps of differentially expressed genes between PC, IC and the novel cell type. (F) Immunofluorescence staining for PARM1 (IC specific) and AQP2 (upper panel) or ATP6V1B1 (lower panel) in the kidney collecting duct. A “double positive” cell is shown at the arrow (G) Ordering single cells along a cell conversion trajectory using Monocle. Three collecting duct cell clusters were used for ordering and plotted on low dimensional space with different colors. The tSNE plots next to the trajectory map show differentially expressed genes in the corresponding cell lineages. (H) Aqp2CremT/mG mouse model used for lineage tracing of AQP2 positive cells. Immunofluorescence staining for GFP, ATP6V1B1 and AQP2. Right panel is the quantification of GFP positive cells (mean ± SD). n=3. Note that AQP2 driven GFP (white) is found in PC (red + white) and IC (green + white) and in transitional cells (not shown) (I) Atp6CremT/mG mouse model used for lineage tracing of ATP6ase positive cells. Immunofluorescence staining for GFP, ATP6V1B1 and AQP2 in Atp6CremT/mG mice. Note that ATP6V1B1 driven GFP (white) is found in PC (red+ white) and IC (green+ white) and in transitional cells (red+ green+ white).
Unexpectedly, our single cell profiling identified a third cell cluster. This cell cluster (cluster 8: Trans) expressed markers of both IC and PC cells (“double positive cells” Fig. 3, A and B) and additional cell type-specific markers. We performed double immunofluorescence staining and in situ hybridization with probes for Aqp2 and Atp6v1b1 (Fig. 3C) and cell type-specific markers such as Parm1 and Sec23b (Fig. 3, D, E, F and fig. S14) to validate the existence of this cell type.
To further investigate this unexpected cell type, we utilized the Monocle analysis toolkit to perform cell trajectory analysis using pseudotime reconstitution of clusters 6–8 (20). We found that the novel cells were located between PCs and ICs suggesting that cluster 8 is a transitional cell type (Fig. 3G). Transitional cells showed low expression levels of stress response genes and cell cycle genes and these cells were present in all batches of our kidney isolates (fig. S15, S16), excluding the possibility that they were injured cells or a proliferating subtype of collecting duct cells, or an artifact. Furthermore, cell trajectory analysis clearly separated IC into A- and B-IC types, and PC cells into their subtypes (principal cells and what are most likely connecting duct cells) as previously identified (Fig. 3G and fig. S17) (7, 25). These results indicate that the collecting duct contains not only PC and IC cells but a third distinct transitional cell type; this raises the possibility that IC and PC cells represent two ends of a spectrum of cellular phenotypes and that the two cell types may undergo cellular transitions.
Fluorescent lineage tracing confirms collecting duct cell plasticity
We next examined whether transitional cells could be identified by conventional in vivo lineage tracing and match our computational characterization. We generated mice that carry a lineage tag in differentiated principal cells (Aqp2CremT/mG) or in differentiated intercalated cells (Atp6CremT/mG) (Fig. 3, H and I). We performed triple immunofluorescence labeling in these animals by staining for GFP (all cells of a specific marker origin), AQP2 (principal cells) and ATP6V1B1 (intercalated cells). As expected, we found that most of the GFP-positive cells were also AQP2-positive in the Aqp2CremT/mG mice. A subset of the GFP positive cells expressed ATP6V1B1, an IC marker, but not AQP2. A smaller subset was “double positive” for ATP6V1B1 and AQP2. Among the Aqp2CremT/mG GFP-positive cells, 61.6% of the cells were AQP2-positive, 29.2% were ATP6V1B1-positive and 9.2% of the cells were positive for both AQP2 and ATP6V1B1 (Fig. 3H). Similar analyses were performed with the Atp6CremT/mG lineage, which showed that both “double positive” AQP2 and ATP6V1B1-positive transitional cells as well as ATP6V1B1-negative true PC cells can originate from ATP6V1B1-positive IC cells (Fig. 3I).
To determine if cell proliferation might be responsible for this cell plasticity, we calculated the expression levels of cell-cycle regulated genes in the single cell transcriptome and in cell trajectory maps. We found that only cluster 9 and 16 (novel cell types 1 and 2), but not any of the collecting duct clusters, expressed high levels of the cell cycle genes (fig. S18). This suggests that cluster 8 is likely to be a transitional cell population and not a proliferating progenitor cell. Thus, in vivo lineage tagging analysis confirmed transitions of PC and IC cells not only during development (15–17) but also in the adult collecting duct via a newly identified transitional cell type.
Collecting duct cell plasticity, driven by Notch ligand and receptor interactions, results in abnormal cell populations in chronic kidney diseases
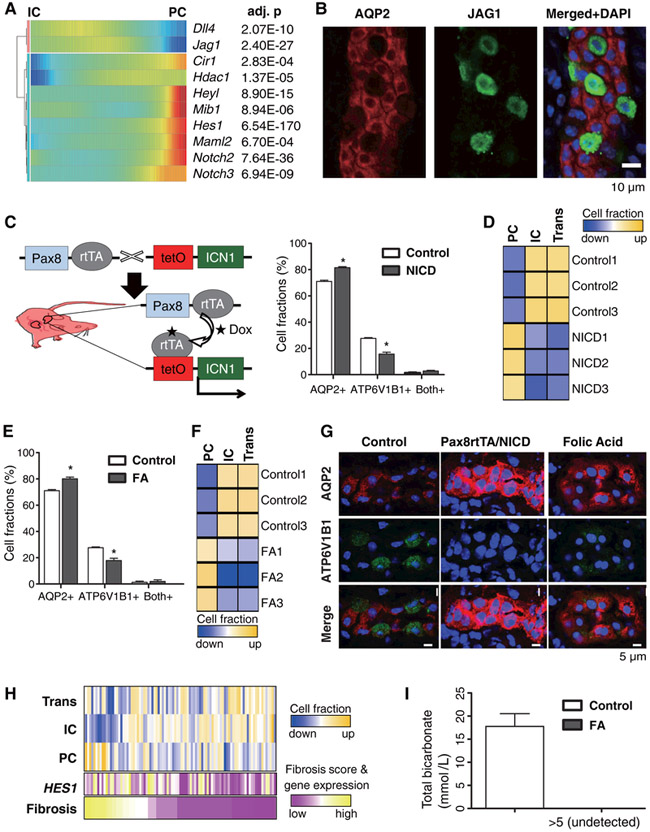
For further analysis of collecting duct cell plasticity, we identified genes whose expression levels change during transitions of PC and IC cells (fig. S19, A and B) (20). PCs showed enriched expression of genes associated with cell adhesion, water homeostasis and salt transport whereas ICs showed enriched expression of genes associated with ATP hydrolysis/synthesis, coupled proton transport and oxidation-reduction processes (fig. S19C). The gene expression patterns revealed that the Notch signaling pathway was activated during the transition of ICs to PCs. Notch regulates the cellular identity of neighboring cells by the expression of either Notch ligands or Notch receptors. Alternating expression of ligands and receptors creates a signal sending cell (Notch-off) and a signal receiving cell (Notch-on). Notch ligands such as Jag1 were highly expressed by ICs whereas their expression levels were low in PCs (Fig. 4A). In contrast, PCs showed high expression levels of Notch2 receptor, and their transcriptional target Hes1 (which is a transcription factor), suggesting that PCs are the Notch signal receiving cells in the collecting duct. Immunofluorescence studies confirmed exclusive expression of the Notch ligand JAG1 in the IC cells (Fig. 4B).
Fig. 4. The IC-to- PC cell transition is driven by Notch ligand and receptor expression.
(A) Transcriptional profiles demonstrating the spectrum of expression of Notch genes in the collecting duct. Cells are ordered in pseudotime and color represents expression levels. (B) Double immunofluorescence staining for AQP2 (red) and JAG1 (green) in the kidney collecting duct. (C) Generation of mice with inducible expression of Notch (ICN1) in kidney tubules. Excess AQP2+ cells and conversely reciprocally decreased ATP6V1B1+ positive cells are found in Pax8rtTA/NICD tubules (mean ± SD). n=3. Asterisk represents significant difference; p-value < 0.01. (D) In silico deconvolution of mouse kidney bulk RNA profiling data. Wild type (WT) and Pax8rtTA/NICD samples were used for analysis. (E) Immunofluorescence quantification of cells labeled with AQP2 and ATP6V1B1 in control and FA mice (mean ± SD). n=3. Asterisk represents significant differences; p-value < 0.01. (F) In silico deconvolution of mouse kidney bulk RNA profiling. Control and kidney samples from FA injected mice were used for analysis. (G) Immunofluorescence staining for AQP2 and ATP6V1B1 in control, Pax8rtTA/NICD and FA collecting ducts. Note the abundance of AQP2+ cells in Pax8rtTA/NICD and FA and conversely the disappearance of ATP6V1B1+ cells (H) In silico deconvolution of bulk RNA profiling data from normal and CKD human samples (n=91). The histological fibrosis scores and HES1 expression levels for the corresponding samples are also shown. (I) Total serum bicarbonate level in control and FA induced kidney fibrosis (mean ± SD). n=5.
To examine whether Notch signaling drives the IC-to-PC cell transition, we generated Pax8rtTA/TRENICD mice, which enables inducible transgenic expression of the conserved Notch intracellular domain portion of the receptor, specifically in differentiated kidney tubule cells (Fig. 4C). This experimental model allowed us to study only the IC to PC transition occurring in adult mice as opposed to those occurring during embryogenesis (15). We found that NOTCH expression disrupted cellular patterning. The number of cells expressing the PC cell marker AQP2 was increased, whereas the number of cells expressing the IC cell marker ATP6V1B1 and the alpha-IC cell marker ADGRF5 was reduced in parallel (Fig. 4C and G, fig. S20). The Notch-mediated transition appeared nearly complete as the cells also expressed multiple PC markers, including AQP3 and HSD11B2 (fig. S21 and S22). Finally, in silico deconvolution analysis of bulk RNA profiling data, examining marker gene expression was performed for control and Pax8rtTA/TRENICD mice (20) to estimate the proportion of ICs and PCs in the collecting duct. The data were consistent with the results of our lineage tracing experiments (Fig. 4D). Collectively, these data indicate that Notch receptor expression and signaling are sufficient to drive the IC-to-PC cell transition in the adult kidney collecting duct.
Because increased Notch expression has been reported in patients and animal models with kidney disease (26, 27), we examined whether disease states disrupt the relative numbers of PC and IC cells. In a mouse model of chronic kidney disease induced by folic acid (FA), which shows structural and functional damage that is similar to human CKD, we found a loss of the typical alternating patterns of IC and PC cells. There was an increase in AQP2+ cells and a decrease in ATP6V1B1+ cells (also ADGRF5+ positive A-type IC cells) compared to untreated mice (Fig. 4E and G, fig. S20). Computational cell deconvolution analysis of bulk RNA sequencing and analysis of marker gene expression in control and FA kidney disease models yielded data consistent with a shift from IC to PC cell fate (Fig. 4F and fig. S23). Using cell markers identified in mice, we performed computational deconvolution analysis of kidney biopsy samples from 91 patients with hypertensive and diabetic CKD, as described under fig. S24. Again, we found that in comparison with healthy samples, the diseased tissue samples showed a higher ratio of PC to IC cells (Fig. 4H and fig. S25), consistent with increased Notch signaling and HES1 expression (that is indicative of active Notch signaling) in these samples. The shift toward PC cells did not correlate with increased expression of cell proliferation associated genes in PC cells nor with increased expression of cell death associated genes in IC cells (fig. S25).
Finally, we analyzed whether the increased IC to PC transition that we observed in the mouse model of CKD (Fig. 4C) and in human samples of CKD (Fig. 4H) had a functional consequence. IC cells are uniquely associated with acid secretion in the kidney by exclusively expressing H+ transporting genes. Conversely, mutations of genes encoding acid transporting such as ATP6V1B1 cause metabolic acidosis, an accumulation of acid in many compartments of the body (fig. S19C and Fig. 2A). We found that total blood CO2 levels (composite measure of serum bicarbonate and pCO2) were significantly reduced in the FA-induced mouse kidney disease model, consistent with metabolic acidosis (Fig. 4I). Together these data show that (i) the IC to PC transition is mediated by Notch ligand (IC) and receptor (PC) expression; and (ii) that a shift toward the PC cell fate is the likely cause of metabolic acidosis in mouse models and patients with CKD.
Discussion
Efforts to describe the cell types that make up the kidney date back to the invention of the microscope. Over the past century, a kidney cell annotation has been developed that is based on the organ’s transporting functions of water and different types of salts. Here, we provide a molecular definition of cell types in the mouse kidney by single cell RNA sequencing of 57,979 cells. At this resolution, we distinguished 21 major cell types defined by quantitative gene expression; these cells included almost all previously described cell types, novel transitional cells in the collecting duct and two additional undefined cells (cluster 9 and 16, Fig. 1). Our work complements previous efforts that have applied this technology to the kidney. Single cell sequencing has been used to study fetal mouse kidneys and sorted kidney segments (7, 28, 29) and a recent study identified kidney cell composition changes in patients with lupus nephritis (30).
Our kidney cell atlas provides insight into kidney function and kidney disease pathogenesis. It demonstrates that the expression of monogenic kidney disease genes is restricted to a single cell type. Therefore, most genetic diseases of the kidney can be traced to single cell types. In this light, each cell type appears to make a non-redundant contribution to a specific type of kidney disease. In contrast, previous transcriptomic studies identified changes in multiple cell types using aggregated data from different kidney diseases. Finally, it appears that the single cell transcriptomics data can be used to infer cell type specific function in vivo at the organismal level.
The atlas also highlights the role of the kidney collecting duct system in health and disease. The expression of genes harboring mutations associated with human disorders such as metabolic acidosis, CKD and BP are specifically localized to this kidney segment. One of the most striking results of our analysis of the mouse kidney was the identification of an unexpected cell type related to the well-known IC and PC cells. Computational and lineage tagging analyses indicated that these “double IC/PC” cells are most likely transitional cells and that the number and patterning of IC and PC cells are controlled by Notch signaling in adult mice. This finding suggests that the Notch pathway may play a role in chronic kidney diseases. We speculate that the transitions between PC and IC is a constitutive process that is activated in disease conditions, since our cell trajectory analysis demonstrates that the transitions occur at low frequency in healthy mice. PCs may be irreplaceable as they are responsible for sodium and water balance, and are involved in regulation of serum potassium levels (31). Elevated serum potassium can cause fatal cardiac conduction abnormalities in patients with chronic kidney failure. On the other hand, acid accumulation due to the loss of ICs can be partially compensated by regulating the respiratory rate, and as a result maintain near normal serum pH. Perhaps this rationale explains the preservation or even expansion of PC cells compared to IC cells in disease states.
In summary, we have generated a comprehensive cell atlas of the mouse kidney, identified cell-type specific markers along with novel cell types, and have uncovered unexpected cell plasticity. This information will enhance our understanding of normal kidney function and disease development.
Supplementary Material
P-values and average natural log expression differences were calculated using the Seurat package as described in Materials and Methods. Percent cells 1 and 2 represent the percent of cells expressing the specific gene in the target cell cluster and average of the non-target clusters, respectively.
P-values and average natural log expression differences were calculated using the Seurat package as described in Materials and Methods. Percent cells 1 and 2 represent the percent of cells expressing the specific gene in the target cell cluster and average of the non-target clusters, respectively.
Each column represents one cell group (from this study) and each row represents the expression of a single gene.
Acknowledgments
Funding: Work in the Susztak lab is supported by NIH NIDDK R01 DK076077, DK087635, DK105821 and DP3 DK108220. Dr. Park is supported by American Diabetes Association Training grant #1–17-PDF-036. Dr. Werth and Barasch are supported by NIH 1U54DK104309–01; NIH 2R01DK073462; DK-16–026 (Kidney Precision Medicine Project).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Processed and raw data can be downloaded from NCBI GEO (GSE107585).
References and Notes:
- 1.Kriz W, Bankir L, A standard nomenclature for structures of the kidney. The Renal Commission of the International Union of Physiological Sciences (IUPS). Kidney international 33, 1–7 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Macosko EZ et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng GX et al. , Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozenblatt-Rosen O, Stubbington MJT, Regev A, Teichmann SA, The Human Cell Atlas: from vision to reality. Nature 550, 451–453 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA, Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lein E, Borm LE, Linnarsson S, The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Chen L et al. , Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A 114, E9989–E9998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaiskos N et al. , The Drosophila embryo at single-cell transcriptome resolution. Science, (2017). [DOI] [PubMed] [Google Scholar]
- 9.Cao J et al. , Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrandt F, Genetic kidney diseases. Lancet 375, 1287–1295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menezes LF, Germino GG, Systems biology of polycystic kidney disease: a critical review. Wiley Interdiscip Rev Syst Biol Med 7, 39–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM et al. , Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 21, 37–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JW, Chou CL, Knepper MA, Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol 26, 2669–2677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang-Panesso M, Humphreys BD, Cellular plasticity in kidney injury and repair. Nature reviews. Nephrology 13, 39–46 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Werth M et al. , Transcription factor TFCP2L1 patterns cells in the mouse kidney collecting ducts. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H et al. , Aqp2-expressing cells give rise to renal intercalated cells. J Am Soc Nephrol 24, 243–252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong HW et al. , Inactivation of Notch signaling in the renal collecting duct causes nephrogenic diabetes insipidus in mice. The Journal of clinical investigation 119, 3290–3300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J et al. , Collecting duct-derived cells display mesenchymal stem cell properties and retain selective in vitro and in vivo epithelial capacity. J Am Soc Nephrol 26, 81–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz GJ, Barasch J, Al-Awqati Q, Plasticity of functional epithelial polarity. Nature 318, 368–371 (1985). [DOI] [PubMed] [Google Scholar]
- 20.See supplementary materials.
- 21.Malaga-Dieguez L, Susztak K, ADCK4 “reenergizes” nephrotic syndrome. The Journal of clinical investigation 123, 4996–4999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattaro C et al. , Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nature communications 7, 10023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren HR et al. , Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nature genetics 49, 403–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin SY et al. , An atlas of genetic influences on human blood metabolites. Nature genetics 46, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welling PA, Roles and Regulation of Renal K Channels. Annual review of physiology 78, 415–435 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Bielesz B et al. , Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. The Journal of clinical investigation 120, 4040–4054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweetwyne MT et al. , Notch1 and Notch2 in Podocytes Play Differential Roles During Diabetic Nephropathy Development. Diabetes 64, 4099–4111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Ye Y, Yang Q, Shi S, Single-cell RNA-sequence analysis of mouse glomerular mesangial cells uncovers mesangial cell essential genes. Kidney international 92, 504–513 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Adam M, Potter AS, Potter SS, Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144, 3625–3632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Der E et al. , Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welling PA, Regulation of renal potassium secretion: molecular mechanisms. Seminars in nephrology 33, 215–228 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P-values and average natural log expression differences were calculated using the Seurat package as described in Materials and Methods. Percent cells 1 and 2 represent the percent of cells expressing the specific gene in the target cell cluster and average of the non-target clusters, respectively.
P-values and average natural log expression differences were calculated using the Seurat package as described in Materials and Methods. Percent cells 1 and 2 represent the percent of cells expressing the specific gene in the target cell cluster and average of the non-target clusters, respectively.
Each column represents one cell group (from this study) and each row represents the expression of a single gene.