Abstract
Nicotinamide adenine dinucleotide (NAD) is the redox cofactor of many enzymes, including the vast aldehyde dehydrogenase (ALDH) superfamily. Although the function of NAD(H) in hydride transfer is established, its influence on protein structure is less understood. Herein, we show that NAD+-binding promotes assembly of the ALDH7A1 tetramer. Multi-angle light scattering, small-angle X-ray scattering, and sedimentation velocity all show a pronounced shift of the dimer-tetramer equilibrium toward the tetramer when NAD+ is present. Furthermore, electron microscopy shows that cofactor binding enhances tetramer formation even at the low enzyme concentration used in activity assays, suggesting the tetramer is the active species. Altogether, our results suggest that the catalytically active oligomer of ALDH7A1 is assembled on demand in response to cofactor availability.
Keywords: Enzyme oligomerization, small-angle X-ray scattering, analytical ultracentrifugation, electron microscopy, aldehyde dehydrogenase, nicotinamide adenine dinucleotide
Introduction
Human aldehyde dehydrogenase 7A1 (ALDH7A1) catalyzes the final step in lysine catabolism, the NAD+-dependent oxidation of α-aminoadipate semialdehyde to α-aminoadipate (Fig. 1). Mutations in the ALDH7A1 gene can cause the autosomal recessive metabolic disease pyridoxine-dependent epilepsy (PDE), which is characterized by seizure activity within days of birth [1–3]. Although PDE symptoms can be mitigated with high doses of pyridoxine (vitamin B6) and a lysine-restricted diet, many patients have long-lasting and untreatable cognitive disability. The molecular basis of PDE is thought to be due to a buildup of Δ1-piperideine-6-carboxylate, the cyclic form of the aldehyde substrate of ALDH7A1 (Fig. 1). Evidence suggests Δ1-piperideine-6-carboxylate can form a covalent adduct with pyridoxal-5’-phosphate (PLP), thereby irreversibly inactivating PLP [4]. Because PLP is a cofactor in ~4% of all enzymes [5], depletion of the cellular PLP pool unsurprisingly has deleterious consequences. To date, over 40 mutations in the ALDH7A1 gene have been identified, but few have been characterized biochemically or structurally.
Fig. 1.
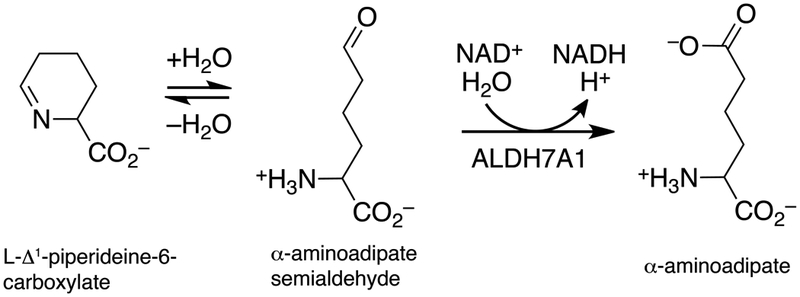
Reactions related to ALDH7A1 biology. ALDH7A1 catalyzes the oxidation of α-aminoadipate semialdehyde to α-aminoadipate, which is in non-enzymatic equilibrium with L-Δ1-piperideine-6-carboxylate.
It is widely assumed that oligomeric structure is intimately linked to catalytic function in ALDHs. Initial structural studies revealed ALDH7A1 forms a dimer-of-dimers tetramer both in crystallo [6–8] and in solution [7]. Many other ALDHs form the same tetramer, including ALDH1 [9–12], ALDH2 [13,14], ALDH5A1 [15], methylmalonate semialdehyde dehydrogenase [16], ALDH21 [17], 2-aminomuconate-6-semialdehyde dehydrogenase [18], betaine aldehyde dehydrogenase [19], lactaldehyde dehydrogenase [20], indole-3-acetaldehyde dehydrogenase [21], α-ketoglutarate-semialdehyde dehydrogenase [22], propionaldehyde dehydrogenase [23], and salicylaldehyde dehydrogenase [24]. Further investigation of the in-solution oligomeric state of ALDH7A1 using analytical ultracentrifugation revealed a concentration-dependent dimer-tetramer equilibrium with a dissociation constant (Kd) of 16 μM [25]. Interestingly, a number of PDE-related mutations map to the oligomeric interfaces of ALDH7A1, and these mutant enzymes are both enzymatically inactive and deficient in their ability to form the tetramer [25]. Additionally, deletion of the C-terminus of ALDH7A1 results in similar defects in both activity and oligomeric state [26]. Also, the disruption of the tetrameric form of ALDH1 by site-directed mutagenesis dramatically decreased catalytic activity [27]. Altogether, these results imply a link between catalytic activity and the ability of ALDH7A1, and presumably other tetrameric ALDHs, to assemble into the tetrameric form.
Despite circumstantial evidence, the true identity of the active oligomeric species of ALDH7A1 remains a conundrum. Although the observation that ALDH7A1 mutants defective in tetramerization are also deficient in catalytic activity suggests the tetramer is the active species, the tetramer-dimer Kd of 16 μM implies that ALDH7A1 is predominantly dimeric in enzyme activity assays where the enzyme concentration typically is on the order of nanomolar. We solve this puzzle by providing evidence that ALDH7A1 tetramerization is dramatically enhanced by the presence of its NAD+ cofactor over a broad range of enzyme concentrations. In-solution analysis of ALDH7A1 by size-exclusion chromatography - multi-angle light scattering - small-angle X-ray scattering (SEC-MALS-SAXS) and sedimentation velocity shows that addition of NAD+ drives the dimer-tetramer equilibrium to strongly favor tetramer formation at protein concentrations near the Kd of the apo enzyme. Further, negative-stain electron microscopy (EM) reveals a dramatic shift toward the tetramer at nanomolar enzyme concentration upon addition of NAD+. Overall, these results suggest that NAD+-cofactor binding drives tetramer formation, and the ALDH7A1 tetramer is the active form of the enzyme.
Materials and methods
ALDH7A1 protein expression and purification
ALDH7A1 was expressed and purified as previously described [25]. The final storage buffer for protein samples - i.e. the SEC mobile phase - contained 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM dithiothreitol (DTT), and 5% (v/v) glycerol. For analytical ultracentrifugation analysis, protein was used without freezing. For all other applications, protein was aliquoted in PCR strip tubes, flash-cooled in liquid nitrogen, and stored at −8000B0030C.
SEC-MALS-SAXS data acquisition and analysis
SAXS experiments were performed at the Advanced Photon Source Sector 18-ID (Argonne National Lab, Chicago, IL, USA) using a 3.5 m camera availing a q range of ~0.005 – 0.38 Å−1. The samples at three nominal protein concentrations ranging from 2 – 8 mg mL−1 (18 – 72 μM by dimer Mr) was loaded onto a Wyatt SEC (1.25 MDa exclusion limit) controlled by an Infinity II HPLC unit (Agilent, Santa Clara, CA, USA). A MALS instrument fitted with a Wyatt DAWN Helios II QELS detector and a Wyatt Optilab T-rEX dRI detector (Wyatt, Goleta, CA, USA) was included in the trajectory between the HPLC UV monitor and the SAXS flow cell. Sample elution and SAXS data acquisition (0.5 s exposures collected every 3 s) occurred simultaneously. MALS and DLS data were processed using the ASTRA software package (Wyatt, Goleta, CA, USA) and the SAXS data were reduced to I(q) vs. q curves (q = 4πsinθ / λ, where 2θ = scattering angle and λ = 1.03 Å). Exposures flanking the elution peak were averaged to generate the I(q) vs. q curve for the buffer, which was then subtracted from the elution peak curves to obtain the sample SAXS curve. Experimental SAXS curves were processed and averaged using ATSAS [28]. We note that each sample eluted as a single SEC peak - i.e. separate dimer and tetramer peaks were not observed. For reference, the SEC-MALS profiles are shown in Fig. S1.
Guinier analysis was performed using PRIMUS [29]. The theoretical scattering curves for the dimeric and tetrameric crystal structure models of ALDH7A1 (PDB ID: 4ZUK) and the goodness-of-fit parameter (χ2) were calculated using FoXS [30]. Theoretical SAXS curves from dimer-tetramer ensemble models were calculated using MultiFoXS [30]. Because of the low signal to noise ratio apparent in the data at q > 0.15 Å−1, only data in the q-range of 0 – 0.15 Å−1 were used for theoretical SAXS curve fitting. The complete experimental SAXS data and Guinier plots are included in Fig. S2 for reference.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a Beckman XL-I analytical ultracentrifuge using an An50Ti rotor at 20°C. Reference buffer and protein samples were loaded into a sedimentation velocity cell containing a two-sector charcoal-Epon centerpiece. The sample was allowed to equilibrate for 2 h prior to centrifugation. The sample was then centrifuged at 82021 g for a total of 300 radial scans spaced at 2 min intervals and acquired using Rayleigh interference optics. Scans 10 – 300 were analyzed and the distribution of apparent sedimentation coefficients, c(s), and distribution of apparent molecular masses, c(M), were determined using SEDFIT [31]. For facile comparison of c(M) distributions, the frictional ratio was fixed at 1.5, which is an approximate average of the refined frictional ratios of each individual sample.
For sedimentation velocity analysis of apo ALDH7A1, a protein sample was dialyzed for overnight against 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM DTT, and 5% (v/v) glycerol. For sedimentation velocity analysis ALDH7A1 in the presence of NAD+, ALDH7A1 was supplemented with 1 mM NAD+ and run over a SEC column with a mobile phase containing 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, and 1 mM NAD+. Final experimental protein concentrations were calculated assuming 1 mg mL−1 is equivalent to 3.33 fringes [32].
Negative-stain scanning electron microscopy
Grids for negative-stain EM were prepared as previously described [33]. Briefly, a 5 μL drop of protein was applied to a glow discharged (Ted Pella, Redding CA USA) carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA, USA) and incubated for 2 min at room temperature. Excess protein was removed by blotting with filter paper and the grids were washed twice in water. The grids were then incubated for 2 minutes in 0.75% (w/v) uranyl formate (UF, Ted Pella, Reading, CA). Excess UF was removed with filter paper, and the grid was air-dried.
Protein particles were visualized using a Tecnai F30 G2 Twin Transmission Electron Microscope (FEI, Hillsboro OR, USA) operated at 200 kV, 3950 microAmp extraction voltage with a gun lens of 5 using low-dose data collection scheme at 59,000x magnification and imaged with an Ultrascan 4000 (Gatan Inc., Warrendale, PA, USA). ALDH7A1 samples, either apo or in the presence of 1 mM NAD+, were diluted in a 10% (v/v) dilution of the protein storage buffer to a concentration of 0.01 mg mL−1 (~90 nM, dimer Mr) and stained with 0.75% w/v UF. CTF correction was performed using CTFFind 4.1.8 [34] using a spherical aberration of 2.0 mm, magnified pixel size of 3.876 Å and an amplitude contrast of 0.6 for apo samples and 0.5 for NAD+ containing micrographs. Automated particle picking (autopicking) was performed using two-dimensional classes generated from ~1000 manually picked particles. A total of 6482 ALDH7A1 apo particles were autopicked using a threshold cutoff of 0.8, a maximum standard deviation of noise was set to 1.1 with a minimum inter-particle distance of 100. A total of 6574 ALDH7A1 + NAD+ particles were autopicked using a threshold cutoff of 1.0 a maximum standard deviation of noise was set to 1.1 with a minimum inter-particle distance of 100. We note that the threshold cutoff was different between the two autopick runs to obtain a similar number of particles for the two independent samples. Data were processed using RELION 2.1 [35]. A total of 30 two-dimensional classes were generated for each ALDH7A1 treatment (apo or + NAD+) using a 130 Å diameter mask.
Results
SEC-MALS-SAXS shows NAD+ shifts the self-association equilibrium of ALDH7A1 to the tetramer
Wild-type ALDH7A1 in the absence of any substrates was subjected to SEC-MALS-SAXS analysis using three nominal loading concentrations in the range of 2 – 8 mg mL−1 (18 – 72 μM by dimer Mr). Results of the MALS experiment show the average in-solution Mr increases from 134 kDa to 158 kDa with increasing protein load concentration (Table 1), consistent with a concentration-dependent dimer-tetramer equilibrium. For reference, the dimer Mr is 111 kDa.
Table 1.
Structural Parameters from MALS and SAXS
| ALDH7A1 apo | ALDH7A1 + NAD+ | |||||
|---|---|---|---|---|---|---|
| Low | Medium | High | Low | Medium | High | |
| MALS Analysis | ||||||
| Rh (Å) | 53 ± 1 | 54 ± 1 | 53 ± 1 | 56 ± 1 | 62 ± 1 | 61 ± 1 |
| Mr (kDa) | 134 ± 1 | 148 ± 1 | 158 ± 1 | 198 ± 2 | 215 ± 1 | 218 ± 2 |
| Guinier Analysis | ||||||
| Rg (Å) | 34.6 ± 0.5 | 37.4 ± 0.3 | 37.4 ± 0.1 | 37.1 ± 0.3 | 38.2 ± 0.2 | 37.9 ± 0.2 |
| qmin (Å−1) | 0.0081 | 0.0054 | 0.0054 | 0.0077 | 0.0069 | 0.0069 |
| qRg range | 0.3–1.3 | 0.2–1.3 | 0.2–1.3 | 0.3–1.3 | 0.3–1.3 | 0.3–1.3 |
| Coefficient of Correlation, R2 | 0.8875 | 0.9709 | 0.9865 | 0.9814 | 0.9876 | 0.9834 |
| P(r) Analysis | ||||||
| Real Space Rg | 35.9 ± 0.3 | 37.0 ± 0.1 | 37.0 ± 0.8 | 37.2 ± 0.1 | 37.8 ± 0.2 | 37.4 ± 0.1 |
| Dmax (Å) | 110 | 118 | 116 | 104 | 119 | 112 |
| P(r) q-range | 0.0081–0.2299 | 0.0054–0.2143 | 0.0054–0.2140 | 0.0078–0.2158 | 0.0069–0.2091 | 0.0069–0.2106 |
| Porod Volume (Å3) | 245,000 | 238,000 | 231,000 | 298,000 | 296,000 | 285,000 |
| FoXS Analysis | ||||||
| Dimer χ2 Value | 2.8 | 16 | 73 | 28 | 37 | 70. |
| Tetramer χ2 value | 1.2 | 1.6 | 3.1 | 1.1 | 1.2 | 1.5 |
| MultiFoXS Analysis | ||||||
| χ2 value (dimer:tetramer) | 1.0 (65:35) | 1.1 (50:50) | 1.6 (40:60) | 1.1 (6:94) | 1.2 (13:87) | 1.6 (12:88) |
| SASBDB Code | SASDD29 | SASDD39 | SASDD49 | SASDD59 | SASDD69 | SASDD79 |
The theoretical Mr values of the dimer and tetramer are 111 and 222 kDa, respectively. The column designations Low, Medium, and High refer to the protein load concentration for the SEC-SAXS experiment.
SAXS data for the apo enzyme are consistent with the MALS results (Fig. 2). Theoretical SAXS curves calculated from the dimer and tetramer crystal structure models (PDB ID: 4ZUK) were fit to the experimental data using FoXS [30]. As expected for an equilibrium mixture containing both dimer and tetramer, neither individual oligomer model provided the best fit to the data (Table 1). As protein concentration increases, single-body fits to the crystallographic dimer statistically worsen, suggesting the dimer character of the sample diminishes with increasing protein concentration (Table 1). Application of a two-body dimer-tetramer ensemble with MultiFoXS [30] improved the χ2 values for each concentration compared to the χ2 value obtained to the single-body fit to the crystallographic tetramer (Table 1). Additionally, qualitative analysis of the fits to the experimental data shows improvement in the q = 0.075 – 0.125 Å−1 range with a dimer-tetramer ensemble fit compared to the tetramer alone (Fig. 2). Further, the composition of the ensemble fit shows increasing amounts of tetramer as protein load concentration increases: from 65:35 dimer:tetramer at the lowest concentration to 40:60 at the highest concentration (Table 1). Overall, these results confirm the presence of an ALDH7A1 dimer-tetramer equilibrium in the absence of substrates, in agreement with earlier studies [25].
Fig. 2.
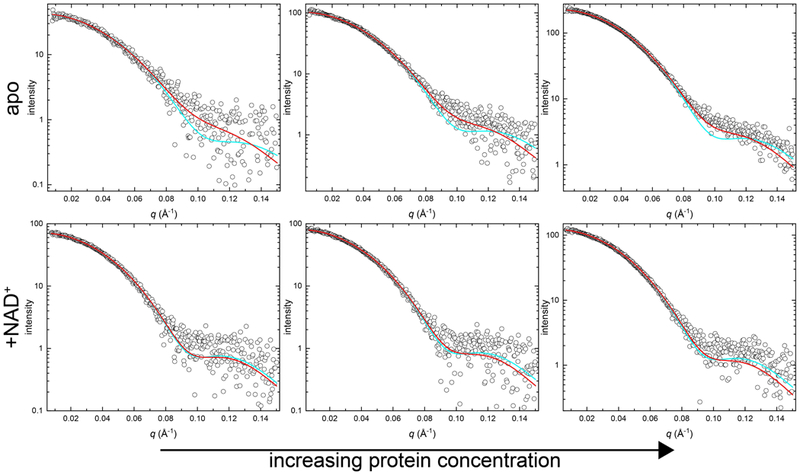
SEC-SAXS analysis of ALDH7A1 in the presence and absence of NAD+. Each panel shows SEC-SAXS experimental data (open circles) and the corresponding FoXS single-body fit to the ALDH7A1 crystallographic tetramer (PDB ID: 4ZUK; cyan curve) or the MultiFoXS two-body ensemble fit to a theoretical mixture of ALDH7A1 crystallographic dimer and tetramer (PDB ID: 4ZUK; red curve). The top three panels display three increasing nominal concentrations (from left to right) of apo ALDH7A1. The bottom three panels display three increasing nominal concentrations (from left to right) of ALDH7A1 in the presence of 1 mM NAD+. The complete experimental SAXS data and Guinier plots are included in Fig. S2 for reference.
To determine whether the presence of NAD+ influences the proportion of tetramer in solution at similar nominal protein load concentrations, an identical SEC-MALS-SAXS experiment was performed in the presence of a saturating concentration of NAD+ (1 mM). MALS analysis reveals a range of in-solution Mr from 198 kDa to 218 kDa with increasing protein load concentration (Table 1). These Mr values are within 11% and 2%, respectively, of the theoretical tetramer Mr (222 kDa). Additionally, the hydrodynamic radius (Rh) increased by ~6 – 17% compared to samples lacking NAD+ (Table 1). In summary, the MALS results show that NAD+ increases the average in-solution Mr and particle size compared to the apo samples, consistent with enhanced tetramerization.
SAXS data are also consistent with NAD+ promoting tetramerization. For example, the Porod volumes observed in the presence of NAD+ show an increase of ~16 – 29% compared to the samples in the absence of NAD+ (Table 1). Further, with NAD+ present, excellent single-body fits to the tetramer were obtained at all three enzyme concentrations, and the two-body dimer-tetramer ensemble afforded no perceptible improvement in χ2 (Fig. 2; Table 1). In fact, use of the two-body ensemble increased χ2 for the highest protein concentration, indicating overfitting of the data. Taken together, these data suggest that inclusion of NAD+ shifted the dimer-tetramer equilibrium substantially toward the tetramer.
Sedimentation velocity shows NAD+ promotes tetramer formation at micromolar enzyme concentrations
Previous studies of ALDH7A1 in the absence of any cofactor or substrate uncovered a dimer-tetramer equilibrium with a Kd of 16 μM [25,26]. To determine whether NAD+ is capable of driving ALDH7A1 tetramerization at concentrations below the reported Kd, sedimentation velocity experiments were conducted in the absence and presence of NAD+ with the enzyme at 9 μM (by dimer Mr).
First, apo ALDH7A1 was analyzed by sedimentation velocity at 9 μM. The c(s) distribution displays two major, overlapping peaks at apparent sedimentation coefficients of 4.5S and 6.5S, as well as a minor peak at 2.7S (Fig. 3A). The major peaks correspond to Mr of 110 kDa and 184 kDa (Fig. 3B), which are both below the theoretical Mr of the tetramer (222 kDa), consistent with an intermediate exchange regime in which free dimer exists in solution simultaneously with a dimer-tetramer equilibrium. The minor species at 2.7S (54 kDa) likely represents a monomer (theoretical Mr of 56 kDa). This peak has been observed previously in ALDH7A1 sedimentation velocity experiments and likely represents a monomeric conformation incapable of assembly into higher order oligomers [25,26].
Fig. 3.
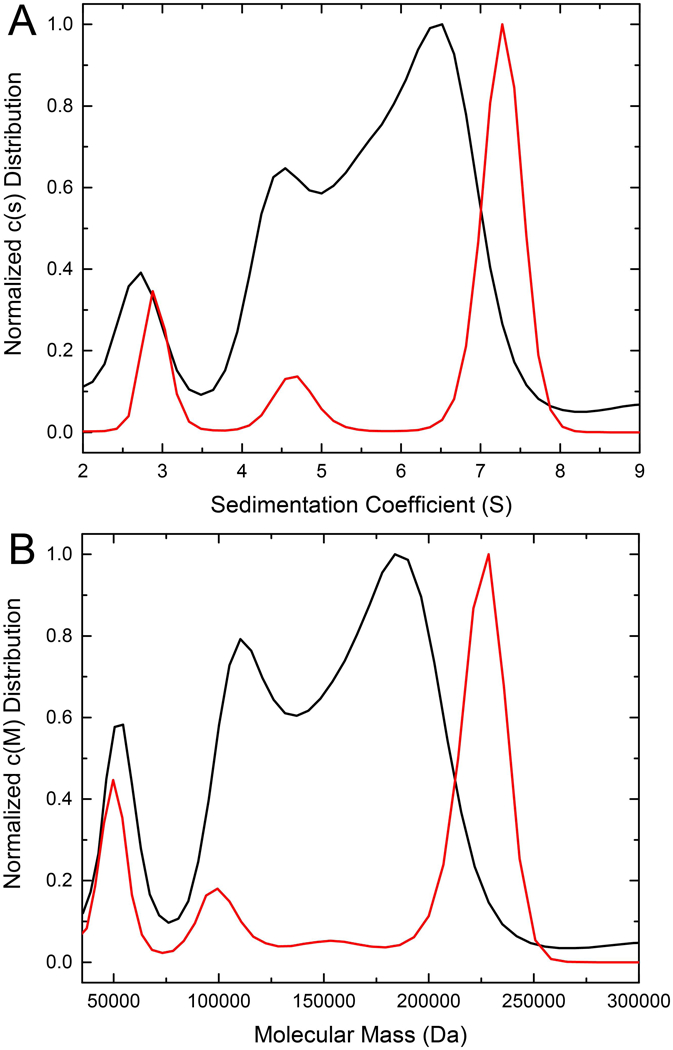
Sedimentation velocity analysis of ALDH7A1 in the absence (black) and presence (red) of NAD+. (A) The distribution of sedimentation coefficients, c(s). (B) The distribution of molecular masses, c(M). In both panels, the black curve depicts apo ALDH7A1 at 9 μM (dimer Mr), and the red curve depicts ALDH7A1 in the presence of 1 mM NAD+ at 9 μM (dimer Mr).
The addition of NAD+ dramatically changes the c(s) distribution. ALDH7A1 at 9 μM in the presence of 1 mM NAD+ has a sharp, major peak at an apparent sedimentation coefficient 7.3S and minor peaks at 4.7S and 2.9S (Fig. 3A). The major peak corresponds to apparent Mr of 228 kDa, which is very close to the theoretical tetramer Mr of 222 kDa (Fig. 3B). Importantly, the major peak at 7.3S is in good agreement with the previously observed apparent sedimentation coefficient of the ALDH7A1 tetramer (7.1S) obtained when using the apo enzyme at 40 μM, i.e. over 4 times the concentration used here. These results suggest the presence of NAD+ promotes tetramer formation at an enzyme concentration below the reported dimer-tetramer Kd of the apo enzyme.
Electron microscopy (EM) evidence for NAD+ promoting tetramerization of ALDH7A1 at nanomolar enzyme concentration
Results to this point indicate that NAD+ enhances ALDH7A1 tetramerization at micromolar enzyme concentrations. An outstanding question, however, is what is the oligomeric state of ALDH7A1 in typical enzyme activity assays, where the enzyme concentration is orders of magnitude lower than micromolar. Addressing this question provides insight into the true identity of the active oligomeric state of ALDH7A1.
Negative-stain EM was employed to assay oligomeric state at an enzyme concentration below the range of typical biophysical methods. First, an apo ALDH7A1 sample was visualized by negative-stain EM at ~90 nM (by dimer Mr). Images revealed a mixture of particles, which were subjected to automated particle picking (autopicking) in Relion 2.1 [35]. The 6482 autopicked particles generated 30 two-dimensional classes. Each class was then compared visually to the structures of the ALDH7A1 dimer and tetramer down-sampled to 20 Å resolution (PDB ID: 4ZUK; Fig. 4A). Of these 30 classes, 20 classes resembled the dimer, whereas only 3 classes resembled the tetramer (Fig. 4B). The remaining 7 classes did not conclusively resemble either the dimer or tetramer. The 20 dimer classes accounted for 5965 particles (92%), while the 3 tetramer classes accounted for 276 particles (4%). These results show that apo ALDH7A1 is overwhelmingly dimeric at 90 nM, as anticipated from the known in-solution dimer-tetramer Kd of 16 μM [25].
Fig. 4.
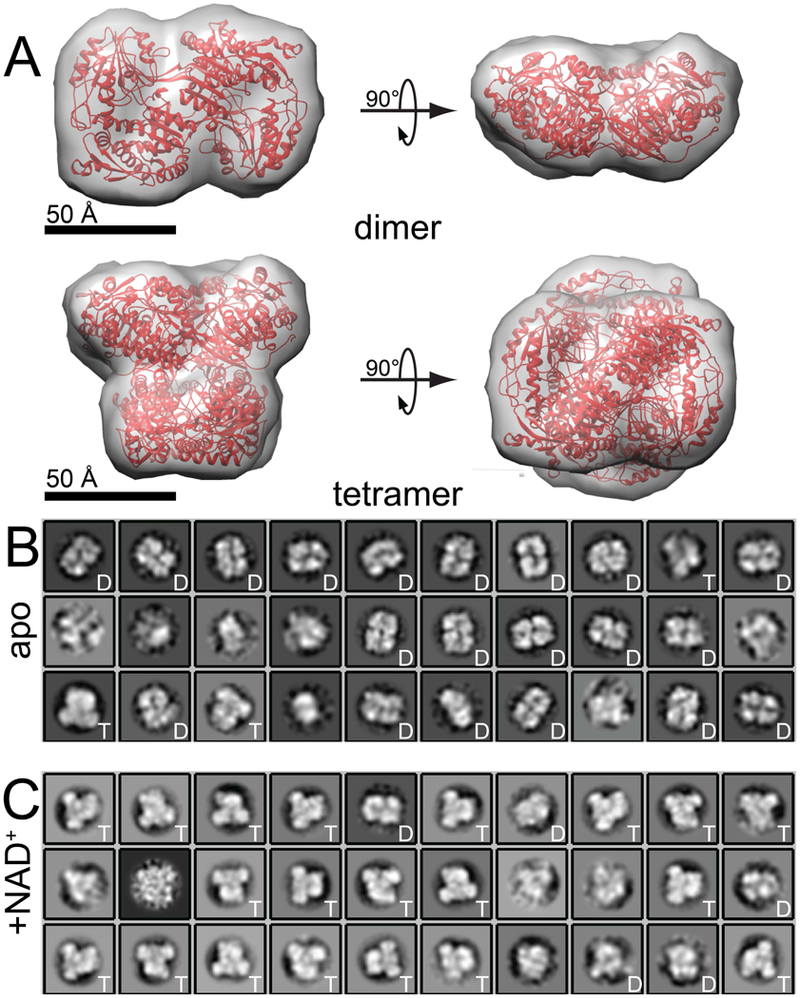
Negative-Stain EM of ALDH7A1 in the presence and absence of NAD+. (A) The crystal structure of ALDH7A1 (PDB ID: 4ZUK) shown as a ribbon in red with an overlaid envelope representing a 20 Å resolution view of both the dimer and tetramer. The 20 Å resolution envelope was generated using Chimera [49]. (B) Two-dimensional class averages for apo ALDH7A1 at 90 nM. (C) Two-dimensional class averages for 90 nM ALDH7A1 in the presence of 1 mM NAD+. For panels B and C, the letters ‘D’ or ‘T’ in the inset of each class average indicate classes that visually resemble the dimer or tetramer, respectively. Classes that lack a letter in the inset were not clearly dimeric or tetrameric.
To determine the effect of NAD+ on oligomeric state, an ALDH7A1 sample containing NAD+ was also visualized by negative-stain EM at ~90 nM (by dimer Mr). Autopicking resulted in 6574 particles, and 30 two-dimensional classes were generated. Of these 30 classes, 20 classes strongly resemble the shape of the tetramer, whereas only 5 classes resembled the dimer (Fig. 4C). The remaining 5 classes could not be unambiguously assigned as dimer or tetramer. The 20 tetramer classes accounted for 5813 particles (88%), while the 5 dimer classes accounted for 262 particles (4%). Thus, the apparent ratio of tetramer to dimer particles exceeds 20. Taken together, the EM results indicate that NAD+-bound ALDH7A1 is predominantly tetrameric under conditions similar to those used in activity assays, consistent with the tetramer being the active form of the enzyme.
To rule out the possibility that the manually-selected particles that were used to seed autopicking biased the results, the autopicking template particles obtained from the +NAD+ micrographs were used to autopick the apo micrographs, and vice versa. Applying this “reverse” autopicking scheme to the apo micrographs resulted in 8135 particles. Generation of 30 two-dimensional classes resulted in 23 dimer classes (7,405 particles, 91%), 3 tetramer classes (452 particles, 6%), and 4 classes that did not resemble the dimer or tetramer (278 particles, 4%) (Fig. S3). Thus, the apo sample appears predominantly dimeric. Autopicking the NAD+ micrographs using apo template resulted in 6701 particles. Generation of 30 two-dimensional classes resulted in 3 dimer classes (262 particles, 4%), 14 tetramer classes (5,375 particles, 80%), and 13 classes that did not resemble the dimer or tetramer (1,064 particles, 16%) (Fig. S3). Thus, the addition of NAD+ favors the tetramer over the dimer. Taken together, these results indicate the two-dimensional classification analyses were not biased by the initial manual particle selection.
Discussion
Herein, we presented evidence suggesting the NAD+ cofactor of human ALDH7A1 dramatically enhances tetramerization of the enzyme. To our knowledge, this is the first observation of NAD+ promoting oligomerization of an ALDH superfamily enzyme. A wide body of research indicates substrate-induced oligomerization is a mechanism of controlling the activity of enzymes [36]. The current work described an example of a dinucleotide redox cofactor influencing the oligomeric state of an enzyme. Other examples of this phenomenon include: FAD induces lipoamide dehydrogenase dimer formation [37,38]; butyryl-CoA dehydrogenase favors the active tetramer in the presence of FAD and CoA [39]; a vanillyl-alcohol oxidase mutant forms an octamer in the presence of its FAD cofactor [40]; NAD(H) influences both homo-tetramerization of the human C-terminal Binding Protein family proteins [41,42], as well as hetero-dimerization with adenovirus protein E1A [42,43]; NAD+ stabilizes the quaternary structure of S-adenosyl-L-homocysteine hydrolase [44]; and mycothione reductase favors tetramer formation in the presence of NADPH [45].
Our results also support the idea of the tetramer as the active form of ALDH7A1. Further, cellular concentrations of NAD+ have been reported in the range of 0.3 – 0.8 mM depending on tissue or organ type [46]. This observation suggests ALDH7A1 is tetrameric in vivo, further implicating the tetramer as the active ALDH7A1 oligomer. Although it may be assumed that the tetramer is the active oligomer of tetrameric ALDHs, direct evidence is rare because biophysical methods for determining oligomeric state are performed at orders of magnitude higher enzyme concentration than is typically used in enzyme assays. In the current report, we circumvented this problem by using negative stain EM, which allows assessment of molecular shape, and hence oligomeric state, at enzyme concentrations similar to those used in activity assays (10 – 100 nM). It would be interesting to investigate the oligomeric structures of other ALDHs similarly.
The high level of structural conservation within the ALDH superfamily implies that the phenomenon of cofactor-enhanced self-association may not be limited to ALDH7A1. We suggest that NAD+ could help promote tetramerization in other ALDH superfamily members. All studied ALDHs have the same protein fold and use an NAD(P)(H) cofactor, and most ALDHs form the dimer-of-dimers tetramer present in ALDH7A1. Our results suggest the potential for a conserved mode of enzyme activation through cofactor-mediated oligomeric assembly amongst tetrameric ALDH superfamily members.
The precise mechanism by which the binding of NAD+ enhances tetramer formation remains to be determined. One possibility is NAD+-binding stabilizes the tetramer. Although the crystal structures of apo (PDB ID: 4ZUK) and NAD+-bound (PDB ID: 4ZVW) ALDH7A1 do not exhibit any major differences, PDBePISA analysis indicates NAD+ binding increases the interface area within the tetramer by ~5%, consistent with the cofactor stabilizing the tetramer [47]. Another possibility is that the binding of NAD+ to the dimer induces structural changes that promote tetramer formation. Elucidating the nature of these conformational changes is challenging, because tetrameric ALDHs invariably crystallize as tetramers, and thus the structure of the free dimer is elusive. Future studies utilizing cryo-EM to study the high-resolution structure of the apo and NAD+-bound enzymes at low concentration may allow for determination of what, if any, conformational changes occur in response to NAD+-binding.
Data deposition
The averaged SAXS curves have been deposited in the SASBDB [48] under the accession codes SASDD29, SASDD39, SASDD49 (apo ALDH7A1 low - high) and SASDD59, SASDD69, SASDD79 (ALDH7A1 + NAD+ low - high).
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the NIGMS of the National Institutes of Health under award number R01GM093123, as well as an administrative supplement to R01GM065546 (Collaborative Supplements for Cryo-Electron Microscopy Technology Transfer). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by grant 9 P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- [1].van Karnebeek CD et al. (2016). Pyridoxine-dependent epilepsy: An expanding clinical spectrum. Pediatr Neurol 59, 6–12. [DOI] [PubMed] [Google Scholar]
- [2].Stockler S et al. (2011). Pyridoxine dependent epilepsy and antiquitin deficiency: clinical and molecular characteristics and recommendations for diagnosis, treatment and follow-up. Mol. Genet. Metab 104, 48–60. [DOI] [PubMed] [Google Scholar]
- [3].Mills PB et al. (2010). Genotypic and phenotypic spectrum of pyridoxine-dependent epilepsy (ALDH7A1 deficiency). Brain 133, 2148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mills PB et al. (2006). Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat. Med 12, 307–9. [DOI] [PubMed] [Google Scholar]
- [5].Mozzarelli A and Bettati S (2006). Exploring the pyridoxal 5’-phosphate-dependent enzymes. Chem Rec 6, 275–87. [DOI] [PubMed] [Google Scholar]
- [6].Luo M, Gates KS, Henzl MT and Tanner JJ (2015). Diethylaminobenzaldehyde is a covalent, irreversible inactivator of ALDH7A1. ACS Chem. Biol 10, 693–7. [DOI] [PubMed] [Google Scholar]
- [7].Luo M and Tanner JJ (2015). Structural basis of substrate recognition by aldehyde dehydrogenase 7A1. Biochemistry 54, 5513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brocker C et al. (2010). Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J. Biol. Chem 285, 18452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Morgan CA and Hurley TD (2015). Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1. Chem Biol Interact 234, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tsybovsky Y and Krupenko SA (2011). Conserved catalytic residues of the ALDH1L1 aldehyde dehydrogenase domain control binding and discharging of the coenzyme. J Biol Chem 286, 23357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen Y, Zhu JY, Hong KH, Mikles DC, Georg GI, Goldstein AS, Amory JK and Schonbrunn E (2018). Structural Basis of ALDH1A2 Inhibition by Irreversible and Reversible Small Molecule Inhibitors. ACS Chem Biol 13, 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Moretti A, Li J, Donini S, Sobol RW, Rizzi M and Garavaglia S (2016). Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD(+) and retinoic acid. Sci Rep 6, 35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Steinmetz CG, Xie P, Weiner H and Hurley TD (1997). Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5, 701–11. [DOI] [PubMed] [Google Scholar]
- [14].Koncitikova R, Vigouroux A, Kopecna M, Andree T, Bartos J, Sebela M, Morera S and Kopecny D (2015). Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7. Biochem. J 468, 109–23. [DOI] [PubMed] [Google Scholar]
- [15].Kim YG, Lee S, Kwon OS, Park SY, Lee SJ, Park BJ and Kim KJ (2009). Redox-switch modulation of human SSADH by dynamic catalytic loop. EMBO J 28, 959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Do H et al. (2016). Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii. J Microbiol 54, 114–21. [DOI] [PubMed] [Google Scholar]
- [17].Kopecna M et al. (2017). The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase. Plant J 92, 229–243. [DOI] [PubMed] [Google Scholar]
- [18].Huo L et al. (2015). Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action. Nat Commun 6, 5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Johansson K, El-Ahmad M, Ramaswamy S, Hjelmqvist L, Jornvall H and Eklund H (1998). Structure of betaine aldehyde dehydrogenase at 2.1 A resolution. Protein Sci 7, 2106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Di Costanzo L, Gomez GA and Christianson DW (2007). Crystal structure of lactaldehyde dehydrogenase from Escherichia coli and inferences regarding substrate and cofactor specificity. J Mol Biol 366, 481–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM and Kunkel BN (2018). Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog 14, e1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Son HF, Park S, Yoo TH, Jung GY and Kim KJ (2017). Structural insights into the production of 3-hydroxypropionic acid by aldehyde dehydrogenase from Azospirillum brasilense. Sci Rep 7, 46005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zarzycki J, Sutter M, Cortina NS, Erb TJ and Kerfeld CA (2017). In Vitro Characterization and Concerted Function of Three Core Enzymes of a Glycyl Radical Enzyme - Associated Bacterial Microcompartment. Sci Rep 7, 42757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Coitinho JB, Pereira MS, Costa DM, Guimaraes SL, Araujo SS, Hengge AC, Brandao TA and Nagem RA (2016). Structural and Kinetic Properties of the Aldehyde Dehydrogenase NahF, a Broad Substrate Specificity Enzyme for Aldehyde Oxidation. Biochemistry 55, 5453–63. [DOI] [PubMed] [Google Scholar]
- [25].Korasick DA, Tanner JJ and Henzl MT (2017). Impact of disease-Linked mutations targeting the oligomerization interfaces of aldehyde dehydrogenase 7A1. Chem Biol Interact 276, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Korasick DA, Wyatt JW, Luo M, Laciak AR, Ruddraraju K, Gates KS, Henzl MT and Tanner JJ (2017). Importance of the C-Terminus of Aldehyde Dehydrogenase 7A1 for Oligomerization and Catalytic Activity. Biochemistry 56, 5910–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rodriguez-Zavala JS and Weiner H (2002). Structural aspects of aldehyde dehydrogenase that influence dimer-tetramer formation. Biochemistry 41, 8229–37. [DOI] [PubMed] [Google Scholar]
- [28].Franke D et al. (2017). ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr 50, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Konarev PV, Volkov VV, Sokolova AV, Koch MHJ and Svergun DI (2003). PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr 36, 1277–1282. [Google Scholar]
- [30].Schneidman-Duhovny D, Hammel M, Tainer JA and Sali A (2016). FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res 44, W424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J 78, 1606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cole JL, Lary JW, T PM and Laue TM (2008). Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol 84, 143–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rames M, Yu Y and Ren G (2014). Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy. J Vis Exp, 90, e51087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mindell JA and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–47. [DOI] [PubMed] [Google Scholar]
- [35].Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Selwood T and Jaffe EK (2012). Dynamic dissociating homo-oligomers and the control of protein function. Arch Biochem Biophys 519, 131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].van Berkel WJ, Benen JA and Snoek MC (1991). On the FAD-induced dimerization of apo-lipoamide dehydrogenase from Azotobacter vinelandii and Pseudomonas fluorescens. Kinetics of reconstitution. Eur J Biochem 197, 769–79. [DOI] [PubMed] [Google Scholar]
- [38].Lindsay H, Beaumont E, Richards SD, Kelly SM, Sanderson SJ, Price NC and Lindsay JG (2000). FAD insertion is essential for attaining the assembly competence of the dihydrolipoamide dehydrogenase (E3) monomer from Escherichia coli. J Biol Chem 275, 36665–70. [DOI] [PubMed] [Google Scholar]
- [39].Van Berkel WJ, Van den Berg WA and Muller F (1988). Large-scale preparation and reconstitution of apo-flavoproteins with special reference to butyryl-CoA dehydrogenase from Megasphaera elsdenii. Hydrophobic-interaction chromatography. Eur J Biochem 178, 197–207. [DOI] [PubMed] [Google Scholar]
- [40].Tahallah N, Van Den Heuvel RH, Van Den Berg WA, Maier CS, Van Berkel WJ and Heck AJ (2002). Cofactor-dependent assembly of the flavoenzyme vanillyl-alcohol oxidase. J Biol Chem 277, 36425–32. [DOI] [PubMed] [Google Scholar]
- [41].Bellesis AG, Jecrois AM, Hayes JA, Schiffer CA and Royer WE Jr. (2018). Assembly of human C-terminal binding protein (CtBP) into tetramers. J Biol Chem 293, 9101–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Madison DL, Wirz JA, Siess D and Lundblad JR (2013). Nicotinamide adenine dinucleotide-induced multimerization of the co-repressor CtBP1 relies on a switching tryptophan. J Biol Chem 288, 27836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Balasubramanian P, Zhao LJ and Chinnadurai G (2003). Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett 537, 157–60. [DOI] [PubMed] [Google Scholar]
- [44].Hohman RJ, Guitton MC and Veron M (1984). Purification of S-adenosyl-L-homocysteine hydrolase from Dictyostelium discoideum: reversible inactivation by cAMP and 2’-deoxyadenosine. Arch Biochem Biophys 233, 785–95. [DOI] [PubMed] [Google Scholar]
- [45].Kumar A, Subramanian Manimekalai MS and Gruber G (2018). Substrate-induced structural alterations of Mycobacterial mycothione reductase and critical residues involved. FEBS Lett 592, 568–585. [DOI] [PubMed] [Google Scholar]
- [46].Stein LR and Imai S (2012). The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 23, 420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Krissinel E and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–97. [DOI] [PubMed] [Google Scholar]
- [48].Valentini E, Kikhney AG, Previtali G, Jeffries CM and Svergun DI (2015). SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 43, D357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


