Abstract
Due to the high genetic heterogeneity of hearing loss, current clinical testing includes sequencing large numbers of genes, which often yields a significant number of novel variants. Therefore, the standardization of variant interpretation is crucial to provide consistent and accurate diagnoses. The Hearing Loss Variant Curation Expert Panel was created within the Clinical Genome Resource to provide expert guidance for standardized genomic interpretation in the context of hearing loss. As one of its major tasks, our Expert Panel has adapted the ACMG/AMP guidelines for the interpretation of sequence variants in hearing loss genes. Here, we provide a comprehensive illustration of the newly specified ACMG/AMP hearing loss rules. Three rules remained unchanged, four rules were removed, and the remaining twenty-one rules were specified. These rules were further validated and refined using a pilot set of 51 variants assessed by curators and expert opinion. Of the 51 variants evaluated in the pilot, 37% (19/51) changed category based upon application of the expert panel specified rules and/or aggregation of evidence across laboratories. These hearing loss-specific ACMG/AMP rules will help standardize variant interpretation, ultimately leading to better care for individuals with hearing loss.
Keywords: Variant interpretation, hearing loss, deafness, ClinGen, ACMG/AMP guidelines, genetic diagnosis
INTRODUCTION
Sensorineural hearing loss (SNHL) is the most common congenital sensory deficit, with an estimated prevalence of 2–3 cases per 1000 individuals (Morton & Nance, 2006). Approximately half of SNHL in children is due to genetic causes, with 70% being nonsyndromic and 30% syndromic (Marazita et al., 1993; Smith, Bale, & White, 2005). Although variation in the GJB2 gene is the most common cause of nonsyndromic genetic SNHL in many populations, there is high genetic and allelic heterogeneity, with over 100 implicated genes (Hereditary Hearing Loss Homepage http://hereditaryhearingloss.org) (Abou Tayoun et al., 2016; Alford et al., 2014).
Given the substantial genetic contribution to SNHL, a clinical genetics evaluation, including comprehensive sequencing, is recommended as part of a standard of care diagnostic work-up. Results of genetic testing can inform clinical management, particularly if a genetic syndrome is identified before the onset of additional clinical manifestations (Alford et al., 2014). The interpretation of sequence variants is a critical component of an accurate genetic diagnosis, and discrepancies in variant interpretation and classification have been well-documented and can have serious implications for patient care (Amendola et al., 2016; Booth et al., 2015; Booth, Kahrizi, et al., 2018; Harrison et al., 2017). The large number of genes sequenced during hearing loss genetic testing routinely yield a large number of novel variants, exacerbating the interpretation challenge. Illustrating the extent of discrepancies in variant interpretation, as of 04/12/2018, 257 (8.1%) of the 3162 sequence variants in the 9 major hearing loss genes represented in our variant pilot (USH2A, GJB2, SLC26A4, MYO7A, KCNQ4, TECTA, MYO6, COCH, and CDH23) had conflicting interpretations in ClinVar.
In an effort to foster accurate interpretation and to reduce discrepancies across laboratories, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published recommendations and guidelines for the interpretation of sequence variants (Richards et al., 2015). The ACMG/AMP guidelines contain several types of evidence that are weighted and categorized. Although these guidelines were intended to be used universally for all Mendelian disorders, certain criteria require gene- or disease-specific knowledge, a lack of which has been shown to contribute to discrepancies in variant interpretation (Pepin et al., 2016). In response to these issues, the NIH-funded Clinical Genome Resource (ClinGen, http://www.clinicalgenome.org) was launched as a centralized resource to provide guidance and tools for defining the clinical validity of gene and variant contributions to disease. Specifically, the ClinGen Hearing Loss Clinical Domain Working Group (HLWG) was established in 2016 to form Expert Panels to evaluate gene-disease associations, as well as standardize variant interpretation in hereditary hearing loss and related syndromes.
Here, we document the work of the Hearing Loss Variant Curation Expert Panel, hereafter referred to as the HL-EP. We adapted the ACMG/AMP guidelines for variant interpretation in the context of hearing loss. These specified rules were developed using the most common genes and syndromes that contribute to genetic hearing loss (HL), with the intent that most specifications could be used broadly across the many associated genes. These hearing loss-specific guidelines will be used in the future by the HL-EP to submit variant interpretations to ClinVar as an “expert panel” submitter, with the goal of resolving discrepancies, moving VUSs towards Benign or Pathogenic and increasing the confidence in HL variant classifications in ClinVar.
MATERIALS AND METHODS
Hearing Loss Clinical Domain Working Groups
As a ClinGen Clinical Domain Working Group (CDWG), the Hearing Loss CDWG aims to create a comprehensive, standardized knowledge base of genes and variants relevant to syndromic and nonsyndromic HL. Members were identified and recruited based on their expertise in hearing loss, and are representative of diverse institutions worldwide, spanning Asia, Australia, Europe, and North America. Members include otolaryngologists, clinical geneticists, molecular geneticists, ClinGen biocurators, clinical researchers, and genetic counselors from over 15 institutions. The Hearing Loss CDWG has so far formed two major efforts defined by ClinGen as a Gene Curation Expert Panel, and a Variant Curation Expert Panel (HL-EP).
Specifications of the ACMG/AMP Guidelines
A smaller task team utilized biweekly conference calls in addition to email correspondence to review, specify, and reach consensus for each of the rules within the ACMG/AMP guidelines. Typically, 15–20 members from the Laboratory for Molecular Medicine (LMM), Children’s Hospital of Philadelphia (CHOP), ARUP Laboratories, Molecular Otolaryngology and Renal Research Laboratories (MORL), and Mayo Clinic participated on each call. Members with specific areas of expertise (e.g. Pendred syndrome) were also contacted for relevant topics. The chairs, ClinGen biocurators, and coordinator of the HL-EP also utilized weekly conference calls for additional planning and preparation. For the larger CDWG, a quarterly conference call was used to present specifications finalized by the smaller group and to obtain final approval from all members. Upon specifying the ACMG/AMP rules, conference calls and email correspondence were used for the variant pilot using a set of 51 variants from nine common HL genes that represent the varied inheritance patterns, evidence types and phenotypic spectrum seen across hearing loss cohorts (see Variant Pilot methods below). The final set of specified rules is shown in Table 1.
Table 1:
ACMG Criteria with HL-EP Specifications
| Pathogenic Criteria | |
|---|---|
| Rule | Rule Description |
| PVS1 | Null variant in a gene with established LOF as a disease mechanism; see PVS1_Strong, PVS1_Moderate, PVS1_Supporting for reduced evidence applications |
| PVS1_ Strong |
See PVS1 flow chart for PVS1_Strong variants in gene where LOF is a known mechanism of disease |
| PVS1_ Moderate |
See PVS1 flowchart for PVS1_Moderate variants in gene where LOF is a known mechanism of disease |
| PVS1_ Supporting |
See PVS1 flowchart for PVS1_Supporting variants in gene where LOF is a known mechanism of disease |
| PS1 | Same amino acid change as an established pathogenic variant; OR splice variants at same nucleotide and with similar impact prediction as previously reported pathogenic variant |
| PS2 | 2 points per tables 5a and 5b: Examples: 1 proven de novo occurrence; OR 2 assumed de novo occurrences |
| PS2_ VeryStrong |
4 points per tables 5a and 5b: Examples: 2 proven de novo occurrences; OR 1 proven + 1 assumed de novo occurrences; OR 4 assumed de novo occurrences |
| PS2_ Moderate |
1 point per tables 5a and 5b: Examples: 1 proven de novo occurrence (phenotype consistent but not specific to gene); OR 1 assumed de novo occurrence; OR 2 assumed de novo occurrences (phenotype/gene not specific) |
| PS2_ Supporting |
0.5 points per tables 5a and 5b: Example: 1 assumed de novo occurrence (phenotype/gene not specific) |
| PS3 | Knock-in mouse model demonstrates the phenotype |
| PS3_ Moderate |
Validated functional studies show a deleterious effect (predefined list) |
| PS3_ Supporting |
Functional studies with limited validation show a deleterious effect |
| PS4 | Fisher Exact or Chi-Squared analysis shows statistical increase in cases over controls, OR Autosomal dominant: ≥15 probands with variant, and variant meets PM2 |
| PS4_ Moderate |
Autosomal dominant: ≥6 probands with variant, and variant meets PM2 |
| PS4_ Supporting |
Autosomal dominant: ≥2 probands with variant, and variant meets PM2 |
| PM1 | Mutational hot spot or well-studied functional domain without benign variation (KCNQ4
pore-forming region; Gly residues in Gly-X-Y motifs of COL4A3/4/5) |
| PM2 | Absent/Rare in population databases (absent or ≤0.00007 (0.007%) for autosomal recessive, ≤0.00002 (0.002%) for autosomal dominant) |
| PM2_ Supporting |
Low MAF in population databases (<0.0007 (0.07%) for autosomal recessive, |
| PM3 | 1 point awarded from tables 7a and 7b Example: Detected in trans with a pathogenic variant (recessive) |
| PM3_ VeryStrong |
4 points awarded from tables 7a and 7b Example: Detected in trans in ≥4 probands with a pathogenic variant (recessive) |
| PM3_ Strong |
2 points awarded from tables 7a and 7b Example: Detected in trans in 2 probands with a pathogenic variant (recessive) |
| PM3_ Supporting |
0.5 points awarded from tables 7a and 7b Examples: Two variants that meet PM2_Supporting detected in trans; OR a homozygous variant meeting PM2_Supporting |
| PM4 | Protein length change due to an in-frame deletion or insertion that are not located in repetitive regions |
| PM5 | Missense change at same codon as another pathogenic missense variant |
| PM5_ Strong |
Missense change at same codon as two different pathogenic missense variants |
| PM6 | See PS2 above |
| PP1 | Segregation in one affected relative for recessive and two affected relatives for dominant |
| PP1_ Strong |
Segregation in three affected relatives for recessive and five affected relatives for dominant |
| PP1_ Moderate |
Segregation in two affected relatives for recessive and 4 affected relatives for dominant |
| PP3 | REVEL score ≥0.7, or predicted impact to splicing using MaxEntScan |
| PP4 | Patient’s phenotype highly specific for gene or fully sequenced gene set (see specifications in Table 7) |
| Benign Criteria | |
| BA1 | MAF of ≥0.005 (0.5%) for autosomal recessive; MAF of ≥0.001 (0.1%) for autosomal dominant |
|
BS1
|
MAF of ≥0.003 (0.3%) for autosomal recessive; MAF of ≥0.0002 (0.02%) for autosomal dominant. Likely benign, provided there is no conflicting evidence. |
| BS1_ Supporting |
MAF of ≥0.0007 (0.07%) for autosomal recessive. No BS1_Supporting criteria for autosomal dominant. |
| BS2 | Observation of variant (biallelic with known pathogenic variant for recessive) in controls inconsistent with disease penetrance. |
| BS3_ Supporting |
Functional study shows no deleterious effect (predefined list) |
| BS4 | Non-segregation with disease |
| BP2 | Observed in trans with a dominant variant/observed in cis with a pathogenic variant (use with caution) |
| BP3 | In-frame indels in repeat region without known function |
| BP4 | Computational evidence suggests no impact; REVEL score ≤0.15 or no impact to splicing in MaxEntScan. |
| BP5 | Variant in an autosomal dominant gene found in a patient with an alternate explanation |
| BP7 | Silent variant with no predicted impact to splicing |
Strikethrough indicates rule was removed or not applicable. Abbreviations: MAF = minor allele frequency; Indels = insertion/deletions.
Pathogenic allele frequency for recessive hearing loss
The frequencies of the most common pathogenic variants in HL were determined using a cohort of 3,673 patients tested at the LMM. The three most common sequence variants were NM_004004.5:c.35delG (p.Gly12Valfs), NM_004004.5:c.109G>A (p.Val37Ile), and NM_004004.5:c.101T>C (p.Met34Thr) in GJB2, while the fourth variant was NM_206933.2:c.2299delG (p.Glu767Serfs) in the USH2A gene (Table 2). Test-based cohorts for the GJB2 (n=1375 probands) or USH2A (n=1887 probands) genes were used to calculate the disease population allele frequency of each one of those variants.
Table 2.
Most common pathogenic variants in hearing loss.
| Variant | LMM (Allele Frequency; Allele Ratio) | MORL (Allele Frequency) |
|---|---|---|
| GJB2, c.35delG | 7.2% (198/2750) | 4.77% (197/4132) |
|
GJB2, c.109G>A p.(Val37Ile) |
4.4% (120/2750) | 2.01% (83/4132) |
|
GJB2, c.101T>C p.(Met34Thr) |
2.1% (57/2750) | 1.91% (79/4132) |
| USH2A, c.2299delG | 1.2% (46/3774) | 0.65% (27/4132) |
LMM: Partners Laboratory for Molecular Medicine; MORL: Iowa Molecular Otolaryngology and Renal Research Laboratories. The higher frequency for each of the above grey shaded variants was used to specify the population frequency rules. RefSeq transcripts are NM_004004.5 for GJB2 and NM_206933.2 for USH2A.
The allele frequencies were confirmed using another HL cohort (n=2066) tested at the Molecular Otolaryngology and Renal Research Laboratories (MORL; Iowa City, IA). Although their frequencies were slightly different, the rankings of the four most common pathogenic variants were recapitulated in this cohort (Table 2). The frequency differences can be attributed to the fact that the MORL cohort excluded patients with positive findings upon GJB2 prescreening. Therefore, the higher allele frequency (from the LMM) for each variant was conservatively used in calculating BA1 and BS1 for recessive HL. Our calculations were made under the assumption of Hardy-Weinberg equilibrium and can be obtained using a recently published web application (https://www.cardiodb.org/allelefrequencyapp/) (Whiffin et al., 2017).
Performance analysis of functional studies
To assess the performance characteristics (sensitivity, specificity, positive predictive value, negative predictive value) of the functional assays in each of GJB2, SLC26A4, and COCH, the number of true and false variant calls were determined as follows. Variants classified in ClinVar and included in functional assays were reviewed for classification accuracy and then used for informing performance. Pathogenic and likely pathogenic variants that had abnormal assay readout, defined as statistically significant deviation from wild type variant(s), were labeled as true positives. Normal assay readouts for pathogenic or likely pathogenic variants were considered false negatives. Similarly, benign and likely benign variants with normal assay readouts (i.e. does not significantly deviate from wild type variants) were counted as true negatives. False positives were benign or likely benign variants with abnormal assay readouts. A summary of true and false variant calls by five functional assays in the above three genes is shown in Supp. Table S1.
Variant Pilot
Following the specification of the ACMG/AMP guidelines, the rules were refined by interpreting a set of 51 variants in GJB2, SLC26A4, USH2A, MYO7A, CDH23, COCH, KCNQ4, MYO6, and TECTA. Each variant was assessed independently by two variant curators, and the variant classification and rules applied were reviewed on a conference call to resolve discrepancies and reach consensus. Curators utilized ClinGen’s Variant Curation Interface (https://curation.clinicalgenome.org/) to assess and document the applicable rules for each variant.
RESULTS
Summary of Specifications
The HL-EP recommended specifications for 21 ACMG/AMP rules (Table1). Four rules had general recommendations on the application of the rule (PS1, PP3, BS4, BP4, and BP5). Seven rules had gene or disease based specifications (PS3, PM1, PM2, PP4, BA1, BS4, BP2). Seven rules had strength-level specifications (PVS1, PS2, PM3, PM5, PM6, PP1, BS3). Three rules had both gene/disease based specifications and strength-level specifications (PS4, BS1, BS2). No changes were recommended for three rules (PM4, BP3, and BP7), and four rules were considered not applicable (PP2, PP5, BP1, and BP6).
Two changes to the ACMG/AMP recommendations were made on how to combine criteria to classify sequence variants. The first is that a likely pathogenic classification can be reached if a variant in a gene associated with autosomal recessive HL meets PVS1 and PM2_Supporting. The second change was that a variant can be classified as likely benign if a variant meets BS1 without valid conflicting evidence that would suggest a pathogenic role.
It should be noted that we do not discuss the technical challenges associated with sequencing some of HL genes, such as homology, GC-rich and/or repetitive regions, which may affect variant calling and interpretation. False positive variants in these regions can exist in population or disease databases and may result in erroneous classifications. Laboratories should carefully assess the technical quality of any variant before interpretation. A full list of technically challenging regions in 109 HL genes was recently characterized (DiStefano et al., 2018).
Population data (BA1, BS1, PM2)
Recessive and dominant forms of HL were distinguished with respect to the interpretation of variant frequency data from the general population. This is mainly due to differences in prevalence, penetrance, and gene contribution. Evidence-based estimates of these attributes are critical for establishing recessive and dominant allele frequency thresholds, at which a benign or pathogenic criterion might be assigned for a given variant. The minor allele frequencies required to apply these criteria are shown in Figure 1. For this work, the ExAC and gnomAD population databases were used, though other databases with a minimum of 2000 alleles are also sufficient. Caution should be used if the cutoffs specified below are applied on an uncharacterized population or a population with minimal representation in ExAC or gnomAD.
Figure 1.
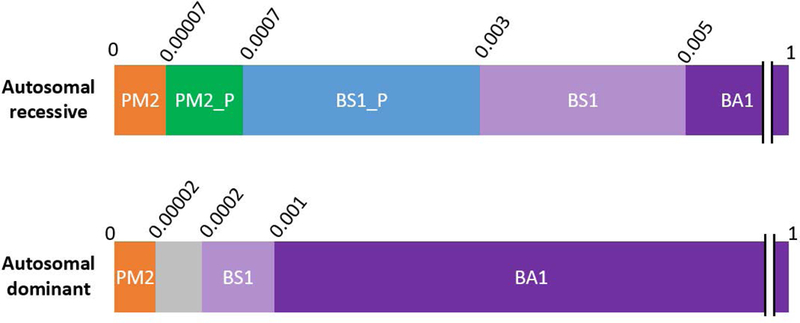
Population data criteria for autosomal recessive and autosomal dominant hearing loss. Minor allele frequencies are represented in decimals above each bar. Abbreviations:PM2_P = PM2_Supporting, BS1_P = BS1_Supporting.
Recessive hearing loss.
Based on a comprehensive literature search, a prevalence of 1 in 200 was used to specify the BA1 rule and to derive the BS1 and PM2 values for recessive HL. This was a highly conservative prevalence estimate from different ethnic groups and accounted for congenital and childhood onset HL (up to 19 years of age) with suspected genetic etiology (Lin, Niparko, & Ferrucci, 2011; Morton & Nance, 2006). Although HL prevalence increases with age, other contributing genetic (mostly dominant) and non-genetic (environmental, such as noise or drug related) factors may become more relevant in late-onset HL.
Penetrance was considered complete for recessive HL, based on the Expert Panel’s experience, with the exception of two variants, GJB2 p.Met34Thr and p.Val37Ile. To derive conservative genetic and allelic heterogeneity values, we calculated the allele frequency of the most common pathogenic variants in HL patient cohorts tested at the LMM and MORL. The same four pathogenic variants were identified as most common among the two cohorts (See Methods and Table 2). The higher frequency value for each variant was used to derive a conservative minor allele frequency (MAF) threshold for BA1, BS1, and BS1_Supporting.
BA1 and BS1.
Using the recently published allele frequency web app, (https://www.cardiodb.org/allelefrequencyapp/), the MAF threshold for BA1 was set at ≥ 0.5% for autosomal recessive HL. This was calculated by using a prevalence of 1/200, complete penetrance (100%), and a maximum allelic heterogeneity contribution of 7.2% based on the most common pathogenic variant (NM_004004.5:c.35delG in GJB2 Table 2). Genetic heterogeneity was set to 1 given that the 7.2% allele frequency used was derived across all genetic causes of hearing loss. We then reviewed all reportedly pathogenic variants with frequencies near or higher than this threshold and developed a BA1 exclusion list which includes two low penetrance variants, GJB2 p.Met34Thr and p.Val37Ile, as well as several founder mutations (Supp. Table S2). Any variant exceeding 0.5% in any subpopulation, aside from variants on the exclusion list (Supp. Table S2), would default to a benign classification. A 95% confidence interval was used to develop the MAF thresholds. Therefore, in practice we recommend using the filtering allele frequency in ExAC, or for other population databases, a 95% confidence interval should be applied using the web application referenced above.
The MAF threshold for BS1 was set at ≥ 0.3% for recessive HL. This was calculated by using a maximum allelic heterogeneity of 4.4%, based upon the second most common pathogenic variant, p.Val37Ile in GJB2 (Table 2), while maintaining the same prevalence, penetrance, genetic heterogeneity, and statistical correction used for the recessive BA1 derivation. Variants exceeding this frequency and having no other conflicting evidence for pathogenicity meet the “Likely Benign” classification, provided they are not the exclusion list in Supp. Table S2.
The BA1 and BS1 cutoffs were validated using an independent approach, where an extensive curation of the literature was performed to estimate disease attributes (prevalence, gene contribution, pathogenic allele frequencies) of the most common causes of HL (GJB2, SLC26A4, USH2A, and MYO7A) across several ancestral groups globally (Supp. Methods and Supp. Tables S3–6). This approach validated the MAF thresholds determined above.
BS1_Supporting, PM2_Supporting and PM2.
To enable broader use of population allele frequencies, we defined supporting strength levels of BS1 and PM2. BS1_Supporting was set at a MAF threshold of ≥ 0.07% using the same metrics as BA1 and BS1 (Table 3), but 1% for the allelic heterogeneity, which is representative of the most common pathogenic variant in the second most common recessive gene in our hearing loss patient cohorts (NM_206933.2:c.2299delG in USH2A - Table 2).
Table 3.
Allele frequency cutoffs for dominant and recessive hearing loss disorders.
| ACMG-AMP Criteria | MAF | Preval. | Allelic Het.† | Penetrance | |
|---|---|---|---|---|---|
|
AUTOSOMAL RECESSIVE |
BA1 |
≥0.005 (0.5%) |
1/200 |
7.2% |
100% |
| BS1 | ≥0.003 (0.3%) |
1/200 |
4.4% | 100% | |
| BS1_Supporting |
≥0.0007 (0.07%) |
1/200 |
1.0% |
100% | |
| PM2_Supporting |
<0.0007 (0.07%) |
Can apply PM2_Supporting if MAF is < BS1_Supporting (0.07%) |
|||
| PM2 | ≤0.00007 (0.007%) |
Can apply PM2_moderate if MAF is an order of magnitude below BS1_Supporting (ie ≤0.007%); |
|||
|
AUTOSOMAL
DOMINANT |
BA1 |
≥0.001 (0.1%) |
1/30 |
5% |
80% |
| BS1 |
≥0.0002 (0.02%) |
1/150 |
5% | 80% | |
| PM2 | ≤0.00002 (0.002%) |
Can apply PM2_Moderate if MAF is an order of magnitude below BS1 (ie ≤0.002%); |
|||
Abbreviations: Preval. = Prevalence. Allelic Het. - Allelic heterogeneity.
Allelic heterogeneity encompassed both genetic and allelic heterogeneity because allele frequencies were derived from hearing loss cases spanning all genetic etiologies.
PM2 was set by selecting a MAF that was an order of magnitude lower than the BS1_Supporting value. Therefore PM2 for recessive disorders is ≤ 0.007% (Table 3). Variants with frequencies between 0.07% and 0.007% can be awarded supporting evidence for pathogenicity (PM2_Supporting) (Table 3).
Dominant hearing loss.
HL prevalence generally increases with age but with decreasing monogenetic contribution (Cunningham & Tucci, 2017). Most dominant HL forms are postlingual and therefore are more difficult to distinguish from non-genetic forms (Shearer, Hildebrand, & Smith, 1993). We chose prevalence in individuals aged 0–49 years to minimize those affected by age related HL. This age range is consistent with the age of onset of the major HL genes, including TECTA (Alloisio et al., 1999; Mustapha et al., 1999; Verhoeven et al., 1998), DFNA5 (Bischoff et al., 2004; Booth, Azaiez, et al., 2018; Van Laer et al., 1998), COCH (Nagy, Horvath, Trexler, Repassy, & Patthy, 2004; Robertson et al., 1998; Street et al., 2005), and WFS1 (Eiberg et al., 2006; Fukuoka, Kanda, Ohta, & Usami, 2007; Hogewind et al., 2010). Based on an NHANES study (n=7490 patients), 1/15 individuals are expected to have HL by 49 years of age (Lin et al., 2011). For dominant BA1 derivation, we conservatively assumed that 50% of those individuals have a genetic cause, and calculated a prevalence of 1/30 (Table 3). This is a deliberate overestimation given that 50% is the estimated genetic contribution to congenital HL, which is the highest contribution among all age groups, and is therefore expected to be lower in adults.
Due to a lack of published data on dominant HL penetrance, we used an estimated penetrance of 80%, based on the collective research and clinical experience of the HL-EP members. The relative contributions of known genes to dominant forms of HL were also assessed in a cohort of 2000 patients tested at MORL. Four genes, TECTA, WFS1, KCNQ4, and COL11A2, were the most common accounting for 20%, 13%, 13%, and 7%, respectively, of all dominant pathogenic variants. Furthermore, among all nonsyndromic dominant genes tested at the LMM (n=3076 probands), KCNQ4 contributed the most for a total of four pathogenic and likely pathogenic variants in one proband each. Therefore, we conservatively assumed the maximum allelic heterogeneity for a dominant HL gene is 25%. Based on these data, we safely estimated that no dominant variant could contribute more than 5% of all HL in the 0–49 year age range. This is based on the assumption that the maximum contribution of one gene would be 20% (TECTA) and the maximum contribution of any one variant to a gene’s HL would be 25% (KCNQ4) (i.e. 20% x 25% = 5%). Our literature search also confirmed a maximum allelic heterogeneity of 5% in any given gene (Hildebrand et al., 2011; Iwasa, Nishio, & Usami, 2016; T. Naito et al., 2013).
Using the above conservative estimates, we set the dominant BA1 cutoff at ≥ 0.1% (Figure 1 and Table 3). To derive the BS1 value, we used a less conservative estimate of dominant prevalence by assuming only 10% of all HL between ages 0–49 years is genetic. Therefore, a prevalence of 1/150 (1/15 × 10%) was used, while keeping all other values as in BA1, leading to a ≥ 0.02% BS1 cutoff. An allele frequency an order of magnitude less than this cutoff (i.e. ≤ 0.002%) was then considered sufficient to be awarded moderate evidence of pathogenicity (PM2) (Table 3). Again, we recommend using the filtering allele frequency in ExAC or applying a 95% confidence interval in the cardiodb.org allele frequency app when applying these thresholds.
PM2_Supporting and BS1_Supporting thresholds were not created for autosomal dominant hearing loss. This was for several reasons. We could not replicate the model used for the development of recessive thresholds for BA1, BS1, and BS1_Supporting, which was partly based on the allele frequency of common pathogenic variants, as no variants are common causes of dominant hearing loss. The value used for recessive BS1_Supporting was also the basis for the development of recessive PM2 and PM2_Supporting. Without the ability to use allele frequencies of common dominant alleles, we were unable to create MAF thresholds that had significant enough gaps between the different benign and pathogenic criteria. Furthermore, the value for dominant BS1 was set very low at 0.02%, and therefore, the difference of a single allele could have a significant effect on the odds impact (Tavtigian et al., 2018). Finally, for dominant hearing loss, we do not expect variants to be present in the general population at an appreciable allele frequency as we would for recessive pathogenic variants, even with the potential for decreased or age-related penetrance in the former. As described above, in setting BA1 and BS1, we lowered the dominant HL penetrance to 80% which, along with the conservative prevalence, allelic and genetic heterogeneity estimates, allows us to account for pathogenic variants with reduced penetrance in the general population.
It is important to note the phenotypic heterogeneity is exhibited by several of the HL genes. Genes known to cause both dominant and recessive forms of HL, such as TECTA, COL11A2, GJB2 and MYO7A should be treated per the more conservative recessive rules for applying BA1 and BS1 rules because the frequency cutoffs for recessive inheritance are higher than those for dominant. This will help to avoid erroneously misclassifying variants as benign or likely benign.
However, when considering pathogenicity evidence, the PM2 rule that matches the proposed form of HL (dominant vs recessive) should be used according to the available evidence about the gene, the segregation data, or the family history information for the proband being assessed. For example, the vast majority of variants reported for GJB2 and MYO7A are associated with autosomal recessive hearing loss and Usher syndrome, respectively. Variants causing autosomal dominant hearing loss in either GJB2 or MYO7A are rare in comparison. Therefore, the autosomal recessive MAF threshold should be utilized for the PM2 rule for these genes. However, for genes that do not have a majority of variants associated with one inheritance pattern, such as TECTA and COL11A2, we recommend using the more conservative dominant rules for PM2, unless there is convincing evidence supporting recessive inheritance.
X-linked forms of HL were not specifically addressed by the HL-EP. However, based on their relative rarity and the fact that most X-linked genes are not strictly recessive or dominant, with carrier females often displaying milder disease, the recessive thresholds for BA1, BS1, and BS1_Supporting can be applied for both X-linked recessive and X-linked dominant HL.
PS4, PS4_Moderate, PS4_Supporting: Prevalence in affected individuals statistically increased over controls
Published case-control studies demonstrating a statistically increased presence of a variant in affected individuals compared to race and ancestry-matched controls can be used as strong evidence towards pathogenicity, as outlined in the ACMG/AMP guidelines. For many variants, published case-control studies may not exist, but a case-control comparison to ancestry-matched controls in gnomAD can still provide critical evidence. This is particularly true for recessive variants with relatively high frequency in the general population. The following recommendations were made for variants in which a published case-control study does not exist.
A Chi-squared or Fisher’s Exact test using a 2×2 contingency table can be performed to determine if the MAF of a variant is statistically higher in cases compared to the general population. This analysis can be used as strong evidence if the MAF of the variant is significantly higher (i.e. the p-value of the Chi-squared or Fisher’s Exact test is ≤ 0.05) in cases than in the general population. It is important to note that to do this comparison, the case and population alleles should be adequately ancestry-matched, the total numbers of positive and negative alleles should be known for both the case and the population cohorts, and the relevant gene must be definitively or strongly associated with hearing loss per the ClinGen Gene Curation framework (Strande et al., 2017).
Large general population databases, such as ExAC or gnomAD, are not true control cohorts and likely contain individuals with HL, given that there is no exclusion for HL. As such, absence of statistical significance should not be used as evidence against pathogenicity.
The ACMG/AMP guidelines allow for the counting of probands as evidence for PS4 in the absence of case-control studies. For autosomal recessive HL, we recommend counting probands through PM3 (see below) where allelic data can be taken into account. For autosomal dominant HL, we adopted the specifications previously published by the ClinGen Cardiomyopathy Expert Panel (CMP-EP) given that dominant HL has similar attributes to cardiomyopathy in terms of penetrance and phenocopies (Kelly et al., 2018). These specifications state that if a variant is absent or extremely rare in the general population (i.e. PM2 is met), identification of ≥ 2, ≥ 6, or ≥ 15 probands with the same variant would reach supporting (PS4_Supporting), moderate (PS4_Moderate), or strong (PS4) evidence, respectively. The rule PM2 must be met to use these PS4 specifications. As noted above, PM2 for autosomal dominant HL was set at ≤ 0.002%, which is slightly lower than the PM2 value of ≤ 0.004% set by the CMP-EP.
COMPUTATIONAL AND PREDICTIVE DATA
Loss of function (LOF) variants (PVS1, PVS1_Strong, PVS1_Moderate, PVS1_Supporting)
The HL-EP and the ClinGen Sequence Variant Interpretation Working Group (SVI) jointly refined this rule to be used for all recessive and haploinsufficient genes across most diseases including HL. This refinement included a detailed guideline for interpreting the PVS1 criteria across all LOF variant types (nonsense, frameshift, canonical splice site variants (+/−1,2), deletions, duplications, and initiation codon variants) while accounting for variant, exon and transcript-specific considerations and the varied evidence weights. This guideline is available in an accompanying paper in this issue (Abou Tayoun et al., this issue) and will not be discussed further here.
Variants affecting the same amino acid residue (PS1, PM5, PM5_Strong)
The ACMG/AMP guideline considers the presence of a known pathogenic variant at the same amino acid residue as either strong or moderate evidence of pathogenicity. This PS1 rule is applied if the variant is a novel nucleotide change but results in the same amino acid change as the known pathogenic variant, and the PM5 rule is applied if the variant is a novel missense change at the same amino acid residue as a known pathogenic missense variant. It should be noted the ACMG/AMP guidelines recommend assessing whether the variants in question could have an impact at the DNA level, such as through an impact on splicing, before applying PS1 or PM5. The HL-EP added a strength modification to the PM5 rule (PM5_Strong) so that a strong level of evidence can be applied if a novel missense variant is located at an amino acid residue with at least two other pathogenic variants at the same residue.
The HL-EP also specified that PS1 can be applied for DNA variants located at the same nucleotide in the splice consensus sequence as a known pathogenic variant. Based on the most conserved regions known to impact splicing, we defined the splice consensus sequence here as the first base and last three bases of the exon, the −12 through −1 positions in the intron, and the +1 through +6 positions in the intron. The +/−1,2 positions were excluded, given that these positions should be evaluated using PVS1 rules. In addition, splice prediction algorithms must predict a similar or greater effect than the comparator known pathogenic variant.
Computational predictive tools (PP3, BP4, BP7)
Computational tools are commonly used to inform variant interpretation. For missense variants, the HL-EP recommends using REVEL, a computational tool that combines predictions from commonly used algorithms for missense variants (Ioannidis et al., 2016). REVEL was selected based on recently published work from members of ClinGen as well as evaluation within the HL-EP and the SVI (Ghosh, Oak, & Plon, 2017). Furthermore, we recommend specific REVEL scores are reached to apply supportive evidence of pathogenicity (PP3) or supportive evidence of benign (BP4). PP3 can be applied with a REVEL score of ≥ 0.7, given that 95% of benign variants are excluded at this score (Ioannidis et al., 2016). BP4 can be applied with a REVEL score of ≤ 0.15, given that 95% of pathogenic variants are excluded at this score (Ioannidis et al., 2016). Variants with a REVEL score between 0.15–0.7 will not receive either rule (PP3 or BP4).
The HL-EP recommends using MaxEntScan when assessing potential splicing impacts at the DNA level for missense, silent, and splice consensus sequence variants outside of the canonical ± 1 or 2 sites. For variants at the canonical splice sites, PP3 cannot be applied if PVS1 is used (Abou Tayoun et al., this issue).
Computational and predictive data rules with no changes (PM4, BP3, BP7)
The HL-EP did not further specify the rules pertaining to protein length changing variants (PM4, BP3). However, the PVS1 flowchart mentioned above should be carefully followed for large in-frame deletions or insertions, particularly those affecting one or more exons. Silent variants with no predicted splicing impact can have BP7 applied, with no specified changes.
FUNCTIONAL DATA
Functional studies (PS3, PS3_Supporting, BS4_Supporting)
Based on systematic assessment of the literature, the HL-EP determined that no functional assays met a strong level of evidence for or against pathogenicity, except for a variant-specific knock-in mouse model. This is mainly due to the lack of well-established functional assays with enough data to assess their validity in predicting variant pathogenicity in most HL genes. Except for the functional assays in the COCH, GJB2 and SLC26A4 genes described below, a PS3_Supporting or BS3_Supporting can be appropriately used for evaluating variants in other genes not specified here if the assay is well-validated with positive and negative controls and the results are consistent with a known protein function.
GJB2.
This gene, which encodes the gap junction protein, connexin 26, is the most common cause of genetic HL and has been well studied. Several research groups have measured the impact of genetic variants on its function primarily using electrical coupling or dye diffusion assays in Xenopus oocytes or mammalian cell systems (Bicego et al., 2006; Choung, Moon, & Park, 2002; D’Andrea et al., 2002; Lee, Derosa, & White, 2009; Mani et al., 2009). In both settings, a mutant and wildtype control cDNA are first transfected into separate oocytes or cell lines. In the electrical coupling assay, connexin 26 (encoded by GJB2) protein function is examined by altering the voltage in one cell and then measuring a current change in a neighboring cell through patch clamping or ion injection and dye sequestering (Ambrosi et al., 2013; Bruzzone et al., 2003; Haack et al., 2006; Mese, Londin, Mui, Brink, & White, 2004; Zhang et al., 2005). In the dye diffusion assay, movement of fluorescent dyes between neighboring cells is quantified as a measure of connexin 26 protein function. Variants disrupting connexin 26 function are expected to significantly reduce electrical coupling or dye transfer compared to control cDNA.
We searched the literature for variants that were assessed by these two assays, and that were validly classified as pathogenic, likely pathogenic, benign, or likely benign. We then determined the performance characteristics for each assay as described in the methods section and shown in Supp. Table S7. Because both assays demonstrated a high sensitivity and positive predictive value, the HL-EP recommends using PS3_Moderate for any novel GJB2 variant resulting in a statistically significant decrease or absence of current or dye transfer compared to wild-type run under the same experimental conditions. On the other hand, given the limited number of benign variants used as negative controls (Supp. Table S1), the HL-EP was less confident regarding the ability of either assay to accurately predict benign effects, and in such cases, using BS3_Supporting is more appropriate.
It should be noted that in dye diffusion assays, a negative control (e.g. non-transfected or water injected control) should not display dye diffusion. In electrical coupling assays, water injected controls should display negligible currents at all potentials.
SLC26A4.
This gene encodes an anion transporter, and its function has been assayed by measuring transport of radioactive anion isotopes (e.g. iodide or chloride) in cells expressing normal or mutant protein subunits (Bizhanova, Chew, Khuon, & Kopp, 2011; B. Y. Choi et al., 2009; Dossena, Bizhanova, et al., 2011; Dossena, Vezzoli, et al., 2006; Gillam et al., 2004; Ishihara et al., 2010; Palos et al., 2008; Reimold et al., 2011; Scott et al., 2000; Yuan et al., 2012). Alternatively, other groups have used halide-sensitive fluorescent probes to measure anion transport (Cirello et al., 2012; Dossena, Bizhanova, et al., 2011; Dossena, Nofziger, et al., 2011; Dossena, Rodighiero, et al., 2006; Fugazzola et al., 2007; Jang et al., 2014; Pera et al., 2008). An assay readout is considered abnormal if it is statistically different than the wild-type vector in the same experiment where a negative control (empty vector) shows absent immunofluorescent staining compared to wild type. In both radioactive and fluorescence based assays, several cell lines usually including Xenopus oocytes, mammalian, or human cells can be used to express relevant SLC26A4 variants. As with GJB2, we determined performance characteristics of both assays by reviewing the results from valid pathogenic/likely pathogenic and benign/likely benign variants in ClinVar (Supp. Tables S1 and S7). Again, there was a limited number of benign variants to assess the ability of both assays to predict benign effects (Supp. Table S1). Furthermore, compared to GJB2, the two assays had lower positive predictive values (Supp. Table S7). Therefore, we recommend using PS3_Supporting or BS3_Supporting if a pathogenic or a benign effect, respectively, was suggested by either assay.
COCH.
This gene encodes cochlin, which is normally secreted into the extracellular matrix of the inner ear (Fransen et al., 1999; Robertson et al., 2006; Robertson et al., 1998). Immunofluorescence and western blotting techniques have been used to assay cochlin secretion, localization, and post-translational modifications subsequent to expression in bacterial cells such as E. coli or mammalian cell lines (Bae et al., 2014; Cho et al., 2012; Grabski et al., 2003; Jung, Kim, Lee, Yang, & Choi, 2015). Compared to wild-type cochlin, abnormal patterns include absence of extracellular deposition (i.e., no secretion), intracellular aggregation (i.e. in the endoplasmic reticulum and Golgi), and stable dimerization due to inappropriate disulfide bond formation interfering with protein secretion and/or function (Cho et al., 2012; Nagy et al., 2004; Street et al., 2005; Yao, Py, Zhu, Bao, & Yuan, 2010). Based on the high sensitivity and positive predictive value (Supp. Tables S1 and S7) of these protein assays, we recommend using PS3_Moderate if a variant exhibits an abnormal secretion, dimerization, or localization pattern compared to wild-type protein. On the other hand, due to a dearth of benign variants tested by these assays, we recommend using only BS3_Supporting if a variant shows a comparable pattern to wild-type cochlin using these protein assays. No criteria should be applied if multiple assay results do not agree.
Mutational hot-spots or functional domains (PM1)
The HL-EP identified one gene, KCNQ4, in the variant pilot set that harbored a functional domain for which PM1 can be applied. Missense variants located in the pore forming region (amino acids 270 – 297) of KCNQ4 are eligible for PM1, based on this region’s extreme intolerance to variation in the general population and significant enrichment of pathogenic missense variants observed in individuals with dominant sensorineural HL (T. Naito et al., 2013).
However, a systematic review of mutational hotspots or functional domains for all genes associated with hearing loss was not performed by the HL-EP. We acknowledge that PM1 can likely be applied to other such regions, such as variants impacting glycine residues in the Gly-X-Y motifs of the collagen genes (COL11A2, COL4A3, COL4A4, and COL4A5), which are essential for the triple helical structure formation (Chakchouk et al., 2015; I. Naito, Kawai, Nomura, Sado, & Osawa, 1996; Savige et al., 2016; Tryggvason, Zhou, Hostikka, & Shows, 1993).
SEGREGATION DATA (PP1, PP1_Moderate, PP1_Strong, BS4)
Segregation with disease is used as evidence towards pathogenicity, and with an increasing number of segregations, stronger evidence can be applied. Given that segregation evidence is not specific to HL, the HL-EP worked in conjunction with the SVI to develop guidelines that further specify PP1. Three levels of evidence were recommended for both autosomal dominant and autosomal recessive segregations. Each strength level was based upon likelihood ratios of 4:1 (LOD 0.6), 16:1 (LOD 1.2), and 32:1 (LOD 1.5) to count as supporting, moderate, and strong evidence, respectively (Table 4a). For autosomal recessive segregations, unaffected individuals with equal probability of inheriting the variant(s) in question, typically siblings of a proband, can be taken into account, as shown in Table 4b.
Table 4a:
Recommendations for PP1 (segregation evidence)
Table 4b:
Recommendations for autosomal recessive segregation evidence (PP1)
| General Recommendations (Phenocopy not an issue) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unaffected Recessive Segregations | ||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Affected segregations | 0 | 0 | 0.12 | 0.25 | 0.37 | 0.5 | 0.62 | 0.75 | 0.87 | 1 | 1.12 | 1.25 |
| 1 | 0.6 | 0.73 | 0.85 | 0.98 | 1.1 | 1.23 | 1.35 | 1.48 | 1.6 | 1.73 | 1.85 | |
| 2 | 1.2 | 1.33 | 1.45 | 1.58 | 1.7 | 1.83 | 1.95 | 2.08 | 2.2 | 2.33 | 2.45 | |
| 3 | 1.81 | 1.93 | 2.06 | 2.18 | 2.31 | 2.43 | 2.56 | 2.68 | 2.81 | 2.93 | 3.06 | |
| 4 | 2.41 | 2.53 | 2.66 | 2.78 | 2.91 | 3.03 | 3.16 | 3.28 | 3.41 | 3.53 | 3.66 | |
| 5 | 3.01 | 3.14 | 3.26 | 3.39 | 3.51 | 3.63 | 3.76 | 3.88 | 4.01 | 4.13 | 4.26 | |
| 6 | 3.61 | 3.74 | 3.86 | 3.99 | 4.11 | 4.24 | 4.36 | 4.49 | 4.61 | 4.74 | 4.86 | |
| 7 | 4.21 | 4.34 | 4.46 | 4.59 | 4.71 | 4.84 | 4.96 | 5.09 | 5.21 | 5.34 | 5.46 | |
| 8 | 4.82 | 4.94 | 5.07 | 5.19 | 5.32 | 5.44 | 5.57 | 5.69 | 5.82 | 5.94 | 6.07 | |
| 9 | 5.42 | 5.54 | 5.67 | 5.79 | 5.92 | 6.04 | 6.17 | 6.29 | 6.42 | 6.54 | 6.67 | |
| 10 | 6.02 | 6.15 | 6.27 | 6.4 | 6.52 | 6.65 | 6.77 | 6.9 | 7.02 | 7.15 | 7.27 | |
Affected segregations are counted in rows and unaffected segregations in columns. Affected segregations are affected family members in whom biallelic compound heterozygous or homozygous variants segregates. Unaffected segregations are defined as unaffected family members, typically siblings, who are at risk to inherit the two variants identified in the proband. These individuals should be either wild-type for both variants identified in the proband, or a heterozygous carrier for a single variant. Unaffected, carrier parents DO NOT count as unaffected segregations. There may be scenarios where individuals other than siblings could be counted as segregations, such as in families where one parent is affected with the autosomal recessive disorder, in large families with multiple branches, or in consanguineous families.
Each cell shows the LOD score of each combination of affected and unaffected segregations. LOD scores were calculated using a simplified LOD score formula, as described in Strande et al., 2017.
Non-segregations can be used as strong benign evidence (BS4), however this should be done with caution. For family members that have HL, but are negative for the variant(s) in question (phenotype-positive, genotype-negative), there is a possibility for a phenocopy. Indications of a potential phenocopy could include differences in phenotype, such as HL age of onset, severity, and the audiogram shape.
For family members who are positive for the variant(s) in question, but have normal hearing (genotype-positive, phenotype-negative), confounding factors that must be taken into consideration include age-related penetrance and variable expression. It is recommended that BS4 is only applied for genotype-positive, phenotype-negative scenarios for genes expected to have full penetrance, including most autosomal recessive genes. Furthermore, there must be confidence that the family member is truly unaffected. Diagnostic audiometric data, including ear specific audiograms or an auditory brainstem response (ABR) should be obtained. If there is any evidence for reduced penetrance, variable expression, or an age of onset later than the age of the unaffected person, BS4 should not be applied.
DE NOVO OCCURRENCE (PS2, PS2_VeryStrong, PS2_Moderate, PS2_Supporting and PM6):
As defined by the 2015 ACMG guidelines, PM6 stands for “de novo, maternity and paternity not confirmed” and PS2 stands for “de novo, paternity and maternity confirmed”. Approximately 80% of nonsyndromic HL is inherited in an autosomal recessive pattern with de novo occurrences being rare (Alford et al., 2014). However, de novo occurrences have been reported, particularly for genes associated with autosomal dominant or X-linked HL (Alvarez et al., 2003; J. W. Choi et al., 2015; Moteki et al., 2015; Yuan et al., 2009). Given that de novo strength-level specifications are not specific to HL genes, the HL-EP worked in conjunction with the SVI and adapted the current recommendations from SVI which are posted on the ClinGen website (https://www.clinicalgenome.org/working-groups/sequence-variant-interpretation/). Different levels of strength are applied to de novo occurrences depending on how many are observed, whether the phenotype is specific, and whether maternity and paternity are confirmed (Tables 5a and 5b).
Table 5a:
Points awarded per de novo occurrence(s)
| Phenotypic consistency | Points per Proband | |
|---|---|---|
| Confirmed de novo | Assumed de novo | |
| Phenotype highly specific for gene | 2 | 1 |
| Phenotype consistent with gene but not highly specific |
1 | 0.5 |
| Phenotype consistent with gene but not highly specific and high genetic heterogeneity† |
0.5 | 0.25 |
| Phenotype not consistent with gene | 0 | 0 |
Maximum allowable value of 1 may contribute to overall score
Table 5b:
Recommendation for determining the appropriate ACMG/AMP evidence strength level for de novo occurrence(s)
| Supporting (PS2_Supporting or PM6_Supporting) |
Moderate (PS2_Moderate or PM6) |
Strong (PS2 or PM6_Strong) |
Very Strong (PS2_VeryStrong or PM6_VeryStrong) |
|---|---|---|---|
| 0.5 points | 1.0 points | 2.0 points | 4.0 points |
ALLELIC DATA (PM3, BS2)
PM3, PM3_Strong, PM3_VeryStrong: In trans with a pathogenic variant (recessive disorders)
For recessive disorders, identifying a variant in trans with a pathogenic variant on the second allele in an affected individual is considered evidence towards pathogenicity. This was previously defined in the ACMG/AMP guidelines as moderate evidence (PM3) (Richards et al., 2015). However, if the variant under consideration is found in multiple probands with pathogenic variants on the other allele, the strength of that evidence should be considered greater. Therefore, in conjunction with the SVI, we developed a scoring system to account for the number of compound heterozygous and homozygous probands identified with a variant, as shown in Table 6. Each instance that a variant is found in a proband, either in homozygosity or compound heterozygosity with a second variant, is given a specified number of points, and the total points corresponds to the strength of PM3 that can be applied. Other scenarios, such as homozygous variants in consanguineous families, and variants of unknown phase, were also taken into account.
Table 6a:
Default points for scoring variants that are observed in trans (PM3 rules)
| Classification/zygosity of other variant | Points per proband |
|
|---|---|---|
| Known in trans | Phase unknown | |
| Pathogenic/Likely pathogenic | 1.0 | 0.5 |
| Homozygous occurrence (Max points from homozygotes=1.0) |
0.5 | N/A |
| Rare uncertain significance variant on other allele, OR Homozygous occurrence due to consanguinity, (Max point= 0.5) |
0.25 | N/A |
Furthermore, if a variant has been observed in many probands, each with a different pathogenic variant on the other allele, the likelihood that the variants are in trans is higher even if phasing has not been performed in every case, assuming the gene has been fully sequenced to detect all variants. In these cases, point values can be adjusted based on confidence of in trans occurrence.
BS2: Observation in controls inconsistent with disease penetrance
BS2 can be utilized when a variant is observed in an adult with normal hearing tests when the hearing loss is expected to be fully penetrant from birth or prelingually. For recessive hearing loss, this would be applicable if the variant is observed in trans with a known pathogenic variant or if the variant occurs in the homozygous state. However, we do not recommend using BS2 if the known pathogenic variant observed in the unaffected individual is a hypomorphic or low penetrance allele, such as the NM_004004.5:p.(Val37Ile) or NM_004004.5:p.(Met34Thr) variants in GJB2. By definition, these hypomorphic or low penetrance alleles could result in normal or low normal hearing thresholds.
For autosomal dominant hearing loss, the variant would be heterozygous. However, BS2 should not be implemented when variable onset and severity of hearing loss has been associated to the gene in which the variant occurs.
BP2: Observation in trans with a pathogenic variant for dominant disorders or observation of a variant in cis with a pathogenic variant
The identification of a variant in cis with a pathogenic variant is supportive evidence for a benign interpretation. However, caution is recommended for the application of this rule for a case with an in trans observation of a dominant pathogenic variant. Careful assessment should include whether an earlier onset or more severe phenotype is possible, which could be consistent with an in trans pathogenic variant.
PHENOTYPIC DATA (PP4 AND BP5)
The HL-EP developed recommendations for PP4, which, in the ACMG/AMP guideline, was intended as supporting evidence of pathogenicity when the patient’s phenotype is highly specific for a single gene. The HL-EP selected a set of syndromes and genes where clinical features in addition to hearing loss are observed in which PP4 can be applied. Isolated hearing loss is not sufficient to apply PP4. The HL-EP applied this rule to HL syndromes if all known causative genes have been sequenced and the detection rate at least doubles when the added clinical feature(s) is present in the proband or one or more prior reported cases (Table 7). The rationale for using a doubling is based on a Bayesian model of the ACMG/AMP guideline whereby any supporting piece of evidence is equivalent to a doubling of likelihood (Tavtigian et al., 2018). The specific clinical features in addition to HL that are required for application of PP4 are shown in Table 7.
Table 7:
Recommendations for specification of PP4
| Gene | Syndrome |
Phenotypes observed in proband or prior reported cases required to apply PP4 |
Detection rate in unselected HL |
Detection rate with specified phenotype |
|---|---|---|---|---|
| SLC26A4 | Pendred syndrome |
Hearing loss with enlarged vestibular aqueduct (EVA) and/or Mondini malformation (incomplete partitioning type 2) |
2.6% (Sloan-Heggen et al., 2016) |
50% for a single mutation (Albert et al., 2006; Azaiez et al., 2007; Chattaraj et al., 2017; B. Y. Choi, Madeo et al., 2009; Pryor et al., 2005) |
|
MYO7A, USH1C, CDH23, PCDH15, USH1G |
Usher syndrome Type I |
Moderately-severe to profound hearing loss and retinitis pigmentosa (onset typically in first decade), +/− vestibular dysfunction |
4.3% (Sloan-Heggen et al., 2016) |
78.7% for 2 mutations (Le Quesne Stabej et al., 2012) |
|
USH2A, ADGRV1 (GPR98), WHRN (DFNB31) |
Usher syndrome Type II |
Mild to severe hearing loss and retinitis pigmentosa (onset typically in first or second decade). |
2.9% (Sloan-Heggen et al., 2016) |
60.3% (Le Quesne Stabej et al., 2012) |
| CLRN1 | Usher syndrome Type III |
Progressive hearing loss with retinitis pigmentosa (variable onset) and vestibular dysfunction |
<1% (Sloan-Heggen et al., 2016) |
50% (2/4) for 2 mutations (Le Quesne Stabej et al., 2012) |
| WFS1 | LF – SNHL | Low frequency autosomal dominant sensorineural hearing loss | 0.4% (Sloan-Heggen et al., 2016) |
28.5% (2/7 cases, LMM unpublished data) |
|
EYA1, SIX1 |
Branchio- oto-renal syndrome |
3 major – or – 2 major + 2 minor – or – 1 major + a 1st degree
relative meeting criteria MAJOR: Branchial anomalies, hearing loss/deafness, preauricular pits, renal anomalies MINOR: External ear anomalies, middle ear anomalies, inner ear anomalies, preauricular tags, other (facial asymmetry, palate abnormalities) (Chang et al., 2004) |
0.2% (Sloan-Heggen et al., 2016) |
45% Testing must include deletion/duplication analysis (Smith, 1993) |
|
OTOF, DFNB59 |
ANSD | Auditory neuropathy spectrum disorder | 1% (Sloan-Heggen et al., 2016) |
9–50% (Matsunaga et al., 2012; Rodriguez-Ballesteros et al., 2008; Varga et al., 2006) |
| MYH9 | MYH9- related disorders |
Congenital macrothrombocytopenia and platelet macrocytosis (Pecci et al., 2014) NOTE: Patient must also be tested for DIAPH1 with no DIAPH1 variants identified |
<0.1% (Sloan-Heggen et al., 2016) |
>90% (Pecci et al., 2014; Pecci et al., 2008; Savoia & Pecci, 1993) |
| DIAPH1 | AD hearing loss with macro- thrombo- cytopenia |
Macrothrombocytopenia (Neuhaus et al., 2017; Stritt et al., 2016) NOTE: Patient must also be tested for MYH9 with no MYH9 variants identified All truncating variants associated to AD occur in exon 27. Truncating variants in other exons have been associated with AR microcephaly. |
Detection rate is unknown. Rare syndrome, prevalence <1/1000000 (DIAPH1-related sensorineural hearing loss- thrombocytopeniasyn drome, n.d.) |
Unknown |
|
PAX3, SOX10, MITF, EDN3, EDNRB |
Waardenbu rg syndrome |
Two or more of the following: 1. Congenital SNHL 2. Pigmentary disturbances of iris 1. Complete heterochromia (two eyes of different color); 2. Partial or segmental heterochromia 3. Hypoplastic blue eyes – characteristic brilliant blue in both eyes 3. Pigmentary disturbances of hair 1. White forelock from birth or in teens 2. Premature graying before age 20 4. A first or second degree relative with 2 or more of criteria 1–3 5. Dystopia canthorum (PAX3 variants) 6. Hirschsprung disease (SOX10, EDN3, EDNRB) Modified from (Liu, Newton, & Read, 1995) |
Detection rate is unknown. Rare syndrome, prevalence of 9/100000 (Pingault et al. 2010; Pingault, 2015) |
>75% for WS1 and WS3 30% for WS2 50% for WS4 (Pingault et al., 2010) Testing must include deletion/duplication analysis |
|
POU3F4 |
X-linked recessive deafness |
Hearing loss + Incomplete partitioning type III (IAC dilation, fistulous connection between basal turn of cochlea and the IAC, cochlear hypoplasia, stapes fixation) |
Detection rate unknown. Prevalence is unknown. Inner ear abnormalities are pathognomonic. (X-linked mixed deafness with perilymphatic gusher, n.d.). |
≥26.7% Testing must include deletion/duplication analysis including 5’ promotor region. |
| GPSM2 | Chudley- McCullough syndrome |
Congenital severe to profound hearing loss with brain abnormalities including frontal polymicrogyria, grey matter heterotopia, cerebellar dysplasia, ventriculomegaly with small frontal horns, agenesis of the corpus callosum, arachnoid cysts (Doherty et al., 2012). |
Detection rate unknown. Rare. Prevalence is 1 / 1 000 000 (Chudley-McCullough syndrome, n.d.) |
>90% (Diaz-Horta et al., 2012; Doherty et al., 2012) |
|
KCNE1, KCNQ1 |
Jervell and Lange- Nielsen syndrome |
Congenital sensorineural hearing loss (must be bilateral and severe to profound) and a reproducible prolonged QTc interval ≥ 480 msec Notes: - GeneReviews states most patients reported to date have QTc ≥500msec - Schwartz Score: ≥3.5 points = high probability of LQTS; QTc ≥ 480 msec (3 points) plus congenital deafness (0.5 points) - Upper limit of normal QTc is 440msec for males and 460msec for post-pubertal females. (Alders, Bikker, & Christiaans, 1993) |
Detection rate unknown. Rare. Prevalence is 1–9 / 1 000 000 (Celano et al. 2009) |
≥90% (Tranebjaerg, Samson, & Green, 1993) |
Abbreviations: LF-SNHL: = Low frequency sensorineural hearing loss. WS1 = Waardenburg syndrome, type 1; WS2 = Waardenburg syndrome, type 2; WS3 = Waardenburg syndrome, type 3; WS4 = Waardenburg syndrome, type 4.
The HL-EP further specified the BP5 rule, which the ACMG/AMP guidelines defined as supporting evidence of benign when a variant is identified in a case with an alternate cause. The HL-EP recommends that for autosomal recessive genes, BP5 is only invoked when the variant is in the homozygous or compound heterozygous state. It is not recommended to be used for heterozygous observations of variants in autosomal recessive genes, given that affected individuals are frequently identified as carriers of other pathogenic recessive variants in genes unrelated to the cause of their HL.
REMOVED AND NOT APPLICABLE RULES
The HL-EP recommends that PP2 (missense variants in genes with low rate of benign missense variation) and BP1 (missense variants in genes where only LOF variants cause disease) are not used. These are not applicable to any known genes associated with HL. In addition, the HL-EP supports the SVI’s decision to remove the two rules pertaining to variant classifications from reputable sources without evidence available (PP5 and BP6) based on the published rationale (Biesecker & Harrison, 2018).
COMBINING CRITERIA RULES
The HL-EP recommends following the rules for combining criteria as outlined in the ACMG/AMP guidelines, with the addition of two modifications.
First, the HL-EP specified that PVS1 and PM2_Supporting can be combined to reach a variant classification of likely pathogenic. Note that the ACMG/AMP criteria require combining at least one moderate or two supporting criteria with a very strong criteria to reach a likely pathogenic classification. This modification was decided given that the HL-EP adopted the stringent criteria for evaluating predicted LOF variants, as described above under PVS1, such that we feel that rare variants meeting at least PM2_Supporting have at least a 90% chance of being pathogenic. However, this modification is not applicable in dominant HL since the HL-EP did not create a MAF threshold for PM2_Supporting in the dominant form of the disease (see above).
The second HL-EP modification was that variants meeting BS1 with no conflicting evidence can be classified as Likely Benign. This is consistent with approaches taken by the RASopathy and Cardiomyopathy Expert Panels (Gelb et al., 2018; Kelly et al., 2018).
Finally, per the new PVS1 rule recommendation (Abou Tayoun et al., this issue), we recommend against combining PVS1 with PP3 for the canonical ±1 or 2 splice bases given that PP3 is essentially applying predictive assumptions already built into PVS1.
VARIANT PILOT OF THE HEARING LOSS SPECIFIED RULES
A set of 51 variants in the GJB2, MYO7A, USH2A, CDH23, MYO6, KCNQ4, COCH, TECTA, and SLC26A4 genes were selected to test and refine the above specifications set by the HL-EP. The majority of variants were in GJB2, USH2A, and SLC26A4 (Figure 2). These variants were either present in ClinVar, on the exclusion list for BA1 and BS1, in a HL-EP’s internal laboratory data, or identified via literature search. All the major rule types supporting pathogenicity were tested, except for the rules specifying de novo occurrences which are extremely rare within HL (Figure 3). Dual curation was performed, and then consensus was reached following discussion for all classifications and their supporting rules among the HL-EP.
Figure 2.

Distribution of pilot variants (n=51) by gene.
Figure 3. Frequency of rules applied during the pilot.
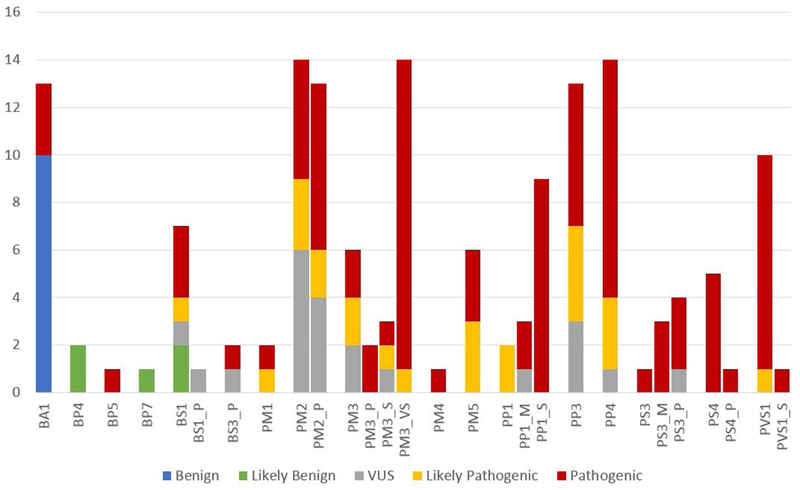
The frequency with which ACMG/AMP rules were applied during the variant pilot and the final classification of the variant when the rule was applied. Rules applied with a modified strength are denoted by the rule followed by _P for Supporting, _M for Moderate, _S for Strong, and _VS for Very Strong.
Of note, the most commonly utilized rules in our pilot were PM2 and PM3 and their modified strength levels (supporting through very strong) (Figure 4). PM2 and PM2_Supporting were used almost equally, with PM2 being applied for 14 variants and PM2_Supporting applied for 13 variants. In addition, PM3 or its modified strengths were used for 25 variants. PP1, PP3, PP4, and PVS1 were also commonly utilized. Pathogenic or likely pathogenic variants where either BA1 or BS1 was applied were only for variants listed on our exclusion list. Furthermore, the single variant of uncertain significance which had BS1 applied was the NM_004004.5:c.−22–2A>C variant in GJB2, which had conflicting data.
Figure 4. Final variant classifications of 51 pilot variants compared to initial classifications in ClinVar.

The total height of each bar represents the total number of variants that were given each classification the HL-EP. The colored segments of each bar represent variants whose ClinVar classification differed from the HL-EP classification. Codes listed in the colored segments of the bars indicate the criteria used to classify the variants that changed category during the variant pilot. Codes were only listed once if they were used for multiple variants in the same category.
The number of variants that were classified as benign, likely benign, uncertain, likely pathogenic and pathogenic, as well as the number of variants that changed from their initial classification in ClinVar to a new classification are shown in Figure 4. Of the 51 variants evaluated in the pilot, 37% (19/51) changed category based upon application of the expert panel specified rules and/or aggregation of evidence across laboratories. We did observe that the number of variants classified as uncertain significance (VUS) decreased as shown in Figure 4. Four variants listed in ClinVar as VUS were reclassified to either likely benign or benign due to the application of either BA1 or BS1 specified by the HL-EP. One variant that was reclassified from VUS to likely pathogenic (NM_004700.3:c.853G>A (p.Gly285Ser) in KCNQ4) did so because of the application rules that were specified by the HL-EP, including PS4_Supporting, PP1, and PM1.
One variant listed in ClinVar as pathogenic was reclassified as uncertain significance. This was the NM_00400.5:c.−22–2A>C variant in GJB2. This variant was present at a frequency sufficient to reach BS1, but several probands were reported in the literature. The HL-EP contacted the ClinVar submitters for clinical information on these probands and determined that the internal laboratory evidence was not sufficient to reach a classification of pathogenic, particularly the identification of one patient who had the NM_00400.5:c.35delG pathogenic variant in GJB2 in trans, yet had normal hearing.
One variant previously classified in ClinVar as likely benign was reclassified to VUS. This variant, NM_000260.3:c.1007G>A p.(p.Arg336His) in MYO7A, had been classified as likely benign based on its frequency, but it only met the BS1_Supporting threshold set by the HL-EP.
Five variants listed previously in ClinVar as Likely Pathogenic were reclassified to Pathogenic. Of note, one of these variants, NM_000441.1: c.349C>T p.(p.Leu117Phe) in SLC26A4, required internal laboratory data from one of the HL-EP members to reach a pathogenic classification.
Variants that had conflicting interpretations of pathogenicity in ClinVar (n=5) were reclassified to either Likely Pathogenic or Pathogenic. The NM_000441.1:c.706C>G p.(p.Leu236Val) variant in SLC26A4 was resolved using expert judgement upon review of phenotypic information of one of the probands reported by a HL-EP member lab. The NM_004004.5:c.109G>A (p.Val37Ile) and NM_004004.5:c.101T>C (p.Met34Thr) variants in GJB2 were also resolved by the HL-EP based on the application of PS4, PM3_VS, PP1_S, PM5, and PS3_M for both of these variants. Of note, the specification of PS4 by the HL-EP aided the classification of these variants, as the high MAF frequency and frequent observation in individuals with hearing loss may have been previously interpreted as conflicting evidence by some ClinVar submitters. The last two variants with conflicting interpretations in ClinVar, NM_004004.5:c.34G>T (p.Gly12Cys) and NM_004004.5:c.167del in GJB2, had multiple submitters that submitted either a pathogenic or likely pathogenic classification, and only one submitter for each variant caused the conflict in ClinVar.
In addition, the consensus discussions performed during the variant pilot led to the identification of several rules that required further clarification to ensure proper application, including PP1, PS4, PP3, BP4, and BP1_Supporting. The clarifications were the result of conversation with the HL-EP as a whole, versus an empirical assessment of the variant pilot. A summary of the genes, their associated phenotype(s) and inheritance pattern(s), and gene-specific rules are shown in Table 8. The list of variants included in the pilot and their classifications and supporting rules are shown in Supp. Table S8.
Table 8:
Variant pilot genes and applicable gene-specific ACMG/AMP criteria
| Gene |
Disease,
Inheritance |
PVS1
Applicable |
PM1: Mutational
hot spot or well- studied functional domain |
Functional Assays |
Phenotype
(PP4) Applicable |
| CDH23 | Usher syndrome, AR |
Yes | N/A | N/A | Yes |
| COCH | Nonsyndromic HL, AD |
N/A | N/A | Localization, secretion, and dimerization studies performed using immunofluorescence and Western blotting techniques |
N/A |
| GJB2 | Nonsyndromic SNHL, AR |
Yes | N/A | Electrical coupling assays, dye transfer assays |
N/A |
| KCNQ4 | Nonsyndromic SNHL, AD |
amino acids 271- 292 |
N/A | N/A | |
| MYO6 | Nonsyndromic SNHL, AD |
Yes | N/A | N/A | N/A |
| MYO7A | Usher syndrome, AR |
Yes | N/A | N/A | Yes |
| SLC26A4 | Pendred syndrome, AR |
N/A | Radio isotope and fluorescence assays |
Yes | |
| TECTA | Nonsyndromic SNHL, AD |
N/A | N/A | N/A | N/A |
| Nonsyndromic SNHL, AR |
Yes | N/A | N/A | N/A | |
| USH2A | Usher syndrome, AR |
Yes | N/A | N/A | Yes |
Abbreviations: AR = autosomal recessive; AD = autosomal dominant; N/A = not applicable
DISCUSSION
Specifying the ACMG/AMP guidelines for HL posed unique challenges, which arose from high prevalence, high genetic heterogeneity, and multiple inheritance patterns. While published specifications exist for cardiomyopathy and RASopathy, these specifications focus on autosomal dominant disorders that typically have pathogenic missense variants as the causal variants. In contrast, HL is frequently autosomal recessive with LOF as the primary disease mechanism, and therefore, the HL-EP specifications addressed and utilized rules that have not been previously refined for more quantitative and nuanced use, including PM3 and PVS1. These rules were some of the most commonly used rules in the variant pilot, particularly for the classification of likely pathogenic and pathogenic variants, and are likely to be frequent among all recessive disorders (Figure 3). As such, these specifications could be applied or serve as a groundwork for other autosomal recessive disorders that rely heavily on biallelic proband observations and predicted LOF variants.
The MAF thresholds for BA1, BS1, and PM2 were some of the most challenging rules to develop for the HL-EP. The MAF thresholds had to meet the “Goldilocks principle”. The thresholds for BA1 and BS1 could not be so high that they would never be reached and they could not be so low that they were over-utilized, as either scenario would lead to variant mis-classification. MAF thresholds had to be developed separately for recessive and dominant hearing loss, given the different attributes for each form. Furthermore, variants for recessive disorders are often expected to be present in the general population at low frequency, which poses a challenge to setting hard MAF thresholds for recessive HL. The addition of BS1_Supporting and PM2_Supporting aided the classification of some variants during the pilot project. Particularly, PM2_Supporting was used to support the classification of two Likely Pathogenic and seven Pathogenic variants, whereas the PM2 rule was used at a similar number of times to support three Likely pathogenic and five Pathogenic variants (Figure 3).
Furthermore, a new combining rule for addressing novel LOF variants was created to address “under-classifying” predicted loss of function variants that the HL-EP believed should be classified as likely pathogenic. By adding a rule whereby PM2_Supporting and PVS1 can be combined to reach a likely pathogenic variant classification, the HL-EP allowed convincing LOF variants with MAFs consistent with pathogenicity to be classified as likely pathogenic while remaining conservative for other potential rule combinations. Although we would consider it appropriate to use this rule combination for both autosomal recessive and autosomal dominant hearing loss, we did not create MAF thresholds for PM2_Supporting or BS1_Supporting for dominant hearing loss, and therefore this rule combination cannot be used for dominant hearing loss.
Our pilot variant project demonstrated the impact of our new rules on HL variant specification (Figure 4). We acknowledge that the pilot project is small (n=51) and the selected variants were biased, as we needed variants to test as many rule specifications as possible. However, we hope that implementation of the HL-EP’s specifications will continue to reduce the number of VUS and lead to fewer conflicts within ClinVar. Indeed, the next steps of the HL-EP will be to provide expert classification on many variants in ClinVar prioritizing medically significant conflicting interpretations (P/LP vs VUS/LB/B) as well as VUSs submitted by multiple labs for which aggregation of data might lead to a shift in classification.
The HL-EP expects that these specifications will evolve as they are utilized by diagnostic laboratories, and as more information on the genetic basis of HL is identified, such as additional autosomal dominant loci, adult-onset HL, and founder mutations in under-studied cohorts. Furthermore, ongoing general refinements to the ACMG/AMP guidelines may be made by the ClinGen SVI which will need to be addressed.
The HL-EP is in the process of submitting for full Expert Panel status for its work on variant interpretation, with the goal of submitting expert variant classifications to ClinVar. These classifications will resolve discrepancies, move VUSs towards Benign or Pathogenic and increase the review status of hearing loss variant classifications present in ClinVar.
Supplementary Material
Table 6b:
Recommendation for determining the appropriate ACMG/AMP evidence strength level for in trans occurrence(s)
| SUPPORTING (PM3_Supporting) |
Moderate (PM3) |
Strong (PM3_Strong) |
Very Strong (PM3_Very Strong) |
|---|---|---|---|
| 0.5 points | 1.0 points | 2.0 points | 4.0 points |
Acknowledgments
The HL-EP would like to thank Steven Harrison, Tina Pesaran, Leslie G. Biesecker and the additional members from the SVI for their feedback on these specifications, and their contributions to the development of the PVS1, PM3, and PP1 rules.
The authors would also like to acknowledge additional members of the HLWG for their contributions, including Sonia Abdelhak, John Alexander, Zippora Brownstein, Rachel Burt., Byung Yoon Choi, Lilian Downie,Thomas Friedman, Anne Giersch, John Greinwald, Jeffrey Holt, Makoto Hosoya, Un-Kyung Kim, Ian Krantz, Suzanne Leal, Saber Masmoudi, Tatsuo Matsunaga, Matías Morín, Cynthia Morton, Hideki Mutai, Arti Pandya, Richard Smith, Mustafa Tekin, Shin-Ichi Usami, Guy Van Camp, Kazuki Yamazawa, Hui-Jun Yuan, Elizabeth Black-Zeigelbein, and Keijan Zhang.
GRANT SPONSOR: Research reported in this publication was supported by the National Human Genome Research Institute (NHGRI) under award number U41HG006834.
REFERENCES
- Abou Tayoun AN, Al Turki SH, Oza AM, Bowser MJ, Hernandez AL, Funke BH (2016). Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med, 18(6), 545–553. 10.1038/gim.2015.141 [DOI] [PubMed] [Google Scholar]
- Abou Tayoun A, Pesaran T, DiStefano M, Oza A, Rehm H, Biesecker L, Harrison S, ClinGen Sequence Variant Interpretation Working Group. (2018). Recommendations for Interpreting the Loss of Function PVS1 ACMG/AMP Variant Criterion. Hum Mutat, 39(this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Blons H, Jonard L, Feldmann D, Chauvin P, Loundon N (2006). SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur J Hum Genet, 14(6), 773–779. 10.1038/sj.ejhg.5201611 [DOI] [PubMed] [Google Scholar]
- Alders M, Bikker H, & Christiaans I (1993). Long QT Syndrome doi:NBK1129 [bookaccession] [Google Scholar]
- Alford RL, Arnos KS, Fox M, Lin JW, Palmer CG, Pandya A (2014). American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med, 16(4), 347–355. 10.1038/gim.2014.2 [DOI] [PubMed] [Google Scholar]
- Alloisio N, Morle L, Bozon M, Godet J, Verhoeven K, Van Camp G (1999). Mutation in the zonadhesin-like domain of alpha-tectorin associated with autosomal dominant non-syndromic hearing loss. Eur J Hum Genet, 7(2), 255–258. 10.1038/sj.ejhg.5200273 [DOI] [PubMed] [Google Scholar]
- Alvarez A, del Castillo I, Pera A, Villamar M, Moreno-Pelayo MA, Moreno F (2003). De novo mutation in the gene encoding connexin-26 (GJB2) in a sporadic case of keratitis-ichthyosis-deafness (KID) syndrome. Am J Med Genet A, 117A(1), 89–91. 10.1002/ajmg.a.10851 [DOI] [PubMed] [Google Scholar]
- Ambrosi C, Walker AE, Depriest AD, Cone AC, Lu C, Badger J (2013). Analysis of trafficking, stability and function of human connexin 26 gap junction channels with deafness-causing mutations in the fourth transmembrane helix. PLoS One, 8(8), e70916 10.1371/journal.pone.0070916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD (2016). Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet, 99(1), 247 10.1016/j.ajhg.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaiez H, Yang T, Prasad S, Sorensen JL, Nishimura CJ, Kimberling WJ (2007). Genotype-phenotype correlations for SLC26A4-related deafness. Hum Genet, 122(5), 451–457. 10.1007/s00439-007-0415-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Robertson NG, Cho HJ, Morton CC, Jung DJ, Baek JI (2014). Identification of pathogenic mechanisms of COCH mutations, abolished cochlin secretion, and intracellular aggregate formation: genotype-phenotype correlations in DFNA9 deafness and vestibular disorder. Hum Mutat, 35(12), 1506–1513. 10.1002/humu.22701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicego M, Beltramello M, Melchionda S, Carella M, Piazza V, Zelante L (2006). Pathogenetic role of the deafness-related M34T mutation of Cx26. Hum Mol Genet, 15(17), 2569–2587. 10.1093/hmg/ddl184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, & Harrison SM (2018). The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 10.1038/gim.2018.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff AM, Luijendijk MW, Huygen PL, van Duijnhoven G, De Leenheer EM, Oudesluijs GG (2004). A novel mutation identified in the DFNA5 gene in a Dutch family: a clinical and genetic evaluation. Audiol Neurootol, 9(1), 34–46. 10.1159/000074185 [DOI] [PubMed] [Google Scholar]
- Bizhanova A, Chew TL, Khuon S, & Kopp P (2011). Analysis of cellular localization and function of carboxy-terminal mutants of pendrin. Cell Physiol Biochem, 28(3), 423–434. 10.1159/000335105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth KT, Azaiez H, Kahrizi K, Simpson AC, Tollefson WT, Sloan CM (2015). PDZD7 and hearing loss: More than just a modifier. Am J Med Genet A, 167A(12), 2957–2965. 10.1002/ajmg.a.37274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth KT, Azaiez H, Kahrizi K, Wang D, Zhang Y, Frees K (2018). Exonic mutations and exon skipping: Lessons learned from DFNA5. Hum Mutat, 39(3), 433–440. 10.1002/humu.23384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth KT, Kahrizi K, Babanejad M, Daghagh H, Bademci G, Arzhangi S (2018). Variants in CIB2 cause DFNB48 and not USH1J. Clin Genet, 93(4), 812–821. 10.1111/cge.13170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R, Veronesi V, Gomes D, Bicego M, Duval N, Marlin S (2003). Loss-of-function and residual channel activity of connexin26 mutations associated with non-syndromic deafness. FEBS Lett, 533(1–3), 79–88. [DOI] [PubMed] [Google Scholar]
- Celano G, Crotti L, Dagradi F, Schwartz P (2009). Jervell and Lange-Nielsen syndrome. Orphanet Encyclopaedia, Retrieved from http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=12056&Disease_Disease_Search_diseaseGroup=90647&Disease_Disease_Search_diseaseType=ORPHA&Disease(s)/group%20of%20diseases=Jervell-and-Lange-Nielsen-syndrome&title=Jervell-and-Lange-Nielsen-syndrome&search=Disease_Search_Simple
- Chakchouk I, Grati M, Bademci G, Bensaid M, Ma Q, Chakroun A (2015). Novel mutations confirm that COL11A2 is responsible for autosomal recessive non-syndromic hearing loss DFNB53. Mol Genet Genomics, 290(4), 1327–1334. 10.1007/s00438-015-0995-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EH, Menezes M, Meyer NC, Cucci RA, Vervoort VS, Schwartz CE (2004). Branchio-oto-renal syndrome: the mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat, 23(6), 582–589. 10.1002/humu.20048 [DOI] [PubMed] [Google Scholar]
- Chattaraj P, Munjal T, Honda K, Rendtorff ND, Ratay JS, Muskett JA (2017). A common SLC26A4-linked haplotype underlying non-syndromic hearing loss with enlargement of the vestibular aqueduct. J Med Genet, 54(10), 665–673. 10.1136/jmedgenet-2017-104721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Park HJ, Trexler M, Venselaar H, Lee KY, Robertson NG (2012). A novel COCH mutation associated with autosomal dominant nonsyndromic hearing loss disrupts the structural stability of the vWFA2 domain. J Mol Med (Berl), 90(11), 1321–1331. 10.1007/s00109-012-0911-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Madeo AC, King KA, Zalewski CK, Pryor SP, Muskett JA (2009). Segregation of enlarged vestibular aqueducts in families with non-diagnostic SLC26A4 genotypes. J Med Genet, 46(12), 856–861. 10.1136/jmg.2009.067892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Stewart AK, Madeo AC, Pryor SP, Lenhard S, Kittles R (2009). Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms? Hum Mutat, 30(4), 599–608. 10.1002/humu.20884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Min B, Kim A, Koo JW, Kim CS, Park WY (2015). De novo large genomic deletions involving POU3F4 in incomplete partition type III inner ear anomaly in East Asian populations and implications for genetic counseling. Otol Neurotol, 36(1), 184–190. 10.1097/MAO.0000000000000343 [DOI] [PubMed] [Google Scholar]
- Choung YH, Moon SK, & Park HJ (2002). Functional study of GJB2 in hereditary hearing loss. Laryngoscope, 112(9), 1667–1671. 10.1097/00005537-200209000-00026 [DOI] [PubMed] [Google Scholar]
- Cirello V, Bazzini C, Vezzoli V, Muzza M, Rodighiero S, Castorina P (2012). Molecular and functional studies of 4 candidate loci in Pendred syndrome and nonsyndromic hearing loss. Mol Cell Endocrinol, 351(2), 342–350. 10.1016/j.mce.2012.01.013 [DOI] [PubMed] [Google Scholar]
- Cunningham LL, & Tucci DL (2017). Hearing Loss in Adults. N Engl J Med, 377(25), 2465–2473. 10.1056/NEJMra1616601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea P, Veronesi V, Bicego M, Melchionda S, Zelante L, Di Iorio E (2002). Hearing loss: frequency and functional studies of the most common connexin26 alleles. Biochem Biophys Res Commun, 296(3), 685–691. [DOI] [PubMed] [Google Scholar]
- Diaz-Horta O, Sirmaci A, Doherty D, Nance W, Arnos K, Pandya A (2012). GPSM2 mutations in Chudley-McCullough syndrome. Am J Med Genet A, 158A(11), 2972–2973. 10.1002/ajmg.a.35636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStefano M, Hemphill S, Cushman B, Bowser M, Hynes E, Grant A (2018). Curating clinically relevant transcripts for the interpretation of sequence variants. bioRxiv. 10.1101/276287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty D, Chudley AE, Coghlan G, Ishak GE, Innes AM, Lemire EG (2012). GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am J Hum Genet, 90(6), 1088–1093. 10.1016/j.ajhg.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossena S, Bizhanova A, Nofziger C, Bernardinelli E, Ramsauer J, Kopp P (2011). Identification of allelic variants of pendrin (SLC26A4) with loss and gain of function. Cell Physiol Biochem, 28(3), 467–476. 10.1159/000335108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossena S, Nofziger C, Brownstein Z, Kanaan M, Avraham KB, & Paulmichl M (2011). Functional characterization of pendrin mutations found in the Israeli and Palestinian populations. Cell Physiol Biochem, 28(3), 477–484. 10.1159/000335109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossena S, Rodighiero S, Vezzoli V, Bazzini C, Sironi C, Meyer G (2006). Fast fluorometric method for measuring pendrin (SLC26A4) Cl-/I- transport activity. Cell Physiol Biochem, 18(1–3), 67–74. 10.1159/000095164 [DOI] [PubMed] [Google Scholar]
- Dossena S, Vezzoli V, Cerutti N, Bazzini C, Tosco M, Sironi C (2006). Functional characterization of wild-type and a mutated form of SLC26A4 identified in a patient with Pendred syndrome. Cell Physiol Biochem, 17(5–6), 245–256. 10.1159/000094137 [DOI] [PubMed] [Google Scholar]
- Eiberg H, Hansen L, Kjer B, Hansen T, Pedersen O, Bille M (2006). Autosomal dominant optic atrophy associated with hearing impairment and impaired glucose regulation caused by a missense mutation in the WFS1 gene. J Med Genet, 43(5), 435–440. 10.1136/jmg.2005.034892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, Verstreken M, Verhagen WI, Wuyts FL, Huygen PL, D’Haese P (1999). High prevalence of symptoms of Meniere’s disease in three families with a mutation in the COCH gene. Hum Mol Genet, 8(8), 1425–1429. [DOI] [PubMed] [Google Scholar]
- Fugazzola L, Cirello V, Dossena S, Rodighiero S, Muzza M, Castorina P (2007). High phenotypic intrafamilial variability in patients with Pendred syndrome and a novel duplication in the SLC26A4 gene: clinical characterization and functional studies of the mutated SLC26A4 protein. Eur J Endocrinol, 157(3), 331–338. 10.1530/EJE-07-0263 [DOI] [PubMed] [Google Scholar]
- Fukuoka H, Kanda Y, Ohta S, & Usami S (2007). Mutations in the WFS1 gene are a frequent cause of autosomal dominant nonsyndromic low-frequency hearing loss in Japanese. J Hum Genet, 52(6), 510–515. 10.1007/s10038-007-0144-3 [DOI] [PubMed] [Google Scholar]
- Gelb BD, Cave H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H (2018). ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet Med. 10.1038/gim.2018.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Oak N, & Plon SE (2017). Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol, 18(1), 225 10.1186/s13059-017-1353-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam MP, Sidhaye AR, Lee EJ, Rutishauser J, Stephan CW, & Kopp P (2004). Functional characterization of pendrin in a polarized cell system. Evidence for pendrin-mediated apical iodide efflux. J Biol Chem, 279(13), 13004–13010. 10.1074/jbc.M313648200 [DOI] [PubMed] [Google Scholar]
- Grabski R, Szul T, Sasaki T, Timpl R, Mayne R, Hicks B (2003). Mutations in COCH that result in non-syndromic autosomal dominant deafness (DFNA9) affect matrix deposition of cochlin. Hum Genet, 113(5), 406–416. 10.1007/s00439-003-0992-7 [DOI] [PubMed] [Google Scholar]
- Haack B, Schmalisch K, Palmada M, Bohmer C, Kohlschmidt N, Keilmann A (2006). Deficient membrane integration of the novel p.N14D-GJB2 mutant associated with non-syndromic hearing impairment. Hum Mutat, 27(11), 1158–1159. 10.1002/humu.9464 [DOI] [PubMed] [Google Scholar]
- Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S (2017). Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med, 19(10), 1096–1104. 10.1038/gim.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand MS, Morin M, Meyer NC, Mayo F, Modamio-Hoybjor S, Mencia A (2011). DFNA8/12 caused by TECTA mutations is the most identified subtype of nonsyndromic autosomal dominant hearing loss. Hum Mutat, 32(7), 825–834. 10.1002/humu.21512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogewind BF, Pennings RJ, Hol FA, Kunst HP, Hoefsloot EH, Cruysberg JR (2010). Autosomal dominant optic neuropathy and sensorineual hearing loss associated with a novel mutation of WFS1. Mol Vis, 16, 26–35. [PMC free article] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S (2016). REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet, 99(4), 877–885. 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- INSERM. (n.d.). “Chudley-McCullough syndrome.” Orphanet Encyclopaedia, Retrieved April 30, 2018 from http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=21500&Disease_Disease_Search_diseaseGroup=314597&Disease_Disease_Search_diseaseType=ORPHA&Disease(s)/group%20of%20diseases=Chudley-McCullough-syndrome&title=Chudley-McCullough-syndrome&search=Disease_Search_Simple
- INSERM. (n.d.). “DIAPH1-related sensorineural hearing loss-thrombocytopenia syndrome.” Orphanet Encyclopaedia, Retrieved April 30, 2018 from http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=25676&Disease_Disease_Search_diseaseGroup=494444&Disease_Disease_Search_diseaseType=ORPHA&Disease(s)/group%20of%20diseases=DIAPH1-related-sensorineural-hearing-loss-thrombocytopenia-syndrome&title=DIAPH1-related-sensorineural-hearing-loss-thrombocytopenia-syndrome&search=Disease_Search_Simple
- INSERM. (n.d.). “X-linked mixed deafness with perilymphatic gusher.” Orphanet Encyclopaedia, Retrieved April 30, 2018 from http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=157&Disease_Disease_Search_diseaseGroup=383&Disease_Disease_Search_diseaseType=ORPHA&Disease(s)/group%20of%20diseases=X-linked-mixed-deafness-with-perilymphatic-gusher&title=X-linked-mixed-deafness-with-perilymphatic-gusher&search=Disease_Search_Simple
- Ishihara K, Okuyama S, Kumano S, Iida K, Hamana H, Murakoshi M (2010). Salicylate restores transport function and anion exchanger activity of missense pendrin mutations. Hear Res, 270(1–2), 110–118. 10.1016/j.heares.2010.08.015 [DOI] [PubMed] [Google Scholar]
- Iwasa YI, Nishio SY, & Usami SI (2016). Comprehensive Genetic Analysis of Japanese Autosomal Dominant Sensorineural Hearing Loss Patients. PLoS One, 11(12), e0166781 10.1371/journal.pone.0166781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JH, Jung J, Kim AR, Cho YM, Kim MY, Lee SY (2014). Identification of novel functional null allele of SLC26A4 associated with enlarged vestibular aqueduct and its possible implication. Audiol Neurootol, 19(5), 319–326. 10.1159/000366190 [DOI] [PubMed] [Google Scholar]
- Jung J, Kim HS, Lee MG, Yang EJ, & Choi JY (2015). Novel COCH p.V123E Mutation, Causative of DFNA9 Sensorineural Hearing Loss and Vestibular Disorder, Shows Impaired Cochlin Post-Translational Cleavage and Secretion. Hum Mutat, 36(12), 1168–1175. 10.1002/humu.22855 [DOI] [PubMed] [Google Scholar]
- Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM (2018). Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med, 20(3), 351–359. 10.1038/gim.2017.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quesne Stabej P, Saihan Z, Rangesh N, Steele-Stallard HB, Ambrose J, Coffey A (2012). Comprehensive sequence analysis of nine Usher syndrome genes in the UK National Collaborative Usher Study. J Med Genet, 49(1), 27–36. 10.1136/jmedgenet-2011-100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Derosa AM, & White TW (2009). Connexin mutations causing skin disease and deafness increase hemichannel activity and cell death when expressed in Xenopus oocytes. J Invest Dermatol, 129(4), 870–878. 10.1038/jid.2008.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Niparko JK, & Ferrucci L (2011). Hearing loss prevalence in the United States. Arch Intern Med, 171(20), 1851–1852. 10.1001/archinternmed.2011.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Newton VE, & Read AP (1995). Waardenburg syndrome type II: phenotypic findings and diagnostic criteria. Am J Med Genet, 55(1), 95–100. 10.1002/ajmg.1320550123 [DOI] [PubMed] [Google Scholar]
- Mani RS, Ganapathy A, Jalvi R, Srikumari Srisailapathy CR, Malhotra V, Chadha S (2009). Functional consequences of novel connexin 26 mutations associated with hereditary hearing loss. Eur J Hum Genet, 17(4), 502–509. 10.1038/ejhg.2008.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, & Nance WE (1993). Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am J Med Genet, 46(5), 486–491. 10.1002/ajmg.1320460504 [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Mutai H, Kunishima S, Namba K, Morimoto N, Shinjo Y (2012). A prevalent founder mutation and genotype-phenotype correlations of OTOF in Japanese patients with auditory neuropathy. Clin Genet, 82(5), 425–432. 10.1111/j.1399-0004.2012.01897.x [DOI] [PubMed] [Google Scholar]
- Mese G, Londin E, Mui R, Brink PR, & White TW (2004). Altered gating properties of functional Cx26 mutants associated with recessive non-syndromic hearing loss. Hum Genet, 115(3), 191–199. 10.1007/s00439-004-1142-6 [DOI] [PubMed] [Google Scholar]
- Morton CC, & Nance WE (2006). Newborn hearing screening--a silent revolution. N Engl J Med, 354(20), 2151–2164. 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- Moteki H, Shearer AE, Izumi S, Kubota Y, Azaiez H, Booth KT (2015). De novo mutation in X-linked hearing loss-associated POU3F4 in a sporadic case of congenital hearing loss. Ann Otol Rhinol Laryngol, 124 Suppl 1, 169S-176S. 10.1177/0003489415575042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS (1999). An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet, 8(3), 409–412. [DOI] [PubMed] [Google Scholar]
- Nagy I, Horvath M, Trexler M, Repassy G, & Patthy L (2004). A novel COCH mutation, V104del, impairs folding of the LCCL domain of cochlin and causes progressive hearing loss. J Med Genet, 41(1), e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito I, Kawai S, Nomura S, Sado Y, & Osawa G (1996). Relationship between COL4A5 gene mutation and distribution of type IV collagen in male X-linked Alport syndrome. Japanese Alport Network. Kidney Int, 50(1), 304–311. [DOI] [PubMed] [Google Scholar]
- Naito T, Nishio SY, Iwasa Y, Yano T, Kumakawa K, Abe S (2013). Comprehensive genetic screening of KCNQ4 in a large autosomal dominant nonsyndromic hearing loss cohort: genotype-phenotype correlations and a founder mutation. PLoS One, 8(5), e63231 10.1371/journal.pone.0063231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus C, Lang-Roth R, Zimmermann U, Heller R, Eisenberger T, Weikert M (2017). Extension of the clinical and molecular phenotype of DIAPH1-associated autosomal dominant hearing loss (DFNA1). Clin Genet, 91(6), 892–901. 10.1111/cge.12915 [DOI] [PubMed] [Google Scholar]
- Palos F, Garcia-Rendueles ME, Araujo-Vilar D, Obregon MJ, Calvo RM, Cameselle-Teijeiro J (2008). Pendred syndrome in two Galician families: insights into clinical phenotypes through cellular, genetic, and molecular studies. J Clin Endocrinol Metab, 93(1), 267–277. 10.1210/jc.2007-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecci A, Klersy C, Gresele P, Lee KJ, De Rocco D, Bozzi V (2014). MYH9-related disease: a novel prognostic model to predict the clinical evolution of the disease based on genotype-phenotype correlations. Hum Mutat, 35(2), 236–247. 10.1002/humu.22476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecci A, Panza E, Pujol-Moix N, Klersy C, Di Bari F, Bozzi V (2008). Position of nonmuscle myosin heavy chain IIA (NMMHC-IIA) mutations predicts the natural history of MYH9-related disease. Hum Mutat, 29(3), 409–417. 10.1002/humu.20661 [DOI] [PubMed] [Google Scholar]
- Pepin MG, Murray ML, Bailey S, Leistritz-Kessler D, Schwarze U, & Byers PH (2016). The challenge of comprehensive and consistent sequence variant interpretation between clinical laboratories. Genet Med, 18(1), 20–24. 10.1038/gim.2015.31 [DOI] [PubMed] [Google Scholar]
- Pera A, Dossena S, Rodighiero S, Gandia M, Botta G, Meyer G (2008). Functional assessment of allelic variants in the SLC26A4 gene involved in Pendred syndrome and nonsyndromic EVA. Proc Natl Acad Sci U S A, 105(47), 18608–18613. 10.1073/pnas.0805831105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault V, (2015). “Waardenburg syndrome.” Orphanet Encyclopaedia, http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=663&Disease_Disease_Search_diseaseGroup=3440&Disease_Disease_Search_diseaseType=ORPHA&Disease(s)/group%20of%20diseases=Waardenburg-syndrome&title=Waardenburg-syndrome&search=Disease_Search_Simple
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, & Bondurand N (2010). Review and update of mutations causing Waardenburg syndrome. Hum Mutat, 31(4), 391–406. 10.1002/humu.21211 [DOI] [PubMed] [Google Scholar]
- Pryor SP, Madeo AC, Reynolds JC, Sarlis NJ, Arnos KS, Nance WE (2005). SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet, 42(2), 159–165. 10.1136/jmg.2004.024208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold FR, Heneghan JF, Stewart AK, Zelikovic I, Vandorpe DH, Shmukler BE (2011). Pendrin function and regulation in Xenopus oocytes. Cell Physiol Biochem, 28(3), 435–450. 10.1159/000335106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson NG, Cremers CW, Huygen PL, Ikezono T, Krastins B, Kremer H (2006). Cochlin immunostaining of inner ear pathologic deposits and proteomic analysis in DFNA9 deafness and vestibular dysfunction. Hum Mol Genet, 15(7), 1071–1085. 10.1093/hmg/ddl022 [DOI] [PubMed] [Google Scholar]
- Robertson NG, Lu L, Heller S, Merchant SN, Eavey RD, McKenna M (1998). Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat Genet, 20(3), 299–303. 10.1038/3118 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ballesteros M, Reynoso R, Olarte M, Villamar M, Morera C, Santarelli R (2008). A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat, 29(6), 823–831. 10.1002/humu.20708 [DOI] [PubMed] [Google Scholar]
- Savige J, Storey H, Il Cheong H, Gyung Kang H, Park E, Hilbert P (2016). X-Linked and Autosomal Recessive Alport Syndrome: Pathogenic Variant Features and Further Genotype-Phenotype Correlations. PLoS One, 11(9), e0161802 10.1371/journal.pone.0161802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoia A, & Pecci A (1993). MYH9-Related Disorders doi:NBK2689 [bookaccession] [Google Scholar]
- Scott DA, Wang R, Kreman TM, Andrews M, McDonald JM, Bishop JR (2000). Functional differences of the PDS gene product are associated with phenotypic variation in patients with Pendred syndrome and non-syndromic hearing loss (DFNB4). Hum Mol Genet, 9(11), 1709–1715. [DOI] [PubMed] [Google Scholar]
- Shearer AE, Hildebrand MS, & Smith RJH (1993). Hereditary Hearing Loss and Deafness Overview doi:NBK1434 [bookaccession] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL (2016). Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet, 135(4), 441–450. 10.1007/s00439-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Bale JF Jr., & White KR (2005). Sensorineural hearing loss in children. Lancet, 365(9462), 879–890. 10.1016/s0140-6736(05)71047-3 [DOI] [PubMed] [Google Scholar]
- Smith RJH (1993). Branchiootorenal Spectrum Disorders doi:NBK1380 [bookaccession] [PubMed] [Google Scholar]
- Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS (2017). Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet, 100(6), 895–906. 10.1016/j.ajhg.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street VA, Kallman JC, Robertson NG, Kuo SF, Morton CC, & Phillips JO (2005). A novel DFNA9 mutation in the vWFA2 domain of COCH alters a conserved cysteine residue and intrachain disulfide bond formation resulting in progressive hearing loss and site-specific vestibular and central oculomotor dysfunction. Am J Med Genet A, 139A(2), 86–95. 10.1002/ajmg.a.30980 [DOI] [PubMed] [Google Scholar]
- Stritt S, Nurden P, Turro E, Greene D, Jansen SB, Westbury SK (2016). A gain-of-function variant in DIAPH1 causes dominant macrothrombocytopenia and hearing loss. Blood, 127(23), 2903–2914. 10.1182/blood-2015-10-675629 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM (2018). Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med. 10.1038/gim.2017.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L, Samson RA, & Green GE (1993). Jervell and Lange-Nielsen Syndrome doi:NBK1405 [bookaccession] [PubMed] [Google Scholar]
- Tryggvason K, Zhou J, Hostikka SL, & Shows TB (1993). Molecular genetics of Alport syndrome. Kidney Int, 43(1), 38–44. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Huizing EH, Verstreken M, van Zuijlen D, Wauters JG, Bossuyt PJ (1998). Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet, 20(2), 194–197. 10.1038/2503 [DOI] [PubMed] [Google Scholar]
- Varga R, Avenarius MR, Kelley PM, Keats BJ, Berlin CI, Hood LJ (2006). OTOF mutations revealed by genetic analysis of hearing loss families including a potential temperature sensitive auditory neuropathy allele. J Med Genet, 43(7), 576–581. 10.1136/jmg.2005.038612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I (1998). Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet, 19(1), 60–62. 10.1038/ng0598-60 [DOI] [PubMed] [Google Scholar]
- Whiffin N, Minikel E, Walsh R, O’Donnell-Luria AH, Karczewski K, Ing AY (2017). Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med, 19(10), 1151–1158. 10.1038/gim.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Py BF, Zhu H, Bao J, & Yuan J (2010). Role of protein misfolding in DFNA9 hearing loss. J Biol Chem, 285(20), 14909–14919. 10.1074/jbc.M110.106724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Guo W, Tang J, Zhang G, Wang G, Han M (2012). Molecular epidemiology and functional assessment of novel allelic variants of SLC26A4 in non-syndromic hearing loss patients with enlarged vestibular aqueduct in China. PLoS One, 7(11), e49984 10.1371/journal.pone.0049984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Huang D, Yu F, Zhu X, Kang D, Yuan H (2009). A de novo GJB2 (connexin 26) mutation, R75W, in a Chinese pedigree with hearing loss and palmoplantar keratoderma. Am J Med Genet A, 149A(4), 689–692. 10.1002/ajmg.a.32461 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tang W, Ahmad S, Sipp JA, Chen P, & Lin X (2005). Gap junction-mediated intercellular biochemical coupling in cochlear supporting cells is required for normal cochlear functions. Proc Natl Acad Sci U S A, 102(42), 15201–15206. 10.1073/pnas.0501859102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


