Abstract
“BRCAX” refers breast cancers occurring in women with a family history predictive of being a BRCA1/2 mutation carrier, but BRCA1/2 genetic screening has failed to find causal mutations. In this study, we report the findings of the genetic architecture of BRCAX with novel and redefined candidate loci and their potential impacts on preventive strategy. We performed a genome-wide association study involving 1,469 BRCAX cases from the Korean Hereditary Breast Cancer study, and high-risk breast cancer cases (1,482 Asians and 9,902 Europeans) from the Breast Cancer Association Consortium. We also evaluated the previously reported susceptibility loci for their roles in the high-risk breast cancers. We have identified three novel loci (PDE7B, UBL3, and a new independent marker in CDKN2B-AS1) associated with BRCAX, and replicated previously reported SNPs (24 of 92) and moderate/high-penetrance (seven of 23) genes for Korean BRCAX. For the novel candidate loci, evidence supported their roles in regulatory function. We estimated that the common low-penetrance loci might explain a substantial part of high-risk breast cancer (39.4% for Koreans and 24.0% for Europeans). Our study findings suggest that common genetic markers with lower penetrance constitute a part of susceptibility to high-risk breast cancers, with potential implications for a more comprehensive genetic screening test.
Introduction
Breast cancer is the most common female cancer worldwide, and its incidence rate is increasing rapidly in Asian populations including South Korea1,2. The National Cancer Screening Program in Korea provides mammography tests for screening breast cancer for women aged 40–69 years3. The effectiveness of this screening program, however, is controversial mainly because the validity of mammography-based test is lower for Asian women than for Caucasians4–8 About 4.8% and 11.1% of breast cancers were diagnosed before age 40 and 45, respectively, in the United States between 2005 and 2014. The proportions of these early-onset cases almost tripled for Korean women during the same period (13.9% and 29.8%), although overall incidence rate is less than a half of that for Western women9,10. Early-onset breast cancer cases are generally more likely to have higher pathologic grade and to be receptor-negative tumors11–14. Given the low predictive value of mammography for Asian women, ultrasonographic breast examination might be introduced to improve the validity but at greater cost. An effective genetic screening test, if available, might be able to identify women at high-risk of breast cancers for personalized prevention programs. So far, to our knowledge, the genetic etiology of high-risk breast cancers is understudied particularly for Asian women with more burdens from high-risk breast cancers.
Mutations in BRCA1 and BRCA2 (hereafter referred to as BRCA1/2), the most important genetic susceptibility markers, account for up to 15% and 30% of the familial recurrence risk (FRR) of overall and high-risk breast cancers15. Clinical practice guidelines are well-established for BRCA1/2 mutation carriers16. Conversely, lack of proper knowledge has left the BRCA1/2 mutation non-carriers’ genetic counseling largely unattended, although these non-carriers with strong family history show similar clinical presentations predictive of BRCA1/2 mutation carriers (“BRCAX” cases), with four-fold or greater lifetime risk of breast cancer than the general population17,18. In Korea, breast cancer cases with early-onset (age < 40) or other indications suggesting a strong genetic burden are entitled to BRCA1/2 mutation testing covered by Korean National Health Insurance. The test has found clinically significant BRCA1/2 mutations for only 15.7% of eligible patients (22.3% of examinees indicated by strong family history, and 8.8% by early-onset)19, to make the understanding of “BRCAX genes” a priority. Several studies have been conducted to elucidate the genetic architecture of BRCAX cancers20–22. Studies do not have replicated the novel loci from each other, suggesting that susceptibility loci for BRCAX might be population-specific, at least partly. Asian women might have specific genetic susceptibility for the high-risk breast cancers, but, to our knowledge, no studies have been conducted toward this end.
We conducted a genome-wide search for BRCAX genes involving 1,469 Korean female breast cancer cases who were eligible for the BRCA1/2 test but negative for causal mutations. Then, we replicated our findings using the Breast Cancer Association Consortium (BCAC) data. We also estimated the role of previously reported susceptibility loci on the risk of BRCAX occurrence. Finally, we evaluated the impacts and estimated risk of those associated markers on high-risk breast cancers for Asians and Europeans.
Results
For the discovery phase, a total of 1,478 Korean BRCAX cases and 5,979 controls were selected with 3,378,933 markers (flowchart is shown in Supplementary Fig. S1). Characteristics of the participants were described in Table 1. Mean age of cases (40.2) was approximately 15 years younger compared to controls (55.1), but allele frequencies of associated markers were not significantly different between age group (Supplementary Table S1). Among 1,469 cases after quality control, 169 and 288 women had one BRCA1 or BRCA2 mutation with unverified significance. For BRCA2 gene, we found 42 variants significantly which are more frequent in the cases compared to the controls (Supplementary Table S2). However, the ClinVar database showed that 40 of these were reported to be benign and only two variants (rs2126042 and rs766173) was reported to be benign or have uncertain significance.
Table 1.
Characteristics of breast cancer cases (n = 1,469).
| Variables | No. (%) | |
|---|---|---|
| Age (years old), mean ± sda | 40.24 ± 9.13 | |
| BRCA1/2 mutationb | BRCA1–neg & BRCA2–uv | 288 (19.6%) |
| BRCA1–uv & BRCA2–neg | 169 (11.5%) | |
| Both negative | 1012 (68.9%) | |
| Age of breast cancer onset | ≤40 | 1037 (70.6%) |
| 40–45 | 143 (9.7%) | |
| >45 | 289 (19.7%) | |
| Family history of breast cancer in the first or second degree relatives | no history | 908 (61.8%) |
| 1 | 492 (38.2%) | |
| 2 | 60 (4.1%) | |
| 3 | 9 (0.6%) | |
| Family history of ovarian cancer in the first or second degree relatives | no history | 1399 (95.2%) |
| 1 | 68 (4.6%) | |
| 2 | 2 (0.1%) | |
| Disease history of cancers other than breast cancer | 72 (4.9%) | |
| Bilateral breast cancer | 122 (8.3%) | |
| Receptor status | ||
| Estrogen receptor | negative | 465 (31.7%) |
| positive | 891 (60.7%) | |
| unknown | 113 (7.7%) | |
| Progesterone receptor | negative | 496 (33.8%) |
| positive | 879 (59.8%) | |
| unknown | 94 (6.4%) | |
| HER2 receptor | negative | 924 (62.9%) |
| positive | 275 (18.7%) | |
| equivocalc | 139 (9.5%) | |
| unknown | 131 (8.9%) | |
| Triple-negative | 223 (15.2%) | |
aSd: standard deviation.
bNeg: negative, uv: unverified mutation.
cImmunohistochemistry (IHC) 2+, Fluorescence In Situ Hybridization (FISH) unknown.
Being indicated by early-onset (≤age 40) comprised the largest proportion of the case composite (70.6%), followed by having a family history of breast cancer (42.9%).
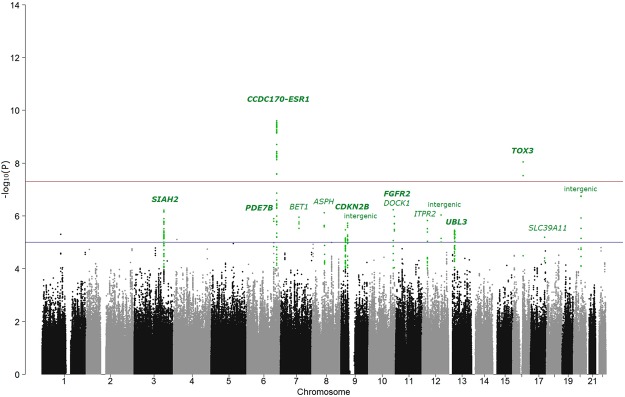
From the genome-wide association study (GWAS), the genomic inflation factor (ʎGC) was 1.09 and p-values were corrected for the inflation factor. Figure 1 and Supplementary Fig. S2 show the Manhattan plot and the quantile-quantile plot. Initially, 15 independent loci of single nucleotide polymorphisms (SNPs) showed p-values < 10−5. Two SNPs reached genome-wide significance level in the first phase analysis, and both were reported before: rs9383936 (p = 1.01 × 10−10, OR = 1.35) on the intron of CCDC170 (near ESR1) and rs4784227 (p = 9.11 × 10−9, OR = 1.31) on the intron of TOX3. Three previously reported loci (SIAH2, FGFR2, PGAM1P5/NTN4) and nine new loci also comprised the initial 15 loci. One variant on CDKN2B-AS1 (rs1011970) was previously associated with breast cancer risk for European descendants23,24, but our study found another variant in the proximal regions of the CDKN2B-AS1 gene, rs78545330. These two SNPs constituted separate haplotype blocks (r2 = 0.026 for East Asians), and we considered rs78545330 as independent.
Figure 1.
Manhattan plot. The red horizontal line represents the genome-wide significance threshold of p-value = 5.0 × 10−8 and the blue horizontal line represents the suggestive significance threshold of p-value = 1.0 × 10−5. For significantly associated regions, SNPs with p-value less than 10-3 are highlighted in green and the replicated genes are marked in bold.
The replication set consisted of 1,482 cases (1,194 early-onset cases and/or 390 women with a family history breast cancer in first-degree relatives, Asian high-risk cases) and 3,612 control women without family history of breast cancer. We compared the characteristics and composite of high-risk breast cancer cases of different populations (Supplementary Table S3). Among the initial 15 candidate loci, six variants were statistically significant (at p-value < 0.05) including two previously known (CCDC170/ESR1, and TOX3) and four novel susceptibility loci. Among four novel loci, rs11154838, located on 6q23.3 (intron of PDE7B), showed associations at genome-wide significance level (p-values: 2.27 × 10−8). Other two novel candidate loci (rs78545330 in the intron of CDKN2B-AS1 and rs278050 on 13q12.3, 73 Kb 5′ of UBL3), while replicated in Asian high-risk cases at nominal p-value, did not reach genome-wide significance in the meta-analysis (p-values: 8.56 × 10−8 and 1.63 × 10−6). The other novel variant, rs4969001 on SLC39A11, was associated in the opposite direction; thus, it was not considered for further analysis. We summarize findings of these 15 candidate loci in Table 2. For European high-risk breast cancers, all three novel susceptibility loci did not show meaningful associations (p-values = 0.34, 0.24 and 0.50).
Table 2.
Genomic loci associated with breast cancer (P-value < 10−5).
| SNP | Chr | Position (hg19) | Nearest Genes (distance to gene) | Function | Risk Allele (fwd) | RAF in 1 kG Phase3a | Korean BRCAX cases (initial) | Asian high-risk cases (replication) | Meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAS | EUR | RAF in casesa | P-value | OR (95% CI)b | RAF in casesa | P-value | OR (95% CI)b | P-value | OR (95% CI)b | P hetero c | ||||||
| rs60538652 | 3 | 150473606 | SIAH2 | Intron | A | 0.61 | 0.98 | 0.65 | 6.24.E-07 | 1.26 (1.15–1.38) | 0.73 | 0.072 | 1.13 (0.99–1.28) | 3.25.E-07 | 1.21 (1.13–1.31) | 0.16 |
| rs11154838 | 6 | 136290942 | PDE7B | Intron | C | 0.72 | 0.65 | 0.75 | 1.28.E-06 | 1.27 (1.15–1.40) | 0.83 | 0.005 | 1.26 (1.07–1.49) | 2.27.E-08 | 1.27 (1.17–1.38) | 0.96 |
| rs9383936 | 6 | 151944614 | 3′ of CCDC170 (2.3 Kb), 5′ of ESR1 (3.3 Kb) | Intergenic | A | 0.31 | 0.08 | 0.34 | 1.01.E-10 | 1.35 (1.23–1.48) | 0.34 | 1.05.E-06 | 1.3 (1.17–1.44) | 6.35.E-16 | 1.33 (1.24–1.42) | 0.58 |
| rs10953105 | 7 | 93622247 | BET1 | 3′ UTR | G | 0.86 | 0.97 | 0.88 | 1.13.E-06 | 1.37 (1.21–1.56) | 0.91 | 0.192 | 1.14 (0.94–1.38) | 1.81.E-06 | 1.30 (1.16–1.44) | 0.11 |
| rs2350923 | 8 | 62605259 | ASPH | Intron | T | 0.72 | 0.70 | 0.76 | 7.75.E-07 | 1.28 (1.16–1.42) | 0.27 | 0.750 | 1.02 (0.92–1.13) | 0.250 | 1.14 (0.91–1.44) | 0.00 |
| rs78545330 | 9 | 21995941 | CDKN2B-AS1 | Intron | A | 0.21 | 0.08 | 0.26 | 3.41.E-06 | 1.26 (1.14–1.39) | 0.20 | 5.73.E-03 | 1.19 (1.05–1.35) | 8.56.E-08 | 1.23 (1.14–1.33) | 0.47 |
| rs10814070 | 9 | 34129839 | 3′ of DCAF12 (3.1 Kb) | Intergenic | T | 0.71 | 0.25 | 0.75 | 1.93.E-06 | 1.26 (1.15–1.39) | 0.65 | 0.469 | 0.96 (0.87–1.07) | 0.465 | 1.10 (0.85–1.44) | 0.00 |
| rs2912774 | 10 | 123348662 | FGFR2 | Intron | T | 0.38 | 0.43 | 0.42 | 5.96.E-07 | 1.25 (1.14–1.36) | 0.43 | 0.052 | 1.11 (1.00–1.21) | 5.60.E-07 | 1.18 (1.10–1.25) | 0.06 |
| rs9418690 | 10 | 128809949 | DOCK1 | Intron | C | 0.73 | 0.76 | 0.76 | 1.08.E-06 | 1.28 (1.16–1.41) | 0.82 | 0.819 | 1.02 (0.85–1.23) | 0.183 | 1.16 (0.93–1.44) | 0.04 |
| rs4964006 | 12 | 26770889 | ITPR2 | Intron | T | 0.95 | 0.69 | 0.98 | 1.52.E-06 | 1.96 (1.49–2.58) | 0.95 | 0.804 | 0.97 (0.76–1.24) | 0.366 | 1.38 (0.69–2.75) | 0.00 |
| rs67129489 | 12 | 96016957 | 5′ of PGAM1P5 (26.0 Kb), 5′ of NTN4 (34.6 Kb) | Intergenic | G | 0.13 | 0.32 | 0.15 | 9.31.E-07 | 1.36 (1.20–1.53) | 0.06 | 0.976 | 1.00 (0.77–1.32) | 0.226 | 1.20 (0.89–1.60) | 0.05 |
| rs278050 | 13 | 30436968 | 5′ of UBL3 (73 Kb) | Intergenic | C | 0.19 | 0.23 | 0.21 | 3.89.E-06 | 1.28 (1.15–1.42) | 0.19 | 0.050 | 1.13 (1.00–1.28) | 1.64.E-06 | 1.22 (1.12–1.32) | 0.14 |
| rs4784227 | 16 | 52583143 | TOX3 | Intron | T | 0.26 | 0.25 | 0.32 | 9.11.E-09 | 1.31 (1.19–1.44) | 0.29 | 6.24.E-05 | 1.25 (1.12–1.39) | 3.26.E-12 | 1.28 (1.20–1.38) | 0.49 |
| rs4969001 | 17 | 70980127 | SLC39A11 | Intron | T | 0.89 | 0.96 | 0.86 | 6.53.E-06 | 1.32 (1.17–1.49) | 0.89 | 0.041 | 0.85 (0.73–0.99) | 0.780 | 1.06 (0.69–1.64) | 0.00 |
| rs73107564 | 20 | 36263775 | — | Intergenic | A | 0.89 | 0.81 | 0.89 | 1.81.E-07 | 1.43 (1.25–1.63) | 0.87 | 0.541 | 0.96 (0.83–1.10) | 0.432 | 1.17 (0.79–1.73) | 0.00 |
aRAF: risk allele frequency, EAS: East Asian, EUR: European.
bOR: odds ratio, CI: confidence interval.
cPhetero: P-value for heterogeneity. Fixed effect model was used if p-value for heterogeneity exceeds 0.05; otherwise random effect model was used.
When we conducted a meta-analysis by the receptor-status, four loci of the initial 15 candidate loci from the GWAS showed receptor-status specific associations with even smaller sample sizes. We only focused on findings where smaller subgroup resulted in the stronger association, to reduce the influence of sample size variation. rs60538652, on SIAH2, rs2912774 on FGFR2 and rs67129489 near PGAM1P5/NTN4 were more strongly associated with ER+ or PR+ cases, while rs9383936 near ESR1 was more strongly associated with ER− or PR− breast cancers (Pheterogeneity <0.05). It is noteworthy that markers at SIAH2, FGFR2, and PGAM1P5/NTN4 showed an increased level of significance in subgroup analyses despite smaller sample sizes. None of the variants identified from the GWAS were differentially associated according to HER2 receptor status or being TNBC. More details about the receptor-status-wise analyses were described in Supplementary Table S4.
Three novel candidate regions were visualized using LD plots and regional association plots (Supplementary Fig. S3) and annotated for regulatory elements (Supplementary Table S5). Two markers (rs6905776 and rs12174235), in the same LD block with rs11154838 on PDE7B, were annotated for regulatory elements (GWAS p-values: 1.63 × 10−6 and 2.72 × 10−5). rs78545330 on the intron of CDKN2B-AS1 and correlated markers showed strong evidence of regulatory function from multiple tissues including breast tissues, while rs1011970 (previously reported SNP) was not annotated for regulatory elements. East Asians have a risk allele of rs78545330 (A, allele frequency = 0.21) three times more frequently than Europeans (0.08). We presented the haplotype structures of this region between Asian and Europeans (Supplementary Fig. S4). For East Asians, multiple LD blocks were more distinct, and the LD between rs78545330 and rs1011970 was weak in East Asians (r2 = 0.02), indicating that rs78545330 is a novel candidate variant. When we examined the genetic regions harboring candidate variant near UBL3, the region consisted of two independent LD blocks, and rs278050 and correlated SNPs showed enhancer-related histone marks in several tissues including breasts and mammary tissues. Also, rs73444211 in the other LD block near UBL3 participated in diverse regulatory functions.
Results from eQTL analyses for three candidate loci also showed evidence of associations with gene-expression levels of different tissues (Supplementary Table S6). The risk allele T of rs11154838 was associated with PDE7B gene-expression level in blood vessels (p-value = 0.001), but not with other tissues. rs78545330 on CDKN2B showed strong associations with gene expressions in multiple tissues. The risk allele substantially decreases the expression of CDKN2B, a tumor suppressor gene (p-value = 1.7 × 10−13, effect size = −0.44). rs278050 near UBL3 also altered cis-gene regulation in several tissues.
When we replicated findings from the previous studies, a part (24 for Korean BRCAX, 55 for European and 14 for Asian high-risk breast cancers) of 92 loci from the GWAS catalog showed meaningful associations (p-value < 0.05 with the same direction of association as in the previous reports, Supplementary Table S7). Generally, replicated variants showed larger risk for BRCAX or high-risk cancers than the previous reports for all breast cancers. Since the majority of known susceptibility markers were reported from studies of European populations, the number of replicated markers was larger for European cases compared with Korean or Asian cases. However, the sum of %FRR (Σ%FRR) of each replicated marker was largest for Korean BRCAX cases (29.8%), followed by European high-risk cases (24.0%) and Asian high-risk cases (10.6%). Three novel candidate variants accounted for additional 9.6% and 3.0% FRR for Koreans and Asians. It is noteworthy that three replicated variants on FGFR2, TOX3, and ESR1 have been reported in previous searches for BRCAX genes20–22. These variants were also associated with overall breast cancer risk in multiple studies25. Other candidate loci reported from previous BRCAX studies were not replicated in this study20–22. Finally, among the breast cancer predisposition genes with moderate/high-penetrance16,26, seven genes (BMPR1A, PTEN, CDH1, NF1, RAD51C, BRIP1, and CHEK2) were replicated for Korean BRCAX (eight for Asian and 15 genes for European high-risk breast cancers, Supplementary Table S8). The estimated contributions from the moderate/high-penetrance variants varied by the populations, accounting for 4.2%, 13.2%, and 7.9% of overall high-risk breast cancers for Koreans, Asians, and Europeans.
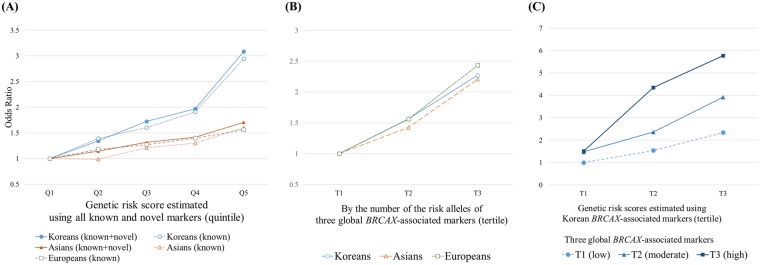
We estimated the increase in risk with increase in genetic risk score (GRS) for each population (Fig. 2A). Predicted risk (OR) by GRS was highest for Korean BRCAX cases, and the OR increased upon adding newly identified markers for both Koreans (OR: from 2.95 to 3.09) and Asians (OR: from 1.60 to 1.71). Also, we estimated the genetic risk of BRCAX according to genotypes of “shared between populations (FGFR2/TOX3/ESR1, reported from multiple BRCAX studies including ours)”. The estimated increases in the risk of BRCAX or high-risk breast cancers from the variants on FGFR2, TOX3, and ESR1 were quite similar across populations (ORs for highest compared to lowest tertile = 2.2 to 2.4, Fig. 2B). When we stratified and compared the estimated risk of GRSs calculated from the three novel and 21 replicated (after exclusion of three markers on FGFR2/TOX3/ESR1 among 24 replicated variants) “Asian-specific BRCAX” markers according to “shared between population” (FGFR2/TOX3/ESR1) genotype status, those with multiple risk alleles of FGFR2/TOX3/ESR1 showed steeper increases in risk for breast cancers, compared with those in none or one allele groups (Fig. 2C).
Figure 2.
Predicted risks by genetic risk scores. (A) Relative risks by genetic risk scores (GRSs) across the population. For two different types of markers sets (“BRCAX-known”: markers replicated for Korean BRCAX cases among previously reported breast cancer-associated markers from the GWAS catalog; “BRCAX-known + novel”: three novel candidate markers added to replicated markers), a predictive performance by GRSs was estimated for Korean BRCAX, Asian, and European high-risk cases. For BRCAX-known markers, predictive performance by GRSs was highest for Korean BRCAX followed by European and Asian high-risk cases. When three novel markers were added, however, prediction improved for Asians compared to Europeans. (B) Predicted risks of breast cancer according to three risk groups using the number of risk alleles of three consistently reported markers from three previous GWAS of BRCAX across the population: FGFR2 (rs2912774), TOX3 (rs4784227), and ESR1 (rs9383936). (C) Predicted BRCAX risks by GRSs using BRCAX-known markers by tertile groups of three global BRCAX-associated markers (on FGFR2, TOX3, and near ESR1): circle symbols are for low-risk group (T1), triangle symbols are for moderate risk group (T2), and square symbols are for high-risk group (T3).
Discussion
From a genome-wide search, three candidate loci were newly identified with suggestive evidence of functional involvement. The three novel loci identified by this study might tag the closely-located functional variants based on the downstream analyses. The marker itself, or markers within the LD block, have marks for regulatory elements, suggesting these SNPs themselves or variants tagged by the SNPs might be associated with the pathophysiology of BRCAX breast cancer development. PDE7B (phosphodiesterase 7B) encodes a cyclic adenosine monophosphate (cAMP)-specific phosphodiesterase and CDKN2B (cyclin-dependent kinase 4 inhibitor B) is a known tumor suppressor gene27. The risk allele of the top hit (rs78545330) on CDKN2B-AS1 showed strong evidence of reducing gene expression level of this tumor suppressor gene.
When we examined the association of newly identified markers with overall Asian cases including late-onset and sporadic breast cancers of BCAC, both rs11154838 (PDE7B) and rs78545330 (CDKN2B-AS1) showed stronger level of significance but with smaller effect sizes (ORs: 1.16 and 1.14 for all breast cancers, 1.26 and 1.19 for high-risk breast cancers), but rs278050 (UBL3) was not associated (p = 0.2). For European cases, rs78545330 was replicated for overall cases only (p = 0.017, OR = 1.04; high-risk breast cancer: p = 0.2), but other two novel candidate loci did not show significant associations. One possible explanation for this population-specific manner of association is that genetic susceptibility to BRCAX might comprise multiple genetic loci which are only partly shared between populations. The newly identified candidate loci might be Asian-specific and also more associated with the early-onset type of high-risk cancers. When we estimated OR according to GRS, the addition of novel variants to known markers increased the risk for Korean and Asian, but not for European high-risk breast cancers.
It is interesting that some of the markers shared between high-risk and general breast cancers showed augmented impacts on the BRCAX cancers. For example, rs12628403 on the intron of APOBEC3A alone explained 6.5% of FRR (OR = 1.49), compared with OR = 1.17 for overall breast cancers. Also, rs10069690 on the intron of TERT was replicated for BRCAX with higher risk estimate (OR = 1.24) than the OR (1.10) from the original studies. These findings are in line with the findings from the colorectal cancer study28: some SNPs show stronger associations with high-risk cases with larger impacts. Our findings suggest that role of some variants might not be same between in high-risk and in general breast cancers, although they are associated with both types. It might be logical to infer that a substantial part of the genetic risk of high-risk breast cancers comprises common variants overlapping with general types of breast cancers but at augmented risks.
Among replicated markers, FGFR2 (rs2912774), TOX3 (rs4784227), and ESR1 (rs9383936) deserve a special remark. All three previous GWAS for BRCAX replicated FGFR2, and multiple studies replicated TOX3 and ESR120–22. Our study also replicated these three variants for high-risk breast cancers in all population data. When we compared the estimated risk of these three variants, the estimated risk was quite similar between three populations (Korean BRCAX, Asian and European high-risk cancer, Fig. 2B). This finding suggests that shared genetic susceptibility toward high-risk breast cancer might exist across the population. These three variants might be only the tip of the iceberg. Our study did not examine sequence level variation which might also be shared across populations. At least, our findings support that a part of common variants associated with low-risk breast cancers also has their roles in the susceptibility of high-risk breast cancer.
In our study, about 62% of Korean BRCAX cases, 48% and 25% of Asian and European high-risk cancers were non-familial early-onset cases. Europeans have more familial cancers probably due to higher prevalence than Asians or inclusion criteria of each study. It is not clear whether two major indications of being a “high-risk” type, i.e., early-onset and familial cases have differences in their genetic constitutions. In our study, differences in the association and effect size between populations for some variants might stem from the differences in this case composites.
When GRS was re-estimated according to the risk allele counts of FGFR2/TOX3/ESR1, the estimated risk from GRS not only was larger but also showed a steeper increase suggesting an effect modification. The risk modification according to FGFR2/TOX3/ESR1 was more evident for BRCAX (Korean), partly because of its strict case definitions (Fig. 2C).
We also evaluated the impact of known and novel susceptibility loci for high-risk breast cancers. The estimated Σ%FRR from replicated markers was 29.8% for Korean BRCAX with 24 markers. The findings from replicated markers (29.8% FRR) might not be attributed to so-called “winner’s curse” bias, an inflated impact estimation in the discovery set itself, because the Korean BRCAX served as a replication set. Three novel loci accounted for 9.6% of the FRR for Korean BRCAX. The impact of three novel loci on Korean BRCAX cases, however, might be inflated for having used the discovery set on itself. Novel candidate loci added 3.0% to Asian, but none to European high-risk cancers. When the %FRR of three novel loci was estimated from all types of Asian breast cancers, the estimate decreased to 0.75%, suggesting that these loci mainly account for the genetic risk of high-risk breast cancers.
Sum of FRR (Σ%FRR) from common variants are considerably compatible between populations: 29.8–39.4% for Koreans, and 24.0% for Europeans, and 10.6–13.6% for other Asians. One reason why Σ%FRR is smaller for Asians might be explained their heterogenic ethnic compositions (In this study, Asian high-risk breast cancer cases consisted of Japanese 28%, Chinese 30%, Asian American 16%, and others 25%). If associated markers have not been identical between ethnic groups, some variants more specifically associated with certain populations might be missed. Moreover, replication was conducted using 92 markers, most of them were reported from studies of European ancestry. Also, a smaller sample size could be another reason for fewer replicated markers in Asians. Wen et al.’s study replicated 44 of 78 previously reported loci involving 11,760 overall breast cancer cases of East Asians29. When we performed an analysis including late-onset breast cancer cases (6,130 cases and 6,371 controls), we could have replicated 24 loci (10 consistent, 12 added and two removed loci). Whether the difference is attributable to the power or the differences in the genetic susceptibility between high-risk and late-onset breast cancers is not conclusive. The six additional loci, however, did not show any clues of possible association with high-risk breast cancers (p > 0.3), and we believe the differences in the genetic architecture between sporadic/low-risk and familial/high-risk breast cancers might explain a part of the discrepancy. Korean data might have shown compatible Σ%FRR of high-risk breast cancers with Europeans, partly due to their size and stricter definition for cases. We believe it might be reasonable to interpret the findings that common variants explain as much Σ%FRR of high-risk cancers as BRCA1/2 mutations. We also examined the associations and their impacts on breast cancer predisposition genes with moderate/high-penetrance. Although imputed markers covered most of the known variants, our approach might have materially underestimated their impacts, and cannot replace the findings from the sequence-based approach.
A proper genetic screening program for BRCA/BRCAX, if implemented, might contribute to detecting high-risk women for providing personalized screening program such as ultrasonography. Additionally, it will provide information for consulting the relatives of BRCAX cases. Findings from our study support the necessity and possibility of developing a new genetic screening for BRCAX. However, our study alone does not provide other important information necessary for effective genetic screening, in particular, estimates of penetrance and predictive values of the test. Estimating sensitivity and specificity of BRCAX-associated markers are required first in larger data. Also, this study considered the genetic markers associated with the occurrence of high-risk breast cancer, and their potential implications for detecting high-risk populations. To provide evidence that support a new genetic screening program, further data about a reduced mortality rate from breast cancer might be required, but these are beyond scope of ours.
We believe that this study confers some advantages over previous BRCAX studies. First, the number of test-proven BRCA-negative cases was not modest (n = 1,469), and more than 11,300 high-risk breast cancer cases were included, larger than previous studies20–22. Second, all the BRCA1/2 mutation status was more accurately identified by standard sequencing analysis of each BRCA1 and BRCA2 exon. Third, we selected high-risk cases with genetic burdens using the similar indications as BRCA1/2 mutation test: age of onset, family history, history of other malignancy, and bilateral cancers. Applying these strict indications might have enabled us to select cases with high genetic burdens. Fourth, we attempted to re-evaluate the findings from our own GWAS study and previous studies jointly including both common variants in the GWAS catalog, and moderate/high-penetrance genes. This approach might have provided more comprehensive views on the genetic architecture of BRCAX and high-risk breast cancers. Similarly, this approach enabled us to estimate the impacts of associated markers on the occurrence of BRCAX for different populations and context. The impact estimates from our study will confer implications for genetic screening test of high-risk women. Finally, to our knowledge, this is the first Asian study focused on BRCAX cases, where epidemiologic features suggest unique susceptibility might exist.
This study also has several limitations. First, exact information about BRCA1/2 mutation status was scarce, and not available for most studies of the BCAC. Applying the criteria for high-risk cancers only which were not confirmed to be BRCA1/2-negative might have decreased the power of detecting associated markers. Nevertheless, this limitation might not have influenced toward increasing false-positive findings in the study. Second, Asian cases in the BCAC data were limited so that the criteria for early-onset age were relaxed to the age 45, while the same criteria for having family history was applied. These relaxed criteria in the replication data might have reduced the power to detect susceptibility loci more specific to the BRCAX cases. When we estimated the associations for replicated and novel loci using different age criteria for Asian high-risk cases (938 cases for replication set when age 40 was used), direction and strength of associations were largely same with slightly less statistical power: the number of replicated markers and Σ%FRR of them declined from 14 (10.6%) to eight (9.0%). However, ORs of three novel markers increased and Σ%FRR increased from 3.0% to 3.8% accordingly. Third, even with the sample size of 1,469 confirmed BRCAX cases, our study has limitations in power to newly detect relatively rare alleles (0.01 < MAF < 0.05) because of the scarcity of high-risk cancer cases. We estimated that the relative risk should exceed 2.5 to be detected as novel with our sample size30. Fourth, although this not being the main purpose of our study, when we attempted to replicate the “moderate/high-penetrance” variants, the effects might have been underestimated because some of the variants were not exactly identified in our imputed genotype data. We believe the estimated impacts from these genes should have been underestimated. The estimated impact of the common variants, however, is not likely affected by this limitations, because the frequency of the common risk alleles is independent of the rare alleles. Fifth, the replication analysis of previously reported loci might have inflated as the replication set could include data of the discovery set. Finally, as the mean age was different by affection status in the discovery set (cases: 40.2, controls: 55.1), the age was not used as a covariate, because we did not observe any difference in allele frequencies between controls (older) and those in the population with same age with cases (younger) (Supplementary Table S1). If there are some susceptibility loci which are expressed in an age-dependent manner, those markers could not have considered in this study. However, we believe older controls might be more appropriate in part due to they have lower chance of developing breast cancer compared to younger controls with similar mean age with the cases (40 years old).
In conclusion, we have identified novel common susceptibility loci from GWAS and replicated some of both common/low-penetrance and rare/high-penetrance variants which were reported previously. We also evaluated the contribution of those susceptibility loci including ours, to high-risk breast cancers of multiple populations. Our findings also suggest that high-risk breast cancers, particularly for Asians, might consist of multiple layers with similar importance, including BRCA1/2, moderate/high-penetration genes, and selected common variants with augmented effects. Our study might add information to consider prevention strategy of early-onset and high-risk breast cancers in Asia, and also to the genetic counseling for relatives of the high-risk breast cancer patients without a mutation in BRCA1/2 genes.
Methods
Study design and participants of the discovery set
We conducted a case-control GWAS. Breast cancer patients were selected from the Korean Hereditary Breast Cancer (KOHBRA) study. Breast cancer patients with one of following five conditions and family members of BRCA1/2 mutation carriers were eligible for BRCA1/2 gene test provided by the Korean National Health Insurance - family history of breast or ovarian cancer, diagnosed at age 40 or younger, bilateral breast cancer, male cases, or diagnosed with another primary malignancy. The KOHBRA study enrolled those who provided informed consent among the “BRCA1/2 gene test eligible” cases and their family members. The detailed information of the KOHBRA study has been described previously31 and in Supplementary Notes. From the KOHBRA participants, unrelated women (estimated identity by descent < 0.125) who met the following criteria were included in this study:
-
(i)
Breast cancer patients who were tested to be free of clinically significant BRCA1/2 gene mutation (clinically unverified mutation cases were included)
AND at least one of the following
-
(ii)
Diagnosed as breast cancer before or at age 40; OR having one or more relative(s) with breast/ovarian cancer; OR bilateral cases; OR previously diagnosed as other cancer.
We used data from Korean Genome Epidemiology Study (KoGES) as control. The profiles of KoGES consortium was described elsewhere32 and in Supplementary Notes. Genetically unrelated (estimated identity by descent <0.125) Korean women without family history of breast or ovarian cancer were included. All participants gave informed written/oral consent, and this study was approved by the Institutional Review Board of Asan Medical Center, Seoul, Republic of Korea. This study was conducted in accordance with the World Medical Association Declaration of Helsinki.
Genotyping and imputation methods
Cases were genotyped using Illumina OncoArray-500K Beadchip at the University of Southern California Epigenome Center, CA, USA. Peripheral blood samples (20 ml) were collected at baseline, and they were divided, treated and stored within 24 hours of sampling. We excluded individuals with low call rate (<95%) and SNPs with genotype call rate <95%, minor allele frequency (MAF)<1%, evidence for deviation from Hardy-Weinberg equilibrium at p-value < 10−9, or poor cluster plots. Genotype data were phased and imputed using SHAPEIT and IMPUTE2 software respectively, and East Asian data from the 1000 Genomes Project (1 kG) Phase 3 were used as reference33–36. From imputed markers, SNPs with info score greater than 0.95 were included. We genotyped cases and controls with four different platforms of dense genome-wide SNP arrays and conducted standard quality control and imputation process. And the batch effect detected through association analysis of control data genotyped from different platforms was removed (p < 0.01). For post-imputation quality control, markers with high missing rate >10%, minor allele frequency (MAF)<1%, evidence for deviation from Hardy-Weinberg equilibrium at p-value < 10−6 were excluded. More detailed information was described in Supplementary Notes.
BRCA1 and BRCA2 genomic mutation analysis
Genomic DNA was extracted from the participants’ peripheral blood samples. For BRCA1/2 mutation testing in cases, 22 coding exons of BRCA1 and 26 coding exons of BRCA2 were scanned through fluorescence-based conformation sensitive gel electrophoresis (F-CSGE) and denaturing high-performance liquid chromatography (DHPLC). For a subset of PCR products with aberrant patterns, direct sequencing was performed on an ABI3100 or ABI3700 (Applied Biosystems, CA) or a MegaBACE500 (GE Healthcare, UK) genetic analyzer. In this study, the definition of a genetic mutation is restricted to the protein-truncating mutation and the missense mutation, which are known to be associated with the disease. This study also includes the participants with a clinically unverified mutation in BRCA1/2 genes. More detailed information about BRCA1/2 mutation analysis has been described before31,37. Since this study also includes the cases with unverified mutation on BRCA1/2, we examined whether non-pathogenic variants on these genes are more frequent in the cases compared to controls at p-value of 0.05. And we searched them at the ClinVar database.
Statistical analyses for the genome-wide search
GWAS was carried out using logistic regression by trends test of PLINK version 1.0738. For visualization of the results, Manhattan plot and quantile-quantile (QQ) plot were generated using R package ‘qqman’39. Principal component analysis (PCA) was conducted using EIGENSTRAT, and the first three principal components were used for adjustment40. Since most cases were early-onset breast cancer and cohorts for control recruited adults with a limited age range, mean age was much younger in cases than controls. Thus, allele frequencies of significant markers were presented by affection status and age groups, rather than adjusting for age.
Replication and meta-analysis of newly identified loci
For the replication of the initial candidate loci, imputed data of Asian population from the BCAC were used. The overall description about the BCAC was previously reported41 and in Supplementary Notes. Since most of the cases in the BCAC lack BRCA1/2 mutation information, we included high-risk breast cancers defined as: breast cancer cases diagnosed before or at age 45 or cases with family history of breast cancer in first degree relatives. For controls, women without family history of breast cancer were included among the BCAC control data.
For newly identified loci, we conducted meta-analyses using fixed or random effect model according to evidence of heterogeneity (fixed effect model was used if p-value for heterogeneity exceeds 0.05). Also, we conducted subgroup analyses by three receptor status (estrogen, progesterone, and HER2 receptor). P-values for heterogeneity by receptor status were calculated using a Cochran’s Q test, and a p-value of 0.05 was used to determine heterogeneity.
Downstream analyses of newly identified loci
For newly identified loci which were replicated using Asian high-risk cases from the BCAC, downstream analyses were performed. These regions were visualized by linkage disequilibrium (LD) plots and regional association plots. LD plots were created using control data with ‘LDheatmap’ package in R42. And regional association plots were created by Locuszoom using East Asian population of the 1 kG Phase 3 as reference43.
Functional information of novel loci was annotated using Encyclopedia of DNA elements (ENCODE) project and HaploReg version 4.144,45. Novel SNPs and correlated markers with r2 > 0.8 from East Asian population were explored. Results of expression quantitative trait loci (eQTL) analyses were found from the Genotype Tissue Expression (GTEx) portal46. Associations between novel markers and expression of the nearest gene of each SNP for all available tissues and associations between these markers and nearby genes with a window size of 1 Mb for the tissue showing the most significant associations were retrieved.
Replicating susceptibility loci previously associated with breast cancers
We estimated associations between previously reported breast cancer susceptibility loci and BRCAX cases and how the significance of associations and their effect sizes differ. First, we selected markers associated with breast cancer at p-value < 10−5 from the NHGRI-EBI GWAS catalog25. We also examined the candidate loci from three previous BRCA1/2-negative cancer GWAS20–22. Finally, the variants on genes associated with increased risk of breast or ovarian cancer listed in the guideline for genetic screening of high-risk breast cancers (suggested by the National Comprehensive Cancer Network, NCCN) or breast cancer predisposition genes used in a recent article were evaluated in this study16,26. Seventeen genes listed on the NCCN guideline were ATM, CDH1, CHEK2, MSH2, MLH1, MSH6, PMS2, EPCAM, NBN, NF1, PALB2, PTEN, RAD51C, RAD51D, STK11, and TP53, except for BRCA1/2; six additional genes suggested from the study by Wong et al. were BARD1, BMPR1A, FANCC, SMAD4, VHL, and XRCC2. Variants in gene regions ± 10 kB were tested for association. In this analysis, the markers with MAF < 1%, which were excluded during quality control, were also included. Independent (r2 > 0.3) SNPs with p-value < 0.01 were included for impact estimation.
To assess the impact of reported loci in the occurrence of BRCAX cases, we estimated the FRRs from Korean BRCAX women, Asian and European high-risk women using the BCAC data (cases with age of onset <45 or with a family history of breast cancer in first-degree relatives, for both Asian and European data). The fraction of FRR (%FRR) explained by markers was calculated as ln(λ)/ln(λ0) where lambda (λ) is the FRR to a relative of an affected individual and λ0 is the overall FRR. λ is calculated as (pr2 × q)/(pr + q)2 where p is the frequency of the risk allele in the general population, q equals 1 − p, and r is the per-allele OR. And λ0 is assumed to be 1.8 as reported from the previous article47,48. For comparison of FRR explained by each SNP, we applied the same allele frequencies from the East Asian or European data from the 1 kG Phase 3 and different odds ratios (ORs) from each analysis.
Impact analysis for novel and replicated loci
To compare predictive power among different groups, we estimated ORs according to GRSs. GRSs were calculated as a weighted sum of the number of risk alleles, in which the weight was taken as the natural log of the OR from association analyses. ORs by quintiles of GRS were estimated using the first quintile as the reference. We compared the predictive power of BRCAX by the different set of markers for different populations (Korean BRCAX, Asian and European high-risk cases).
We re-evaluated the ORs from polygenic GRS, by stratifying subjects according to their risk alleles associated with BRCAX in multiple studies20–22. GRSs were calculated using markers replicated for Korean BRCAX among previously reported markers in breast cancer from the GWAS catalog (“BRCAX-known”), and with novel candidate markers from the GWAS added to replicated markers (“BRCAX-known + novel”).
Electronic supplementary material
Acknowledgements
This work was supported by the Post-Genome Technology Development Program (10050164, Developing Korean Reference Genome) funded by the Ministry of Trade, Industry and Energy (MOTIE), Republic of Korea. This study was also funded by a grant (grant no. 2012-0510/2013-0510) from the Asan Institute for Life Science, Asan Medical Center, Seoul, Republic of Korea and supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (No. 1020350). We thank the BCAC and KoGES consortium for access to the data. Also, we thank all investigators of the KOHBRA Study: Beom Seok Kwak, Byeong-Woo Park, Byung Ho Son, Byung-In Moon, Cha Kyong Yom, Chan Heun Park, Chan Seok Yoon, Chang Hyun Lee, Dae Sung Yoon, Dong-Young Noh, Doo Ho Choi, Eundeok Chang, Eun-Kyu Kim, Eunyoung Kang, Hae Kyung Lee, Hai-Lin Park, Hyde Lee, Hyeong-Gon Moon, Hyun-Ah Kim, Il-Kyun Lee, Jeong Eon Lee, Jihyoun Lee, Jong Won Lee, Jong-Han Yu, Joon Jeong, Jung Han Yoon, Jung-Hyun Yang, Keumhee Kwak, Ki-Tae Hwang, Ku Sang Kim, Lee Su Kim, Min Hee Hur, Min Ho Park, Min Hyuk Lee, Myung Chul Chang, Nam Sun Paik, Sang Ah Han, Sang Seol Jung, Sang Uk Woo, Se Jeong Oh, Sehwan Han, Sei Joong Kim, Sei-Hyun Ahn, Seok-Jin Nam, Seung Sang Ko, Sung Hoo Jung, Sung Soo Kang, Sung Yong Kim, Sung-Won Kim, Tae Hyun Kim, Tae Wan Won, Tae Woo Kang, Wonshik Han, Woo-Chul Noh, Yong Lai Park, Yongsik Jung, Young Jin Suh, Young Tae Bae, Young Up Cho, Young-Ik Hong, Sue K. Park, Yoon Joo Jung, Su Yun Choi, Young Bum Yoo, and Soo-Jung Lee.
Author Contributions
J.-Y.L. performed all analyses and wrote the manuscript. S.-W.K., J.W.L., S.K.P., S.H.A., M.H.L. are principal investigators of KOHBRA study and they participated in recruitment and maintenance of this cohort with J.K., S.B.L., Y.J.S., D.-Y.N., B.H.S. and Y.U.C. J.K., J.S. and J.L.H. made substantial contributions to acquisition of KOHBRA, KoGES, and BCAC data, respectively. J.-Y.L., J.S. J.L.H. and J.K. contributed to writing and editing of the manuscript. J.W.L., J.L.H. and J.S. supervised the research. All authors approved the final version of the manuscript.
Data Availability
The datasets generated and/or analysed during the current study are available in the Open Science Framework repository, https://osf.io/a7kn5/. The data that support the findings of this study are available from BCAC but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the BCAC group.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Joo-Yeon Lee and Jisun Kim contributed equally
Jong Won Lee, John L. Hopper and Joohon Sung jointly supervised this work.
Contributor Information
Jong Won Lee, Email: jjjongwr@hanmail.net.
John L. Hopper, Email: j.hopper@unimelb.edu.au
Joohon Sung, Email: jsung@snu.ac.kr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31859-8.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, et al. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2014. Cancer research and treatment: official journal of Korean Cancer Association. 2017;49:292–305. doi: 10.4143/crt.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korea National Cancer Information Center, Breast Cancer Screening Guideline. Available at: http://www.cancer.go.kr. Accessed April 26th, 2017.
- 4.Yoo KB, et al. Is mammography for breast cancer screening cost-effective in both Western and asian countries?: results of a systematic review. Asian Pacific journal of cancer prevention: APJCP. 2013;14:4141–4149. doi: 10.7314/APJCP.2013.14.7.4141. [DOI] [PubMed] [Google Scholar]
- 5.del Carmen MG, et al. Mammographic breast density and race. AJR Am J Roentgenol. 2007;188:1147–1150. doi: 10.2214/AJR.06.0619. [DOI] [PubMed] [Google Scholar]
- 6.El-Bastawissi AY, White E, Mandelson MT, Taplin S. Variation in mammographic breast density by race. Ann Epidemiol. 2001;11:257–263. doi: 10.1016/S1047-2797(00)00225-8. [DOI] [PubMed] [Google Scholar]
- 7.Mandelson MT, et al. Breast density as a predictor of mammographic detection: comparison of interval- and screen-detected cancers. Journal of the National Cancer Institute. 2000;92:1081–1087. doi: 10.1093/jnci/92.13.1081. [DOI] [PubMed] [Google Scholar]
- 8.Carney PA, et al. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann Intern Med. 2003;138:168–175. doi: 10.7326/0003-4819-138-3-200302040-00008. [DOI] [PubMed] [Google Scholar]
- 9.Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2016 Sub (1973-2014 varying) - Linked To County Attributes - Total U.S., 1969–2015 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2017, based on the November 2016 submission.
- 10.KOrean Statistical Information Service (Ministry of Health and Welfare, Korea, Cancer Registration Statistics). Breast Cancer Incident Cases for Women by Age Groups, 2005–2014. Available at: http://kosis.kr. Accessed April 26th, 2017.
- 11.Gabriel CA, Domchek SM. Breast cancer in young women. Breast cancer research: BCR. 2010;12:212. doi: 10.1186/bcr2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredholm H, et al. Breast cancer in young women: poor survival despite intensive treatment. PloS one. 2009;4:e7695. doi: 10.1371/journal.pone.0007695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narod SA. Breast cancer in young women. Nat Rev Clin Oncol. 2012;9:460–470. doi: 10.1038/nrclinonc.2012.102. [DOI] [PubMed] [Google Scholar]
- 14.Anders CK, Johnson R, Litton J, Phillips M, Bleyer A. Breast cancer before age 40 years. Semin Oncol. 2009;36:237–249. doi: 10.1053/j.seminoncol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343:1466–1470. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Comprehensive Cancer Network, Genetic/Familial High-Risk Assessment: Breast and Ovarian (Version 2. 2017). Available at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf. Accessed April 26th, 2017.
- 17.Metcalfe KA, et al. Breast cancer risks in women with a family history of breast or ovarian cancer who have tested negative for a BRCA1 or BRCA2 mutation. British journal of cancer. 2009;100:421–425. doi: 10.1038/sj.bjc.6604830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotsopoulos J, et al. Prospective study of high-risk, BRCA1/2-mutation negative women: the ‘negative study’. BMC Cancer. 2014;14:221. doi: 10.1186/1471-2407-14-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang E, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast cancer research and treatment. 2015;151:157–168. doi: 10.1007/s10549-015-3377-4. [DOI] [PubMed] [Google Scholar]
- 20.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palomba G, et al. Genome-wide association study of susceptibility loci for breast cancer in Sardinian population. BMC Cancer. 2015;15:383. doi: 10.1186/s12885-015-1392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinella ES, et al. Genetic variants associated with breast cancer risk for Ashkenazi Jewish women with strong family histories but no identifiable BRCA1/2 mutation. Human genetics. 2013;132:523–536. doi: 10.1007/s00439-013-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics. 2013;45(353–361):361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nature genetics. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The NHGRI-EBI Catalog of published genome-wide association studies. Available at: www.ebi.ac.uk/gwas. Accessed March 1st, 2017. [DOI] [PMC free article] [PubMed]
- 26.Wong ESY, et al. Inherited breast cancer predisposition in Asians: multigene panel testing outcomes from Singapore. NPJ Genom Med. 2016;1:15003. doi: 10.1038/npjgenmed.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins MA, et al. Quantifying the utility of single nucleotide polymorphisms to guide colorectal cancer screening. Future Oncol. 2016;12:503–513. doi: 10.2217/fon.15.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen W, et al. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast cancer research: BCR. 2016;18:124. doi: 10.1186/s13058-016-0786-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11:773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han SA, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clinical oncology. 2011;23:434–441. doi: 10.1016/j.clon.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Han BG, Ko GES. g. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. International journal of epidemiology. 2016 doi: 10.1093/ije/dyv316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. 2011;G3(1):457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 37.Son BH, et al. Prevalence of BRCA1 and BRCA2 mutations in non-familial breast cancer patients with high risks in Korea: the Korean Hereditary Breast Cancer (KOHBRA) Study. Breast cancer research and treatment. 2012;133:1143–1152. doi: 10.1007/s10549-012-2001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner, S. D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots., 10.1101/005165.
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Breast Cancer Association Consortium. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. Journal of the National Cancer Institute98, 1382–1396, 10.1093/jnci/djj374 (2006). [DOI] [PubMed]
- 42.Shin, J.-H., Blay, S., McNeney, B. & Graham, J. L. Dheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. Journal of statistical software16, Code Snippet 3 (2006).
- 43.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74, 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collaborative Group on Hormonal Factors in Breast Cancer. Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet358, 1389–1399, 10.1016/S0140-6736(01)06524-2 (2001). [DOI] [PubMed]
- 48.Zheng W, et al. Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Human molecular genetics. 2013;22:2539–2550. doi: 10.1093/hmg/ddt089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Open Science Framework repository, https://osf.io/a7kn5/. The data that support the findings of this study are available from BCAC but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the BCAC group.