Abstract
Migratory birds are an increasing focus of interest when it comes to infection dynamics and the spread of avian influenza viruses (AIV). However, we lack detailed understanding migratory birds’ contribution to local AIV prevalence levels and their downstream socio-economic costs and threats.
To explain the potential differential roles of migratory and resident birds in local AIV infection dynamics, we used a susceptible-infectious-recovered (SIR) model. We investigated five (mutually non- exclusive) mechanisms potentially driving observed prevalence patterns: 1) a pronounced birth pulse (e.g. the synchronised annual influx of immunologically naïve individuals), 2) short-term immunity, 3) increase of susceptible migrants, 4) differential susceptibility to infection (i.e. transmission rate) for migrants and residents, and 5) replacement of migrants during peak migration.
SIR models describing all possible combinations of the five mechanisms were fitted to individual AIV infection data from a detailed longitudinal surveillance study in the partially migratory mallard duck (Anas platyrhynchos). During autumn and winter, the local resident mallard community also held migratory mallards that exhibited distinct AIV infection dynamics.
Replacement of migratory birds during peak migration in autumn was found to be the most important mechanism driving the variation in local AIV infection patterns. This suggests that a constant influx of migratory birds, likely immunological naïve to locally circulating AIV strains, is required to predict the observed temporal prevalence patterns and the distinct differences in prevalence between residents and migrants.
Synthesis and applications. Our analysis reveals a key mechanism that could explain the amplifying role of migratory birds in local avian influenza virus infection dynamics; the constant flow and replacement of migratory birds during peak migration. Aside from monitoring efforts, in order to achieve adequate disease management and control in wildlife - with knock-on effects for livestock and humans, - we conclude that it is crucial, in future surveillance studies, to record host demographical parameters such as population density, timing of birth and turnover of migrants.
Keywords: Avian influenza, Migratory birds, SIR, Migratory connectivity, Mallard, Host-pathogen interactions, Epidemiology, Immunity
Introduction
Highly pathogenic avian influenza (e.g. H5N1, H5N8) is a prime example of an emerging infectious disease with rapid rise in incidences in poultry, and potentially in humans and wild birds (Alexander 2007). When it comes to the spread and local amplification of avian influenza viruses (AIV), herewith threatening global economies and public health, migratory birds are increasingly thought to play a central role (Lycett et al. 2016). Several characteristics of migratory species make them seemingly perfect vectors for a variety of pathogens (Altizer, Bartel & Han 2011). During their migratory journey migrants may encounter a broad range of parasite species and strains, thereby increasing the likelihood of transmitting novel parasites to resident communities they encounter en-route (Waldenström et al. 2002). Moreover, the physiological challenges migrants face during their migration, leading to potential trade-offs with their immune function, may increase their susceptibility to infection (Buehler et al. 2008). Finally, many migrants aggregate in large numbers at so-called stop-over sites leading to further enhancement of pathogen transmission (Krauss et al. 2010; Fritzsche McKay & Hoye 2016). However, despite the pervasive support for the role of migrants in pathogen dispersal (e.g. Fourment, Darling & Holmes 2017), conceptual support of clearly documented and quantified examples of their role in infection dynamics are surprisingly rare (Altizer, Bartel & Han 2011). This significantly constrains our ability to design strategies to recognise and mitigate potential disease threats coming from wildlife populations, which in turn could minimize the risk of spill-over to domestic animals and humans.
In this study, we built on a unique dataset from a detailed study on mallard ducks (Anas platyrhynchos); van Dijk et al., (2014) described AIV infection dynamics at a small spatial scale, a single duck decoy, over a full annual cycle. During part of the year (i.e. autumn and winter), the local mallard community consisted of resident and migratory birds and by characterizing the majority of individuals as either migratory or resident, van Dijk et al. (2014) showed that the major peak of AIV infection in autumn coincided with the arrival of susceptible migratory mallards. Here, we use a Susceptible-Infectious-Recovered (SIR) modelling framework, aiming to explore multiple mechanisms that are suggested to drive local AIV infection dynamics in wild birds, quantify their relative importance, and identify the differential role of migrants and residents within the population. We start with a very basic demographic and epidemiological model, gradually increasing the complexity by adding and combining the following five, non-mutually exclusive, mechanisms:
1 – Birth pulse
The synchronised hatching of chicks can be a major factor influencing seasonal changes to the density of susceptible individuals. The vast majority of animal populations show marked seasonal variation in the timing of birth, resulting in a pulsed influx of immunologically naïve juveniles (Hosseini, Dhondt & Dobson 2004; Begon et al. 2009). Such seasonal birth pulses have been shown to both precede annual peaks in infection prevalence in wildlife (Hinshaw et al. 1985; Peel et al. 2014; Avril et al. 2016), and be fundamental to producing these dynamics in empirically validated models (Hosseini, Dhondt & Dobson 2004; He 2005; Begon et al. 2009).
2 – Short-term immunity
The vast majority of theoretical AIV infection studies assume long-term or even permanent immunity (e.g. Galsworthy et al. 2011; Nickbakhsh et al. 2014). In fact, the immune response to AIV within the host appears to be sufficient to attenuate the duration and the intensity of subsequent infections (Fereidouni et al. 2010; Jourdain et al. 2010). However, the relatively weak antibody response may be short-term and antibodies might be detectable for a few months only (Kida, Yanagawa & Matsuoka 1980; Hoye et al. 2011; Samuel et al. 2015).
3 – Increase of susceptible migrants
A key feature that put migrants into the spotlight of infectious disease dynamics is the fact that they visit disparate locations throughout their annual cycle (Altizer, Bartel & Han 2011). In combination with the rather strain-specific immune response to AIV infections (Jourdain et al. 2010), migrants may thus be generally more susceptible to local AIV strains once they arrive at a new location (Verhagen et al. 2014).
4 – Differential susceptibility
The physiological challenges associated with migratory journeys may result in a trade-off with immune functioning, leading to a reduced immunocompetence in migrants compared to residents (Altizer, Bartel & Han 2011). These differences in immune status may translate into a higher likelihood of migrants becoming infected with AIV after contact with an infectious individual.
5 – Replacement of migrants
In the context of infectious disease dynamics, like with AIV, a yet understudied part of migration is the diversity in migratory strategy. At one extreme all individuals of a population may have an identical spatial-temporal pattern in their migration (e.g. Orell et al. 2007; Stanley et al. 2012), while at the other end of the spectrum, individuals migrate within a broad time window and may not necessarily follow the same route. These differences in migration timing and arrival can have profound effects on the local composition, density and turnover of individuals at breeding, wintering and staging sites, and therewith on the local host-pathogen dynamics (Møller & Szep 2011; Bauer, Lisovski & Hahn 2016).
Materials and Methods
Study Species
Mallards are one of the most common and numerous waterfowl species around the world, with an estimated population size of 19 million individuals (Delany & Scott 2006). The species is also considered to be the major AIV reservoir in the wild (Webster et al. 1992). Mallards are partially migratory, meaning that the population consists of both migratory and resident birds. Birds breeding in western Europe (e.g. the Netherlands) are mainly sedentary, and northern breeding birds (e.g. Scandinavia, the Baltic, north-west Russia) migrate in autumn to overwinter between Denmark, northern France and Britain (Scott & Rose 1996).
Study Site and Sampling
Detailed description of the sampling diagnostic methods can be found in van Dijk et al. (2014). In short, mallards were caught using swim-in traps of a duck decoy (Payne-Gallwey 1886) located near Oud Alblas (4°42’26’’E, 51°52’38’’N), the Netherlands. Sampling took place from March 2010 until February 2011. On average, the duck decoy was visited six times per month, capturing approximately 15 individuals per visit, resulting in a total of 1109 samples being collected. For detection of current AIV infection, both cloacal and oropharyngeal samples were taken and analysed. To determine the origin of the individuals, thus distinguishing between migrants and residents, stable hydrogen isotope (δ2H) ratios were measured within freshly moulted feather collected from the individuals caught between August and December. The origin of 319 out of 458 individuals could be classified (van Dijk, Meissner & Klaassen 2014).
Modelling
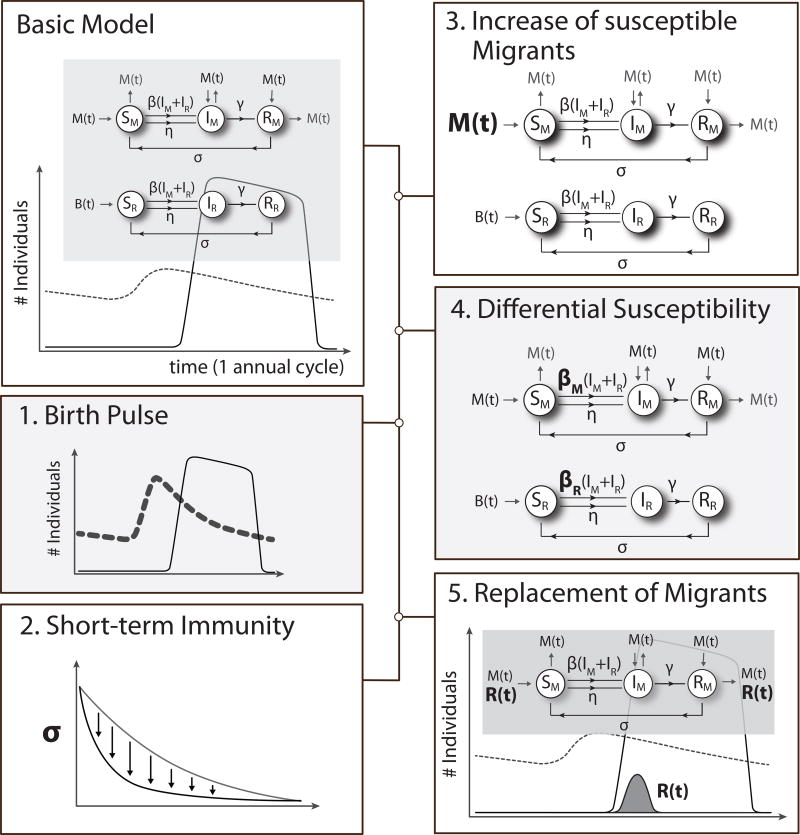
All five potential mechanisms (“modifications” hereafter) had an identical underlying “basic” model (Figure 1), describing mallard demography – birth, death, migration – at the study site and the basic AIV infection dynamics.
Figure 1.
The structure of the basic model and the main differences of the modifications that are based on five potential mechanisms that may drive local AIV infection dynamics in wild birds. For the basic model, the grey box shows the flowchart of the movement of migrant (M) and resident (R) individuals between the susceptible (S), infectious (I) and recovered (R) compartments as described by the model equations 1–6. Natural mortality (m) is not depicted, but is assumed to occur within all three compartments and at the same rate for all individuals (i.e. resident and migrant). The graph below the grey box shows the general annual demographic dynamics of resident mallards (dashed line) and migratory mallards (solid line) visiting the study site during the annual cycle. For the 1st model modification, the bold dashed line shows the potential dynamics of the resident population with a more pronounced birth pulse B(t). The 2nd model modification assumes a reduced immune rate (σ) and thus a faster loss of immunity against AIV infections. In the 3rd model modification, relatively more migrants enter into the pool of susceptible (S) individuals. In the 4th model modification, the transmission rate (β) is modelled separately and has different values for migrants (βM) and residents (βR). For the 5th model modification, the R(t) curve describes the amount and the shape at which migrants within the infectious (I) and the recovered (R) pool are replaced by new susceptible migrants.
Basic model structure
The AIV infection dynamics were modelled using a SIR model with the components Susceptible (S), Infectious (I) and Recovered (R) (Figure 1). Infections are generally thought to be density dependent (McCallum, Barlow & Hone 2001), and modelled with a transmission term β, describing the rate at which susceptible birds (S) become infected through direct or indirect contact with infectious individuals (I). For AIV, indirect transmission is considered to be of crucial importance and follow a faecal-oral route via water (Webster et al. 1992). In addition to the β term for transmission, we allowed for background transmission (η) to account for virus persistence in water (e.g. Stallknecht et al. 1990), because environmental transmission can potentially occur after infectious individuals have left the site. However, background transmission was set to a low value given that Nazir et al. (2010) found persistence of AIV to range between days to a couple of weeks depending on temperature. Most importantly background transmission allows occasional re-introduction of the virus in the absence of infectious birds and is a crucial mechanism enabling the persistence of pathogens, particularly within small wildlife communities that are below the critical community size where epidemics cannot be sustained by direct transmission only (Breban et al. 2009). Birds that recovered from AIV infection at rate γ, were moved from the infectious compartment (I) to the recovered compartment (R). Loss of immunity occurred at rate σ, transferring individuals from the recovered (R) to the susceptible (S) compartment. Arriving migrants were allocated across the susceptible (S), infectious (I) and recovered (R) compartments in the same proportions as the resident population.
The demography was modelled as an integral part of the SIR model, with separate differential equations describing the migrant and resident population. The basic model assumed a resident population of 700 adult individuals, reflecting the approximate number of residents observed at the study site (van Dijk et al. 2014). Birth rate (B(t)) was modelled for residents only and followed a normal distribution, defined by mean day of birth (Bmean) and its standard deviation (Bsd), which was multiplied by the number of breeding pairs (0.5*Npop, i.e. half the resident population size) and a fixed number of hatchlings per pair (Nhatch) to derive a daily number of hatchlings that enter the population. All individuals (i.e. residents and migrants) experienced natural mortality at rate m. The arrival of migrants at rate M(t) was also modelled following a normal distribution defined by mean arrival date (Amean) and its standard deviation (Asd), which was multiplied by the resident population size (Npop) and the ratio of migrants to residents (Prmig). Departure of migrants was modelled by setting the migratory population to 0 at day 92 (1st of April), when migrants were expected to have left the area for spring migration (Cramps & Simmons 1977).
These assumptions formed the basic model that consisted of six ordinary differential equations:
Resident population
| eq. 1 |
| eq. 2 |
| eq. 3 |
Migrant population
| eq. 4 |
| eq. 5 |
| eq. 6 |
Model modifications
For the 1st modification birth pulse we divided the resident population into adults (>10 months old) and juveniles (<10 months old), with 10 months being the period between hatching and sexual maturity in juveniles. Separation in age class allowed for differential mortality rates. At the end of the annual cycle (defined at day 92, April 1st) all juveniles were transferred into the pool of adults. The 2nd modification, short-term immunity, did not require a structural change of the basic model. In the 3rd modification, increase of susceptible migrants, relatively more migrants (b) enter into the susceptible (S) compartment instead of being distributed across the three compartments according to the proportions of the resident population. In the 4th modification, differential susceptibility, we allowed for separate transmission rates for residents and migrants (βR and βM, respectively). To model the 5th modification, replacement of migrants, a function describing this replacement was added R(t), which was modelled using a symmetric double logistic function with parameters mean, amplitude, slope and kurtosis (i.e. Rmean, Ramp, Rslope, Rkurt respectively). All model equations can be found in supplementary material S1.
Model parameterisation
Only few model parameters could be fixed at a value derived from literature or personal observations (Table 1). For most parameters, we lack data since these are often difficult or even impossible to measure. For ‘non-fixed’ parameters we defined a likely range (Table 1) over which they were allowed to vary during model simulations. Simulations were conducted with 32 different model scenarios, where each scenario consisted of a combination of the basic model and one or more of the five model modifications.
Table 1.
The parameters of the basic model and model modifications. If the value of a parameter is a single integer, the parameter was hold fixed during the Markov Chain Monte Carlo (MCMC) simulation. All other parameters were optimised within the given range. Large numbers on the left group parameters that are unique to a certain model modification. See methods for references and validation of the defined parameter ranges.
| Symbol | Definition | Value/Range | Units | |
|---|---|---|---|---|
| β | transmission rate | 0.1x10−4 to 0.4x10−3 | bird−1 day−1 | |
|
|
||||
| γ | recovery rate | 1/12 to 1/3 | day−1 | |
|
|
||||
| σ | immune rate | 0.0013 to 0.013 | bird−1 day−1 | |
|
|
||||
| η | background transmission rate | 10−5 | day−1 | |
|
|
||||
| Bmean | mean day of birth | 135 to 220 | day of the year | |
|
|
||||
| Bsd | standard deviation of birth | 0.5 to 25 | days | |
|
|
||||
| Nhatch | number of hatchlings per pair | 0.63 | individuals | |
|
|
||||
| Prmig | ratio of migrants to residents | 0.5 to 4 | proportion | |
|
|
||||
| Amean | mean arrival day of migrants | 240 to 335 | day of the year | |
|
|
||||
| Asd | standard deviation of arrival of migrants | 0.5 to 50 | days | |
|
|
||||
| m | mortality rate | 0.315/365 | bird−1 day−1 | |
|
|
||||
| 1 | Npulse | number of hatchlings per pair | 4 | individuals |
|
| ||||
| mjuv | juvenile mortality rate | estimated | bird−1 day−1 | |
|
|
||||
| 2 | σs | short-term immunity rate | 0.0013 to 0.066 | bird−1 day−1 |
|
|
||||
| 3 | b | susceptible migrants | 5 to 75 | percent |
|
|
||||
| 4 | βM | transmission rate in migrants | 0.1x10−4 to 0.3x10−2 | bird−1 day−1 |
|
| ||||
| βR | transmission rate in residents | 0.1x10−4 to 0.4x10−3 | bird−1 day−1 | |
|
|
||||
| 5 | Rmean | mean of migratory replacement | 240 to 288 | day of the year |
|
| ||||
| Ramp | proportion of migratory replacement | 0.05 to 0.6 | proportion | |
|
| ||||
| Rslope | slope of migratory replacement | 2 to 25 | days | |
|
| ||||
| Rkurt | kurtosis of migratory replacement | 2 to 3 | days | |
In the basic model, birth was modelled using a fixed number of 0.63 hatchlings per pair (Nhatch). This value ensured a stable population size over time given a natural daily mortality rate (m) of 8.63×10−5, which was based on a life expectancy of 2.27 years for mallards (Schekkerman & Slaterus 2008). AIV transmission rates (β) in wildlife populations are largely unknown, therefore we chose a broad range for β, making sure that the basic reproduction number of the virus (R0) ranges from 0.8 to 8.0. The background infection rate (η) was set to a fixed value of η = 10−5, which gives a probability of 1% for a single mallard to become infected in a mean lifetime of 828 days (Galsworthy et al. 2011). The mean recovery rate (γ) and the immune loss rate (σ) can vary between host-species, their immunological history and between AIV strains (Costa et al. 2010; Fereidouni et al. 2010; Jourdain et al. 2010; Curran 2012). A mild strain may cause longer periods of virus excretion (γ≈1/12: Kida, Yanagawa & Matsuoka 1980; Costa et al. 2010; Jourdain et al. 2010), whereas a more severe strain may have a short generation time of approximately three days (γ≈1/3: Ng & Higgins 1986; van der Goot et al. 2008; Latorre-Margalef et al. 2009). The immune rate (σ) was set to a range resulting in a mean loss of immunity within 75 to 730 days. Mean autumn migration (Amean) was set from August 27th to November 30th, depicting the period that migratory mallards may arrive at the wintering grounds in Western Europe. This period reflects the observed bird migration window (Fransson & Petersson 2001; Bakken, Runde & Tjorve 2003).
For the 1st modification birth pulse, a larger number of hatchlings was chosen. Mallards are known to produce large clutches with an average clutch size of 9 to 13 eggs (Cramp & Simmons 1977), that potentially result in 4 instead of 0.63 hatchlings per pair (Npulse) that enter the resident population (Figure 1). Mortality rates for juveniles (mjuv) were estimated prior to each simulation to ensure a stable population size (N = 700), taking the Bmean, Bsd, and m into account. The parameter boundaries describing the replacement of migrants (R(t)) in the 5th modification replacement of migrants was defined to allow the replacement of individuals at the start of the migratory period (August 27th) until mid-October, depicting the peak of migration. Migrants with a current AIV infection were not subject to replacement, since this would not change the number of infected individuals and the infection dynamic. Although in reality, infected individuals might also be subjected to replacement. Empirical studies in mallards and other waterfowl have shown that AIV infection may hamper migration and movements (van Gils et al. 2007; van Dijk et al. 2015; Hoye et al. 2016), suggesting that infected individuals may remain stationary during the course of an infection.
Simulation and model fit
To allow demographic and infection patterns to stabilize, all models were run over ten annual cycles, where the last cycle was used for comparison with the empirical patterns observed in the field. All possible model scenarios (n = 32) were written in C++ and compiled, as well as integrated using the ‘ode’ method in R Package deSolve (Soetaert, Petzoldt & Setzer 2010). To estimate parameter values, their relative importance and uncertainties, we used a Markov Chain Monte Carlo (MCMC) simulation with a delayed adaptive Metropolis algorithm (Haario et al. 2006), implemented in the function MCMCmod from R Package FME (Soetaert & Petzoldt 2010). This MCMC algorithm uses a multivariate proposal distribution that is automatically adapted to allow for posterior corrections between parameters and identifies the direction of principal change along the ridges in the posterior landscape. The acceptance rate is improved by the delayed rejection part of the algorithm where, instead of immediately advancing the chain following rejection of a parameter set, a second proposal is made that depends on both the current position of the chain and the rejected parameter set. Ultimately, the DRAM algorithm produces posterior distributions of the parameters by minimizing the so-called ‘model costs’ defined as the negative sum of the log-binomial densities (binomial response of the number of infected and non-infected individuals).
For all possible model scenarios, we simulated 25 independent MCMC chains using 10,000 iterations with an update of the covariance matrix after every 50th iterations and one delayed rejection step. Initial parameters were chosen randomly from the parameter ranges for each MCMC chain. The possible parameter space for all non-fixed parameters is shown in Table 1. The last 2,500 iterations were used to describe the posterior distribution of each of the non-fixed parameters as well as to calculate the confidence interval of the model predication and the goodness of fit used to compare model scenarios based on Watanabe-Akaike information criterion (WAIC), a pointwise out-of-sample prediction accuracy (Watanabe 2010; Gelman et al. 2013). Within the DRAM optimization routine, observations for residents and migrants were compared separately with the respective model output (i.e. number of infected and non-infected individuals in residents and migrants). Sampling results of individuals with unknown migration status were also included in the model and compared with the pooled predicted prevalence in both residents and migrants. Thus, the best fitting model is based on the sum of three (i.e. resident, migrant, unknown status) separate ‘model costs’ (or the sum of three log-binomial density distributions). We used median WAIC value for each model scenario to rank their potential in predicting the observed AIV infection dynamic.
We investigated potential collinearity of model parameters for each of the scenarios by calculating the correlation matrix of the parameters across the final 2,500 iterations.
Results
Model scenarios
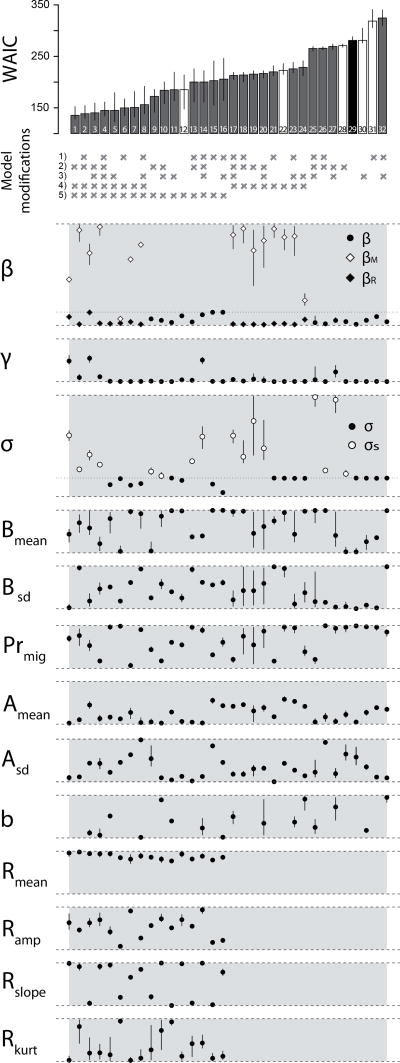
The basic model in combination with the five different model modifications led to 32 possible scenarios (Figure 2). The median WAIC values of the model fit for these 32 scenarios ranged from 135.5 to 324.6, with lower values indicating better model fits and a higher potential to predict the observed AIV infection dynamic. Although their annual AIV infection dynamics appeared slightly different, the best fitting three scenarios (rank 1 to 3) were within a small WAIC range of 4.8, and could be considered similarly good (Figure 2). In fact, the first 8 scenarios differed in their ability to predict the observed infection dynamic from the remaining scenarios with median WAIC values from 135.46 to 156.17 compared to > 172.3 in the ranks from 9 to 32. These 16 best ranked scenarios all included the 5th modification, the replacement of migrants. In addition to the 5th modification, modification 4 was found to increase the fit in the 8 best-ranked scenarios (Figure 2). This modification also contributed the 8 scenarios missing the 5th most important modification (rank 17 to 24).
Figure 2.
Ranked model scenarios based on median WAIC (‘model cost’) of the model fit. White bars indicate scenarios with a single modification on top of the basic model. The black bar shows the basic model without any modification. Models with lower WAIC represent better model fits. The bars represent the median WAIC over 25 independent MCMC chains with random selection of initial priors within the range of the respective parameter. Error bars indicate 95% confidence intervals. The inclusion of the five model modifications to the basic model are shown below the bar plot, with a cross indicating that the particular modification was included in the scenario. The model scenarios are 1) Birth Pulse, 2) Short-term immunity, 3) Increase of Susceptible Migrants, 4) Differential Susceptibility and 5) Replacement of Migrants. Below, all estimated parameter values across the 32 scenarios are shown with the 50% (circle) and the 10% and 90% percentiles of the posterior distributions (10,000 iterations). The grey areas between the dashed lines indicate the pre-set parameter ranges. In case of the transmission rate (β) and immune rate (σ), the dotted lines indicate the reduced boundaries of the parameter for the 4th differential susceptibility and the 2nd short-term immunity modifications.
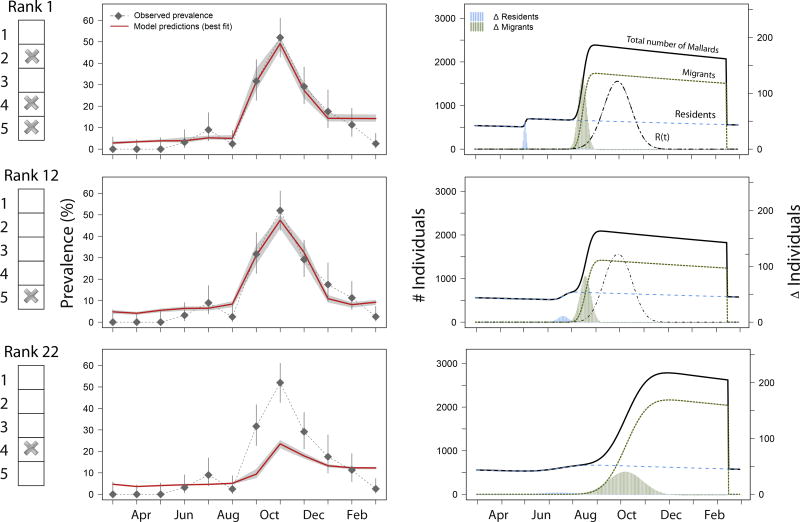
The replacement of migrant modification was the best modification in explaining the observed WAIV prevalence pattern in isolation with the basic model (rank 12, Figure 2 and Figure 3). However, in isolation, the 4th modification resulted in a relatively poor fit (rank 22, Figure 3). No eminent pattern was found in the distribution of the other three modifications across the 32 scenarios ranked by their WAIC value (Figure 2).
Figure 3.
Results of three model scenarios: the best-ranked scenario (with 3rd, 4th and 5th modification, Rank 1), the scenario with the most influential modification only (5th modification, Rank 12), and the scenario with the second most influential modification only (4th modification; Rank 22). The left column shows which of the model modifications were included in the respective scenario. The middle column shows the observed AIV prevalence levels (±95% CI) at the study site (dashed line with diamond symbols), and the model prediction (bold line) of the best fitting model with the respective modifications. Grey area around model predictions indicate the sensitivity range of the fit (e.g. ±95% CI). The underlying demography for each depicted scenario is shown in the right column with absolute numbers of individuals (black line) consisting of migrants (dotted line) and residents (dashed line). The replaced migrants (R(t)) is also shown as the absolute number of individuals replaced at time t (in days). Additionally, the rate of change in individuals (∆ Individuals) is shown for residents indicating birth (light grey bars) and migrants indicating initial arrival (dark grey bars).
Parameter estimates
Looking at the best parameter combinations across all 32 scenarios (Figure 2), once the transmission rates were modelled separately for migrants and residents (thus including the 4th differential susceptibility modification), βM was estimated higher than βR except for rank 6 that was the only scenarios where βM was chosen within the boundaries of βR. The recovery rates (γ) were almost always at the lower end of the pre-defined parameter range: AIV infected individuals recovered after an average of approximately 12 days. The immune rates (σ) were highly variable across the different scenarios and thus unrelated to the goodness of fit (WAIC value) of the models. However, in all scenarios that included short-term immunity, the rate (σs) was higher than it would have been in the absence of the 2nd short-term immunity modification. This was notably true for scenarios missing the 4th and the 5th modification when short-term immunity was the mechanism that could elevate the prevalence levels during the late summer and autumn period. In almost all scenarios, the ratio of migrants to residents (Prmig) was highly skewed towards migrants: 3–4 times the resident population.
In general, across the first 16 scenarios that included the 5th replacement of migrants modification, the arrival peak of migrants (Amean) occurred relatively early (low Amean) with respect to the pre-set window (early August). In contrast, without this modification, peak arrival dates occurred in mid-October. The shape of the arrival curve (Asd) was consistently in the lower half of the pre-set parameter range and was particularly low, i.e. reflecting a quick and synchronised arrival, in the best fitting scenarios. Not much variation was found in the timing of replacement (Rmean), i.e. the start was always at the upper boundary and therefore as early as possible. The amplitude of the replacement curve (Ramp) varied, but was consistent within the upper half of the parameter range (0.35–0.6).
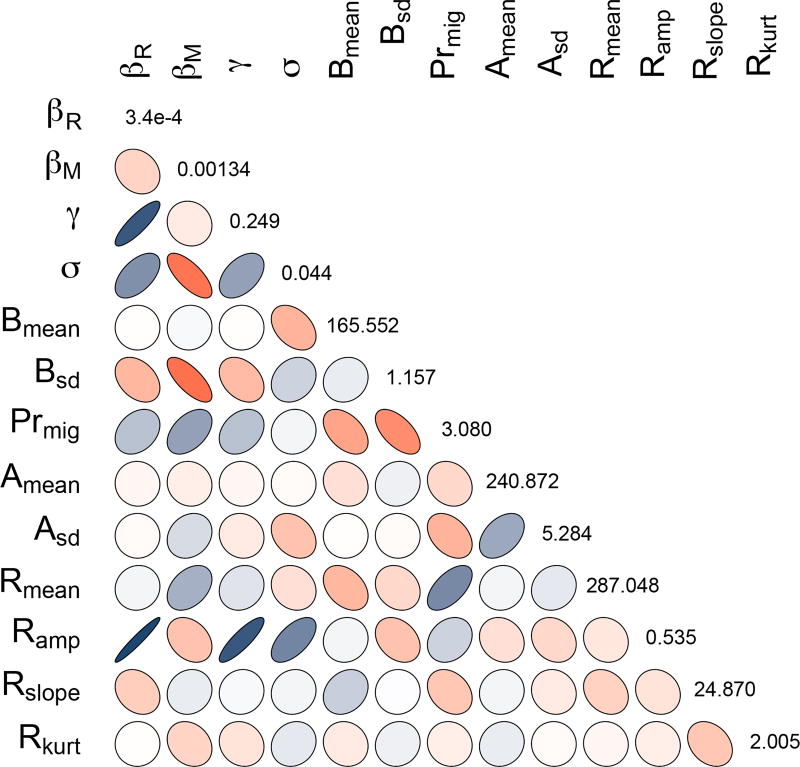
Parameter correlation
We used the posterior distributions of each parameter from the last 2,500 MCMC iterations to illustrate the potential correlation between parameters. In the best ranked model scenario (rank 1) that included the 2rd short-term immunity, the 4th differential susceptibility and the 5th replacement of migrants modification, many parameters showed high correlation (indicating parameter identification problem), e.g. the recovery rate (γ) was positively correlated with the transmission rate of the resident population (βR). Whereas the immune rate (σ) was negatively correlated with the transmission rate of the migrant population (βM). This means that all four parameters could contribute to higher prevalence levels during the major peak. Additionally, the shape of the replacement of migrants distribution (Rsd) was found to be correlated with the transmission rates of residents (γ) and the immune rate. However, correlations between parameters different immensely between the scenarios (supplementary material S2).
Discussion
Mathematical modelling has great potential to probe the complex dynamics of infectious diseases and identify the mechanisms of transmission. Therewith, it may also indicate approaches for prevention and control that may help shape national and international public health policy (Heesterbeek et al. 2015). In this study, we used a mathematical modelling approach to evaluate several mechanisms that have been suggested to drive local AIV dynamics in wild bird populations. To evaluate these mechanisms, we fitted the predicted infection dynamics to a unique sampling dataset of a year-round, small-scale AIV surveillance study in the key European AIV host species, the mallard (van Dijk et al. 2014). We found that one particular mechanism, the local replacement of migrants during the peak migration, contributed most significantly towards better model predictions.
There is a general perception that animal migration plays a central role in wildlife disease dynamics by enhancing the global spread of pathogens (Altizer, Bartel & Han 2011). Notably with respect to AIV there are a number of high-profile studies stressing this case (Verhagen, Herfst & Fouchier 2015; Hill et al. 2016; Lycett et al. 2016). In addition, we show that migrants may importantly facilitate local AIV infection dynamics. Our models provide strong indications that the role of migrants in infection dynamics is not simply determined by the presence of migrants, but critically relies on how migration takes place over time. Migratory birds within a population may migrate highly synchronised and visit stop-over sites all at once, or they may differ in their timing leading to several waves of migrants and extended periods during which migrants arrive at and depart from stop-over sites (Bauer, Lisovski & Hahn 2016). In most species, including the mallard (Fransson & Petersson 2001; Bakken, Runde & Tjorve 2003), the migratory season protracts over several weeks up to a few months. This is likely to lead to the mechanisms behind migratory replacement used as a modification in our model: arriving migrants stay in the area for a limited period of time after which they move on and are replaced by newly arriving individuals. As a consequence, individuals that have acquired some degree of protection against re-infection by means of AIV specific antibodies (either due to AIV exposure prior to arrival or at the study site itself), are replaced by potentially susceptible individuals that may perpetuate or even invigorate local transmission dynamics.
Besides the strong effect of the replacement of migrants, the models with better predicting power also included the modification differential susceptibility with higher transmission rates in migrants compared to residents. This is in line with the observed data showing that the major AIV prevalence peak coincided with the arrival of migrants (van Dijk et al. 2014). There are several non-mutually exclusive mechanisms that may explain why the better predicting models had an increased susceptibility in individual migrants. Empirical studies have shown that the physiological challenges accompanied with migration, including a potential trade-off with the immune system, can reduce their immunocompetence and render migrants more susceptible (Buehler et al. 2008). It has also been suggested that migrants are generally more susceptible, since their immune system is less specialised but adapted to cope with the exposure of different and disparate environments and their pathogens (Waldenström et al. 2002).
Combining the findings from the model and the empirical study, we can conclude that migrants may not exclusively affect the AIV infection dynamics of resident populations through their often suggested role in the introduction of new virus strains (e.g. Verhagen, Herfst & Fouchier 2015; Lycett et al. 2016) and the general increase in host-densities (Gaidet et al. 2012, Hill et al. 2012), but may have an additional role in the amplification of the virus (Yin et al. 2017). Since migration is a large-scale multi-species phenomenon, this might more generally be the case and it questions the hypothesis that migrants only have a role in dispersing and introducing AIVs and thereby affecting resident populations.
Most northern hemisphere AIV surveillance studies in wild bird populations show similar patterns of pronounced late summer - early autumn infection peaks (e.g. Munster et al. 2007; Hénaux et al. 2013; Lisovski, Hoye & Klaassen 2017). Clearly, our model scenarios are ranked by their ability to capture this pronounced feature within the entire annual infection dynamic. While allowing for differential susceptibility to infection for migrants and residents seems to strengthen the predictions of the AIV infection peak, the overall importance of this parameter (i.e. transmission rate (β)) reduces the informative power of the remaining evaluated mechanisms, like the birth pulse (B(t)), the short-term immunity (σs) and the epidemiological state at which migrants enter a resident population (M(t)). However, those mechanisms might still be crucial. Indeed, the correlation matrix (Figure 4) shows that the immune rate (σ) and the transmission rate (β) could be parameters of importance and are highly correlated with parameters that significantly influence the autumn infection peak, like the ratio of migrants to residents (Prmig) and the amplitude of migratory replacement (Ramp). However, the correlation between such potentially important parameters also indicates that we require more information to narrow their potential range, otherwise the estimates become rather uninformative.
Figure 4.
The parameters of the best ranked model scenario (matrix diagonal) and the correlation matrix of all parameters from the last 2.500 MCMC iterations. The intensity of grey and shape of the ellipsoids represent the strength (dark colour and narrow ellipse represent correlation coefficients close to 1 or −1) and direction of the correlation.
Birth pulses have previously been shown to be of importance in AIV infection dynamics in wild waterfowl (Hénaux et al. 2013), and appeared to be fundamental to produce annual infection peaks in empirically validated disease models (Hosseini, Dhondt & Dobson 2004; He 2005; Begon et al. 2009). Avril et al. (2016) showed that migratory juvenile mallards had a consistently higher risk of getting infected with AIV compared to adults. Hénaux et al. (2013) in particular showed that the early autumn AIV infection peak in a major host species across the North American continent, the blue-winged teal (Anas discors), was mainly due to infections in immunologically naïve juveniles, and that only a small proportion of adults were within the susceptible pool and contributed to the transmission dynamics. Although knowledge on the number of migrants among adults was not included in that study, our results raise questions whether accounting for the underlying geography of the locations at which the birds were sampled, including associated differences in their migration strategies, could lead to other conclusions. Migration routes across the North American continent are grouped into four major north-south stretching flyways, and phylogenetic analysis of AIV indeed indicate that those flyways represent corridors for gene flow with more restricted east-west gene flow, suggesting that migration of waterfowl occurs on a large scale from north to south with little longitudinal mixing (Lam et al. 2012; Fourment, Darling & Holmes 2017, but see Krauss et al. 2010 for bottleneck in wader migration within North America). The preferentially north-south migration across a broad east-west front may restrict the origin and number of migrants within wetlands along the different migratory routes and reduce their influence on the local AIV infection dynamics. In contrast, the wetlands in central-northern Europe (e.g. the Netherlands) are within a bottleneck of the East-Atlantic Flyway in which most routes of waterfowl from a vast geographical origin merge (Scott & Pose 1996).
In our effort to construct relatively simple models allowing for comprehensive testing of a set of hypothesised drivers for local AIV infection dynamics, we had to make a considerable number of assumptions. Possibly the most important one is that we ignored the existence of viral subtypes, and that AIV infection might elicit subtype specific immunity against further infections (Latorre-Margalef et al. 2017). However, ducks may show limited immune responses to AIV infection contrasting findings from e.g. chickens (Kida, Yanagawa & Matsuoka 1980) and can quickly be re-infected with the same AIV subtype (Chaise et al. 2014) providing support for our simplifying approach to ignore potential antigenic variation in AIV infected mallards. Furthermore, like most SIR(S) modelling approaches, we ignore individual variation and the potentially important role of transmission heterogeneity and “superspreaders” in the infection dynamics (e.g. Lloyd-Smith et al. 2005). However, although we consider this of great importance, empirical data to support the existence of such transmission heterogeneity for AIV among conspecifics within wildlife populations is thus far lacking. Finally, the nature of the empirical data set that we used, e.g. one annual cycle at one location for one bird population, might limit our ability to extrapolate our findings. However, the temporal pattern and amplitude of the epizootic is comparable to what has been found in other studies in north-western Europe (e.g. Munster et al. 2007) and temperate areas in North America (e.g. Lisovski, Hoye & Klaassen 2017). Interestingly, despite some profound geographical variation in AIV infection patterns (Gaidet et al. 2012; Lisovski, Hoye & Klaassen 2017), globally the drivers for those patterns in AIV prevalence appear to be the same.
Management Implications
Avian influenza viruses are currently an increasing threat to the global poultry production sector and to public health (Hien, de Jong & Farrar 2004). The poultry industry and trade is an important part of this problem, with some highly pathogenic AIV strains being endemic in poultry in several countries in Africa and Asia, where also low pathogenic AIV is sometimes more readily circulating among domestic than wild birds (Hassan et al. 2017). Irrespectively, wild birds, and notably birds of the order Anseriformes (ducks and geese), are the ancestral reservoir host for AIV (Caron, Capelle & Gaidet 2017) and remain of key importance for global AIV diversity (Alexander 2007), notably in the face of readily reassorting high pathogenic AIV virus such as H5 clade 2.3.4.4 (Lee et al. 2017). Anseriformes also play a major role in the dispersal of AIV (Alexander 2007), including dispersal of highly pathogenic strains (e.g. Lycett et al. 2016); the latter most likely through spill-back from poultry (Messenger, Barnes & Gray 2014). It is for these reasons that an understanding of the ecology and transmission of AIV in wildlife, and migratory waterbirds in particular, is required to assist its management and control in livestock and humans in the future (Kuiken et al. 2005, Coker et al. 2011). Mathematical models allow probing the complex dynamics of host-pathogen interactions to help identify the mechanisms of transmission, enabling prediction and possibly prevention of outbreaks. In spite of AIV infection dynamics being arguably one of the best studied avian-wildlife host-pathogen systems, our study highlights that we typically lack crucial information allowing identifying and quantifying the major mechanisms that lead to high prevalence levels in cases where migrants are involved; i.e., data on the number and turnover of migrants in a bird population. Thus, besides maintaining efforts in virus sampling and growing our understanding of virus diversity and evolution, their temporal occurrence and host range (Olsen et al. 2006; Munster et al. 2007; Caron, Capelle & Gaidet 2017) we additionally need increased efforts in recording of host demography. Given the here revealed importance of migration, and notably the timing and strategy of migration, it appears crucial to extend the wildlife disease surveillance database with demographic features such as the timing of birth and density, as well as turnover. Turnover can be estimated by flow models (Nolet & Drent 1998; Drever & Hrachowitz 2015), by combining counts with concurrent (re)sightings of marked individuals (e.g. Frederiksen et al. 2001) or by behavioural-based simulations of stopover site use (Nolet et al. 2016; Stillman et al. 2015). Knowledge of these parameters would allow us to use more in depth mathematical models allowing the estimation of e.g. transmission-, recovery- and immune rates, the key processes in host-pathogen interactions (McCallum 2000). Therefore, we believe that besides unravelling the mechanistic understanding of infection dynamics of wildlife diseases, understanding demographic patterns, especially for systems that involve considerable numbers of migratory individuals, are as important as pathogen detection.
Supplementary Material
Acknowledgments
We thank Hans Heesterbeek, Bethany Hoye, Elaine Ferguson and Fränzi Korner-Nievergelt for valuable discussions. Three anonymous referees and Bred Elderd helped to significantly improve the manuscript. SL was partly funded by the Australian Animal Health Laboratory (AAHL), Geelong. This study was supported by the Australian Research Council (DP130101935). The empirical study was funded by the Netherlands Organization for Scientific Research (NWO; Grant 820.01.018) and the National Institute of Health (NIH; contract NIAID HHSN266200700010C).
Footnotes
Authors’ contributions
MK, JvD, BN, DK, RF and SL conceived the research. SL analysed the data. SL and MK wrote the paper and all authors contributed significantly to the drafts and gave final approval for publication.
Data accessibility
Virus and antibody information, CT-values and hydrogen stable isotope measurements are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.j855b (van Dijk et al. 2013). The code is available from Zenodo. DOI: 10.5281/zenodo.1203836 (Lisovski et al. 2018).
References
- Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Altermatt F. Tell me what you eat and I'll tell you when you fly: diet can predict phenological changes in response to climate change. Ecology Letters. 2010;13:1475–1484. doi: 10.1111/j.1461-0248.2010.01534.x. [DOI] [PubMed] [Google Scholar]
- Altizer S, Bartel R, Han BA. Animal Migration and Infectious Disease Risk. Science. 2011;331:296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Seasonality and the dynamics of infectious diseases. Ecology Letters. 2006;9:467–484. doi: 10.1111/j.1461-0248.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- Avril A, Grosbois V, Latorre-Margalef N, Gaidet N, Tolf C, Olsen B, Waldenstrom J. Capturing individual-level parameters of influenza A virus dynamics in wild ducks using multistate models. Journal of Applied Ecology. 2016;53:1289–1297. [Google Scholar]
- Bakken V, Runde O, Tjorve E. Norwegian bird ringing atlas. Stavanger: Stavanger Museum; 2003. [Google Scholar]
- Bauer S, Lisovski S, Hahn S. Timing is crucial for consequences of migratory connectivity. Oikos. 2016;125:605–612. [Google Scholar]
- Begon M, Telfer S, Burthe S, Lambin X, Smith MJ, Paterson S. Effects of abundance on infection in natural populations: Field voles and cowpox virus. Epidemics. 2009;1:35–46. doi: 10.1016/j.epidem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Breban R, Drake JM, Stallknecht DE, Rohani P. The Role of Environmental Transmission in Recurrent Avian Influenza Epidemics. Plos Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler DM, Piersma T, Matson KD, Tieleman BI. Seasonal Redistribution of Immune Function in a Migrant Shorebird: Annual-Cycle Effects Override Adjustments to Thermal Regime. American Naturalist. 2008;172:783–796. doi: 10.1086/592865. [DOI] [PubMed] [Google Scholar]
- Caron A, Capelle J, Gaidet N. Challenging the conceptual framework of maintainance host for influenza A virus in wild birds. Journal of Applied Ecology. 2017;54:681–690. [Google Scholar]
- Chaise C, Lalmanach A-C, Marty H, Soubies SM, Croville G, Loupias J, Marc D, Quéré P, Guérin J-L. Protection Patterns in Duck and Chicken after Homo- or Hetero-Subtypic Reinfections with H5 and H7 Low Pathogenicity Avian Influenza Viruses: A Comparative Study. Plos One. 2014;9:e105189. doi: 10.1371/journal.pone.0105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker R, Rushton J, Mounier-Jack S, Karimuribo E, Lutumba P, Kambarage D, Pfeiffer DU, Stärk K, Rweyemamu M. Towards a conceptual framework to support one-health research for policy on emerging zoonoses. Lancet Infectious Diseases. 2011;11:326–331. doi: 10.1016/S1473-3099(10)70312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TP, Brown JD, Howerth EW, Stallknecht DE. Effect of a Prior Exposure to a Low Pathogenic Avian Influenza Virus in the Outcome of a Heterosubtypic Low Pathogenic Avian Influenza Infection in Mallards (Anas platyrhynchos) Avian Diseases. 2010;54:1286–1291. doi: 10.1637/9480-072210-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramp S, Simmons KEL. Handbook of the Birds of Europe, the Middle East, and North Africa: The Birds of the Western Palearctic, Vol. 1: Ostrich to Ducks. Oxford University Press; Oxford: 1977. [Google Scholar]
- Curran JM. The surveillance and risk assessment of wild birds in northern Australia for highly pathogenic avian influenza H5N1 virus. Doctor of Philosophy. Murdoch University; 2012. [Google Scholar]
- Delany S, Scott D. Waterbird population estimates. 4. Wetlands International; Wageningen, The Netherlands: 2006. [Google Scholar]
- Drever MC, Hrachowityz M. Migration as flow: using hydrological concepts to estimate the residence time of migrating birds from the daily counts. Methods in Ecology and Evolution. 2017;9:1146–1157. [Google Scholar]
- Fereidouni SR, Grund C, Haeuslaigner R, Lange E, Wilking H, Harder TC, Beer M, Starick E. Dynamics of Specific Antibody Responses Induced in Mallards After Infection by or Immunization with Low Pathogenicity Avian Influenza Viruses. Avian Diseases. 2010;54:79–85. doi: 10.1637/9005-073109-Reg.1. [DOI] [PubMed] [Google Scholar]
- Fourment M, Darling AE, Holmes EC. The impact of migratory flyways on the spread of avian influenza virus in North America. Bmc Evolutionary Biology. 2017 doi: 10.1186/s12862-017-0965-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson T, Petersson J. Svensk ringmaerkningsatlas [Swedish bird ringing atlas] Ljungfieretagen Tryckeri AB; Orebro: 2001. [Google Scholar]
- Frederiksen M, Fox AD, Madsen J, Colhoun K. Estimating the total number of birds using a staging site. Journal of Wildlife Management. 2001;65:282–289. [Google Scholar]
- Fritzsche McKay A, Hoye BJ. Are migratory animals superspreaders of infection? Integrative and Comparative Biology. 2016;56 doi: 10.1093/icb/icw054. [DOI] [PubMed] [Google Scholar]
- Gaidet N, Caron A, Cappelle J, Cumming GS, Balanca G, Hammoumi S, Cattoli G, Abolnik C, de Almeida RS, Gil P, Fereidouni SR, Grosbois V, Tran A, Mundava J, Fofana B, El Mamy ABO, Ndlovu M, Mondain-Monval JY, Triplet P, Hagemeijer W, Karesh WB, Newman SH, Dodman T. Understanding the ecological drivers of avian influenza virus infection in wildfowl: a continental-scale study across Africa. Proceedings of the Royal Society B-Biological Sciences. 2012;279:1131–1141. doi: 10.1098/rspb.2011.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galsworthy SJ, ten Bosch QA, Hoye BJ, Heesterbeek JAP, Klaassen M, Klinkenberg D. Effects of Infection-Induced Migration Delays on the Epidemiology of Avian Influenza in Wild Mallard Populations. Plos One. 2011;6 doi: 10.1371/journal.pone.0026118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. Bayesian Data Analysis. CRC Press; Boca Raton: 2013. [Google Scholar]
- Haario H, Laine M, Mira A, Saksman E. DRAM: efficient adaptive MCMC. Statistics and Computing. 2006;16:339–354. [Google Scholar]
- Hassan MM, Hoque A, Debnath NC, Yamage M, Klaassen M. Are poultry or wild birds the main reservoirs for Avian influenza in Bangladesh? Ecohealth. 2017 doi: 10.1007/s10393-017-1257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. Deriving a neutral model of species abundance from fundamental mechanisms of population dynamics. Functional Ecology. 2005;19:187–193. [Google Scholar]
- Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, Eames KTD, Edmunds WJ, Frost SDW, Funk S, Hollingsworth TD, House T, Isham V, Klepac P, Lessler J, Lloyd-Smith JO, Metcalf CJE, Mollison D, Pellis L, Pulliam JRC, Roberts MG, Viboud C, Collaboration INII. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347 doi: 10.1126/science.aaa4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hénaux V, Parmley J, Soos C, Samuel MD. Estimating transmission of avian influenza in wild birds from incomplete epizootic data: implications for surveillance and disease spread. Journal of Applied Ecology. 2013;50:223–231. [Google Scholar]
- Hien TT, de Jong M, Farrar J. Avian influenza - A challenge to global health care structures. New England Journal of Medicine. 2004;351:2363–2365. doi: 10.1056/NEJMp048267. [DOI] [PubMed] [Google Scholar]
- Hill NJ, Ma EJ, Meixell BW, Lindberg MS, Boyce WM, Runstadler JA. Transmission of influenza reflects seasonality of wild birds across the annual cycle. Ecology Letters. 2016;19:915–925. doi: 10.1111/ele.12629. [DOI] [PubMed] [Google Scholar]
- Hinshaw VS, Wood JM, Webster RG, Deibel R, Turner B. Circulation of Influenza-Viruses and Paramyxoviruses in Waterfowl Originating from two Different Areas of North-America. Bulletin of the World Health Organization. 1985;63:771–719. [PMC free article] [PubMed] [Google Scholar]
- Hosseini PR, Dhondt AA, Dobson A. Seasonality and wildlife disease: how seasonal birth, aggregation and variation in immunity affect the dynamics of Mycoplasma gallisepticum in house finches. Proceedings of the Royal Society B-Biological Sciences. 2004;271:2569–2577. doi: 10.1098/rspb.2004.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye BJ, Munster VJ, Huig N, de Vries P, Oosterbeek K, Tijsen W, Klaassen M, Fouchier RAM, van Gils JA. Hampered Performance of Migratory Swans: Intra- and Inter-Seasonal Effects of Avian Influenza Virus. Integrative and Comparative Biology. 2016;56:317–329. doi: 10.1093/icb/icw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye BJ, Munster VJ, Nishiura H, Fouchier RAM, Madsen J, Klaassen M. Reconstructing an annual cycle of interaction: natural infection and antibody dynamics to avian influenza along a migratory flyway. Oikos. 2011;120:748–755. [Google Scholar]
- Jourdain E, Gunnarsson G, Wahlgren J, Latorre-Margalef N, Brojer C, Sahlin S, Svensson L, Waldenstrom J, Lundkvist A, Olsen B. Influenza Virus in a Natural Host, the Mallard: Experimental Infection Data. Plos One. 2010;5 doi: 10.1371/journal.pone.0008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H, Yanagawa R, Matsuoka Y. Duck Influenza Lacking Evidence of Disease Signs and Immune-Response. Infection and Immunity. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG. Coincident ruddy turnstone migration and horseshoe crab spawning creates an ecological 'hot spot' for influenza viruses. Proceedings of the Royal Society B-Biological Sciences. 2010;277:3373–3379. doi: 10.1098/rspb.2010.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Leighton FA, Fouchier RAM, LeDuc JW, Peiris JSM, Schudel A, Stöhr K, Osterhaus ADME. Pathogen surveillance in animals. Science. 2005;309:1680–1681. doi: 10.1126/science.1113310. [DOI] [PubMed] [Google Scholar]
- Lam TTY, Ip HS, Ghedin E, Wentworth DE, Halpin RA, Stockwell TB, Spiro DJ, Dusek RJ, Bortner JB, Hoskins J, Bales BD, Yparraguirre DR, Holmes EC. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecology Letters. 2012;15:24–33. doi: 10.1111/j.1461-0248.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N, Brown JD, Fojtik A, Poulson RL, Carter D, Franca M, Stallknecht DE. Competition between influenza A virus subtypes through heterosubtypic immunity modulates re-infection and antibody dynamics in the mallard duck. Plos Pathogens. 2017;13:e1006419. doi: 10.1371/journal.ppat.1006419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N, Gunnarsson G, Munster VJ, Fouchier RAM, Osterhaus ADME, Elmberg J, Olsen B, Wallensten A, Haemig PD, Fransson T, Brudin L, Waldenstrom J. Effects of influenza A virus infection on migrating mallard ducks. Proceedings of the Royal Society B-Biological Sciences. 2009;276:1029–1036. doi: 10.1098/rspb.2008.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. Journal of Veterinary Science. 2017;18:269–280. doi: 10.4142/jvs.2017.18.S1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisovski S, Hoye BJ, Klaassen M. Geographic variation in seasonality and its influence on the dynamics of an infectious disease. Oikos 2017 [Google Scholar]
- Lisovski S, van Dijk JGB, Klinkenberg D, Nolet BA, Fouchier RAM, Klaassen M. Code for: The roles of migratory and resident birds in local avian influenza infection dynamics. Zenodo. 2018 doi: 10.5281/zenodo.1203837. [DOI] [PMC free article] [PubMed]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett SJ, Bodewes R, Pohlmann A, Banks J, Banyai K, Boni MF, Bouwstra R, Breed AC, Brown IH, Chen HL, Dan A, DeLiberto TJ, Diep N, Gilbert M, Hill S, Ip HS, Ke CW, Kida H, Killian ML, Koopmans MP, Kwon JH, Lee DH, Lee YJ, Lu L, Monne I, Pasick J, Pybus OG, Rambaut A, Robinson TP, Sakoda Y, Zohari S, Song CS, Swayne DE, Torchetti MK, Tsai HJ, Fouchier RAM, Beer M, Woolhouse M, Kuiken T, Related GCHN. Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354:213–217. doi: 10.1126/science.aaf8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H. Population parameters: Estimation for Ecological Models. Blackwell Science; Oxford: 2000. [Google Scholar]
- McCallum H, Barlow N, Hone J. How should pathogen transmission be modelled? Trends in Ecology & Evolution. 2001;16:295–300. doi: 10.1016/s0169-5347(01)02144-9. [DOI] [PubMed] [Google Scholar]
- Messenger AM, Barnes AN, Gray GC. Reverse Zoonotic Disease Transmission (Zooanthroponosis): A Systematic Review of Seldom-Documented Human Biological Threats to Animals. Plos One. 2014;9 doi: 10.1371/journal.pone.0089055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller AP, Szep T. The role of parasites in ecology and evolution of migration and migratory connectivity. Journal of Ornithology. 2011;152:141–150. [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus ADME, Fouchier RAM. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. Plos Pathogens. 2007;3:630–638. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir J, Haumacher H, Ike A, Stumpf P, Böhm R, Marschang RE. Long-Term Study on Tenacity of Avian Influenza Viruses in Water (Distilled Water, Normal Saline, and Surface Water) at Different Temperatures. Avian Diseases. 2010;54:720–724. doi: 10.1637/8754-033109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Ng PLK, Higgins DA. Bile Immunoglobulin of the Duck (Anas-Platyrhynchos) Developmental and Comparative Immunology. 1986;10:100. [PMC free article] [PubMed] [Google Scholar]
- Nickbakhsh S, Matthews L, Reid SWJ, Kao RR. A metapopulation model for highly pathogenic avian influenza: implications for compartmentalization as a control measure. Epidemiology and Infection. 2014;142:1813–1825. doi: 10.1017/S0950268813002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolet BA, Drent RH. Bewick's Swans refuelling on pondweed tubers in the Dvina Bay (White Sea) during their spring migration: first come, first served. Journal of Avian Biology. 1998;29:574–581. [Google Scholar]
- Nolet BA, Gyimesi A, van Krimpen RRD, de Boer WF, Stillman RA. Predicting effects of water regime changes on waterbirds: insights from staging swans. PLoS ONE. 2016;11:e0147340. doi: 10.1371/journal.pone.0147340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus ADME, Fouchier RAM. Global patterns of influenza A virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Orell P, Erkinaro J, Svenning MA, Davidsen JG, Niemela E. Synchrony in the downstream migration of smolts and upstream migration of adult Atlantic salmon in the subarctic River Utsjoki. Journal of Fish Biology. 2007;71:1735–1750. [Google Scholar]
- Payne-Gallwey R. The book of duck decoys, their construction, management, and history. Jvan Voorst; London, UK: 1886. [Google Scholar]
- Peel AJ, Pulliam JR, Luis AD, Plowright RK, O'Shea TJ, Hayman DT, Wood JL, Webb CT, Restif O. The effect of seasonal birth pulses on pathogen persistence in wild mammal populations. Proc Biol Sci. 2014;281 doi: 10.1098/rspb.2013.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MD, Hall JS, Brown JD, Goldberg DR, Ip H, Baranyuk VV. The dynamics of avian influenza in Lesser Snow Geese: implications for annual and migratory infection patterns. Ecological Applications. 2015;25:1851–1859. doi: 10.1890/14-1820.1. [DOI] [PubMed] [Google Scholar]
- Schekkerman H, Slaterus R. Avian influenza and wildfowl populations. British Trust for Ornithology (BTO); Thetford, Norfolk, UK: 2008. Population dynamics and prevalence of influenza A viruses in mallard, mute swan and other wildfowl. [Google Scholar]
- Scott D, Rose PM. Atlas of Anatidae populations in Africa and Western Eurasia, Wetlands International publication no. 41. Wetlands International; Wageningen, The Netherlands: 1996. [Google Scholar]
- Scott DA, Pose PM. Atlas of Anatidae Populations in Africa and Western Eurasia. Wetlands International, Wageningen, The Netherlands. 1996;41 [Google Scholar]
- Soetaert K, Petzoldt T. Inverse Modelling, Sensitivity and Monte Carlo Analysis in R Using Package FME. Journal of Statistical Software. 2010;33:1–28. [Google Scholar]
- Soetaert K, Petzoldt T, Setzer RW. Solving Differantial Equations in R: Package deSolve. Journal of Statistical Software. 2010;33:1–25. [Google Scholar]
- Stallknecht DE, Shane SM, Kearney MT, Zwank PJ. Persistence of Avian Influenza-Viruses in Water. Avian Diseases. 1990;34:406–411. [PubMed] [Google Scholar]
- Stanley CQ, MacPherson M, Fraser KC, McKinnon EA, Stutchbury BJM. Repeat Tracking of Individual Songbirds Reveals Consistent Migration Timing but Flexibility in Route. Plos One. 2012;7 doi: 10.1371/journal.pone.0040688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman RA, Wood KA, Gilkerson W, Elkinton E, Black JM, Ward DH, Petrie M. Predicting effects of environmental change on a migratory herbivore. Ecosphere. 2015;6:art114. [Google Scholar]
- Tian HY, Zhou S, Dong L, Van Boeckel TP, Cui YJ, Wu YR, Cazelles B, Huang SQ, Yang RF, Grenfell BT, Xu B. Avian influenza H5N1 viral and bird migration networks in Asia. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:172–177. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot JA, van Boven M, Stegeman A, de Water SGPV, de Jong MCM, Koch G. Transmission of highly pathogenic avian influenza H5N1 virus in Pekin ducks is significantly reduced by a genetically distant H5N2 vaccine. Virology. 2008;382:91–97. doi: 10.1016/j.virol.2008.08.037. [DOI] [PubMed] [Google Scholar]
- van Dijk JGB, Hoye BJ, Verhagen JH, Nolet BA, Fouchier RAM, Klaassen M. Data from: Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. Dryad Digital Repository. 2013 doi: 10.5061/dryad.j855b. [DOI] [PMC free article] [PubMed]
- van Dijk JGB, Hoye BJ, Verhagen JH, Nolet BA, Fouchier RAM, Klaassen M. Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. Journal of Animal Ecology. 2014;83:266–275. doi: 10.1111/1365-2656.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk JGB, Kleyheeg E, Soons MB, Nolet BA, Fouchier RAM, Klaassen M. Weak negative associations between avian influenza virus infection and movement behaviour in a key host species, the mallard Anas platyrhynchos. Oikos 2015 [Google Scholar]
- van Dijk JGB, Meissner W, Klaassen M. Improving provenance studies in migratory birds when using feather hydrogen stable isotopes. Journal of Avian Biology. 2014;45:103–108. [Google Scholar]
- van Gils JA, Munster VJ, Radersma R, Liefhebber D, Fouchier RAM, Klaassen M. Hampered Foraging and Migratory Performance in Swans Infected with Low-Pathogenic Avian Influenza A Virus. Plos One. 2007;2 doi: 10.1371/journal.pone.0000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen JH, Herfst S, Fouchier RAM. How a virus travels the world. Science. 2015;347:616–617. doi: 10.1126/science.aaa6724. [DOI] [PubMed] [Google Scholar]
- Verhagen JH, van Dijk JGB, Vuong O, Bestebroer T, Lexmond P, Klaassen M, Fouchier RAM. Migratory Birds Reinforce Local Circulation of Avian Influenza Viruses. Plos One. 2014;9 doi: 10.1371/journal.pone.0112366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Molecular Ecology. 2002;11:1545–1554. doi: 10.1046/j.1365-294x.2002.01523.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S. Asymptotic equivalence of Bayes cross validation and widely applicable information criterionin singular learning theory. The Journal of Machine Learning Research. 2010;11:3571–3594. [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and Ecology of Influenza-a Viruses. Microbiological Reviews. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Kleijn D, Müskens GJDM, Fouchier RAM, Verhagen JH, Glazov PM, Si Y, Prins HHT, F dBW. No evidence that migratory geese disperse avian influenza viruses from breeding to wintering ground. Plos One. 2017;12:e0177790. doi: 10.1371/journal.pone.0177790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.