Abstract
Medical genomics research performed in diverse population facilitates a better understanding of the genetic basis of developmental disorders, with regional implications for community genetics. Autosomal recessive mitochondrial complex I deficiency (MCID) accounts for a constellation of clinical features, including encephalopathies, myopathies, and Leigh Syndrome. Using whole-exome sequencing, we identified biallelic missense variants in NDUFV1 that encodes the 51-kD subunit of complex I (NADH dehydrogenase) NDUFV1. Mapping the variants on published crystal structures of mitochondrial complex I demonstrate that the novel c.1118T > C (p.(Phe373Ser)) variant is predicted to diminish the affinity of the active pocket of NDUFV1 for FMN that correlates to an early onset of debilitating MCID symptoms. The c.1156C > T (p.(Arg386Cys)) variant is predicted to alter electron shuttling required for energy production and correlate to a disease onset in childhood. NDUFV1 c.1156C > T (p.(Arg386Cys)) represents a founder variant in South Asian populations that have value in prioritizing this variant in a population-specific manner for genetic diagnostic evaluation. In conclusion, our results demonstrate the advantage of analyzing population-specific sequences to understand the disease pathophysiology and prevalence of inherited risk variants in the underrepresented populations.
Introduction
Aggregation of large-scale sequencing data and increasing clinical use of whole-exome sequencing (WES) for genetic diagnosis, is uncovering genetic variation that aids in annotation and interpretation of disease alleles. Increased inclusion of WES data from diverse populations, underrepresented in genomic databases uncovers clinically significant genetic variation that advances our understanding of disease alleles and underlying biology. Elucidating rare genetic diversity and how it contributes to Mendelian inherited disorders has implications for community genetics, where a better understanding of the ancestry-specific genomics can direct carrier testing, presymptomatic diagnosis, and potential interventions to delay the onset of symptoms in a community-specific manner.
Autosomal recessive mitochondrial complex I deficiency (MCID) accounts for approximately 23% of childhood respiratory chain deficiency cases [1]. Mitochondrial complex I is composed of 44 structural subunits and over 10 assembly factors, which underscores its diverse clinical manifestations that diverge based on severity and age of onset [2]. The clinical presentations range from infant onset subacute necrotizing encephalomyelopathy (Leigh syndrome [MIM: 256000]) to adult-onset exercise-induced myopathy. Genetic studies have identified disease-causing variants in 11 nuclear-encoded complex I genes [MIM: 252010], including NDUFV1 [3–13].
In the present study, we report genetic findings from two unrelated probands from the South Asian population (Country of origin: India, region: Southern India) that presented with divergent features of MCID. Proband 1 exhibited symptoms of MCID at 6 months of age and presented with a novel homozygous c.1118T > C (p.(Phe373Ser)) missense variant in exon 8 of NDUFV1. Proband 2 was homozygous for the previously reported c.1156C > T (p.(Arg386Cys)) variant, also in exon 8 of NDUFV1 (Figs. 1c, 2a), but did not exhibit symptoms until 6 years of age [14–16]. The discrepancy in clinical presentation and age of onset between these cases is supported by the molecular impact as modeled on published crystal structures of mitochondrial complex I. The NDUFV1 p.(Arg386Cys) variant exhibits higher frequency in South Asian population suggesting a founder effect with implication for community genetics [14].
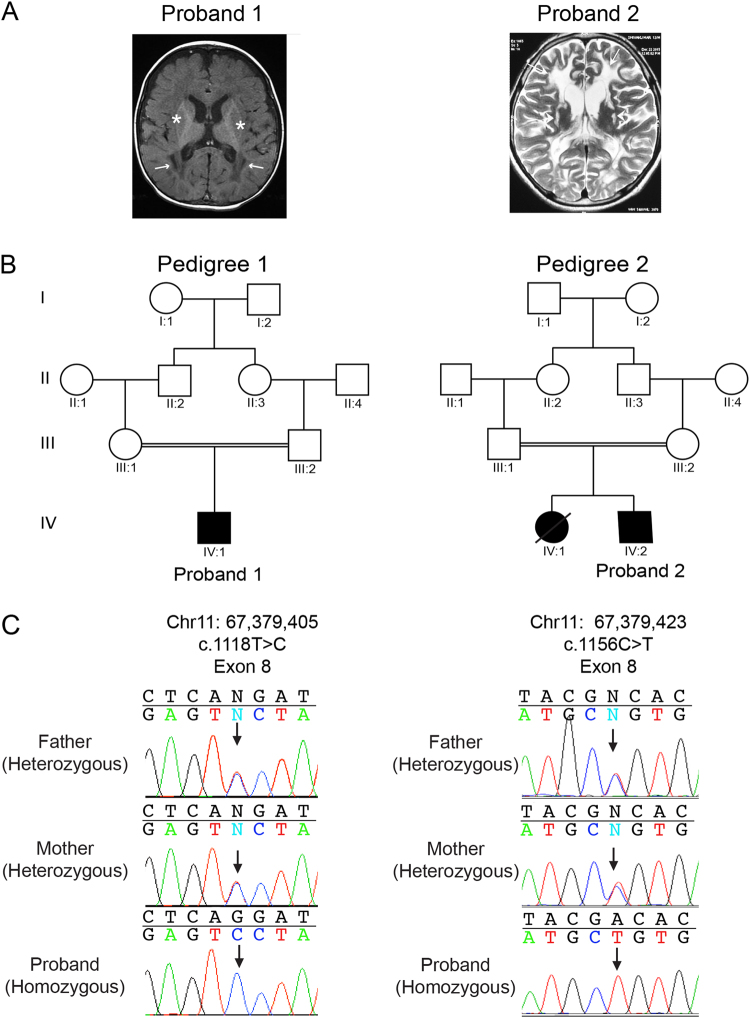
Fig. 1.
Brain imaging and NDUFV1 variants in Proband 1 and 2. a T1-weighted axial brain MRI of proband 1 at 6 months demonstrates pachy diffusion involving centrum semiovale, corona radiate (arrows), and periventricular external capsule (stars). Proband 2 T2 weighted axial brain MRI at 6 years of age shows diffuse white matter demyelination with gliotic areas and atrophy. b Pedigrees for family 1 and family 2, both of South Asian population. Filled symbols denote individuals affected with the MCID. Double lines denote consanguinity. c Sequence chromatograms showing the biallelic inheritance of NDUFV1 missense variants (black arrows) in proband 1 and 2 (lower panel, proband) consistent with consanguinity in the family
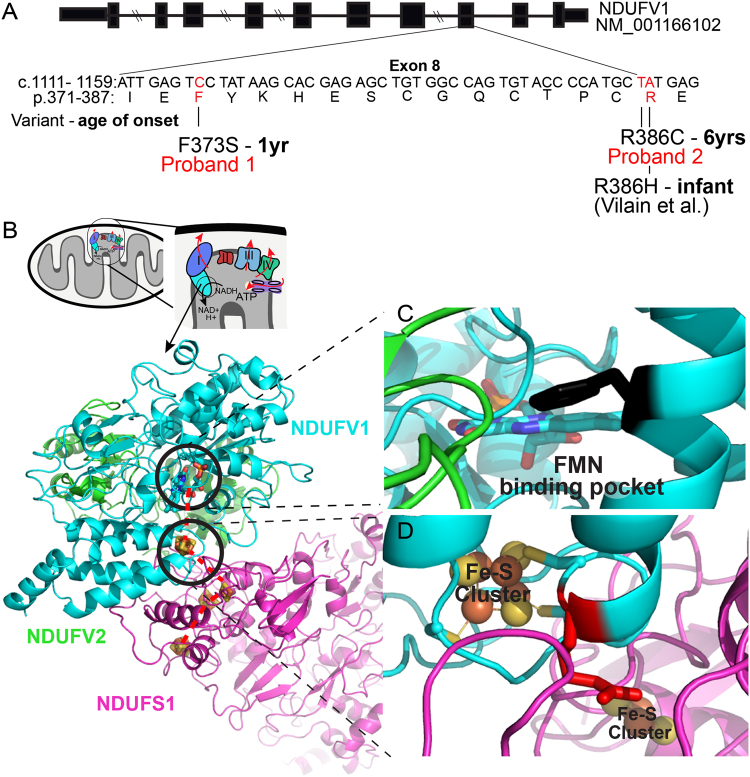
Fig. 2.
a Schematic showing the intron-exon structure of human NDUFV1. The cDNA sequence from position 1111 (NM_007103.3) and the corresponding peptide sequence from Ile371 to Glu387 are shown. Genetic variants that affect function or amino acid substitutions identified in mitochondrial complex I deficiency patient are marked in red. b Structure of mammalian mitochondria and close-up depicting membrane embedded respiratory chain complexes. The circled region shows the N module of complex I, composed of NDUFV1, NDUFV2, and NDUFS1 proteins. Crystal structure of N module derived from T. thermophilus (3IAM), where colors denote distinct proteins (blue = NDUFV1, pink = NDUFS, and green = NDUFV2). Colored elements: Fe–S cluster = red and orange, FMN binding pocket = blue and green, and missense variants identified in proband 1 and 2 are encircled with bold circle. Dashed red lines denote the trajectory of shuttled electrons. c Magnified image of substituted amino acid close to FMN binding pocket in proband 1. The amino acid in black protruding out from alpha helix is the phenylalanine at position 373. The variation p.(Phe373Ser) disrupts the FMN binding to the NDUFV1. (d) Magnification of amino acids close to Fe–S cluster disrupted by proband 2 substitution. Arginine at position 386 is shown in red. The p.(Arg386Cys) substitution will alter buffering of Fe–S clusters and electrons transfer across NDUFV1
Clinical reports
Proband 1
One-year-old proband 1, the first born of third-degree consanguineous parents (Fig. 1b), was referred for developmental delay. He was born at full term by normal vaginal delivery. He weighed 2.5 kg at birth. An excessive cry was noted. Seizures were first observed at the age of 6 months. His development was delayed mildly, with momentary loss of head control, and roll over at 7 months of age. He was diagnosed with myopia. He started smiling and reaching for objects after the use of spectacles. On clinical examination at the age of 1 year, height was 78.5 cm (−0.9 SD), head circumference was 49 cm (+1 SD), and weight was 9.8 kg (−1 SD). He was observed to have bilateral lower set ears, nystagmus, mosaic pigmentary anomalies, hepatomegaly, and spasticity in lower limbs, extreme plantar responses and brisk deep tendon reflexes. Electroencephalogram and fundus examinations were found to be normal. Brain imaging sequences show diffuse hyperintensity in the cerebral white matter, cerebellar white matter and brainstem white matter, and small cystic areas in the periventricular white matter (Fig. 1a). Magnetic resonance spectrometry showed reduced N-acetyl aspartate and elevated choline levels and an inverted double peak of lactate.
Proband 2
Proband 2, second born of third-degree consanguineous parents (Fig. 1b), developed normally until the 6 years of age at which time he presented with neuroregression, mild cognitive decline with regressive speech deficiencies, bilateral optic atrophy, and marked motor decline. Proband 2 was of normal height and weight at 137 cm and 25 kg. History of seizures was noted. Sibling with similar clinical features died at the age of 15 years. Physical examination showed a height of 137 cm (−1 SD), head circumference of 46 cm (−4 SD), and weight 25 kg (−3 SD) with spasticity in all four limbs, clonus, and nystagmus. Optic atrophy was reported on ophthalmology evaluation. However, blood lactate and pyruvate levels were within normal range. Brain imaging displayed a diffuse white matter demyelination with cystic areas consistent with neurodegeneration (Fig. 1a). Based on these features, proband 2 was provided with a diagnosis of leukodystrophy.
Material and methods
Ethics statement and clinical sample collection
The present study is a part of an ongoing combined clinical/research project (Ethical approval number: Indo-Foreign/Neuro/154/2015) that started in the year 2013 to recruit individuals with inherited neurodevelopmental disorders. A total of 450 consanguineous families from Southern India have been recruited to date. The parents provided written informed consent for WES. DNA was isolated from peripheral blood by standard procedures [17].
Whole-exome sequencing
Exome libraries of affected and unaffected genomic DNA were generated using the Illumina TruSeq DNA Sample Prep kit, following the manufacturer’s instructions. Parents-proband WES was performed on family 1. Coding sequences were prepared and captured with the Agilent SureSelect All Exon kit-v4 and sequenced on an Illumina HiSeq 2500 instrument as described previously [18]. Only proband 2 underwent WES in family 2, as previously described [19].
Variant calling and filtering
WES data was processed using GATK callers and SeqMule. ANNOVAR was used to functionally annotate the detected genetic variants [20]. Variants were further filtered against public databases such as 1000 Genomes Project phase 3, ExAC, National Heart, Lung, and Blood Institute and Exome Sequencing Project Exome Variant Server (ESP6500SI-V2). Variants flagged as low quality or putative false positives (Phred quality score < 20 and low quality by depth < 20) and minor allele frequency > 1% was excluded from the analysis.
Protein modeling
Mitochondrial complex I crystal structure of bacterial [T. thermophilus (PDB:3IAM)] and bovine [Bos Taurus (PDB:5lc5)] was obtained from Protein Data Bank (http://www.rcsb.org/pdb/). Sequences were mapped to Homo sapiens using ClustalW. Pymol software was used to model the structure of N-modules of bacterial and bovine mitochondrial complex I.
Results
Variant prioritization
In Proband 1, seven rare variants, including those in NDUFV1 (NM_007103.3) were prioritized as candidates for the clinical presentation. Among these, only one was responsible for neurodevelopmental disorders; homozygous missense variant in NRXN2 was ruled out because of its prevalence in public databases and inconsistencies in associated phenotypic outcomes (Supplemental Table 1). Alternatively, the novel homozygous missense c.1118T > C (p.(Phe373Ser)) NDUFV1 variant accounts for the MCID phenotype that overlaps with the clinical features of proband 1. The NDUFV1 c.1118T > C (p.(Phe373Ser)) variant was not detected in ExAC browser. The variant was submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/431452/; Submission Accession: SCV000588197.1).
However, in proband 2, three rare variants were prioritized in the genes NDUFV1, KIRREL3, and TFG. Among these, the biallelic NDUFV1 variant c.1156C > T (p.(Arg386Cys)) located in exon 8 (numbered as in NG_013353.1; NM_007103.3) was deemed to affect function in silico by PolyPhen-2 http://genetics.bwh.harvard.edu/pph2/) supported by a NDUFV1 c.1157G > A (p.(Arg386His)) substitution previously associated with severe MCID and onset of symptoms in infancy (Fig. 2a) [21]. Variant p.(Arg386Cys) is a previously published variant in additional unrelated individuals from the South Asian population and its submission accession is SCV000566902.2. The parents were heterozygous for both the variants identified in Proband 1 and 2, as confirmed by Sanger sequencing and consistent with consanguinity (Fig. 1c)
Protein modeling analysis
We modeled the functional impact of proband 2 homozygous NDUFV1 p.(Arg386Cys) variant relative to the previously characterized p.(Arg386His) variant that alters the same amino acid Arg386, on the bacterial and bovine crystal structure of mitochondrial complex I [22–25] (Fig. 2b). The amino acid substitution p.(Arg386Cys) is predicted to disrupt the protein–protein interactions that facilitate Fe–S cluster buffering. In the three-dimensional conformation of complex I, Arg386 is in close proximity to Cys385, which participates in Fe–S cluster stabilization within the complex and buffering (Fig. 2d). The volume of the Cys side chain is small compared to that of His as observed in amino acid substitution p.(Arg386His). Increased bulkiness of His may perturb local interactions between Fe–S binding motifs of NDUFV1. However, the functional consequence of having two consecutive Cys at the Fe–S binding motif, as would be the case with the p.(Arg386Cys) substitution is predicted to be less disruptive to the Fe–S binding motif, and further compensate for this substitution by facilitating Fe–S cluster buffering.
Protein modeling and crystal structure analysis of p.(Phe373Ser) suggests that the variant is located in FMN/NADH binding site of complex I and is involved in FMN binding. Substitution of the highly hydrophobic Phe to a small and polar Ser is predicted to diminish the affinity for FMN to the active pocket of NDUFV1, altering the first step of electron transfer that promotes the redox activity of complex I. We surveyed the published variants in NDUFV1 relative to the functions ascribed by yeast genetic screens to demonstrate the predictive value of evaluating pathogenicity relative to the available mammalian crystal structure (Fig. 2c). These findings are summarized in supplemental Table 2.
Variant interpretation: ACMG guidelines
According to the variant classification guidelines of ACMG, genetic variant p.(Phe373Ser) is categorized as “likely pathogenic” whereas variant p.(Arg386Cys) is categorized as “pathogenic” (Supplemental Table 3).
Discussion
Mitochondrial complex I of the respiratory chain functions to liberate and transfer electrons from NADH to ubiquinone for ATP production. Missense variants in NDUFV1 can disrupt three important functions of complex I: (1) binding of FMN and NADH, (2) transfer of electrons between iron–sulfur clusters, and (3) structural integrity required to maintain the interactions between complex I subunits. Yeast functional studies have proven useful for assaying deleterious alleles in NDUFV1, but do not differentiate between alleles corresponding to the clinical outcome of differing severity. Modeling the impact of missense substitutions in NDUFV1 relative to these functions, using the bacterial and bovine complex I crystal structures, provide evidence to support the molecular impact of deleterious amino acid substitutions. Using this approach, we provide a rationale for how substitutions of the same amino acid residue, i.e., p.(Arg386Cys) and p.(Arg386His) result in MCID with different age of onset and clinical outcomes. Genetic variants p.(Arg386Cys) and p.(Arg386His) were both classified as strongly functionally impacted variants in the yeast genetic screens [24]. Based on the bacteria and bovine crystal structure, the p.(Arg386His) substitution results in an amino acid side chain that is bulkier than the p.(Arg386Cys) substitution. The bulkier His side chain is predicted to more severely disrupt protein–protein interaction between NDUFV1 and NDUFS1 and buffering between the Fe–S clusters through complex I than the Cys, providing a molecular rationale for the less severe later onset MCID associated with the p. (Arg386Cys) variant, as compared with p. (Arg386His) that is associated with early onset of severe clinical manifestations followed by death in infancy [21].
Functional validation of our novel p.(Phe373Ser) variant has not been performed in yeast. Modeling studies suggest that p.(Phe373Ser) lies in the close proximity to Arg88, Lys111, Ala117 and Glu246, NDUFV1 amino acids shown to be deleterious in functional assays when mutated to clinically relevant MCID substitutions. These amino acids maintain the conformation of the FMN/NADH binding pocket, implicating a role for variant p.(Phe373Ser) in complex I function. This analysis approach predicts that p.(Phe373Ser) affects complex I function and the severity of clinical outcomes can be traced back to the nature of the amino acid substitution, with those most dissimilar to the wildtype amino acid negatively impacting FMN binding kinetics and clinical outcomes.
Heterozygous NDUFV1 c.1156C > T (p.(Arg386Cys)) was detected at a frequency of 0.0001 in the ExAC database. While rare, it is noteworthy that 13 of the 15 c.1156C > T (p.(Arg386Cys)) alleles described were observed in South Asian populations and accentuates a pervious report that associated this allele to late-onset MCID, in unrelated South Asian families [14]. Additionally, our inherited neurodevelopmental disorder cohort of 450 consanguineous families also identified NDUFV1 c.1156C > T (p.(Arg386Cys)) in one of the two families. Altogether, the recessive inheritance of c.1156C > T, independent of consanguinity, suggests this allele is a founder variant, yet defining enrichment in this population is complicated by the underrepresentation of this population in ExAC or related sequence databases [16]. Our results demonstrate the value of including genetically diverse populations in genomic medicine research. Consequently, first-tier genetic screening of c.1156C > T (p.(Arg386Cys)) may prove to have high molecular diagnostic yield for South Asian children that present with cardinal features of late-onset MCID.
In conclusion, our results provide a rationale for how substitutions of the same amino acid residue can be associated with different ages of onset. We also demonstrate the value of including ancestrally diverse population in genomic medicine and clinical research studies, to improve the reliability of molecular diagnosis and reduce global health disparities.
Electronic supplementary material
Candidate genes pointed by whole-exome sequencing of proband 1 and proband 2
Acknowledgments
We thank the patient and all the family members for their participation. The study was supported by the grant from National Institute of Health (1R21NS094047-01).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0209-0) contains supplementary material, which is available to authorized users.
References
- 1.Hoefs SJ, Skjeldal OH, Rodenburg RJ, et al. Novel mutations in the NDUFS1 gene cause low residual activities in human complex I deficiencies. Mol Genet Metab. 2010;100:251–6. doi: 10.1016/j.ymgme.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Alston CL, Compton AG, Formosa LE, et al. Biallelic mutations in TMEM126B cause severe complex I deficiency with a variable clinical phenotype. Am J Hum Genet. 2016;99:217–27. doi: 10.1016/j.ajhg.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benit P, Beugnot R, Chretien D, et al. Mutant NDUFV2 subunit of mitochondrial complex I causes early onset hypertrophic cardiomyopathy and encephalopathy. Hum Mutat. 2003;21:582–6. doi: 10.1002/humu.10225. [DOI] [PubMed] [Google Scholar]
- 4.Benit P, Slama A, Cartault F, et al. Mutant NDUFS3 subunit of mitochondrial complex I causes Leigh syndrome. J Med Genet. 2004;41:14–17. doi: 10.1136/jmg.2003.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirby DM, Salemi R, Sugiana C, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest. 2004;114:837–45. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeffen J, Elpeleg O, Smeitink J, et al. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol. 2001;49:195–201. doi: 10.1002/1531-8249(20010201)49:2<195::AID-ANA39>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Loeffen J, Smeitink J, Triepels R, et al. The first nuclear-encoded complex I mutation in a patient with Leigh syndrome. Am J Hum Genet. 1998;63:1598–608. doi: 10.1086/302154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuelke M, Smeitink J, Mariman E, et al. Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat Genet. 1999;21:260–1. doi: 10.1038/6772. [DOI] [PubMed] [Google Scholar]
- 9.Triepels RH, van den Heuvel LP, Loeffen JL, et al. Leigh syndrome associated with a mutation in the NDUFS7 (PSST) nuclear-encoded subunit of complex I. Ann Neurol. 1999;45:787–90. doi: 10.1002/1531-8249(199906)45:6<787::AID-ANA13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.van den Heuvel L, Ruitenbeek W, Smeets R, et al. Demonstration of a new pathogenic mutation in human complex I deficiency: a 5-bp duplication in the nuclear gene encoding the 18-kD (AQDQ) subunit. Am J Hum Genet. 1998;62:262–8. doi: 10.1086/301716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bénit P, Chretien D, Kadhom N, et al. Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am J Hum Genet. 2001;68:1344–52. doi: 10.1086/320603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O. Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol. 2008;63:405–8. doi: 10.1002/ana.21332. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Moreira D, Ugalde C, Smeets R, et al. X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol. 2007;61:73–83. doi: 10.1002/ana.21036. [DOI] [PubMed] [Google Scholar]
- 14.Marin SE, Mesterman R, Robinson B, Rodenburg RJ, Smeitink J, Tarnopolsky MA. Leigh syndrome associated with mitochondrial complex I deficiency due to novel mutations In NDUFV1 and NDUFS2. Gene. 2013;516:162–7. doi: 10.1016/j.gene.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Breningstall GN, Shoffner J, Patterson RJ. Siblings with leukoencephalopathy. Semin Pediatr Neurol. 2008;15:212–5. doi: 10.1016/j.spen.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girisha KM, Shukla A, Trujillano D, et al. A homozygous nonsense variant in IFT52 is associated with a human skeletal ciliopathy. Clin Genet. 2016;90:536–9. doi: 10.1111/cge.12762. [DOI] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilain C, Rens C, Aeby A, et al. A novel NDUFV1 gene mutation in complex I deficiency in consanguineous siblings with brainstem lesions and Leigh syndrome. Clin Genet. 2012;82:264–70. doi: 10.1111/j.1399-0004.2011.01743.x. [DOI] [PubMed] [Google Scholar]
- 22.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–8. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–6. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 24.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varghese F, Atcheson E, Bridges HR, Hirst J. Characterization of clinically identified mutations in NDUFV1, the flavin-binding subunit of respiratory complex I, using a yeast model system. Hum Mol Genet. 2015;24:6350–60. doi: 10.1093/hmg/ddv344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Candidate genes pointed by whole-exome sequencing of proband 1 and proband 2