Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by social communication deficits and restricted, repetitive patterns of behavior. For more than a decade, genetically-modified, risk factor-induced, as well as naturally occurring rodent models for ASD have been used as the most predominant tools to dissect the molecular and circuitry mechanisms underlying ASD. However, the apparent evolutionary differences in terms of social behavior and brain anatomy between rodents and humans have become an issue of debate regarding the translational value of rodent models for studying ASD. More recently, genome manipulation of non human primates using lentivirus-based gene expression, TALEN and CRISPR/Cas9 mediated gene editing techniques, has been reported. Genetically modified non-human primate models for ASD have been produced and characterized. While the feasibility, value, and exciting opportunities provided by the non-human primate models have been clearly demonstrated, many challenges still remain. Here, we review current progress, discuss the remaining challenges, and highlight the key issues in the development of non-human primate models for ASD research and drug development.
Keywords: non-human primate, autism, social behavior, CRISPR/Cas9
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by impaired social interactions and restricted, repetitive behaviors. Research has shown that ASD involves both genetic and environmental factors and that genetic causes have been identified in approximately 25% of the patients [Iossifov et al., 2014; De Rubeis et al., 2014; Bourgeron, 2015; Krumm et al., 2015]. Whole exome or genome sequencing, targeted resequencing, and genome copy number analysis of ASD patients have confirmed the significance of deleterious mutations in approximately 70 genes or loci [Sanders et al., 2011, 2015]. The molecular heterogeneity of ASD, including the large number of low penetrant alleles relating to ASD, has created a significant challenge to those who have attempted to create animal models [McCarroll & Hyman, 2013]. Apart from genetic causes, epidemiological evidence also suggests that non-genetic factors, such as maternal use of anti-epilepsy drug of valproic acid (VPA) [Moore et al., 2000; Rasalam et al., 2005; Bromley et al., 2013] and maternal infection during pregnancy, may represent risk factors for ASD [Ashwood, Wills, & Van de Water, 2006; Atladottir et al., 2009; Lee et al., 2015].
Postmortem neuropathological and brain imaging studies of ASD patients have suggested several brain regions underlying the pathogenesis of ASD [Uddin, Supekar, & Menon, 2013; Di Martino et al., 2014; Lainhart, 2015]. Indeed, the examination of ASD postmortem brains has revealed neuroinflammation [Vargas, Nascimbene, Krishnan, Zimmerman, & Pardo, 2005; Gesundheit et al., 2013] and abnormalities in specific regions that are related to the core symptoms of ASD, such as the amygdala, prefrontal cortex, and striatum. The amygdala, which shows abnormal organization in ASD patients, is associated with emotion and motivation, and processes both fearful and rewarding stimuli [Kemper & Bauman, 1993]. Magnetic resonance imaging (MRI) has revealed enlargement of the cortical surface area in ASD infants [Shen et al., 2013; Hazlett et al., 2017]. However, these studies do not support the causality between the brain regions and ASD behaviors because of the apparent technical limitations in human studies. Rodents have been widely used to study the pathogenesis of ASD. However, the evolutionary differences in brain anatomy and social behavior between humans and rodents have posed the interesting debate of whether rodent models are of value in studying the pathological mechanisms underlying ASD. Recently, a few non-human primate models have been created to facilitate the study of ASD (Fig. 1 and Table 1) [Liu et al., 2016a; Chen et al., 2017; Zhao et al., 2017]. Here, we review the findings from these new non-human primate models for ASD and discuss the value, as well as the challenges, of using these models in translational research.
Figure 1.
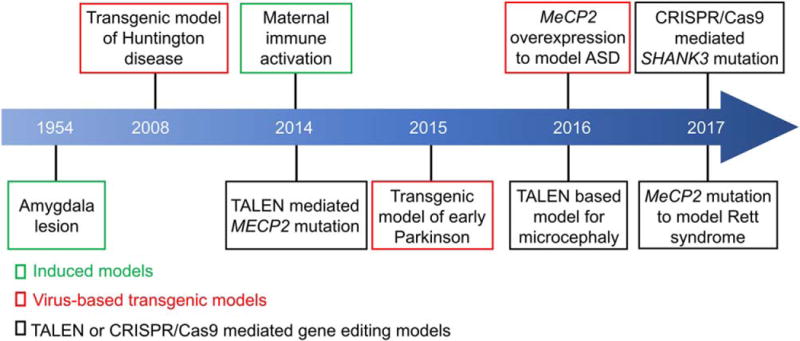
Timeline of brain disease models developed in non-human primates.
Table 1.
Phenotype Analysis of Non-Human Primate Models for ASD
| Autism models | Methods | Phenotypes
|
References | ||
|---|---|---|---|---|---|
| Behavioral | Cognitive | Imaging | |||
| Rett syndrome | TALEN | Fragmented sleep, increased sensory threshold, less social contact and more stereotypical behaviors | Defects in recognizing emotional expressions in eye-tracking test | Changes of cortical and sub-cortical volume by MRI | Chen et al., 2017 |
| MECP2 duplication | Overexpression | Repetitive circular routing, impaired social interaction and increased anxiety | Largely normal in WGTA assay | ND | Liu et al., 2016a |
| Maternal immune activation | a Poly IC injection | Increased repetitive behaviors, self-directed behaviors and abnormal social interactions | Abnormal gaze patterns to salient social information in eye-tracking test | Altered dendritic morphology |
Bauman et al., 2014; Machado et al., 2015; Rose et al., 2017 |
| b IgG-ASD injection | Inappropriate approach behavior | ND | Enlarged brain volume in males by MRI | Bauman et al., 2013 | |
| SHANK3 related autism | CRISPR/Cas9 | ND | ND | Fewer neurons and reduced spine density | Zhao et al., 2017 |
Pregnant rhesus macaques were injected with polyinosinic:polycytidylic acid (poly I:C), a double stranded RNA analog (viral mimic).
Pregnant rhesus macaques were injected with IgG isolated from mothers of children with ASD (IgG-ASD), which target fetal brain tissue.
Note: ND, not determined; MRI, magnetic resonance imaging.
Rodent Models for ASD
Rodent models recapitulating the molecular genetic defects associated with ASD in human patients serve as valid tools to dissect the molecular and cellular mechanisms underlying ASD. Numerous rodent models have been developed for known ASD risk genes and environmental risk factors; the comprehensive information can be found in the Autism Database (AutDB, http://autism.mindspec.org/autdb/AM_About.do and https://gene.sfari.org/) [Kumar et al., 2011; Abrahams et al., 2013]. Genetic models are developed based on well-studied monogenic or syndromic genes associated with ASD, such as MECP2, FMR1, NLGN3, TSC1/2, SHANK1/2/3, and UBE3A [Bey & Jiang, 2014; Hulbert & Jiang, 2016; Kumar et al., 2011]. Naturally occurring BTBR mice also exhibit a spectrum of autistic-like behaviors [McFarlane et al., 2008; Han, Tai, Jones, Scheuer, & Catterall, 2014]. Other rodent models for ASD are risk factors induced such as maternal VPA-exposure [Rodier, 2002; Rinaldi, Kulangara, Antoniello, & Markram, 2007], maternal autoantibodies, and maternal immune activation (MIA) [Camacho et al., 2014; Choi et al., 2016; Kim et al., 2017].
Evolutionary Differences between Rodents and Primates: molecular Pathways, Brain Structure, and Behavior
While rodent models have been widely used to model human genetic disorders over the last two decades, the evolutionary differences between these species have also been the focus for extensive discussion in terms of translational potential. Specifically, there are considerable differences between the mouse and human in many aspects, such as brain anatomy, cognitive capacity, and behavioral repertoire [Watson & Platt, 2012; Izpisua Belmonte et al., 2015; Jennings et al., 2016]. Several brain regions including the amygdala, the frontal lobe, the parietal cortex and the superior temporal cortex are implicated in social behaviors through brain lesion and imaging studies in humans [Adolphs, 2001; Insel & Fernald, 2004]. The brain regions of the orbitofrontal cortex and caudate nucleus are associated with the restricted, repetitive patterns of behavior of ASD patients [Amaral, Schumann, & Nordahl, 2008; Estes et al., 2011]. The key brain regions implicated in social behavior are remarkably similar between humans and non-human primates [Bauman & Schumann, 2018], but divergent in rodents.
Because of their strong similarity to humans in terms of brain anatomy, non-human primates are considered to be better models for ASD than rodents. Old World monkeys, mainly Macaca fascicularis and Macaca mulatta, are widely used in basic and translational biomedical research, for many reasons. Old World monkeys share a common ancestor with humans about 23 million years ago, while rodents and humans underwent divergence 70 million years ago [Norgren, 2013]. The ratio of encephalization quotient (a rough measure of brain size allometrically scaled to body size) ranges from 7 to 9 in humans and macaques, but is only 1 in rodents [Dorus et al., 2004]. The primate brain shows an enlarged prefrontal cortex and increased cortical folding compared with rodents. The prefrontal cortex plays an important role in decision-making, emotional regulation, attention control, and working memory. These differences may limit the translational value of knowledge learned from mouse models to be directly applied in human patients.
Transcriptome and epigenome studies have shown that differences dominate similarities between humans and mice [Lin et al., 2014]. The transcript isoforms repertoire varies markedly between humans and mice [Barbosa-Morais et al., 2012; Merkin, Russell, Chen, & Burge, 2012]. For instance, the G Protein-Coupled Receptor 56 (GPR56) splicing pattern differs extensively in mice and humans; the noncoding GPR56 exon 2 is only present in non-human primates and humans [Bae et al., 2014]. GPR56 regulates lateral cortical development in gyrencephalic mammals [Bae et al., 2014]. Furthermore, reduced neocortical NOS1 expression was found specifically in human fragile X syndrome patients, but not in FMRP deficient mice [Kwan et al., 2012]. This provides a partial explanation for the failure of mutant mice to reproduce key cognitive features of the patients with fragile X syndrome.
Non-human primates exhibit unique social behaviors and cognitive functions, which are in line with those of humans [Phillips et al., 2014]. Deficits in social information processing and emotional regulation are important aspects of ASD [Endevelt-Shapira et al., 2018]. Genetically-engineered mouse models frequently fail to recapitulate the social and communicative impairments of ASD patients [Hyman, 2014], because the social and environmental contexts that shaped human behavior are highly divergent from that of rodents during evolution. For instance, monkeys have forward-looking eyes to process social stimuli, while rodents rely more on olfaction. Abnormal eye gaze and reduced visual contact are common features of ASD patients [Guastella, Mitchell, & Dadds, 2008]. Monkeys also have the ability to recognize individuals from photos or videos, and the focus of eye gaze can be quantitatively analyzed [Phillips et al., 2014]. Thus, non-human primate models can provide more valuable and unique information than rodent models to understand how ASD-associated genetic mutations influence human brain development and behavior.
New Genome Editing Tools for Non-Human Primates
Lentivirus-based transgenic methods have been successfully used to generate transgenic monkey models. For example, in 2001, a retroviral vector injection method was used to create a transgenic rhesus monkey (Macaca mulatta) expressing green fluorescence protein (GFP) [Chan, Chong, Martinovich, Simerly, & Schatten, 2001]. This seminal work opened up a new era and eventually led to the creation of several lines of transgenic monkeys which can be used to model different brain disorders. Yang et al., generated the first nonhuman primate model for Huntington disease, in which the Huntingtin (HTT) mutant gene, responsible for the Huntington disease, was transfected into monkey zygotes by lentivirus (Fig. 1) [Yang et al., 2008]. These monkeys showed signs of neuronal death and motor defects. More recently, transgenic monkeys expressing mutant a-synuclein (A53T) were generated to mimic familial Parkinsonism [Niu et al., 2015], while transgenic monkeys overexpressing Rett syndrome-associated MECP2 protein exhibited defective social interactions [Liu et al., 2016a].
We have now entered a new era of non-human primate research, facilitated by the development of highly efficient and precise genome editing tools, such as TALEN (transcription activator-like (TAL) effector nucleases) and CRISPR/Cas9 gene editing systems (Fig. 2). Cynomolgus monkeys are more often used than rhesus monkeys for gene editing, as they have menstrual cycles throughout the entire year, while rhesus monkeys reproduce only seasonally. Niu and colleagues first used Cas9 mRNA and sgRNA to edit genes in cynomolgus monkeys [Niu et al., 2014]. Subsequently, Chen et al. used CRISPR/Cas9 to target the monkey dystrophin gene to mimic Duchenne muscular dystrophy (DMD) [Chen et al., 2015], while Zhao et al., generated three mutant offspring containing deleterious SHANK3 mutations by using CRISPR/Cas9 [Zhao et al., 2017]. In other studies, MECP2 (methyl-CpG binding protein 2) and MCPH1 (Microcephalin 1) were targeted by TALEN in cynomolgus monkeys to model Rett syndrome and microcephaly, respectively [Liu et al., 2014a; Liu et al., 2014b; Ke et al., 2016].
Figure 2.
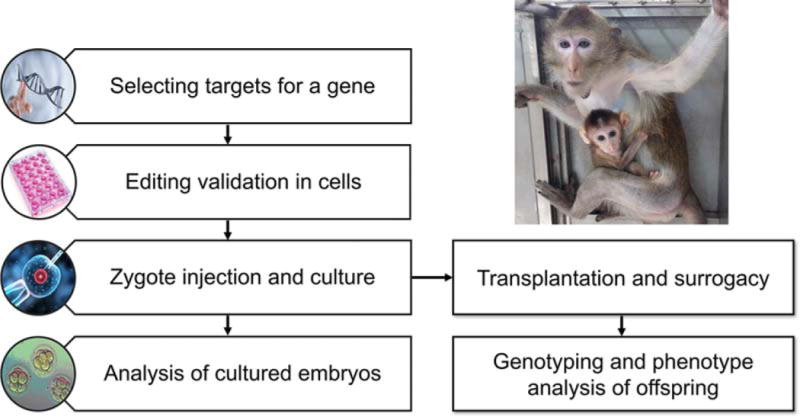
Diagram of TALEN- and CRISPR/Cas9-mediated gene editing to make ASD monkey models.
Monkey Models for ASD
Genetic and induced non-human primate models have been produced and characterized to allow us to investigate the pathological mechanisms of ASD (Fig. 1). Naturally-occurring genetic variations in non-human primates offer an excellent opportunity to exploit genotype-phenotype association, although relatively few studies have been reported thus far [Haus et al., 2014]. Madlon-Kay et al. reported naturally occurring variants of SHANK3 among a large free-ranging feral population of rhesus macaques on Cayo Santiago Island [Madlon-Kay et al., 2015]. It would be interesting to further explore the relationships between these genetic variants and social and cognitive performance in these monkeys.
The administration of MIA and VPA treatment during pregnancy were reported to induce ASD-related abnormalities in monkey offspring [Machado, Whitaker, Smith, Patterson, & Bauman, 2015; Yasue et al., 2015]. During pregnancy, maternal IgG antibodies that react to fetal brain proteins represent risk factors for ASD [Bauman et al., 2013]. Monkeys that are prenatally-exposed to antibodies from the mothers of autistic patients exhibit stereotyped behaviors and are more hyperactive compared with controls [Bauman et al., 2013]. Prenatal MIA has also been linked to altered brain development and behavior abnormalities in humans and animal models [Estes & McAllister, 2016; Kim et al., 2017]. Rhesus monkeys exposed to MIA show abnormal gaze to salient social information [Machado, Whitaker, Smith, Patterson, & Bauman, 2015], repetitive behaviors, and abnormal social interactions [Bauman et al., 2014]. Other studies have shown that VPA-treated marmosets do not recognize social interactions between human caregivers while untreated controls do [Yasue et al., 2015]. The progress of induced monkey models for ASD has been recently reviewed by Bauman and Schumann [Bauman & Schumann, 2018].
Genes implicated in ASD, such as MECP2 and SHANK3, have been targeted to produce non-human primate models of ASD [Liu et al., 2016a; Chen et al., 2017; Zhao et al., 2017]. MECP2 duplication syndrome, characterized by ASD, intellectual disability, and anxiety, is caused by a common genomic rearrangement. Monkeys overexpressing MECP2 mediated by lentivirus infection exhibit characteristic features associated with MECP2 duplication in humans such as abnormal social and repetitive circulating behavior, as well as increased anxiety [Liu et al., 2016a]. Rett syndrome is a severe form of ASD caused by mutations in MECP2. MECP2 deficient monkeys, induced by TALEN-mediated gene editing, show defects in social behavior, sleep, and cognitive function [Chen et al., 2017]. Heterozygous SHANK3 mutations and deletions are one of the most common genetic mutations found in ASD [Leblond et al., 2014]. Furthermore, SHANK3 deficit monkeys, induced by CRISPR/Cas9, show reduced levels of postsynaptic PSD95 protein and a subset of glutamate receptors, reduced spine density, and fewer mature neurons in the prefrontal cortex [Zhao et al., 2017].
Phenotyping Tools for Non-Human Primates
Similar to disease models in other species, phenotype analyses for mutant monkey models of ASD typically include biochemical, molecular, cellular, behavioral assays, and brain imaging. There are extensively validated social behavior experimental paradigms [Sclafani et al. 2016] and ethograms for non-human primates (Table 1). Robust and comprehensive methods to phenotype non-human primates are essential for the successful investigation of models for human ASD. A list of the currently available phenotype assays, focusing on behavior and cognitive analysis, and brain imaging, are shown in Table 1.
Behavioral analysis in monkeys is typically performed by scoring video-recorded behavioral data in both natural and designed experimental settings. The behaviors frequently examined include active social contacts, such as grooming and sitting together, environmental exploration, aggressive behavior, and stereotypical behavior. Stereotypical behaviors are repetitive and involuntary movements, such as hand flapping, body-rocking, and head-rolling. Monkeys with amygdala lesions show more self-directed stereotypies and spend less time in social interactions [Rosvold, Mirsky, & Pribram, 1954; Bauman, Toscano, Babineau, Mason, & Amaral, 2008]. MECP2 hypomorphic mutant monkeys exhibit less social interaction and more stereotypical behavior without aggressive behavior [Chen et al., 2017]. Many ASD patients have comorbid symptoms such as sleep disturbance, obsessive compulsive disorder, anxiety, and sensory processing problems. Increased sensory threshold and fragmented sleep have also been observed in MECP2 mutant monkeys [Chen et al., 2017]. Transgenic MECP2 duplication monkeys show defects in social interaction, increased repetitive circular locomotion, but no difference in the total time spent in locomotion [Liu et al., 2016a].
Eye tracking assays are useful in assessing the neural circuitry activity of the non-human primate brain. Visual attention is the ability to focus onto relevant aspects of the visual world but simultaneously ignore distracting aspects. Abnormal eye attention is considered to be one of the key features of ASD [Guastella, Mitchell, & Dadds, 2008]. In a manner similar to humans, non-human primates allow a measure of social information processing and facial expression recognition with an eye tracking system, which is not applicable in rodents [Gothard, Erickson, & Amaral, 2004; Machado, Whitaker, Smith, Patterson, & Bauman, 2015]. In vivo physiological assays record a small number of neurons in various regions of the brain in freely-moving monkeys [Moore & Armstrong, 2003]. Non-invasive electroencephalograms (EEGs) can also be used to record the network activity of cortical neurons in freely-moving monkey [Gil-da-Costa, Stoner, Fung, & Albright, 2013].
Brain imaging such as MRI/fMRI and diffusion tensor imaging (DTI) are routinely used to analyze brain structures and for longitudinal follow-up studies in humans [Pardini et al., 2009; Schumann et al., 2010]. Functional MRI (fMRI) is often used to assess changes in brain activity when performing a certain task. The longitudinal assessment of normal brain growth patterns using MRI has been reported in monkeys [Scott et al., 2016]. Studies of brain structures in ASD patients using MRI have also revealed abnormal brain growth during early childhood [Piven et al., 1995; Courchesne et al., 2001; Schumann & Nordahl, 2011]. MECP2-deficient monkeys exhibit abnormal brain development including reduced gray matter volume and cortical thickness in specific brain regions [Chen et al., 2017]. DTI can also provide unique contrast, quantitative diffusion metrics, and diffusion tractography to allow the investigation of white matter organization in monkey brains [Calabrese et al., 2015]. However, DTI and fMRI have not yet been applied to non-human primate models for ASD.
Aside from behavioral and imaging assays, molecular, cellular, and ultrastructural analysis techniques are available for rodents and can be readily used in studies involving non-human primates. For example, cellular analysis, using various neuronal and glial markers, has discovered fewer mature neurons and more astrocytes in SHANK3 deficit monkey brains, which have not been observed in Shank3 mutant mice [Zhao et al., 2017].
Challenges and Perspectives
Our ability to genetically manipulate non-human primates has certainly provided an unprecedented opportunity to model neurological problems in humans, particularly neuropsychiatric disorders. Successful generation of several non-human primate models for genetic diseases provide considerable support for the feasibility of gene manipulations in non-human primates. Although the feasibility of several other molecular systems has been demonstrated, we predict that CRISPR/Cas9 will be the predominant technique of the future. While the power of CRISPR/Cas9 gene-editing in non-human primates is clearly evident, this technique is still associated with several limitations in the manipulation of non-human primate genes such as inadequate gene editing efficiency, mosaicism, and off-target effects in the founder animal [Chen, Niu, & Ji, 2016; Zhao et al., 2017]. Variable targeting efficiency and low pregnancy rates have also been documented [Niu et al., 2014, Zhao et al., 2017]. Whether these issues are gene specific remains to be seen. The solution to resolve mosaicism and off-target mutations is to breed the founder animals to obtain germline transmission for the targeted mutation, but segregate out the off-targeted mutations. This is biologically feasible and simple in other species such as rodents, but can be challenging from a practical point-of-view in non-human primates. The frequency of germline transmission of mutations mediated by CRISPR/Cas9 remains to be established. The long reproductive cycle and the monotocous nature of breeding in macaques make this even more difficult, if not impossible, because of our inability to obtain a large colony in a short time period. On average, it will take almost 5 years (4 years for female sexual maturation and 5 months for pregnancy) for a founder animal to produce its first offspring through a natural reproduction cycle. A more rapid acquisition of monkey offspring, using testis xenograft techniques (testis tissues from juvenile male monkey are xenografted in nude mouse), was achieved in transgenic MECP2 mutants produced by lentivirus technology [Liu et al., 2016a,b]. The xenograft technique can shorten the F1 generation time to 2.5 years. Developing ASD models in marmosets, a New World monkey species, is another option because of their quick sexual maturity (about 14 months) and high fertility (80% chance to have twins) [Miller et al., 2016].
In addition to the limitations associated with the CRISPR/Cas9 technique, there remain other challenges. First, cost inevitably remains a daunting hurdle to overcome. The inability to practically produce a sufficient number of animals due to the long reproduction cycle may pose a significant challenge for experimental design and data analyses in some experiments. Second, the limited availability of phenotype analysis tools is likely to limit the value of non-human primate mutants in dissecting the pathophysiology of human disease, at least in the short term. Third, ethical concerns may be encountered; the use of non-human primates rather than other animal models requires greater ethical consideration of potential animal suffering and benefits of the research. Lastly, similar to behavioral experiment in other species, it is expected that a range of nongenetic factors, including food supply, living environment, and health of the experimental animals may exert a significant confounding factor on behavioral performances.
Despite these issues, a steady growth in the use of non-human primates as a model for ASD and other neuropsychiatric disorders is anticipated. The repeated failure to advance psychiatric drug development from studies on rodent models clearly supports the need for alternative models, and the non-human primate is certainly a logical option [Seok et al., 2013]. The partnership between academic and industrial entities is likely to be critically important in this endeavor. Modeling human ASD is a high priority for investment. It is likely that researchers will learn a great deal from characterizing monkey models for ASD. These monkey models will allow investigators to design new experimental paradigms with which to use non-human primates to better model ASD, and other human brain disorders.
Lay Summary.
Over the last two decades, genetically modified rat and mouse models have been used as the most predominant tools to study mechanisms underlying autism spectrum disorder (ASD). However, the apparent evolutionary differences between rodents and humans limit the translational value of rodent models for studying ASD. Recently, several non-human primate models for ASD have been established and characterized. Here, we review current progress, discuss the challenges, and highlight the key issues in the development of non-human primate models for ASD research and drug development.
Acknowledgments
We are grateful to Dr. David Amaral and Dr. Xiao-jiang Li for introducing us to non-human primates. We thank Qiqi Wang for discussion and critical reading of the manuscript. The study was supported by grants from the Ministry of Science and Technology (2014CB942803) and the National Natural Science Foundation of China (31490592) to Y. Q. Zhang, and National Institute for Health grants (MH098114, MH104316, HD087795) to Y.H. Jiang.
Contributor Information
Hui Zhao, State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China; University of Chinese Academy of Sciences, Beijing, China.
Yong-Hui Jiang, Department of Pediatrics and Department of Neurobiology, Duke University, Durham, North Carolina 27710.
Yong Q. Zhang, State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Molecular Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs R. The neurobiology of social cognition. Current Opinion in Neurobiology. 2001;11:231–239. doi: 10.1016/s0959-4388(00)00202-6. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends in Neurosciences. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Ashwood P, Wills S, Van-de-Water J. The immune response in autism: A new frontier for autism research. Journal of Leukocyte Biology. 2006;80:1–15. doi: 10.1189/jlb.1205707. [DOI] [PubMed] [Google Scholar]
- Atladottir HO, Pedersen MG, Thorsen P, Mortensen PB, Deleuran B, Eaton WW, Parner ET. Association of family history of autoimmune diseases and autism spectrum disorders. Pediatrics. 2009;124:687–694. doi: 10.1542/peds.2008-2445. [DOI] [PubMed] [Google Scholar]
- Bae BI, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, Walsh CA. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bauman MD, Schumann CM. Advances in nonhuman primate models of autism: Integrating neuroscience and behavior. Experimental Neurology. 2018;299:252–265. doi: 10.1016/j.expneurol.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman MD, Iosif AM, Ashwood P, Braunschweig D, Lee A, Schumann CM, Amaral DG. Maternal antibodies from mothers of children with autism alter brain growth and social behavior development in the rhesus monkey. Translational Psychiatry. 2013;3:e278. doi: 10.1038/tp.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, Patterson PH. Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biological Psychiatry. 2014;75:332–341. doi: 10.1016/j.biopsych.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman MD, Toscano JE, Babineau BA, Mason WA, Amaral DG. Emergence of stereotypies in juvenile monkeys (Macaca mulatta) with neonatal amygdala or hippocampus lesions. Behavioral Neuroscience. 2008;122:1005–1015. doi: 10.1037/a0012600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey AL, Jiang YH. Overview of mouse models of autism spectrum disorders. Curr Protoc Pharmacol. 2014;66:56661–656626. doi: 10.1002/0471141755.ph0566s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature Reviews Neuroscience. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- Bromley RL, Mawer GE, Briggs M, Cheyne C, Clayton-Smith J, Garcia-Finana M, Baker GA. The prevalence of neurodevelopmental disorders in children prenatally exposed to antiepileptic drugs. Journal of Neurology Neurosurgery and Psychiatry. 2013;84:637–643. doi: 10.1136/jnnp-2012-304270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage. 2015;117:408–416. doi: 10.1016/j.neuroimage.2015.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho J, Jones K, Miller E, Ariza J, Noctor S, de Water JV, Martínez-Cerdeño V. Embryonic intraventricular exposure to autism-specific maternal autoantibodies produces alterations in autistic-like stereotypical behaviors in offspring mice. Behavioural Brain Research. 2014;266:46–51. doi: 10.1016/j.bbr.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- Chen Y, Niu Y, Ji W. Genome editing in nonhuman primates: approach to generating human disease models. Journal of Internal Medicine. 2016;280:246–251. doi: 10.1111/joim.12469. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, Sun YE. Modeling Rett syndrome using TALEN-edited MECP2 mutant cynomolgus monkeys. Cell. 2017;169:945–955. doi: 10.1016/j.cell.2017.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Li XJ. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Human Molecular Genetics. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Huh JR. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, Courchesne RY. Unusual brain growth patterns in early life in patients with autistic disorder: An MRI study. Neurology. 2001;57:245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Milham MP. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Endevelt-Shapira Y, Perl O, Ravia A, Amir D, Eisen A, Bezalel V, Sobel N. Altered responses to social chemosignals in autism spectrum disorder. Nature Neuroscience. 2018;21:111–119. doi: 10.1038/s41593-017-0024-x. [DOI] [PubMed] [Google Scholar]
- Estes A, Shaw DW, Sparks BF, Friedman S, Giedd JN, Dawson G, Dager SR. Basal ganglia morphometry and repetitive behavior in young children with autism spectrum disorder. Autism Research. 2011;4:212–220. doi: 10.1002/aur.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes ML, McAllister AK. Maternal immune activation: Implications for neuropsychiatric disorders. Science. 2016;353:772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesundheit B, Rosenzweig JP, Naor D, Lerer B, Zachor DA, Prochazka V, Ashwood P. Immunological and autoimmune considerations of autism spectrum disorders. Journal of Autoimmunity. 2013;44:1–7. doi: 10.1016/j.jaut.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Gil-da-Costa R, Stoner GR, Fung R, Albright TD. Nonhuman primate model of schizophrenia using a noninvasive EEG method. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15425–15430. doi: 10.1073/pnas.1312264110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, Erickson CA, Amaral DG. How do rhesus monkeys (Macaca mulatta) scan faces in a visual paired comparison task? Animal Cognition. 2004;7:25–36. doi: 10.1007/s10071-003-0179-6. [DOI] [PubMed] [Google Scholar]
- Guastella AJ, Mitchell PB, Dadds MR. Oxytocin increases gaze to the eye region of human faces. Biological Psychiatry. 2008;63:3–5. doi: 10.1016/j.biopsych.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Han S, Tai C, Jones CJ, Scheuer T, Catterall WA. Enhancement of inhibitory neurotransmission by GABAA receptors having alpha2,3-subunits ameliorates behavioral deficits in a mouse model of autism. Neuron. 2014;81:1282–1289. doi: 10.1016/j.neuron.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haus T, Ferguson B, Rogers J, Doxiadis G, Certa U, Rose NJ, Roos C. Genome typing of nonhuman primate models: Implications for biomedical research. Trends in Genetics. 2014;30:482–487. doi: 10.1016/j.tig.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, Piven J. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542:348–351. doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SW, Jiang YH. Monogenic mouse models of autism spectrum disorders: Common mechanisms and missing links. Neuroscience. 2016;321:3–23. doi: 10.1016/j.neuroscience.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE. How far can mice carry autism research? Cell. 2014;158:13–14. doi: 10.1016/j.cell.2014.06.032. [DOI] [PubMed] [Google Scholar]
- Insel TR, Fernald RD. How the brain processes social information: Searching for the social brain. Annual Review of Neuroscience. 2004;27:697–722. doi: 10.1146/annurev.neuro.27.070203.144148. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izpisua-Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Zhang F. Brains, genes, and primates. Neuron. 2015;86:617–631. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings CG, Landman R, Zhou Y, Sharma J, Hyman J, Movshon JA, Feng G. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nature Neuroscience. 2016;19:1123–1130. doi: 10.1038/nn.4362. [DOI] [PubMed] [Google Scholar]
- Ke Q, Li W, Lai X, Chen H, Huang L, Kang Z, Xiang AP. TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Research. 2016;26:1048–1061. doi: 10.1038/cr.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper TL, Bauman ML. The contribution of neuropathologic studies to the understanding of autism. Neurologic Clinics. 1993;11:175–187. [PubMed] [Google Scholar]
- Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Huh JR. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Eichler EE. Excess of rare, inherited truncating mutations in autism. Nature Genetics. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Wadhawan R, Swanwick CC, Kollu R, Basu SN, Banerjee-Basu S. Animal model integration to AutDB, a genetic database for autism. BMC Medical Genomics. 2011;4:15. doi: 10.1186/1755-8794-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rasin MR, Sestan N. Species-dependent post-transcriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainhart JE. Brain imaging research in autism spectrum disorders: in search of neuropathology and health across the lifespan. Current Opinion in Psychiatry. 2015;28:76–82. doi: 10.1097/YCO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, Bourgeon T. Meta-analysis of SHANK mutations in autism spectrum disorders: A gradient of severity in cognitive impairments. PLoS Genetics. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Magnusson C, Gardner RM, Blomstrom A, Newschaffer CJ, Burstyn I, Dalman C. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behavior and Immunity. 2015;44:100–105. doi: 10.1016/j.bbi.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Snyder MP. Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Ji W. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell. 2014a;14:323–328. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, Qiu Z. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. 2016a;530:98–102. doi: 10.1038/nature16533. [DOI] [PubMed] [Google Scholar]
- Liu Z, Nie YH, Zhang CC, Cai YJ, Wang Y, Lu HP, Sun Q. Generation of macaques with sperm derived from juvenile monkey testicular xenografts. Cell Research. 2016b;26:139–142. doi: 10.1038/cr.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou X, Zhu Y, Chen ZF, Yu B, Wang Y, Qiu Z. Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neuroscience Bulletin. 2014b;30:381–386. doi: 10.1007/s12264-014-1434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CJ, Whitaker AM, Smith SE, Patterson PH, Bauman MD. Maternal immune activation in nonhuman primates alters social attention in juvenile offspring. Biological Psychiatry. 2015;77:823–832. doi: 10.1016/j.biopsych.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlon-Kay S, Bey A, Passman R, Brent L, Watson K, Skene P, Jiang Y. 2015 Neuroscience Meeting Planner. Chicago, IL: Society for Neuroscience; 2015. SHANK3 single nucleotide variant associated with social behavior in rhesus macaques. Online Program No. 306. 03. [Google Scholar]
- McCarroll SA, Hyman SE. Progress in the genetics of polygenic brain disorders: significant new challenges for neurobiology. Neuron. 2013;80:578–587. doi: 10.1016/j.neuron.2013.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral pheno-types in BTBR T1tf/J mice. Genes Brain and Behavior. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Freiwald WA, Leopold DA, Mitchell JF, Silva AC, Wang X. Marmosets: A neuroscientific model of human social behavior. Neuron. 2016;90:219–233. doi: 10.1016/j.neuron.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SJ, Turnpenny P, Quinn A, Glover S, Lloyd DJ, Montgomery T, Dean JC. A clinical study of 57 children with fetal anticonvulsant syndromes. Journal of Medical Genetics. 2000;37:489–497. doi: 10.1136/jmg.37.7.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Armstrong KM. Selective gating of visual signals by microstimulation of frontal cortex. Nature. 2003;421:370–373. doi: 10.1038/nature01341. [DOI] [PubMed] [Google Scholar]
- Niu Y, Guo X, Chen Y, Wang CE, Gao J, Yang W, Li XJ. Early Parkinson’s disease symptoms in alpha-synuclein transgenic monkeys. Human Molecular Genetics. 2015;24:2308–2317. doi: 10.1093/hmg/ddu748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Norgren RB., Jr Improving genome assemblies and annotations for nonhuman primates. ILAR Journal. 2013;54:144–153. doi: 10.1093/ilar/ilt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardini M, Garaci FG, Bonzano L, Roccatagliata L, Palmieri MG, Pompili E, Emberti GL. White matter reduced streamline coherence in young men with autism and mental retardation. European Journal of Neurology. 2009;16:1185–1190. doi: 10.1111/j.1468-1331.2009.02699.x. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, T Hart BA, Voytko ML. Why primate models matter. American Journal of Primatology. 2014;76:801–827. doi: 10.1002/ajp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piven J, Arndt S, Bailey J, Havercamp S, Andreasen NC, Palmer P. An MRI study of brain size in autism. American Journal of Psychiatry. 1995;152:1145–1149. doi: 10.1176/ajp.152.8.1145. [DOI] [PubMed] [Google Scholar]
- Rasalam AD, Hailey H, Williams JH, Moore SJ, Turnpenny PD, Lloyd DJ, Dean JC. Characteristics of fetal anticonvulsant syndrome associated autistic disorder. Developmental Medicine and Child Neurology. 2005;47:551–555. doi: 10.1017/s0012162205001076. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Kulangara K, Antoniello K, Markram H. Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13501–13506. doi: 10.1073/pnas.0704391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM. Converging evidence for brain stem injury in autism. Developmental Psychopathology. 2002;14:537–557. doi: 10.1017/s0954579402003085. [DOI] [PubMed] [Google Scholar]
- Rose DR, Careaga M, Van-de-Water J, McAllister K, Bauman MD, Ashwood P. Long-term altered immune responses following fetal priming in a non-human primate model of maternal immune activation. Brain Behavioral and Immunity. 2017;63:60–70. doi: 10.1016/j.bbi.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvold HE, Mirsky AF, Pribram KH. Influence of amygdalectomy on social behavior in monkeys. Journal of Comparative and Physiological Psychology. 1954;47:173–178. doi: 10.1037/h0058870. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, State MW. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, State MW. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Nordahl CW. Bridging the gap between MRI and postmortem research in autism. Brain Research. 2011;1380:175–186. doi: 10.1016/j.brainres.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Bloss CS, Barnes CC, Wideman GM, Carper RA, Akshoomoff N, Courchesne E. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. Journal of Neuroscience. 2010;30:4419–4427. doi: 10.1523/JNEUROSCI.5714-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani V, Del Rosso LA, Seil SK, Calonder LA, Madrid JE, Bone KJ, Parker KJ. Early predictors of impaired social functioning in male rhesus macaques (Macaca mulatta) PLoS One. 2016;11:e0165401. doi: 10.1371/journal.pone.0165401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Grayson D, Fletcher E, Lee A, Bauman MD, Schumann CM, Amaral DJ. Longitudinal analysis of the developing rhesus monkey brain using magnetic resonance imaging: birth to adulthood. Brain Structure & Function. 2016;221:2847–2871. doi: 10.1007/s00429-015-1076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MD, Nordahl CW, Young GS, Wootton-Gorges SL, Lee A, Liston SE, Amaral DJ. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain. 2013;136:2825–2835. doi: 10.1093/brain/awt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Menon V. Reconceptualizing functional brain connectivity in autism from a developmental perspective. Frontiers in Human Neuroscience. 2013;7 doi: 10.3389/fnhum.2013.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Annals of Neurology. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- Watson KK, Platt ML. Of mice and monkeys: Using non-human primate models to bridge mouse- and human-based investigations of autism spectrum disorders. Journal of Neurodevelopmental Disorders. 2012;4:21. doi: 10.1186/1866-1955-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, Chan AW. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasue M, Nakagami A, Banno T, Nakagaki K, Ichinohe N, Kawai N. Indifference of marmosets with prenatal valproate exposure to third-party non-reciprocal interactions with otherwise avoided non-reciprocal individuals. Behavioural Brain Research. 2015;292:323–326. doi: 10.1016/j.bbr.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tu Z, Xu H, Yan S, Yan H, Zheng Y, Zhang YQ. Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Research. 2017;27:1293–1297. doi: 10.1038/cr.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]


