Abstract
Objective
We performed small-scale mutation and large genomic rearrangement (LGR) analysis of BRCA1/2 in ovarian cancer patients to determine the prevalence and the characteristics of the mutations.
Methods
All ovarian cancer patients who visited a single institution between September 2015 and April 2017 were included. Sanger sequencing, multiplex ligation-dependent probe amplification (MLPA), and long-range polymerase chain reaction (PCR) were performed to comprehensively study BRCA1/2. The genetic risk models BRCAPRO, Myriad, and BOADICEA were used to evaluate the mutation analysis.
Results
In total, 131 patients were enrolled. Of the 131 patients, Sanger sequencing identified 16 different BRCA1/2 small-scale mutations in 20 patients (15.3%). Two novel nonsense mutations were detected in 2 patients with a serous borderline tumor and a large-cell neuroendocrine carcinoma. MLPA analysis of BRCA1/2 in Sanger-negative patients revealed 2 LGRs. The LGRs accounted for 14.3% of all identified BRCA1 mutations, and the prevalence of LGRs identified in this study was 1.8% in 111 Sanger-negative patients. The genetic risk models showed statistically significant differences between mutation carriers and non-carriers. The 2 patients with LGRs had at least one blood relative with breast or ovarian cancer.
Conclusion
Twenty-two (16.8%) of the unselected ovarian cancer patients had BRCA1/2 mutations that were detected through comprehensive BRCA1/2 genetic testing. Ovarian cancer patients with Sanger-negative results should be considered for LGR detection if they have one blood relative with breast or ovarian cancer. The detection of more BRCA1/2 mutations in patients is important for efforts to provide targeted therapy to ovarian cancer patients.
Keywords: Mutation, Genes, Neoplasms, Ovary, Korea
INTRODUCTION
Ovarian cancer, which is the most fatal gynecologic malignancy in the world, is a heterogeneous disease with multiple histologic subtypes. It is the fourth most common cancer in females aged 0–34, and its prevalence has been increasing in Korea [1]. The majority (90%) of ovarian cancers are epithelial ovarian cancers [2]. The most common histologic subtype of epithelial ovarian cancer is high-grade serous ovarian cancer, accounting for about 70% of cases, a majority of which are diagnosed at an advanced stage [3].
High-penetrance ovarian cancer susceptibility genes BRCA1 and BRCA2 are tumor suppressor genes located at 17q21.31 and 13q13.1, respectively. In a recent prospective cohort study, the cumulative risk of ovarian cancer by age 80 was 44% for BRCA1 mutation carriers and 17% for BRCA2 mutation carriers [4]. In May 2013, Angelina Jolie, an actress from the United States, publicized her story of having a BRCA1 mutation and undergoing risk-reducing surgery. After her announcement, referrals to genetic services and risk-reducing surgeries increased, the so-called “Angelina effect.” Moreover, In Korea, the National Health Insurance system promoted a strategy of BRCA1/2 testing coverage for epithelial ovarian cancer patients in May 2012. Both the “Angelina effect” and Health insurance coverage are thought to be behind the gradual increase in BRCA testing in Korea [5].
The majority of BRCA1/2 mutations are small-scale mutations (point mutations, small deletions, or insertions), resulting in protein truncation, disruption of messenger RNA processing, or amino acid substitutions that have a significant impact on protein function. Such mutations occur throughout the whole coding sequence or at the splice junctions of both genes. These mutations are readily detected by standard Sanger sequencing methods using polymerase chain reaction (PCR)-amplified gene segments [6]. Another mechanism of BRCA1 and BRCA2 inactivation are large genomic rearrangements (LGRs), which are responsible for a variable but significant proportion of BRCA1/2 mutations [7]. Because Sanger sequencing is incapable of detecting LGRs, other techniques, such as multiplex ligation-dependent probe amplification (MLPA) or next-generation sequencing, are required. MLPA is a semi-quantitative technique that is the most commonly used method for detecting LGRs in BRCA1/2. A high prevalence of LGRs in BRCA1 or BRCA2 has been demonstrated in several populations, including the Dutch, Northern Italian, Danish, and Portuguese [8,9,10,11]. In contrast, studies in some populations, including the French-Canadian and Sri Lankan, found no LGR mutations in BRCA1/2, suggesting that LGRs are probably very rare in such populations [12,13]. In populations with rare LGRs, a cost-effective screening strategy for LGR detection is necessary to identify all patients that may benefit from targeted therapies, such as a poly ADP-ribose polymerase inhibitor.
According to a recent study in Korea, 24.6% of 232 epithelial ovarian patients had small-scale mutations of BRCA1/2 that were detected using Sanger sequencing [14]. A few LGRs in primary breast cancer patients were also reported in Korea [15,16,17], but the prevalence of LGRs in primary ovarian cancer patients has not yet been reported in Korea. Additionally, most previous BRCA1/2 studies in ovarian cancer have been focused on epithelial subtypes, but BRCA1/2 mutations in various other ovarian cancer histologic subtypes have also been reported [18,19].
Therefore, we performed small-scale mutation and LGR analysis without selection in multiple ovarian cancer subtypes. We also evaluated the usefulness of several mutation carrier prediction algorithms for predicting BRCA1/2 mutations, including LGRs. Here, we report the results of a comprehensive BRCA1/2 genetic study that determined the prevalence and the characteristics of mutations among unselected ovarian cancer patients.
MATERIALS AND METHODS
1. Study population
All ovarian, primary peritoneal, primary fallopian tube cancer patients who visited the Department of Obstetrics and Gynecology at Dongsan Medical Center of Keimyung University for genetic testing between September 2015 and April 2017 were included in this study. The family histories, medical record, and tumor pathology of the probands and their family members were detailed through genetic counseling and/or review of the patient's medical record. All participants provided informed consent, and this study was approved by the Institutional Review Board (IRB)/Ethics Committee of Dongsan Medical Center (IRB No. 2016-05-002).
2. Sanger sequencing, MLPA, and long-range PCR
Sanger sequencing was performed in all ovarian cancer patients to detect small-scale mutations. Genomic DNA was isolated from the peripheral blood leukocytes using the QIAmp DNA Mini Kit (Qiagen, Hamburg, Germany). Sanger sequencing was performed on a 3500xL DNA Analyzer with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and analyzed using Sequencer 5.0 software (Gene Codes Corporation, Ann Arbor, MI, USA). Exon numbering and DNA sequence variant descriptions of BRCA1/2 were based on NM_007294.3 (NG_005905.2) and NM_000059.3 (NG_012772.3) as reference sequences, obtained from the NCBI RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/rsg/). All BRCA1/2 variants were categorized into pathogenic, variants of uncertain significance (VUS), or neutral. Pathogenic variants were defined as previously reported pathogenic variants and novel nonsense or frameshift variants that resulted in protein truncation. Neutral variants were those that were clearly not pathogenic or were unlikely to be pathogenic. Other variants were classified as VUS.
MLPA was performed on Sanger sequencing-negative patients. MLPA probe mixes P002 and P045 were used for the screening of LGRs in BRCA1 and BRCA2, respectively, and P087 and P077 were used for confirmation, according to the manufacturer's recommendations (MRC-Holland, Amsterdam, The Netherlands). MLPA was performed as previously described [15]. Genemarker v1.91 software (Softgenetics, State College, PA, USA) was used for fragment analysis. After normalization of raw data, a deletion or duplication was identified when the peak height ratio was below 0.70 or more than 1.40, respectively. We also used Sanger sequencing of the probe binding and ligation sites to detect any variants that may have led to a false positive result [6,20].
To characterize the LGRs detected in this study, long-range PCR was performed using primers Int20-3′ F 5′-CCTGGGAGAACCCCAGAGT-3′ and Int20-3′ R 5′-CTGGTCCTGGAGGAGGAGTT-3′ and an Expand Long Range dNTPack (Roche Diagnostics GmbH, Mannheim, Germany). The PCR cycling conditions were as follows: an initial denaturation at 92°C for 2 minutes; 10 cycles at 92°C for 10 seconds, 60°C for 15 seconds, and 68°C for 15 minutes; 25 cycles at 92°C for 10 seconds, 60°C for 15 seconds, and 68°C for 15 minutes (increase of 20 seconds per cycle); and final extension at 68°C for 15 minutes.
To determine the exact breakpoint of LGR, the target band was cut from the agarose gel and was employed to purify the target fragment using SolGent UB solution (SolGent, Daejeon, Korea). The product was sequenced with a series of additional primers to successfully narrow down the breakpoint regions. The primer that eventually allowed for characterization of the exon 21–23 deletion breakpoint was Internal1 F 5′-GAGGTCGAGTCTCTGTTGCC-3′. The breakpoint of LGR was described according to the Human Genome Variation Society (HGVS) nomenclature criteria (http://varnomen.hgvs.org/recommendations/). To explore the mechanism underpinning LGR, L78833.1 was used as a reference sequence to search for the location of Alu sequences.
3. Mutation carrier prediction algorithms for BRCA1/2
We used the genetic risk models BRCAPRO, Myriad, and BOADICEA to compute a probability or a score for the likelihood of carrying a BRCA1/2 mutation.
BRCAPRO is a statistical model that uses Mendelian inheritance and Bayesian analysis based on all available data, including current age/age at death, age of diagnosis, ethnicity, personal/familial history of cancer, cancer markers, and genetic test results [21]. BRCAPRO scores were calculated using the latest version of the software program, CancerGene 6.0 (available at http://www4.utsouthwestern.edu/breasthealth/cagene/).
The Myriad BRCA1 and BRCA2 prevalence tables provide the probability of detecting BRCA1/2 mutations and are based on observations of deleterious mutations by Myriad Genetic Laboratories through its clinical testing service [22]. We used the latest version of the table, which was based on 162,914 tests in individuals without an Ashkenazi ancestry (available at https://new.myriadpro.com/products/bracanalysis-overview/#1479845138516-ee8a82d1-427c).
BOADICEA is a computer program that is used to calculate the risk of breast or ovarian cancer in women and the probability of being a BRCA1/2 mutation carrier based on family history [23]. BOADICEA scores were calculated using the latest version of software, BOADICEA Web Application v3 (available at https://pluto.srl.cam.ac.uk/cgi-bin/bd3/v3/bd.cgi).
4. Statistical analyses
Categorical variables were compared using a χ2 test or Fisher's exact test. Continuous variables were compared using the Mann-Whitney U test. To determine the performance of the mutation carrier prediction algorithms, receiver operating characteristics (ROC) curves were estimated, and the area under the ROC curve and 95% confidence interval (CI) were calculated. SPSS version 20 (IBM Corp., Armonk, NY, USA) was used. A p value of <0.05 was considered statistically significant.
RESULTS
In total, 131 patients were enrolled. All patients were of Korean ethnicity, and the mean age at diagnosis was 52 years. Of the 131 patients, 126 had ovarian cancer and 5 had primary peritoneal or fallopian tube cancer. Five patients had a personal history of breast cancer, and 15 patients had close blood relatives (including first-, second-, or third-degree relatives) with BRCA-associated cancer (breast, ovarian, primary peritoneal, fallopian tube, pancreatic, or prostate cancer). Histology of the tumors revealed that 87 patients (66.4%) had serous type and 44 (33.6%) patients had a non-serous type. Sixty-nine patients (52.7%) had advanced-stage (III, IV) cancer. Ninety-nine patients (75.6%) had a high-grade cancer (Table 1).
Table 1. Baseline characteristics of the patients (n=131).
| Characteristic | Value | |
|---|---|---|
| Age at diagnosis (yr) | 52 (20–74) | |
| Type of cancer | ||
| Ovarian | 126 (96.2) | |
| Primary peritoneal | 2 (1.5) | |
| Primary fallopian tube | 3 (2.3) | |
| Personal history of breast cancer | 5 (3.8) | |
| Family history (1st– 3rd degree relatives) of BRCA-associated cancer* | 15 (11.5) | |
| Tumor histology | ||
| Serous | 87 (66.4) | |
| Clear cell | 18 (13.7) | |
| Mucinous | 9 (6.9) | |
| Endometrioid | 9 (6.9) | |
| Seromucinous | 2 (1.5) | |
| Serous borderline tumor | 1 (0.8) | |
| Squamous cell | 2 (1.5) | |
| Sertoli-Leydig cell | 1 (0.8) | |
| Carcinosarcoma | 1 (0.8) | |
| Large cell neuroendocrine | 1 (0.8) | |
| Stage | ||
| I–II | 62 (47.3) | |
| III–IV | 69 (52.7) | |
| Tumor grade | ||
| 1–2 | 32 (24.4) | |
| 3 | 99 (75.6) | |
Data are described as the mean (range) or number (%).
*BRCA-associated cancers include breast, ovarian, primary peritoneal, fallopian tube, pancreatic, and prostate.
Of the 131 patients, Sanger sequencing identified 10 different BRCA1 small-scale mutations in 12 patients (9.2%) and 6 BRCA2 small-scale mutations in 8 patients (6.1%). Of the 16 BRCA1/2 small-scale mutations, 2 BRCA1 mutations (c.3655G>T and c.4253delT) were novel. One BRCA1 mutation (c.5080G>T, n=3) and 2 BRCA2 mutations (c.1399A>T, n=2; c.7480C>T, n=2) were recurrent (Table 2). One BRCA2 mutation (c.3096_3110delinsT) was found in a patient with primary fallopian tube cancer.
Table 2. Deleterious BRCA1/2 gene mutations found in this study.
| Gene | Exon/intron | Nucleotide change* | Protein change* | Mutation type | NCBI SNP ID number | No. |
|---|---|---|---|---|---|---|
| BRCA1 | Exon6 | c.390C>A | p.Tyr130Ter | Nonsense | rs80356888 | 1 |
| Exon10 | c.1354delG | p.Val452Ter | Nonsense | rs886039946 | 1 | |
| c.1831delC | p.Leu611Ter | Nonsense | rs397508913 | 1 | ||
| c.3296delC | p.Pro1099Leufs | Frameshift | rs80357815 | 1 | ||
| c.3627dupA | p.Glu1210Argfs | Frameshift | rs80357729 | 1 | ||
| c.3655G>T | p.Glu1219Ter | Nonsense | Novel | 1 | ||
| Exon12 | c.4253delT | p.Leu1418Ter | Nonsense | Novel | 1 | |
| c.4327C>T | p.Arg1443Ter | Nonsense | rs41293455 | 1 | ||
| Exon17 | c.5080G>T | p.Glu1694Ter | Nonsense | rs80356896 | 3 | |
| Exon23 | c.5496_5506delinsA | p.Val1833Serfs | Frameshift | rs273902775 | ||
| Exon1–2 | c.(?_-232)_(c.80+1_81-1)del | LGR | - | 1 | ||
| Exon21–23 | c.5332+558_*6809del | LGR | 1 | |||
| BRCA2 | Exon10 | c.1399A>T | p.Lys467Ter | Nonsense | rs80358427 | 2 |
| Exon11 | c.3096_3110delinsT† | p.Lys1032Asnfs | Frameshift | rs397507655 | 1 | |
| c.6553delG | p.Ala2185Leufs | Frameshift | rs80359603 | 1 | ||
| c.7480C>T | p.Arg2494Ter | Nonsense | rs80358972 | 2 | ||
| Exon23 | c.9117G>A | p.Pro3039= | Splice site | rs28897756 | 1 | |
| Exon24 | c.9253delA | p.Thr3085Glnfs | Frameshift | rs397508041 | 1 |
NCBI, National Center for Biotechnology Information; SNP, single nucleotide polymorphism; HGVS, Human Genome Variation Society.
*All variants were described according to the recommended HGVS nomenclature; †This variant was found in a patient with primary fallopian tube cancer.
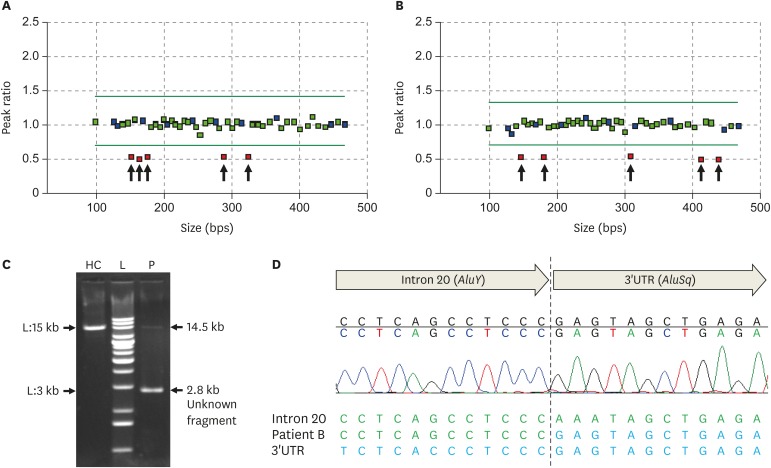
MLPA analysis of BRCA1/2 in Sanger-negative patients revealed 2 LGRs: a deletion of BRCA1 exons 1–2 in patient A (Fig. 1A) and a deletion of BRCA1 exons 21–23 in patient B (Fig. 1B). The long-range PCR in patient B revealed a 2.8-kb-sized unknown fragment with a 14.5-kb-sized normal fragment, showing an approximately 11.7-kb-sized deletion of the BRCA1 gene (Fig. 1C). Finally, the subsequent Sanger sequencing of this unknown fragment revealed the exact breakpoint of the BRCA1 LGR in patient B. The patient had an 11,637-bp-sized deletion that ranged from intron 20 to the 3′ untranslated region (UTR). The LGR in patient B was described as NG_005905.2(NM_007294.3):c.5332+558_*6809del. Because the deletion breakpoint of BRCA1 LGR in patient A was uncharacterized, the description of the LGR was NG_005905.2(NM_007294.3):c.(?_-232)_(c.80+1_81-1)del according to the Human Genome Variation Society nomenclature criteria. This LGR fuses the AluY sequence in intron 20 with the AluSq sequence of the 3′ UTR (Fig. 1D). In total, including the two BRCA1 LGR cases, 22 (16.8%) of the 131 enrolled patients had BRCA1/2 mutations. The LGRs consisted of 14.3% of all identified BRCA1 mutations, and the prevalence of LGRs identified in this study was 1.8% in 111 Sanger-negative patients and 1.5% in all enrolled patients (Table 2).
Fig. 1. MLPA analysis of patient A (A) and B (B). The red squares indicate the deleted BRCA1 probes. (C) Long-range PCR results for patient B showing an 11.7 kb-sized deletion of the BRCA1 gene. (D) Sanger sequencing of the abnormal band revealed the LGR breakpoint: NG_005905.2:g.167479_179115del11637. This LGR fuses the AluY sequence with the AluSq sequence.
MLPA, multiplex ligation-dependent probe amplification; PCR, polymerase chain reaction; LGR, large genomic rearrangement; HC, healthy control; L, DNA extension ladder; P, patient B; UTR, untranslated region.
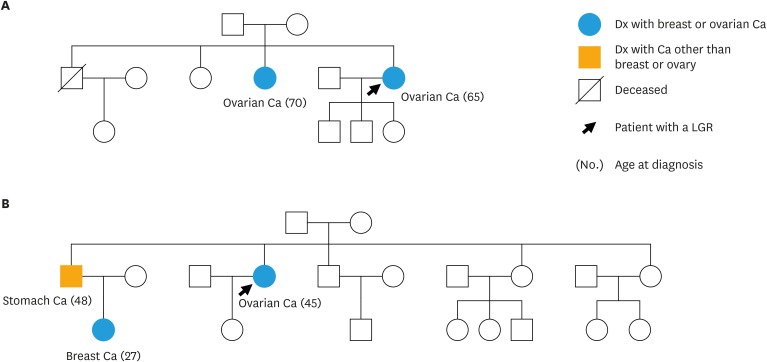
The pedigrees of the 2 patients with LGRs are shown in Fig. 2. Patient A, who was diagnosed with stage III high-grade serous ovarian cancer at age 65, had one first-degree blood relative with ovarian cancer (Fig. 2A). Patient B, who was diagnosed with stage III high-grade serous ovarian cancer at age 45, had one second-degree blood relative with breast cancer and one first-degree blood relative with stomach cancer (Fig. 2B).
Fig. 2. Pedigree of the 2 patients with LGR mutations in this study. (A) Family pedigree of patient A with a deletion of BRCA1 exons 1–2. (B) Family pedigree of patient B with a deletion of BRCA1 exons 21–23.
LGR, large genomic rearrangement; Ca, cancer; Dx, diagnosed.
The characteristics of the patients according to BRCA1/2 mutation status are shown in Table 3. Patients with BRCA1/2 mutations were more likely to have a personal history of breast cancer (p=0.033) and a family history (including third-degree blood relatives) of BRCA-associated cancer (p<0.001). Among the 22 patients with BRCA1/2 mutations, 18 (81.8%) had serous histologic type and two patients (9.1%) had endometrioid histologic type. Two patients (9.0%) with BRCA1/2 mutations had serous borderline tumor and large-cell neuroendocrine carcinoma (LCNC) types. The BRCAPRO, Myriad, and BOADICEA scores were significantly higher among patients with BRCA1/2 mutations (p=0.006, p<0.001, and p=0.040, respectively).
Table 3. Characteristics of the patients according to BRCA1/2 mutation status.
| Characteristic | BRCA mutation positive (n=22) | BRCA mutation negative (n=109) | p value | |
|---|---|---|---|---|
| Age at diagnosis (yr) | 57.1 (41–73) | 51.5 (20–74) | ||
| Type of cancer | ||||
| Ovarian | 21 (95.5) | 105 (96.3) | ||
| Primary peritoneal | 0 | 2 (1.8) | ||
| Primary fallopian tube | 1 (4.5) | 2 (1.8) | ||
| Personal history of breast cancer | 3 (13.6) | 2 (1.8) | 0.033 | |
| Family history (1st–3rd degree relatives) of BRCA-associated cancer | 9 (40.9) | 6 (5.5) | <0.001 | |
| Tumor histology | ||||
| Serous | 18 (81.8) | 69 (63.3) | ||
| Clear cell | 0 | 18 (16.5) | ||
| Mucinous | 0 | 9 (8.3) | ||
| Endometrioid | 2 (9.1) | 7 (6.4) | ||
| Seromucinous | 0 | 2 (1.8) | ||
| Serous borderline tumor | 1 (4.5) | 0 | ||
| Squamous cell | 0 | 2 (1.8) | ||
| Sertoli-Leydig cell | 0 | 1 (0.9) | ||
| Carcinosarcoma | 0 | 1 (0.9) | ||
| Large cell neuroendocrine | 1 (4.5) | 0 | ||
| Stage | 0.509 | |||
| I–II | 9 (40.9) | 53 (48.7) | ||
| III–IV | 13 (59.1) | 56 (51.3) | ||
| Tumor grade | 0.197 | |||
| 1–2 | 3 (13.6) | 29 (26.6) | ||
| 3 | 19 (86.4) | 80 (73.4) | ||
| BRCAPRO score | 0.113 (0.011–0.802) | 0.032 (0.003–0.300) | 0.006 | |
| Myriad score | 0.105 (0.077–0.263) | 0.080 (0.077–0.147) | <0.001 | |
| BOADICEA score | 0.081 (0.005–0.511) | 0.029 (0.002–0.291) | 0.040 | |
Data are described as the mean (range) or number (%).
The estimated ROC curves for all patients are shown in Supplementary Fig. 1. The areas under the ROC curves for predicting the probability of BRCA1/2 mutations were 0.685 (95% CI=0.538–0.831), 0.660 (95% CI=0.518–0.803), and 0.639 (95% CI=0.486–0.793) for the BRCAPRO, Myriad, and BOADICEA scores, respectively. The suggested cutoff point of BRCAPRO by the closest value to the left upper corner was 3.04%, and the sensitivity and specificity values at the cutoff points were 0.636 and 0.789, respectively. The suggested cutoff point of Myriad by the closest value to the left upper corner was 9.9%, and the sensitivity and specificity values at the cutoff point were 0.364 and 0.954, respectively. The suggested cutoff point of BOADICEA by the closest value to the left upper corner was 3.5%, and the sensitivity and specificity values at the cutoff point were 0.545 and 0.807, respectively. Using a 10% cutoff, the traditional cutoff for offering a BRCA1/2 genetic testing, 16 mutations (72.7%) in BRCAPRO, 14 mutations (63.6%) in Myriad, and 16 mutations (72.7%) in BOADICEA would have been missed. Even if we had used each suggested cutoff point from the ROC curves, all of which were lower than 10%, the total number of missed mutations would be 8 (36.4%) using BRCAPRO, 14 (63.6%) using Myriad, and 10 (45.5%) using BOADICEA.
DISCUSSION
Of the 131 unselected Korean ovarian (including primary peritoneal or fallopian tube) cancer patients evaluated in this study, BRCA1/2 mutations, including the two LGRs, were detected in 22 (16.8%). This is a lower mutation prevalence than found by a previous study on Korean patients (24.6%) [14], and it may have resulted from our unselective inclusion of all ovarian cancer patients. The BRCAPRO, Myriad, and BOADICEA scores were significantly higher for patients with BRCA1/2 mutations.
BRCAPRO, Myriad, and BOADICEA are the most widely used models for estimating the likelihood that a BRCA1/2 mutation is present based on family history. BRCAPRO and Myriad models incorporate the effects of breast and ovarian cancer. The BOADICEA model incorporates information on breast, ovarian, pancreatic, and prostate cancer. The BRCAPRO, Myriad, and BOADICEA scores were significantly higher in BRCA1/2 mutation carriers compared to non-carriers; however, many patients classified as low risk by these models were much more likely than predicted to have BRCA1/2 mutations. These findings support a universal testing strategy for all ovarian cancer patients regardless of family history as assessment of family history by validated prediction models cannot effectively target testing to a high-risk ovarian cancer patient population [24].
Interestingly, 2 novel nonsense mutations were detected in 2 patients with a serous borderline tumor and LCNC. Ovarian borderline tumors, otherwise known as ovarian tumors of low malignant potential (LMP), are a histologic subtype of epithelial ovarian cancer. According to a recent report, aggressive tumor characteristics were suggested as a common feature in ovarian LMP tumors with BRCA1/2 mutations [25]. In this study, the patient had a stage III serous borderline tumor and was diagnosed with hepatic metastasis at age 47. This patient died from hepatic failure. Therefore BRCA1/2 testing should be considered in patients with ovarian LMP tumors who exhibit aggressive tumor characteristics. LCNC, which is an aggressive cancer with a tendency to present at advanced stages, has no standard treatment. LCNC is a rare cancer, and only a few cases have been reported in the literature [26]. Only one previous study reported a BRCA2 germline mutation in a patient with LCNC of the ovary [19], and, to the best of our knowledge, this is the first report of a BRCA1 germline mutation carrier with LCNC of the ovary. Therefore, even ovarian cancer patients with non-epithelial histological subtypes, especially LCNC, should be considered for BRCA1/2 genetic testing.
The LGRs consisted of 9.1% of all detected BRCA1/2 mutations, and the prevalence of LGRs identified in this study was 1.8% in 111 Sanger-negative patients and 1.5% in all enrolled patients. The major mechanisms of LGRs found in BRCA1/2 are Alu-mediated unequal homologous recombination and non-homologous events, such as Alu/non-Alu or non-Alu/non-Alu. The breakpoint analysis of LGR in patient B revealed a fusion between the AluY sequence in intron 20 and the AluSq sequence of the 3′ UTR, which is suggestive of an unequal homologous recombination event between these two Alu sequences that share 84% homology. Until recently, only BRCA1 LGR cases had been reported in Korea [15,16,17,27], and our two LGR cases also involved BRCA1. As no LGRs in BRCA2 have been identified, these mutations must be rare in our population. Of the reported BRCA1/2 LGR cases in other populations, BRCA1 LGRs were in the majority [10,28]. BRCA1 contains more Alu sequences than BRCA2, which may explain why more LGRs have been reported in BRCA1 [28].
In populations with a low BRCA1/2 LGR prevalence, LGR screening is rarely performed in routine genetic laboratories. Therefore, patients with BRCA1/2 LGR mutations who may benefit from targeted therapies would be missed by routine genetic testing. For such populations, an effective screening strategy for BRCA1/2 LGR detection is necessary to enable a more efficient and lower-cost mutational screening approach. In Korea, only three ovarian patients with BRCA1 LGRs, including the 2 patients in this study, have been reported (Table 4). All three patients had high-grade serous ovarian cancer, which was diagnosed at ages 45, 35, and 65 years. Each patient had at least one blood relative with breast or ovarian cancer, and this finding is similar to that of a previous large-scale study of BRCA1/2 LGRs [29]. Additionally, in our study, most BRCAPRO, Myriad, and BOADICEA scores were higher in patients carrying BRCA1/2 mutations. These results partially support a previous study that showed a positive correlation between the proportion of mutations due to a LGR and the BRCAPRO score. Therefore, Sanger-negative ovarian cancer patients should be considered for LGR testing if they have one blood relative with breast or ovarian cancer. Additionally, an above-average score on the BRCAPRO, Myriad, or BOADICEA assessments could also be an indication for LGR testing.
Table 4. Characteristics of ovarian cancer patients with BRCA1 LGRs reported in Korea.
| Exon rearrangement | Age at Dx (yr) | FHx of BRCA-associated cancer | Tumor histology | Tumor grade | Stage | BRCAPRO score | Myriad score | BOADICEA score | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Deletion of exon 1–2 | 45 | 1 BC | serous | 3 | III | 0.199 | 0.143 | 0.164 | This study |
| Duplication of exon 4–6 | 35 | 1 BC, 1OC | serous | 3 | III | 0.730 | 0.344 | 0.398 | [17] |
| Deletions of exon 21–23 | 65 | 1OC | serous | 3 | III | 0.089 | 0.147 | 0.024 | This study |
LGR, large genomic rearrangement; Dx, diagnosis; FHx, family history; BC, breast cancer; OC, ovarian cancer.
This study had several limitations. First, as this was a single-institution study performed over 18 months, the number of non-epithelial ovarian cancer or ovarian LMP tumors was small (n=6). Additionally, more detailed data on the characteristics of BRCA1/2 LGR mutations in Korea could be gathered through larger studies. Second, some family histories were taken from medical records, which may have introduced bias. Despite these limitations, the current study identified 2 mutations in patients with a serous borderline tumor and LCNC. We also found two BRCA1 LGRs in patients with a family history of BRCA-associated cancer. These mutations accounted for 9.1% of all detected BRCA1/2 mutations and were found in 1.8% of Sanger-negative patients. A larger multicenter study would improve the statistical significance of research in this area.
In conclusion, 16.8% of unselected ovarian cancer patients had BRCA1/2 mutations detected through a comprehensive BRCA1/2 genetic study. We identified two rare cases of BRCA1 mutations in patients with an ovarian LMP tumor and LCNC. Therefore, patients with ovarian LMP tumors with aggressive characteristics and LCNC of the ovary should be considered for BRCA1/2 genetic testing. We also found 2 cases with BRCA1 LGRs. Based on these results, ovarian cancer patients who are Sanger-negative should be considered for LGR detection if they have one blood relative with breast or ovarian cancer. Additionally, an above-average score on the BRCAPRO, Myriad, or BOADICEA assessments may also indicate the need for LGR testing in Sanger-negative patients. A cost-efficient mutation testing strategy will detect more BRCA1/2 mutations in ovarian cancer patients in populations with a low LGR prevalence. This is essential for the ability to provide targeted therapies to and thereby increase the survival rate of patients with ovarian cancer.
ACKNOWLEDGMENTS
We thank K. S. Park, K. S. Lee, T. Y. Park, S. H. Woo, of Dongsan Medical Center, for their technical assistance with this manuscript.
Footnotes
Funding: This work was supported by the research promoting grant from the Keimyung University Dongsan Medical Center in 2016.
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: K.D.H., H.J.S.
- Data curation: K.D.H., K.S.Y., H.J.S.
- Formal analysis: K.D.H.
- Funding acquisition: H.J.S.
- Investigation: L.W.
- Methodology: K.D.H., H.J.S.
- Project administration: H.J.S.
- Resources: C.C.H., K.S.Y.
- Software: K.D.H.
- Supervision: J.D.S.
- Validation: K.D.H., K.S.Y.
- Visualization: R.N.H.
- Writing - original draft: K.D.H.
- Writing - review & editing: H.J.S.
SUPPLEMENTARY MATERIAL
ROC curve for the BRCAPRO, Myriad, and BOADICEA scores. The areas under the ROC curves were 0.685 (95% CI=0.538–0.831), 0.660 (95% CI=0.518–0.803), and 0.639 (95% CI=0.486–0.793) for the BRCAPRO (blue line), Myriad (green line), and BOADICEA (yellow line) scores, respectively. ROC, receiver operating characteristics; CI, confidence interval.
References
- 1.Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2014. Cancer Res Treat. 2017;49:292–305. doi: 10.4143/crt.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SI, Lee JW, Lee M, Kim HS, Chung HH, Kim JW, et al. Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecol Oncol. 2018;148:375–382. doi: 10.1016/j.ygyno.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Lee YJ, Lee SW, Kim KR, Jung KH, Lee JW, Kim YM. Pathologic findings at risk-reducing salpingo-oophorectomy (RRSO) in germline BRCA mutation carriers with breast cancer: significance of bilateral RRSO at the optimal age in germline BRCA mutation carriers. J Gynecol Oncol. 2017;28:e3. doi: 10.3802/jgo.2017.28.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 5.Lee J, Kim S, Kang E, Park S, Kim Z, Lee MH, et al. Influence of the Angelina Jolie announcement and insurance reimbursement on practice patterns for hereditary breast cancer. J Breast Cancer. 2017;20:203–207. doi: 10.4048/jbc.2017.20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewald IP, Ribeiro PL, Palmero EI, Cossio SL, Giugliani R, Ashton-Prolla P. Genomic rearrangements in BRCA1 and BRCA2: a literature review. Genet Mol Biol. 2009;32:437–446. doi: 10.1590/S1415-47572009005000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodward AM, Davis TA, Silva AG, Kirk JA, Leary JA kConFab Investigators. Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet. 2005;42:e31. doi: 10.1136/jmg.2004.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 9.Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, et al. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003;12:1055–1061. doi: 10.1093/hmg/ddg120. [DOI] [PubMed] [Google Scholar]
- 10.Hansen T, Jønson L, Albrechtsen A, Andersen MK, Ejlertsen B, Nielsen FC. Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat. 2009;115:315–323. doi: 10.1007/s10549-008-0088-0. [DOI] [PubMed] [Google Scholar]
- 11.Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M, et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol. 2007;25:2027–2034. doi: 10.1200/JCO.2006.06.9443. [DOI] [PubMed] [Google Scholar]
- 12.Moisan AM, Fortin J, Dumont M, Samson C, Bessette P, Chiquette J, et al. No evidence of BRCA1/2 genomic rearrangements in high-risk French-Canadian breast/ovarian cancer families. Genet Test. 2006;10:104–115. doi: 10.1089/gte.2006.10.104. [DOI] [PubMed] [Google Scholar]
- 13.De Silva S, Tennekoon KH, Karunanayake EH, Amarasinghe I, Angunawela P. Analysis of BRCA1 and BRCA2 large genomic rearrangements in Sri Lankan familial breast cancer patients and at risk individuals. BMC Res Notes. 2014;7:344. doi: 10.1186/1756-0500-7-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eoh KJ, Park JS, Park HS, Lee ST, Han J, Lee JY, et al. BRCA1 and BRCA2 mutation predictions using the BRCAPRO and Myriad models in Korean ovarian cancer patients. Gynecol Oncol. 2017;145:137–141. doi: 10.1016/j.ygyno.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Kim DH, Chae H, Jo I, Yoo J, Lee H, Jang W, et al. Identification of large genomic rearrangement of BRCA1/2 in high risk patients in Korea. BMC Med Genet. 2017;18:38. doi: 10.1186/s12881-017-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seong MW, Cho SI, Kim KH, Chung IY, Kang E, Lee JW, et al. A multi-institutional study of the prevalence of BRCA1 and BRCA2 large genomic rearrangements in familial breast cancer patients. BMC Cancer. 2014;14:645. doi: 10.1186/1471-2407-14-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JY, Cho DY, Ahn SH, Choi SY, Shin I, Park HG, et al. Large genomic rearrangement of BRCA1 and BRCA2 genes in familial breast cancer patients in Korea. Fam Cancer. 2014;13:205–211. doi: 10.1007/s10689-014-9704-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Royer R, Li S, McLaughlin JR, Rosen B, Risch HA, et al. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol Oncol. 2011;121:353–357. doi: 10.1016/j.ygyno.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Herold N, Wappenschmidt B, Markiefka B, Keupp K, Kröber S, Hahnen E, et al. Non-small cell neuroendocrine carcinoma of the ovary in a BRCA2-germline mutation carrier: a case report and brief review of the literature. Oncol Lett. 2018;15:4093–4096. doi: 10.3892/ol.2018.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewald IP, Cossio SL, Palmero EI, Pinheiro M, Nascimento IL, Machado TM, et al. BRCA1 and BRCA2 rearrangements in Brazilian individuals with hereditary breast and ovarian cancer syndrome. Genet Mol Biol. 2016;39:223–231. doi: 10.1590/1678-4685-GMB-2014-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 23.Lee AJ, Cunningham AP, Kuchenbaecker KB, Mavaddat N, Easton DF, Antoniou AC, et al. BOADICEA breast cancer risk prediction model: updates to cancer incidences, tumour pathology and web interface. Br J Cancer. 2014;110:535–545. doi: 10.1038/bjc.2013.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniels MS, Babb SA, King RH, Urbauer DL, Batte BA, Brandt AC, et al. Underestimation of risk of a BRCA1 or BRCA2 mutation in women with high-grade serous ovarian cancer by BRCAPRO: a multi-institution study. J Clin Oncol. 2014;32:1249–1255. doi: 10.1200/JCO.2013.50.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo K, Tierney KE, Schneider DM, Mhawech-Fauceglia P, Roman LD, Gershenson DM. Characteristics of ovarian tumors of low malignant potential in BRCA mutation carriers: a case series. Gynecol Oncol Rep. 2015;13:36–39. doi: 10.1016/j.gore.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YS, Im KS, Choi DI. Ovarian large cell neuroendocrine carcinoma in the youngest woman. Eur J Gynaecol Oncol. 2016;37:244–246. [PubMed] [Google Scholar]
- 27.Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer (KOHBRA) study. Breast Cancer Res Treat. 2015;151:157–168. doi: 10.1007/s10549-015-3377-4. [DOI] [PubMed] [Google Scholar]
- 28.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125:325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 29.Engert S, Wappenschmidt B, Betz B, Kast K, Kutsche M, Hellebrand H, et al. MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat. 2008;29:948–958. doi: 10.1002/humu.20723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ROC curve for the BRCAPRO, Myriad, and BOADICEA scores. The areas under the ROC curves were 0.685 (95% CI=0.538–0.831), 0.660 (95% CI=0.518–0.803), and 0.639 (95% CI=0.486–0.793) for the BRCAPRO (blue line), Myriad (green line), and BOADICEA (yellow line) scores, respectively. ROC, receiver operating characteristics; CI, confidence interval.