Autophagy is an evolutionary conserved, degradative process from single-cell eukaryotes, such as Saccharomyces cerevisiae, to higher mammals, such as humans. The regulation of autophagy has been elucidated through the combined study of yeast, Caenorhabditis elegans, mice, Drosophila melanogaster, and humans.
KEYWORDS: AMPK, ATG101, ATG13, autophagy, beclin 1, LC3, MTOR, RB1CC1, ULK1, ULK2
ABSTRACT
Autophagy is an evolutionary conserved, degradative process from single-cell eukaryotes, such as Saccharomyces cerevisiae, to higher mammals, such as humans. The regulation of autophagy has been elucidated through the combined study of yeast, Caenorhabditis elegans, mice, Drosophila melanogaster, and humans. MTOR, the major negative regulator of autophagy, and activating nutrient kinases, such as 5′-AMP-activated protein kinase (AMPK), interact with the autophagy regulatory complex: ULK1/2, RB1CC1, ATG13, and ATG101. The ULK1/2 complex induces autophagy by phosphorylating downstream autophagy complexes, such as the BECN1 PIK3 signaling complex that leads to the creation of LC3+ autophagosomes. We highlight in this review various reports of autophagy induction that are independent of these regulators. We discuss reports of MTOR-independent, AMPK-independent, ULK1/2-independent, and BECN1-PIK3C3-independent autophagy. We illustrate that autophagy induction and the components required vary by the nature of the induction signal and type of cell and do not always require canonical members of the autophagy signaling pathway. We illustrate that rather than thinking of autophagy as a linear pathway, it is better to think of autophagy induction as an interconnecting web of key regulators, many of which can induce autophagy through different requirements depending on the type and length of induction signals.
INTRODUCTION
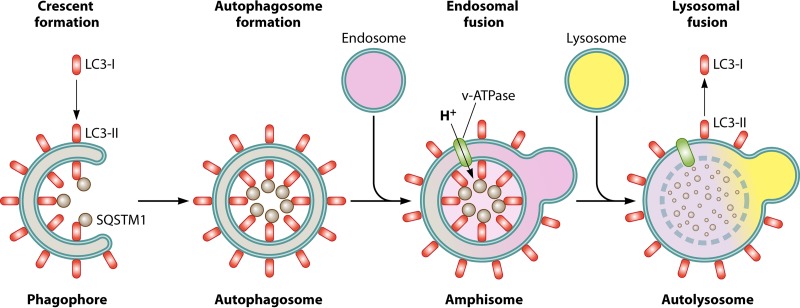
Autophagy is a degradative cellular process that has been characterized since the early 1970s (1). The word autophagosome was coined by Christian de Duve and comes from the Greek words autos, meaning “self,” phagos, meaning “to eat,” and soma, meaning “body” (1). Autophagy can occur in several forms, depending on the induction signal and type of cargo, such as macroautophagy, mitophagy, autophagy of the endoplasmic reticulum (ER-phagy), and lipophagy. Macroautophagy (here referred to as autophagy) is characterized as the bulk degradative process that primarily degrades nonspecific cytosolic cargo. Autophagic induction directs formation of characteristic double-membrane vesicles (autophagosomes) that engulf cytosolic cargo (2). Autophagy occurs in several key steps, starting with the formation of the phagophore/omegasome upon autophagy induction. Next, the phagophore membrane expands, leading to the engulfment of cytosolic cargo, as well as membrane fusion, leading to autophagosome formation. The autophagosome is acidified by fusion with an acidic late endosome, leading to an amphisome. Final degradation of cargo occurs by fusion of an autophagosome or amphisome with the lysosome, leading to the formation of the autolysosome (Fig. 1).
FIG 1.
The major steps in autophagy. Upon signaling induction of autophagy, the preautophagosomal membrane known as the phagophore forms. The LC3 ubiqutin-like lipidation complex takes cytosolic LC3-I and conjugates it to a phosphoethanolamine to generate membrane-associated LC3-II. Autophagy cargo receptors such as SQSTM1 bring cargo into the forming phagophore and bind to LC3 via the LC3 interaction region (LIR) domain. Eventual expansion of the double membrane around cytosolic cargo forms the characteristic double-membrane vesicle known as the autophagosome. Through SNARE-mediated fusion, the autophagosome fuses with an endosome that contains vacuolar ATPases (v-ATPase). This leads to acidification of the autophagosome, forming the amphisome. Upon acidification, the amphisome can then fuse with a lysosome to bring in the degradative enzymes that degrade the cargo, including LC3 and SQSTM1. This forms the autolysosome. LC3-II can be converted back into LC3-I if located on the exterior of the autolysosome.
Autophagy is a basal homeostatic process that can be upregulated during times of stress, such as nutrient starvation or pathogen infection (2). Autophagy also plays a role in several different types of cancer. It can serve as an inhibitory process, through elimination of reactive oxygen species (ROS) generation due to damaged organelles, and prevent cancer formation. Autophagy can also promote cancer progression by providing nutrients to tumors that are in nutrient-poor conditions. Therefore, the impact of autophagy is highly dependent on the cancer model being studied (3).
Autophagy is evolutionarily conserved from single-cell eukaryotes, such as Saccharomyces cerevisiae, to multicellular organisms, such as plants, fungi, and animals (2). Microtubule-associated protein 1 light chain 3 beta (MAP1LC3B/LC3B/LC3-I) is a protein that localizes to both the nascent phagophore and autophagosome membranes upon lipidation with phosphoethanolamine via a ubiquitin-like ligation complex. Recent studies, though, have found that LC3 may have an additional nonautophagy role in phagocytosis, known as LC3-associated phagocytosis (LAP). This process has been characterized to be generally independent of most of the regulatory proteins discussed here (4).
The lipidation-dependent band shift of LC3 on a Western blot is widely used as a marker of autophagy induction (5). Fluorescently tagged LC3 can be used to monitor autophagy using immunofluorescence microscopy through quantification of cytosolic puncta (5, 6). Unfortunately, due to the recycling of LC3-II to cytosolic LC3-I during autophagy (Fig. 1), an increase in LC3-II levels can result from either induction or blockage of degradation, making interpretation of LC3 data difficult. An LC3 flux assay was developed that inhibits lysosomal proteases and allows for a measure of induction, and measuring steady-state levels of autophagic cargo receptors gives insight into active degradation (5, 7). Two early autophagosome markers, WD repeat domain, phosphoinositide interacting 1/2 (WIPI1/2), can be used to monitor autophagy induction via quantification of puncta (5, 8). The combined use of electron microscopy and a proper autophagy flux assay can also be utilized as a measure of autophagy induction. For a more in-depth review on autophagy assays and their interpretation, please refer to the work of Klionsky et al. (5).
S. cerevisiae encodes a master regulator protein known as target of rapamycin (Tor) that negatively regulates autophagy but also regulates many cellular process (9, 10). In an effort to find autophagy-specific regulators, large-scale genetic screens were conducted which led to the discovery of several vital genes that directly impact autophagy (11–15). One of the first genes identified, the autophagy related 1 gene (ATG1), has a serine/threonine kinase domain (11). ATG1 also contains regulatory domains that either enhance binding with yeast scaffold protein autophagy related 13 (Atg13p) or inhibit kinase activity through Tor (13, 15). For the first time since the discovery of autophagy, the field had insight into potential signaling regulators.
The link between yeast to a multicellular organism was found when searching for ATG1 homologs in Caenorhabiditis elegans. ATG1 has significant homology to the C. elegans gene uncoordinated 51 (unc-51), which also impacts autophagy. Furthermore, after genomic comparisons, Iarger eukaryotes were found to have a family of unc-51-like kinases (ULKs) that shared homology to unc-51 (1, 16–19). Of these kinases, ULK1 and ULK2 have been characterized as autophagy-specific regulators with at least partially overlapping autophagy functions (1, 20). Since then, the field has extensively characterized the signaling transduction pathway involved in the generation of LC3+ autophagosomes, though there has been evidence that LC3 lipidation is not an absolute requirement for autophagosome formation (21, 22). Our focus is on complexes downstream of mammalian mechanistic target of rapamycin (MTOR), the mammalian homolog of yeast Tor (Fig. 2). We begin with the ULK1 complex composed of ULK1, autophagy-related 13 (ATG13), RB1-inducible coiled coil 1 (RB1CC1), and autophagy-related 101 (ATG101) (23–28). While it is widely accepted that ULK1/2 are the primary upstream autophagy-specific kinases, there have been reports of autophagy induction that is not dependent on the MTOR-ULK1/2 signaling axis. This review highlights that while ULK1/2 play pivotal roles in autophagy and in development, they are not the only kinases that induce bona fide (degradative) autophagy.
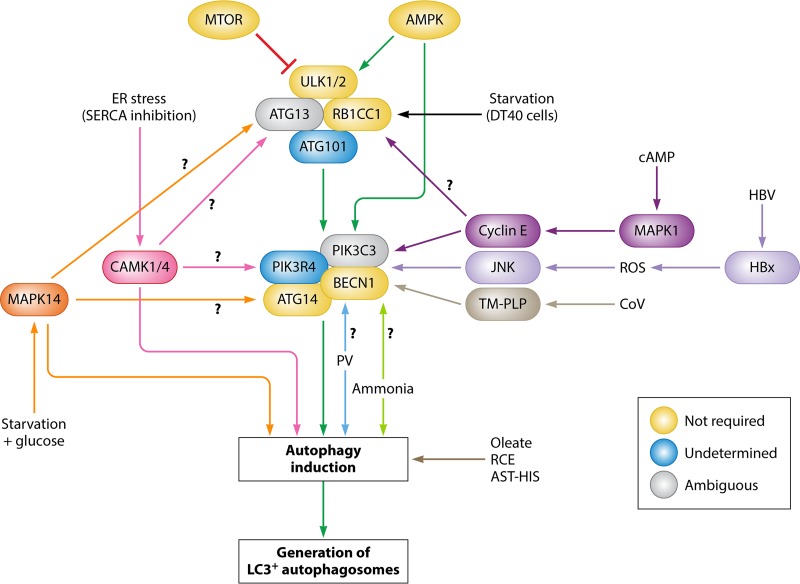
FIG 2.
Map of autophagy induction. The canonical autophagy pathway (green arrows) is depicted down the center, where MTOR negatively regulates and AMPK positively regulates the ULK1/2 complex. These then activate the downstream BECN1 complex, leading to autophagy induction and subsequently the generation of LC3+ autophagosomes. Additionally, AMPK has been shown to directly phosphorylate BECN1. In yellow are complex members that have been shown to be dispensable for autophagy induction. In blue are complex members whose requirement status has not been determined. In gray are complex members whose role is ambiguous, with conflicting reports of induction requirement. ER stress-induced autophagy through SERCA inhibition (pink arrows) activates CAMK1/4, which then induces autophagy but not degradative autophagy. The roles of the ULK1/2 and BECN1 complexes have not been determined. Glucose addition during starvation (orange arrows) activates MAPK14, which induces autophagy, but the roles of the ULK1/2 and BECN1 complexes remain unknown. Starvation in DT40 cells (black arrow) requires RB1CC1 and ATG13 but does not require ULK1/2. Cyclic AMP (dark purple arrows) activates MAPK1 and cyclin E and induces autophagy through BECN1, although it is not certain if this pathway also activates the ULK1/2 complex. Hepatitis B virus X protein (HBx) induces ROS (light purple arrows) which activate JNK signaling and induce incomplete autophagy via BECN1. Human coronavirus transmembrane papain-like proteases (PLPs) interact with BECN1 (gray arrows) to induce incomplete autophagy. Ammonia (lime green arrows) induces autophagy in a ULK1/2-independent manner, although the role of the BECN1 complex was not assessed. Poliovirus (PV) induces autophagic signals (blue arrows) independently of the ULK1/2 complex, although the role of the BECN1 complex has not been assessed. Oleate, Rhus coriaria extract (RCE), and astemizole-histamine (AST-HIS) induce autophagy (brown arrow) in a BECN1-independent manner.
CANONICAL AUTOPHAGY INDUCTION THROUGH MTOR-ULK1/2
ULK1- and ULK2-containing complexes regulate autophagy.
Atg1p forms a complex with Atg13p upon conditions requiring upregulation of autophagy for survival (15). Atg1p and Atg13p form a complex only when Atg1p is active (15). The mammalian homologs ULK1 and ULK2 also form complexes, but unlike Atg1p-Atg13p, the complexes persist under homeostatic (i.e., nutrient-rich) conditions (23, 24, 26, 29). The first ULK1/2 complex member was identified in mice, and then humans, as RB1CC1, originally known as FAK-interacting protein of 200 kDa (FIP200) (25). RB1CC1 coimmunoprecipitates with ULK1 or ULK2 independently of kinase activity, and the C-terminal domain (CTD) of ULK1 is required for interaction with RB1CC1. RB1CC1 colocalizes with ATG5, a known member of the vital LC3 lipidation complex, suggesting a link between the regulatory ULK1/2 complex and downstream autophagy machinery. Loss of RB1CC1 inhibits autophagy in mouse embryonic fibroblasts (MEFs), due to ULK1 instability (25).
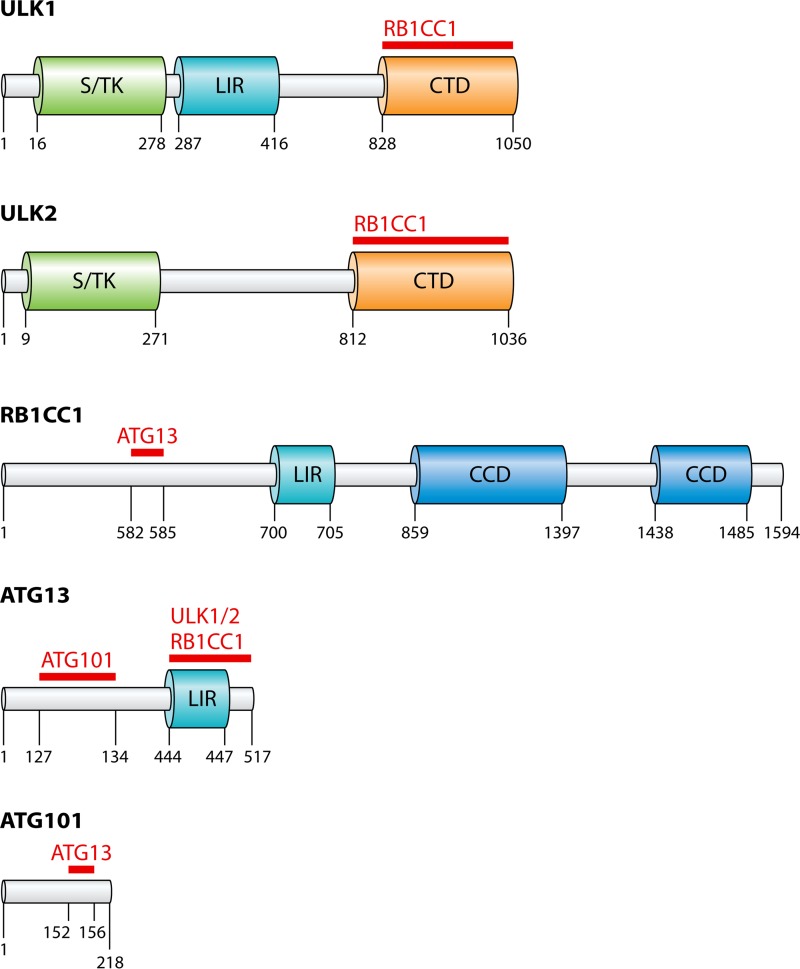
Searches for additional ULK1-interacting partners identified a mammalian gene that shares weak homology with yeast and C. elegans Atg13 (23, 28, 29). Mammalian ATG13 interacts with ULK1, ULK2, and RB1CC1 via the C-terminal domain and localizes to phagophore membranes upon autophagy induction, enabling stabilization of ULK1-RB1CC1 interactions (23, 28–30). Intriguingly, loss of RB1CC1 does not impact ULK1-ATG13 binding, suggesting that ULK1-ATG13 association precedes RB1CC1 binding (22, 27, 28). ATG13 is a vital member of the complex, as deletion or RNA interference (RNAi)-mediated knockdown disrupts autophagy induction and interrupts ULK1 localization to phagophore membranes (23, 28, 29). ULK1 and MTOR regulate ATG13 through hyperphosphorylation, which regulates the ability of ATG13 to promote autophagy. Finally, there is a single unique complex member in larger eukaryotes that is not conserved in yeast. Autophagy-related 101 (ATG101) was found not only to interact with the ULK1 complex but also to be conserved in C. elegans, D. melanogaster, Mus musculus, and Homo sapiens (26). ATG101 interacts with ATG13 and RB1CC1, and the loss of ATG101 inhibits autophagy induction due to instability of the complex, indicating a chaperone-like role for the protein (26, 31). ULK1, ATG13, and RB1CC1 have LC3 interaction regions (LIR) that may play a role in facilitating complex assembly (32–34) (Fig. 3).
FIG 3.
The major domains of the ULK complex. The numbers refer to the amino acid sequence of the protein. The red lines and protein names refer to known sites of interaction with the corresponding protein. ULK1 and ULK2 contain conserved serine/threonine kinase (S/TK) domains and conserved C-terminal domains (CTD) that are crucial for binding with RB1CC1. ULK1, RB1CC1, and ATG13 have been characterized to contain LC3 interaction regions (LIR) that enable binding to LC3 and the GABARAP family members. RB1CC1 contains two coiled-coil domains (CCD) which may play a role in complex formation as well as non-autophagy-related functions. ATG13 and ATG101 interact via the HORMA domain.
Nutrient-sensing kinases MTOR and AMPK regulate the ULK1 complex.
After the discovery of ULK1 and ULK2, MTOR was shown to phosphorylate the kinases to prevent autophagy induction under nutrient-rich conditions (27). MTOR forms a complex, known as MTORC1, composed of regulatory-associated protein of MTOR complex 1 (RPTOR), MTOR-associated protein LST8 (MLST8), AKT1 substrate 1 (AKT1S1), and DEP domain containing MTOR-interacting protein (DEPTOR) (35–37). MTORC1 directly phosphorylates ULK1 (27). Another major nutrient-sensing regulatory kinase, which is known to directly phosphorylate ULK1/2 during times of glucose starvation, is 5′-AMP-activated protein kinase (AMPK) (27, 38). AMPK has been characterized to have multiple functions aside from the general regulation of cellular metabolism, including, but not limited to, diabetes and β cell biology, regulation of the NLRP3 inflammasome during aging, and mitochonrdrial homeostasis (39–41). AMPK activates ULK1 by phosphorylating serine 317 and 777 (27, 38). ULK1 phosphorylation at serine 757 by MTORC1 disrupts ULK1-AMPK interactions (27). MTORC1 phosphorylates ATG13 at serine 258 to further inhibit ULK1/2 (42). AMPK activates chaperones that inhibit MTORC1, as well as directly inactivating MTOR. Phosphorylation of ATG13 at serine 224 by AMPK inhibits starvation-induced autophagy (42). ULK1 and ULK2 phosphorylate both MTOR and AMPK; consequences include a negative-feedback loop with AMPK, phosphorylating and inhibiting AMPK activity (43, 44). ULK1 phosphorylates RPTOR at multiple sites and interferes with MTORC1 ATP loading, inactivating MTOR activity under nutrient starvation conditions (43).
ULK1 regulates the activity of the phosphatidylinositol 3-kinase complex composed of BECN1, PIK3C3, ATG14, and PIK3R4.
The major downstream target of the ULK1/2 complex is the beclin 1 (BECN1) complex, composed of BECN1, autophagy-related 14 (ATG14), phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3), and phosphatidylinositol 3-kinase regulatory subunit 4 (PIK3R4) (Fig. 2, green arrows) (2). PIK3C3 exists in multiple complexes under homeostatic conditions, with many of those complexes having nonautophagic functions (32–36). PIK3C3 activity is regulated by the interactions with the other complex members (32–36). One of these complexes, the BECN1 complex, promotes the generation of phosphatidylinositol 3-phosphate (PI3-P) lipids that are vital to the formation of the initial phagophore structure (2). ULK1 phosphorylates BECN1 at serines 15 and 30 and causes dissociation of BECN1 from the apoptosis regulatory protein BCL2 (45–47). Additionally, ULK1 phosphorylates ATG14 at serine 29, which enhances ATG14:BECN1 binding and promotes the formation of the autophagy PIK3C3 complexes (47). Upon generation of PI3-P lipids, WIPI2 binds and recruits the LC3 conjugation complex: autophagy-related 5 (ATG5), autophagy-related 12 (ATG12), and autophagy-related 16 (ATG16) (48). LC3-I is converted to membrane-bound LC3-II, and the phagophore begins expansion into a closed autophagosome.
MTOR- AND AMPK-INDEPENDENT AUTOPHAGY INDUCTION
MTOR-independent autophagy induction.
There are several reports of autophagy induction that does not require MTOR inhibition or in which autophagy is induced despite MTOR activity. When J16 Jurkat human T lymphocytes or chicken DT40 B lymphocyte cells are treated with thapsigargin (TG), a sarcoplasmic ER Ca2+ ATPase (SERCA) pump inhibitor, autophagic signals (i.e., LC3 lipidation, formation of LC3 puncta, and formation of WIPI1 puncta) are detected after 1 h (49). Addition of a divalent cation chelator inhibits the formation of LC3 and WIPI1 puncta, suggesting that ER Ca2+ influx induces autophagy. Intriguingly, phosphorylation levels of p70S6K, an MTOR target, are slightly decreased in TG-treated cells but not to the same extent as cells treated with rapamycin, an MTOR inhibitor, suggesting MTOR-independent induction (49). TG treatment triggers the unfolded protein response in the ER, inducing what has been referred to as ER-phagy (50, 51). It was not until after publication of reference 49 that TG was found to be a potent inhibitor of autophagosome-lysosome fusion and therefore not an inducer of bona fide autophagy (49, 52). TG is hypothesized to work as an inhibitor of flux by inhibiting SERCA, which was shown to be vital for the fusion of the autophagosome to the lysosome (53). This new information, coupled with the results obtained with J16 Jurkat cells and DT40 cells, suggests that TG promotes upstream autophagy induction signals but does not induce degradative autophagy, mimicking findings for various viral and bacterial infections.
NIH 3T3 cells starved by deprivation of amino acids, glucose, and serum for 2 to 4 h respond to addition of glucose by inducing autophagic signals, as observed by monitoring LC3 lipidation and the use of LC3 flux assays, that are MTOR independent (54). Phosphorylation of 4E-BP1, an MTOR target, increases upon glucose addition under starvation conditions, but other targets, such as p70S6K and ULK1, are not phosphorylated, suggesting target-specific inhibition of MTORC1. Pretreatment of starved cells with rapamycin fails to increase LC3-II levels in the presence or absence of glucose addition, suggesting that glucose addition induces MTOR-independent autophagy. Importantly, inhibition of glycolysis, via sodium oxamate, blocks glucose-induced autophagy in starved cells (54). These reports suggest that in the case of ER stress or glucose influx, autophagy is induced in an MTOR-independent mechanism.
Autophagy is induced not only during times of starvation but also during times of cellular stress. Mouse oocytes induce bona fide autophagy upon fertilization (55). Additionally, treatment of oocytes with torin and PP242, which are MTOR inhibitors, fails to induce autophagy in these cells. Fertilized oocytes that are treated with cycloheximide, which activates MTOR, continue autophagy progression (55, 56). The authors determined that PIK3 signaling is required for fertilization-induced autophagy (55). Another group uncovered a different MTOR-independent autophagy induction mechanism in nucleus pulposus (NP) cells under hypoxic conditions (57). The authors demonstrated that hypoxia triggers autophagy induction but not degradative autophagy: LC3 puncta form and LC3-II levels increase, but autophagic degradation does not increase, as measured by a lack of change in protein level of autophagic cargo receptor sequestesome 1 (SQSTM1) (57). Furthermore, levels of the LC3 lipidation machinery do not change, nor are there any changes seen in the phosphorylation states of ULK1, suggesting no MTOR inhibition. NP cells, placed under hypoxic conditions, do not respond further to rapamycin treatment, suggesting MTOR-independent autophagy induction. Additionally, NP cells under basal conditions treated with torin fail to induce autophagy (57). Could AMPK be responsible for some of these data? MTOR-independent autophagy during fertilization of mouse oocytes may be explained by altered AMPK signaling, as the study did not investigate the role of AMPK. However, AMPK activation was not seen during hypoxia (57). In summary, MTOR signaling can be bypassed under conditions that are not directly related to starvation or metabolite addition. In the case of TG, glucose, and hypoxia, altered AMPK signaling was not observed, as discussed in the following section.
AMPK-independent autophagy induction.
The role of AMPK regulation of autophagy has been controversial. AMPK has been shown to both induce autophagy under low-glucose conditions and inhibit autophagy under other conditions, such as amino acid starvation (27, 38, 58). AMPKα1/AMPKα2 knockout MEFs are still able to induce autophagy when treated with TG (49). Another group found similar results: U2OS cells expressing a stable green fluorescent protein (GFP)-WIPI1 were screened for WIPI1 puncta and LC3 puncta as markers of autophagic induction against a library of chemicals known to disrupt calcium transport (59). Inhibitors of calcium transport induced autophagic signals in both studies, while calcium chelators inhibited autophagic signals. This held true in AMPKα1/AMPKα2 knockout MEFs, despite a decrease in numbers of basal WIPI1 puncta (59). As mentioned above, TG does not induce bona fide autophagy; therefore these results suggest that TG-induced ER stress causes the formation of autophagosomes in an AMPK-independent manner but that ultimately these vesicles are unable to fuse with lysosomes and proceed to degradation.
In regard to glucose starvation, AMPKα1/AMPKα2 knockout MEFs induce autophagic signals, as measured by LC3 lipidation, LC3 flux assays, and quantification of LC3 puncta, under low-glucose conditions before inducing apoptosis (60). This type of autophagy induction was shown to be sensitive to 3-methyladenine (3-MA) treatment, which inhibits PIK3 signaling, autophagy, and apoptosis (60). Starved NIH 3T3 cells that were fed glucose subsequently induce autophagic signals without inducing the phosphorylation of AMPKα2 threonine 172, a known activating event. Additionally, under these conditions phosphorylation of acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), an AMPK target, is diminished (54). A similar trend is observed in NP cells under hypoxic conditions, in which phosphorylation of ULK1 serine 777, a target of AMPK, does not change despite autophagy induction (57). A recent study investigating the role of AMPK in either amino acid or glucose starvation in multiple cell lines found that amino acid starvation, but not glucose starvation, induces bona fide autophagy (58). AMPK, while active under glucose starvation, does not significantly increase LC3-II levels or LC3 punctum formation. Furthermore, AMPK inhibits ULK1 by phosphorylating ULK1 at serine 555 under starvation conditions, as well as inhibiting lysosome activity and autophagy flux (58). In conclusion, it appears that AMPK is not an absolute requirement for autophagy induction in response to ER stress or glucose addition, suggesting that the role of autophagy induction of AMPK may not be as all-encompassing as once thought.
Autophagy induction through CAMK1 (CaMKI), MAPK1 (ERK)-cyclin E-BECN1, and MAPK14 (p38/MAPK) signaling.
As discussed previously, ER stress through the use of TG induces autophagic signals that are both MTOR and AMPK independent (Fig. 2, pink arrows). The literature provides clues to the mechanism of this induction. RNAi-mediated knockdown of calcium/calmodulin-dependent protein kinase kinase 1/2 (CAMKK1/2), calcium/calmodulin-dependent protein kinase 1 (CAMK1), or CAMK1/4, prevents induction of autophagic signals by TG and other calcium transporter inhibitors, suggesting a role for the CAMK pathway (49, 59). Mesenchymal stem cells (MSCs) are known to utilize cyclic AMP as differentiation signals (61). Treatment with prostaglandin E2 (PGE2), which increases intracellular cAMP levels, induces autophagic signals as measured by LC3 lipidation, LC3 flux assays, and LC3 punctum quantification (62). Addition of the cAMP analog 8-CPT provides the same result. Addition of 3-MA to PGE2-treated MSCs inhibits autophagic signals, suggesting a role for PIK3 signaling (Fig. 2, dark purple arrows). Additionally, knockdown of BECN1, cyclin E, or mitogen-activated protein kinase 1 (MAPK1)/extracellular signal-regulated kinase (ERK) inhibits cAMP-induced autophagic signals, suggesting a role for MAPK1-cyclin E signaling inducing autophagy through interactions with BECN1 (62). The PIK3 inhibitor LY294002 abolished autophagy in starved NIH 3T3 cells that were fed glucose. This indicates that glucose induction of autophagy in this model is dependent on PIK3 signaling (54). Addition of glucose increases the phosphorylation of mitogen-activated protein kinase 14 (MAPK14/p38), cAMP-responsive element binding protein 1 (CREB1), and activating transcription factor 2 (ATF2). This phosphorylation was not dependent on mitogen-activated protein kinase kinase 3/6 (MAP2K3/MAP2K6), the kinases known to phosphorylate MAPK14. Inhibition of MAPK14 signaling inhibits autophagic signaling, suggesting a role for MAPK14 in autophagy induction (54) (Fig. 2, orange arrows). These three studies did not investigate the role of the ULK1/2 complex, leaving the possibility that CAMK1, MAPK1, and MAPK14 interact with the ULK1/2 complex to induce autophagy. In summary, while MTOR is the master negative regulator of autophagy, it is evident that multiple upstream kinases can initiate autophagic induction, depending on the type of induction signal.
ULK1/2-INDEPENDENT AUTOPHAGY INDUCTION
ULK1/2-independent autophagy induction under nutrient starvation.
The current understanding of autophagy regulation places ULK1/2 as the most upstream, autophagy-specific kinases; as such, they serve as the nexus for signal inputs in the cell. However, an increasing number of reports have illustrated that while ULK1/2 may induce autophagy, they are not absolute requirements.
Amino acid starvation fails to induce autophagy in ULK1/2 knockout MEFs, but glucose starvation still induces autophagy in these cells (Fig. 2, lime green arrows). This suggests that autophagy induction by glucose starvation is ULK1/2 independent (63). Ammonia metabolism was shown to be important in this autophagy induction pathway: glucose starvation increases intracellular levels of ammonia in MEFs, while treatment with methyl pyruvate, which eliminates ammonia, results in an inhibition of autophagy induction (63). Additionally, p70S6K phosphorylation levels do not change after addition of ammonia, suggesting active MTOR signaling during autophagy induction (63).
Another study reported that ULK1/2 is dispensable for starvation-induced autophagy in chicken DT40 cells cultured in Earle's balanced salt solution (EBSS) for 2 h (64). Loss of ATG13 inhibits autophagy, and LC3-II levels fail to increase with treatment with bafilomycin A1, a vacuolar-ATPase inhibitor. Knockout of ULK1/2 does not inhibit starvation-induced autophagy. Furthermore, knockdown of ULK3, another homolog of ULK1 and ULK2, fails to inhibit autophagy in DT40 cells (64). Surprisingly, MTOR inhibition through rapamycin or torin fails to induce autophagy under basal conditions in DT40 cells (64). Finally, ULK1/2-independent autophagy induction in DT40 cells requires ATG13-RB1CC1 binding: expression of ATG13 splice variants that lack RB1CC1 binding domains in ATG13KO DT40 cells fails to rescue autophagy induction (64) (Fig. 2, black arrow). U2OS cells or mice treated with palmitate or oleate fatty acids induce autophagy in vitro and in vivo (65). Surprisingly, ULK1 and ULK2 knockdown affects palmitate induction of autophagy but not oleate, suggesting that oleate induces ULK1/2-independent autophagy (65) (Fig. 2, brown arrow).
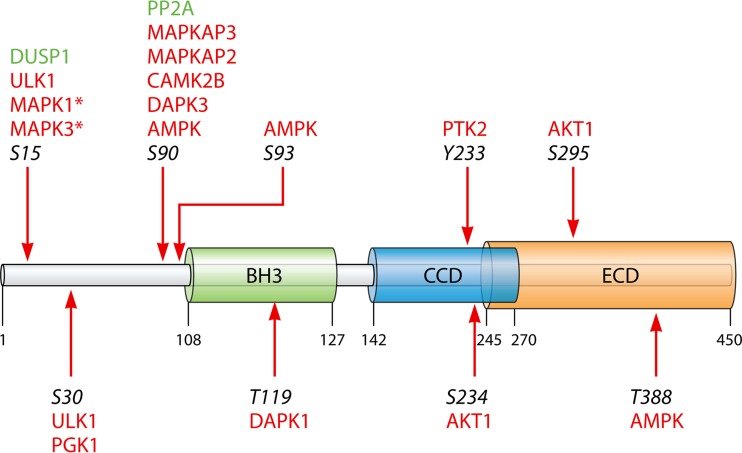
One possible mechanism of ULK1/2 bypass would require direct interaction with downstream autophagy complexes, such as the BECN1 complex. AMPK regulates PIK3C3 complexes during nutrient starvation (66). Glucose starvation was shown to increase the formation of proautophagy PIK3C3 complexes, containing PIK3C3, BECN1, ATG14, and PIK3R4. This equilibrium shift is inhibited in AMPKα1/AMPKα2 knockout MEFs (66). Furthermore, overexpression of kinase-dead AMPKα in AMPKα1/AMPKα2 knockout MEFs fails to rescue induction of autophagy by glucose starvation, suggesting a requirement of active AMPK. AMPK directly phosphorylates BECN1 at serines 91 and 94 and is required for stable ATG14 interactions for the proautophagy activation of the BECN1 complex (66). Similar results were found for HEK-293T cells (67). Cells that were treated with AICAR, an AMPK activator, induced autophagic signals, as measured by LC3 lipidation and LC3 punctum quantification, to the same extent as glucose starvation. Treatment with compound C (dorsomorphin), an AMPK inhibitor, or knockdown of PRKAA1 (human AMPKα1) inhibits autophagy under glucose starvation conditions (67). AMPK directly phosphorylates BECN1 at threonine 388, and this phosphorylation is required for glucose starvation autophagy induction (67). These two findings may suggest a mechanism of ULK1/2 bypass under rapid response to starvation conditions (66, 67). It is worth noting, however, that AMPK does not constitute the only possible bypass of the ULK1 complex. For example, BECN1 is phosphorylated by multiple kinases, many of which may have an impact on autophagy signaling (Fig. 4) (68–75).
FIG 4.
Schematic diagram of BECN1 with the known Bcl2 homology domain (BH3), coiled-coil domain (CCD), and evolutionarily conserved domain (ECD), which is conserved between mammals and yeast. Known phosphorylation sites are reflected with arrows and italicized numbers of the cognate residues. In red are the kinases that have been reported to phosphorylate that site. In green are phosphatases that are known to act on those specific phosphoresidues. The asterisk reflects a tentative connection of MAPK1/3 to S15 of BECN1 based on the dephosphorylation activity of DUSP1.
ULK1/2-independent autophagy induction in the context of infection.
While autophagy is essential under basal and starvation conditions, pathogens hijack this process for their own replication (76–78). Here we report on three different types of viruses utilizing autophagy for their own benefit in a ULK1/2-independent manner: coronaviruses, hepatitis B virus (HBV), and poliovirus (PV) (79–81). The transmembrane papain-like proteases (PLPs) in human coronavirus are able to induce autophagic signals, measured by LC3 lipidation, but not degradation (79). Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and porcine epidemic diarrhea virus (PEDV) encode PLPs that induce a similar response that is independent of the protease activity of PLPs (Fig. 2, gray arrows). These PLPs inhibit the ability of autophagosomes to acidify, and they enhance BECN1 interactions with transmembrane protein 173 (TMEM173), formally known as STING, which is an antiviral host signaling protein. Knockdown of BECN1 inhibits PLP-induced autophagic signals, as well as inhibiting viral replication of PEDV (79). Furthermore, PLPs directly interact with BECN1, suggesting a ULK1/2 bypass mechanism that upregulates autophagy induction without proceeding to degradation (79). The promotion of immature autophagosomes is hypothesized to help play a role in coronavirus replication as well as inhibiting antiviral responses during infection. Hepatitis B virus X (HBx) protein induces nondegradative autophagy through BECN1 (80). Overexpression of HBx leads to increased MTOR activity, but rapamycin did not increase LC3-II levels. Knockdown of BECN1 or PIK3C3 inhibits HBx induction of autophagy, as does treatment with 3-MA, suggesting a requirement of the BECN1 PIK3 signaling for autophagy (80). This study also found that loss of Jun N-terminal protein kinase (JNK) signaling, by knockdown or inhibition, prevents HBx-induced autophagy, demonstrating another potential bypass of the MTOR-ULK1/2 signaling for autophagy induction. ROS were found to be increased during HBx overexpression or during HBV infection, suggesting that ROS serve as the induction signal for JNK to signal for autophagy through the BECN1 complex (80) (Fig. 2, light purple arrows). HBV may utilize HBx to promote viral replication, limit antiviral activity in the infected cell, and interfere with the utilization of autophagy as a defense mechanism during infection. A recent report from our group found that poliovirus (PV) induces autophagic signals, as measured by LC3 lipidation and WIPI1 puncta, independently of ULK1/2 (81). Knockdown of ULK1, ULK2, and RB1CC1 does not impact viral replication or autophagic induction, as seen by increased levels of LC3-II. Surprisingly, we found that PV mediates instability of the ULK1/2 complex, as the protein levels of all ULK complex members decrease during infection (81). Whether PV induces bona fide autophagy has yet to be determined; PV and other picornaviruses cleave SQSTM1 during infection, confounding our ability to measure autophagic flux (81–83). PV also does not significantly alter the levels and phosphorylation states of MTOR and AMPK, suggesting that PV-mediated autophagic induction is independent of the MTOR-ULK1/2 signaling axis (81) (Fig. 2, blue arrows). While ULK1/2 are developmentally vital, their requirement in autophagy induction varies in the context of starvation, nutrient availability, and infection.
BECN1-INDEPENDENT AUTOPHAGY INDUCTION
Autophagy induction does not always require MTOR inhibition, AMPK activation, or ULK1/2 signaling. Mechanistically, we can reconcile this disparity in regard to autophagy induction and regulation on the idea that all signaling pathways must eventually converge into conserved autophagy machinery that is proximal to membrane formation, such as the LC3 lipidation complex. Surprisingly, there have been reports of BECN1- and PIK3C3-independent autophagy induction, suggesting that even conserved downstream autophagy proteins may not be required for autophagy induction. MCF-7 cells treated with astemizole-histamine (AST-HIS) undergo not only apoptosis but also induction of autophagic signals, as measured by LC3 lipidation (84) (Fig. 2, brown arrow). SQSTM1 levels dropped with AST-HIS treatment, suggesting that this was genuine degradative autophagy. Surprisingly, knockdown of BECN1 in MCF-7 cells treated with AST-HIS fails to inhibit autophagy, suggesting a BECN1-independent pathway of autophagy induction (84). The role of PIK3C3 or PIK3 signaling in general was not assessed, so there is the potential that PIK3 signaling is not impacted by knockdown of BECN1. Another study found that knockdown of BECN1, PIK3C3, and ATG14 inhibits palmitate-induced, but not oleate-induced, autophagy. This suggests that oleate-induced autophagy is independent of the entire BECN1 complex (65) (Fig. 2, brown arrow). Treatment of U2OS cells with 3-MA also has no effect on oleate induction of autophagy. TSC2 knockout MEFs, in which MTOR is constitutively active, have lower levels of palmitate- and oleate-induced autophagy, suggesting that while ULK1/2 and BECN1 are not required, MTOR is still able to inhibit fatty acid-induced autophagy (65). Finally, oleate-induced, BECN1-independent autophagy was replicated in S. cerevisiae and C. elegans, suggesting an evolutionarily conserved pathway for oleate and similar fatty acids (65). The differential requirements of autophagy induction dependent on the fatty acid substrate may have consequences as to the lipid makeup or composition of cellular membranes.
Proliferation of the colon cancer cell lines HT-29 and Caco-2 is inhibited by the extract of Rhus coriaria (RCE), more commonly known as the Sicilian sumac (85). RCE treatment induces bona fide autophagy in HT-29 and Caco-2 cells (85). Intriguingly, BECN1 levels decrease, in a dose-dependent manner, in these cells, and knockdown of BECN1 has no effect on RCE induction of autophagy. RCE does not impact BECN1 transcript levels but targets BECN1, MTOR, AKT, TP53, and caspase 3 (CASP3) for proteasomal degradation (85). There is a temporal association between autophagy and apoptosis. Autophagy occurs within 12 h of treatment with RCE, while apoptosis is detected at 48 h posttreatment (85). Inhibition of autophagy is shown to prevent apoptosis in HT-29 and Caco-2 cells treated with RCE. This suggests that RCE induces BECN1-independent autophagy, leading to apoptosis in colon cancer cells (85) (Fig. 2, brown arrow). The BECN1-independent autophagy pathway has not been as well characterized as the MTOR-AMPK-ULK1/2 independent pathways; it will be interesting to uncover what core autophagy proteins and complexes are required for degradative, bona fide autophagy induction.
CONCLUDING REMARKS
Since the discovery of autophagy several decades ago, much progress has been made to elucidate the regulatory mechanism behind this vital cellular process. While MTOR, AMPK, and ULK1/2 play significant roles in the regulation and induction of autophagy under many circumstances, an increasing number of reports have shown that not all of the components of the autophagy pathway are required all of the time. It is our hope that this review has illustrated that autophagy induction and required components vary depending on the type of induction signal and the types of cells utilized (Table 1). There is a difference between amino acid, glucose, and serum deprivation. Different small molecules and compounds, such as palmitate and oleate, elicit different autophagy responses. Even pathogens that benefit from the autophagy pathway have different requirements for autophagy induction. As we proceed in characterizing the vast network of interactions and regulation mechanisms of autophagy, it is vital that researchers define and classify which type of autophagy induction is being tested, which components are required, and which components are bypassed altogether. Throughout the literature, the length and nature of starvation or induction treatment varies, and the speed at which proautophagy signals are fed into the autophagy pathway may be a major factor in determining which components are bypassed in order to allow for cell survival under those conditions. We must change our understanding of autophagy induction as a linear pathway to a web of different key players, that all can induce autophagy, differing only on the type and length of induction signal.
TABLE 1.
Summary of the studies cited in this reviewa
| Induction condition | Cells | Dispensable protein(s) | Required protein(s) | Degradative autophagy | Reference(s) |
|---|---|---|---|---|---|
| ER stress (SERCA inhibition) | J16 T lymphocytes (HU), DT40 B lymphocytes (CK) | MTOR, AMPK | ULK1/2 complex?, BECN1 complex, CAMK1/4 | No | 49, 59 |
| Starvation + glucose addition | NIH 3T3 (MS) | MTOR, AMPK | ULK1/2 complex?, BECN1 complex?, MAPK14 | Yes | 54 |
| Fertilization | Oocytes (MS) | MTOR | AMPK?, ULK1/2 complex?, BECN1 complex | ? | 55 |
| Hypoxia | Nucleus-pulposus cells (RT, HU) | MTOR, AMPK | ULK1/2 complex? BECN1 complex? | No | 57 |
| Glucose starvation | Embryonic fibroblasts (MS) | AMPK | ULK1/2 complex?, BECN1 complex | ? | 60 |
| cAMP | Mesenchymal stem cells (HU) | AMPK | ULK1/2 complex?, BECN1, MAPK1, cyclin E | ? | 62 |
| Ammonia | Embryonic fibroblasts (MS) | ULK1/2, MTOR | BECN1 complex?, AMPK? | ? | 63 |
| Amino acid starvation | DT40 B lymphocytes (CK) | ULK1/2/3, MTOR | ATG13, RB1CC1, BECN1 complex? | ? | 64 |
| Oleate | U2OS (HU), heart, liver, muscle tissue (MS) | ULK1/2, BECN1, PIK3C3 | ATG5, ATG7, MTOR | Yes | 65 |
| Coronavirus infection | HEK-293T (HU), HeLa (HU), MCF-7 (HU) | ULK1/2 complex | BECN1, PIK3C3? | No | 79 |
| Hepatitis B virus infection | HepG2 (HU) | ULK1/2 complex | JNK, BECN1, PIK3C3? | No | 80 |
| Poliovirus infection | HEK-293T (HU), H1-HeLa (HU) | ULK1/2 complex | BECN1 complex? | No | 81 |
| Astemizole-histamine | MCF-7 (HU) | BECN1 | ULK1/2 complex?, PIK3C3? | Yes | 84 |
| Rhus coriaria extract | HT-29 (HU), Caco-2 (HU) | BECN1, MTOR | ULK1/2 complex?, PIK3C3? | Yes | 85 |
Cell types utilized in each study are listed as either human (HU), chicken (CK), mouse (MS), or rat (RT). Autophagy proteins found to be dispensable or required and whether each induction condition induced degradative autophagy are noted. Question marks indicate that the requirement for the marked proteins or whether the induction condition led to degradative autophagy was not assessed.
ACKNOWLEDGMENTS
A.F.C.V. was supported by NIAID training grant AI007540. The work was supported by NIAID grant AI104928 to W.T.J.
We thank Abigail K. Corona for extensive suggestions and revisions and the members of the Jackson and Frieman labs for enlightening discussions. W.T.J. thanks the Grateful Dead for inspiring the title.
We declare that we have no conflict of interest.
REFERENCES
- 1.Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res 24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanida I. 2011. Autophagy basics. Microbiol Immunol 55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- 3.White E, Mehnert JM, Chan CS. 2015. Autophagy, metabolism, and cancer. Clin Cancer Res 21:5037–5046. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, Ziegler U, Roesler J, Dengjel J, Reichenbach J, Münz C. 2013. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol 203:757–766. doi: 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky DJ, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura S, Fujita N, Noda T, Yoshimori T. 2009. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 7.Jiang P, Mizushima N. 2015. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods 75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Thost A-K, Dönnes P, Kohlbacher O, Proikas-Cezanne T. 2015. Fluorescence-based imaging of autophagy progression by human WIPI protein detection. Methods 75:69–78. doi: 10.1016/j.ymeth.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Noda T, Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 10.Chan TF, Carvalho J, Riles L, Zheng XF. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc Natl Acad Sci U S A 97:13227–13232. doi: 10.1073/pnas.240444197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245–250. doi: 10.1016/S0378-1119(97)00084-X. [DOI] [PubMed] [Google Scholar]
- 12.Kametaka S, Matsuura A, Wada Y, Ohsumi Y. 1996. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178:139–143. doi: 10.1016/0378-1119(96)00354-X. [DOI] [PubMed] [Google Scholar]
- 13.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. 1997. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192:207–213. doi: 10.1016/S0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 14.Papinski D, Kraft C. 2014. Atg1 kinase organizes autophagosome formation by phosphorylating Atg9. Autophagy 10:1338–1340. doi: 10.4161/auto.28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang Y-Y, Neufeld TP. 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20:2004–2014. doi: 10.1091/mbc.e08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. 1998. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun 246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 17.Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T. 1998. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics 51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- 18.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. 1999. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 19.Chan EYW, Kir S, Tooze SA. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 20.Lee E-J, Tournier C. 2011. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7:689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. 2014. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol 16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215:857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Mizushima N. 2009. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 5:85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 25.Hara T, Takamura A, Kishi C, Iemura S-I, Natsume T, Guan J-L, Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. 2009. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Kundu M, Viollet B, Guan K-L. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan J-L, Oshiro N, Mizushima N. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20:1981–1991. doi: 10.1091/mbc.e08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003. doi: 10.1091/mbc.e08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Wang C, Yeo S, Liang C-C, Okamoto T, Sun S, Wen J, Guan J-L. 2016. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev 30:856–869. doi: 10.1101/gad.276428.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi S, Kim DJ, Stjepanovic G, Hurley JH. 2015. Structure of the human Atg13-Atg101 HORMA heterodimer: an interaction hub within the ULK1 complex. Structure 23:1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Øvervatn A, Kirkin V, Johansen T. 2012. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. 2000. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res 85:1–12. doi: 10.1016/S0169-328X(00)00218-7. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H, Tabata K, Morita E, Kawasaki M, Kato R, Dobson RCJ, Yoshimori T, Wakatsuki S. 2014. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure 22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 36.Kim D-H, Sarbassov DD, Ali SM, Latek RR, Guntur KVP, Erdjument-Bromage H, Tempst P, Sabatini DM. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 11:895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 37.Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Lee JW, Park S, Takahashi Y, Wang H-G. 2010. The association of AMPK with ULK1 regulates autophagy. PLoS One 5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rourke JL, Hu Q, Screaton RA. 2018. AMPK and friends: central regulators of β cell biology. Trends Endocrinol Metab 29:111–122. doi: 10.1016/j.tem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Cordero MD, Williams MR, Ryffel B. 2018. AMP-activated protein kinase regulation of the NLRP3 inflammasome during aging. Trends Endocrinol Metab 29:8–17. doi: 10.1016/j.tem.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Herzig S, Shaw RJ. 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19:121–135. doi: 10.1038/nrn.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente C, Hendrickson RC, Jiang X. 2016. Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J Biol Chem 291:6026–6035. doi: 10.1074/jbc.M115.689646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. 2011. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 7:737–747. doi: 10.4161/auto.7.7.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Löffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. 2011. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 7:696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- 45.Russell RC, Tian Y, Yuan H, Park HW, Chang Y-Y, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fogel AI, Dlouhy BJ, Wang C, Ryu S-W, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. 2013. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol 33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park J-M, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, Higgins L, Kim D-H. 2018. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14:584–597. doi: 10.1080/15548627.2017.1422851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. 2014. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grotemeier A, Alers S, Pfisterer SG, Paasch F, Daubrawa M, Dieterle A, Viollet B, Wesselborg S, Proikas-Cezanne T, Stork B. 2010. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell Signal 22:914–925. doi: 10.1016/j.cellsig.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP. 1990. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A 87:2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernales S, McDonald KL, Walter P. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganley IG, Wong P-M, Gammoh N, Jiang X. 2011. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauvezin C, Nagy P, Juhász G, Neufeld TP. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moruno-Manchón JF, Pérez-Jiménez E, Knecht E. 2013. Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway. Biochem J 449:497–506. doi: 10.1042/BJ20121122. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto A, Mizushima N, Tsukamoto S. 2014. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol Reprod 91:7. doi: 10.1095/biolreprod.113.115816. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe-Asano T, Kuma A, Mizushima N. 2014. Cycloheximide inhibits starvation-induced autophagy through mTORC1 activation. Biochem Biophys Res Commun 445:334–339. doi: 10.1016/j.bbrc.2014.01.180. [DOI] [PubMed] [Google Scholar]
- 57.Choi H, Merceron C, Mangiavini L, Seifert EL, Schipani E, Shapiro IM, Risbud MV. 2016. Hypoxia promotes noncanonical autophagy in nucleus pulposus cells independent of MTOR and HIF1A signaling. Autophagy 12:1631–1646. doi: 10.1080/15548627.2016.1192753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nwadike C, Williamson LE, Gallagher LE, Guan J-L, Chan EYW. 2018. AMPK inhibits ULK1-dependent autophagosome formation and lysosomal acidification via distinct mechanisms. Mol Cell Biol 38:e00023-18. doi: 10.1128/MCB.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfisterer SG, Mauthe M, Codogno P, Proikas-Cezanne T. 2011. Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol Pharmacol 80:1066–1075. doi: 10.1124/mol.111.071761. [DOI] [PubMed] [Google Scholar]
- 60.Williams T, Forsberg LJ, Viollet B, Brenman JE. 2009. Basal autophagy induction without AMP-activated protein kinase under low glucose conditions. Autophagy 5:1155–1165. doi: 10.4161/auto.5.8.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yazawa T, Imamichi Y, Miyamoto K, Khan MRI, Uwada J, Umezawa A, Taniguchi T. 2016. Induction of steroidogenic cells from adult stem cells and pluripotent stem cells. Endocr J 63:943–951. doi: 10.1507/endocrj.EJ16-0373. [DOI] [PubMed] [Google Scholar]
- 62.Ugland H, Naderi S, Brech A, Collas P, Blomhoff HK. 2011. cAMP induces autophagy via a novel pathway involving ERK, cyclin E and Beclin 1. Autophagy 7:1199–1211. doi: 10.4161/auto.7.10.16649. [DOI] [PubMed] [Google Scholar]
- 63.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. 2011. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A 108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alers S, Löffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, Stork B. 2011. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 7:1423–1433. doi: 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Mariño G, Cianfanelli V, Ben-Younès A, Troncoso R, Markaki M, Sica V, Izzo V, Chaba K, Bauvy C, Dupont N, Kepp O, Rockenfeller P, Wolinski H, Madeo F, Lavandero S, Codogno P, Harper F, Pierron G, Tavernarakis N, Cecconi F, Maiuri MC, Galluzzi L, Kroemer G. 2015. Unsaturated fatty acids induce non-canonical autophagy. EMBO J 34:1025–1041. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan K-L. 2013. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D, Wang W, Sun X, Xu D, Wang C, Zhang Q, Wang H, Luo W, Chen Y, Chen H, Liu Z. 2016. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy 12:1447–1459. doi: 10.1080/15548627.2016.1185576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Wu X-Q, Deng R, Li D-D, Tang J, Chen W-D, Chen J-H, Ji J, Jiao L, Jiang S, Yang F, Feng G-K, Senthilkumar R, Yue F, Zhang H-L, Wu R-Y, Yu Y, Xu X-L, Mai J, Li Z-L, Peng X-D, Huang Y, Huang X, Ma N-F, Tao Q, Zeng Y-X, Zhu X-F. 2017. CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nat Commun 8:1159. doi: 10.1038/s41467-017-01272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian X, Li X, Lu Z. 2017. Protein kinase activity of the glycolytic enzyme PGK1 regulates autophagy to promote tumorigenesis. Autophagy 13:1246–1247. doi: 10.1080/15548627.2017.1313945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng Z, Zhu Q, Dee R, Opheim Z, Mack CP, Cyr DM, Taylor JM. 2017. Focal adhesion kinase-mediated phosphorylation of beclin1 protein suppresses cardiomyocyte autophagy and initiates hypertrophic growth. J Biol Chem 292:2065–2079. doi: 10.1074/jbc.M116.758268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujiwara N, Usui T, Ohama T, Sato K. 2016. Regulation of Beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3). J Biol Chem 291:10858–10866. doi: 10.1074/jbc.M115.704908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. 2015. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 4:e05289. doi: 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J, Zhou J-Y, Kho D, Reiners JJ, Wu GS. 2016. Role for DUSP1 (dual-specificity protein phosphatase 1) in the regulation of autophagy. Autophagy 12:1791–1803. doi: 10.1080/15548627.2016.1203483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. 2012. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Birmingham CL, Higgins DE, Brumell JH. 2008. Avoiding death by autophagy: interactions of Listeria monocytogenes with the macrophage autophagy system. Autophagy 4:368–371. doi: 10.4161/auto.5594. [DOI] [PubMed] [Google Scholar]
- 77.Alirezaei M, Flynn CT, Wood MR, Harkins S, Whitton JL. 2015. Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo. Autophagy 11:1389–1407. doi: 10.1080/15548627.2015.1063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackson WT. 2015. Viruses and the autophagy pathway. Virology 479-480:450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Wang K, Xing Y, Tu J, Yang X, Zhao Q, Li K, Chen Z. 2014. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell 5:912–927. doi: 10.1007/s13238-014-0104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong L, Shu W, Dai W, Gao B, Xiong S. 2017. Reactive oxygen species-mediated c-Jun NH2-terminal kinase activation contributes to hepatitis B virus X protein-induced autophagy via regulation of the Beclin-1/Bcl-2 interaction. J Virol 91:e00001-17. doi: 10.1128/JVI.00001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corona Velazquez A, Corona A, Klein KA, Jackson WT. 2018. Poliovirus induces autophagic signaling independent of the ULK1 complex. Autophagy 14:1201–1213. doi: 10.1080/15548627.2018.1458805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corona AK, Saulsbery HM, Corona Velazquez AF, Jackson WT. 2018. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep 22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J, Li X, Jan E, Luo H. 2013. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 9:1591–1603. doi: 10.4161/auto.26059. [DOI] [PubMed] [Google Scholar]
- 84.Jakhar R, Paul S, Bhardwaj M, Kang SC. 2016. Astemizole-histamine induces Beclin-1-independent autophagy by targeting p53-dependent crosstalk between autophagy and apoptosis. Cancer Lett 372:89–100. doi: 10.1016/j.canlet.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 85.Athamneh K, Hasasna HE, Samri HA, Attoub S, Arafat K, Benhalilou N, Rashedi AA, Dhaheri YA, AbuQamar S, Eid A, Iratni R. 2017. Rhus coriaria increases protein ubiquitination, proteasomal degradation and triggers noncanonical Beclin-1-independent autophagy and apoptotic cell death in colon cancer cells. Sci Rep 7:11633. doi: 10.1038/s41598-017-11202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]