Influenza virus infection remains a major public health problem. Utilizing a recombinant green fluorescent protein-expressing influenza virus, we compared the in vivo transcriptomes of directly infected and uninfected bystander cells from infected mouse lungs and discovered many pathways uniquely regulated in each population. The Wnt signaling pathway was downregulated in directly infected cells and was shown to affect virus but not interferon production. Our study is the first to discern the in vivo transcriptome changes induced by direct viral infection compared to mere exposure to the lung inflammatory milieu and highlight the downregulation of Wnt signaling. This downregulation has important implications for understanding influenza virus pathogenesis, as Wnt signaling is critical for lung epithelial stem cells and lung epithelial cell differentiation. Our findings reveal a mechanism by which influenza virus may affect host lung repair and suggest interventions that prevent damage or accelerate recovery of the lung.
KEYWORDS: alveolar epithelial cells, transcriptome, Wnt signaling, influenza
ABSTRACT
Influenza virus outbreaks remain a serious threat to public health. A greater understanding of how cells targeted by the virus respond to the infection can provide insight into the pathogenesis of disease. Here we examined the transcriptional profile of in vivo-infected and uninfected type 2 alveolar epithelial cells (AEC) in the lungs of influenza virus-infected mice. We show for the first time the unique gene expression profiles induced by the in vivo infection of AEC as well as the transcriptional response of uninfected bystander cells. This work allows us to distinguish the direct and indirect effects of infection at the cellular level. Transcriptome analysis revealed that although directly infected and bystander AEC from infected animals shared many transcriptome changes compared to AEC from uninfected animals, directly infected cells produce more interferon and express lower levels of Wnt signaling-associated transcripts, while concurrently expressing more transcripts associated with cell death pathways, than bystander uninfected AEC. The Wnt signaling pathway was downregulated in both in vivo-infected AEC and in vitro-infected human lung epithelial A549 cells. Wnt signaling did not affect type I and III interferon production by infected A549 cells. Our results reveal unique transcriptional changes that occur within infected AEC and show that influenza virus downregulates Wnt signaling. In light of recent findings that Wnt signaling is essential for lung epithelial stem cells, our findings reveal a mechanism by which influenza virus may affect host lung repair.
IMPORTANCE Influenza virus infection remains a major public health problem. Utilizing a recombinant green fluorescent protein-expressing influenza virus, we compared the in vivo transcriptomes of directly infected and uninfected bystander cells from infected mouse lungs and discovered many pathways uniquely regulated in each population. The Wnt signaling pathway was downregulated in directly infected cells and was shown to affect virus but not interferon production. Our study is the first to discern the in vivo transcriptome changes induced by direct viral infection compared to mere exposure to the lung inflammatory milieu and highlight the downregulation of Wnt signaling. This downregulation has important implications for understanding influenza virus pathogenesis, as Wnt signaling is critical for lung epithelial stem cells and lung epithelial cell differentiation. Our findings reveal a mechanism by which influenza virus may affect host lung repair and suggest interventions that prevent damage or accelerate recovery of the lung.
INTRODUCTION
In the United States, 5 to 20% of the population suffers from seasonal influenza virus outbreaks each year (1). The severity of the disease is caused by the cytopathic effect of the virus in conjunction with lung infiltration of proinflammatory cells and inflammatory cytokine production. Thus far, three types of influenza viruses have been identified, types A, B, and C (2). The most common, influenza A virus (IAV), infects cells lining the respiratory tract, typically alveolar epithelial cells (AEC) and alveolar macrophages. Infection of type 2 AEC leads to productive replication and budding of new infectious virions, allowing the viral infection to spread within the lung (3). Previous studies have shown that in vitro infection of human lung epithelial cells with IAV leads to changes in the RNA transcriptome (4) and proteome (5). Studies conducted in vivo examining lung tissue from IAV-infected patients, mice, and birds have also shown changes in lung gene expression induced by infection (6–10). These observed alterations in gene transcripts from whole lung tissue, however, are the result of the combination of IAV-infected cells, bystander uninfected cells, and infiltrating immune cells. Thus, the changes between infected and bystander cells in vivo remain to be investigated.
Previous studies have indicated the important role of Wnt signaling in lung development and disease. Even single Wnt ligand manipulation was shown to have detrimental effects on lung development, as deletion of Wnt7b leads to perinatal death, which was attributed to respiratory failure, with early developing lungs demonstrating hypoplasia (11). Other studies show the importance of Wnt signaling, as both the complete lack of Wnt signaling as well as augmented Wnt signaling are capable of affecting lung morphology and AEC differentiation. Wnt5a deletion alone drastically altered lung development and decreased differentiation of AEC (12). The opposite effect was achieved through Wnt5a overexpression, resulting in increased differentiation of AEC (13). The role of Wnt signaling in adults is less well characterized. Wnt signaling may be necessary for adult lung homeostasis, as expressions of many Wnt signaling components are detected in transplant lung tissue (14). Previous studies have demonstrated that Wnt signaling controls stem cell niches, and AEC turnover occurs in normal homeostasis; therefore, these fluctuations in Wnt pathway expression may reflect cell turnover and responses to lung injury (15). Recently, Wnt signaling has been shown to maintain adult lung epithelial stem cell niches, and downregulation is necessary for differentiation of type II to type I AEC, highlighting the delicate balance that Wnt signaling plays in lung homeostasis and injury (16). However, how Wnt signaling is affected during lung infections such as influenza virus infection and what role it may play during infection remain unknown.
To determine directly what in vivo changes in RNA expression are induced in IAV-infected AEC and what changes occur indirectly in bystander uninfected AEC during IAV infection, we performed RNA sequencing (RNA-seq) on infected and uninfected bystander type 2 AEC isolated from lungs of mice infected with a recombinant green fluorescent protein (GFP)-expressing influenza virus (17) and type 2 AEC from uninfected mice. Our results reveal a number of unique differentially expressed genes and pathways within influenza virus-infected as well as bystander uninfected epithelial cells. Many pathways involved in an antiviral immune response were among the pathways most well represented in both GFP-positive (GFP+) directly infected AEC and GFP-negative (GFP−) bystander AEC transcriptomes. Uniquely, directly infected AEC exhibited reduced Wnt signaling and many pathways associated with cellular organization and polarity while demonstrating increased cell death pathways and apoptosis compared to bystander AEC. These results provide evidence for unique transcriptional expression profiles in directly infected AEC compared to bystander AEC that could be utilized to target virally infected cells in order to reduce influenza virus-induced morbidity and mortality.
RESULTS
Directly IAV-infected and bystander AEC have distinct transcriptomes.
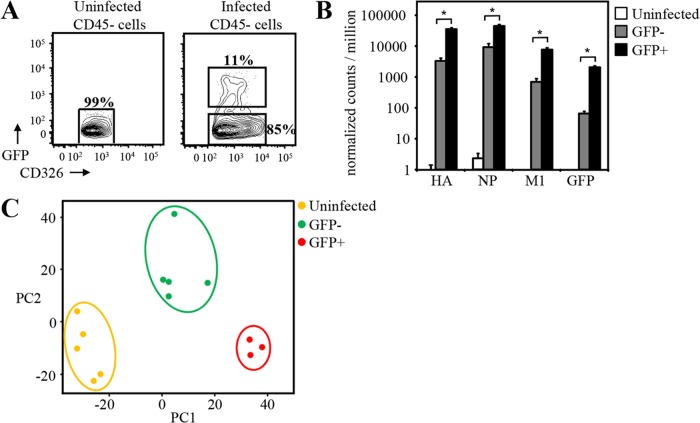
In order to determine the differences in the transcriptomes between cells directly infected by influenza virus and bystander cells exposed to the inflammatory milieu of a virally infected lung, C57BL/6 mice were intranasally infected with GFP-expressing A/Puerto Rico/8/1934 IAV (PR8-GFP) (17). On day 3 after infection with PR8-GFP, directly infected alveolar epithelial cells (CD45− CD326+ GFP+) as well as bystander uninfected cells (CD45− CD326+ GFP−) were fluorescence-activated cell sorter (FACS) sorted from the lungs of infected mice (Fig. 1A). Type 2 AEC (CD45− CD326+) from uninfected animals served as controls. RNA was isolated from these cells, and RNA-seq was performed. From these separate sorted populations, we first performed an initial screening of both influenza virus genes as well as GFP. As would be expected, the levels of these transcripts were highest in infected cells and undetectable in uninfected cells. Bystander cell population sequencing, however, showed some influenza virus transcripts. Although present in the bystander cells, we found that levels of IAV RNA and GFP RNA reads were 10- to 20-fold lower in bystander AEC than in infected AEC, and this corresponded to ∼5 to 10% contamination of infected cells in the bystander cell samples (Fig. 1B). These infection-associated transcripts can be explained by newly infected cells (GFP protein negative and viral RNA positive) being sorted with the GFP− bystander population.
FIG 1.
GFP and viral RNA expression by IAV-infected and bystander type 2 AEC sorted from GFP-flu-infected mice. (A) GFP expression in uninfected and GFP-flu-infected mice at day 3 postinfection (CD326+ CD45− cells are shown). Infected (CD326+ CD45− GFP+) and uninfected bystander (CD326+ CD45−) type 2 AEC were clearly distinguished by GFP expression and sorted. Type 2 AEC from uninfected mice were sorted as controls (CD326+ CD45−). (B) RNA-seq confirmed the infection state of cells sorted based on GFP expression. IAV hemagglutinin (HA), nuclear protein (NP), matrix protein (M1), and GFP RNA reads were determined by RNA-seq in bystander (GFP−), infected (GFP+), and uninfected AEC (*, adjusted P value of <0.05 by an unpaired t test). (C) Principal-component (PC) analysis of type 2 AEC (n = top 1,000 genes). (GFP+ refers to infected GFP+ AEC, and GFP− refers to bystander GFP− AEC.)
Principal-component analysis (PCA) of the transcriptomes of the different AEC showed a clear segregation based on their infection status (Fig. 1C). These findings revealed a unique and distinct transcriptome that characterizes AEC directly infected by IAV and those AEC that remained uninfected but were exposed to the inflammatory pulmonary milieu that results from IAV infection. Both of these AEC transcriptional profiles were distinct from each other and from that of AEC isolated from uninfected lungs (Fig. 1C).
The interferon response and antiviral transcripts are more abundant in directly infected AEC.
Analysis of the relative abundances of genes in the infected and bystander AECs revealed significant differences in how these cells respond to IAV infection (Fig. 2A to D). When comparing the magnitudes of relative mRNA abundances with respect to the comparisons of infected to uninfected AEC and bystander to uninfected AEC, the degree of change in mRNA abundance is more pronounced in the comparison of infected to uninfected AEC (Fig. 2A and B). As would be expected, the top 25 most abundant transcripts detected within infected compared to uninfected AEC were heavily enriched in antiviral and interferon-associated genes. Interferon beta 1 (Ifnb1) was the most abundantly represented gene, being 1,842-fold induced, and the abundances of other genes associated with interferon signaling were also increased, including interferon-stimulated gene 15 (Isg15) (433-fold), IFN-λ3 (Ifnl3) (360-fold), MX dynamin-like GTPase 1 (Mx1) (136-fold), 2′-5′-oligoadenylate synthetase 3 (Oas3) (128-fold induction), IFN-λ2 (Ifnl2) (120-fold), and interferon-induced protein with tetratricopeptide repeats 1 (Ifit1) (113-fold). Other transcripts with increased abundances in infected AEC have different functional properties for the anti-influenza immune response, including chemokine ligand 5 (Ccl5) (1,237-fold), which acts as a chemoattractant for CD8+ T cells in the lung (18). Apolipoproteins (Apol9a and Apol9b [245-fold and 214-fold, respectively]) have been suggested to have roles in tissue repair in addition to antiviral activity (19, 20). Z-DNA binding protein 1 (Zbp1) (159-fold) acts as a cytosolic DNA immune sensor for type I IFN production, while viperin, also known as radical S-adenosylmethionine domain containing 2 (Rsad2) (154-fold), is an interferon-inducible iron-sulfur cluster binding protein that is capable of inhibiting viral replication (21). Proteoglycan 2 (Prg2) (101-fold), also known as natural killer cell activator, was first purified from a T cell hybridoma and shown to augment NK cell function (22). When the transcriptome of bystander AEC was compared to that of uninfected AEC, levels of interferons and antiviral genes were also increased albeit to a lesser extent (Fig. 2B). From the data described above, it is apparent that the most abundantly represented genes in both infected and bystander AEC involve the interferons, interferon response genes, and other antiviral genes.
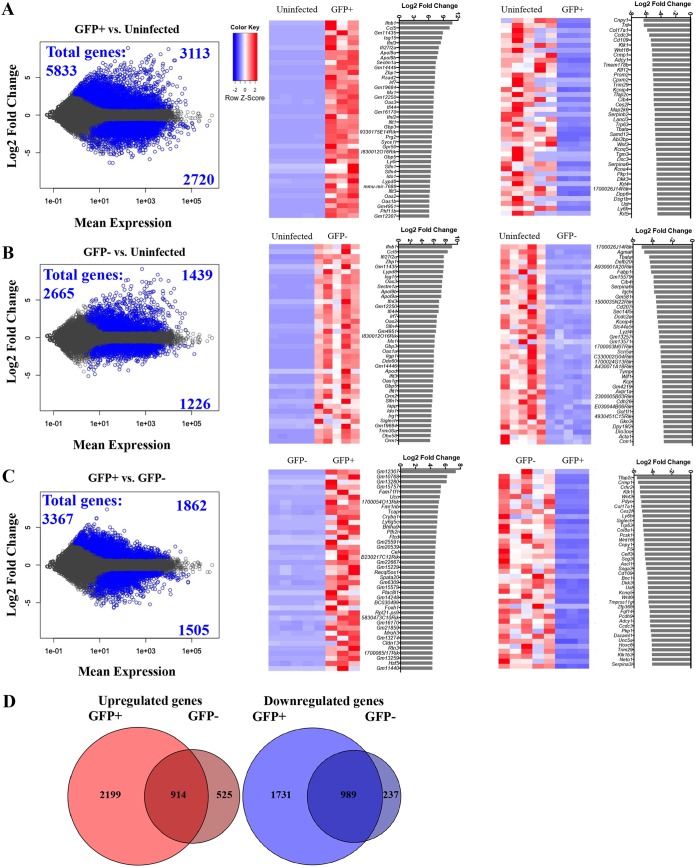
FIG 2.
Pairwise comparisons of in vivo influenza virus-infected, uninfected bystander, and uninfected type 2 AEC. (A to C) Genes that show differential expression (adjusted P value of <0.05) and a change of >2-fold between different infection statuses of AEC are shown in blue in plots on the left for comparisons of infected versus bystander cells (A), infected versus uninfected cells (B), and bystander versus uninfected cells (C). Infected cells refer to GFP+ infected AEC, and bystander cells refer to GFP− bystander AEC, both sorted from influenza virus-infected mice; uninfected cells refer to AEC sorted from uninfected animals. Heat maps show the 40 genes most differentially expressed under the different conditions. Normalized read count values of each library are centered and scaled for each gene. Bar graphs show the log2 fold changes (DESeq2) of the most increased and decreased genes for each comparison. (D) Venn diagrams of all unique and shared increased and decreased transcripts of infected and bystander type 2 AEC compared to uninfected type 2 AEC (GFP+ refers to infected GFP+ AEC, and GFP− refers to bystander GFP− AEC.)
As mentioned above, antiviral genes and the interferon response are enriched in epithelial cells following influenza virus infection. However, when the GFP+ infected AEC are compared to the GFP− bystander AEC, it becomes more evident which genes are most affected by direct influenza virus infection or by the lung inflammatory milieu of infection (Fig. 2C, left). Among these there are certain genes with a common defined function that are clearly represented in infected AEC (Fig. 2C, right). Many genes involved in Wnt signaling (Wnt3, Wnt16, Wnt6, and Dkk3) are among the most decreased transcripts in GFP+ infected AEC. A number of genes involved in cell adhesion and morphology can also be found in this list, including Fgf14 (fibroblast growth factor 14), Pcdh9 (protocadherin 9), Dscaml1 (Down syndrome cell adhesion molecule 1), Serpina3i (serpin peptidase inhibitor), Cnpy1 (canopy homologue 1), Col8a1 (collagen type VIII alpha chain 1), and Col17a1 (collagen type XVII alpha chain 1), among others. These data suggest that direct infection with influenza virus plays a role in altering Wnt signaling as well as cellular morphology and adhesion during infection.
Distinct signaling pathways are affected by direct influenza virus infection of AEC.
Compared to uninfected AEC, infected AEC showed the largest number of increased or decreased RNA transcripts, with 2- to 3-fold-higher abundances than in the bystander AEC. Infected and bystander AEC shared 914 genes that were significantly increased and 989 genes that were significantly decreased by more than 2-fold compared to uninfected AEC (Fig. 2D). However, there were also 2,199 genes that were uniquely increased in infected AEC and 525 genes that increased only in bystander AEC (Fig. 2D). Infected AEC also had 1,731 genes that were uniquely decreased, while in bystander AEC, 237 genes were uniquely decreased (Fig. 2D). Thus, direct infection of AEC with influenza virus results in much larger numbers of genes being affected during influenza virus infection when comparing the uniquely increased transcripts of infected AEC to those of bystander AEC.
In an effort to understand how the differentially expressed genes fit into canonical cellular pathways, totals of 5,833, 3,367, and 2,665 genes from the pairwise comparisons of infected versus uninfected AEC, infected versus bystander AEC, and bystander versus uninfected AEC, respectively, were analyzed using Ingenuity Pathway Analysis (IPA). These genes were selected based on criteria of a >2-fold change, >5 normalized counts per kilobase of gene (NCPKG), and an adjusted P value of <0.05. Using IPA to further enrich the analysis for the pathways that were affected by influenza virus infection, a combination of trend and score (Z-score for activation or inhibition) assigned by IPA was used to rank pathways based on differentially expressed genes from each of the pairwise comparisons (Fig. 3A). Pathways expected to be increased during an immune response were among the most affected pathways in both directly infected and bystander AEC and included interferon signaling, role of pattern recognition receptors (PRR) of bacteria and viruses, interleukin-8 (IL-8) signaling, NF-κB signaling, role of RIG-I-like receptors in antiviral innate immunity, and activation of interferon regulatory factor (IRF) by cytosolic pattern recognition receptors (Fig. 3A). Pathways such as death receptor signaling and apoptosis signaling were also increased in both directly infected and bystander AEC compared to uninfected AEC (Fig. 3A). These pathways most likely represent the response of AEC to the inflammatory milieu of the influenza virus-infected lung.
FIG 3.
Top canonical signaling pathways modulated by influenza virus infection. (A) IPA was used to generate a Treemap image depicting those biological processes affected by influenza virus infection in infected (GFP+) versus bystander (GFP−) AECs. Blue and orange represent negative and positive Z-scores, respectively, indicating that the biological process is increased or decreased based on transcripts contained in each category. PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-activated protein kinase; TGF-β, transforming growth factor β; DARPP32, dopamine- and cAMP-regulated neuronal phosphoprotein; eNOS, endothelial nitric oxide synthase; HMGB1, high-mobility group box 1; PTEN, phosphatase and tensin homolog; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; CREB, cAMP response element binding protein; GNRH, gonadotropin-releasing hormone; PCP, planar cell polarity; IRF, interferon regulatory factor; IL-1, interleukin-1; LPS, lipopolysaccharide; RXR, retinoid X receptor; MAPK, mitogen activated protein kinase; NFAT, nuclear factor of activated T cells; GDNF, glial cell line-derived neurotrophic factor; NF-κB, nuclear factor κB; PPAR, peroxisome proliferator-activated receptor; UVA, ultraviolet A; NRF2, nuclear factor erythroid 2-related factor 2. (B) The most significant canonical pathways based on the score and direction of change of positive correlation for bystander AEC were used to generate a heat map based upon each pairwise comparison. Blue and orange represent negative and positive Z-scores, respectively. (GFP+ refers to infected GFP+ AEC, and GFP− refers to bystander GFP− AEC.)
To visualize those biological trends affected by direct influenza virus infection, Treemap images were constructed based upon pathway activation or inhibition in combination with a Z-score and a P value generated by IPA based upon the differentially expressed individual profiles in the pairwise comparison of directly infected and bystander AEC. In accordance with the Venn diagrams, which suggest a reduced abundance of transcripts in the directly infected AEC (Fig. 2D), the Treemap image constructed based upon the comparison of GFP+ directly infected AEC compared to GFP− bystander AEC illustrated that direct influenza virus infection had a negative impact on many cellular functions and signaling pathways based on the Z-score and the P value (Fig. 3B; see also Table S1 in the supplemental material). Among the biological processes reduced were those involved in cell morphology, movement, assembly and organization, and growth and proliferation, while the associated functions were related to differentiation of cells, organization of the cytoskeleton, organization of the cytoplasm, and cell viability. Only a few pathways were positively impacted by influenza virus infection, and these pathways were cell death and survival and organismal injury and abnormalities, which are all related to cell death, necrosis, and apoptosis (Fig. 3B and Table S1). These data suggest that the directly infected AEC are effectively shutting down cellular functions and initiating programmed cell death.
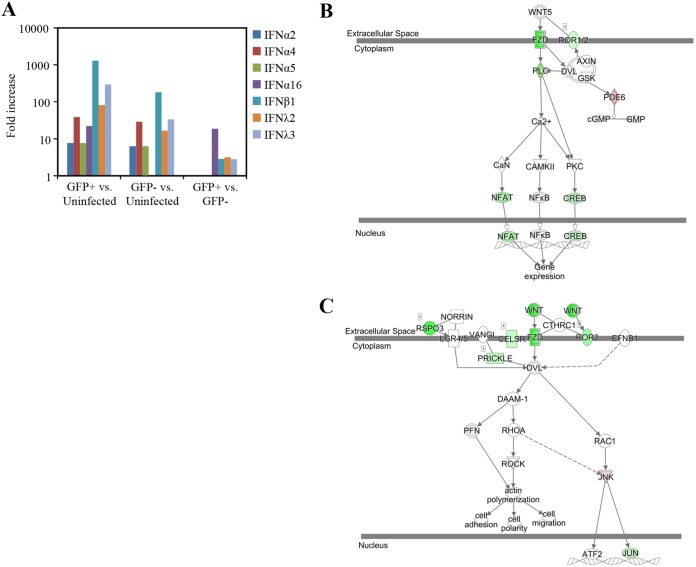
As mentioned above, the IFN pathway genes were among the most increased in AEC from infected animals. Within the 10 most abundant transcripts in the infected AEC compared to uninfected AEC, we observed a 1,842-fold increase in the Ifnb1 mRNA abundance, along with a 360-fold increase in the Ifnl3 abundance and a 152-fold increase in IRF7 transcript expression (Fig. 4A) (GFP-Influenza database). In contrast, in bystander AEC, a more modest 551-fold increase in Ifnb1 transcripts, a 118-fold increase in Ifnl3 transcripts, and a 91-fold increase in Irf7 transcripts were found (Fig. 4A) (GFP-Influenza database). Uniquely, the abundance of IFN-α16 (also known as IFN-α6T) is increased in the infected AEC, with a 42-fold increase over the abundance in uninfected cells (Fig. 4A). The abundance of IFN-α16 was not increased in bystander AEC. It is not known whether the IFN-α16 gene is responsive to viral infection, as previous studies have shown that the promoter sequences of IFN-α16 contain mutations in the predicted IRF binding motifs, and thus, this IFN′s promoter may not be bound by IRF7 (23). These results suggest that infected cells, on a per-cell basis, are producing more antiviral cytokines than bystander cells.
FIG 4.
Interferons and Wnt pathways are differentially expressed in infected type 2 AEC. (A) Type I and III interferons are more abundant in influenza virus-infected type 2 AEC cells. All comparisons were statistically significant. (B and C) Pathway images from IPA illustrating genes associated with the Wnt/Ca2+ noncanonical signaling pathway (B) and the planar cell polarity pathway (C) that showed differential expression are highlighted in color. The color intensity indicates the degree of upregulation (red) or downregulation (green) within the pathway. (GFP+ refers to infected GFP+ AEC, and GFP− refers to bystander GFP− AEC.)
Direct influenza virus infection of AEC selectively decreases the Wnt pathway.
Among the pathways that were identified by IPA to be differentially regulated in directly infected AEC compared to bystander AEC was the Wnt signaling pathway. Both the Wnt/Ca+ pathway (Fig. 4B) and the planar cell polarity (PCP) pathway (Fig. 4C) were decreased in directly infected AEC (see Table S1 in the supplemental material) compared to bystander AEC. These pathways were not affected in bystander AEC when they were compared to those in uninfected AEC (Table S1). Previous in vitro research has shown that the Wnt signaling pathway intersects with viral infection and that different Wnt ligands can have different effects on interferon production (24, 25). Two Wnt-related signaling pathways were decreased in infected AEC compared to bystander AEC, the Wnt/Ca+ pathway and the PCP pathway (Fig. 4B and C), while CTNNB1 (which encodes β-catenin) was identified as one of the top upstream regulators inhibited (Table S2). The top upstream regulator analysis identifies a common transcription factor or signaling molecule that could explain the changes observed in global gene transcription. Within these pathways, ligands, receptors, and activators were among the genes with decreased abundances within GFP-positive directly infected cells compared to uninfected bystander cells. When considering only Wnt ligands, many were found to have decreased abundances within infected AEC compared to bystander AEC, including Wnt3 (39-fold), Wnt16 (30-fold), Wnt6 (23-fold), Wnt10a (9-fold), and Wnt4 (2.6-fold). In addition, the abundances of known receptors for Wnt ligands, including Frizzled 10 (Fzd10) (26) and Ror2 (27), were also reduced 17-fold and 4-fold in infected AEC compared to bystander AEC, respectively.
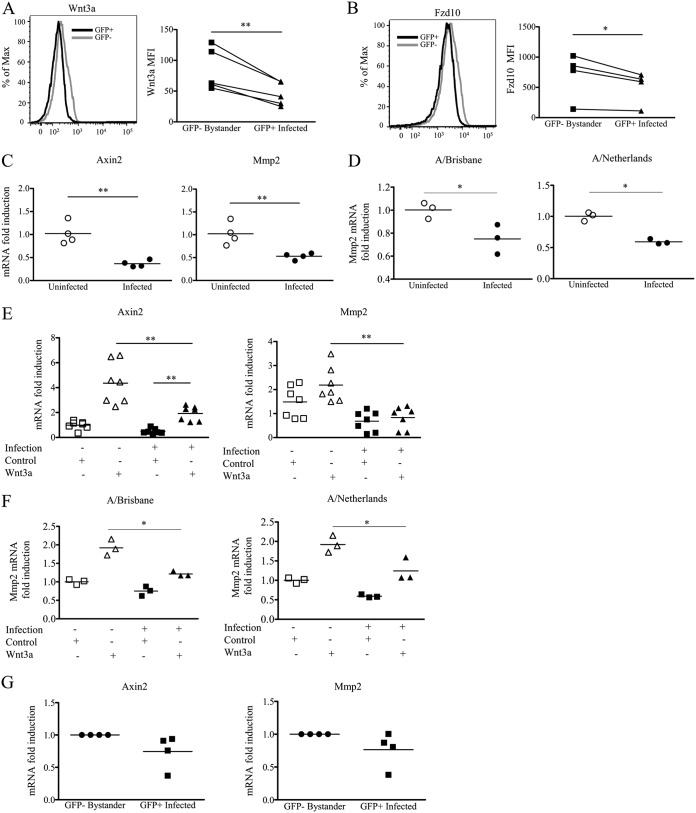
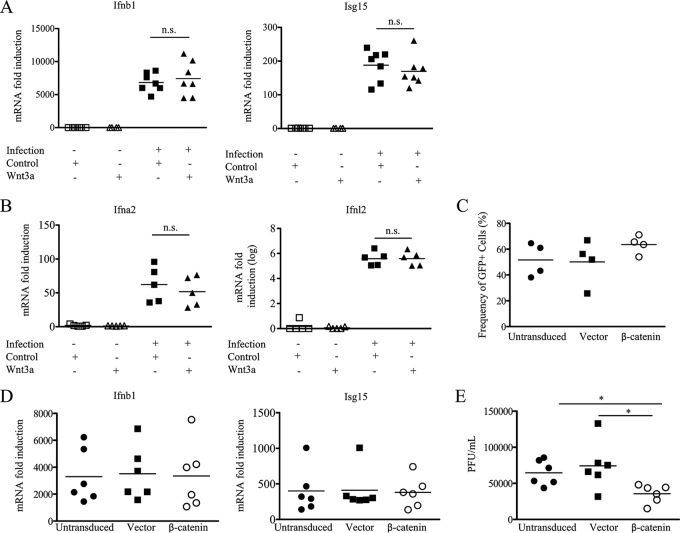
To confirm these findings in vitro and in a human model of influenza virus infection, we infected A549 human lung epithelial cells with PR8-GFP influenza virus. Using flow cytometry to quantitate protein expression, we examined PR8-GFP-infected A549 cell cultures for the expression of the Wnt ligand Wnt3a and the Wnt receptor Fzd10. GFP-positive infected cells compared to GFP-negative uninfected cells from the same cultures showed reduced protein levels for Wnt3a (Fig. 5A) and Fzd10 (Fig. 5B), confirming our transcriptome results. Reduced Wnt signaling in these cells was indicated by the lower fold induction of downstream Wnt-associated transcripts, axin 2 (Axin2) and matrix metalloprotease 2 (Mmp2), in infected A549 cells (Fig. 5C). As PR8-GFP is a laboratory-adapted strain, we also infected A549 cells with primary seasonal influenza virus isolates and found similar downregulation of Wnt-dependent transcripts following infection (Fig. 5D). To further confirm the active downregulation of Wnt signaling, we treated PR8-GFP-infected A549 cells with conditioned medium containing Wnt3a. The addition of exogenous Wnt3a to uninfected cells upregulated the Wnt/β-catenin-associated transcripts Axin2 and Mmp2 (Fig. 5E and F). However, when exogenous Wnt3a was added to A549 cells infected with PR8-GFP or primary seasonal influenza virus isolates, infected cells failed to upregulate the Wnt/β-catenin-associated transcripts Axin2 and Mmp2 to the same extent as their uninfected counterparts (Fig. 5E and F). Additionally, in order to confirm the active Wnt signaling downregulation in influenza virus-infected A549 cells, infected cell cultures were sorted based on GFP expression to determine whether infected cells have fewer β-catenin-dependent transcripts. Sorted GFP-positive directly infected A549 cells demonstrated a trend toward lower levels of Wnt-dependent transcripts at 12 h postinfection than sorted GFP-negative A549 cells from the same cultures. A limitation of this assay is that newly infected cells may not express GFP at 12 h postinfection and thus could significantly contaminate the truly GFP-negative noninfected population (Fig. 5G). It should be noted that the GFP-negative and GFP-positive A549 cells in these cultures had significant downregulation of Wnt-dependent transcripts, which were ∼40% less abundant than those in A549 cells from uninfected cultures (data not shown). Previous studies have shown that transfection of the pandemic 1918 virus PB2 protein and, to a lesser extent, of an avian PB2 protein was capable of reducing the induction of Wnt/β-catenin-associated transcripts after the addition of a recombinant Wnt3a (10). This observation together with previous literature that demonstrated that Wnt signaling could potentiate the interferon response (28) led previously to the proposition that influenza virus may modulate type I IFN production via the Wnt pathway as an evasion mechanism (10, 28). To test this, we examined Ifnb1 and Isg15 transcript levels after Wnt3a treatment of A549 cells. However, we found no difference in Ifnb1 or Isg15 transcripts after exogenous Wnt3a addition to infected cultures (Fig. 6A). This observation was also true for other type I interferons and type III interferons (Fig. 6B). Thus, any effects of Wnt signaling during influenza virus infection appear to be IFN independent. This result could be due to downregulation of the Wnt receptor, and in order to circumvent this problem, we retrovirally expressed a constitutively active β-catenin molecule in A549 cells to activate the canonical Wnt signaling pathway. When we infected these cells with influenza virus, we found no difference between groups in terms of infectivity as determined by the frequency of GFP-positive cells in infected cultures where more than 60% of the cells were GFP positive (Fig. 6C), nor did we find any difference in Ifnb1 transcripts or the induction of interferon-stimulated genes (Isg15) (Fig. 6D). However, cells transduced with the constitutively active β-catenin molecule produced less infectious virus than both control transduced and untransduced A549 cells, as determined by plaque assays on the supernatants of infected cell cultures (Fig. 6E). How β-catenin signaling affects infectious virus production, however, remains to be elucidated (29). The above-described observations suggest that Wnt signaling negatively impacts virus production independently of an effect on Ifnb1. From these studies and the studies of others (24, 28), it is clear that Wnt signaling is decreased after influenza virus infection.
FIG 5.
Wnt signaling within infected epithelial cells is reduced compared to that in bystander uninfected epithelial cells. (A and B) Representative histograms of reduced expression of Wnt3a (A) and Fzd10 (B) in PR8-GFP-infected (GFP+) A549 cells compared to bystander GFP− cells. Dot plots display the mean fluorescence intensity (MFI) 48 h after infection with influenza virus after a single-round infection. Each dot represents data from a separate independent experiment (n ≤ 4 experiments; paired t test). (C and D) Wnt-stimulated gene transcripts from infected A549 cell cultures 16 h after infection with PR8-GFP (C) or the seasonal influenza viruses A/Brisbane/10/2007 (A/Brisbane) and A/Netherlands/602/209 (A/Netherlands) (D). (E and F) Wnt-dependent transcript downregulation was also observed in the presence of exogenous Wnt3a 12 to 16 h after infection with PR8-GFP (E) and seasonal influenza viruses (F). (G) Real-time PCR analysis of Wnt-dependent transcripts from A549 cells infected with PR8-GFP and sorted at 12 h postinfection (n ≥ 3 experiments) (*, P ≤ 0.05; **, P ≤ 0.01 [by an unpaired t test or a Mann-Whitney U test]). (GFP+ refers to infected GFP+ AEC, and GFP− refers to bystander GFP− AEC.)
FIG 6.
Wnt signaling does not affect IFN gene induction in infected cells. (A and B) Induction of Ifnb1 and Isg15 transcripts (A) and Ifna2 and Ifnl2 transcripts (B) assessed after treatment of PR8-GFP influenza virus-infected cells with exogenous Wnt3a 12 to 16 h after infection of A549 cells after a single-round infection. Fold induction was determined by comparison to uninfected cell cultures. (C and D) Constitutive expression of the active β-catenin molecule in A549 cells does not change the frequency of GFP+ cells when infected with PR8-GFP influenza virus at 24 h postinfection (C) or the induction of Ifnb1 and Isg15 in PR8-GFP influenza virus-infected cells (D). (E) MDCK plaque assays quantifying the number of PFU produced in the cell supernatants of infected cell cultures per milliliter collected 48 h after infection with PR8-GFP at a MOI of 1 (n ≥ 3 experiments) (*, P ≤ 0.05; n.s. or no asterisk, nonsignificant result [by a paired t test]).
DISCUSSION
Our studies have determined the transcriptome of infected AEC and bystander uninfected AEC from an in vivo mouse IAV infection, showing for the first time that unique gene expression profiles are induced by direct IAV infection. The transcriptome profile of AEC isolated from infected lungs showed universally increased transcript abundances for pathways involved in the early immune response to IAV, including interferon signaling, chemokine signaling, and cell death. The inflammatory milieu and viral products in the lung induce gene expression changes in bystander uninfected cells, but the presence of virus in infected AEC induced more potent and unique changes.
Of particular interest among the pathways affected by influenza virus were the interferon, Wnt, and cell death pathways. These pathways seemed to be differentially affected by influenza virus infection between GFP-negative bystander cells and GFP-positive infected cells. The interferon signaling pathway showed mainly increases in many interferon transcripts, most likely due to PRR activation within the cytoplasm by viral products that augmented the production of interferons. Interestingly, unique interferon induction, like that of IFN-α16, within directly infected AEC suggested that different interferon subtypes could play a role in tempering the immune response in infected cells, as different interferon alpha subtypes have been shown to possess different capacities to stimulate interferon-regulated genes (30).
Although the above-mentioned pathways related to the antiviral response were increased within infected AEC, the majority of pathways were decreased compared to those in bystander uninfected AEC. Among these decreased pathways were the Wnt and NF-κB signaling pathways. Wnt signaling is a pathway involved in many aspects of cellular development, proliferation, structure, polarization, and differentiation (31). Wnt signaling includes numerous molecules and transcriptional targets but is essentially divided into three pathways: the canonical Wnt signaling pathway (which is β-catenin dependent) and two noncanonical pathways, the Wnt/calcium pathway and the PCP pathway (32). Stimulation of cells with Wnt3a is the most common method used to signal through the canonical pathway. The interplay between influenza virus and the Wnt pathway remains unclear (24, 28, 33). Using a cell reporter system to quantify interferon-stimulated response element (ISRE) promoter activity, Shapira et al. showed that pretreatment with Wnt3a could potentiate the interferon response when cells were transfected with viral RNA or infected with an NS1-deleted influenza virus (24). Other studies using A549 cells demonstrated that when cells were transiently transfected with a constitutively active β-catenin molecule, viral progeny were reduced when infected with an avian influenza virus (28). In contrast, Karlas et al. showed that with small interfering RNA (siRNA)-mediated knockdown of Wnt ligands, there was strong inhibition of influenza virus replication (33). Those studies, however, did not address how Wnt signaling is affected by influenza virus infection in an in vivo infection. Our data show, for the first time, that influenza virus-infected cells uniquely downregulate Wnt receptors and ligands compared to bystander cells for the same infected lungs. Our in vitro studies further confirmed that infection of A549 cells downregulates Wnt pathway signaling. In addition, our studies confirm that infection of A549 cells that express a constitutively active β-catenin produce less virus than control cell. This occurred despite interferon transcription and induction of interferon-stimulated genes remaining unaffected by constitutive Wnt signaling. As all three Wnt signaling pathways were downregulated with in vivo infection of AEC, this could suggest a more complex response to influenza virus infection.
Although previous research has shown that Wnt signaling is affected during viral infection and could possibly affect the antiviral response (10, 24, 25, 28), most of those studies focused on canonical Wnt signaling, but we are the first to show in an in vivo model the decreased PCP and Wnt/Ca+ pathways in influenza virus infection. Fzd10 has been reported to be expressed in the airway epithelium (26), but its role remains unclear, while another receptor, Ror2, is a receptor tyrosine kinase that has been implicated in modulating the balance between canonical and noncanonical Wnt signaling in the lungs (27). A number of studies have suggested that Wnt signaling enhances type I IFN signaling, and influenza virus has been reported to suppress Wnt signaling (10, 24, 28). Thus, the diminished expression of the Wnt pathway found specifically within influenza virus-infected AEC was proposed to reflect a strategy employed by IAV to mitigate IFN signaling and its antiviral effect. Our studies, however, using cells with constitutively active β-catenin and Wnt3a-treated cells suggest that Wnt signaling is not affecting type I IFN production and signaling during influenza infection. Wnt signaling, particularly the Wnt/calcium pathway and the PCP pathway, is responsible for the internal structure and organization of the cell. Loss of these pathways can compromise the cell's orientation and function and result in Jun N-terminal protein kinase (JNK)-induced cell death (34, 35). In addition, junctional protein expression and function have been shown to play a major role in cellular apoptosis, with some proteins (e.g., connexins) being found to be in close association with apoptosis-associated proteins (36). Activation of this cell death pathway is prevented in normal cells by simultaneous activation of NF-κB signaling (34). Interestingly, we find that NF-κB signaling is also downregulated in the GFP-positive AEC compared to the GFP-negative AEC, suggesting that the directly infected cells are initiating a programmed cell death pathway. Indeed, among the few biological functions increased during infection were those involved in cell death and apoptosis. As PR8 infection models a severe influenza virus infection, the lung damage induced in the alveolar epithelia is considerable, and this downregulation of Wnt signaling may play a role in the pathogenicity of influenza virus, as seen by others (37). Downregulation of all three Wnt signaling pathways, involved in maintaining the structure and positioning of cells, would be expected to compromise the function of the lung as well as epithelial repair and likely contributes to the pathogenicity observed in the lung during influenza virus infection. In line with our conclusions, previous studies also found that augmentation of β-catenin signaling by lithium chloride resulted in reduced viral progeny, and inversely, reduced epithelial repair was observed due to downregulation of fibroblast growth factor receptor 2b (fgfr2b) in epithelial cells (37). Indeed, in our data set, fgfr2b was significantly downregulated by ∼1.6-fold in influenza virus-infected AEC (37). Thus, in addition to the inhibitory effect that Wnt signaling may have on virus production, it also plays a role in epithelial repair. Wnt signaling downregulation may on the one hand benefit the virus but also affect tissue repair and damage. Targeting Wnt signaling therefore may serve as a means to mitigate damage and inhibit viral replication.
Recent studies have suggested that Wnt signaling in subsets of type 2 AEC maintains their stem cell-like qualities, and reduction of this signaling promotes their differentiation into type 1 AEC (16). Thus, the downregulation of Wnt signaling that we observe with influenza virus infection may also be a host response in order to promote the differentiation of type 2 AEC to type 1 AEC to replenish the pool of cells compromised during influenza virus infection. Interestingly, studies performed in a mouse model of chronic obstructive pulmonary disease found that Wnt5a impaired lung healing (38), which is one of the main Wnt proteins that we find to be downregulated in our data set (Fig. 4B). The downregulation of this pathway, specifically within infected cells, may promote influenza pathogenicity, as Wnt signaling is required for the renewal of type 2 AEC (16). Therefore, Wnt signaling downregulation by influenza virus may in the short term replenish type 1 AEC but in the long term impair host lung repair programs by depleting the renewal of type 2 AEC. Given the complex effects of Wnt signaling on AEC and the lung, additional studies are needed to address whether Wnt administration can improve or accelerate lung recovery from influenza virus.
Our in vivo transcriptome studies give further insight into the host response to IAV infection, allowing us to isolate many of the individual contributions of directly infected and bystander AEC in the lungs of infected animals. Our findings identify signaling pathways and genes that are affected in influenza virus-infected cells and may provide targets of intervention to reduce the morbidity and mortality associated with IAV infections.
MATERIALS AND METHODS
Animals.
All mice were housed at Drexel University in certified barrier facilities in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care (AAALAC). All animal work was reviewed and approved by the Drexel University Institutional Animal Care and Use Committee (IACUC). Animal research with mice was conducted under the approved IACUC protocol 210108. The use of 9-day-old embryonated chicken eggs was exempt from review by the IACUC. Embryonated specific-pathogen-free (SPF) eggs were purchased from a commercial vendor (B&E Eggs, PA). All animal work was carried out in compliance with U.S. government regulations, including the Animal Welfare Act and Public Health Service Policy on Humane Care and Use of Laboratory Animals (39).
Virus and cells.
The recombinant A/Puerto Rico/8/34/GFP influenza virus (GFP-flu H1N1 strain, from A. García-Sastre) was propagated in 9-day-old chicken eggs and titrated by a plaque assay as previously described (40). Seasonal influenza virus strains A/Brisbane/10/2007 and A/Netherlands/602/2009 were generous gifts from Guus Rimmelzwaan (Department of Virology, Erasmus MC, University Medical Center Rotterdam). The human lung epithelial line A549 (ATCC, Manassas, VA) was propagated in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco), 2 mM glutamine (Gibco), 100 U/ml penicillin (Gibco), and 100 U/ml streptomycin (Gibco) at 37°C in 5% CO2. Influenza viral infections were performed at a multiplicity of infection (MOI) of 1, and cells were harvested for downstream analysis at the indicated time points.
Animal infections and cell sorting.
Specific-pathogen-free female C57BL/6 mice were purchased from Jackson Laboratories. Mice were anesthetized with 2,2,2-tribromoethanol (250 mg/kg of body weight, injected intraperitoneally) (Avertin; Acros) and infected intranasally with a sublethal dose (1 × 106 PFU) of GFP-flu. At 3 days postinfection, lungs of infected and uninfected control mice were harvested and processed into single-cell suspensions. Cells were panned twice on anti-CD16/32 (Fc Block, 2.4G2; BD Biosciences) and anti-CD45 (30-F11; BD Biosciences) antibody-coated petri dishes to enrich for type 2 AEC. Panned cells were then stained as previously described (41), using monoclonal anti-CD45.2 (104; eBioscience) and anti-CD326 (G8.8; eBioscience) antibodies before sorting on a FACSAria instrument (BD Biosciences). Sorted type 2 AEC were then frozen in TRIzol LS reagent (Life Technologies) at −80°C.
Type 2 AEC RNA extraction, library preparation, and sequencing.
RNA was extracted from sorted type 2 AEC and quantified by UV spectrophotometry (NanoDrop). A bioanalyzer (Agilent) was used to determine the integrity of the extracted RNA. Barcoded sequencing libraries were generated using a TruSeq stranded mRNA kit (Illumina). Quality of libraries was assessed via the bioanalyzer. Kapa quantitative PCR (qPCR) was used to quantify fragments with adapters on both ends. Samples with >5% adapter dimer content were repurified using solid phase reversible immobilization (SPRI) beads. Samples were then diluted and shipped to Beijing Genomics Institute (BGI) (Hong Kong, China). Library quality and integrity were reassessed via a bioanalyzer and Kapa qPCR before cluster generation and 100-bp single-end sequencing. Libraries were sequenced with HTSeq2000 Illumina technology.
Processing and genomic mapping of RNA-seq libraries.
RNA-seq libraries were trimmed from library preparation adapters and low-quality ends with Trim Galore (version 0.3.5 [http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/]) using default parameters. Mapping against the mouse mm10 genomic version (Mus_musculus.GRCm38) was carried out using Tophat2 (42) (v2.0.11, with —p 6 —g 1 parameters), providing the Mus_musculus.GRCm38.70.gtf annotation.
Analysis of differential transcript abundance, normalization of read counts by gene size, and downstream analyses.
Mapped reads were counted along genes with HTSeq (29), using default parameters and the —s reverse option. Mus_musculus.GRCm38.75.gtf annotation was used for counting. Differential expression analysis and normalization of read counts of all the libraries together were done with DESeq2 (v 1.4.5) (43). The results for the different pairwise comparisons were extracted with the contrast argument. Normalized read counts of each gene were corrected by the gene size (sum of all the exons in the Mus_musculus.GRCm38.75.gtf annotation) using R and the GenomicFeatures (44) library and expressed as NCPKG (considering only exonic regions). The NCPKG was used to filter out genes with low expression (<5 NCPKG). Statistically significant gene lists for each pairwise comparison (adjusted P value of <0.05 with a fold change of >2 and >5 NCPKG) were used for the selection of significant canonical pathways and molecular networks using Ingenuity Pathway Analysis (IPA; Qiagen, USA). Pathway enrichment P values (Fisher's exact test) and activation Z-scores were calculated by IPA and used to rank the significant pathways.
A549 infections.
A549 cells were plated on 6-well plates (Corning Life Sciences) at a concentration of 1.0 × 106 cells per well. For multiple-round infections, cells were incubated with recombinant PR8-GFP influenza or seasonal influenza virus at a multiplicity of infection (MOI) of 1 for 2 h at 37°C, washed, and incubated with serum-free medium in the presence of trypsin-EDTA (Fisher Scientific) for 48 h. For single-round infections, cells were infected in the same manner as described above, with the exception being that after infection, serum-containing medium was replaced, or cells were incubated with conditioned medium containing Wnt3a or control conditioned medium until the indicated time point. Cells were subsequently resuspended in TRIzol reagent (Life Technologies).
RNA extraction and real-time PCR.
Primary mouse type 2 AEC or A549 cell RNA was extracted using the RNeasy minikit (Qiagen). Extracted RNA was reverse transcribed using a high-capacity cDNA kit (Applied Biosystems) and analyzed using a 7900HT fast real-time PCR machine (Applied Biosystems). TaqMan gene expression assays for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (assay Hs00610344_m1), Axin-2 (Axin2) (assay Hs00610344_m1), matrix metalloproteinase 2 (Mmp2) (assay Hs01548727_m1), IFN-β (Ifnb1) (assay Hs01077958_s1), IFN-α2 (Ifna2) (assay Hs00265051_s1), IFN-λ2 (Ifnl2) (assay Hs00820125_g1), and interferon-stimulated gene 15 (Isg15) (assay Hs01921425_s1) were obtained from ThermoFisher Scientific. Statistical analysis was performed using GraphPad Prism software.
A549 cell staining and analysis.
Cells were stained as previously described (41). For annexin V staining, all buffers contained 2.5 mM CaCl2. Cells were permeabilized according to the eBioscience IC fixation buffer protocol and incubated with either of the following monoclonal antibodies: anti-Wnt3a (clone 217804; R&D Systems) or a rat IgG2A allophycocyanin (APC) isotype control (clone 54447; R&D Systems). Alternatively, cells were incubated with anti-Frizzled 10 (Fzd10) (rabbit polyclonal, catalogue number sc-368103; Santa Cruz). Cells were washed with permeabilization buffer and incubated with a goat anti-rabbit IgG phycoerythrin (PE)-conjugated cross-adsorbed secondary antibody (catalogue number 4052-09; Southern Biotech). Cells were washed after all incubations, fixed in 1% paraformaldehyde (PFA) with 2.5 mM CaCl2, and analyzed by using an LSR Fortessa instrument. All data were analyzed with FlowJo software (TreeStar).
MDCK plaque assays.
Supernatants were collected from infected cell cultures and frozen at −80°C prior to quantification. Madin-Darby canine kidney (MDCK) cells (ATCC) were seeded at a concentration of 3.0 × 106 cells per well in 6-well plates on the day prior to infection. Cells were washed with serum-free medium and incubated with dilutions of infected supernatants for 1 h with mixing. The medium containing virus was removed, and cells were layered with a prewarmed mixture of 30 ml of a 1.8% agarose mixture; 30 ml of Lonzo Leibovitz L-15 modified medium (catalogue number 21083-027; Life Technologies) supplemented with sodium bicarbonate (catalogue number 25080060; Life Technologies), HEPES buffer (catalogue number 15630-056; Life Technologies), and penicillin-streptomycin (catalogue number 15070063; Life Technologies); and 120 μl of a 0.l% trypsin–tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) (catalogue number LS003740; Worthington Biochemical)-treated solution. Cells were then incubated at 37°C for 72 h prior to fixation with 10% paraformaldehyde (catalogue number 15710; Brunschwig Chemie) for 30 min and staining with a 0.5% crystal violet (catalogue number 204330050; ThermoFisher Scientific)–methanol solution for 30 min. Plaques were then quantified, and PFU were back-calculated based upon the dilution.
Accession number(s).
RNA-seq data were deposited in the Gene Expression Omnibus (GEO) under accession number GSE119123.
Supplementary Material
ACKNOWLEDGMENTS
E.M.-C., S.L., and M.T. were supported by the Biotechnology and Biological Sciences Research Council. This work was partially supported by NIAID grant U19AI106754 and by CRIP (Center for Research in Influenza Pathogenesis), an NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS) (contract number HHSN272201400008C) (to A.G.-S.). This work was supported by funds from the Drexel University College of Medicine and Erasmus MC, University Medical Center Rotterdam (to P.D.K.).
Primary influenza virus isolates and help with infections were kindly provided by the Department of Virology at Erasmus MC, University Medical Center Rotterdam, specifically Theo Bestebroer and Guus Rimmelzwaan. Wnt3a conditioned medium was a generous gift from Tokameh Mahmoudi, Elise de Crignis, and Shahla Romal (Department of Biochemistry, Erasmus MC, University Medical Center Rotterdam).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01325-18.
REFERENCES
- 1.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Chau NVV, Khanh TH, Dong VC, Qui PT, Van Cam B, Ha DQ, Guan Y, Peiris JSM, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrhardt C, Ludwig S. 2009. A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell Microbiol 11:863–871. doi: 10.1111/j.1462-5822.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, Mason RJ. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am J Respir Cell Mol Biol 45:582–591. doi: 10.1165/rcmb.2010-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti A, Vipat V, Mukherjee S, Singh R, Pawar S, Mishra A. 2010. Host gene expression profiling in influenza A virus-infected lung epithelial (A549) cells: a comparative analysis between highly pathogenic and modified H5N1 viruses. Virol J 7:219. doi: 10.1186/1743-422X-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroeker AL, Ezzati P, Halayko AJ, Coombs KM. 2012. Response of primary human airway epithelial cells to influenza infection: a quantitative proteomic study. J Proteome Res 11:4132–4146. doi: 10.1021/pr300239r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Brahmakshatriya V, Lupiani B, Reddy S, Soibam B, Benham A, Gunaratne P, Liu H, Trakooljul N, Ing N, Okimoto R, Zhou H. 2012. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genomics 13:278. doi: 10.1186/1471-2164-13-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pommerenke C, Wilk E, Srivastava B, Schulze A, Novoselova N, Geffers R, Schughart K. 2012. Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses. PLoS One 7:e41169. doi: 10.1371/journal.pone.0041169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parnell G, McLean A, Booth D, Armstrong N, Nalos M, Huang S, Manak J, Tang W, Tam O-Y, Chan S, Tang B. 2012. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong H, Morrison J, Ferris MT, Gralinski LE, Whitmore AC, Green R, Thomas MJ, Tisoncik-Go J, Schroth GP, Pardo-Manuel de Villena FF, Baric RS, Heise MT, Peng X, Katze MG. 2014. Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression. G3 (Bethesda) 4:1429–1444. doi: 10.1534/g3.114.011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forero A, Tisoncik-Go J, Watanabe T, Zhong G, Hatta M, Tchitchek N, Selinger C, Chang J, Barker K, Morrison J, Berndt JD, Moon RT, Josset L, Kawaoka Y, Katze MG. 2015. The 1918 influenza virus PB2 protein enhances virulence through the disruption of inflammatory and Wnt-mediated signaling in mice. J Virol 90:2240–2253. doi: 10.1128/JVI.02974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu W, Jiang YQ, Lu MM, Morrisey EE. 2002. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development 129:4831–4842. [DOI] [PubMed] [Google Scholar]
- 12.Rieger ME, Zhou B, Solomon N, Sunohara M, Li C, Nguyen C, Liu Y, Pan JH, Minoo P, Crandall ED, Brody SL, Kahn M, Borok Z. 2016. p300/beta-catenin interactions regulate adult progenitor cell differentiation downstream of WNT5a/protein kinase C (PKC). J Biol Chem 291:6569–6582. doi: 10.1074/jbc.M115.706416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Hu L, Xiao J, Chen H, Li JT, Bellusci S, Delanghe S, Minoo P. 2005. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol 287:86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 14.Konigshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. 2008. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One 3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotton DN, Morrisey EE. 2014. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med 20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabhan A, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. 1 February 2018. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galkina E, Thatte J, Dabak V, Williams MB, Ley K, Braciale TJ. 2005. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J Clin Invest 115:3473–3483. doi: 10.1172/JCI24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L, Miyoshi H, Origanti S, Nice TJ, Barger AC, Manieri NA, Fogel LA, French AR, Piwnica-Worms D, Piwnica-Worms H, Virgin HW, Lenschow DJ, Stappenbeck TS. 2015. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe 17:85–97. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreit M, Vertommen D, Gillet L, Michiels T. 2015. The interferon-inducible mouse apolipoprotein L9 and prohibitins cooperate to restrict Theiler's virus replication. PLoS One 10:e0133190. doi: 10.1371/journal.pone.0133190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo JY, Yaneva R, Cresswell P. 2011. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10:534–539. doi: 10.1016/j.chom.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimatsu K, Ohya Y, Shikata Y, Seto T, Hasegawa Y, Tanaka I, Kawamura T, Kitoh K, Toyoshima S, Osawa T. 1992. Purification and cDNA cloning of a novel factor produced by a human T-cell hybridoma: sequence homology with animal lectins. Mol Immunol 29:537–546. doi: 10.1016/0161-5890(92)90012-M. [DOI] [PubMed] [Google Scholar]
- 23.van Pesch V, Lanaya H, Renauld J-C, Michiels T. 2004. Characterization of the murine alpha interferon gene family. J Virol 78:8219–8228. doi: 10.1128/JVI.78.15.8219-8228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapira SD, Gat-Viks I, Shum BOV, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baril M, Es-Saad S, Chatel-Chaix L, Fink K, Pham T, Raymond VA, Audette K, Guenier AS, Duchaine J, Servant M, Bilodeau M, Cohen E, Grandvaux N, Lamarre D. 2013. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog 9:e1003416. doi: 10.1371/journal.ppat.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Shu W, Lu MM, Morrisey EE. 2005. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol 25:5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Chen H, Hu L, Xing Y, Sasaki T, Villosis MF, Li J, Nishita M, Minami Y, Minoo P. 2008. Ror2 modulates the canonical Wnt signaling in lung epithelial cells through cooperation with Fzd2. BMC Mol Biol 9:11. doi: 10.1186/1471-2199-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillesheim A, Nordhoff C, Boergeling Y, Ludwig S, Wixler V. 2014. β-Catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-κB signaling. Cell Commun Signal 12:29. doi: 10.1186/1478-811X-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francois C, Bernard I, Castelain S, Charleston B, Fray MD, Capiod JC, Duverlie G. 2005. Quantification of different human alpha interferon subtypes and pegylated interferon activities by measuring MxA promoter activation. Antimicrob Agents Chemother 49:3770–3775. doi: 10.1128/AAC.49.9.3770-3775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staal FJ, Luis TC, Tiemessen MM. 2008. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 32.Clevers H, Nusse R. 2012. Wnt/beta-catenin signaling and disease. Cell 149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Lin A. 2005. Role of JNK activation in apoptosis: a double-edged sword. Cell Res 15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- 35.Royer C, Lu X. 2011. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ 18:1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giardina SF, Mikami M, Goubaeva F, Yang J. 2007. Connexin 43 confers resistance to hydrogen peroxide-mediated apoptosis. Biochem Biophys Res Commun 362:747–752. doi: 10.1016/j.bbrc.2007.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadasz I, Mayer K, Gattenloehner S, Fink L, Matrosovich M, Li X, Seeger W, Lohmeyer J, Bellusci S, Herold S. 2016. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog 12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baarsma HA, Skronska-Wasek W, Mutze K, Ciolek F, Wagner DE, John-Schuster G, Heinzelmann K, Günther A, Bracke KR, Dagouassat M, Boczkowski J, Brusselle GG, Smits R, Eickelberg O, Yildirim AÖ, Königshoff M. 2017. Noncanonical WNT-5A signaling impairs endogenous lung repair in COPD. J Exp Med 214:143–163. doi: 10.1084/jem.20160675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Institutes of Health. 2002. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 40.Tobita K, Sugiura A, Enomote C, Furuyama M. 1975. Plaque assay and primary isolation of influenza A viruses in an established line of canine kidney cells (MDCK) in the presence of trypsin. Med Microbiol Immunol 162:9–14. doi: 10.1007/BF02123572. [DOI] [PubMed] [Google Scholar]
- 41.Dolfi DV, Duttagupta PA, Boesteanu AC, Mueller YM, Oliai CH, Borowski AB, Katsikis PD. 2011. Dendritic cells and CD28 costimulation are required to sustain virus-specific CD8+ T cell responses during the effector phase in vivo. J Immunol 186:4599–4608. doi: 10.4049/jimmunol.1001972. [DOI] [PubMed] [Google Scholar]
- 42.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.