Abstract
On the basis of an initial molecular modeling study suggesting the favorable binding of the “privileged” fragment 8-hydroxyquinoline with IN at the IN-LEDGF/p75 interface, we developed a set of modified 8-hydroxyquinoline fragments demonstrating micromolar IC50 values for inhibition of the IN-LEDGF/p75 interaction, but significant cytotoxicity was associated with these initial compounds. Diverse modifications at the C5 and C7 carbons of the 8-hydroxyquinoline core improved potency, but reduction of diversity to only modifications at the C5 position ultimately yielded potent inhibitors with low cytotoxicity. Two of these particular compounds, 5-((p-tolylamino)methyl)quinolin-8-ol and 5-(((3,4-dimethylphenyl)amino)methyl)quinolin-8-ol, inhibited viral replication in MT-4 cells with low micromolar EC50. This is the first study providing evidence for 8-hydroxyquinolines as novel inhibitors of the IN-LEDGF/p75 interaction. Our lead compounds are drug-like, have low molecular weights, and are amenable to various substitutions suitable for enhancing their potency and selectivity.
INTRODUCTION
Currently, over 30 FDA approved drugs, targeting various stages of the viral life cycle, are available to treat HIV-1 infection,1 but drug resistance2 and reservoirs of latent viral infection3, 4 have prevented total eradication of the virus. HIV-1 integrase (IN) is an essential viral enzyme responsible for integrating the provirus into the host chromosomes.5 After several unsuccessful attempts in developing clinically safe and efficacious IN inhibitors, only Raltegravir has shown remarkable safety and selectivity profiles and gained accelerated approval from the FDA in 2007.6 However, viral resistance has necessitated the continued development of novel IN inhibitors.7 The discovery of the interaction between IN and the host protein LEDGF/p758 has opened the possibility for the design of allosteric inhibitors selectively targeting this interaction. LEDGF/p75 is an important cofactor of viral replication, plays a crucial role in tethering IN to the host chromosome,9 and stimulates concerted integration of the HIV-1 provirus.10 Several small-molecule inhibitors of this virus-host interaction have recently been developed that not only allosterically inhibit HIV-1 integration, but also exhibit antiviral activity.11–17
Fragment-based drug design (FBDD) is becoming a popular complement to traditional high-throughput screening efforts.18, 19 In FBDD, successful scaffolds from FDA-approved or otherwise clinically advanced drugs may be used as starting points for inhibitor development. This technique can significantly reduce the amount of compounds screened relative to traditional high-throughput screening and can also increase the success rate for lead development. FBDD has been used to study ligand-binding mechanisms for HIV-1 IN.20–22 Quinoline has functioned as a “privileged”23 scaffold of several FDA-approved drugs, including mefloquine, quinine, quinidine, montelukast, chloroquine, amodiaquine, primaquine, and dibucaine,24–31 and along with quinolone derivatives was also explored in the development of IN inhibitors over the last 10 years.32–35 Several quinolines, quinolones, and quinines possess antifungal, antimicrobial, and anti-malarial properties, respectively,36–38 demonstrating a potential utility for these scaffolds in drug discovery. In this study we present a novel class of 8-hydroxyquinoline inhibitors of the IN-LEDGF/p75 interaction. 8-hydroxyquinoline analogues have been previously explored for anti-IN activity39–42 (examples in Figure 1), but to date there exist no reports of 8-hydroxyquinoline inhibition of IN-LEDGF/p75.
Figure 1.
8-hydroxyquinolines as catalytic inhibitors of HIV-1 IN.
RESULTS
8-Hydroxyquinoline-Based Fragments Inhibit the IN-LEDGF/p75 Interaction
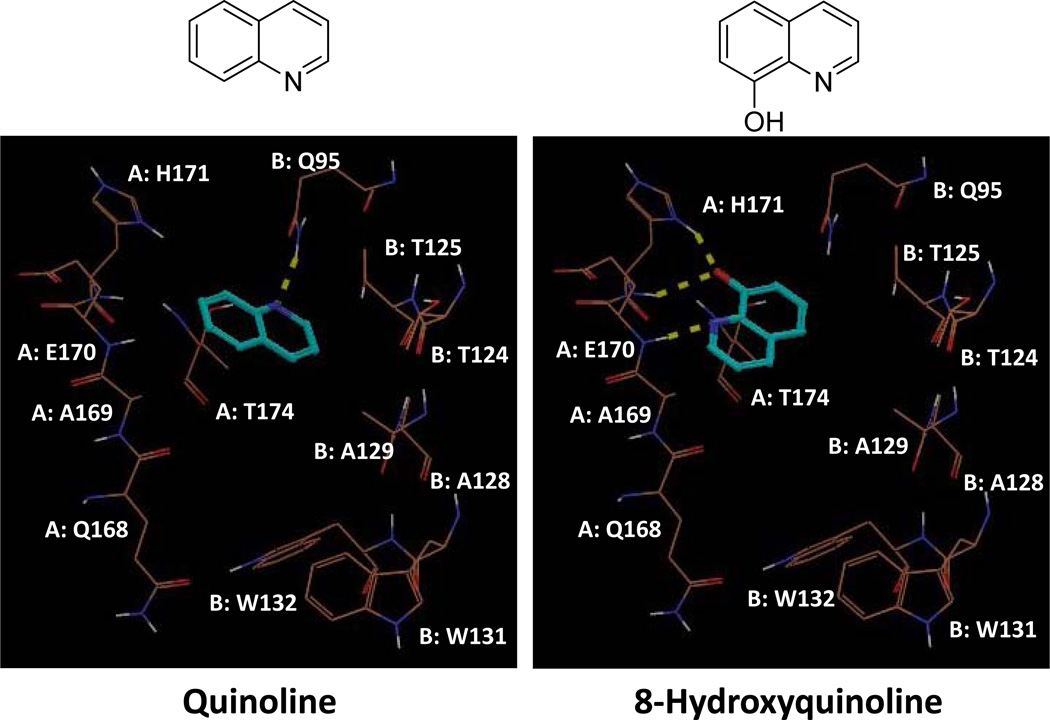
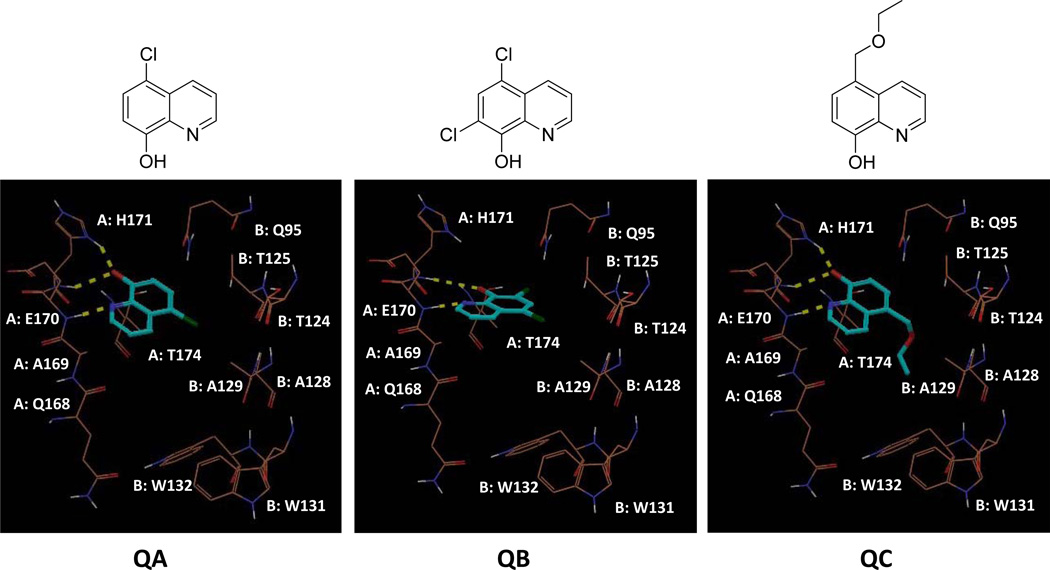
Using Grid-based Ligand Docking from Energetics (Glide), we performed molecular docking of quinoline at the IN-LEDGF/p75 interface (PDB ID: 2B4J). While quinoline formed hydrogen bond (H-bond) interactions with only the side chain NH of Gln 95 from IN chain B, 8-hydroxyquinoline formed several H-bonds, specifically with His 171 and Glu 170 (Figure 2). We subsequently began to assay 8-hydroxyquinoline fragments for their potency in inhibiting the IN-LEDGF/p75 interaction. We used an AlphaScreen protein-protein interaction assay that has been previously validated for efficacy in identifying specific IN-LEDGF/p75 inhibitors.12, 16, 43 The AlphaScreen luminescence assay is distinct from the IN enzymatic and cell-based antiviral assays. As shown in Table 1, the 8- hydroxyquinoline inhibitors of the IN-LEDGF/p75 interaction lack inhibitory activity in the IN enzymatic assay. Compounds with minor structural additions to the 8-hydroxyquinoline core (Figure 3) exhibited favorable potency. 5-chloroquinolin-8-ol, 5,7-dichloroquinolin-8-ol, and 5-(ethoxymethyl)quinolin-8-ol (QA, QB, and QC) inhibited IN-LEDGF/p75 with IC50 values of 3.6 ± 0.5, 4.3 ± 0.2, and 2.4 ± 0.4 µM, respectively (Table 1). Molecular docking with Glide (Figure 3) suggested a similar IN-binding pattern for each of the three compounds. The quinoline nitrogen of each compound formed an H-bond with the backbone NH of Glu 170. Furthermore, the 8-olate ions of QA and QC formed two H-bonds with the backbone and side chain NHs of His 171, while that of QB formed only one H-bond with backbone NH of His 171. However, the ethoxymethyl group of QC was additionally predicted to position itself within the hydrophobic pocket formed by Ala 128, Ala 129, Glu 170, Thr 124, Thr 125, Trp 131 and Trp 132. The additional hydrophobic interactions of QC may explain its increased potency compared to the other two fragments, QA and QB. Initial MTT analysis in LNCaP cells revealed significant cytotoxicity associated with all three of these fragments (Table 1), which led us to extend our analysis to more divergent and bulky additions to the core fragment.
Figure 2.
Docking poses of simple quinoline versus 8-hydroxyquinoline into the LEDGF/p75 binding site of HIV-1 IN (PDB ID: 2B4J). Tan colored lines represent important residues from the two monomers (A and B) of IN, while cyan colored sticks are the ligand. Yellow dotted lines are H-bonds between protein and ligands.
Table 1.
Biological activities of 8-hydroxyquinoline derivatives.
| IN- LEDGF/p75 |
MTT Lncap |
IN 3'-P |
IN ST |
Quench | MTT/MT-4 | |||
|---|---|---|---|---|---|---|---|---|
| (IC50, µM)a | CC50 (µM)b | IC50 (µM)c |
IC50 (µM)d |
% Inhib.e | EC50f | CC50g | SIh | |
| QA | 3.6 ± 0.5 | 5 | >20 | >20 | <50 | ND | ND | |
| QB | 4.3 ± 0.2 | 2.5 | >20 | >20 | <50 | ND | ND | |
| QC | 2.4 ± 0.4 | 2 | >20 | >20 | <50 | ND | ND | |
| Q1-1 | 1.4 ± 0.2 | 0.68 ± 0.03 | >20 | >20 | <50 | ND | ND | |
| Q1–2 | 2.0 ± 1 | 0.58 ± 0.02 | >20 | >20 | <50 | ND | ND | |
| Q1–3 | 2.0 ± 1 | 0.58 ± 0.03 | >20 | >20 | <50 | ND | ND | |
| Q1–4 | 2.6 ± 0.5 | >10 | >20 | >20 | <50 | >10 | 10 | |
| Q1–5 | 3.0 ± 1 | 0.74 ± 0.06 | >20 | >20 | <50 | ND | ND | |
| Q1–6 | 3.5 ± 1.6 | 5 ± 0.6 | >20 | >20 | <50 | ND | ND | |
| Q1–7 | 3.9 ± 1.8 | >10 | >20 | >20 | <50 | >42 | 42 | |
| Q1–8 | 4.0 ± 0 | 0.72 ± 0.1 | >20 | >20 | <50 | ND | ND | |
| Q1–9 | 4.0 ± 0 | 3 ± 0.08 | >20 | >20 | <50 | ND | ND | |
| Q1–10 | 4.0 ± 1 | 0.62 ± 0.03 | >20 | >20 | <50 | ND | ND | |
| Q1–11 | 4.0 ± 3 | 0.62 ± 0.03 | >20 | >20 | <50 | ND | ND | |
| Q1–12 | 4.7 ± 3.4 | 2.4 ± 0.1 | >20 | >20 | <50 | ND | ND | |
| Q2-1 | 1.4 ± 0.1 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q2-2 | 2.0 ± 0.4 | 0.1 ± 0.02 | >20 | >20 | <50 | ND | ND | |
| Q2–3 | 2.1 ± 0.3 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q2–4 | 2.8 ± 0.9 | >10 | >20 | >20 | <50 | >2 | 2 | |
| Q2–5 | 2.9 ± 0.5 | >10 | >20 | >20 | <50 | >10 | 10 | |
| Q2–6 | 3.0 ± 0.3 | >10 | >20 | >20 | <50 | >13 | 13 | |
| Q2–7 | 3.2 ± 1.3 | 5 ± 0.2 | >20 | >20 | <50 | ND | ND | |
| Q2–8 | 3.3 ± 1.3 | >10 | >20 | >20 | <50 | 15.41 | 36.5±16.5 | 2 |
| Q2–9 | 3.5 ± 1.2 | >10 | >20 | >20 | <50 | >11 | 11 | |
| Q2–10 | 3.7 ± 2.0 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q2–11 | 4.0 ± 1.3 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q2–12 | 4.0 ± 1.7 | >10 | >20 | >20 | <50 | >11 | 11 | |
| Q2–13 | 4.6 ± 1.2 | >10 | >20 | >20 | <50 | 17.85 | 33.5±7.5 | 2 |
| Q2–14 | 4.8 ± 0.3 | >10 | >20 | >20 | <50 | >34 | 34 | |
| Q3-1 | 0.4 ± 0.1 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q3-2 | 0.8 ± 0.6 | >10 | >20 | >20 | <50 | >35 | 35 | |
| Q3-3 | 1.2 ± 0.5 | >10 | >20 | >20 | <50 | >42 | 42 | |
| Q3–4 | 1.5 ± 0.4 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q3–5 | 1.9 ± 0.4 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q3–6 | 2.0 ± 0.2 | >10 | >20 | >20 | <50 | >9 | 9 | |
| Q3–7 | 2.1 ± 1.0 | 5 ± 0.8 | >20 | >20 | <50 | ND | ND | |
| Q3–8 | 2.2 ± 0.4 | >10 | >20 | >20 | <50 | ND | ND | |
| Q3–9 | 2.2 ± 0.4 | >10 | >20 | >20 | <50 | >11 | 11 | |
| Q4-1 | 1.7 ± 0.4 | >10 | >20 | >20 | <50 | >2 | 2 | |
| Q5-1 | 3.0 ± 1 | 0.8 ± 0.06 | >20 | >20 | <50 | >2 | 2 | |
| Q5-2 | 3.0 ± 2 | >10 | >20 | >20 | <50 | ND | ND | |
Concentration required to inhibit the in vitro protein-protein interaction by 50%.
Cytotoxic concentration reducing Lncap cell viability by 50%.
Concentration required to inhibit IN 3’-processing catalysis by 50%.
Concentration required to inhibit IN strand transfer catalysis by 50%.
Percent inhibition exerted by compounds in quench counter-screen when tested at a concentration of 10 µM.
Effective concentration required to reduce HIV-1 induced cytopathic effect by 50% in MT-4 cells.
Cytotoxic concentration reducing MT-4 cell viability by 50%.
Selectivity index: ratio CC50/EC50.
Figure 3.
Docking poses of our initial 8-hydroxyquinoline fragments, QA, QB, and QC into the LEDGF/p75 binding site of HIV-1 IN (PDB ID: 2B4J). Tan colored lines represent important residues from the two monomers (A and B) of IN, while cyan colored sticks are the ligand. Yellow dotted lines are H-bonds between protein and ligands.
Structure-Based Pharmacophore Study
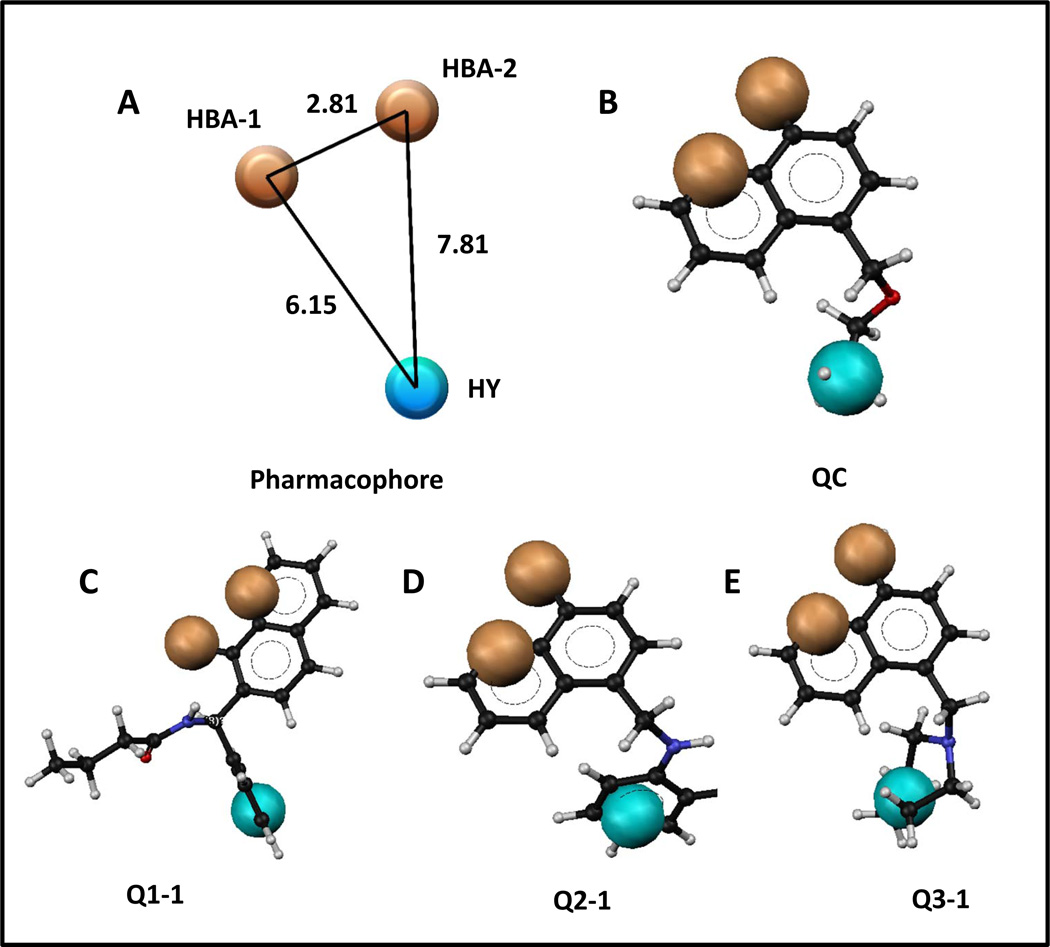
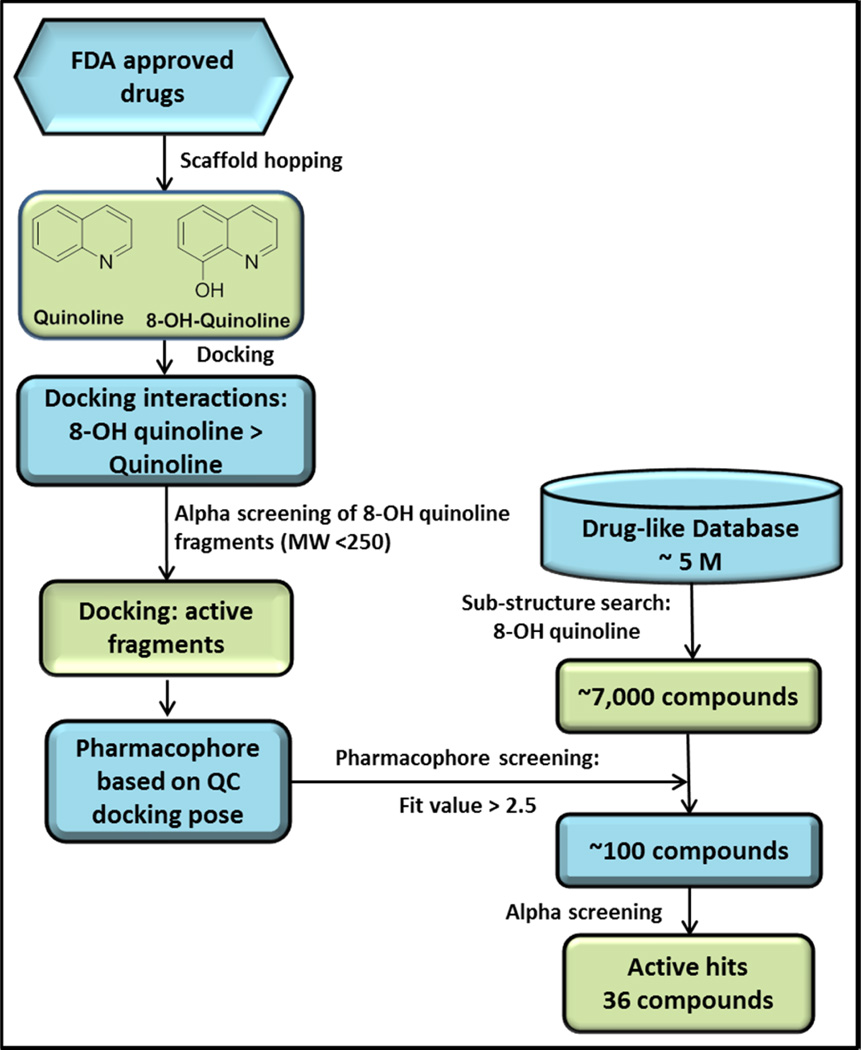
A structure-based pharmacophore was generated on the basis of the docked poses of most potent fragment, QC. The pharmacophore consisted of two H-bond acceptors (HBA) and a hydrophobic feature (HY, Figure 4A). The two HBAs represent three distinct H-bond interactions. Specifically, HBA-1 represents two H-bonds between the QC 8-olate group and the backbone and side chain NHs of His 171, and HBA-2 represents an H-bond between the QC quinoline nitrogen and the backbone NH of Glu170. The HY represents the hydrophobic interactions of the QC ethoxymethyl group within hydrophobic pocket formed by Ala128, Ala129, Glu170, Thr124. Thr125, Trp131 and Trp132. Figure 4B–E also details the phamacophore mapped onto initial fragment compound QC and top compounds Q1-1, Q2-1, and Q3-1, which are discussed below. This pharmacophore was used to screen approximately 7,000 compounds resulting from a substructure search of 5 million drug-like commercially available compounds using 8-hydroxyquinoline as a query. The pharmacophore screening resulted in approximately 100 8-hydroxyquinoline compounds with greater than 2.5 fit values. We systematically evaluated these compounds through clustering into different subsets of compounds, and the complete flow chart of our approach is depicted in Figure 5. Although the pharmacophore identified the neutral form of compounds from the database, we have used the ionized form for the purpose of pharmacophore mapping, as we consider these compounds to be ionized in the context of receptor binding.
Figure 4.
A) Pharmacophore generated based on docking interactions between IN and QC; B) pharmacophore mapped onto QC; C) pharmacophore mapped onto Q1-1; D) pharmacophore mapped onto Q2-1 ; E) pharmacophore mapped onto Q3-1. Tan colored balls represent hydrogen bond acceptors (HBA-1 and HBA-2), while the cyan colored ball represents a hydrophobic (HY) feature. Black, white, blue, and red represent elemental colors of compounds as carbon, hydrogen, nitrogen, and oxygen, respectively. Distances between features are in angstroms (Å).
Figure 5.
Flow-chart of rational drug design of IN inhibitors at the IN-LEDGF/p75 interaction interface. After initial molecular docking of 8-hydroxyquinoline, we found that fragments based on this scaffold exhibited micromolar inhibition of the IN-LEDGF/p75 interaction in vitro. Following an 8-hydroxyquinoline substructure search of a 5-million compound industrial database, virtual screening using a pharmacophore based on the top fragment yielded around 100 compounds. These compounds were subjected to AlphaScreening, ultimately yielding 36 active hits.
Expansion of Chemical Diversity of 8-Hydroxyquinolines
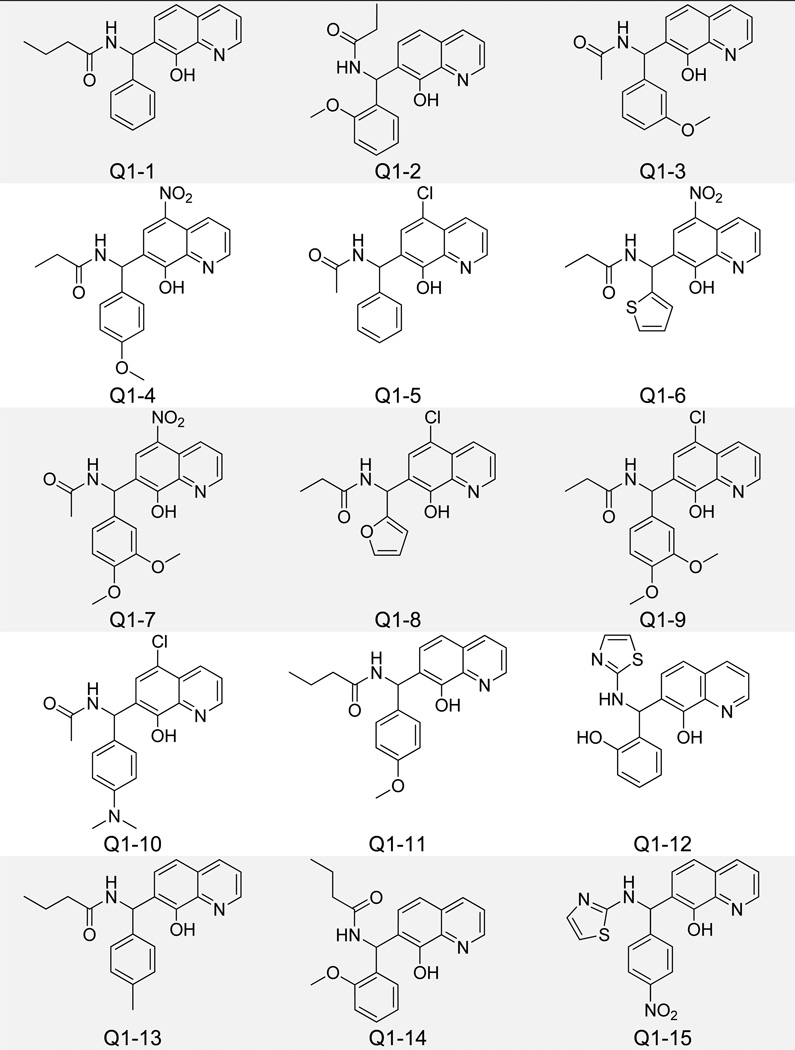
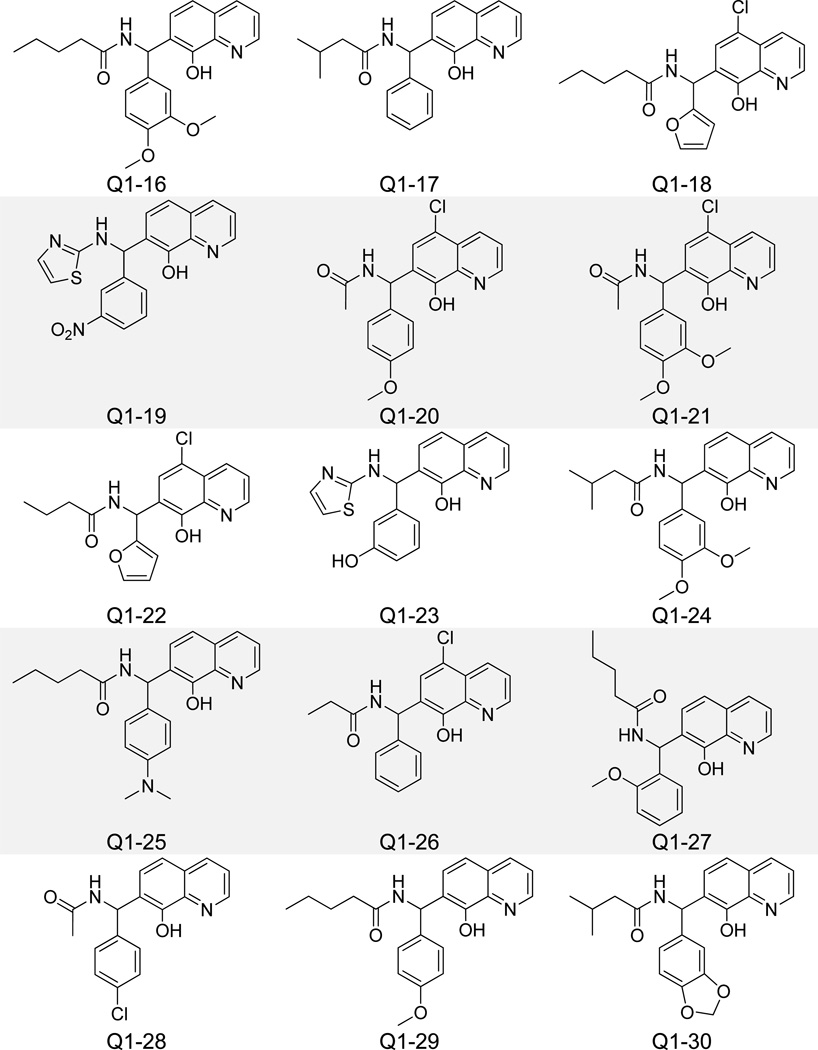
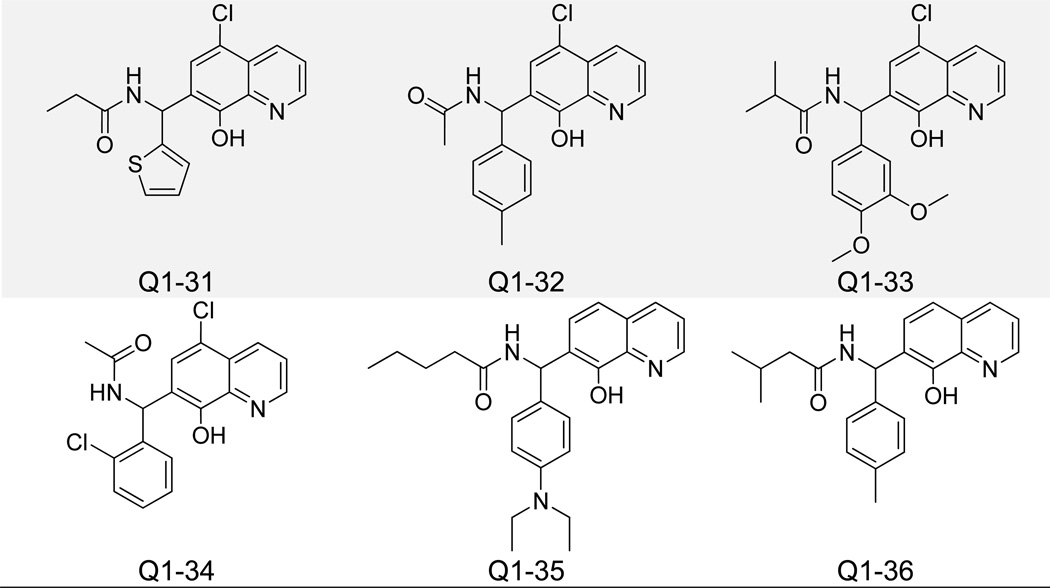
Pharmacophore-identified compounds containing substitutions at both the C5 and C7 carbons of the quinoline core were next investigated (Figure 6). Only compounds with IC50 values less than 5 µM for IN-LEDGF/p75 interaction inhibition are listed in Table 1, while all compounds with IC50 values above 5 µM are listed in Supplementary Table 1. Propan-2-one, butan-2-one, and pentan-2-one were the favored moieties at the C5 carbon position, while longer aliphatic hexan-2-ones and divergent 2-methylthiazole groups decreased activity. The C7 carbon position generally contained a modified phenyl ring for all compounds tested, and an additional lack of bulk was associated with increased potency in this group. Structures of inactive (IC50 > 20 µM) analogues of this and other clusters of 8-hydroxyquinolines can be found in Supplementary Figure 1. Molecular docking revealed an alternative spatial orientation for compounds Q1-1 and Q1–2 relative to the fragments QA-C (Supplementary Figure 2). More specifically, the quinoline nitrogen from Q1-1 and Q1–2 formed an H-bond with the side chain NH of His 171 on IN, while the 8-olate group formed an H-bond with the backbone NH of His 171. The C7 carbon-attached phenyl moieties of these two compounds packed into the hydrophobic pocket formed by Ala 128, Ala 129, Gln 168, Trp 131, and Trp 132. Hydrogen bond distances between all compounds detailed in this study and IN residues are listed in Supplementary Table 2. Though the top compounds Q1-1 and Q1–2 showed reasonable potency for inhibiting the IN-LEDGF/p75 interaction in vitro (IC50 = 1.4 and 2.0 µM, respectively), both exhibited notable cytotoxicity in the MTT LNCaP assay (Table 1).
Figure 6.
Structures of 8-Hydroxyquinoline Analogues Containing C5 and C7 Carbon Substitutions.
Non-Cytotoxic 8-Hydroxyquinoline Analogues
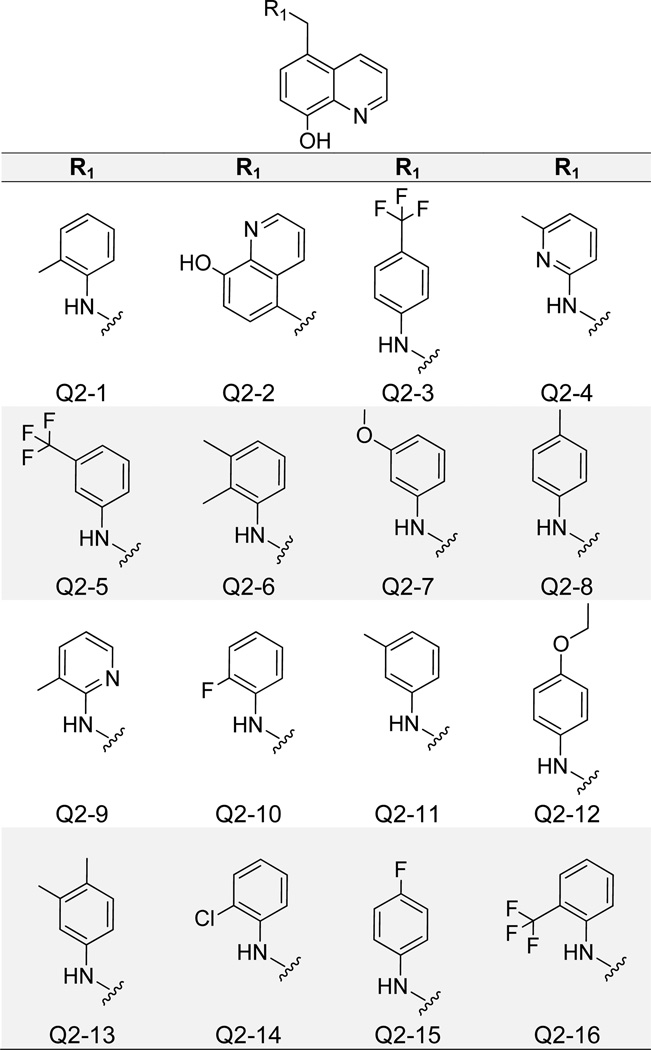
Consistent cytotoxicity of 8-hydroxyquinoline derivatives was eventually overcome with compounds containing only modifications at the core scaffold C5 carbon, rather than both C5 and C7 (Figure 7). Most compounds in this group (Q2-1 – Q2–14) inhibited the IN-LEDGF/p75 interaction with IC50 values below 5 µM, and many were non-cytotoxic (Table 1). Top compound Q2-1 exhibited an IN-LEDGF/p75 IC50 value of 1.4 µM and displayed 0% inhibition of cell growth in the MTT LNCaP assay. However, quinolin-8-ol and 3-methoxyaniline modifications at the R1 position (Q2-2 and Q2–7) did confer notable cytotoxicity. Docking poses of top two compounds Q2-1 and Q2-2 from this cluster were similar to QC (Supplementary Figure 2), as the quinoline nitrogen formed an H-bond with the backbone NH of Glu 170, and the 8-olate group formed two H-bonds with side chain and backbone NHs of His 171. The toluene moiety of Q2-1 and the second quinoline moiety of Q2-2 packed into the hydrophobic pocket formed by Ala 128, Ala 129, Gln 168, Trp 131, and Trp 132, similar to the R2 phenyl moieties of compounds from the Q1 cluster.
Figure 7.
Structures of 8-Hydroxyquinoline Analogues Containing Only C5 Carbon Substitutions.
Enhanced Potency with Piperidine and Piperizine Substitutions
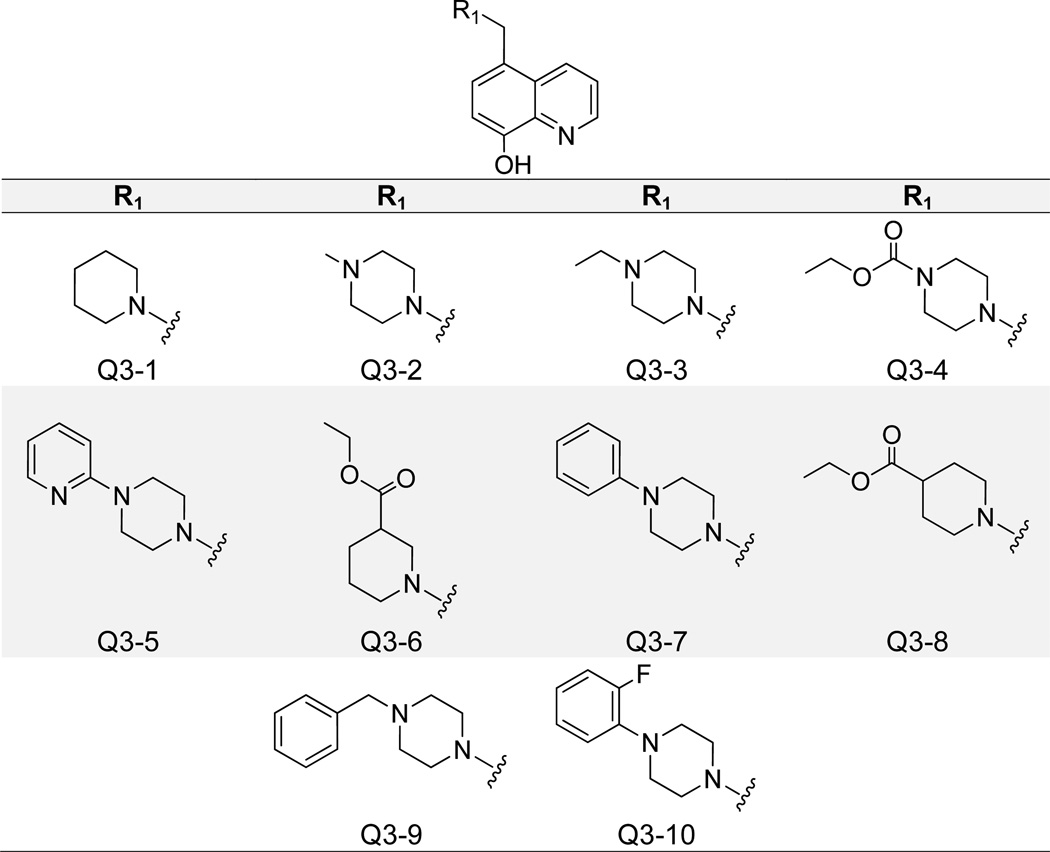
Replacement of the C5 phenyl substitutions with piperidine- and piperizine-based groups generated a class of compounds with increased potency and reduced cytotoxicity (Figure 8). The best compound in this series, Q3-1, contained a piperidine moiety and inhibited the IN-LEDGF/p75 interaction with an IC50 value of 400 nM. Extension to a 1-methylpiperizine (Q3-2) maintained decent activity at 800 nM, but further added bulk was generally associated with a significant decrease in potency (Table 1). Q3-1 and Q3-2 shared similar binding modes, as molecular docking revealed that the quinoline nitrogens formed an H-bond with the backbone nitrogen of Glu 170, and the 8-olate group formed two H-bonds with the side chain and backbone NHs of His 171 (Supplementary Figure 2). Q3-1 piperidine and Q3-2 methylpiperazine moieties both packed into the hydrophobic pocket formed by Ala 128, Ala 129, Gln 168, Trp 131, and Trp 132. Many of the assayed compounds in this class maintained low cytotoxicity (Table 1). Only the 1-phenylpiperazine (Q3–7) modification yielded a compound with notable cell-growth inhibition at 10 µM in the MTT LNCaP assay. Close analogue 1-(2-fluorophenyl)piperizine (Q3–10) showed minimal cytotoxicity but also was less potent against IN-LEDGF/p75 with an IC50 of 12.5 µM.
Figure 8.
Structures of 8-Hydroxyquinoline Analogues Containing Piperidine- and Piperizine-Based C5 Substitutions.
Replacement of the 8-Hydroxy Component Maintains Inhibitory Potency
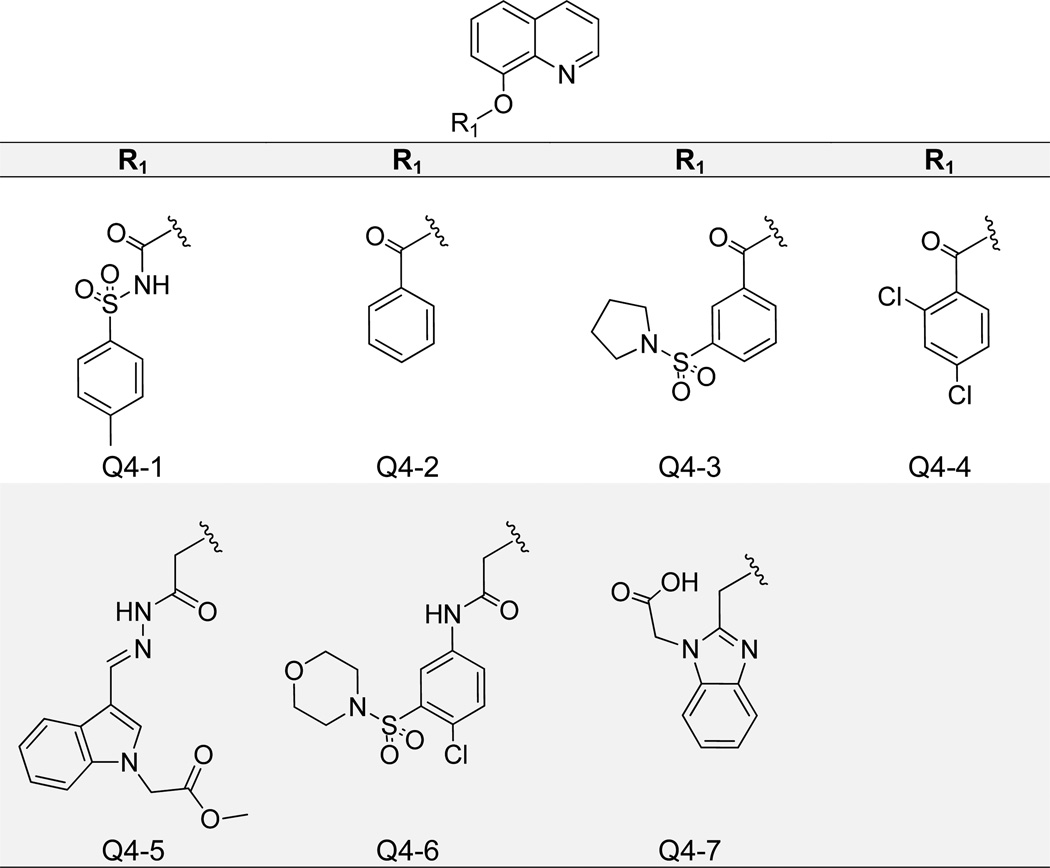
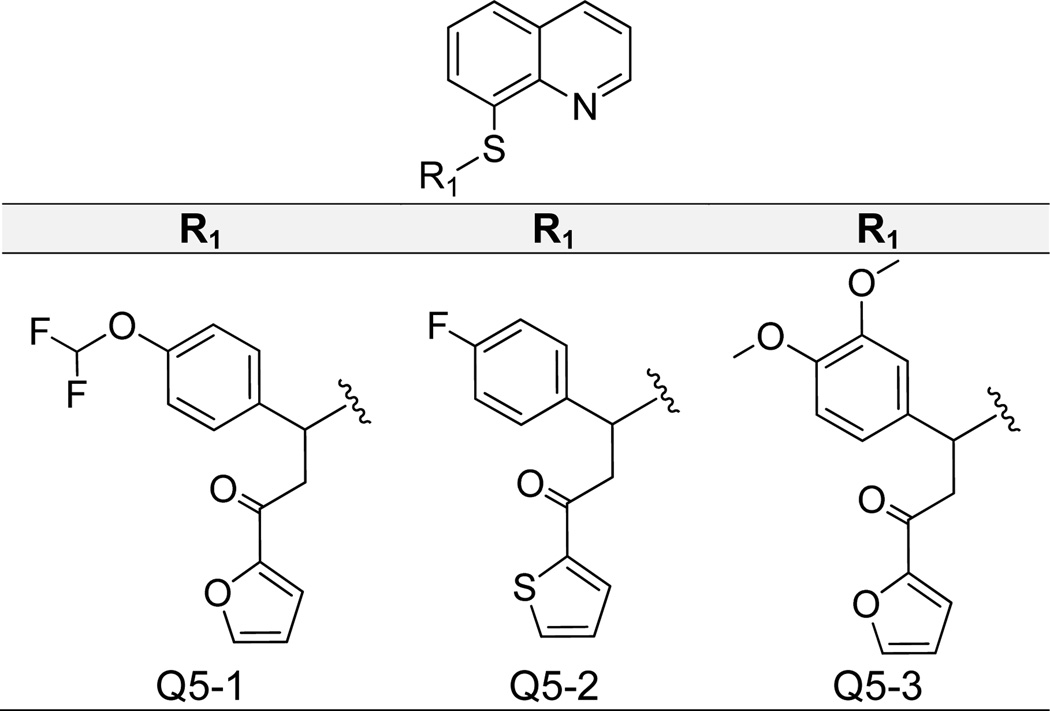
Finally, we found that the 8-hydroxy substituent itself is not essential for IN-LEDGF/p75 inhibition, as bulky additions (Figure 9) and replacement of the oxygen with sulfur (Figure 10) still yielded active compounds. In Figure 9 we show that modification of the 8-hydroxy with an added N-tosylformamide generated a compound (Q4-1) with IN-LEDGF/p75 IC50 of 1.7 µM and low cytotoxicity, with 13% cell growth inhibition at the tested concentration of 10 µM (Table 1). Ligand binding mirrored prior actives, as the quinoline nitrogen formed an H-bond with Glu 170, and the phenyl moiety packed into the applicable hydrophobic pocket (Supplementary Figure 2). Additionally, the carbonyl substituent formed two H-bonds with His 171 backbone and side chain NHs. However, more diverse additions at this point on the molecule resulted in reduced potency, as the second best compound with an added benzaldehyde (Q4-2) showed an IC50 value of over 5 µM. Figure 10 presents data for three quinolin-8-thiol derivatives. Interestingly, both 3-(4-(difluoromethoxy)phenyl)-1-(furan-2-yl)propan-1-one (Q5-1) and 3-(4-fluorophenyl)-1-(thiophen-2-yl)propan-1-one (Q5-2) maintained similar IN-LEDGF/p75 activity with IC50 values of 3 µM, but Q5-1 exhibited markedly increased cytotoxicity with 73% inhibition of cell growth, compared to 21% by Q5-2 (Figure 10). Molecular docking revealed slightly different binding modes for these compounds, presumably due to varied hydrophobic substituent bulkiness and length, but both formed hydrogen bond interactions with Glu 170 and/or His 171 (Supplementary Figure 2). The additional Q5-1 furyl oxygen may potentially contribute to off-target effects associated with cytotoxicity. An analogous 3-(3,4-dimethoxyphenyl)-1-(furan-2-yl)propan-1-one (Q5-3) substitution generated a compound that was slightly less active, with an IN-LEDGF/p75 IC50 of 5 µM.
Figure 9.
Structures of 8-Hydroxyquinolines Containing 8-Hydroxy Substitutions.
Figure 10.
Structures of Quinoline-8-Thiol Analogues.
Counter-Screening and Antiviral Activity
All of the compounds listed herein that showed IC50 values below 5 µM for IN-LEDGF/p75 inhibition (Table 1) were subjected to counter-screening for validation of specificity. This counter-screen is an AlphaScreen-based assay utilizing a Flag-6x-histidine construct that is capable of binding to both donor and acceptor AlphaScreen beads (for detailed description of AlphaScreen technology, see 44). Since the single construct binds to both beads, the assay is able to identify compounds that are 1) nonspecific binders, 2) competing with Flag or histidine for bead binding, or 3) quenching transfer of luminescent signal. All of the compounds assayed in this study proved inactive in this counter-screen assay (Table 1). We next screened all active compounds with an IC50 for inhibition of the IN-LEDGF/p75 interaction below 5 µM in an IN enzymatic assay to probe effect on IN catalysis. Again none of the compounds tested were active in this assay, and therefore neither inhibited the 3’processing, nor the strand transfer activity of IN (Table 1).
Finally, non-cytotoxic compounds inhibiting IN-LEDGF/p75 with IC50 < 5 µM were subjected to antiviral activity testing in a classical MT-4 cellular phenotypic assay where cytotoxicity in these cell lines was evaluated in parallel. While some compounds proved more cytotoxic (i.e Q2–4, Q4-1, Q5-1) in MT-4 cells than in LNCaP, most compounds showed CC50s around or above 10 µM. All compounds were screened for reduction of HIV-induced cytopathic effect in MT-4 cells at their CC50 concentration, and two (Q2–8 and Q2–13) displayed moderate but reproducible antiviral activity with EC50 values, at 15 and 18 µM, respectively.
DISCUSSION
Recent advances in anti-HIV drug discovery have validated the IN-LEDGF/p75 interaction as a novel drug target. We therefore aimed at developing molecules based on a novel chemical scaffold as potent inhibitors of this virus-host interaction in vitro and in cell culture. Molecular docking data demonstrated that 8-hydroxyquinoline formed multiple H-bonds with IN residues at the LEDGF/p75 binding interface, which led us to perform structure-activity relationship analysis to better understand their mechanism of binding. Initial fragments containing simple additions to the 8-hydroxyquinoline core exhibited low micromolar activity against IN-LEDGF/p75, but these compounds were also cytotoxic in the MTT assay. Modification at both the C5 and C7 carbons yielded similar in vitro potency to the initial fragments, but significant cytotoxicity was still apparent. Simplification of the structure to include only modifications at the C5 carbon position yielded compounds with identical in vitro activity, but far reduced cellular toxicity. Variation of these C5 additions to include piperidine-and piperizine-based moieties yielded the most potent compounds, Q3-1 and Q3-2, which showed negligible inhibition of cell growth in MTT assay and nanomolar IC50 values for the inhibition of the IN-LEDGF/p75 interaction. This represents a 7-fold increase in potency from initial fragments to our best compounds, accompanied with more than 90% reduced inhibition of cell growth. 8-hydroxyquinolines have a broad spectrum of cellular targets, and off-target effects imparted by our initial classes of compounds could have contributed to their cytotoxicity. Reduction in nonspecific binding by certain modifications (i.e. those inherent in our Q2 and Q3 classes of compounds) could explain the decrease in cell growth inhibition observed through the course of our 8-hydroxyquinoline optimization.
The 8-hydroxy component of our core scaffold initially seemed important for binding to IN, as normal quinoline did not dock well. However, with our Q4 and Q5 compound classes, we found that the 8-hydroxy component could be modified with bulky additions or substituted to form quinolin-8-thiol derivatives. The latter exhibited slightly increased cytotoxicity, presumably due to the added sulfur. Nonetheless, this data clearly validates the potential for further structural optimization toward increased potency of IN-LEDGF/p75 inhibition. Analogous substitutions of p-toluidine and 3,4-dimethylaniline (Q2–8 and Q2–13) resulted in measurable antiviral activity in vitro. However, these were not the most potent compounds in their respective cluster in terms of IN-LEDGF/p75 in vitro inhibition. In fact Q2-1 showed an AlphaScreen potency 3-fold higher than Q2–8, but cytotoxicity in MT-4 cells limited antiviral analysis of Q2-1 to a maximum concentration of 9 µM. Similarly, the 2,3-dimethylaniline addition (Q2–6) generated a structure almost identical to Q2–8 and Q2–13, but it was three-fold more toxic in MT-4 cells. Therefore, the maximum concentration of Q2–6 amenable to antiviral testing was 13 µM, and no antiviral activity could be detected at this concentration. Further chemical optimization based upon the structure-activity relationship laid out in this initial report should yield 8-hydroxyquinoline derivatives with reduced cytotoxicity and increased potency for IN-LEDGF/p75 inhibition in cells.
All of the compounds reported in this study lacked allosteric inhibition of IN enzymatic activity, unlike recently reported LEDGINs, which were described to inhibit IN enzymatic activity likely through multimeric stabilization and disruption of the enzyme’s multimerization dynamics.15, 16 Some of these LEDGINs exhibit a quinoine-based scaffold, but modifications are based mainly on the C3 and C4 carbons, which may explain a divergent mode of interaction with IN residues. X-ray crystallization revealed that an initial lead LEDGIN 2-(quinolin-3-yl)acetic acid derivative formed H-bonds with IN residues Glu 170 and His 171 and packed directly against IN residue A128.12 Our antiviral compounds Q2–8 and Q2–13 differed slightly in plausible binding, as their extended toluene moieties at the C5 position were responsible for A128 packing, which differed from the LEDGIN chlorine modification at the C6 position. Such minor alterations in binding mode could explain more large-scale effects, such as the absence of IN catalysis inhibition evoked by our top compounds.
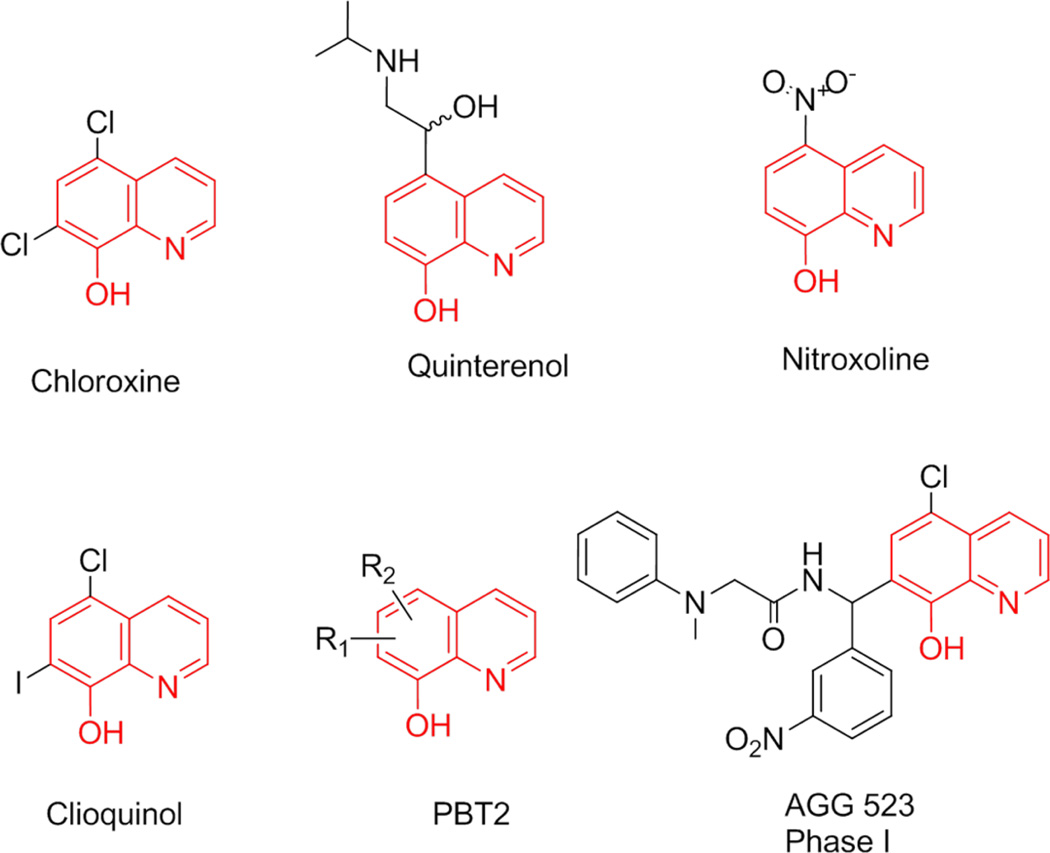
Multiple drugs used in the treatment of different diseases contain 8-hydroxyquinoline (Figure 11). For example, chloroxine, which is identical to our initial fragment QB, is used topically for dandruff and seborrheic dermatitis.24 Nitroxoline is an antibiotic used for the treatment of urinary tract infections25 and has recently been reported to exert antiangiogenic activity.26 Quinterenol is a beta adrenergic stimulant used in asthma.27 Clioquinol is an antifungal and antiprotozoal drug29 but was recently withdrawn from the market due to possible neurotoxicity. However, this compound was separately shown to halt cognitive decline in Alzheimer’s disease, and clioquinol analogues were developed for this purpose.30 PBT2 is one of these analogues, which lacks neurotoxicity and is currently being evaluated in phase II clinical trials for Alzheimer’s disease.45 And finally, AGG 523 is an aggrecanase inhibitor in phase I clinical trials for osteoarthritis.31 Our nontoxic 8-hydroxyquinoline inhibitors of the IN-LEDGF/p75 interaction with nanomolar activity are drug-like and warrant further optimization. More potent congeners of antiviral compounds Q2–8 and Q2–13 will be necessary to enable profiling their precise mechanism of action on their desired target, the LEDGF/p75-IN interaction.
Figure 11.
FDA-approved drugs and drugs in clinical trials containing an 8-hydroxyquinoline moiety.24–31 The 8-hydroxyquinoline core is highlighted in red, with various modifications in black.
CONCLUSIONS
From initial molecular modeling data suggesting a favorable interaction of 8-hydroxyquinoline with HIV-1 IN, we have developed a panel of potent inhibitors of the IN-LEDGF/p75 interaction. Initial cytotoxicity was overcome by concerted substitution of various regions of the core scaffold. Through this optimization, we achieved an approximately 7-fold increase in potency from initial 8-hydroxyquinoline fragments to most potent lead compound. Two compounds, 5-((p-tolylamino)methyl)quinolin-8-ol and 5-(((3,4-dimethylphenyl)amino)methyl)quinolin-8-ol (Q2–8 and Q2–13), exhibited low micromolar EC50 values for inhibition of viral replication in MT-4 cells. The structural components of IN principally responsible for its interaction with LEDGF/p75 are the backbone conformation of residues 168–171 of one monomer and a hydrophobic patch from α-helices 1 and 3 of the second monomer. Molecular docking analysis confirmed that the plausible binding mode of our most potent antiviral compound included the formation of an H-bond with His 171 and packed into the hydrophobic pocket involving IN residue A128, effectively abrogating LEDGF/p75 binding. This novel class of IN-LEDGF/p75 inhibitors is promising for further preclinical and clinical development, as they are nontoxic and possess low molecular weights.
EXPERIMENTAL SECTION
IN-LEDGF/p75 AlphaScreen
The AlphaScreen assay was performed according to the manufacturer’s protocol (PerkinElmer, Benelux). Reactions were performed in a 25 µl final volume in 384-well Optiwell™ microtiter plates (PerkinElmer). The reaction buffer contained 25 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.01% (v/v) Tween-20 and 0.1% (w/v) bovine serum albumin. His6-tagged integrase (300 nM final concentration) was incubated with compound at a final concentration of 20 µM for 30 minutes at 4 °C. FLAG-tagged LEDGF/p75 protein was then added at 300 nM final concentration, and the reaction was incubated for an additional 60 minutes at 4 °C. Subsequently, 5 µL Ni-chelate-coated donor beads and 5 µL anti-FLAG antibody coated acceptor beads were added to a final concentration of 20 µg/mL of both beads. Proteins and beads were incubated for 1 h at 30 °C in order to allow association to occur. Exposure of the reaction to direct light was omitted as much as possible and the emission of light from the acceptor beads was measured in the EnVision plate reader (PerkinElmer) and analyzed using the EnVision manager software.
Quench CounterScreen
Quench counterscreening was performed identically to IN-LEDGF/p75 AlphaScreen, but a Flag-6xhistidine construct was used in place of HIV-1 IN and LEDGF/p75.
Cell Culture
The LNCaP cell line was kindly provided by Dr. Alan Epstein (University of Southern California, Los Angeles, CA). The cell line was grown in RPMI 1640 medium (Cellgro, Mediatech Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS; Gemini-Bioproducts, Woodland, CA) at 37 °C in a humidified atmosphere of 5% CO2. To remove the adherent cells from the flask, cells were washed with 1 x PBS, incubated with 0.05% trypsin-EDTA solution (Gemini-Bioproducts, Woodland, CA) at 37 °C for 5 minutes, collected in fresh culture medium and centrifuged. After removal of supernatant, the cell pellet was resuspended in fresh culture medium for subculture and experiments. All experiments were performed with cells in exponential growth phase.
Cytotoxicity Assay
Potential cytotoxicity of compounds was evaluated with 3-(4,5-dimethylthiazol-2-yl) −2,5-diphenyltetrazolium bromide (MTT) assay.46, 47 LNCaP cells were seeded in 96-well tissue culture plates at the concentration of 2,500 cells/well. After overnight attachment, cells were treated with a continuous exposure to compounds or DMSO (0.1% vol/vol) for 72 h. MTT solution (3 mg/mL in PBS) was then added to each well for a final concentration of 0.3mg/mL MTT. After 4h incubation at 37 °C, media from each well was removed, and DMSO was added to dissolve the formazan crystals formed by live cells. Absorbance was measured at 570 nm using a microplate reader (Molecular devices, Sunnyvale, CA). Cytotoxicity of compounds was presented as percentage inhibition of cell proliferation against DMSO treated controls:
% Inhibition of cell proliferation = 1 - (ODcompound - ODblank) / (ODcontrol - ODblank)
IC50 for the cytotoxic compounds was then determined from a plot of percentage inhibition of cell proliferation versus log compound concentration.
Antiviral Assay
The inhibitory effect of potentially antiviral drugs on the HIV-induced cytopathic effect in MT-4 cell culture was determined by the MTT assay as previously described.12 In short the 50% cell culture infective dose of the HIV strains was determined by titration of the virus stock using MT-4 cells. For the drug susceptibility assays, MT-4 cells were infected with 100 to 300 50% cell culture infective doses (CCID50) of the HIV strains in the presence of fivefold serial dilutions of the antiviral drugs. The concentration of the compound achieving 50% protection against the CPE of HIV, which is defined as the 50% effective concentration (IC50), was determined. The concentration of the compound killing 50% of the MT-4 cells, which is defined as the 50% cytotoxic concentration (CC50), was determined as well.
Molecular Docking
Molecular docking was performed using the Glide (Grid-based Ligand Docking from Energetics) program (version 5.7, Schrodinger, LLC, New York, 2011).48–50 The crystal structure of HIV-1 IN protein catalytic core domain complexed with the IN binding domain of LEDGF/p75 (PDB ID: 2B4J) was used for the docking of ligands. The LEDGF/p75 protein and water molecules were deleted from the crystal structure, and the same method was used in protein preparation wizard. The wizard uses OPLS 2005 force field to minimize the protein to 0.30 Å rmsd by adding hydrogen and adjusting bond orders. Prior to docking, compounds were minimized, and all possible combinations of stereo isomers and conformers were generated with LigPrep from Schrodinger using OPLS-2005 force field. Within LigPrep, Epik module was used to generate all possible ionization forms of ligands at pH 7.0 ± 0.2. Compounds with their all possible combinations of stereoisomers and conformers were docked into the LEDGF/p75 binding site of the prepared IN protein using the extra precision (XP) docking mode of Glide. Glide performs an exhaustive search of the positional, orientational, and conformational space available to the docked ligand using a series of hierarchical filters, followed by energy optimization. Finally, the conformations were further refined via a Monte Carlo sampling that examined nearby torsional minima.50
Pharmacophore Study
A pharmacophore was generated using Catalyst (Accelrys, Inc.).51 The docked conformation of compound QC was imported into Catalyst, and pharmacophoric features were generated based on docking interactions to recreate specific ligand-protein interactions within the pharmacophore. Using the merge hypotheses features tool, all these features were merged into a pharmacophore hypothesis, which was then used to screen a 3D database that stores a diverse sampling of all the energetically accessible conformations of five million drug-like compounds from various commercial sources including Asinex, Enamine, and VitasM Lab.52–54 Compounds used in this studies were purchased from Asinex and Enamine as solid powder with >95 purity.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a research grant from an NIH/NIAID (R21 AI081610) grant and the Campbell foundation. ES was a recipient of USC Oakley Fellowship. We thank Hyungju Kim for assistance with high-throughput screening.
ABBREVIATIONS USED
- IN
HIV-1 integrase
- LEDGF/p75
lens epithelium-derived growth factor/p75
- FBDD
fragment-based drug design
- Glide
Grid-based Ligand Docking from Energetics
- H-bond
hydrogen bond
- IC50
50% inhibitory concentration
- EC50
50% effective concentration
- CC50
50% cytotoxic concentration
- CCID50
cell culture infective dose
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supplementary Figures 1–2 and Supplementary Tables 1–2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Broder S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral Res. 2010;85:1–18. doi: 10.1016/j.antiviral.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis. 2011;203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandegraaff N, Engelman A. Molecular mechanisms of HIV integration and therapeutic intervention. Expert Rev Mol Med. 2007;9:1–19. doi: 10.1017/S1462399407000257. [DOI] [PubMed] [Google Scholar]
- 6.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 7.Serrao E, Odde S, Ramkumar K, Neamati N. Raltegravir, elvitegravir, and metoogravir: the birth of "me-too" HIV-1 integrase inhibitors. Retrovirology. 2009;6:25. doi: 10.1186/1742-4690-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 9.Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 2007;3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey KK, Sinha S, Grandgenett DP. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J Virol. 2007;81:3969–3979. doi: 10.1128/JVI.02322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Luca L, Barreca ML, Ferro S, Christ F, Iraci N, Gitto R, Monforte AM, Debyser Z, Chimirri A. Pharmacophore-based discovery of small-molecule inhibitors of protein-protein interactions between HIV-1 integrase and cellular cofactor LEDGF/p75. ChemMedChem. 2009;4:1311–1316. doi: 10.1002/cmdc.200900070. [DOI] [PubMed] [Google Scholar]
- 12.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 13.De Luca L, Ferro S, Morreale F, De Grazia S, Chimirri A. Inhibitors of the interactions between HIV-1 IN and the cofactor LEDGF/p75. ChemMedChem. 2011;6:1184–1191. doi: 10.1002/cmdc.201100071. [DOI] [PubMed] [Google Scholar]
- 14.Fan X, Zhang FH, Al-Safi RI, Zeng LF, Shabaik Y, Debnath B, Sanchez TW, Odde S, Neamati N, Long YQ. Design of HIV-1 integrase inhibitors targeting the catalytic domain as well as its interaction with LEDGF/p75: a scaffold hopping approach using salicylate and catechol groups. Bioorg Med Chem. 2011;19:4935–4952. doi: 10.1016/j.bmc.2011.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem. 2012;287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. Small-Molecule Inhibitors of the LEDGF/p75 Binding Site of Integrase Block HIV Replication and Modulate Integrase Multimerization. Antimicrob Agents Chemother. 2012;56:4365–4374. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peat TS, Rhodes DI, Vandegraaff N, Le G, Smith JA, Clark LJ, Jones ED, Coates JA, Thienthong N, Newman J, Dolezal O, Mulder R, Ryan JH, Savage GP, Francis CL, Deadman JJ. Small molecule inhibitors of the LEDGF site of human immunodeficiency virus integrase identified by fragment screening and structure based design. PLoS One. 2012;7:e40147. doi: 10.1371/journal.pone.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat Rev Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 19.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 20.Majerz-Maniecka K, Musiol R, Skorska-Stania A, Tabak D, Mazur P, Oleksyn BJ, Polanski J. X-ray and molecular modelling in fragment-based design of three small quinoline scaffolds for HIV integrase inhibitors. Bioorg Med Chem. 2011;19:1606–1612. doi: 10.1016/j.bmc.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DI, Peat TS, Vandegraaff N, Jeevarajah D, Le G, Jones ED, Smith JA, Coates JA, Winfield LJ, Thienthong N, Newman J, Lucent D, Ryan JH, Savage GP, Francis CL, Deadman JJ. Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design. Antivir Chem Chemother. 2011;21:155–168. doi: 10.3851/IMP1716. [DOI] [PubMed] [Google Scholar]
- 22.Wielens J, Headey SJ, Deadman JJ, Rhodes DI, Le GT, Parker MW, Chalmers DK, Scanlon MJ. Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1. ChemMedChem. 2011;6:258–261. doi: 10.1002/cmdc.201000483. [DOI] [PubMed] [Google Scholar]
- 23.DeSimone RW, Currie KS, Mitchell SA, Darrow JW, Pippin DA. Privileged structures: applications in drug discovery. Comb Chem High Throughput Screen. 2004;7:473–494. doi: 10.2174/1386207043328544. [DOI] [PubMed] [Google Scholar]
- 24.Negosanti M, Bettoli V, Valenti R, Patrone P, Celasco G. [Clinical evaluation on the usefulness of 5,7-dichloro-8-hydroxyquinoline (chloroxine) in association with betamethasone 17-benzoate in the topical treatment of infected cortisone-sensitive dermopathies] G Ital Dermatol Venereol. 1985;120:XVII–XXIII. [PubMed] [Google Scholar]
- 25.Bergogne-Berezin E, Berthelot G, Muller-Serieys C. [Present status of nitroxoline] Pathol Biol (Paris) 1987;35:873–878. [PubMed] [Google Scholar]
- 26.Shim JS, Matsui Y, Bhat S, Nacev BA, Xu J, Bhang HE, Dhara S, Han KC, Chong CR, Pomper MG, So A, Liu JO. Effect of nitroxoline on angiogenesis and growth of human bladder cancer. J Natl Cancer Inst. 2010;102:1855–1873. doi: 10.1093/jnci/djq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bleecker E, McKinney W, Lyons H, Steen SN. Bronchodilator effects of Quinterenol sulfate--a preliminary clinical study. Anesth Analg. 1969;48:7–9. [PubMed] [Google Scholar]
- 28.Bleecker ER, Siler T, Owen R, Kramer B. Bronchodilator efficacy and safety of indacaterol 150 mug once daily in patients with COPD: an analysis of pooled data. Int J Chron Obstruct Pulmon Dis. 2011;6:431–438. doi: 10.2147/COPD.S21073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gholz LM, Arons WL. Prophylaxis and Therapy of Amebiasis and Shigellosis with Iodochlorhydroxyquin. Am J Trop Med Hyg. 1964;13:396–401. doi: 10.4269/ajtmh.1964.13.396. [DOI] [PubMed] [Google Scholar]
- 30.Adlard PA, Cherny RA, Finkelstein DI, Gautier E, Robb E, Cortes M, Volitakis I, Liu X, Smith JP, Perez K, Laughton K, Li QX, Charman SA, Nicolazzo JA, Wilkins S, Deleva K, Lynch T, Kok G, Ritchie CW, Tanzi RE, Cappai R, Masters CL, Barnham KJ, Bush AI. Rapid restoration of cognition in Alzheimer's transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Abeta. Neuron. 2008;59:43–55. doi: 10.1016/j.neuron.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Chockalingam PS, Sun W, Rivera-Bermudez MA, Zeng W, Dufield DR, Larsson S, Lohmander LS, Flannery CR, Glasson SS, Georgiadis KE, Morris EA. Elevated aggrecanase activity in a rat model of joint injury is attenuated by an aggrecanase specific inhibitor. Osteoarthritis Cartilage. 2011;19:315–323. doi: 10.1016/j.joca.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Hazuda D, Iwamoto M, Wenning L. Emerging pharmacology: inhibitors of human immunodeficiency virus integration. Annu Rev Pharmacol Toxicol. 2009;49:377–394. doi: 10.1146/annurev.pharmtox.011008.145553. [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, Ikeda S, Kodama E, Matsuoka M, Shinkai H. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem. 2006;49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 34.Dayam R, Al-Mawsawi LQ, Zawahir Z, Witvrouw M, Debyser Z, Neamati N. Quinolone 3-carboxylic acid pharmacophore: design of second generation HIV-1 integrase inhibitors. J Med Chem. 2008;51:1136–1144. doi: 10.1021/jm070609b. [DOI] [PubMed] [Google Scholar]
- 35.Mekouar K, Mouscadet JF, Desmaele D, Subra F, Leh H, Savoure D, Auclair C, d'Angelo J. Styrylquinoline derivatives: a new class of potent HIV-1 integrase inhibitors that block HIV-1 replication in CEM cells. J Med Chem. 1998;41:2846–2857. doi: 10.1021/jm980043e. [DOI] [PubMed] [Google Scholar]
- 36.Wiles JA, Bradbury BJ, Pucci MJ. New quinolone antibiotics: a survey of the literature from 2005 to 2010. Expert Opin Ther Pat. 2010;20:1295–1319. doi: 10.1517/13543776.2010.505922. [DOI] [PubMed] [Google Scholar]
- 37.Musiol R, Jampilek J, Buchta V, Silva L, Niedbala H, Podeszwa B, Palka A, Majerz-Maniecka K, Oleksyn B, Polanski J. Antifungal properties of new series of quinoline derivatives. Bioorg Med Chem. 2006;14:3592–3598. doi: 10.1016/j.bmc.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D'Alessandro U. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar J. 2011;10:144. doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu L, Zhang S, He X, Luo Z, Wang X, Liu W, Qin X. Design and synthesis of novel beta-diketo derivatives as HIV-1 integrase inhibitors. Bioorg Med Chem. 2012;20:177–182. doi: 10.1016/j.bmc.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Rabaaoui S, Zouhiri F, Lancon A, Leh H, d'Angelo J, Wattel E. Inhibitors of strand transfer that prevent integration and inhibit human T-cell leukemia virus type 1 early replication. Antimicrob Agents Chemother. 2008;52:3532–3541. doi: 10.1128/AAC.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H, Cai RZ, Schacherer L, Jabri S, Tsiang M, Fardis M, Chen X, Chen JM, Kim CU. Design, synthesis, and SAR studies of novel and highly active tri-cyclic HIV integrase inhibitors. Bioorg Med Chem Lett. 2006;16:3989–3992. doi: 10.1016/j.bmcl.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Majerz-Maniecka K, Musiol R, Nitek W, Oleksyn BJ, Mouscadet JF, Le Bret M, Polanski J. Intermolecular interactions in the crystal structures of potential HIV-1 integrase inhibitors. Bioorg Med Chem Lett. 2006;16:1005–1009. doi: 10.1016/j.bmcl.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 43.De Luca L, Ferro S, Gitto R, Barreca ML, Agnello S, Christ F, Debyser Z, Chimirri A. Small molecules targeting the interaction between HIV-1 integrase and LEDGF/p75 cofactor. Bioorg Med Chem. 2010;18:7515–7521. doi: 10.1016/j.bmc.2010.08.051. [DOI] [PubMed] [Google Scholar]
- 44.Serrao E, Thys W, Demeulemeester J, Al-Mawsawi LQ, Christ F, Debyser Z, Neamati N. A Symmetric Region of the HIV-1 Integrase Dimerization Interface Is Essential for Viral Replication. PLoS One. 2012;7:e45177. doi: 10.1371/journal.pone.0045177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenche VB, Barnham KJ. Alzheimer's disease & metals: therapeutic opportunities. Br. J. Pharmacol. 2011;163:211–219. doi: 10.1111/j.1476-5381.2011.01221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 47.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 48.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 49.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 50.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 51.Catalyst v 4.11. San Diego, CA: Accelrys Software Inc; 2005. http://www.accelrys.com. [Google Scholar]
- 52. http://www.asinex.com.
- 53. http://www.enamine.net.
- 54. http://www.vitasmlab.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.