Abstract
DNA methylation is a conserved epigenetic mark that is critical for many biological processes in plants and mammals. In Arabidopsis, the antagonistic activities of RNA-directed DNA methylation (RdDM) and ROS1-dependent active DNA demethylation are key for the dynamic regulation of locus-specific DNA methylation. However, the molecular factors that coordinate RdDM and active demethylation are largely unknown. Here we report that CLSY4 and its three paralogous SWI2/SNF2-type chromatin-remodeling proteins function in both RdDM and DNA demethylation in Arabidopsis. We initially identified CLSY4 in a genetic screen for DNA demethylation factors and subsequently demonstrated that it also is important in RdDM. Comprehensive genetic analyses using single and high order mutants of CLSY family proteins revealed their roles as double agents in the balance between methylation and demethylation reactions. The four CLSY proteins collectively are necessary for the canonical RdDM pathway; at the same time, each CLSY likely mediates DNA demethylation at specific loci where DNA methylation depends on RdDM. These results indicate that the four chromatin-remodeling proteins have dual functions in regulating genomic DNA methylation, and thus provide new insights into the dynamic regulation of DNA methylation in a model multicellular eukaryotic organism.
Introduction
DNA methylation is an important epigenetic mark that confers transcriptional regulation of genes and transposable elements1–4. Aberrant DNA methylation can disrupt normal developmental processes and cause disease symptoms in plants and mammals5,6. A specific DNA methylation pattern is an outcome of dynamic regulation by methylation establishment and maintenance, together with methylation removal activities.
In plants, de novo DNA methylation can be established by the RNA-directed DNA methylation (RdDM) pathway, in which complementary pairing between 24-nt siRNAs and nascent scaffold RNAs, together with protein–protein interactions, recruits the DNA methyltransferase DRM2 for DNA methylation4,7–10. Production of nearly all 24-nt siRNAs depends on the plant-specific RNA polymerase Pol IV, which generates single-stranded non-coding RNAs that serve as templates for downstream processing by RDR2 (RNA-dependent RNA polymerase 2) and DCL3 (Dicer-like 3) to produce 24-nt siRNAs11–17. Pol IV is recruited to RdDM targets in part by a SAWADEE domain-containing protein, SHH1/DTF1, which binds via its Tudor domain to histone H3 with methylated H3K918,19. Both Pol IV and DTF1/SHH1 associate with CLSY118,19, which is a chromatin-remodeling protein involved in Pol IV-dependent siRNA production20. However, it appears that CLSY1 only affects a part of Pol IV-dependent RdDM21, and it is unclear whether other chromatin-remodeling proteins are involved in Pol IV-dependent siRNA production and DNA methylation.
Active DNA demethylation counteracts the establishment and maintenance of DNA methylation to prevent DNA hypermethylation at specific loci22. In plants, a family of bifunctional 5-mC DNA glycosylases initiates active DNA demethylation through a base excision repair pathway23–26. ROS1 is a major DNA demethylase in Arabidopsis and is particularly important for counteracting DNA methylation established by the RdDM pathway, although it also demethylates RdDM-independent methylation at some genomic regions22,23,27,28. The ROS1 gene promoter contains a short DNA sequence termed MEMS (DNA MEthylation Monitoring Sequence). DNA methylation is positively correlated with ROS1 gene expression and is co-regulated by the RdDM pathway, by the CG methylation maintenance methyltransferase MET1, and by ROS1 itself;29–31 thus MEMS serves as an indicator of general methylation and demethylation activities and enables the coordination of DNA methylation and active DNA demethylation through transcriptional regulation of ROS1. Although it is evident that DNA methylation is dynamically regulated by methylation and demethylation processes, the underlying mechanisms of this fine dynamic regulation, besides the transcriptional regulation of ROS1, remain to be discovered.
In this study, our genetic screen for Arabidopsis DNA demethylation factors led to the isolation of a clsy4 mutant. Investigations on single, triple, and quadruple mutants of CLSY1, CLSY2, CLSY3, and CLSY4 revealed that these four chromatin-remodeling proteins not only redundantly function in Pol IV-dependent RdDM, but also individually mediate active DNA demethylation at specific genomic regions including RdDM targets. Our results show that the four CLSY proteins collectively are necessary for Pol IV-dependent siRNA production and DNA methylation in the canonical RdDM pathway. Importantly, each CLSY also mediates DNA demethylation at loci where DNA methylation depends on one or more of the other three CLSY proteins, thereby providing a novel mechanism underlying the dynamic regulation of DNA methylation in Arabidopsis.
Results
Identification of CLSY4/CHR40 as a DNA demethylation factor
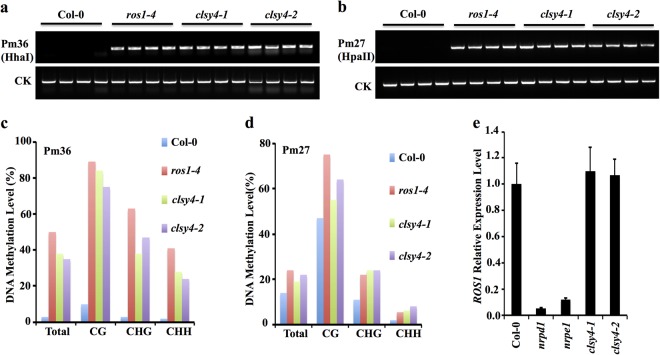
In searching for molecular factors involved in active DNA demethylation, we conducted a genetic screen in a T-DNA mutant collection (Supplementary Table S1) using Chop-PCR to assess the DNA methylation status of two genomic loci, Pm36 and Pm27 (Supplementary Fig. S1a). Pm36 and Pm27 are located in the 3′ regions of At1g26400 and At1g26390, respectively, and both show DNA hypermethylation in the ros1 mutant and were used successfully as markers to isolate idm1 (increased DNA methylation 1) and idm2 (increased DNA methylation 2) mutants that are defective in the regulation of active DNA demethylation (Supplementary Fig. S1a)32,33. In a T-DNA insertion mutant (SALK_102252) of At3g24340, both Pm36 and Pm27 could be amplified after methylation-sensitive enzyme digestion, suggesting DNA hypermethylation in this mutant (Fig. 1a, b; Supplementary Fig. S1b, c). Moreover, another T-DNA insertion mutant allele for this gene (SALK_003876) also exhibited DNA hypermethylation (Fig.1a, b; Supplementary Fig. S1b, c). At3g24340 encodes Chromatin Remodeling 40 (CHR40), a member of the ATP-dependent chromatin remodeler protein family that is also known as CLSY434. To confirm the DNA hypermethylation phenotype, we performed bisulfite sequencing at the two loci and found that DNA methylation levels in all cytosine contexts (CG, CHG, and CHH, where H represents A, T, and C) were increased in both clsy4/chr40 alleles as is also the case in the ros1 mutant (Fig. 1c, d).
Fig. 1. Identification of CLSY4/CHR40 as a regulator of DNA demethylation.
a DNA methylation analysis at Pm36 by chop-PCR with digestion by the methylation-sensitive enzyme HhaI. Undigested DNA was used as the control (CK). b DNA methylation analysis at Pm27 by chop-PCR with digestion by the methylation-sensitive enzyme HpaII. Undigested DNA was used as the control. c and d Individual locus bisulfite sequencing at Pm36 (c) and Pm27 (d). e qRT-PCR analysis of ROS1 transcript levels. Bars indicate the standard error of three biological replicates
Previous reports showed that the reduced ROS1 expression in various RdDM mutants causes increased DNA methylation at certain loci in the genome22,30,35. Given that CHR40 was previously found to be associated with the Pol IV protein complex34, we suspected that DNA hypermethylation in clsy4/chr40 mutants might be caused indirectly by a decrease in ROS1 expression. However, real-time PCR assays showed that the transcript level of ROS1 was not changed in either of the clsy4/chr40 mutant alleles compared to wild-type plants (Fig. 1e), suggesting that CLSY4/CHR40 is more likely to be directly involved in the ROS1-mediated active DNA demethylation pathway.
In addition, transgenic expression of CLSY4 driven by its native promoter rescued the CHH and CHG hypermethylation phenotype in the clsy4-1/chr40-1 mutant (Supplementary Fig. S1d, e), supporting the inference that the mutations in CLSY4/CHR40 are responsible for DNA hypermethylation at the tested loci.
CLSY4 affects DNA methylation at many genomic regions
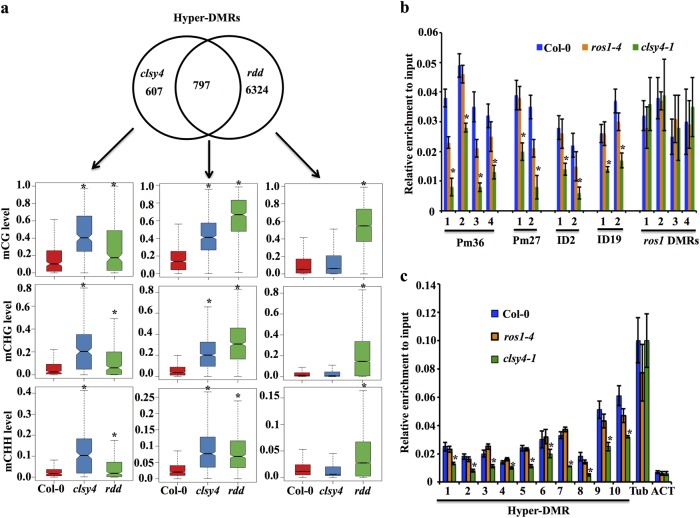
CLSY4/CHR40 belongs to the SWI2/SNF2 family of chromatin-remodeling proteins, and is in the clade of six plant-specific members (Supplementary Fig. S2a) that include CLSY1 and DRD1; DRD1 is a component of the DDR complex required for Pol V function in RdDM36. A previous study reported that CLSY4 could be co-purified with the largest subunit of Pol IV, suggesting its possible role in RdDM34. However, our results above suggested that CLSY4 functions in DNA demethylation. To investigate this inconsistency, we performed high coverage (~60x) whole-genome bisulfite sequencing of the clsy4-1/chr40-1 mutant in order to examine the DNA methylome of clsy4 mutant plants (Supplementary Table S2). We identified 1404 hypermethylated differentially methylated regions (hyper-DMRs) and 314 hypomethylated DMRs (hypo-DMRs) in clsy4-1 mutant plants compared to wild-type plants (Supplementary Fig. S2b). For the hyper-DMRs, DNA methylation in three sequence contexts, including CG, CHG, and CHH, was increased in the clsy4-1/chr40-1 mutant (Fig. 2a). This pattern of increased methylation in all three sequence contexts was also observed in ros1 single and ros1dml2dml3 (rdd) triple mutants32. According to our analysis, about 60% of the hyper-DMRs in clsy4-1/chr40-1 also showed hypermethylation in the rdd mutant (Fig. 2a). Even for the clsy4-specific hyper-DMRs, DNA methylation in CG, CHG, and CHH contexts was also significantly increased in rdd mutant plants, although the increase was less than that in clsy4 (Fig. 2a), possibly due to the remaining 5-mC DNA glycosylase DME in rdd.
Fig. 2. Features of clsy4 hyper-DMRs.
a Venn diagram showing the overlap of hyper-DMRs between clsy4 and rdd mutants, and box plots showing the DNA methylation levels in three sequence contexts at clsy4-specific, rdd-specific, and overlapped DMRs. *Indicates significant difference by Wilcoxon sum test (P < 0.01). b Nucleosome density indicated by histone H3 levels at the Pm36, Pm27, ID2, and ID19 loci with four rdd-specific hyper-DMRs as controls measured by chromatin immunoprecipitation (ChIP). Bars indicate the standard deviation of three biological replicates. *Indicates statistical difference between the wild type and clsy4 according to Student’s t-test (P < 0.01). c Nucleosome density indicated by histone H3 levels at 10 clsy4 hyper-DMRs measured by ChIP. Tubulin 8 and actin 2 served as controls. *Indicates statistical difference between the wild type and clsy4 according to Student’s t-test (P < 0.01). Bars indicate the standard deviation of three biological replicates
Increased DNA methylation levels at two hyper-DMRs, ID2 (increased DNA methylation 2) and ID19, were confirmed by individual locus bisulfite sequencing (Supplementary Fig. S2c, d). The siRNA abundance at Pm36, ID2, and ID19 was not increased in clsy4 mutants, as shown by northern blotting (Supplementary Fig. S2e), indicating that the DNA hypermethylation was not due to increased accumulation of siRNAs. Given that CLSY4 encodes a putative ATP-dependent chromatin-remodeling protein that might regulate nucleosome positioning, we assessed the nucleosome occupancy at the hyper-DMRs37 using anti-histone H3 for ChIP-qPCR assays after MNase digestion. At four genomic regions (Supplementary Fig. S3a) that were confirmed to be hypermethylated by individual locus bisulfite sequencing in both clsy4 and ros1 (Fig. 1c, d; Supplementary Fig. S2c, d), the ChIP signals were significantly lower in clsy4 mutant plants than in wild-type plants, indicating that the nucleosome occupancy at those regions was reduced in clsy4 (Fig. 2b). In contrast, the signals in the wild type and the ros1 mutant were comparable (Fig. 2b), indicating that ROS1 does not affect nucleosome positioning. In addition, at four rdd mutant-specific hyper-DMRs (Supplementary Fig. S3b), the nucleosome occupancy was not deceased in the clsy4 mutant (Fig. 2b), suggesting a direct role of CLSY4 in the regulation of active demethylation. At 10 other randomly selected hyper-DMRs of clsy4 (Supplementary Fig. S4), the clsy4 mutant also showed lower nucleosome occupancy than the wild type and ros1 (Fig. 2c). These results suggest that CLSY4 likely facilitates active DNA demethylation through nucleosome remodeling.
To determine whether the increased DNA methylation in clsy4 affects gene expression, we selected 12 genes near the hyper-DMRs and measured their expression levels (Supplementary Figs. S5, S6). Eight of the 12 selected genes showed reduced transcript levels in the clsy4 mutant (Supplementary Figs. S5, S6). One gene, At3g44910, showed an increased expression in clsy4 and ros1 (Supplementary Fig. S6a), providing a case in which DNA methylation is positively correlated with the transcript level. Taken together, the results show that CLSY4-dependent DNA demethylation contributes to gene regulation.
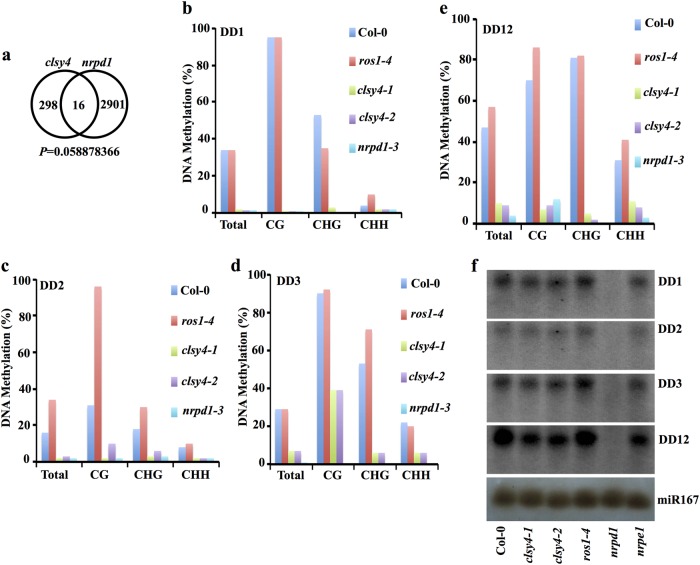
Among the 314 hypo-DMRs identified in the clsy4-1/chr40-1 mutant, only 16 overlapped with nrpd1 hypo-DMRs, suggesting that a large subset of the hypo-DMRs in clys4 mutant are not RdDM target loci (Fig. 3a). Four common hypo-DMRs were selected for validation by individual locus bisulfite sequencing, which showed that DNA methylation was indeed reduced in both the clsy4 and nrpd1 mutants relative to the Col-0 wild type and ros1 mutant (Fig. 3b–e). siRNAs were abolished at the tested loci in the nrpd1 mutant and were not reduced in the clsy4 mutant (Fig. 3f). siRNA accumulation at several known RdDM target loci such as Solo-LTR, AtSN1, and GP1, was unaffected in clsy4 (Supplementary Fig. S7a); DNA methylation at these loci was also unaffected in clsy4 mutant plants (Supplementary Fig. S7b–d). Altogether, our genome-wide analyses of DMRs in clsy4 revealed that CLSY4 is likely involved in DNA demethylation and, to a lesser degree, in Pol IV-dependent RdDM.
Fig. 3. CLSY4 is required for DNA methylation at certain RdDM target loci.
a Venn diagram showing the overlap of hypo-DMRs between clsy4 and nrpd1. b–e The DNA methylation levels at the DD1, DD2, DD3, and DD12 loci as determined by individual locus bisulfite sequencing. f Small RNA northern blots showing siRNA abundance at the indicated hypo-DMRs
The relationship between RdDM and DNA demethylation mediated by CLSY4
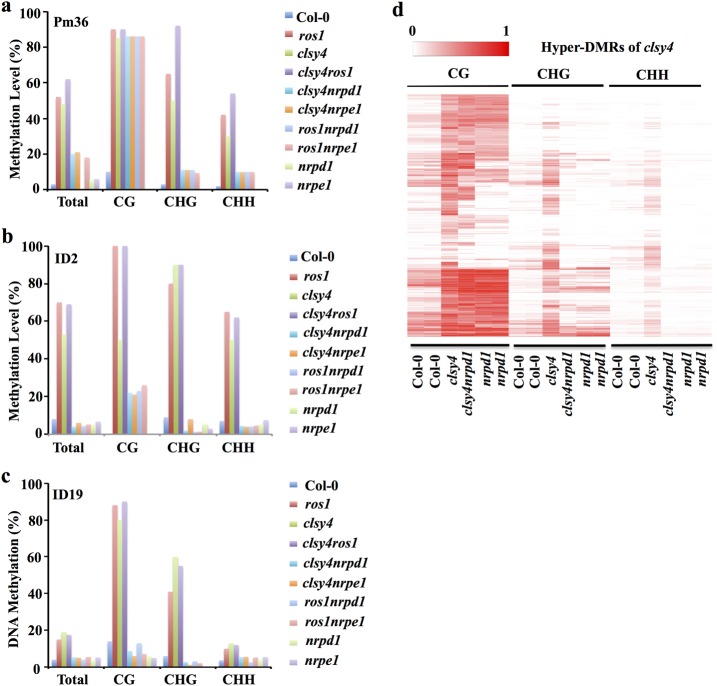
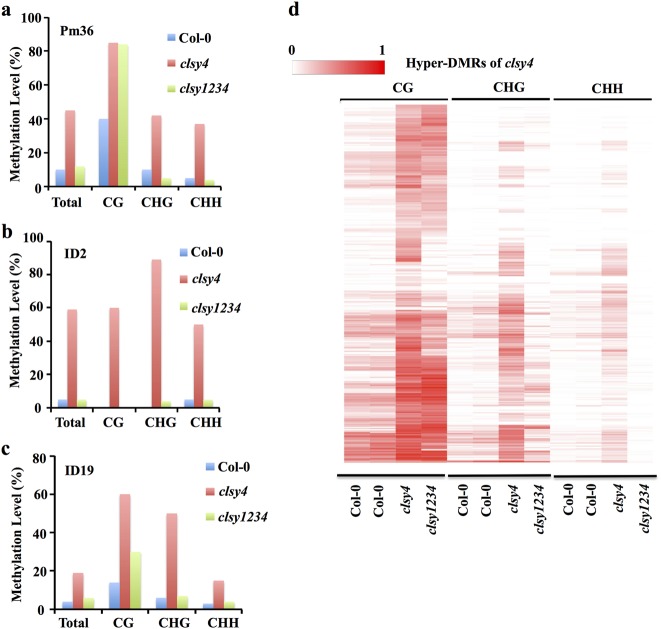
To investigate the relationship between CLSY4-mediated DNA demethylation and RdDM, we carried out genetic analysis in the clsy4-1nrpd1-3 and clsy4-1nrpe1-11 double mutants at three clsy4 hyper-DMRs (Pm36, ID2, and ID19). These tested loci are also ROS1 target loci (Fig. 4a–c). At these loci, the DNA methylation in CHG and CHH contexts was substantially lower in the double mutants clsy4nrpd1, clsy4nrpe1, ros1nrpd1, and ros1nrpe1 than that in the clsy4 and ros1 mutants (Fig. 4a–c), indicating that these loci are RdDM targets. The variation in CG methylation was substantial and included full maintenance at PM36 in clsy4nrpd1, clsy4nrpe1, ros1nrpd1, and ros1nrpe1; partial maintenance at ID2 in ros1nrpd1 and ros1nrpe1; and complete reduction at ID19 in clsy4nrpd1, clsy4nrpe1, ros1nrpd1, and ros1nrpe1 (Fig. 4a–c). These results suggest that the DNA hypermethylation in clsy4 depends on the RdDM pathway. We did not observe any additive effect between clsy4 and ros1 by comparing the DNA methylation levels at these loci in clsy4 and ros1 single mutants to those in the ros1clsy4 double mutant (Fig. 4a–c). These genetic analysis results, together with the data showing that the DNA methylation levels at clsy4 hyper-DMRs were increased in rdd (Fig. 2a), strongly suggest that CLSY4/CHR40 and the ROS1 family DNA demethylases function in the same genetic pathway for DNA demethylation.
Fig. 4. DNA hypermethylation of clsy4 depends on RdDM.
a–c DNA methylation levels at the indicated clsy4 hyper-DMRs in various genotypes as measured by individual locus bisulfite sequencing. d Heatmap analysis of the DNA methylation levels of three types (CG, CHG, and CHH) of clsy4 hyper-DMRs in the indicated genotypes
To further examine the dependence of the clsy4 hyper-DMRs on RdDM, we conducted high coverage (~58x) methylome analysis of the clsy4nrpd1 double mutant using whole-genome bisulfite sequencing (Supplementary Table S2). The cytosine methylation level at clsy4 hyper-DMRs was clearly reduced in the clsy4nrpd1 double mutant (Supplementary Fig. 8). In the CHG and CHH sequence contexts, DNA methylation in the double mutant was significantly lower than that in the Col-0 wild type, but was comparable to that in the nrpd1 mutant, whereas DNA methylation in the CG context was only slightly reduced in the double mutant (Supplementary Fig. S8a–d). Heatmap analysis also showed that the CHH methylation at most clsy4 hyper-DMRs was decreased in the clsy4nrpd1 double mutant (Fig. 4d), while CHG methylation was reduced at the majority of the hyper-DMRs, and CG methylation was reduced at a subset of the hyper-DMRs in the double mutant (Fig. 4d). These results suggest that the hypermethylation of non-CG sequences throughout the genome in clsy4 depends on RdDM.
Four CLSYs redundantly regulate DNA methylation at RdDM target loci
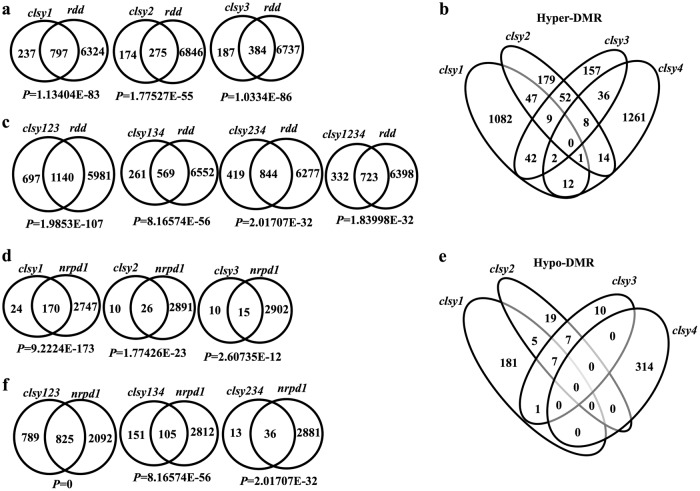
The Arabidopsis CLSY1, CLSY2, CLSY3, and CLSY4 are four closely related chromatin-remodeling proteins and thus may have redundant functions in regulating DNA methylation. To understand their genetic relationships, we crossed their respective mutants and constructed high order mutants including three triple mutants (clsy123 refers to clsy1clsy2clsy3, clsy134 refers to clsy1clsy3clsy4, and clsy234 refers to clsy2clsy3clsy4) and a quadruple mutant (clsy1234 refers to clsy1clsy2clsy3clsy4) (Supplementary Table S3). Whole-genome bisulfite sequencing was conducted in the high order mutants together with the single mutants with high coverage (Supplementary Table S2). Hundreds of hyper-DMRs were identified in clsy1, clsy2, and clsy3 single mutants, with the majority being shared by the rdd mutant, i.e., 77% in clsy1, 61% in clsy2, and 67% in clsy3 overlapped with the hyper-DMRs in rdd (Fig. 5a). Consistent with the hypermethylation in clsy single mutants, the CHG and CHH methylation level of transposons was higher in clsy1, clsy2, and clsy3 than that in the wild type (Supplementary Fig. S9a). The overlapped hyper-DMRs among the four clsy single mutants were very limited (Fig. 5b), suggesting that each CLSY controls DNA demethylation in a locus-specific manner. The majority of the hyper-DMRs in the triple mutants and the quadruple mutant were also shared by rdd, with 62% overlap for clsy123, 69% overlap for clsy134, 67% overlap for clsy234, and 69% overlap for clsy1234 (Fig. 5c).
Fig. 5. Analyses of DMRs in clsy single and triple mutants.
Venn diagram showing the overlap of hyper-DMRs between a rdd and clsy1, clsy2, or clsy3; b clsy single mutants; and c rdd and clsy123, clsy134, clsy234, or clsy1234. Venn diagram showing the overlap of hypo-DMRs between d nrpd1 and clsy1, clsy2, and clsy3; e clsy single mutants; and f. nrpd1 and clsy123, clsy134, and clsy234. P-values are indicated under each overlap (hypergeometric distribution test)
In contrast to the hypo-DMRs in clsy4 (Fig. 3a), the majority of hypo-DMRs in the other three clsy single mutants were shared by nrpd1 (Fig. 5d), indicating important roles of CLSY1, 2, and 3 in RdDM. Again, each CLSY affected DNA methylation in largely distinct genomic regions (Fig. 5e). Even though certain hypo-DMRs were identified in the four clsy single mutants, DNA methylation levels at gene body and transposon regions in the single mutants were not as low as those in the nrpd1 mutant (Supplementary Fig. S9a), suggesting that the four CLSYs are largely redundant in mediating RdDM. The triple mutant clsy123 had the highest number of hypo-DMRs (n = 1614), while clsy134 had an intermediate number (n = 256), and clsy234 had the lowest number (n = 49) (Fig. 5f). The majority of the hypo-DMRs in the three triple mutants were RdDM targets (Fig. 5f). In addition, among the three triple mutants, clsy123 had the lowest and clsy234 had the highest CHH methylation levels at both gene and transposon regions (Supplementary Fig. S9b), suggesting a predominant role of CLSY1 and a minor role of CLSY4 in controlling of DNA methylation at RdDM target regions.
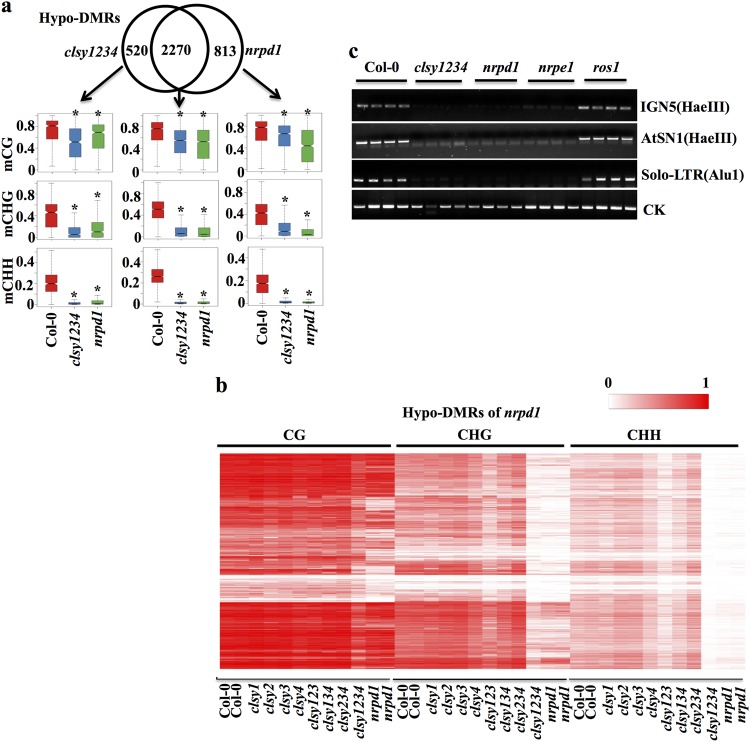
The CHG and CHH methylation levels at gene body regions in the clsy1234 mutant were quite similar to those in nrpd1 (Supplementary Fig. S9b). At transposons, which are the major RdDM targets, the CHG and CHH methylation levels were reduced more in clsy1234 than in nrpd1 (Supplementary Fig. S9b). Thousands of hypo-DMRs were identified in clsy1234, the majority of which were shared by nrpd1 (Fig. 6a). Importantly, the methylation level was also reduced in nrpd1 at clsy1234-specific hypo-DMRs, and vice versa (Fig. 6a). Heatmap analysis showed highly similar patterns of CHG and CHH hypomethylation in clsy1234 and nrpd1 at nrpd1 hypo-DMRs (Fig. 6b). DNA hypomethylation at RdDM target loci was observed in the single mutants clsy1 and clsy4, whereas clsy2 and clsy3 showed little difference compared to the wild type (Fig. 6b). Non-CG methylation was moderately reduced in clsy123 and was less reduced in clsy134, whereas clsy234 showed little reduction in non-CG methylation (Fig. 6b). The methylation analyses of RdDM regions of the single and triple mutants also suggested that CLSY1 predominates among the four CLSY proteins in mediating RdDM, which might explain why CLSY1 but not the other three CLSY proteins has been identified from genetic screens for RdDM factors20. These results demonstrate that the four CLSY members redundantly control DNA methylation at RdDM target loci. DNA methylation at several known RdDM target loci was also examined using chop-PCR. The chop-PCR results showed that IGN5, AtSN1, and Solo-LTR were hypomethylated in clsy1234 as well as in the RdDM mutants nrpd1 and nrpe1 (Fig. 6c), supporting the inference that RdDM was blocked in the quadruple mutant.
Fig. 6. Simultaneous mutations in all four CLSYs eliminate DNA methylation at RdDM target loci.
a Methylation levels of CG, CHG, and CHH at clsy1234-specific, nrpd1-specific, and their common hypo-DMRs. b Heatmap showing the methylation levels in various clsy mutants at nprd1 hypo-DMRs in different cytosine contexts (CG, CHG, and CHH). c Analysis of DNA methylation in the indicated genotypes at IGN5, AtSN1, and Solo-LTR by chop-PCR. Undigested DNA served as the control
clsy mutations impair siRNA production and transposon silencing
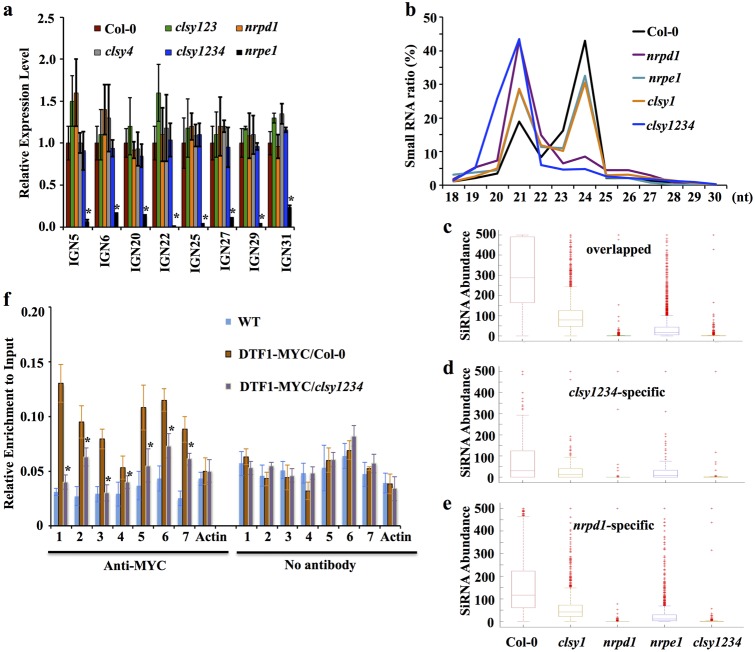
Because DRD1 is a putative chromatin-remodeling protein that is closely related to the CLSYs and that functions to facilitate Pol V transcription36, we tested whether the CLSY proteins might also be involved in Pol V transcription to produce the scaffold RNAs essential for AGO4 targeting38. Quantitative RT-PCR assays showed that the transcript levels of eight Pol V scaffold RNAs in clsy4, clsy123, and clsy1234 were comparable to those in the wild type (Fig. 7a), suggesting that the CLSYs are not involved in Pol V transcription. The four CLSYs proteins are physically associated with Pol IV, which indicates their potential roles in siRNA biosynthesis34. To test this possibility, we conducted small RNA sequencing using clsy1234 mutant plants. As was the case in nrpd1, 24-nt siRNAs were almost eliminated in the quadruple mutant (Fig. 7b). In the clsy1 single mutant, 24-nt siRNAs were slightly reduced, resembling the nrpe1 mutant (Fig. 7b). These patterns suggest that the four CLSYs are collectively required for 24-nt siRNA production. At the clsy1234-specific, nrpd1-specific, and their overlapped hypo-DMRs (Fig. 6a), 24-nt siRNAs were nearly eliminated in clsy1234 and nrpd1 mutants but were moderately reduced in clsy1 and nrpe1 mutants (Fig. 7c–e), supporting the inference that the four CLSYs redundantly regulate siRNA biogenesis at all RdDM target loci. Because DTF1/SHH1 is required for Pol IV function and 24-nt siRNA biogenesis18,19, we examined DTF1/SHH1 enrichment at several RdDM target loci (Supplementary Fig. S10) and found that the chromatin occupancy of DTF1/SHH1 was less in clsy1234 plants than that in wild-type plants (Fig. 7f). This suggested that the four CLSYs are required for proper DTF1/SHH1 binding at the RdDM target loci.
Fig. 7. Pol V transcripts, small RNA abundance, and DTF1/SHH1 enrichment in the clsy1234 mutant.
a Pol V transcript levels in the indicated genotypes. b Length distribution of small RNAs in the indicated genotypes. c siRNA abundance at clsy1234 and nrpd1 overlapped hypo-DMRs. d siRNA abundance at clsy1234-specific hypo-DMRs. e siRNA abundance at nrpd1-specific hypo-DMRs. f Relative ChIP signals for DTF1/SHH1 in the indicated genotypes. *Indicates statistical difference between DTF1-MYC/Col-0 and DTF1-MYC/clsy1234 according to Student’s t-test (P < 0.01)
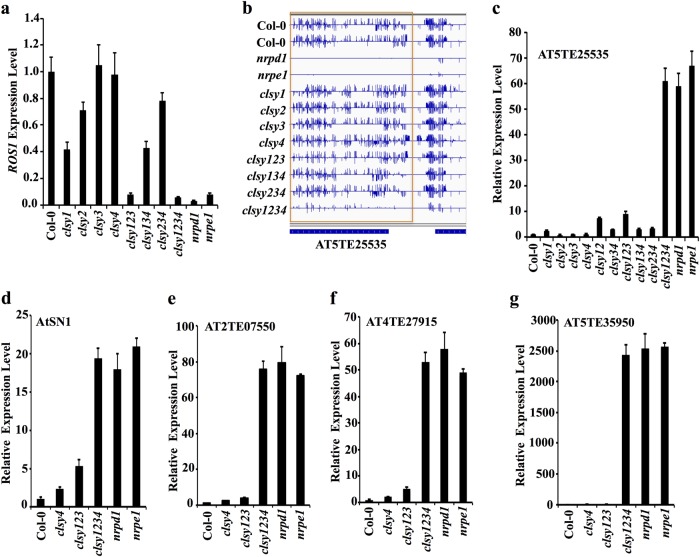
ROS1 expression level is an excellent indicator ofo RdDM activities, because ROS1 expression positively correlates with RdDM-dependent DNA methylation in its promoter region30,39. Like nrpd1 and npre1, clsy1234 showed a drastic reduction in ROS1 transcript levels (Fig. 8a). Among the four single clsy mutants, clsy1 had the lowest ROS1 expression, while clsy123 had the lowest ROS1 expression among the three triple mutants (Fig. 8a). These patterns of ROS1 gene repression are consistent with the DNA hypomethylation phenotypes in the mutants (Fig. 6b).
Fig. 8. The four CLSYs redundantly regulate transposon expression.
a Real-time PCR assay for ROS1 transcript levels in various clsy mutants. b Snapshot showing the DNA methylation of AT5TE25535 in various genotypes. c Real-time PCR assay for AT5TE25535 expression levels in various clsy mutants. d–g The expression levels of the indicated transposons in various mutants
Because RdDM can transcriptionally silence transposons, we selected several TEs that are known to be released in the drd1 mutant40, and examined their DNA methylation levels and transcript levels in various clsy mutants and nrpd1 and nrpe1 mutants. As shown in Fig. 8b, DNA methylation at AT5TE25535 was nearly eliminated in clsy1234 but not in the other clsy mutants (Fig. 8b). Consistent with the DNA methylation patterns, the expression level of AT5TE25535 in clsy1234 was increased by about 60 fold, which was comparable to that in nrpd1 and nrpe1 mutants; in the other clsy mutants, in contrast, AT5TE25535 expression levels were either not changed or were only slightly increased (Fig. 8c). As was the case at AT5TE25535, DNA methylation at the other four tested transposons was eliminated in clsy1234, nrpd1, and nrpe1 mutants, but remained largely unchanged in the other clsy mutants (Supplementary Fig. S11a–d). Their transcript levels were dramatically increased in clsy1234 as well as in nrpd1 and nrpe1 (Fig. 8d–g). Therefore, consistent with their redundant roles in RdDM, the four CLSY proteins have redundant roles in the transcriptional silencing of transposons.
The four CLSY members antagonistically regulate DNA methylation
Because the DNA hypermethylation in clsy4 mutants depends on RdDM, which in turn requires the redundant function of the four CLSY proteins, we suspected that the DNA demethylation function of one CLSY is antagonized by the other CLSY proteins. To examine this possibility, we examined methylation levels of three clsy4 hyper-DMRs using individual locus bisulfite sequencing. At Pm36, the non-CG hypermethylation of clsy4 was suppressed in clsy1234 mutant plants, while the CG hypermethylation did not change in the quadruple mutant (Fig. 9a). At the other two clsy4 hyper-DMRs, ID2 and ID19, hypermethylation was reduced or abolished in all three contexts in the quadruple mutant (Fig. 9b, c). In addition, Integrated Genome Viewer (IGV) examination of randomly selected clsy4 hyper-DMRs from whole-genome bisulfite sequencing results also showed a loss of DNA hypermethylation in clsy1234 compared to clsy4 (Supplementary Fig. S12a–f). These results revealed that, at some RdDM target loci, CLSY4-dependent DNA demethylation counteracts the DNA methylation mediated by the other three CLSY proteins, demonstrating an important role of the CLSY proteins in regulating DNA methylation dynamics.
Fig. 9. DNA hypermethylation in clsy4 depends on the other three CLSYs.
a–c DNA methylation analysis by individual locus bisulfite sequencing of the hyper-DMRs Pm36 (a), ID2 (b), and ID19 (c) in the wild type, clsy4, and clsy1234. d Methylation levels of CG, CHG, and CHH at clsy4 hyper-DMRs
On a whole-genome scale, CHG and CHH methylation at clsy4 hyper-DMRs was significantly reduced in clsy1234 compared to clsy4, as shown by boxplot analysis (Supplementary Fig. S12g–j). Moreover, heatmap analysis showed that the increased CHG and CHH DNA methylation at most clsy4 hyper-DMRs was lost in clsy1234, whereas the CG hypermethylation at clsy4 hyper-DMRs largely remained in clsy1234 (Fig. 9d). These genome-wide patterns further supported the conclusion that CLSY4 antagonizes the other CLSYs in balancing DNA methylation levels at specific genomic loci, although CLSY4 also positively contributes to DNA methylation at some other loci in a way that is genetically redundant with the other CLSYs.
We further investigated the relationship among the four CLSYs by comparing all clsy single mutants with clsy1234. At clsy1, clsy2, or clsy3 hyper-DMRs, DNA hypermethylation was largely abolished in clsy1234, as shown by heatmap analyses (Supplementary Fig. S13a–c). In addition, at these clsy single mutant hyper-DMRs, cytosine methylation levels and especially CHG and CHH methylation levels were reduced in clsy1234 (Supplementary Fig. S14a–c), indicating a general antagonistic relationship among the four CLSY members in regulating DNA methylation dynamics. Consistent with this inference, the hyper-DMRs in clsy123, clsy134, and clsy234 were also largely reduced in the clsy1234 mutant (Supplementary Fig. S13d–f), with CHG and CHH methylation reduced more than CG methylation (Supplementary Fig. S15a–c).
Discussion
Arabidopsis CLSY1 was first identified in a genetic screen for cellular factors required for gene silencing induced by double-stranded RNAs and was demonstrated to function with RDR2 and NRPD1 in the production of 24-nt siRNA at endogenous loci20. However, DNA methylome analysis of multiple RdDM mutants showed that the methylation level at RdDM target loci was only weakly reduced in clsy1 mutants21, suggesting that other chromatin-remodeling proteins likely work together with CLSY1 in siRNA biogenesis. Later, CLSY1 and its three paralogs, CLSY2, CLSY3, and CLSY4, were identified as Pol IV-associated proteins, suggesting that these proteins probably function redundantly in siRNA production in the RdDM pathway34. Here we profiled the DNA methylomes of single, triple, and quadruple clsy mutants and found that the DNA methylation of RdDM target loci is indeed cooperatively controlled by the four CLSY proteins. We found that 24-nt siRNAs were almost eliminated in the clsy1234 mutant. Although the detailed molecular mechanism underlying the involvement of the CLSYs in Pol IV function remains to be elucidated, chromatin enrichment of DTF1/SHH1, the cofactor important for Pol IV recruitment and 24-nt siRNA production at a subset of RdDM loci18,19, was reduced in clsy1234 at several of the tested genomic regions.
In this study, CLSY4 (a close homolog of CLSY1) was originally identified in our genetic screen for DNA demethylation factors. More than 1000 hyper-DMRs were found in clsy4 mutant plants, indicating that CLSY4 is important for genome-wide DNA demethylation. ROS1 expression was not reduced in clsy4 mutants, indicating that CLSY4 does not function indirectly in demethylation by being an RdDM factor that controls ROS1 expression. The biochemical function of CLSY4 in DNA demethylation is unknown, and we have not observed a physical association between ROS1 and any of the four CLSYs (unpublished data). Nevertheless, we found that the nucleosome occupancy was reduced in clsy4 at several tested genomic loci targeted by active DNA demethylation. ATP-dependent chromatin-remodeling proteins generally function in nucleosome sliding, eviction, and histone exchange on chromatin41. It is possible that active DNA demethylation at some genomic loci may somehow involve CLSY-mediated nucleosome movement. In support of this notion of a chromatin remodeling functioning in active DNA demethylation, Ikeda et al. have documented that the FACT histone chaperone component SSRP1 plays an important role in DME-mediated active DNA demethylation and gene imprinting42.
With the use of clsy4nrpd1 and clsy1234 mutants, our genetic analysis demonstrated that DNA hypermethylation in clsy4 depends on the RdDM pathway, a pattern resembling that of the ros1 mutant. It is possible that, besides CLSY4, the other three CLSYs also directly regulate DNA demethylation at discrete genomic regions, because hundreds of hyper-DMRs were identified in the single and triple clsy mutants. Those hyper-DMRs also depended on the RdDM pathway, because DNA hypermethylation was largely abolished in the clsy1234 mutant, in which DNA methylation and siRNA accumulation at RdDM target loci was eliminated. Therefore, the dual function of CLSY proteins and especially of CLSY4, whose mutation results in the highest number of hyper-DMRs, in DNA methylation via RdDM and in active DNA demethylation mediated by the ROS1 family of DNA demethylases represents a new mechanism underlying the dynamic regulation of DNA methylation. DNA methylation and demethylation activities were previously determined to be coordinated by a “methylstat”, a methylation-sensitive cis-element at the ROS1 promoter30,31. The current results suggest that chromatin remodeling by the four CLSYs is another mechanism for the coordination of DNA methylation and demethylation.
Mutations in the CLSYs led to increased DNA methylation at certain genomic sites but deceased DNA methylation at other sites. The opposite DNA methylation phenotypes in the clsy mutants raises the possibility that other epigenetic marks, like histone modifications and pre-existing DNA methylation together with nucleosome occupancy regulated by CLSYs, may collectively determine whether a region undergoes RdDM or active DNA demethylation. The chromatin status regulated by the CLSYs is only one of the factors determining the dominance of RdDM or active DNA demethylation. How RdDM and active DNA demethylation differ in their requirements for chromatin environments warrants additional study.
Materials and methods
Plant materials and growth conditions
All Arabidopsis plants used in this study were in the Col-0 background. The clsy1-3 mutant was reported previously20. All other mutants (Supplementary Table S3) were either ordered from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org) or made in this study. The oligos used for genotyping and making higher order mutants are listed in Supplementary Table S4. Plants that were subjected to DNA methylation and small RNA analyses were 2-week-old seedlings grown on 1/2 strength Murashige Skoog (MS) medium supplemented with 0.7% agar and 1% sucrose. The seedlings were grown in growth chambers (Percival, Inc.) at 22 °C with 16 h of light and 8 h of darkness.
Chop-PCR and genetic screen
The T-DNA mutants (Supplementary Table S1) were grown in soil for 5 weeks. Four plants of each genotype were selected for DNA extraction by the CTAB method. About 2 µg of DNA of each plant was diluted and digested with HhaI or HpaII in a 20-µl reaction mixture for 8 h. The PCR reaction used 1 µl of enzyme-digested DNA as the template. Agarose (1%) gels were used to compare the amplified DNA signal, with the wild type and ros1-4 as controls. Candidate mutants were then further genotyped and confirmed.
Individual locus bisulfite sequencing
After the genomic DNA was isolated from 4-week-old Arabidopsis plants using the Plant DNeasy Mini Kit (Qiagen), about 100 ng of genomic DNA was subjected to sodium bisulfite treatment and purification using the BisulFlash DNA Modification Kit (Epigentek) and following the manufacturer’s protocol. A 1 µl volume of bisulfite-converted DNA was used for PCR reaction with oligos designed for specific target regions (Supplementary Table S4). The PCR products were ligated into the pMD18-T vector (Takara, Japan). The ligation product was transformed into DH5α competent cells, and at least 15 single clones were sequenced for each type of transformant. The sequencing results were aligned and analyzed using CyMate (http://www.cymate.org).
Whole-genome bisulfite sequencing and data analysis
Genomic DNA was extracted from 2-week-old seedlings grown in 1/2 strength MS medium and was sent to the Beijing Genomics Institute (Shenzhen, China) for bisulfite treatment, library construction, and high-throughput sequencing. Clean reads were generated by trimming adaptor and low-quality sequences (q < 20) and then were mapped to the Arabidopsis genome (TAIR 10) using BSMAP (Bisulfite Sequence Mapping Program) and allowing two mismatches. Differentially methylated regions (DMRs) were identified as previously described16. First, the cytosines with a read coverage < 5 were filtered out. The DNA methylation level in every 200-bp window with a step size of 50 bp was compared between control and mutant plants using Fisher’s exact test. P-values calculated from the tests were then adjusted using the Benjamini-Hochberg method to control for the false discovery rate (FDR). Windows with an adjusted P-value < 0.05 were then tested for the number of DMCs (differentially methylated cytosines), which were defined as cytosines with a P-value < 0.01 in Fisher’s exact test. The DMR-identified windows with at least 7 DMCs and ≥ 1.5-fold change in DNA methylation levels were combined to generate the final list of DMRs if the gap length between two windows was ≤ 100 bp.
All of the figures related to DNA methylomes were generated using R (https://www.r-project.org/). The heatmaps were generated using “heatmap.2” from “gplots” library, with the following parameters: Colv = FALSE, dendrogram = “row”, trace = “none”, and labRow = “NA”. The methylation data were not normalized for generating the heatmaps.
Genome-wide small RNA sequencing and analysis
Total RNA was isolated from 2-week-old seedlings with TRIzole reagent (Sigma) and the standard protocol. Total RNA was separated on PAGE gels and < 35-nt fractions were cut and purified for small RNA library preparation and sequencing at BGI (Shenzhen, China). After sequencing, adapter sequences were trimmed, and clean reads from 18 to 30 nt were mapped to the Arabidopsis genome (TAIR10) using Bowtie with parameter “-v 0 –k 10”16 Reads were mapped to annotated structural RNAs including tRNAs, rRNAs, snRNAs, and snoRNAs were excluded. Read counts were normalized to Reads Per Ten Million (RPTM) based on total mapped reads. The “hits-normalized abundance” (HNA) values were calculated by dividing the normalized abundance (in RPTM) for each small RNA hit, where a hit was simply defined as the number of loci at which a given sequence perfectly matched the genome16, and the values were used for boxplot generation.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation was conducted using 2-week-old seedlings of the wild type Col-0, DTF1-MYC/clsy1234, and DTF1-MYC/Col-0 grown in 1/2 strength MS solid medium as previously described43. The antibody used for DTF1 ChIP assays was anti-MYC (16–219, Millipore). The DNA was diluted in 50 μl of TE, and a 1.5 μl volume was used for each qPCR reaction. The relative enrichment to input was calculated from three biological replicates.
Nuclei were extracted from 2-week-old Arabidopsis seedlings grown in 1/2 strength MS medium as previously described38 and were digested with Micrococcal Nuclease (MNase; NEB) before chromatin immunoprecipitation was conducted using antibodies against H3 (Abcam, ab1791) as previously reported44.
Small RNA extraction and northern blotting
After total RNA was extracted from 2-week-old Arabidopsis seedlings with TRIzole reagent (Sigma) and the standard protocol, the small RNA fraction was precipitated by the PEG method. In brief, equal volumes of a PEG8000 solution (20% PEG 8000, 1 M NaCl) were added to the total RNA. After centrifugation at 16000 g at 4 °C, an equal volume of isoproponal and 0.1 volume of 3 M NaAC were added to the supernatant. Following another centrifugation at 16000 g at 4 °C, the small RNA pellet was re-suspended in DEPC-treated water. For each sample, small RNAs from about 100 µg of total RNA were separated on a 17% polyacrylamide gel, which was electrotransferred to a Hybond N+ membrane (GE Lifesciences). Membranes were cross-linked, incubated for 2 h at 80 °C, and hybridized overnight at 38 °C with 32P-labelled DNA probes or oligonucleotides (listed in Supplementary Table S4) in PerfectHyb buffer (Sigma). The washed membranes were then exposed in the presence of X-ray film at −80 °C for 7 days.
Data access
The sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm. nih.gov/geo/) and are accessible through accession number GSE110338.
Electronic supplementary material
Acknowledgements
We thank David C. Baulcombe for the seeds of the clsy1-3 mutant. This work was supported by the Chinese Academy of Sciences to J.-K. Z., and by the National Science Foundation of China (31671340), the Natural Science Foundation of Jiangsu (BK20170027), the Innovative Project of State Key Laboratory of Crop Genetics and Germplasm Enhancement (ZZ2017009), Fundamental Research Funds for the Central Universities (KYZ201503, KJJQ201801), and the Jiangsu Collaborative Innovation Center for Modern Crop Production to D.-L. Y.
Author contributions
Experimental design and data analysis: D.L.Y. and J.K.Z. Experimental performance: D.L.Y., G.Z., D.X., D.M. Bioinformation analysis: L.Y., L.Z., K.T., and L.W. Contribution of reagents/materials/analysis tools: J.L., C.G.D., and C.D. Manuscript writing: D.L.Y, H.Z., and J.K.Z.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong-Lei Yang, Email: dlyang@njau.edu.cn.
Jian-Kang Zhu, Email: jkzhu@sibs.ac.cn.
Electronic supplementary material
Supplementary Information accompanies the paper at (10.1038/s41421-018-0056-8).
References
- 1.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 2.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant. Biol. 2007;10:528–533. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 3.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 6.Lang Z, et al. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA. 2017;114:E4511–E4517. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr. Opin. Plant. Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb. Symp. Quant. Biol. 2012;77:205–212. doi: 10.1101/sqb.2013.77.014803. [DOI] [PubMed] [Google Scholar]
- 9.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 10.Cuerda-Gil D, Slotkin RK. Non-canonical RNA-directed DNA methylation. Nat. Plants. 2016;2:16163. doi: 10.1038/nplants.2016.163. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, et al. Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol. Biol. Evol. 2015;32:1788–1799. doi: 10.1093/molbev/msv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai J, et al. A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell. 2015;163:445–455. doi: 10.1016/j.cell.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blevins T, et al. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife. 2015;4:e09591. doi: 10.7554/eLife.09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang DL, et al. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 2016;26:66–82. doi: 10.1038/cr.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye R, et al. A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell. 2016;61:222–235. doi: 10.1016/j.molcel.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang K, Lang Z, Zhang H, Zhu JK. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants. 2016;2:16169. doi: 10.1038/nplants.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/S0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 24.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 27.He XJ, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Lei M, et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:3553–3557. doi: 10.1073/pnas.1502279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams BP, Pignatta D, Henikoff S, Gehring M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS. Genet. 2015;11:e1005142. doi: 10.1371/journal.pgen.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian W, et al. Regulation of active DNA demethylation by an alpha-crystallin domain protein in Arabidopsis. Mol. Cell. 2014;55:361–371. doi: 10.1016/j.molcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS. Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:11425–11430. doi: 10.1073/pnas.1208557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law JA, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan DP, Owen-Hughes T. Snf2-family proteins: chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 2011;15:649–656. doi: 10.1016/j.cbpa.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda Y, et al. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev. Cell. 2011;21:589–596. doi: 10.1016/j.devcel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008;3:1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.