Growth differentiation factor 15 (GDF15) is a secreted protein with pleotropic functions from the transforming growth factor β (TGF-β) family. GDF15 is synthesized as a precursor and undergoes proteolytic cleavage to generate mature GDF15.
KEYWORDS: GDF15, PCSK, heart disease, cardiomyocytes, cardiac endocrinology
ABSTRACT
Growth differentiation factor 15 (GDF15) is a secreted protein with pleotropic functions from the transforming growth factor β (TGF-β) family. GDF15 is synthesized as a precursor and undergoes proteolytic cleavage to generate mature GDF15. The strong appetite-suppressing effect of mature GDF15 makes it an attractive therapeutic agent/target for diseases such as obesity and cachexia. In addition, clinical studies indicate that circulating, mature GDF15 is an independent biomarker for heart failure. We recently found that GDF15 functions as a heart-derived hormone that inhibits liver growth hormone signaling and postnatal body growth in the pediatric period. However, little is known about the mechanism of GDF15 maturation, in particular the enzymes that mediate GDF15 precursor cleavage. We investigated which candidate proteases can cleave GDF15 precursor and generate mature GDF15 in cardiomyocytes in vitro and mouse hearts in vivo. We discovered that three members of the proprotein convertase, subtilisin/kexin-type (PCSK) family, namely, PCSK3, PCSK5, and PCSK6, can efficiently cleave GDF15 precursor, therefore licensing its maturation both in vitro and in vivo. Our studies suggest that PCSK3, -5, and -6 mediate a crucial step of GDF15 maturation through proteolytic cleavage of the precursor. These results also reveal new targets for therapeutic application of GDF15 in treating obesity and cachexia.
INTRODUCTION
Growth differentiation factor 15 (GDF15; also known as MIC-1, NAG-1, PTGFB, PDF, and PLAB) is a distant member of the transforming growth factor β (TGF-β) family (1–7). Basal tissue expression of Gdf15 is low except in the placenta and prostate, but its expression can be induced in pathological conditions (5, 8). In particular, an elevated serum GDF15 level has been found in heart disease patients and is associated with increased morbidity and mortality. GDF15 is therefore used as an independent biomarker for heart disease, especially heart failure (9, 10). We recently discovered GDF15 as a heart-derived hormone that regulates postnatal body growth (11). Cardiac Gdf15 expression is highly induced in pediatric heart disease, and circulating GDF15 acts on the liver to inhibit growth hormone signaling and body growth. Plasma GDF15 is increased in children with concomitant heart disease and failure to thrive (FTT), providing a potential explanation for the well-established clinical observation that children with heart diseases often develop FTT. In addition, it was recently shown that GDF15 suppresses appetite via activating its receptor GFRAL in the brain stem (12–17). GDF15 is currently being explored as a potential therapeutic agent or target for a variety of indications, including obesity, metabolic disease, anorexia, and cancer cachexia.
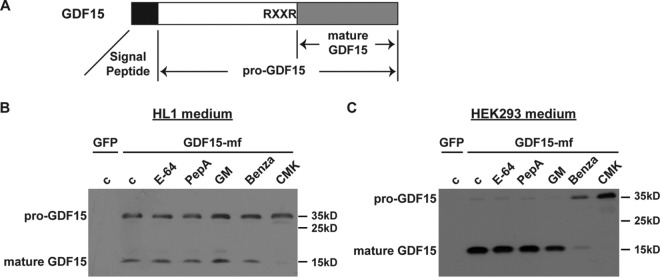
So far, most studies on GDF15 have focused on its biological function and disease biomarker implications. The biochemical properties of the GDF15 protein itself remain little understood. Like many other protein hormones, GDF15 protein is synthesized as a biologically inactive precursor protein (pre-pro-GDF15), with an N-terminal signal peptide important for its intracellular trafficking and secretion (Fig. 1A). After removal of the signal peptide, the remaining GDF15 propeptide (pro-GDF15; ∼30 kDa) undergoes proteolytic cleavage to yield the C-terminal mature GDF15 (∼13 kDa), which forms a homodimer as the major secreted form of GDF15 found in serum (18). However, the enzymes responsible for the proteolytic processing and generation of mature GDF15 remain elusive.
FIG 1.
pro-GDF15 is cleaved by the PCSK family of proteases. (A) Schematic diagram of pre-pro-GDF15 with its canonical RXXR cleavage site shown. (B and C) Representative Western blot images showing pro-GDF15 and mature GDF15 in media of HL1 cardiomyocytes (B) and HEK293 cells (C) transfected with GDF15-mf plasmid and treated with different protease inhibitors. GDF15-mf, C-terminal myc-FLAG-tagged pre-pro-GDF15; c, control; PepA, pepstatin A; GM, GM-6001; Benza, benzamidine; CMK, decanoyl-RVKR-CMK.
The mammalian proprotein convertase, subtilisin/kexin-type (PCSK) proteins are a family of serine proteinases that share homology with bacterial enzyme subtilisin and yeast enzyme kexin (19, 20). Among the nine mammalian PCSK enzymes, PCSK9 is well known for its role in cleaving and degrading low-density lipoprotein receptor (LDLR), which is crucial in cholesterol metabolism (21). Antibodies targeting PCSK9 (Praluent and Repatha) constitute the latest therapy for hypercholesterolemia and heart disease by significantly reducing serum LDL and the risk of cardiovascular events (22). The substrates of PCSK8 (also known as SKI-1/S1P) include sterol-regulated element binding proteins (SREBPs), brain-derived neurotrophic factor (BDNF), and somatostatin. PCSK1 to -7 recognize and cleave right after the basic residues Arg/Lys-X2n-Arg/Lys↓ (R/KX2nR/K, where X is any amino acid and n = 0 to 3). They are essential for the cleavage and maturation of many secreted proteins, including important hormones (i.e., insulin, glucagon, and adrenocorticotropic hormone [ACTH]). Intriguingly, PCSK3 (also known as furin) has been reported as the major enzyme for TGF-β family proteins TGF-β and bone morphogenetic protein 10 (23, 24). In addition, GDF15 contains the canonical RXXR sequence right before the cleavage site, which can be recognized by PCSK1 to -7. These observations raised the possibility that one or more of the PCSK enzymes are responsible for the cleavage and maturation of GDF15.
In this study, we investigated the mechanisms of GDF15 maturation in cardiomyocytes, in which it is highly induced under the condition of heart disease. We found that PCSK3, PCSK5, and PCSK6 can efficiently cleave pro-GDF15 for maturation both in vitro and in vivo. These results revealed the long-sought identity of the enzymes responsible for GDF15 maturation and uncovered novel means for therapeutic application of GDF15 in treating metabolic disease, anorexia, and cachexia.
RESULTS
PCSK3, -5, and -6 cleave pro-GDF15 into the mature form in vitro.
To determine the enzyme class that cleaves pro-GDF15, we expressed mouse C-terminal myc-FLAG-tagged pre-pro-GDF15 (GDF15-mf) in HL1 mouse cardiomyocytes or HEK293 human embryonic kidney cells. These cells expressed little endogenous GDF15, making them a good system for analysis. We then treated the cells with different classes of protease inhibitors: cysteine protease inhibitor E-64, aspartyl protease inhibitor pepstatin, broad-spectrum matrix metalloprotease inhibitor GM-6001, serine protease inhibitor benzamidine, and pan-PCSK inhibitor decanoyl-RVKR-CMK. Western blot analysis revealed that both benzamidine and CMK could inhibit the processing of pro-GDF15 (decreased ratio of mature GDF15 to pro-GDF15), while the other classes of protease inhibitors had no effect (Fig. 1B and C). These results suggest that GDF15 is cleaved by one or more PCSK family members in these cells.
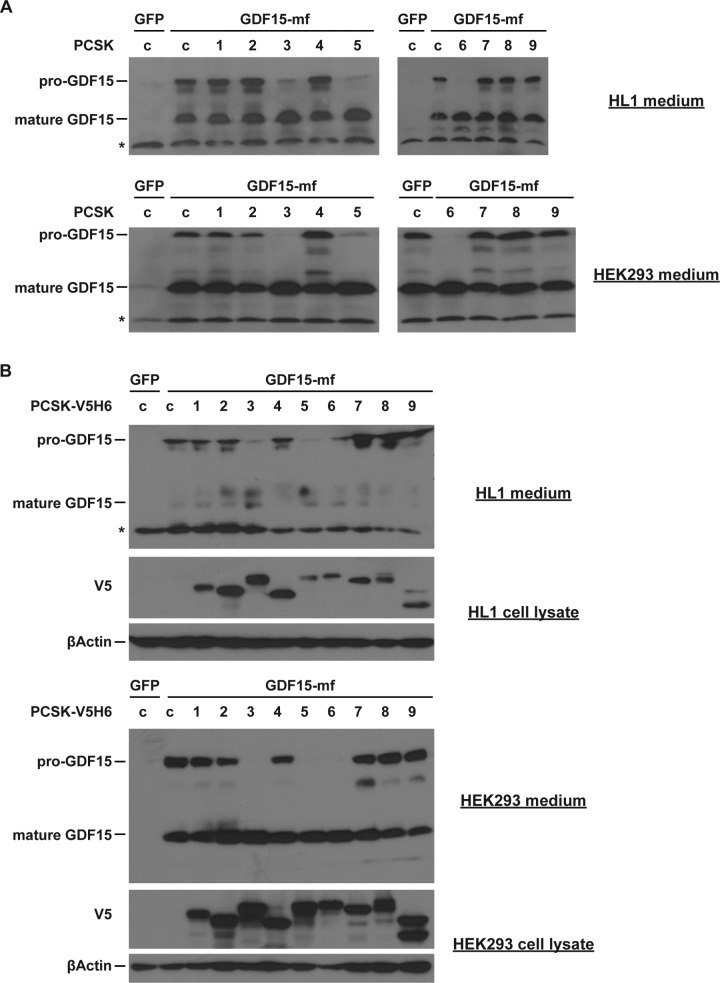
To determine the specific PCSK protein that cleaves GDF15, we expressed individual PCSK proteins together with pre-pro-GDF15 in HL1 or HEK293 cells. Western blot analysis showed that only PCSK3, PCSK5, and PCSK6 (also known as PACE4) effectively processed pro-GDF15 into mature GDF15 (Fig. 2A). All the other PCSK members exhibited no GDF15-processing capability. To ensure that all PCSK proteins are expressed at comparable levels, we also performed these experiments using tagged PCSK vectors, and we obtained the same results (Fig. 2B). These results suggest that PCSK3, PCSK5, and PCSK6 are cell-type-independent enzymes that process pro-GDF15 into the mature form.
FIG 2.
PCSK3, -5, and -6 cleave pro-GDF15 in vitro. Representative Western blot images show pro-GDF15 and mature GDF15 in media of HL1 cardiomyocytes and HEK293 cells transfected with GDF15-mf and different PCSK plasmids. (A) Nontagged PCSK; (B) V5H6-tagged PCSK. In panel B, cellular PCSK protein levels were also determined by Western blotting using V5 antibody. β-Actin served as a loading control. *, nonspecific bands.
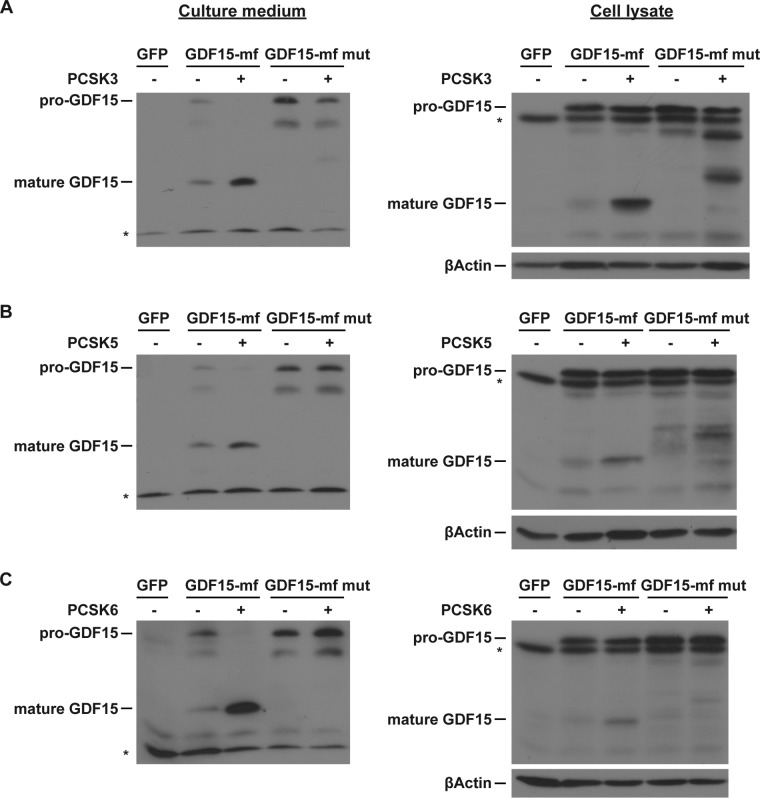
We next investigated whether the PCSK recognition sequence RXXR in pro-GDF15 (ending at position 188 in mouse and position 193 in human pre-pro-GDF15) is essential for its processing. We mutated the last Arg to Gln (R188Q) to disrupt this RXXR motif. While coexpression of PCSK3, PCSK5, or PCSK6 could cleave wild-type (WT) pro-GDF15 into mature GDF15 in both HL1 cell lysate and culture medium, all three PCSK proteins were ineffective against this RXXR motif mutant pro-GDF15 (Fig. 3). These results suggest that PCSK3, PCSK5, and PCSK6 cleave pro-GDF15 at its canonical RXXR sequence.
FIG 3.
RXXR motif of pro-GDF15 is crucial for PCSK3, -5, and -6-mediated cleavage. Representative Western blot images of HL1 medium and cell lysates are shown. HL1 cardiomyocytes were transfected with WT or R188Q mutant GDF15-mf, with or without an equal amount of plasmid encoding full-length PCSK3 (A), PCSK5 (B), or PCSK6 (C). β-Actin served as a loading control.
PCSK3, -5, and -6 catalytic activities are essential for pro-GDF15 cleavage.
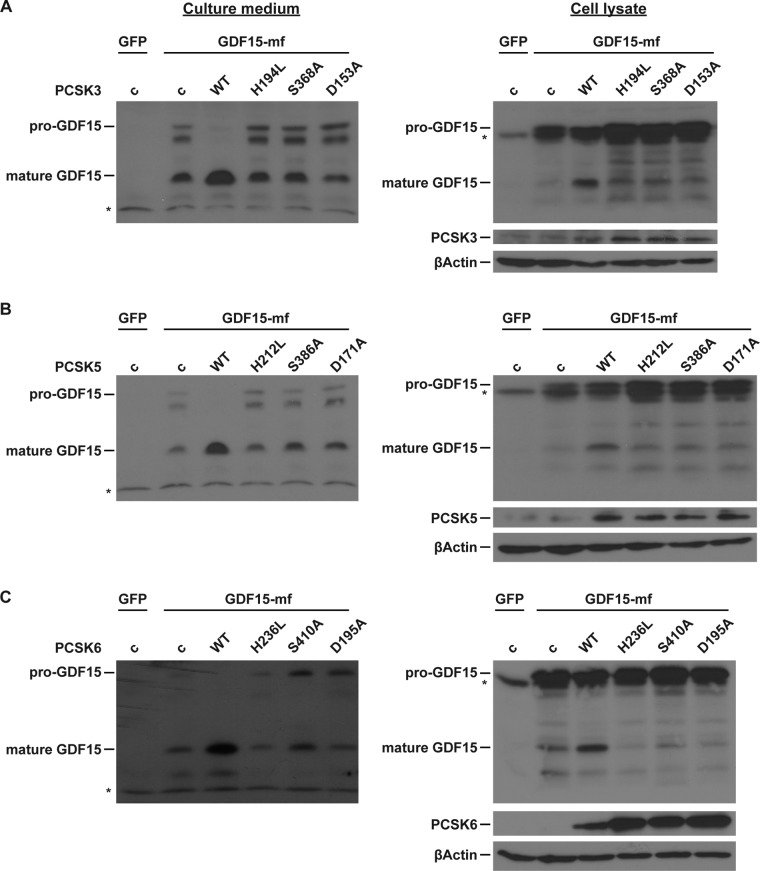
The so-called catalytic triad Asp-His-Ser forms the catalytic active sites of PCSK enzymes and is conserved from bacterial subtilisin and yeast kexin to mammal PCSK (19, 20). We next investigated whether the catalytic triad of PCSK3, -5, and -6 is essential for pro-GDF15 cleavage. We mutated the individual essential amino acid (Asp to Ala, His to Lys, and Ser to Ala) in the catalytic triad of PCSK3, PCSK5, and PCSK6. These mutations seem to increase the stability of PCSK3 and PCSK6 but not PCSK5 (Fig. 4). While WT PCSK3, PCSK5, or PCSK6 could cleave pro-GDF15 into mature GDF15 in both HL1 cell lysate and culture medium, none of the mutant forms were able to cleave pro-GDF15 (Fig. 4). These results suggest that the catalytic activities of PCSK3, -5, and -6 are essential for pro-GDF15 processing.
FIG 4.
PCSK3, -5, and -6 catalytic activities are essential for pro-GDF15 cleavage. Representative Western blot images of HL1 medium and cell lysates are shown. HL1 cardiomyocytes were transfected with WT GDF15-mf and an equal amount of plasmids encoding WT or mutant PCSK3 (A), PCSK5 (B), or PCSK6 (C). β-Actin served as a loading control.
PCSK3, -5, and -6 license GDF15 maturation in vivo.
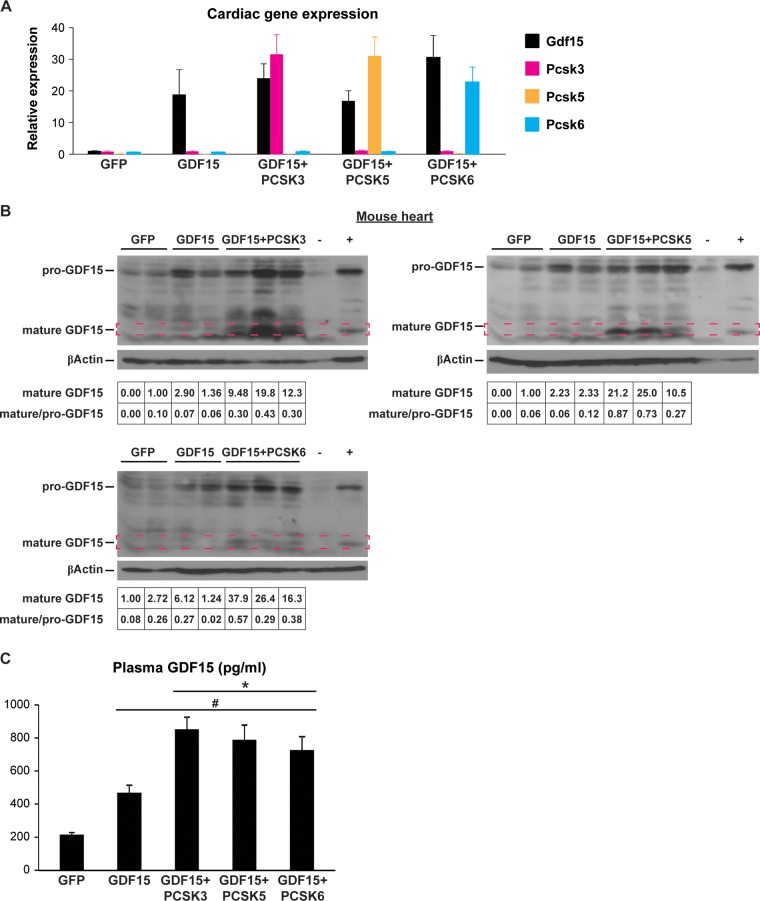
We next investigated whether PCSK3, PCSK5, or PCSK6 was able to cleave pro-GDF15 in the mouse heart in vivo. We generated and performed pericardial injection of adeno-associated virus 9 (AAV9) encoding a cardiac troponin (cTnt) promoter-driven pre-pro-GDF15, in combination with AAV9-cTnt-green fluorescent protein (GFP) (as a control), PCSK3, PCSK5, or PCSK6 into postnatal day 3 to 5 C57BL/6 WT mice and collected the hearts 10 days after injection. We and others have previously shown that pericardial injection of AAV9 achieves stable and cardiac tissue-specific expression of transgenes (11, 25). Quantitative PCR (qPCR) results confirmed the expression of individual genes in the mouse heart (Fig. 5A). GDF15 overexpression alone resulted in increased pro-GDF15 but little change of mature GDF15 protein in the WT mouse hearts (Fig. 5B). In contrast, coexpression of PCSK3, PCSK5, or PCSK6 significantly increased both pro-GDF15 and mature GDF15, and importantly, the ratio of mature/pro-GDF15. Furthermore, the presence of PCSK3, PCSK5, or PCSK6 doubled the amount of circulating, mature GDF15 (18) (Fig. 5C). Together, these results suggest that PCSK3, PCSK5, and PCSK6 license GDF15 maturation in vivo.
FIG 5.
PCSK3, -5, and -6 license GDF15 maturation in vivo. (A) Cardiac Gdf15, Pcsk3, Pcsk5, and Pcsk6 mRNA levels in mice receiving AAV9 pericardial injection. (B) Representative Western blot images of pro-GDF15 and mature GDF15 (framed) in AAV-injected heart lysates. −, negative control (GDF15 KO heart lysate); +, positive control (heart lysates from cardiac ERRα/γ KO mice). β-Actin served as a loading control. Quantification of mature GDF15 band density (normalized to β-actin) and mature versus pro-GDF15 ratio is shown at the bottom. (C) Plasma GDF15 levels in AAV9-injected mice (n = 12 to 18 per group). #, P < 0.0001 versus AAV9-GFP; *, P < 0.01 versus AAV9-GDF15.
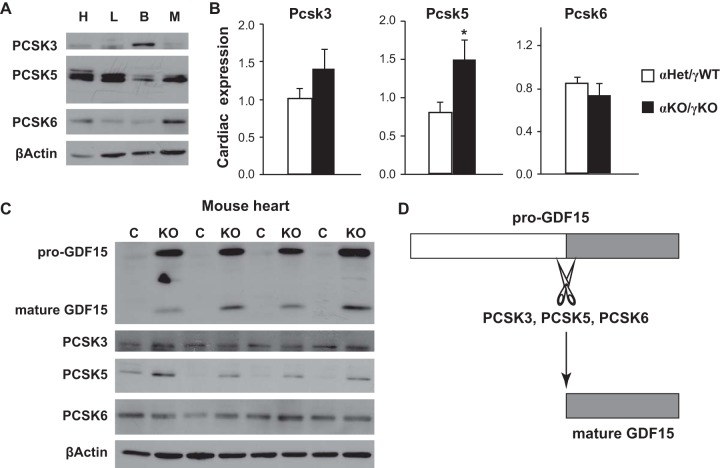
We last examined PCSK3, -5, and -6 expression in a mouse model of heart disease. We have previously shown that cardiac loss of two transcription factors essential for normal cardiac metabolism and function, estrogen-related receptor alpha (ERRα) and gamma (ERRγ), results in postnatal cardiomyopathy, heart failure, and FTT (11, 26). Gdf15 transcription is highly induced in cardiac ERRα/γ knockout (KO) mice, resulting in increased pro-GDF15 and mature GDF15 production in the heart. Circulating GDF15, in turn, inhibits liver growth hormone signaling and pediatric body growth, thereby coordinating body growth rate with cardiac function. We found that PCSK3, -5, and -6 proteins are present in multiple tissues important in metabolism (heart, liver, brown fat, and gastrocnemius muscle [Fig. 6A]). Although PCSK3 and PCSK6 abundance were little changed, both the RNA and protein levels of PCSK5 were significantly increased in cardiac ERRα/γ KO mouse hearts, further supporting significant GDF15 maturation (Fig. 6B and C). Although further investigations are required to determine whether the activities of these PCSK enzymes alter under other physiological or pathological conditions, these results suggest that PCSK3, -5, and -6 can support GDF15 maturation in heart disease in vivo.
FIG 6.
PCSK3, -5, and -6 expression in mouse models of heart disease. (A) Representative Western blot images of PCSK3, PCSK5, and PCSK6 proteins in the heart (H), liver (L), brown fat (B), and gastrocnemius muscle (M) of 16-day-old control αHet/γWT mice. (B) Expression of Pcsk3, Pcsk5, and Pcsk6 in the hearts of control αHet/γWT and cardiac ERRα/γ KO (αKO/γKO) mice (n = 6 to 8). *, P < 0.05. (C) Representative Western blot images of GDF15, PCSK3, PCSK5, and PCSK6 proteins in the hearts of littermate control αHet/γWT and cardiac ERRα/γ KO mice (n = 4). (D) Diagram summarizing findings of pro-GDF15 cleavage by PCSK3, -5, and -6. β-Actin served as a loading control in panels A and C.
DISCUSSION
Like most hormones and many other TGF-β family member proteins, GDF15 is synthesized as a precursor protein and undergoes proteolytic processing to release the mature form as a secreted hormone. However, little is known about the protein enzymes that mediate this critical cleavage step. This issue becomes more important considering the current strong therapeutic interest in GDF15 for treating obesity and cachexia and its implication in cardiovascular disease. Although a recent report that focused on PCSK6 in prostate cancer touched on GDF15 as one of the substrates of PCSK6 (27), this study took a more systematic approach and identified three enzymes that effectively process pro-GDF15 to maturation. PCSK3, -5, and -6 (but not other PCSK proteins) can all recognize and cleave the canonical substrate sequence of pro-GDF15 in vitro and in vivo (Fig. 6D). We also showed that the catalytic triad Asp-His-Ser of PCSK3, -5, and -6 is essential for the members' enzymatic activities and GDF15 maturation. Together, these studies provided novel mechanistic insights into our understanding of GDF15 biology.
We showed that PCSK3, -5, and -6 can all cleave pro-GDF15 in multiple cell types in vitro and in the mouse heart in vivo. Whether they are essential for GDF15 in vivo maturation remains unclear at this moment, as inactivation of all three PCSK enzymes is likely needed to definitively address this question. It remains possible that one particular PCSK is critical for pro-GDF15 processing in certain cell types or tissues due to its much higher abundance than other PCSKs. In addition, it is also possible that in a complex organ (such as the heart) composed of many cell types, different PCSKs are essential for pro-GDF15 processing in distinct cell types. Future studies will address these possibilities. On the other hand, how cellular PCSK3, -5, and -6 activities are regulated remains little understood. It is possible that certain physiological stimuli or pathological conditions promote specific PCSK abundance and/or activity, thereby regulating mature GDF15 availability, as we showed for PCSK5 in heart disease.
MATERIALS AND METHODS
Animal studies and AAV injection.
All animal studies were approved by and performed under the guidelines of the Institutional Animal Care and Use Committee of the Children's Hospital of Philadelphia (CHOP) and comply with the ARRIVE guidelines. The WT C57BL6/J mice were from the Jackson Laboratory. Cardiac ERRα/γ KO mice in the C57BL6/J background were previously described (26). Mice were maintained in a pathogen-free environment on a 12/12-h light/dark cycle with ad libitum access to food and water. Both sexes were included in studies. The mouse pups and dams were fed a breeder diet (LabDiet 5058), while weaned mice were fed a standard chow diet (LabDiet 5L0D). AAV9-cTnt-GFP, mouse pre-pro-GDF15, mouse PCSK3, human PCSK5, and mouse PCSK6 were generated at the CHOP Research Vector Core. Pericardial injection of mice at 3 to 5 days of age with 3 × 1011 vector genomes of AAV9 was performed as previously described (11). Mice were returned to their home cage after AAV injection and maintained for an additional 10 days before being euthanized.
Cell culture, plasmids, and transfection.
HEK293 cells were cultured in Dulbecco's modified Eagle medium (DMEM) with l-glutamine, 4.5 g/liter of glucose, sodium pyruvate (Corning; 10013CV), and 10% fetal bovine serum (FBS; Sigma; F2442). HL1 cardiomyocytes were maintained in Claycomb medium (Sigma; 51800C) supplemented with FBS (Sigma; F2442), 0.1 mM l-(−)-norepinephrine bitartrate (Cayman Chemical; 16673), and 2 mM l-glutamine (Gibco; 25030) as previously described (26). Plasmid encoding mouse pre-pro-GDF15 was from Origene (MR204247). Plasmids encoding full-length mouse PCSK1 to -4 and PCSK7 to -9 were generated by PCR cloning from mouse cDNAs into the pcDNA3.1 vector. Plasmid encoding full-length human PCSK5 was from GeneCopoeia (Ex-T3161-M98). Plasmid encoding full-length mouse PCSK6 with FLAG tag was from Nanopro Biosciences (751729-1). The V5H6-tagged PCSK1 to -9 vectors were generated by PCR cloning into the pcDNA3.1D-V5H6 vector. The mutant plasmids GDF15 R188Q, PCSK3 H194L, S368A, and D153A, PCSK5 H212L, S386A, and D171A, and PCSK6 H236L, S410A, and D195A were generated by site-directed mutagenesis as previously described (26, 28, 29). HEK293 and HL1 cells were transfected using 1.5:1 and 5:1 ratios of FUGENE 6 (Promega), respectively.
Protease inhibitors and Western blot analysis.
Protease inhibitors used in this study include E-64 (10 μM; Cayman Chemical; 10007963), pepstatin A (10 μM; Cayman Chemical; 9000469), GM-6001 (50 μM; Enzo Life Sciences; BML-EI300), benzamidine (5 mM; Sigma; B6506), and decanoyl-RVKR-CMK (25 μM; Cayman Chemical; 14965). Twenty-four hours after transfection, cells were treated with aforementioned protease inhibitors for 24 h. The culture medium was then collected and briefly centrifuged to remove cell debris; cells were lysed with radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (Roche; 4693159001). Western blot analysis was performed as previously described (26, 28, 29) using the following antibodies: GDF15 (Abcam; ab189358), PCSK3 (Santa Cruz; sc-133142), PCSK5 (Thermo Fisher; PA5-42378), PCSK6 (Abcam; AB151562), FLAG (Sigma; F1804), V5 (Thermo Fisher; MA1-81617), and β-actin (Cell Signaling; 4967). Image quantification was performed using ImageJ from films scanned at 1,200 dots per inch (dpi) in TIFF format by selecting uniform rectangles that enclose the bands. We separated peaks corresponding to different bands using ImageJ, which allowed accurate calculation of mature and pro-GDF15 band densities from closely located, nonspecific bands. Relative ratio was calculated by dividing the value of each band by that of its corresponding control band.
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed as previously described (26, 30). Primers sequences are listed in Table 1.
TABLE 1.
qPCR primer sequences
| Gene | Species | Forward | Reverse |
|---|---|---|---|
| Pcsk1 | Mouse | AGTTGGAGGCATAAGAATGCTG | GCCTTCTGGGCTAGTCTGC |
| Pcsk2 | Mouse | AGAGAGACCCCAGGATAAAGATG | CTTGCCCAGTGTTGAACAGGT |
| Pcsk3 | Mouse | TAGCAGGCAATTATGACCCTGG | TAAGCTACACCTACGCCACAG |
| Pcsk4 | Mouse | CTTTCGCTCTCCTACAGCCG | AGTTGTTGTAGCCTTGGCCG |
| Pcsk5 | Human | GAGGGACCCACAGTTTCATTTC | TGGGCACGACTGAAGTCATAA |
| Pcsk6 | Mouse | CAGGCGCGAAGTGACTCTC | GACCGACAGCGACTGTTCTT |
| Pcsk7 | Mouse | AGAGAGTCTGACGCAACAGG | TATGCCCAGTAGGCTGGACAA |
| Pcsk8 | Mouse | CTGGTGGTTTTGCTCTGTGG | GGCTGTGAAGTATCCGTTGAAAG |
| Pcsk9 | Mouse | GAGACCCAGAGGCTACAGATT | AATGTACTCCACATGGGGCAA |
| Gdf15 | Mouse | CTGGCAATGCCTGAACAACG | GGTCGGGACTTGGTTCTGAG |
| 36b4 | Mouse | AGATGCAGCAGATCCGCAT | GTTCTTGCCCATCAGCACC |
ELISA.
Mouse blood was collected using a lithium heparin-coated microvette (Sarstedt; CB300LH) and centrifuged at 3,000 × g for 5 min at 4°C. Plasma GDF15 levels were measured with an enzyme-linked immunosorbent assay (ELISA) kit (R&D; DY6385) by following the manufacturer's instructions.
Statistics.
Data are presented means plus standard errors of the means (SEMs). Two-tailed, two-sample unequal-variance Student's t test was used to determine the statistical significance. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Douglas Wallace, Mitchell Lazar, Matthew Weitzman, Amita Sehgal, Michael Marks, and Mark Kahn for critical discussion of the project.
L.P. conceived and directed the project. J.J.L., J.L., and K.L. performed most experiments. X.L. and L.Z. generated AAV vectors. J.J.L. and L.P. wrote the manuscript, and all authors reviewed or edited the manuscript.
We and this work were supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer Reviewed Medical Research Program under award no. W81XWH-16-1-0400 and by NIH grant DK111495 (L.P.). The AAV vector work was supported by the CHOP RVC internal operation fund.
We declare no conflict of interest.
REFERENCES
- 1.Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, Zhang HP, Donnellan M, Mahler S, Pryor K, Walsh BJ, Nicholson RC, Fairlie WD, Por SB, Robbins JM, Breit SN. 1997. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci U S A 94:11514–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hromas R, Hufford M, Sutton J, Xu D, Li Y, Lu L. 1997. PLAB, a novel placental bone morphogenetic protein. Biochim Biophys Acta 1354:40–44. doi: 10.1016/S0167-4781(97)00122-X. [DOI] [PubMed] [Google Scholar]
- 3.Lawton LN, Bonaldo MF, Jelenc PC, Qiu L, Baumes SA, Marcelino RA, de Jesus GM, Wellington S, Knowles JA, Warburton D, Brown S, Soares MB. 1997. Identification of a novel member of the TGF-beta superfamily highly expressed in human placenta. Gene 203:17–26. doi: 10.1016/S0378-1119(97)00485-X. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama-Kobayashi M, Saeki M, Sekine S, Kato S. 1997. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J Biochem 122:622–626. doi: 10.1093/oxfordjournals.jbchem.a021798. [DOI] [PubMed] [Google Scholar]
- 5.Paralkar VM, Vail AL, Grasser WA, Brown TA, Xu H, Vukicevic S, Ke HZ, Qi H, Owen TA, Thompson DD. 1998. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J Biol Chem 273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 6.Böttner M, Laaff M, Schechinger B, Rappold G, Unsicker K, Suter-Crazzolara C. 1999. Characterization of the rat, mouse, and human genes of growth/differentiation factor-15/macrophage inhibiting cytokine-1 (GDF-15/MIC-1). Gene 237:105–111. doi: 10.1016/S0378-1119(99)00309-1. [DOI] [PubMed] [Google Scholar]
- 7.Baek SJ, Kim KS, Nixon JB, Wilson LC, Eling TE. 2001. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol Pharmacol 59:901–908. doi: 10.1124/mol.59.4.901. [DOI] [PubMed] [Google Scholar]
- 8.Unsicker K, Spittau B, Krieglstein K. 2013. The multiple facets of the TGF-beta family cytokine growth/differentiation factor-15/macrophage inhibitory cytokine-1. Cytokine Growth Factor Rev 24:373–384. doi: 10.1016/j.cytogfr.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Baggen VJ, van den Bosch AE, Eindhoven JA, Schut AW, Cuypers JA, Witsenburg M, de Waart M, van Schaik RH, Zijlstra F, Boersma E, Roos-Hesselink JW. 2017. Prognostic value of N-terminal pro-B-type natriuretic peptide, troponin-T, and growth-differentiation factor 15 in adult congenital heart disease. Circulation 135:264–279. doi: 10.1161/CIRCULATIONAHA.116.023255. [DOI] [PubMed] [Google Scholar]
- 10.Wollert KC, Kempf T, Wallentin L. 2017. Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin Chem 63:140–151. doi: 10.1373/clinchem.2016.255174. [DOI] [PubMed] [Google Scholar]
- 11.Wang T, Liu J, McDonald C, Lupino K, Zhai X, Wilkins BJ, Hakonarson H, Pei L. 2017. GDF15 is a heart-derived hormone that regulates body growth. EMBO Mol Med 9:1150–1164. doi: 10.15252/emmm.201707604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnen H, Lin S, Kuffner T, Brown DA, Tsai VW, Bauskin AR, Wu L, Pankhurst G, Jiang L, Junankar S, Hunter M, Fairlie WD, Lee NJ, Enriquez RF, Baldock PA, Corey E, Apple FS, Murakami MM, Lin EJ, Wang C, During MJ, Sainsbury A, Herzog H, Breit SN. 2007. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat Med 13:1333–1340. doi: 10.1038/nm1677. [DOI] [PubMed] [Google Scholar]
- 13.Mullican SE, Lin-Schmidt X, Chin CN, Chavez JA, Furman JL, Armstrong AA, Beck SC, South VJ, Dinh TQ, Cash-Mason TD, Cavanaugh CR, Nelson S, Huang C, Hunter MJ, Rangwala SM. 2017. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat Med 23:1150–1157. doi: 10.1038/nm.4392. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Chang CC, Sun Z, Madsen D, Zhu H, Padkjaer SB, Wu X, Huang T, Hultman K, Paulsen SJ, Wang J, Bugge A, Frantzen JB, Norgaard P, Jeppesen JF, Yang Z, Secher A, Chen H, Li X, John LM, Shan B, He Z, Gao X, Su J, Hansen KT, Yang W, Jorgensen SB. 2017. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat Med 23:1158–1166. doi: 10.1038/nm.4394. [DOI] [PubMed] [Google Scholar]
- 15.Emmerson PJ, Wang F, Du Y, Liu Q, Pickard RT, Gonciarz MD, Coskun T, Hamang MJ, Sindelar DK, Ballman KK, Foltz LA, Muppidi A, Alsina-Fernandez J, Barnard GC, Tang JX, Liu X, Mao X, Siegel R, Sloan JH, Mitchell PJ, Zhang BB, Gimeno RE, Shan B, Wu X. 2017. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat Med 23:1215–1219. doi: 10.1038/nm.4393. [DOI] [PubMed] [Google Scholar]
- 16.Hsu JY, Crawley S, Chen M, Ayupova DA, Lindhout DA, Higbee J, Kutach A, Joo W, Gao Z, Fu D, To C, Mondal K, Li B, Kekatpure A, Wang M, Laird T, Horner G, Chan J, McEntee M, Lopez M, Lakshminarasimhan D, White A, Wang SP, Yao J, Yie J, Matern H, Solloway M, Haldankar R, Parsons T, Tang J, Shen WD, Alice Chen Y, Tian H, Allan BB. 2017. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550:255–259. doi: 10.1038/nature24042. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Walker K, Min X, Hale C, Tran T, Komorowski R, Yang J, Davda J, Nuanmanee N, Kemp D, Wang X, Liu H, Miller S, Lee KJ, Wang Z, Veniant MM. 2017. Long-acting MIC-1/GDF15 molecules to treat obesity: evidence from mice to monkeys. Sci Transl Med 9:eaan8732. doi: 10.1126/scitranslmed.aan8732. [DOI] [PubMed] [Google Scholar]
- 18.Kempf T, Horn-Wichmann R, Brabant G, Peter T, Allhoff T, Klein G, Drexler H, Johnston N, Wallentin L, Wollert KC. 2007. Circulating concentrations of growth-differentiation factor 15 in apparently healthy elderly individuals and patients with chronic heart failure as assessed by a new immunoradiometric sandwich assay. Clin Chem 53:284–291. doi: 10.1373/clinchem.2006.076828. [DOI] [PubMed] [Google Scholar]
- 19.Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, Prat A. 2008. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol 40:1111–1125. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 20.Seidah NG, Sadr MS, Chretien M, Mbikay M. 2013. The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J Biol Chem 288:21473–21481. doi: 10.1074/jbc.R113.481549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton JD, Cohen JC, Hobbs HH. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci 32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR. 2017. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 23.Dubois CM, Blanchette F, Laprise MH, Leduc R, Grondin F, Seidah NG. 2001. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am J Pathol 158:305–316. doi: 10.1016/S0002-9440(10)63970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Susan-Resiga D, Essalmani R, Hamelin J, Asselin MC, Benjannet S, Chamberland A, Day R, Szumska D, Constam D, Bhattacharya S, Prat A, Seidah NG. 2011. Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J Biol Chem 286:22785–22794. doi: 10.1074/jbc.M111.233577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bish LT, Morine K, Sleeper MM, Sanmiguel J, Wu D, Gao G, Wilson JM, Sweeney HL. 2008. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther 19:1359–1368. doi: 10.1089/hum.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, McDonald C, Petrenko NB, Leblanc M, Giguere V, Evans RM, Patel VV, Pei L. 2015. Estrogen-related receptor alpha (ERRalpha) and ERRgamma are essential coordinators of cardiac metabolism and function. Mol Cell Biol 35:1281–1298. doi: 10.1128/MCB.01156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couture F, Sabbagh R, Kwiatkowska A, Desjardins R, Guay SP, Bouchard L, Day R. 2017. PACE4 undergoes an oncogenic alternative splicing switch in cancer. Cancer Res 77:6863–6879. doi: 10.1158/0008-5472.CAN-17-1397. [DOI] [PubMed] [Google Scholar]
- 28.Pei L, Mu Y, Leblanc M, Alaynick W, Barish GD, Pankratz M, Tseng TW, Kaufman S, Liddle C, Yu RT, Downes M, Pfaff SL, Auwerx J, Gage FH, Evans RM. 2015. Dependence of hippocampal function on ERRgamma-regulated mitochondrial metabolism. Cell Metab 21:628–636. doi: 10.1016/j.cmet.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei L, Leblanc M, Barish G, Atkins A, Nofsinger R, Whyte J, Gold D, He M, Kawamura K, Li HR, Downes M, Yu RT, Powell HC, Lingrel JB, Evans RM. 2011. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat Med 17:1466–1472. doi: 10.1038/nm.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Lupino K, Wilkins BJ, Qiu C, Liu J, Omura Y, Allred AL, McDonald C, Susztak K, Barish GD, Pei L. 2018. Genomic integration of ERRgamma-HNF1beta regulates renal bioenergetics and prevents chronic kidney disease. Proc Natl Acad Sci U S A 115:E4910–E4919. doi: 10.1073/pnas.1804965115. [DOI] [PMC free article] [PubMed] [Google Scholar]