Abstract
Somatic genome instability is a hallmark of cancer genomes and has been linked to aging and a variety of other pathologies. Large-scale cancer genome and exome sequencing have revealed that mutation load and spectra in cancers can be influenced by environmental exposures, the anatomical site of exposures, and tissue type. There is now an abundance of data favoring the hypothesis that a substantial portion of the mutations in cancers originate prior to carcinogenesis in stem-cells of the healthy individual. Rapid advances in sequencing of non-cancer cells from healthy humans have shown that their mutation loads and spectra resemble cancer data. Like cancer genomes, mutation profiles of healthy cells show marked intra-individual variation, thus providing a metric of the various factors—environmental and endogenous— involved in mutagenesis in these individuals. This review focuses on the current methodologies to measure mutation loads and to determine mutation signatures for evaluating the environmental and endogenous sources of DNA damage in human somatic cells. We anticipate that in future, such large-scale studies aimed at exploring the landscapes of somatic mutations across different cell-types in healthy people would provide a valuable resource for designing personalized preventative strategies against diseases associated with somatic genome instability.
Introduction
The adult human body consists of 1014 cells that arose from a single zygote via cell division. During the divisions and in the non-dividing terminally differentiated stage, each cell has the ability to acquire mutations from both endogenous and environmental processes. It has been reported that a cell may encounter more than 70,000 DNA lesions per day (Lindahl and Barnes, 2000; Tubbs and Nussenzweig, 2017). Such lesions may be due to spontaneous cytosine deamination, endogenous oxidative damage, or due to exogenous DNA damaging agents which include ultra-violet radiation, X-rays and tobacco smoke. If left unrepaired or if erroneously repaired, these lesions may result in DNA nucleotide substitutions (also termed as single nucleotide variations – SNVs), small or large insertions and deletions (indels), copy number variations (CNVs) and gross chromosomal rearrangements (GCRs). Moreover, DNA replication, transcription and recombination can destabilize and mutagenize DNA, which further adds to the genome mutation burden. To avoid deleterious outcomes of DNA damage, cells have evolved a large repertoire of DNA repair pathways. Each pathway is specialized for a subset of lesions with defects leading to increased rates of DNA damage and mutagenesis. Thus, the somatic mutation burden, spectra and landscape can collectively act as a lifetime record reflecting the environmental exposures of the individuals and the efficacy of DNA repair processes in their cells.
With the advent of large-scale next generation genome and exome sequencing projects, a vast variety of cancer types have been sequenced. Since cell populations in tumors are highly clonal, most of the somatic mutations are found in high fraction sequence reads making mutation calls reliable and amenable to validation. Analysis of mutational burden and spectra in many thousands of tumor genomes led to the striking discovery that mutation profiles vary amongst cancer types based on cell-type and location in the body, and the known DNA damaging agents the individuals may have been exposed to over time (Alexandrov et al., 2013; Lawrence et al., 2013; Roberts and Gordenin, 2014; Roberts et al., 2013). On the other hand, cell populations in the specimens directly collected from non-cancerous tissues are mostly non-clonal thus not allowing for similar analyses in healthy individuals. As such, the somatic genome mutation loads and spectra from healthy individuals is largely an untapped resource. Advances in single-cell genome sequencing, and newer technologies to isolate single-cell-derived clones are providing unforeseen insights into this field. This review is focused on the current methodologies aimed to measure mutation loads and determine mutation spectra in non-cancerous somatic cells. We also highlight how such data can be used to understand the impact of environmental and endogenous DNA damage in the individual’s cells across the human body and through the individual’s lifetime. Understanding the mutation loads and spectra in cancer-free individuals is essential to providing the base-line for defining norm and pathology in human somatic genome instability.
Somatic Mutations associated with diseases and with aging
The mutator phenotype hypothesis postulates that increased mutation rates in cancer cells provides a fitness advantage by leading to tumor heterogeneity which may further contribute to the generation of resistant cell populations to chemotherapeutic agents (Loeb, 2001). Several examples can be found for factors leading to increased spontaneous or induced mutation rates in cells that are associated with heightened cancer risk and poor prognosis (Aaltonen et al., 1993; Cleaver and Crowley, 2002; Cunningham et al., 1998; Gansmo et al., 2017; Hart et al., 1977; Hecht, 2008; Joo et al., 2016; Loeb et al., 1974; Parsons et al., 1993). Over the last decade, several international consortia such as The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), Pan Cancer Analysis of Whole Genomes (PCAWG) have undertaken efforts toward the sequencing and annotation of mutations in cancer exomes and genomes. Somatic mutation loads in tumors were found to be highly variable across cancer types ranging from ~100 mutations to >106 mutations per genome. This variability could in part be explained by the differences in the mutator phenotypic manifestation in the cancers, as well as the replicative history of the tissue, the age of cancer onset and the source of mutations, such that childhood tumors like Rhabdoid tumors or tumors in neuronal cells tend to have lower number of somatic mutations while, cancers with known strong environmental mutagen exposures like lung and skin tend to have higher mutation loads (Alexandrov et al., 2013; Lawrence et al., 2013). Moreover, mutation spectra in the samples resembled known spectra of the environmental mutagens, such that lung cancers were predominantly C→A changes, characteristic of oxidative damage, while skin melanomas were characterized by a majority of C→T changes, attributable to ultra-violet radiation (Alexandrov et al., 2013; Lawrence et al., 2013). Post zygotic mutations, have also been implicated in the pathology of a variety of diseases, other than cancer ranging from neurodevelopmental disorders, including the autism spectrum disorders, skin abnormalities like incontinentia pigmenti and syndromes with more widespread phenotypes including Proteus syndrome, Klippel–Trenaunay syndrome and Sturge–Weber syndrome (Table 1) and reviewed in (Erickson, 2003, 2010, 2014, 2016).
Table 1.
Examples of diseases other than cancer associated with somatic genome changes and mosaicism
| Type of Instability | Tissue Tested | Examples of Diseases |
|---|---|---|
| Single base substitutions and small insertion deletions | Blood | Chronic infantile neurologic cutaneous articular, CHARGE syndrome, Paroxysmal nocturnal hemoglobinuria, Congential central hypoventilatilation syndrome, Costello syndrome, Hemophilia A and B, Hereditary spastic parapelagia, Hunter disease, Hypocalcemia, Loeys-Dietz, Marfans, MYH9 disorders, Neonatal diabetes, Ornithine transcarbamylase deficiency, Retinoblastoma, Rett syndrome, Skeletal dysplasia, von Hippel-Lindau disease |
| Skin cells and hair roots | Androgen insensitivity, McCune-Albright syndrome, EEC (ectrodactyly, ectodermal dysplasia, and orofacial clefts), Hutchinson-Gilford progeria, Lesch-Nyhan, Loeys-Dietz, MYH9 disorders, Ornithine transcarbamylase deficiency, X-linked hypophosphatemic rickets | |
| Buccal cells | Chronic infantile neurologic cutaneous articular (CINCA), Epidermolysis bullosa simplex, Loeys-Dietz, MYH9 disorders | |
| Cerebral cortex | Alzheimer disease | |
| Skeletal tissue | Osteogenesis imperfecta | |
| Deletions | Blood | Campomelic dysplasia, Hemophilia A and B, Infantile spinal muscular atrophy, Rubinstein-Taybi, X-linked dyskeratosis congenita |
| Skin | Incontinentia pigmenti, Neurofibromatosis 1 and 2 | |
| Nervous system | Autism spectrum disorders, Incontinentia pigmenti, Neurofibromatosis 1 and 2 | |
| Duplications | Blood | X linked mental retardation |
| Repetitive element instability | Blood | Fascioscapular humeral muscular dystrophy |
Table adapted from (Erickson, 2003, 2010, 2014, 2016)
Both environmental and endogenous DNA damaging factors continuously mutagenize the somatic genomes of the human body’s cells over an individual’s lifetime. Increase in mutation loads with age has been documented in both model organisms and human cells from healthy or cancerous tissues. Moreover, defects in DNA repair pathways that could increase mutation load are often associated with diseases that manifest as premature aging (Gregg et al., 2012; Hoeijmakers, 2009; Loeb et al., 1974). Studies with reporter transgenes introduced in mouse liver, brain, heart, small intestine and spleen demonstrated an age-dependent increase in transgene mutation frequencies (Dolle et al., 1997; Dolle and Vijg, 2002). A similar age-associated increase in transgene mutation frequencies was demonstrated for Drosophila melanogaster. Interestingly, increased temperature, which has been shown to decrease the fly lifespan, had a dramatic impact on accelerating the mutation rates in the flies, implying that mutation loads and organismal fitness can be inversely correlated (Garcia et al., 2010), however, one cannot rule out other potential impacts of temperature on lifespan modulation in flies. Analysis of mutations in the HPRT, HLA-A, T-cell receptor and glycophorin A genes in human cells also demonstrated an increase in somatic mutation load with age (Cibulskis et al., 2013; Davies et al., 1992; Koboldt et al., 2012; Kroigard et al., 2016; Van der Auwera et al., 2013). In both mice and humans, the rate of increase in mutation frequencies varied in the tissues implying a dependence of the mutation loads on both the replicative potential of the cells and on varying exposures to DNA damaging agents in the tissues (Reviewed in (Kennedy et al., 2012)). In addition to single base substitutions, increase in other forms of genome instability including large-scale GCRs and CNVs (Dolle et al., 2000; Dolle and Vijg, 2002; Ramsey et al., 1995; Vijg and Dolle, 2002), and increased expression of retrotransposons and elevated L1 copy numbers, likely indicating increased transposition events (Lawrence et al., 2013), have also been seen to be associated with aging and age-associated diseases.
In further support of the association between increased somatic mutation loads and age, are the cancer-based studies of genome instability. Cancer is a known age-associated pathology and the cumulative risk of developing cancer has been shown to increase with age up to 70 years (White et al., 2014). Somatic genome instability was found to correlate with age of the patients at the time of cancer detection (Milholland et al., 2015; Tomasetti et al., 2013), and analyses of mutations in cancer genomes suggested that age-associated mutation loads double every 8 years (Podolskiy et al., 2016). Importantly, tissues with high stem-cell turnover, demonstrate higher cancer risk, likely due to increased probabilities of accumulating potentially deleterious mutations (Tomasetti and Vogelstein, 2015).
Somatic mutation load and spectra in healthy cells
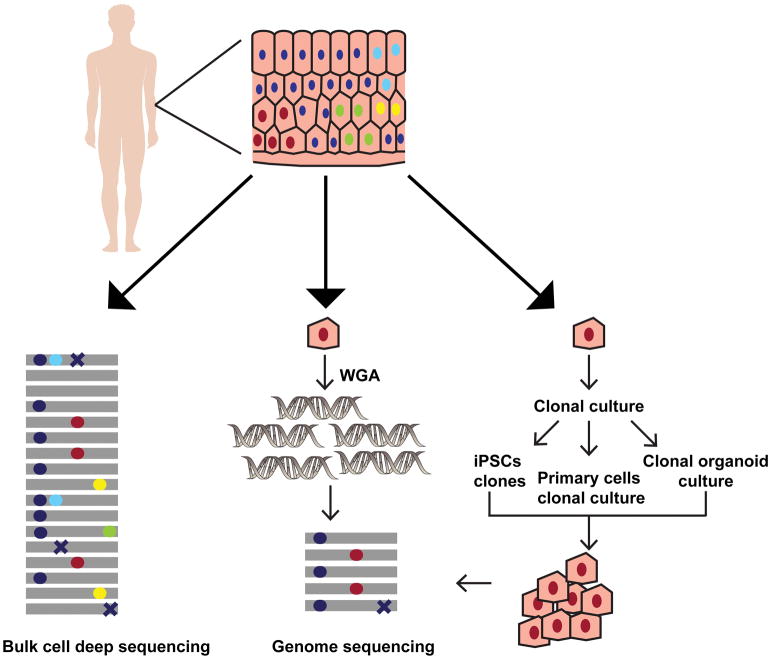
While it is estimated that more than half the mutations in cancer somatic genomes, arise in healthy cells prior to carcinogenesis (Tomasetti et al., 2013), the knowledge about and accurate measurements of somatic mutations load and spectra in non-cancerous human cells are limited. In recent years, novel technological advances in obtaining clonal populations or in sequencing single cells have initiated the progress in the accrual of genomic data for healthy human tissues and cells (Figure 1). Below we describe the methodologies developed for NextGen genome sequencing of healthy somatic cells and tissues.
Figure 1. Overview of the methods for detecting somatic mutations in healthy human cells.
The various approaches to analyze somatic mutations in a heterogenous tissue sample are depicted using skin as an example. The colored circles (red, cyan, yellow and green) in the tissue denote nuclei within the cells with different somatic mutations respectively. The dark blue circles denote nuclei with germline polymorphisms present across all cells in the tissue. DNA extraction from the bulk cells followed by deep sequencing provides a measure of the diverse somatic mutations present in the sample (left panel). The gray lines denote individual reads in NGS, and the colored circles on them imply heterozygous somatic mutations or germline polymorphisms present in the tissue. The somatic mutations in bulk sequencing samples are often present in very low allele fractions. In addition, sequencing errors (crosses) may further confound analysis in deep-sequenced heterogenous samples. To determine the somatic mutations in a given cell whole genome amplification from a single cell followed by WGS may be used (middle panel). On the other hand, pluripotent stem cells may be derived from the tissue and grown clonally to get enough number of cells for WGS. The primary cells may also be either cultured directly, or organoid cultures may be established from the stem cells within the tissue allowing propagation in vitro to obtain DNA for WGS (right panel). In single cell sequencing, and clonal population sequencing somatic mutations are present as high frequency alleles, while errors generated during library preparation and sequencing are usually present in a smaller fraction of reads. Therefore, somatic mutation calling using these approaches is more accurate.
Deep sequencing of bulk cell populations
The heterogeneity of healthy tissue poses a problem for reliable and comprehensive identification of somatic variants in a sample via standard sequencing methods. To detect rare somatic variants, present in low allele fractions in a sample, deep sequencing to ultra-high coverage of either a fraction of the genome or the entire genome may be utilized. Moreover, estimation of allele frequencies of the somatic variants detected can allow for assessment of the clonality of the sample. Deep sequencing of a panel of driver genes from small biopsies obtained from eyelids of patients that underwent blepharoplasty (eyelid lift surgery), revealed that somatic genome changes are abundant in healthy tissue, with no evidence of negative selection of potentially deleterious variants. The majority among the mutations detected by deep sequencing in these skin biopsies were C→T and CC→TT changes and likely rose due to UV-irradiation of skin cells during the lifetime of the donors (Table 2). Interestingly, based on low fraction of reads containing mutations, clonality in these sequenced samples was very low, implying that the somatic mutations were present in very small localized regions of skin with large heterogeneity within the tissue (Martincorena et al., 2015). Sequencing the exomes of the bulk cells from skin biopsies from the forearms of hips demonstrated a similar clonal architecture, wherein each biopsy that was 4mm in size contained at least 10 clonal lineages. The cells from the forearms carried a higher mutation burden, and UV-mutation spectra were prevalent in cells from the forearms (Saini et al., 2016).
Table 2.
Somatic mutation types, loads and spectra detected in healthy human cells.
| Somatic Variation | Tissue type analyzed | Load and spectra of somatic changes | Methodology used | Reference |
|---|---|---|---|---|
| Copy number changes | Brain | Large CNVs > 1Mb can occur in 13–41% of neurons from healthy individuals and hemimegalencephaly patients. | Single cell sequencing and single cell SNP arrays | (Cai et al., 2014; McConnell et al., 2013) |
| Skin | Approximately 30% skin fibroblasts have megabase-scale CNVs | iPSCs sequencing | (Abyzov et al., 2012) | |
| Skin and brain | 8–9% of the cells have at least 1 megabase-scale CNV | Single cell sequencing | (Knouse et al., 2016; Knouse et al., 2014) | |
| Skin | Skin fibroblasts have at least 1 somatic CNV, and ~30% cells have megabase-scale CNVs. Most CNVs are near known fragile genomic regions. | Single-cell-derived clonal lineage sequencing | (Saini et al., 2016) | |
| Structural Variations | Skin | All skin fibroblasts have at least 1 somatic structural variation. Deletions are the most abundant SV, however duplications, inversions and translocations were detected in the cells. Most SVs are near known fragile genomic regions. | Single-cell-derived clonal lineage sequencing | (Saini et al., 2016) |
| Retrotransposition | Brain | <0.6 somatic L1 retrotransposition events detected per neuron | Single cell sequencing | (Evrony et al., 2012) |
| Single base substitutions | Skin | 3760 mutations found across the 234 samples from four individuals. Prevalence of C→T and CC→TT changes characteristic of UV-induced mutations | Deep sequencing of 74 genes from eyelid biopsies | (Martincorena et al., 2015) |
| Brain | ~1500 somatic mutations per neuron. The major mutation signature was C→T changes at CpG motifs. | Single cell sequencing | (Lodato et al., 2015) | |
| Skin | ~600 to 13000 somatic mutations per skin fibroblast obtained from skin biopsies from the hips and forearms. Mutation load in sun-exposed skin is higher, with a prevalence of UV-mutation signature. | Single-cell-derived clonal lineage sequencing | (Saini et al., 2016) | |
| Skin | ~1000 somatic mutations per skin fibroblast obtained from donor underarm skin biopsy. | iPSCs sequencing | (Abyzov et al., 2017) | |
| Skin and Blood | 14 to 28 somatic mutations in protein coding genes and 391 somatic changes in endothelial progenitor cells from one 57 year old individual. | iPSCs and monoclonal EPCs sequencing | (Rouhani et al., 2016) | |
| Brain | 200–400 somatic mutations in neurons from 12–14 week old fetus. C→T changes at CpG motifs and C→A changes characteristic of oxidative damage were the prevalent mutation signatures. | Single-cell-derived clonal lineage sequencing | (Bae et al., 2018) | |
| Colon, small intestine and liver | 1000–3000 mutations per cell. Linear increase in mutation loads with age. C→T changes at CpG motif detected in small intestine and colon cells. Mutation signature attributable to an unknown source in liver cells. | Adult stem cells-derived organoids sequencing | (Blokzijl et al., 2016) |
A major caveat of deep sequencing bulk cells is the limitation of the current NGS technologies to accurately detect very low frequency somatic changes. The ability to detect rare variants is dependent on the number of reads obtained during sequencing, erroneous calls due to sequencing errors and the amount of the starting genetic material. As such, it may often not be feasible or cost effective to ultra-deep sequence genomes using the canonical NGS methods. As an alternative, DNA barcoding strategies have been developed wherein DNA molecules are uniquely barcoded at the ends with specific DNA sequence tags. The barcoded DNA is then PCR amplified and mutations present in >95% of the barcoded samples from the same barcoded molecule are considered true positives (Kinde et al., 2011). Alternatively, the individual strands of DNA may be barcoded with tag sequences incorporated within Illumina sequencing adapters followed by strand-specific PCR. The amplified copies of the barcoded molecules allow for sequencing of each DNA strand repeatedly, thus, generation of consensus read sequences from these drastically reduces errors made during amplification and sequencing. This allows for detection of variants at frequencies as low as 10−5 mutations per base pair (Schmitt et al., 2012). A further improvement to these techniques is the CypherSeq approach which clones the barcoded library into circular vectors that can be enriched using rolling circle amplification of targeted molecules, thus maximizing read depth without off-target amplification. CypherSeq has been shown to accurately detect variants as low as 2.4 × 10−7 mutations per base pair (Gregory et al., 2016). Optimization and usage of these sequencing strategies for high throughput sequencing of panels of targeted genes can permit detection of somatic mutations across populations with high accuracy.
Sequencing single cells
The somatic mutation load present in a single cell represents the mutations accumulated in that cell lineage over an individual’s lifetime. Although sequencing bulk cells provided the first approximation of the mutation load and spectra in non-cancerous cells, such studies cannot address cell-to-cell variability within a tissue and provide the mutation load and spectra within independent cells. A single diploid human cell contains approximately 6pg of DNA, therefore, whole genome amplification is a prerequisite for single cell DNA sequencing (Figure 1). Since the first single cell genome sequencing study by Navin et. al in 2011 (Navin et al., 2011), a variety of methods to amplify and sequence genomes from single cells have become available. Initial attempts to amplify the genome relied on PCR using either degenerate oligonucleotides, or primers targeting either specific sequences in the genomes or adaptors ligated to sheared genomic fragments. However, PCR-based methods were not efficient in yielding uniform coverage across the entire genome. The second generation of whole genome amplification methods including Multiple displacement amplification (MDA) or Multiple Annealing and Looping Based Amplification Cycles (MALBAC), involved isothermal amplification using the phi29 polymerase, which has a very low error-rate and high processivity with the ability to do strand displacement, allowing for large genomic regions to be amplified (Reviewed in (Gawad et al., 2016)). A major challenge in whole genome amplification (WGA) from single cells is the introduction of contaminating DNA from the environment. Performing single-cell WGA in microfluidic devices was found to drastically reduce such contamination (Blainey and Quake, 2011) and increase uniformity of genome coverage during both MDA and MALBAC approaches (Marcy et al., 2007; Yu et al., 2014). More recently, Chen et.al. developed Linear Amplification via Transposon Insertion (LIANTI) to linearly amplify single cell genomes. LIANTI involves DNA fragmentation by Tn5 insertions, and T7 transcription at promoters inserted during the transposition event followed by reverse transcription and cDNA synthesis to linearly amplify the DNA, thus reducing off target priming and the lack of exponential DNA synthesis abrogates amplification biases and artifacts (Chen et al., 2017). These technologies are revolutionizing the field of single cell DNA sequencing and are enabling efficient and accurate measurements in somatic cells that are difficult to culture in vitro.
Single cell DNA sequencing has been instrumental in understanding somatic genome variation in non-cancerous cells, especially in terminally-differentiated cell types that cannot be easily propagated in vitro to obtain sufficient amount of clonal DNA for high coverage genome sequencing. Using MDA or GenomePlex (a PCR-based WGA technique [Sigma Aldrich Cat# WGA4, (Arneson et al., 2008)] followed by either low coverage whole genome sequencing (WGS) (Cai et al., 2014), or a combination of both low coverage WGS and SNP arrays (McConnell et al., 2013), it was established that neuronal genomes often carry large copy number variations (CNV) leading to mosaicism in the brain. With a predisposition to deletions, large-scale CNVs spanning megabases of the chromosomes, and whole chromosome aneuploidies could be detected in 13% to 41% of the cells analyzed (Cai et al., 2014; McConnell et al., 2013). A slightly lower frequency (8–10%) was seen for the presence of large CNVs which were more than 5 Mb in size, in skin keratinocytes and neurons, when the sensitivity and selectivity of the CNV-calling algorithms was improved (Knouse et al., 2016) (Table 2).
Increasing the coverage for WGS studies to 60X allowed for estimation of the number of retrotransposition events and single nucleotide variants in single neurons. L1 retrotransposition was found to be a rare event with a frequency of >0.6 retrotranspositions per genome (Evrony et al., 2012). On the other hand, 1685 to 1793 somatic SNVs were detected per neuron, and upon accounting for false-discovery rate due to amplification errors, the average number of mutations per neuron ranged from 1458 to 1580 (Lodato et al., 2015). Interestingly, mutation loads were found to vary with the expression levels of genes and the predominant mutation types in neurons were C→T at CpG motifs, implying normal cellular transcription and repair input into mutagenesis in neurons. CNVs and SNVs were often also found to be shared between cells from the same individuals, suggesting that these changes may have occurred early during development and neurogenesis thus allowing to track developmental lineages within the tissues (Cai et al., 2014; Evrony et al., 2012; Evrony et al., 2015; Knouse et al., 2016; Lodato et al., 2015).
Combining single-cell sequencing techniques with strand-specific sequencing (StrandSeq) has further enabled detection of structural changes in the genome including inversions, which are otherwise difficult to sequence, with very high accuracy. In this approach, cells are grown in the presence of a thymidine analog 5-bromo-2′-deoxyuridin (BrdU) that is incorporated in the nascent strand during DNA replication. Irradiation by UV leads to nicking and removal of the BrdU-containing strand and allows for strand-specific DNA libraries to be created. Sequencing and alignment of the reads to the plus or minus strands maintains parental DNA directionality and permits identification of structural rearrangements (Falconer et al., 2012; Sanders et al., 2016). StrandSeq can provide information about sequence composition of homologous chromosomes, copy neutral rearrangements including translocations between homologs and between sister chromatids, which are currently unattainable by the conventional NGS approaches (Sanders et al., 2017). Single-cell sequencing of 47 cord blood samples using StrandSeq demonstrated the presence of 111 polymorphic inversions in the genomes with the smallest inversion being ~17kb in length (Sanders et al., 2016).
Overall, single cell WGS has already provided insights into the somatic mutation loads and spectra in healthy human tissues. However, the intrinsic error rates associated with whole genome amplification, and the lack of genetic material to orthogonally validate somatic mutations are still the major impediments preventing accurate measurements of mutations in somatic cells. Despite the advances in WGA techniques, these still lag behind bulk DNA sequencing which can obtain 90% genome coverage with read-depths as low as 4X. MDA on the other hand, only covers 73% of the genome at 25X average read-depth, while MALBAC is limited to approximately 34% of the genome at the same average read-depths (Zong et al., 2012). Such loss in genome coverage lead to high rate of SNV-dropouts and often only one of the alleles are covered in the final data (allelic-dropout). Efficiencies of detecting both alleles of known SNVs range from 91% in MALBAC to as low as 10% in MDA (Zong et al., 2012). LIANTI was found to outperform both methodologies and achieve upto 97% genome coverage at 30X average read depths. LIANTI was found to outperform both MALBAC and MDA and achieve upto 97% genome coverage at 30X average read depth with only a 17% allele dropout rate, compared to upto 65% allele dropout rates by the previous methodologies (Chen et al., 2017; Zong et al., 2012). Furthermore, regions of the genome carrying repetitive DNA and other structural or sequence elements that may prevent amplification are significantly underrepresented in WGA and downstream WGS (Zong et al., 2012). WGA methods also have high SNV false positive rates being ~4 × 10−5 for classical WGA techniques to 5.4 × 10−6 for LIANTI. The false positives could potentially be eliminated by independently sequencing multiple single-kindred cells or kindred clonal lineages without need for amplification and comparing mutation calls (Zong et al., 2012). Alternatively, false positive calls may be ablated by comparison of single-cell sequencing data with unamplified bulk cells from the same population. This method was first applied to reliably detect base substitutions introduced by the activity of the mutagen N-ethyl-N-nitrosourea in Drosophila and mouse cells (Gundry et al., 2012), and subsequently to evaluate concordant mutation calls in whole genome amplified single human somatic cells and bulk cells obtained from the same source (Dong et al., 2017). Furthermore, performing single-cell lysis on ice and treating DNA by Uracil-N-Glycosylase and endonucleases to eliminate temperature-induced cytosine deamination to uracil that may have occurred during DNA preparation and library preparation steps are expected to further decrease artefactual calls (Chen et al., 2017; Dong et al., 2017).
Sequencing clonal lineages
Sequencing single cell-derived clonal lineages that have been cultured in vitro up to sufficient cell numbers to provide DNA for WGS, can bypass the caveats of bulk and single cell genome sequencing mentioned above. Most adult human cell types are terminally differentiated are not amenable to growth in culture media. Reprogramming adult skin cells into induced-pluripotent stem cells (iPSCs), Abyzov et.al. found that almost 30% of skin cells carry large-scale copy number changes (Abyzov et al., 2012). Like the single-neuron WGS, skin cell-derived iPSCs harbor ~1000 somatic mutations per cell (Abyzov et al., 2017). An independent study involving sequencing of protein coding regions from iPSCs derived from skin cells showed the presence of 14 and 28 somatic mutations per cell, with no SNVs shared between the cells further corroborating the conclusion of very high mosaicism in skin. Interestingly, comparisons of iPSCs derived from the same source helped determine the mutation rate and spectra during reprogramming. The process of reprogramming terminally differentiated cells was found to be inherently mutagenic with a prevalence of oxidative damage-induced mutations (Ji et al., 2012; Rouhani et al., 2016).
Although iPSCs allow for sequencing of otherwise unculturable cell types, the increased copy number, structural genome changes, and elevated mutation rates during the process of reprogramming preclude accurate measurements of these changes in somatic cells (reviewed in (von Joest et al., 2016)). Two methodologies have been developed that involve the least manipulations to the cells and therefore provide the most accuracy in downstream mutation calling. The first approach involves directly obtaining single cells from a donor’s tissue, and culturing them in vitro to obtain single cell-derived clones (Figure 1 and(Bae et al., 2018; Saini et al., 2016)). This approach was used to measure mutation loads in fibroblasts obtained from the left and right hips and forearms of two healthy donors. After filtering to remove somatic genome changes that were present in the germline of the donors, or those that may have occurred during culture, each cell analyzed carried at least one structural and copy number variant and from 581 to 12,743 single nucleotide substitutions. The majority of the structural variants were present in the vicinity of known fragile sites, indicating that replication impediments at these loci are the primary cause of gross chromosomal rearrangements in somatic cells (Table 2). Validation by PCR and Sanger sequencing of ~10 exonic mutations from the cells and amplification and Sanger sequencing of the breakpoints of the structural variants, showed a high rate of true positive calls. The aging-associate C→T changes in CpG motifs (Table 3 and (Alexandrov et al., 2015)) were detected in all cells analyzed in the study. Furthermore, overall mutation loads in cells from the forearms exceed those in hips, and UV-signature mutations were prevalent in the sun-exposed samples.
Table 3.
Signatures associated with different mutational mechanisms in cancers and healthy human somatic cells.
| Mutation source | Signature motif | Signature motif (detailed) | Mutation signature | Rationale | References |
|---|---|---|---|---|---|
| CpG deamination | nCga | 5′ [a|t|g|c]C[g] 3′b | nCg→nTg | Measure of C→T mutagenesis in CpG dinucleotides. | (Alexandrov et al., 2013; Saini et al., 2016) |
| rCg | 5′ [a|g]C[g] 3′ | rCg→rTg | Helps differentiate mutagenesis at CpG motifs from UV-signature mutagenesis | (Saini et al., 2016) | |
| UV | yCn | 5′ [t|c]C[a|t|g|c] 3′ | yCn→yTn | Measure of C→T mutagenesis at cyclobutane dimers formed at yC dinucleotides by UV. | (Alexandrov et al., 2013; Berger et al., 2012; Saini et al., 2016) |
| nTt | 5′ [a|t|g|c]T[t] 3′ | nTt→nCt | Measure of T→C mutagenesis at thymine dimers formed by UV. Helps differentiate UV mutated samples from CpG and APOBEC mutated samples. |
(Saini et al., 2016) | |
| rTt | 5′ [a|g]T[t] 3′ | rTt→rCt | More specific than nTt→nCt signature of UV mutagenesis | (Saini et al., 2016) | |
| CC | 5′ CC 3′ | CC→TTc | Measure of mutagenesis at cyclobutane dimers formed at CC dinucleotides by UV. | (Berger et al., 2012; Saini et al., 2016) | |
| APOBEC3 | tCw | 5′[t]C[a|t] 3′ | tCw→tTw or tCw→tGw | Measure of C→T mutagenesis caused by APOBEC3-induced cytidine deamination | (Alexandrov et al., 2013; Roberts et al., 2013) |
| APOBEC3A | ytCad | 5′[t|c][t]C[a] 3′ | ytCa→ytTa or ytCa→ytGa | Measure of C→T mutagenesis caused by APOBEC3A-induced cytidine deamination | (Chan et al., 2015) |
| APOBEC3B | rtCad | 5′[a|g][t]C[a] 3′ | rtCa→rtTa or rtCa→rtGa | Measure of C→T mutagenesis caused by APOBEC3B-induced cytidine deamination | (Chan et al., 2015) |
| AID | wrChe | 5′ [a|t][a|g]C[a|t|c] 3′ | wrCh→wrTh or wrCh→wrGh | Measure of C→T mutagenesis caused by AID-induced cytidine deamination | (Aaltonen et al., 1993; Hart et al., 1977; Maul and Gearhart, 2010; Peled et al., 2008; Rogozin et al., 2016) |
| Temozolomide | nCy | 5′ [a|t|c|g]C[t|c] 3′ | nCy→nTy | Measure of C→T mutagenesis caused by base damage by the alkylating agent temozolomide. | (Johnson et al., 2014) |
| Aristolochic acid | cTr | 5′ [c]T[a|g] 3′ | cTr→cAr | Measure of T→A mutagenesis caused by base damage by aristolochic acid | (Hoang et al., 2013) |
| Replicative DNA polymerase ε | tCt | 5′[t]C[t] 3′ | tCt→tAt | Measure of C→A mutagenesis due to errors made by replicative polymerases | (Palles et al., 2013) |
| tCg | 5′[t]C[g] 3′ | tCg→tTg | Measure of C→T mutagenesis due to errors made by replicative polymerases | (Palles et al., 2013) | |
| DNA polymerase η | nTw | 5′[a|t|g|c]T[a|t] 3′ | nTw→nNw | Measure of T→N mutagenesis due errors made by translesion polymerase η. | (Rogozin et al., 2018; Rogozin et al., 2001) |
| NTHL1 defect | nCn | 5′ nCn 3′ | nCn→nTn | Measure of C→T mutagenesis due to defects in NTHL1 DNA glycosylase. | (Drost et al., 2017) |
| ERCC2 defect | broad spectrum | NA | Broad spectrum, similar to Signature 5 in COSMIC | Measure of mutagenesis due to ERCC2 defects. Mutations attributable to this signature also increase with age. | (Kim et al., 2016) |
Signature motifs are depicted in the trinucleotide context with −1 and +1 flanks exceptions specified below). Mutated residue is capitalized. IUPAC nomenclature is used to denote degenerate nucleotides.
Nucleotides for the flanking positions are shown in brackets; if various nucleotides are possible, then separated by “|”
CC→TT mutation signature is highly specific for UV-induced mutations, flanking nucleotides are not shown.
APOBEC3A and APOBEC3B-specific signatures take into account the −2 positions allowing differentiation between the activities of the two cytosine deaminases.
The AID-signature motif demonstrates specificity in the −2 nucleotide position and is a tetranucleotide motif.
The second approach to analyze clonal-lineages involves in vitro organoid culture from multipotent adult stem cells (Figure 1). Organoids are three dimensional multicellular cultures that mimic in vivo tissue. These can be constructed using either embryonic or induced pluripotent stem cells, or tissue specific adult stem cells. Organoids are suspended in extracellular matrix and cultured in laminin-rich Matrigel with growth factors that induce stem-cell expansion. For example, stem cells obtained from intestinal crypts are grown in the presence of Wnt, noggin and epidermal growth factors, required for maintenance and expansion of Lgr5+ stem cells (Blokzijl et al., 2016; and reviewed in Rookmaaker et al., 2015). Since organoids are derived from a single adult stem cell, this methodology has allowed researchers to obtain clonal cell populations in vitro for whole genome sequencing and tissue-specific mutation analysis while bypassing the need for genome amplification as needed for single cell studies (Behjati et al., 2014; Blokzijl et al., 2016). Whole genome sequencing of organoid cultures from mice stomach, small bowel, large bowel and prostate demonstrated wide variability in mutation loads and spectra in the tissues with mutation loads varying from 100 to 600 mutations per culture (Behjati et al., 2014). This approach was later applied to human tissues, and WGS from organoids derived from the colon, small intestine and liver of individuals ranging from 3 to 87 years demonstrated that each cell carried 1000 to 3000 point mutations per genome with an age dependent increase of approximately 40 mutations per year. The major mutation signature in the colon and small-intestine derived cells was C→T at CpG dincucleotides. An additional signature whose source is currently unknown was detected in liver-derived organoids (Blokzijl et al., 2016) (Table 2).
These studies highlight the impacts of both replication and tissue-specific extrinsic sources of DNA damage in generation of the somatic “mosaicome” (Bae et al., 2018) across the human body. The tissue-specific mutation loads and profiles in the above-mentioned studies clearly demonstrate that somatic genomes can be a dosimeter of the endogenous and exogenous mutagenic processes operative in cells over the lifetime of the donors. Sequencing larger populations of individuals, and wider cell types will provide us with the ranges of somatic genome variability.
Somatic mutation calling
In addition to the methodology used for sequencing somatic cells, accuracy of somatic mutation calls is further dependent on the alignment and mutation calling algorithms used and downstream filtering applied. Currently there are a multitude of pipelines for whole genome sequence alignments and mutation annotation. Many of these somatic mutation callers have been assessed for specificity and sensitivity in comparative studies. It is evident that the balance between sensitivity and accuracy of true positive calls varies between the algorithms. For example, analysis of melanoma and lung tumor samples with matched normal demonstrated that MuTect (Cibulskis et al., 2013) and VarScan2 (Koboldt et al., 2009; Koboldt et al., 2012) were highly sensitive and accurate mutation callers, however, MuTect’s stringent filtering criteria led to the rejection of more somatic SNVs than VarScan2, while VarScan2 reported more false-positives than MuTect. On the other hand, Strelka (Saunders et al., 2012) and SomaticSniper (Larson et al., 2012) identified fewer true positive SNVs however, they also identified fewer false positive SNVs (Wang et al., 2013). Furthermore, analysis of breast cancers by nine different somatic mutation callers, demonstrated MuTect, EBCaller (Shiraishi et al., 2013), Shimmer (Hansen et al., 2013) and Strelka to be the most accurate and reliable algorithms and capable of outperforming VarScan2 (Kroigard et al., 2016). As such, it is preferable to utilize multiple somatic mutation callers to accurately identify somatic variants while minimizing caller-specific false positives.
Our own approach has revolved around using the Genome Analysis Toolkit best practices pipeline (Van der Auwera et al., 2013), followed by mutation calling using the consensus calls from three independent mutation callers (Saini et al., 2016). Specifically, sequencing reads are aligned to the human genome using bwa-mem. The resulting SAM files are converted to the BAM format with addition of read groups needed for GATK downstream processing, PCR duplicates are marked and removed and the resulting BAM files are realigned around indels and the base quality scores are recalibrated. Germline polymorphisms are determined in blood DNA or the matched “normal” DNA using the GATK Haplotype caller and the SNVs are filtered by training on known SNPs present in HapMap, dbSNP and the 1000 genomes project. Somatic mutations are determined in the clone or single cell using three callers – the GATK Haplotype caller which reports all variants in the sample and subsequently the germline variants are subtracted from the mutation list, as well as MuTect and VarScan2 which use data from the matched normal to directly output the clone- or cell-specific mutation calls. Only high-confidence variants called as “somatic” by all three callers are denoted as true-somatic calls.
To obtain high quality somatic mutation calls, further filters may be applied. VariantFiltration from GATK may be used to filter out variants that do not conform to certain criteria such as SNPs with quality (QUAL) scores less than 30 or low depth of coverage. Similar filters can be applied to VarScan2 and MuTect such that mutations present in regions with low depth, variants only detected in one orientation, SNPs next to indels, triallelic events or low-confidence somatic SNPs can be filtered out. Further, variant positions that overlap with known dbSNPs or variants within simple repeat tracts and blacklisted genomic regions (obtained from https://genome.ucsc.edu/cgi-bin/hgTables), are removed from consideration. Finally, for single cell or clonal population sequencing, allele frequency filters should be applied such that SNPs with allele frequencies between 45% and 55% are considered as heterozygous alleles, while those with frequencies >90% are considered homozygous alleles. SNPs with allele frequencies that are not within these ranges likely arose in vitro during cell culture or whole genome amplification and are therefore removed.
Human somatic mutation rates
The assessment of somatic mutation rates in multicellular organisms, especially humans, in situ has been a long standing unanswered question. The underlying cause for this gap in knowledge so far has been the lack of accurate measurements of somatic mutation loads in single human cells. Initially mutations accumulated in single gene reporters or in a small fraction of the genome were used as a proxy to determine somatic mutation rates in humans. These estimates vary dramatically between 10−9 to 10−7 mutations per base per cell division (Araten et al., 2005; Li et al., 2014; Lynch, 2010). Moreover, based on genomic studies in human somatic cells and cancers, it is now clear that mutation rates vary across the genome in a non-random manner. The majority of the mutagenic processes lead to increased mutation loads in late replicating heterochromatic genomic regions in part due to reduced DNA repair capacity in these regions (Koren et al., 2012; Lawrence et al., 2013; Polak et al., 2015; Polak et al., 2014; Sabarinathan et al., 2016; Saini et al., 2016; Schuster-Bockler and Lehner, 2012). As such, measuring mutation rates based on mutation accumulation at a single locus may lead to biased results.
The innovations in single cell sequencing techniques, or single cell derived clones and organoids’ sequencing has provided insights into somatic mutation rates. Single cell sequencing from mice and human cells showed a median mutation rate of 2.8 X 10−7 and 4.4 X 10−7 per bp for human and mouse cells. Accounting for cell divisions based on the number of cells in the body and tissue homeostasis after birth enabled correction of the mutation rates to 2.66 X 10−9 and 8.1 X 10−9 mutations per bp per mitosis in humans and mice (Milholland et al., 2017). On the other hand, estimation of telomere lengths in somatic fibroblast clones provided an estimate of approximately 60 cell divisions in adult skin cells since birth. Using this metric, the somatic mutation rates in hip and forearm fibroblast clones varied from 1.4X10−9 to 3X10−8 mutations per nucleotide per cell division, with higher mutation rates in sun-exposed skin cells carrying a prominent UV-mutation signature (Saini et al., 2016).
Interestingly, studies have also reported elevated accumulation of structural variations during early embryogenesis (Vanneste et al., 2009). Additionally, Ju et.al estimated somatic mutation rates during early embryogenesis to be approximately 2.8 mutations per cell doubling. Similar to the age-associated mutation profile 43% of the mutations generated during early embryogenesis were C→T changes at CpG dinucleotides (Table 2 and Table 3) (Ju et al., 2017). The rate of somatic mutation accumulation in neurons was found to be ~5.1 mutations per day during neurogenesis, which was lowered to ~1.3 mutations per day in early embryos. Moreover, analysis of mutation spectra in these cells demonstrated heightened C→T changes at CpG motifs generated during early neurogenesis coupled with increased C→A changes likely attributable to oxidative damage (Bae et al., 2018).
Somatic mutation spectra as an indicator of environmental or endogenous DNA damage in cancer-free cells
Currently, 30 mutation signatures are defined in the COSMIC database for cancer genomes (Alexandrov et al., 2013; Forbes et al., 2017). A variety of mutation signatures are highly concordant with known mutagenic activities of a variety of DNA damaging agents, including APOBEC3-induced tCw→tTw and tCw→tGw changes (Signatures 2 and 13 in COSMIC), UV-induced C→T changes at pyrimidine dimers (Signature 7 and 11 in COSMIC) and oxidative damage induced C→A changes due to erroneous bypass of 8-oxoguanine residues (Signature 4 in COSMIC); and signatures also vary with defects in DNA repair pathways (Drost et al., 2017; Kim et al., 2016 also Table 3). Based on these observations, it can be hypothesized that mutation signatures attributable to known environmental or endogenous DNA damage in human somatic cells can provide insights into the major mutagenic sources acting upon the tissue types tested. In support of this conjecture, the predominant mutation signature detected in sun-exposed healthy skin cells was of the UV-induced mutations (Abyzov et al., 2017; Martincorena et al., 2015; Saini et al., 2016), while in sun-unexposed skin cells this signature was much lower (Saini et al., 2016). Moreover, the aging-associated ubiquitous nCg→nTg signature has been shown to be present in almost all cell types tested (Blokzijl et al., 2016; Lodato et al., 2015; Saini et al., 2016), demonstrating that deamination at methylated cytosines is active in all cell types across the human body. With further accumulation of the data for healthy human cells, it is possible that similar to cancers, a catalog of mutation signatures may be developed, highlighting the mutational mechanisms that exclusively function in either cancerous tissues or healthy cells.
The caveats of such an undertaking are that a large number of signatures have overlapping components, while for many signatures, the etiology of the processes is unknown. Thus, it is a challenging effort to assign, where possible, the DNA damage and transaction processes to the signatures. Examples of such overlapping mutation signatures include the UV mutation motif (yCn→yTn), AID mutation motif (wrC→wrT), APOBEC mutation motif (tCw→tTw) and the aging-associated mutation motif (nCg→nTg) (Roberts et al., 2013; Rogozin et al., 2016; Saini et al., 2016)(Table 3). One way to annotate specific processes with their signatures is to split mutation signatures into non-overlapping components. Saini et. al. were able to differentiate between UV-induced mutagenesis at yCn motifs from CpG mutation signatures by splitting nCg→nTg to rCg and yCg respectively (Saini et al., 2016) (Table 3). This approach has also been used in determining which of the APOBEC3 enzymes are the major mutagens in cancer genomes. Both the APOBEC3A and 3B enzymes preferentially deaminate cytosines in the tCw motif. Using yeast systems ectopically expressing these enzymes, Chan et.al demonstrated that the preferred moiety for APOBEC3A is ytCa (y is a pyrimidine), while APOBEC3B favored rtCa (r is a purine) (Table 3). This choice of the −2 nucleotide by the enzymes was further corroborated by analysis of cancer genomes, which led to the conclusion that APOBEC3A is responsible for higher mutation loads in cancers than APOBEC3B (Chan et al., 2015). Interestingly, Rogozin et.al. demonstrated that C→T changes at methylated cytosines at CpG residues that are within the AID-mutable motif wrC are often overrepresented. Thus, AID-induced mutagenesis in wrCg motifs was found to be enriched in cancers where based on analysis of the classical AID motif (wrC) no AID activity was detected (Rogozin et al., 2016). Such analyses for other mutation signature motifs will help delineate the various mechanisms that damage DNA and measure their impacts on genome stability in human somatic cells.
Conclusions and the future challenges in somatic mutation analysis
The advances in somatic genome sequencing techniques over the last few years have significantly enhanced our knowledge about the somatic mutation loads and spectra in non-cancerous human tissues. Similar to cancer genomes, the mutation loads and spectra across the body and in different cell types were found to be dependent on the proliferative ability of the cells, their DNA repair capacity and the endogenous or exogenous DNA damaging agents the cells can encounter.
Measuring mutations at the cellular level is essential to understand the impacts of environmental or endogenous factors on genome stability since even deep sequencing of bulk tissue specimens would reflect primarily a few overrepresented mutations or cell lineages. However, studying the majority of the cells across the body is a challenging feat since most terminally differentiated cell types cannot be cultured. To overcome this caveat, induced pluripotent cell lines may be generated, however this process is inherently mutagenic (Ji et al., 2012; Rouhani et al., 2016). On the other hand, techniques involving single cell-derived clones (Saini et al., 2016) or organoids (Behjati et al., 2014; Blokzijl et al., 2016) are reliant on the minor fraction of cells that retain proliferative ability within the tissue. Therefore, single-cell sequencing for measuring mutation loads across most cell types makes this the desired future approach for large-scale studies. The current single-cell sequencing technologies have already allowed to sequence genomes of otherwise inaccessible cell types. However, one of the major caveats of single cell sequencing techniques is the high error-rate of the whole genome amplification techniques. Benchmarking the current technologies using sequenced clonal lineages alongside better-quality bioinformatic algorithms to determine the mutation rates and spectra associated with the different WGA technologies, can drastically improve the rates of calling true-positive mutations in single cells. Overall, we anticipate that advances in single cell sequencing technologies will provide the means for high-throughput profiling of somatic genomes in the cells across the human body.
The current and future studies on somatic mutation loads and spectra in healthy human cells have the potential to build the platform for utilization of these data as a “dosimeter” of environmental and endogenous DNA damage accrued over an individual’s lifetime allowing mutational information in cells to be used as part of cancer prevention screens. However, in order to transition into such screening procedures, a variety of challenges must be addressed. The first and foremost difficulty anticipated is the unavailability of or highly invasive nature to obtain a large variety of cell types from healthy living donors, including neuronal cells which currently can only be accessed post mortem. Hence, development of techniques and bioinformatic tools for sequencing of cell-free DNA from blood and calling somatic mutations (reviewed in (Cai et al., 2015; Heitzer et al., 2016)) may bypass the need to sequence specific cell types, allowing expansion of these studies to a large population at a fraction of the costs. Moreover, we can anticipate that somatic mutation estimations in easily accessible cell types like blood cells or buccal swabs of individuals who may have been exposed to carcinogens, would become common practice in measuring the impacts of the exposure to their health outcomes. Such measurements across populations, including individuals carrying known deleterious mutations in DNA repair genes or pathways predisposing them to accumulation of mutations, will provide the range of somatic mutation loads in healthy people thus informing about the normal and pathological levels of human somatic mutations.
In summary, estimation of somatic mutations in healthy human tissues has the potential to revolutionize our understanding of the causes underlying and the consequences of somatic genome instability on human health. Similar to the burst in cancer genome sequencing studies, with improvements in NGS technologies, and reduction in cost for sequencing genomes we expect large-scale efforts in somatic mutation detection in healthy individuals paving the way to understanding and screening for cancer and other somatic disease predisposition.
Acknowledgments
We are thankful to Dr. S Syed, Dr. M Howard and Dr. S. Vijayraghavan for critically reading this manuscript and providing their feedback. This work was supported by the US National Institute of Health Intramural Research Program Project Z1AES103266 to DAG
References
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino AF, Rosenberg Belmaker LA, Szekely A, Wilson M, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Tomasini L, Zhou B, Vasmatzis N, Coppola G, Amenduni M, Pattni R, Wilson M, Gerstein M, Weissman S, et al. One thousand somatic SNVs per skin fibroblast cell set baseline of mosaic mutational load with patterns that suggest proliferative origin. Genome Res. 2017;27:512–523. doi: 10.1101/gr.215517.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, Luzzatto L. A quantitative measurement of the human somatic mutation rate. Cancer research. 2005;65:8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- Arneson N, Hughes S, Houlston R, Done S. GenomePlex Whole-Genome Amplification. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4920. pdb prot4920. [DOI] [PubMed] [Google Scholar]
- Bae T, Tomasini L, Mariani J, Zhou B, Roychowdhury T, Franjic D, Pletikos M, Pattni R, Chen BJ, Venturini E, et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018;359:550–555. doi: 10.1126/science.aan8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Huch M, van Boxtel R, Karthaus W, Wedge DC, Tamuri AU, Martincorena I, Petljak M, Alexandrov LB, Gundem G, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, Huch M, Boymans S, Kuijk E, Prins P, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A, Walsh CA. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Janku F, Zhan Q, Fan JB. Accessing Genetic Information with Liquid Biopsies. Trends Genet. 2015;31:564–575. doi: 10.1016/j.tig.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, Xie XS. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Crowley E. UV damage, DNA repair and skin carcinogenesis. Front Biosci. 2002;7:d1024–1043. doi: 10.2741/A829. [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer research. 1998;58:3455–3460. [PubMed] [Google Scholar]
- Davies MJ, Lovell DP, Anderson D. Thioguanine-resistant mutant frequency in T-lymphocytes from a healthy human population. Mutat Res. 1992;265:165–171. doi: 10.1016/0027-5107(92)90045-4. [DOI] [PubMed] [Google Scholar]
- Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- Dolle ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc Natl Acad Sci U S A. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle ME, Vijg J. Genome dynamics in aging mice. Genome Res. 2002;12:1732–1738. doi: 10.1101/gr.125502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhang L, Milholland B, Lee M, Maslov AY, Wang T, Vijg J. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson RP. Somatic gene mutation and human disease other than cancer. Mutat Res. 2003;543:125–136. doi: 10.1016/s1383-5742(03)00010-3. [DOI] [PubMed] [Google Scholar]
- Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res. 2010;705:96–106. doi: 10.1016/j.mrrev.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Erickson RP. Recent advances in the study of somatic mosaicism and diseases other than cancer. Curr Opin Genet Dev. 2014;26:73–78. doi: 10.1016/j.gde.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Erickson RP. The importance of de novo mutations for pediatric neurological disease--It is not all in utero or birth trauma. Mutat Res Rev Mutat Res. 2016;767:42–58. doi: 10.1016/j.mrrev.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Hills M, Naumann U, Poon SS, Chavez EA, Sanders AD, Zhao Y, Hirst M, Lansdorp PM. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods. 2012;9:1107–1112. doi: 10.1038/nmeth.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansmo LB, Romundstad P, Hveem K, Vatten L, Nik-Zainal S, Lonning PE, Knappskog S. APOBEC3A/B deletion polymorphism and cancer risk. Carcinogenesis. 2017 doi: 10.1093/carcin/bgx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AM, Calder RB, Dolle ME, Lundell M, Kapahi P, Vijg J. Age- and temperature-dependent somatic mutation accumulation in Drosophila melanogaster. PLoS Genet. 2010;6:e1000950. doi: 10.1371/journal.pgen.1000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- Gregg SQ, Gutierrez V, Robinson AR, Woodell T, Nakao A, Ross MA, Michalopoulos GK, Rigatti L, Rothermel CE, Kamileri I, et al. A mouse model of accelerated liver aging caused by a defect in DNA repair. Hepatology. 2012;55:609–621. doi: 10.1002/hep.24713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MT, Bertout JA, Ericson NG, Taylor SD, Mukherjee R, Robins HS, Drescher CW, Bielas JH. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016;44:e22. doi: 10.1093/nar/gkv915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry M, Li W, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic Acids Res. 2012;40:2032–2040. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen NF, Gartner JJ, Mei L, Samuels Y, Mullikin JC. Shimmer: detection of genetic alterations in tumors using next-generation sequence data. Bioinformatics. 2013;29:1498–1503. doi: 10.1093/bioinformatics/btt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RW, Setlow RB, Woodhead AD. Evidence that pyrimidine dimers in DNA can give rise to tumors. Proc Natl Acad Sci U S A. 1977;74:5574–5578. doi: 10.1073/pnas.74.12.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E, Ulz P, Geigl JB, Speicher MR. Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol Oncol. 2016;10:494–502. doi: 10.1016/j.molonc.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang ML, Chen CH, Sidorenko VS, He J, Dickman KG, Yun BH, Moriya M, Niknafs N, Douville C, Karchin R, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JHJ. DNA Damage, Aging, and Cancer. (vol 361, pg 1475, 2009) New Engl J Med. 2009;361:1914–1914. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Ji J, Ng SH, Sharma V, Neculai D, Hussein S, Sam M, Trinh Q, Church GM, McPherson JD, Nagy A, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J, Yoon KA, Hayashi T, Kong SY, Shin HJ, Park B, Kim YM, Hwang SH, Kim J, Shin A, et al. Nucleotide Excision Repair Gene ERCC2 and ERCC5 Variants Increase Risk of Uterine Cervical Cancer. Cancer Res Treat. 2016;48:708–714. doi: 10.4143/crt.2015.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Martincorena I, Gerstung M, Petljak M, Alexandrov LB, Rahbari R, Wedge DC, Davies HR, Ramakrishna M, Fullam A, et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature. 2017;543:714–718. doi: 10.1038/nature21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133:118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, Rosenberg JE, Van Allen EM, D’Andrea A, Getz G. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Amon A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 2016;26:376–384. doi: 10.1101/gr.198937.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroigard AB, Thomassen M, Laenkholm AV, Kruse TA, Larsen MJ. Evaluation of Nine Somatic Variant Callers for Detection of Somatic Mutations in Exome and Targeted Deep Sequencing Data. PLoS One. 2016;11:e0151664. doi: 10.1371/journal.pone.0151664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Montpetit A, Rousseau M, Wu SY, Greenwood CM, Spector TD, Pollak M, Polychronakos C, Richards JB. Somatic point mutations occurring early in development: a monozygotic twin study. J Med Genet. 2014;51:28–34. doi: 10.1136/jmedgenet-2013-101712. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA. A mutator phenotype in cancer. Cancer research. 2001;61:3230–3239. [PubMed] [Google Scholar]
- Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer research. 1974;34:2311–2321. [PubMed] [Google Scholar]
- Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SM, Quake SR. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ. AID and Somatic Hypermutation. Adv Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milholland B, Auton A, Suh Y, Vijg J. Age-related somatic mutations in the cancer genome. Oncotarget. 2015;6:24627–24635. doi: 10.18632/oncotarget.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milholland B, Dong X, Zhang L, Hao X, Suh Y, Vijg J. Differences between germline and somatic mutation rates in humans and mice. Nat Commun. 2017;8:15183. doi: 10.1038/ncomms15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman ME, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Podolskiy DI, Lobanov AV, Kryukov GV, Gladyshev VN. Analysis of cancer genomes reveals basic features of human aging and its role in cancer development. Nat Commun. 2016;7:12157. doi: 10.1038/ncomms12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Lawrence MS, Haugen E, Stoletzki N, Stojanov P, Thurman RE, Garraway LA, Mirkin S, Getz G, Stamatoyannopoulos JA, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nature Biotechnology. 2014;32:71. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MJ, Moore DH, Briner JF, Lee DA, Olsen LA, Senft JR, Tucker JD. The Effects of Age and Life-Style Factors on the Accumulation of Cytogenetic Damage as Measured by Chromosome Painting. Mutat Res-Dnaging G. 1995;338:95–106. doi: 10.1016/0921-8734(95)00015-x. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Goncearenco A, Lada AG, De S, Yurchenko V, Nudelman G, Panchenko AR, Cooper DN, Pavlov YI. DNA polymerase eta mutational signatures are found in a variety of different types of cancer. Cell cycle. 2018:1–8. doi: 10.1080/15384101.2017.1404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Lada AG, Goncearenco A, Green MR, De S, Nudelman G, Panchenko AR, Koonin EV, Pavlov YI. Activation induced deaminase mutational signature overlaps with CpG methylation sites in follicular lymphoma and other cancers. Sci Rep. 2016;6:38133. doi: 10.1038/srep38133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nature immunology. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H. Development and application of human adult stem or progenitor cell organoids. Nature reviews Nephrology. 2015;11:546–554. doi: 10.1038/nrneph.2015.118. [DOI] [PubMed] [Google Scholar]
- Rouhani FJ, Nik-Zainal S, Wuster A, Li Y, Conte N, Koike-Yusa H, Kumasaka N, Vallier L, Yusa K, Bradley A. Mutational History of a Human Cell Lineage from Somatic to Induced Pluripotent Stem Cells. PLoS Genet. 2016;12:e1005932. doi: 10.1371/journal.pgen.1005932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- Saini N, Roberts SA, Klimczak LJ, Chan K, Grimm SA, Dai S, Fargo DC, Boyer JC, Kaufmann WK, Taylor JA, et al. The Impact of Environmental and Endogenous Damage on Somatic Mutation Load in Human Skin Fibroblasts. PLoS Genet. 2016;12:e1006385. doi: 10.1371/journal.pgen.1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Falconer E, Hills M, Spierings DCJ, Lansdorp PM. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat Protoc. 2017;12:1151–1176. doi: 10.1038/nprot.2017.029. [DOI] [PubMed] [Google Scholar]
- Sanders AD, Hills M, Porubsky D, Guryev V, Falconer E, Lansdorp PM. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 2016;26:1575–1587. doi: 10.1101/gr.201160.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci U S A. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current protocols in bioinformatics. 2013;43:11.10 11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Vijg J, Dolle ME. Large genome rearrangements as a primary cause of aging. Mech Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- von Joest M, Bua Aguin S, Li H. Genomic stability during cellular reprogramming: Mission impossible? Mutat Res. 2016;788:12–16. doi: 10.1016/j.mrfmmm.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Wang Q, Jia P, Li F, Chen H, Ji H, Hucks D, Dahlman KB, Pao W, Zhao Z. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome medicine. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MC, Holman DM, Boehm JE, Peipins LA, Grossman M, Henley SJ. Age and cancer risk: a potentially modifiable relationship. Am J Prev Med. 2014;46:S7–15. doi: 10.1016/j.amepre.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lu S, Huang Y. Microfluidic whole genome amplification device for single cell sequencing. Analytical chemistry. 2014;86:9386–9390. doi: 10.1021/ac5032176. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]