Abstract
GenomeConnect, the NIH-funded Clinical Genome Resource (ClinGen) patient registry, engages patients in data sharing to support the goal of creating a genomic knowledge base to inform clinical care and research. Participant self-reported health information and genomic variants from genetic testing reports are curated and shared with public databases, such as ClinVar. There are four primary benefits of GenomeConnect: 1) sharing novel genomic data - 47.9% of variants were new to ClinVar, highlighting patients as a genomic data source; 2) contributing additional phenotypic information - of the 52.1% of variants already in ClinVar, GenomeConnect provided enhanced case-level data; 3) providing a way for patients to receive variant classification updates if the reporting laboratory submits to ClinVar – 97.3% of responding participants opted to receive such information and 13 updates have been identified; and 4) supporting connections with others, including other participants, clinicians, and researchers to enable the exchange of information and support - 60.4% of participants have opted to partake in participant matching. Moving forward, ClinGen plans to increase patient-centric data sharing by partnering with other existing patient groups. By engaging patients, more information is contributed to the public knowledge base, benefiting both patients and the genomics community.
Keywords: Patient Registry, Genomic Data Sharing, Matchmaking, ClinGen, ClinVar, Variant Interpretation
Background
The expansion and broader availability of genomic testing has allowed for an increasing number of genomic variants to be identified in patients with rare diseases, common disorders, and even in healthy individuals. Interpretation of these variants remains complex, and their impact on health is often unclear, requiring collaborative efforts for genomic and phenotypic data sharing to improve the quality, consistency, and accuracy of variant interpretation and to better inform patient care (Harrison et al. 2017). Because of this, professional societies, funding bodies, and biomedical journals have endorsed data sharing efforts (National Institutes of Health, 2014; National Society of Genetic Counselors, 2015; ACMG Board of Directors, 2017; American Medical Association, 2013; Barsh et al., 2015). Supporting data sharing is also a key goal of the National Institutes of Health (NIH)-funded Clinical Genome Resource (ClinGen) (Rehm et al., 2015; Rehm, 2017; https://www.clinicalgenome.org/). ClinGen is building central resources that define the clinical relevance of genes and genomic variants to inform clinical care and research (Rehm et al., 2015). This effort relies on data sharing, and, as a result, ClinGen actively collaborates with laboratories, researchers, clinicians, patients, and other stakeholders to enable submission of genomic data to the publicly available ClinVar database, a repository of human genomic variants and their relationship to human health within the National Center for Biotechnology Information (NCBI) at the NIH (Landrum et al., 2014, Landrum et al., 2016; Landrum et al., 2018;https://www.ncbi.nlm.nih.gov/clinvar/ Rehm, 2017;).
GenomeConnect
GenomeConnect, the ClinGen online patient registry, was developed and launched in October 2014 to engage patients in data sharing efforts (Kirkpatrick et al., 2015; https://genomeconnect.org). Specifically, GenomeConnect was created to capture individual-level phenotype information, vital to the variant interpretation process, from the patients themselves. Genomic testing laboratories often do not have detailed phenotype information regarding patients in whom rare variants are found. Access to this detailed phenotype information can greatly inform the interpretation process (Riggs et al., 2012).
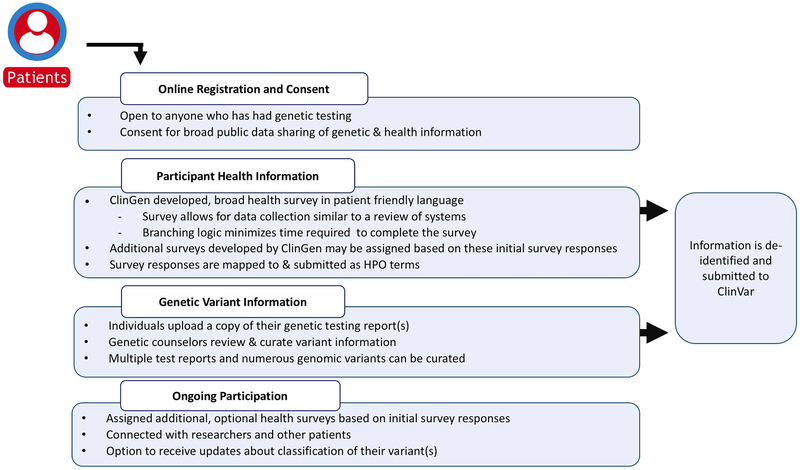
Participation in GenomeConnect is open to anyone who has had genetic testing regardless of genetic test results or diagnosis. Participants consent to have their genetic and health information de-identified and shared with public databases, such as ClinVar. Health information is collected via participant-completed surveys and importantly, genomic data is reviewed and curated from participants’ uploaded genetic test reports by genetic counseling staff to ensure accuracy and consistency of genomic data (Kirkpatrick et al, 2015; Figure 1).
Figure 1.
GenomeConnect participation process. The figure depicts the steps GenomeConnect participants engage in while enrolling in the registry, data collection methods, and ongoing engagement.
As of April 2018, 1,601 participants from 32 countries have enrolled in GenomeConnect. Participants report learning about GenomeConnect in varied ways; the most common sources of referral are healthcare providers (29.4%%, n=471/1601) and information on genetic testing reports (27.1%, n=434/1601). GenomeConnect participants range in age from less than a year to 91 years old. Sixty percent (60.5%, n=968/1601) of participants identify as female, 37.5% (n=601/1601) identify as male, 0.6% (n=10/1601) identified as something else (e.g., Female to Male (FTM)/Transman/Transgender male, Intersex, Other) and 1.4% (n=22/1601) did not provide a response. The majority of participants underwent genetic testing to confirm a diagnosis (20.4%, n=326/1601), evaluate symptoms (42.7%, n=684/1601), or determine if they carry a familial variant (11.9%, n=191/1601). Healthy individuals who have undergone testing to be proactive about their health or to determine reproductive risks have also enrolled in the registry (4.6%, n=74/1601).
Collection of Participant-provided Phenotype Data via Health Survey
Phenotype information is collected through participant health surveys and shared using corresponding Human Phenotype Ontology (HPO) terms (Robinson & Mundlos, 2010; Köhler et al., 2017). The GenomeConnect team collaborated with the ClinGen Phenotype working group to develop an initial health survey and describe Human Phenotype Ontology terms in patient-friendly language (Kirkpatrick et al., 2015). Invited working group members included clinicians, laboratorians, and representatives from HPO and other phenotyping tools. A review of clinical intake and high-level HPO terms informed the structure of this general health survey, designed to serve as an overall “review of systems” for each GenomeConnect participant. For all survey questions, the associated medical term is provided along with example(s) or a patient-friendly description developed by members of the Phenotype working group. Appropriate HPO mappings were determined by the group and programmed into the survey so that survey responses automatically map to HPO terms on the back-end and can be exported using these structured identifiers. The current version of the GenomeConnect general health survey is available online and has been available for review and use since GenomeConnect’s launch in 2014 (https://tinyurl.com/y9v29onh; Supp. Figure S1). Other registries and research groups have adopted all or portions of this survey for use as a general health data collection tool including other registries on the same platform at GenomeConnect through Invitae and the Undiagnosed Disease Network.
For each general body system, the participant is asked whether or not they have any issues related to this system. If they answer “Yes,” they are presented with a brief list of phenotypic features within that body system deemed by the ClinGen Phenotype working group to be potentially indicative of a genetic disorder. This list is purposefully brief and the survey uses branching logic in order to minimize time required to complete the survey. For example, if the participant answered “No” to the high-level question for a given body system (e.g. “Ears and/or Hearing”), they would not be presented with the follow-up questions related to that body system. Currently, the GenomeConnect survey has not been validated against data extracted from patients’ electronic health records, but validation efforts and further survey modifications are being pursued through a Patient-Centered Outcomes Research Institute funded collaboration (https://tinyurl.com/ybl3eb5z).
Any concerns indicated on the initial survey can also be further detailed in subsequent sub-surveys as needed (Kirkpatrick et al. 2015). For example, if the participant indicated that they had a history of cardiomyopathy, they may be invited to participate in a follow-up survey designed to learn more about the specific type, when and how it was diagnosed, treatment modalities, other risk factors, and outcomes. Additional surveys have been developed by the GenomeConnect team in collaboration with ClinGen clinical domain working groups and expert panels, in an effort to collect phenotypic data that would aid their curation efforts (https://tinyurl.com/y89gbka9). Surveys developed to date include cardiomyopathy, congenital heart defect, arrhythmia, and cancer surveys. Responses from these surveys are also mapped to HPO terms to allow for structured data sharing.
Submission of Patient-Derived Data to ClinVar
NCBI’s ClinVar provides a publicly available database of the relationship between genomic variants and phenotypes. ClinVar is a database that relies on submissions from many groups, including clinical genetic testing and research laboratories, clinicians, locus-specific databases, OMIM®, GeneReviews™, expert panels, and practice guidelines (Landrum et al., 2016). ClinVar aggregates submitted interpretations of the clinical significance of variants and allows submitters to provide structured and free text supporting evidence (Landrum et al., 2014).
GenomeConnect facilitates genotype and phenotype data submission from patients. Phenotype information is collected through participant health surveys. Genomic information is obtained from the participant’s genetic testing report by genetic counselor staff. All clinical reports are accepted regardless of testing indication or variant pathogenicity. Research and direct to consumer testing reports are reviewed by the GenomeConnect team but are not be submitted to ClinVar if results were not confirmed in a CLIA laboratory or if there is concern regarding the validity of a result.
This genomic and health data is submitted to ClinVar as a “phenotyping only” submission. This collection method is reserved for variants submitted to ClinVar that provide individual observations with phenotype data without an independent variant interpretation from the submitter (Landrum et al., 2018). This distinct collection method highlights for the user the fact that these variants may also be submitted directly from the reporting laboratory (typically under the collection method “clinical testing”). The reporting laboratory name, that laboratory’s reported interpretation, the report date, segregation data, and detailed phenotype terms are included in GenomeConnect’s “phenotyping only” submissions. However, some of this information is not currently displayed in the ClinVar web display, and is only available in the XML monthly data release (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/xml/).
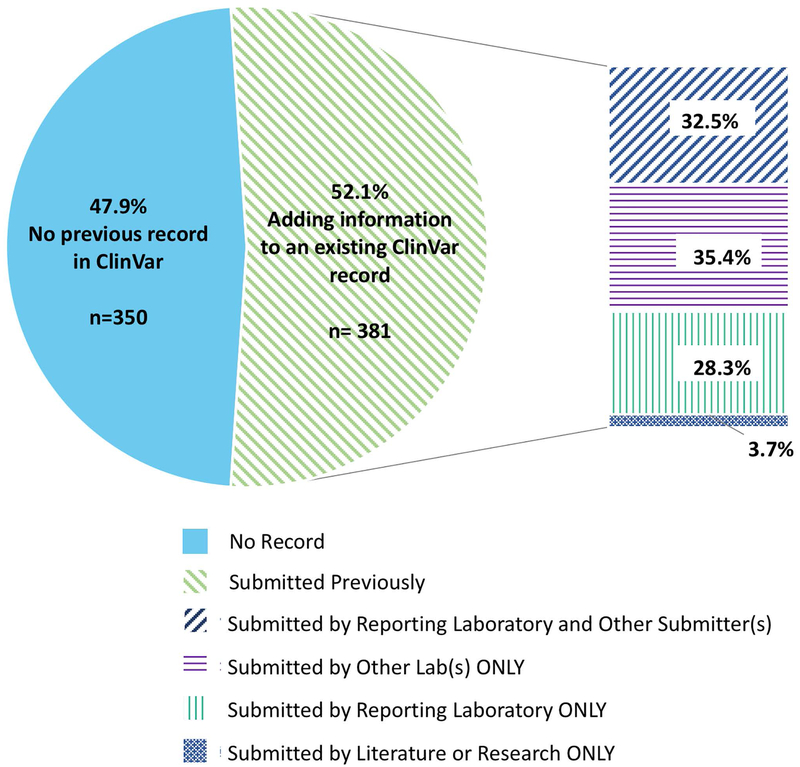
The first GenomeConnect submission to ClinVar was completed in August 2017; a second is in progress and subsequent submissions are planned on a quarterly basis. To assess the impact of patient-derived data sharing, we examined the impact of the registry’s ClinVar submissions to date. Of 731 sequence variants submitted, or prepared for submission, 47.9% (n=350) had not been previously submitted to ClinVar. Also, 67.7% of variants (n=495/731) were not in the Human Gene Mutation Database. Of all submitted sequence variants, 34.8% (n=255/731) were neither in ClinVar or the Human Gene Mutation Database and, are therefore, likely previously unreported in publicly available databases in association with disease. These variants were reported by laboratories that do not regularly submit to ClinVar as well as laboratories that regularly share data. Report dates ranged from 2012–2018 and variants not previously shared included all five major pathogenicity classifications (benign, likely benign, uncertain significance, likely pathogenic, and pathogenic). The remaining 52.1% (n=381/731) of variants had previously been shared with ClinVar by other submitters; 60.9% (n=232/381) of these were submitted by the GenomeConnect participant’s reporting laboratory. Of the variants not submitted by the participant’s reporting laboratory, 35.4% (n=135/381) were submitted by other clinical laboratories, and 3.7% (n=14/381) were submitted from research results or as a citation to the literature (“literature only”) (Figure 2). Although GenomeConnect was originally created to provide a source of enhanced phenotypic information from patients to inform variant interpretation efforts, the registry’s ClinVar variant submissions also demonstrate the importance of patients as a source of novel genomic data.
Figure 2.
Previous ClinVar records of the 731 sequence variants submitted or prepared for submission by GenomeConnect. These 731 variants were shared by 424 GenomeConnect participants.
Facilitating Variant Interpretation Updates
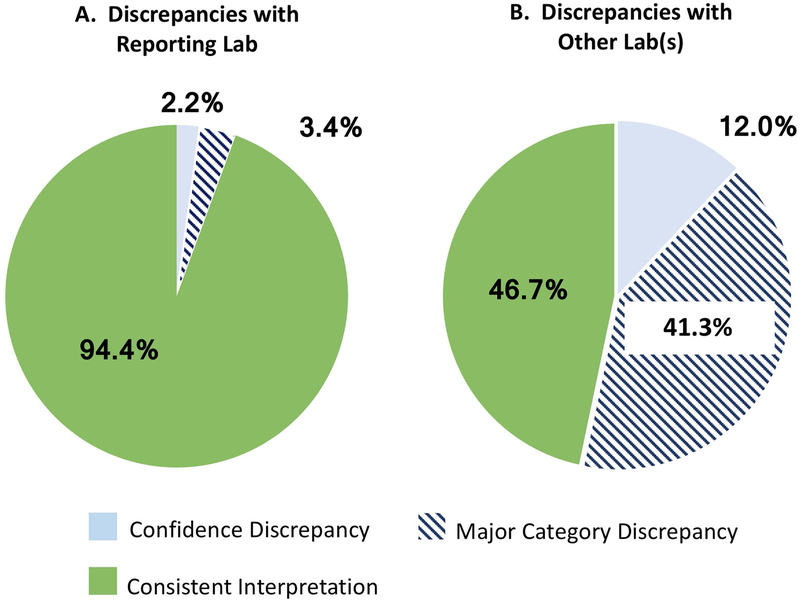
Of the variants previously submitted by the participant’s reporting laboratory (n=232), the classification on the participant’s report was out of date compared to the laboratory’s current ClinVar entry for 13 (5.6%) variants. In all instances, the reporting laboratory had reclassified the variant since the patient’s results were reported. As shown in Figure 3, nine of the variants had a difference between the three major classification levels “pathogenic(P)/likely pathogenic(LP)”, “uncertain significance (VUS)”, and “benign(B)/likely benign(LB),” and the remaining four had a confidence discrepancy with a difference between P and LP (Figure 3).
Figure 3.
GenomeConnect Report and ClinVar Submission Interpretation Discrepancies. A: Discrepancies between the participant’s report and the reporting laboratory. Major category discrepancies are defined as those differences between the three main classification categories 1) “pathogenic (P)/likely pathogenic (LP)”, “uncertain significance (VUS)”, and “benign (B)/likely benign (LB).” Confidence discrepancies are differences between Pathogenic and Likely pathogenic or Benign and Likely benign. B: Discrepancies between the participant’s report and other submitting laboratories.
Evidence supporting or refuting a given variant’s reported interpretation may emerge over time, prompting a laboratory to update their interpretation. Currently, practices pertaining to updating genetic testing reports and contacting ordering healthcare providers concerning variant interpretation modifications vary between testing laboratories. Laboratories may attempt to inform clinicians about updates to variant classifications, but this updated information is not always relayed to the patient for a variety of reasons, including lack of resources and reimbursement as well as difficulties re-contacting the ordering physician or patient. Moreover, as the number of variants detected by a laboratory has grown enormously, the ability of laboratories to provide timely updates to providers has already become unsustainable (Richards et al., 2015).
GenomeConnect is a logical resource for participants to remain up to date about variant reclassifications. If the registry becomes aware of a discrepancy between the classification on the participant’s report and the classification in ClinVar through the registry’s submission to the database, this information can be relayed to participants. Before proceeding with a plan to potentially provide these updates, GenomeConnect surveyed participants regarding their preferences for receiving this information. In September 2016, GenomeConnect distributed a survey invitation by email to current participants to capture preferences regarding receiving updates about their genetic testing results from the registry. The goal of the survey was to facilitate planning and resource allocation by GenomeConnect. The survey was sent to 698 participants or their guardians; 137 participants responded for a response rate of approximately 20%. Participants were provided with a brief summary about genetic testing results and how interpretations may change over time. Survey responses were anonymous, and no demographic data was collected. Participants were then asked up to eight questions regarding whether they would like to learn about these updates and if they would like to learn these updates from GenomeConnect. Of those who completed the survey, 98.5% (n=135/137) indicated that they would want to receive these updates and that they would want GenomeConnect to contact them with this information if the registry became aware of it. In fact, 97.8% (n=134/137) of participants saw the potential of receiving updates about their or their child’s testing as a benefit of participating in GenomeConnect.
The two participants who were unsure if they would like to receive such updates from GenomeConnect were asked why. Responses included that they would prefer to receive updates from their healthcare provider, they felt GenomeConnect would not be able to provide enough information about any updates, and they were unsure or not interested in receiving updates in general.
Some variant interpretation updates might impact medical management, while others may not. For example, an update from VUS to LB for a BRCA1 variant would not likely impact clinical care, while an update from VUS to LP could affect care. Consequently, GenomeConnect asked participants what type of updates they would like to receive. Ninety-two percent of respondents (n=126/137) wanted all possible updates, 7.3% (n=10/137) only wanted those that could impact medical management, and one participant was unsure. In addition to obtaining information on preferences about the type of updates that are provided, participants were asked if receiving updates should be something they should have the option to decline. Eighty-two percent of respondents (n=113/137) indicated that there should be the ability to opt out of receiving such information. Given that survey responses were anonymized, it is unclear how respondents differed from non-respondents.
In summary, by surveying GenomeConnect participants, it became apparent that registry participants want to receive information about potential updates to their genetic testing results. Participants see GenomeConnect as an acceptable intermediary in providing these updates, and they believe that receiving updates is a potential benefit to participation. Based on these survey data, GenomeConnect has implemented a process to provide participants with the option to receive potential updates to their variant interpretation. During the registration process, each participant now elects whether they would like to receive this information. Although the majority of responding participants indicated that they would want to receive updates, the majority also felt that individuals should have the ability to choose not to receive this information. Moreover, it is possible that participants that did not respond to this survey may be less interested in obtaining variant interpretation updates. As such, the GenomeConnect team felt that each participant should have the option to choose to receive updated variant classification information. Participants who registered prior to the implementation of this process have been prompted to update their preferences. To date, 97.3% (n= 727/747) of responding participants have opted to receive updates.
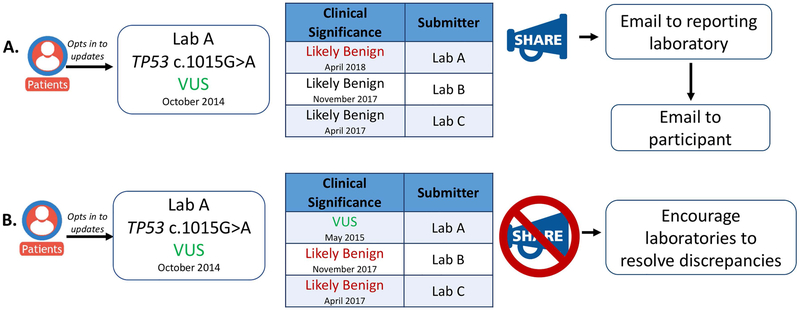
If an update has been identified and the participant has elected to receive this information, GenomeConnect will first alert the reporting genetic testing laboratory via email. During registration, GenomeConnect obtains consent from the participant to communicate with their testing laboratory. If an update is identified, GenomeConnect then uses the accession or laboratory identification number on the report to communicate with the testing laboratory. Because each laboratory’s policy regarding updating genetic test reports varies, alerting the laboratory can prompt report updating, if needed. Next, the GenomeConnect participant is contacted via email. The email includes a general statement that there may be updated information about their genetic test results, suggest the participant contact the ordering healthcare provider or a genetics provider in their area, provides information that would be helpful to share with their provider, and reminds them to upload any updated genetic testing reports they may receive so the GenomeConnect ClinVar submission can reflect the change (Figure 4). GenomeConnect currently does not provide the specific update to the participant given that, as a registry, GenomeConnect is unable to fully assess clinical correlation, recommend appropriate medical management, or request an updated clinical report. Because these activities are best completed as part of a provider-patient relationship, GenomeConnect suggests participants contact a healthcare provider to further review potential updates. It should be mentioned, however, that ClinVar is the source of the updated information and therefore participants are able to access the same information in ClinVar. To date, of the 13 instances in which the classification on the participant’s report was out of date compared to the laboratory’s current ClinVar entry, three updates have been provided to participants and an additional three are in process. The remaining seven updates have not been shared with participants. Despite multiple prompts to update their profiles, these seven participants have not selected their preference regarding the receipt of this information. Moving forward, GenomeConnect will explore mechanisms to automate and scale variant tracking to facilitate variant interpretation updates as the volume increases. The GenomeConnect team will also continue to evaluate other scenarios where interpretation updates could be passed along to participants and will continue gathering experience returning updates from the participants’ reporting laboratory to inform future efforts.
Figure 4.
Informing GenomeConnect participants about variant discrepancies in ClinVar. A: Process of providing updates if the participant’s report is out of date compared to the laboratory’s current ClinVar submission. Updates are provided if the participant opts in to receiving them and the reporting laboratory’s interpretation is out of date on the participant’s report. An email is first sent to the reporting laboratory and then an email is sent to the participant referring them back to their healthcare provider and encouraging them to upload any genetic test results they may receive. B: Updates are not provided if discrepancies are only identified between the participants’ report and other submitters.
Facilitating Discrepancy Resolution
Of the genomic variants previously submitted by a laboratory other than the reporting institution, 41.3% (n=107/259) had a major category difference in variant classification between the participant report and another submission. As shown in Figure 3, 12% (n= 31/259) had a confidence discrepancy between the participant report and another submission. Information about these discrepancies with other laboratories is not currently shared with participants since the participant’s genetic testing report from the clinical testing laboratory may still represent that laboratory’s interpretation (Figure 4). However, although GenomeConnect is not relaying this information to participants, the registry is working with the ClinGen Sequence Variant Inter-Laboratory Discrepancy Resolution group to identify variants that are discrepant in ClinVar and have been reported through a GenomeConnect participants’ testing. The ClinGen Sequence Variant Inter-Laboratory Discrepancy Resolution group then encourages submitting laboratories with these discrepant interpretations to address major category discrepancies (Harrison et al., 2017). GenomeConnect can provide additional participant phenotype data to inform these deliberations.
In conjunction with this working group, all variants with major category discrepancies between the GenomeConnect participant’s report and other ClinVar submitters were reviewed (n=107). From that review, 22 variants were prioritized for outlier reassessment (if at least one submitting laboratory submitted an interpretation that differed from the majority of other submitters). These laboratories were contacted by the working group and prompted to reassess the variants. Of the 22 variants, 13 had medically significant discrepancies (LP/P vs. LB/B/VUS) and 9 were not medically significant (LB/B vs. VUS). Eight discrepancies were resolved completely and three were partially resolved. Those that were partially resolved initially had a majority of laboratories with concordant variant classifications and two or more laboratories with a differing interpretation. In these cases, one laboratory resolved their discrepancy with the majority of other submitters, but at least one other discrepancy remained (S.M. Harrison, personal communication, April 18, 2018). Of the eleven discrepancies resolved or partially addressed, seven resulted in a classification update from the GenomeConnect participants’ reporting laboratory. These seven variant interpretation updates will be shared with participants that opted to receive them. In one instance to date, GenomeConnect participant data indicating the variant was de novo led to resolution of a medically significant discrepancy between two clinical laboratories.
Facilitating Matchmaking
In addition to facilitating genomic data sharing, GenomeConnect also allows participants to connect with other stakeholders, including researchers, clinicians, and other patients and families. Within the registry, participants can search for others based on gene, disease, or US state using the participant matching interface (Kirkpatrick et al., 2015). Currently, 60.4% (n=967/1601) of participants have opted to participate in this feature. If any matches are returned, de-identified information about the match is provided including age, gender, and location. The participant can then elect to contact their match via the portal. If both participants wish to connect, they can then do so outside of the portal. Currently, 12.8% (n=205/1601) of participants can match based on gene, 33.9% (n=543/1601) based on diagnosis, and 89.6% can match on US state (n=1434/1601). In addition to matching based on gene, participants can also request to attempt to match based on specific copy number or sequence variant by emailing the GenomeConnect coordinator team. Given differences in variant nomenclature and potential difficulties searching, this process has not been automated and is facilitated by genetic counselor staff. If a match is identified, the coordinator will send a query on behalf of the participants interested in connecting.
In addition to connecting participants within the registry, GenomeConnect also allows for matchmaking with external clinicians and researchers through Matchmaker Exchange (MME). MME is a federated network of rare disease datasets established to facilitate candidate gene matching (Philippakis et al., 2015; Sobreira et al., 2017). Currently, GenomeConnect actively submits variants in genes of uncertain significance on behalf of our participants to MME through GeneMatcher (Sobreira et al., 2015), one of the connected nodes. To date, 14/16 submitted cases yielded potential matches. All submitters of potential matches were contacted by the GenomeConnect genetic counselors and, in three cases, researchers requested to connect directly with participants.
Matching across MME is currently limited to clinician and researcher-initiated cases. To ensure GenomeConnect participants also match with the patient-initiated cases from MyGene2 (University of Washington Center for Mendelian Genomics, n.d.; http://www.mygene2.org), a web-portal for families with rare genetic conditions who wish to share their genetic and health data, GenomeConnect genetic counselor staff periodically manually query MyGene2 for patient-submitted gene matches and contact registry participants regarding potential matches. If the MyGene2 entry clearly represents the same individual or family enrolled in GenomeConnect, this match is excluded and the GenomeConnect participant is not contacted. Doing so helps increase participants’ ability to find other individuals and families with variants in the same gene to enable support and information exchange. To date, 53 GenomeConnect participants without matches within the registry have been contacted to inform them there may be a match in MyGene2. Individuals and families impacted by rare disease can feel isolated when there is a lack of available information regarding their disease or genetic test results (Zurynski et al., 2008). Facilitating connections between individuals and families can allow for exchange of meaningful informational and emotional support. Our experience reveals the need for a mechanism to automate and scale patient-initiated matching across rare-disease platforms to promote such connections.
GenomeConnect also facilitates matching between patients and clinicians, researchers, and laboratories to advance genomic medicine. If a researcher wishes to connect with a subset of participants based on gene, disease, or variant, GenomeConnect staff review the request and relay this invitation along to appropriate participants. Several researchers and clinicians have contacted GenomeConnect based on the registry’s ClinVar submissions to request additional phenotype data or invite participants to participate in research. By facilitating these connections, GenomeConnect protects patients’ privacy while contributing to the delineation of gene-disease relationships, benefiting the research community, clinicians, and patients.
Broadening Patient-Centered Data Sharing
GenomeConnect was one of the first patient registries to submit patient shared genetic and health information to ClinVar, but many other patient registries and advocacy organizations exist and can empower participants to share data in a similar way. Due to the success of GenomeConnect’s data sharing efforts, ClinGen aims to ensure this opportunity is available to other patient-focused organizations. Moving forward, ClinGen plans to partner with additional sources for patient data sharing, including other gene or condition specific registries and patient support and advocacy groups. Individual patients whether enrolled or not in a gene or condition specific registry can also enroll in GenomeConnect to share their genetic and health information. Gene or condition specific registries built on the same registry platform as GenomeConnect (PatientCrossroads, Inc. doing business as (dba.) AltaVoice, a wholly-owned subsidiary of Invitae Corporation), can elect to have GenomeConnect staff curate and submit data to ClinVar from participants who opt to share. All other features in GenomeConnect are also available to these participants.
Registries on other platforms can discuss data sharing opportunities with ClinGen to determine how such efforts can be supported. Support or advocacy groups that do not yet have a registry can elect to collaborate with ClinGen to enable participants to share data or create a separate registry with Invitae or another platform service. ClinGen is prioritizing engagement of groups that represent a disease area where ClinGen has existing gene and variant curation efforts (e.g. RASopathies and inborn errors of metabolism). Doing so will increase data available for curation efforts and can ultimately allow patients to receive updated variant interpretations that may be produced as a result. ClinGen is working to pilot these data sharing partnerships with several existing registries and advocacy groups interested in creating a disease specific registry.
Conclusions
Broad data sharing of genotypic and phenotypic information is needed to inform variant interpretation, gene-disease relationships, and actionability of genomic information to ultimately improve patient care. Through GenomeConnect and collaborations with external patient groups, ClinGen is partnering with patients in data sharing efforts. GenomeConnect’s experience to date highlights the utility of patient-shared data and the ways in which increasing patient engagement in genomic data sharing benefits both patients and the genomics community.
Supplementary Material
The current version of the GenomeConnect general health survey that displays question format and wording, HPO mappings and survey branching logic.
Acknowledgements
Most authors are clinical service providers noted in their affiliations. Vanessa Rangel Miller, Jud Rhode, and Jo Anne Vidal are employed by and own stock in Invitae.
This work is supported through the following grant: U41HG006834. ClinVar is supported, in part, by the intramural research program of the National Library of Medicine, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Patient data have been obtained in a manner conforming with IRB and/or granting agency ethical guidelines.
The authors would like to acknowledge Annie Niehaus, Tam Sneddon, and the members of the Education Work Group of the ClinGen Resource for their contributions to GenomeConnect and the data shared here as well as the ClinGen Sequence Variant Inter-Laboratory Discrepancy Resolution group and participating laboratories for their discrepancy resolution efforts. Finally, we would like to acknowledge our GenomeConnect participants that make this work possible.
References
- American Medical Association. (2013). Genome Analysis and Variant Identification Policy D-460.971. [Google Scholar]
- American College of Medical Genetics (ACMG) Board of Directors. (2017). Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 19, 721–722. doi: 10.1038/gim.2016.196 [DOI] [PubMed] [Google Scholar]
- Barsh GS, Cooper GM, Copenhaver GP, Gibson G, McCarthy MI, Tang H, … Williams SM (2015). PLOS Genetics Data Sharing Policy: In Pursuit of Functional Utility. PLoS Genetics 11(12), e1005716. doi: 10.1371/journal.pgen.1005716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, … Rehm HL (2017). Clinical Laboratories Collaborate to Resolve Differences in Variant Interpretations Submitted to ClinVar. Genetics in Medicine, 19, 1096–1104. doi: 10.1038/gim.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick BE, Riggs ER, Azzariti DR, Miller VR, Ledbetter DH, Miller DT, … Faucett WA on behalf of the ClinGen Resource. (2015) GenomeConnect: matchmaking between patients, clinical laboratories, and researchers to improve genomic knowledge. Human Mutation, 36, 974–978. doi: 10.1002/humu.22838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Ayme S, … Robinson PN (2017). The human phenotype ontology in 2017. Nucleic Acids Research, 45, D865–D876. doi: 10.1093/nar/gkw1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, & Maglott DR (2014). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research, 42, D980–D985. Doi: 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, … Maglott DR(2016). ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Research, 44, D862–D868. doi: 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JL, Benson M, Brown GR, Chao C, Chitipiralla S.., Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Research, 46(D1), D1062–D1067.doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health. (2014). Genomic Data Sharing. 79 FR 51345. https://www.gpo.gov/fdsys/pkg/FR-2014-08-28/pdf/2014-20385.pdf [Google Scholar]
- National Society of Genetic Counselors. (2015). Clinical Data Sharing. https://www.nsgc.org/p/bl/et/blogaid=330 [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, … Rehm, H.L. (2015). The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Human Mutation, 36, 915–921. doi: 10.1002/humu.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17, 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs ER, Jackson L, Miller DT, & Van Vooren S (2012). Phenotypic information in genomic variant databases enhances clinical care and research: the International Standards for Cytogenomic Arrays Consortium experience. Human Mutation, 33(5), 787–96. 10.1002/humu.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, … Watson MS (2015). ClinGen — The Clinical Genome Resource. New England Journal of Medicine, 372(23), 2235–2242. doi: 10.1056/nejmsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL (2017). A new era in the interpretation of human genomic interpretation. Genetics in Medicine, 19, 1092–1095. doi: 10.1038/gim.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN & Mundlos S (2010). The human phenotype ontology. Clinical Genetics, 77, 525–534. doi: 10.1111/j.1399-0004.2010.01436.x [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schienttecatte F, Valle D, & Hamosh A (2015). GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Human Mutation, 10, 928–930. doi: 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira NL, Arachchi H, Buske OJ, Chong JX, Hutton B, Foreman J, … Rehm HL (2017). Matchmaker Exchange. Current Protocols in Human Genetics. 95(1), 9.31.1–9.31.15. doi: 10.1002/cphg.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- University of Washington Center for Mendelian Genomics. (n.d.). MyGene2. Retrieved April 03, 2018, from https://mygene2.org/MyGene2/
- Zurynski Y, Frith K, Leonard H, & Elliott E (2008). Rare childhood diseases: How should we respond? Archives of Disease in Childhood, 93(12), 1071–1074. doi: 10.1136/adc.2007.134940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The current version of the GenomeConnect general health survey that displays question format and wording, HPO mappings and survey branching logic.