Many flaviviruses are significant human pathogens that pose global threats to public health. Although licensed vaccines are available for yellow fever, Japanese encephalitis, tick-borne encephalitis, and dengue viruses, new approaches are needed to develop improved vaccines. Using dengue virus as a model, we developed a vaccine platform using a virion assembly-defective virus. We show that such an assembly-defective virus could be rescued to higher titers and infect cells for a single round. Mice immunized with the assembly-defective virus were protected from wild-type-virus infection. This vaccine approach could be applicable to other viral pathogens.
KEYWORDS: dengue virus, flavivirus vaccine, NS2A, trans-complementation, viral assembly
ABSTRACT
Dengue virus (DENV) is the most prevalent mosquito-transmitted viral pathogen in humans. The recently licensed dengue vaccine has major weaknesses. Therefore, there is an urgent need to develop improved dengue vaccines. Here, we report a virion assembly-defective DENV as a vaccine platform. DENV containing an amino acid deletion (K188) in nonstructural protein 2A (NS2A) is fully competent in viral RNA replication but is completely defective in virion assembly. When trans-complemented with wild-type NS2A protein, the virion assembly defect could be rescued, generating pseudoinfectious virus (PIVNS2A) that could initiate single-round infection. The trans-complementation efficiency could be significantly improved through selection for adaptive mutations, leading to high-yield PIVNS2A production, with titers of >107 infectious-focus units (IFU)/ml. Mice immunized with a single dose of PIVNS2A elicited strong T cell immune responses and neutralization antibodies and were protected from wild-type-virus challenge. Collectively, the results proved the concept of using assembly-defective virus as a vaccine approach. The study also solved the technical bottleneck in producing high yields of PIVNS2A vaccine. The technology could be applicable to vaccine development for other viral pathogens.
IMPORTANCE Many flaviviruses are significant human pathogens that pose global threats to public health. Although licensed vaccines are available for yellow fever, Japanese encephalitis, tick-borne encephalitis, and dengue viruses, new approaches are needed to develop improved vaccines. Using dengue virus as a model, we developed a vaccine platform using a virion assembly-defective virus. We show that such an assembly-defective virus could be rescued to higher titers and infect cells for a single round. Mice immunized with the assembly-defective virus were protected from wild-type-virus infection. This vaccine approach could be applicable to other viral pathogens.
INTRODUCTION
The four serotypes of dengue virus (DENV) are the most important mosquito-borne viral pathogens in humans, with approximately 390 million human infections each year (1). DENV infection causes dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS). Infection with one DENV serotype elicits lifelong immunity to that particular serotype but confers only partial and transient protection against the other three serotypes. Secondary infections with heterogeneous DENV serotypes increase the risk of severe disease (DHF and DSS), which is mediated at least in part through an antibody-dependent enhancement (ADE) of infection (2). Thus, an ideal dengue vaccine should confer balanced immune protection against all four serotypes. Dengvaxia, the first clinically approved dengue vaccine, has an average efficacy of 30 to 61% (3–6). Unfortunately, Dengvaxia seems to increase the risk of hospitalization over time in children <9 years of age who are seronegative at the time of vaccination, possibly through vaccine-induced antibody enhancement. The World Health Organization recently recommended the use of Dengvaxia only in areas where the disease is highly prevalent (7). Although two other live attenuated dengue vaccines (NIH and Takeda) are currently under late clinical development (8), new approaches are needed for development of improved vaccines for dengue virus and other flaviviruses.
Besides DENV, many other flaviviruses are also important human pathogens, including yellow fever (YFV), West Nile (WNV), Japanese encephalitis (JEV), tick-borne encephalitis (TBEV), and Zika (ZIKV) viruses. The flavivirus genome encodes three structural proteins (capsid [C], premembrane [prM], and envelope [E] proteins) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Structural proteins form virions, whereas nonstructural proteins participate in viral RNA synthesis, virion assembly, and evasion of immune response (9). Virus-like particles (VLPs) represent an attractive vaccine approach, as exemplified by the clinically approved human vaccines for papillomavirus and hepatitis B virus (10). Two forms of VLPs have been reported for flaviviruses: empty VLPs and subgenomic VLPs (also known as pseudoinfectious viruses [PIVs]). Empty VLPs are produced during natural flavivirus infection or from cells expressing viral prM and E proteins (11). Subgenomic VLPs contain viral RNA with a deletion in a structural gene(s); such VLPs are produced by complementation of the subgenomic RNA in helper cells that express the deleted structural protein(s) (12–14, 46). Immunization of mice or rhesus macaques with TBEV, YFV, or WNV subgenomic VLPs protects against wild-type (WT) virus infection (15, 16).
Flavivirus assembly is modulated by NS1 (17), NS2A (18, 19), NS2B (20), and NS3 (21). NS2A is a highly hydrophobic membrane protein located in the endoplasmic reticulum (ER) (22). Different regions of NS2A function in viral RNA synthesis and virion assembly (18): (i) distinct NS2A mutations selectively abolish DENV serotype 2 (DENV-2) assembly without significantly reducing viral RNA synthesis; (ii) the defect of virion assembly could be rescued in helper cells expressing WT NS2A protein, generating PIVNS2As; (iii) the rescued PIVNS2A could initiate single-round infection (23). These findings prompted us to explore the feasibility of developing PIVs as a novel vaccine platform for DENV. Here, we demonstrate the production of high titers of PIVNS2As (>107 infectious-focus units [IFU]/ml) and single-dose efficacy of PIVs in a DENV-2 mouse model. This technology could be applied to the development of other flavivirus vaccines.
RESULTS
Experimental rationale.
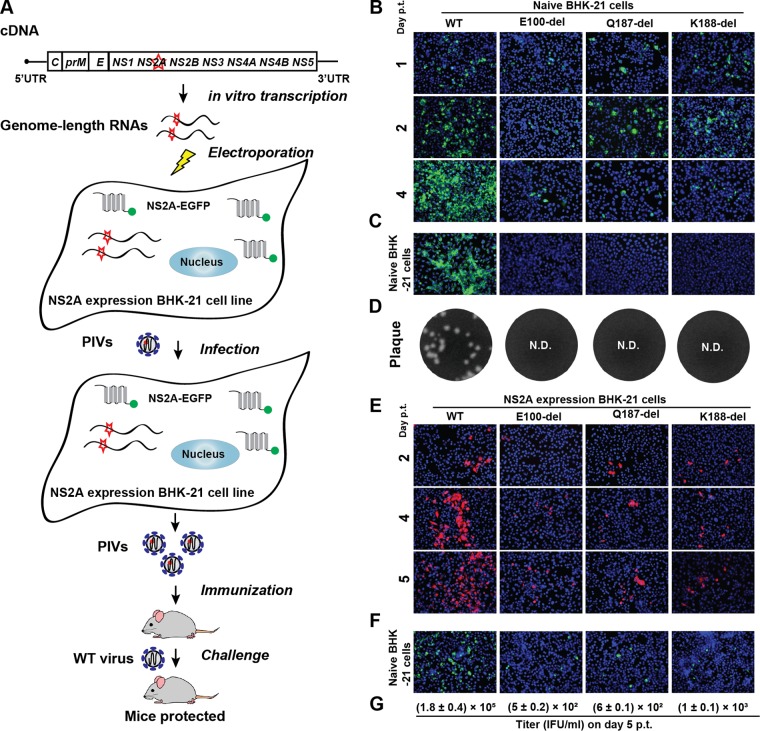
Figure 1A outlines the experimental rationale for PIVNS2A production and efficacy evaluation. An NS2A mutant virus defective in virion assembly (but competent in viral RNA replication) was trans-supplied with WT NS2A protein in a helper cell line, producing PIVNS2As. The NS2A-expressing cell line constitutively expressed the WT NS2A protein fused with a C-terminal green fluorescent protein (GFP) tag (23). The efficiency of PIVNS2A production was improved by selection of adaptive mutations (after multiple rounds of culturing PIVNS2As on the NS2A-expressing helper cells). The PIVNS2As were evaluated for vaccine efficacy in a mouse model.
FIG 1.
Characterization of NS2A E100-del, Q187-del, and K188-del mutants. (A) Experimental rationale. The diagram outlines the production of PIVNS2As and vaccination in mice. The NS2A-expressing cell line constitutively expressed the WT NS2A protein fused with a C-terminal GFP tag (23). The GFP tag (indicated by a green dot) was engineered to facilitate the detection of NS2A expression because no reliable NS2A antibodies are currently available. (B) IFA of naive BHK-21 cells transfected with the WT or NS2A mutant genome-length RNA. Equal amounts (10 μg) of the WT and mutant (E100-del, Q187-del, and K188-del) genome-length RNAs were transfected into BHK-21 cells. From days 1 to 4 p.t., the cells were stained with anti-E protein MAb 4G2 (green). The blue color shows the nuclei of cells stained with DAPI. (C) IFA of naive BHK-21 cells that were infected with the culture fluids harvested from transfected cells from panel B on day 4 p.t. The IFA was performed on day 2 postinfection. (D) Plaque assay. Naive BHK-21 cells were infected with the culture fluids harvested from transfected cells from panel B on day 5 p.t. Plaques developed on day 5 postinfection. N.D., not detectable. (E) IFA of NS2A-expressing BHK-21 cells transfected with the WT or NS2A mutant genome-length RNA. Results from days 2 to 5 p.t. are shown. E protein was detected by MAb 4G2 (red). (F) IFA of naive BHK-21 cells infected with the culture fluids harvested from transfected cells from panel E. The IFA was performed on day 2 postinfection. The expression of E protein is indicated in green. (G) Quantification of the infectious titers of PIVNS2As by IFA. The E-positive cells from panel F were counted to estimate the infectious titers of PIVNS2As. The titers on day 5 p.t. are shown as means ± standard deviations (SD) from three independent experiments.
PIVNS2A production.
Three NS2A amino acids (E100, Q187, and K188), whose mutations were previously shown to selectively abolish virion assembly (23), were selected to generate PIVNS2As for vaccine development. To minimize reversion, individual amino acids were deleted within an infectious cDNA clone of DENV-2 (24), resulting in mutants E100-del, Q187-del, and K188-del. As shown in Fig. 1B, BHK-21 cells transfected with both WT and mutant genome-length RNAs expressed viral E protein, suggesting that the deletion mutants were replication competent; however, from days 1 to 4 posttransfection (p.t.), the number of E-positive cells increased only in the WT-RNA-transfected cells, not in the mutant-RNA-transfected cells, suggesting no virus spreading in the mutant-RNA-transfected cells. Among the three deletion mutants, E100-del generated fewer E protein-positive cells than Q187-del and K188-del. Incubation of naive BHK-21 cells with culture medium from the transfected cells showed that only the WT RNA, not the mutant RNAs, generated infectious virus, as indicated by the E-positive cells (Fig. 1C). These data were corroborated by the plaque assay results showing that no infectious virus was recovered from any of the mutant-RNA-transfected cells (Fig. 1D). Collectively, the results suggest that E100-del, Q187-del, and K188-del are defective in virion assembly and/or release.
To evaluate the effects of the three NS2A deletions on viral RNA synthesis, we performed a transient luciferase replicon assay (Fig. 2). Upon transfection into BHK-21 cells, both the WT and mutant replicons generated equivalent luciferase signals at 2 to 6 h p.t.; different luciferase activities were detected at 24 h p.t. in the following order: WT ≥ K188-del ≥ Q187-del > E100-del. Equivalent luciferase signals were observed for the WT and mutants at 48 h posttransfection. These results indicate that K188-del, Q187-del, and E100-del do not dramatically compromise viral RNA replication.
FIG 2.
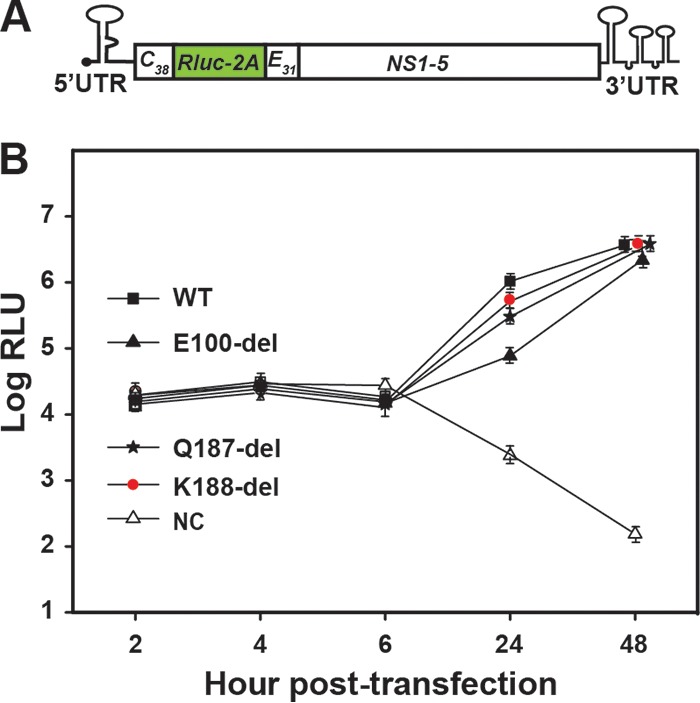
Transient replicon analysis of DENV-2 NS2A mutants. (A) Schematic diagram of a luciferase replicon of DENV-2 (22). Rluc-2A, a Renilla luciferase gene fused with the foot-and-mouth disease virus 2A sequence; C38, nucleotides encoding the first 38 amino acids of C protein; E31, nucleotides encoding the last 31 amino acids of E protein; HDVr, hepatitis delta virus ribozyme sequence; UTR, untranslated region. (B) Transient replicon assay. WT or mutant replicon RNAs containing single or combined mutations were electroporated into naive BHK-21 cells. A nonreplicative NS4B mutant, K143A, was included as a negative control (NC). Each data point shows the average of three independent experiments. The error bars represent standard deviations.
To rescue the defect of virion assembly/release, we transfected the three mutant genome-length RNAs individually into BHK-21 cells that constitutively expressed DENV-2 WT NS2A protein. From days 2 to 5 p.t., an increasing number of viral E-positive cells were observed in the following order: WT > K188-del ≥ Q187-del ≥ E100-del (Fig. 1E). Incubation of naive BHK-21 cells with the culture medium from the transfected cells generated E-positive cells, suggesting that infectious PIVNS2As were produced (Fig. 1F). Quantification of the E-positive cells suggested PIVNS2A titers of 1.8 × 105, 5 × 102, 6 × 102, and 1 × 103 IFU/ml for WT, E100-del, Q187-del, and K188-del viruses, respectively (Fig. 1G). To exclude the possibility that the observed trans-complementation was due to reversion of the NS2A deletions, we extracted the total intracellular RNAs from the infected naive BHK-21 cells shown in Fig. 1F and sequenced the entire viral genome. The sequencing results showed that the engineered NS2A deletions were retained without secondary mutations. A new round of infection of naive BHK-21 cells with culture fluids from cells shown in Fig. 1F did not yield any E-positive cells (data not shown), suggesting that the PIVNS2As could perform only a single-round infection.
To further minimize potential reversion, we generated a double amino acid deletion mutant, Q187K188-del. As shown in Fig. 3, the Q187K188-del mutant was highly defective in viral replication in BHK-21 cells. Therefore, we did not further pursue the double-mutant approach.
FIG 3.
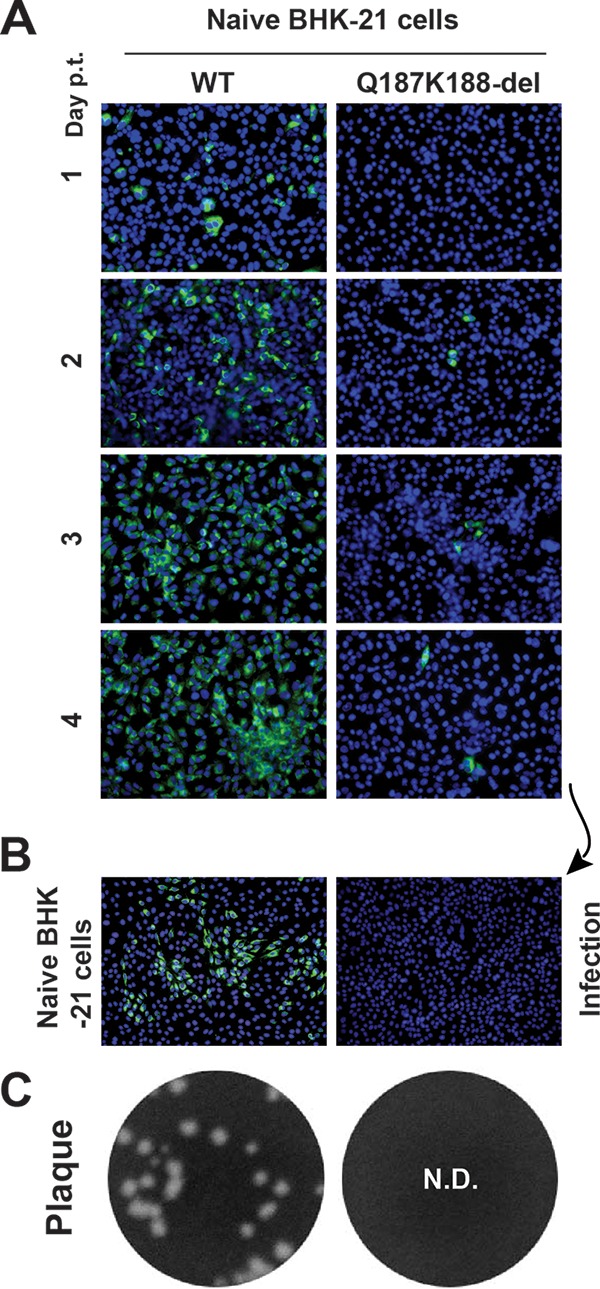
Characterization of Q187K188-del mutants. (A) IFA of naive BHK-21 cells transfected with WT or Q187K188-del genome-length RNA. (B) IFA of naive BHK-21 cells infected with the culture fluids harvested on day 4 p.t. from the transfected cells in panel A. (C) Plaque assay. Culture fluids from the transfected BHK-21 cells on day 5 p.t. were subjected to plaque assay using BHK-21 cells. No detectable (N.D.) plaques were observed for the Q187K188-del mutant.
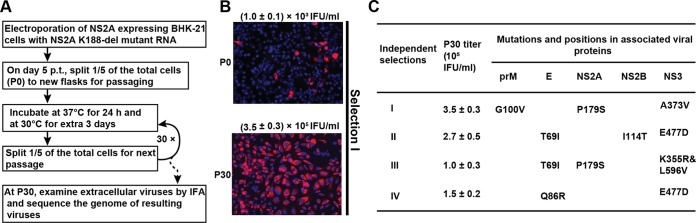
Selection for improved PIVNS2A production.
We performed selection experiments to improve PIVNS2A production (Fig. 4A). Mutant K188-del was chosen for the selection experiment because of its higher PIV titer in the above-mentioned trans-complementation assay (Fig. 1G). Four independent selections were performed. After passaging the NS2A-expressing cells (which were transfected with the K188-del genome-length RNA) for 30 rounds (4 days per round) (Fig. 4A), the E-positive cells increased from 3% at passage 0 (P0) to almost 100% at P30 (Fig. 4B [selection I; selections II to IV not shown]). The P30 PIVNS2A titers were improved to ≥1 × 105 IFU/ml in all four independent selections (Fig. 4C). Sequencing of the P30 PIVNS2A genomic RNAs revealed that, besides the engineered NS2A K188-del, additional mutations accumulated in both structural and nonstructural genes (Fig. 4C). These results suggest that PIVNS2A production could be improved by accumulating adaptive mutations in the K188-del genome.
FIG 4.
Selection for adaptive mutations to improve the yield of K188-del PIVNS2A production. (A) Experiment flowchart. The diagram shows the procedure for continuous passaging of PIVNS2As on NS2A-expressing BHK-21 cells. (B) IFA of NS2A-expressing BHK-21 cells infected with P0 and P30 PIVNS2As. The cells were stained for E protein expression (red) using MAb 4G2. (C) Summary of adaptive mutations. The complete genomes of P30 PIVNS2As were sequenced for each of the four independent selections. The adaptive mutations are indicated by the amino acid positions encoded by individual genes. Consensus sequencing was performed to identify the adaptive mutations. The titers of P30 PIVNS2As are also indicated.
Validation of selection I mutations to confer high PIVNS2A production.
To validate the roles of adaptive mutations, we engineered the mutations into DENV-2 in the presence and absence of an NS2A K188-del background. We chose to focus on the mutations identified from selections I and II because these two selections produced the highest PIVNS2A titers in the NS2A-expressing cells (Fig. 4C).
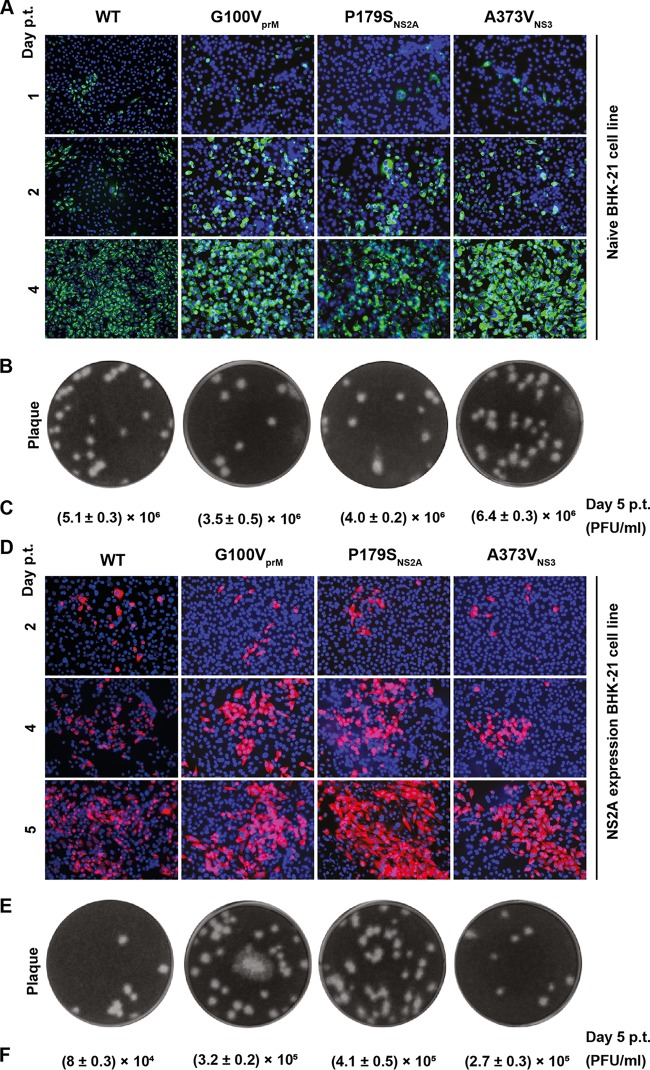
For selection I, we initially examined the effects of three adaptive mutations (G100VPrM, P179SNS2A, and A373VNS3) on viral replication by engineering individual mutations into the WT genome-length RNA. After electroporation into naive BHK-21 cells, the WT and mutant RNAs produced comparable amounts of E-positive cells (Fig. 5A) and generated infectious viruses with similar plaque morphologies (Fig. 5B) and equivalent viral titers on day 5 p.t. (Fig. 5C). We also electroporated the genome-length RNAs into the NS2A-expressing cells. Three mutant RNAs generated slightly more E-positive cells than the WT RNA on day 5 p.t. (Fig. 5D), the WT and mutant viruses showed similar plaque morphologies on naive BHK-21 cells (Fig. 5E), and the mutant RNAs generated more infectious viruses than the WT RNA on day 5 p.t. (Fig. 5F). Interestingly, both the WT and mutant RNAs generated ≥10-fold more infectious viruses on the naive cells than on the NS2A-expressing cells (compare Fig. 5C and F), which might have been caused by the competition for limited cellular factors between the exogenous and endogenous NS2A proteins.
FIG 5.
Effects of selection I mutations on viral replication in the absence of the K188-del mutation. (A) IFA of naive BHK-21 cells transfected with WT or mutant (G100VprM, P179SNS2A, and A373VNS3) genome-length RNA. Viral E protein expression (green) was detected using MAb 4G2. (B) Plaque morphology. The culture fluids harvested from the transfected cells from panel A on day 5 p.t. were subjected to plaque assay using BHK-21 cells. (C) Viral titers on day 5 from the transfected cells from panel A. (D) IFA of NS2A-expressing BHK-21 cells transfected with the WT or mutant genome-length RNA. E protein expression (red) was detected using MAb 4G2. (E) Plaque morphology. The culture fluids harvested on day 5 p.t. from cells in panel D were used to perform a plaque assay on BHK-21 cells. (F) Viral titers on day 5 p.t. from the transfected cells in panel D. Means ± standard deviations from three independent experiments are presented.
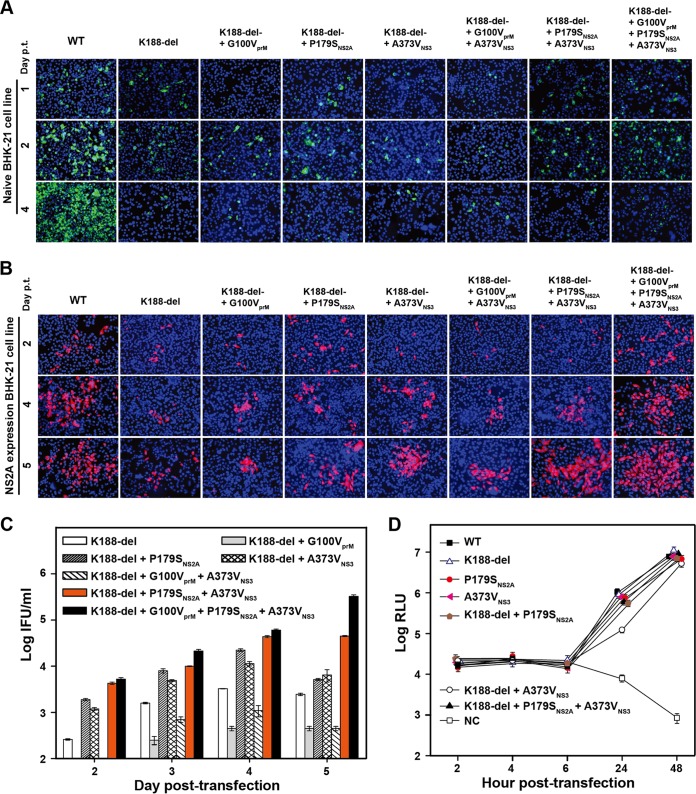
Next, we analyzed the effects of selection I mutations on viral replication in the context of NS2A K188-del genome-length RNA. In naive BHK-21 cells, addition of adaptive mutations (individually or in combination) did not rescue the virion assembly defect of K188-del RNA, as evidenced by lack of increase in E-positive cells from days 1 to 4 p.t. (Fig. 6A) and lack of infectious virus on day 5 p.t. (data not shown). The results demonstrate that the adaptive mutations could not rescue the defect of virion assembly in naive BHK-21 cells. Remarkably, in NS2A-expressing cells, the addition of P179SNS2A and A373VNS3, alone or in combination, improved virion assembly, as indicated by E-positive cells (Fig. 6B) and PIVNS2A titers (Fig. 6C). Interestingly, the G100VprM mutation reduced PIVNS2A production in the context of K188-del plus G100VprM or 188-del plus G100VprM plus A373VNS3; however, the mutation did enhance PIVNS2A production when combined with all selection I mutations (188-del plus G100VprM plus P179SNS2A plus A373VNS3). These results demonstrate that selection I mutations contribute to improved PIV production.
FIG 6.
Improvement of PIVNS2A production through adaptive mutations from selection I. (A) IFA of naive BHK-21 cells transfected with the genome-length RNA of the WT or mutants that contain NS2A K188-del and the mutations derived from selection I. The transfected cells were monitored for E protein expression (green) using MAb 4G2. (B) IFA of NS2A-expressing BHK-21 cells transfected with the genome-length RNA of the WT or mutants that contain NS2A K188-del and different mutations rescued from selection I. The E protein was stained in red using MAb 4G2. (C) Quantification of PIVNS2A titers generated from NS2A-expressing BHK-21 cells transfected with the indicated genome-length RNAs. The detection limit of IFA quantification was 10 IFU/ml. Mean values from three independent experiments are shown, and the error bars represent standard deviations. (D) Replicon analysis. WT or mutant replicon RNAs containing single or combined mutations were electroporated into naive BHK-21 cells. An NS4B lethal mutation, K143A (45), was included as a negative control (NC). The means and standard deviations from the results of three independent experiments are presented.
To exclude the possibility that the above-mentioned observation was due to a mutational effect on viral RNA synthesis, we examined the selection I mutations alone or in combination with K188-del in a transient replicon assay in naive BHK-21 cells. As shown in Fig. 6D, only replicon 188-del plus A373VNS3 attenuated viral replication at 24 h p.t.; all other replicons replicated to the WT level. The results suggest that selection I mutations alone do not affect viral RNA synthesis.
Validation of selection II mutations for high PIVNS2A production.
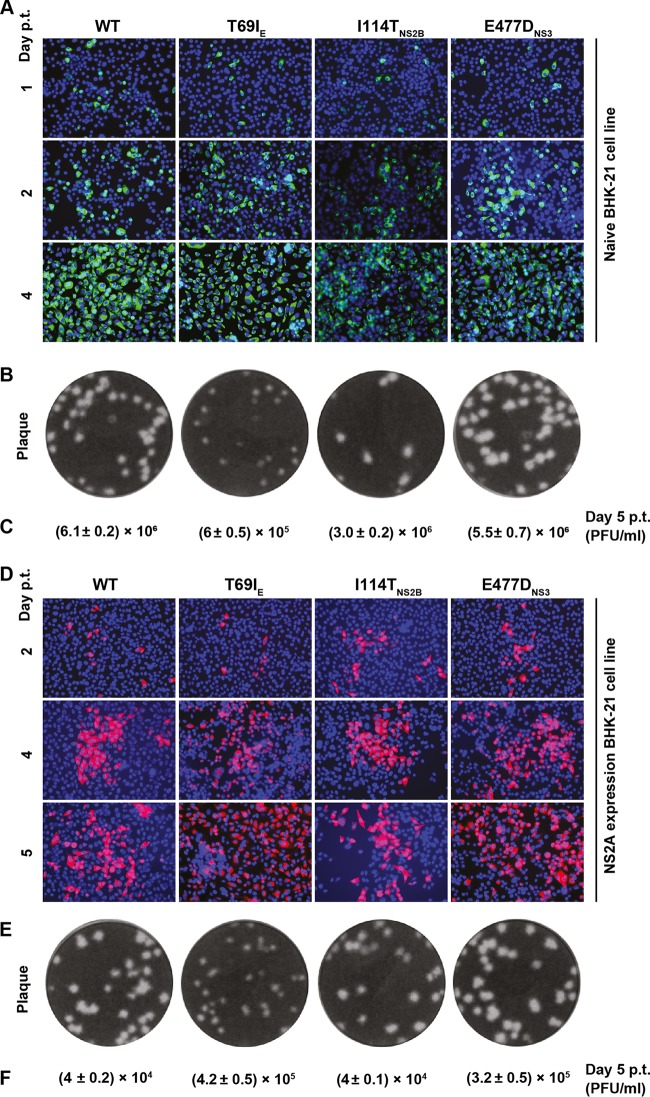
We took a similar approach to analyze selection II mutations. In the absence of K188-del, genome-length RNA encoding a T69I mutation in the envelope protein (T69IE) produced 10-fold less infectious virus than the WT RNA upon transfection into naive BHK-21 cells, whereas mutation I114TNS2B or E477DNS3 did not affect viral replication (Fig. 7A to C). In NS2A-expressing cells, the mutant RNAs replicated to levels equivalent to (I114TNS2B) or higher than (T69IE or E477DNS3) those of the WT RNA (Fig. 7D to F).
FIG 7.
Effects of selection II mutations on viral replication in the absence of the K188-del mutation. (A) IFA of naive BHK-21 cells transfected with WT or mutant (T69IE, I114TNS2B, and E477DNS3) genome-length RNA. E-positive cells strained by 4G2 are shown in green. (B) Plaque morphology. Culture fluids harvested from the transfected cells from panel A on day 5 p.t. were subjected to plaques assay using BHK-21 cells. (C) Viral titers on day 5 from the transfected cells from panel A. (D) IFA of NS2A-expressing BHK-21 cells transfected with WT or mutant genome-length RNA. E-positive cells stained with MAb 4G2 are shown in red. (E) Plaque morphology. The culture fluids harvested on day 5 p.t. from panel D were used to perform a plaque assay on naive BHK-21 cells. (F) Viral titers on day 5 from the transfected cells in panel D. The data are presented as means ± standard deviations from the results of three independent experiments.
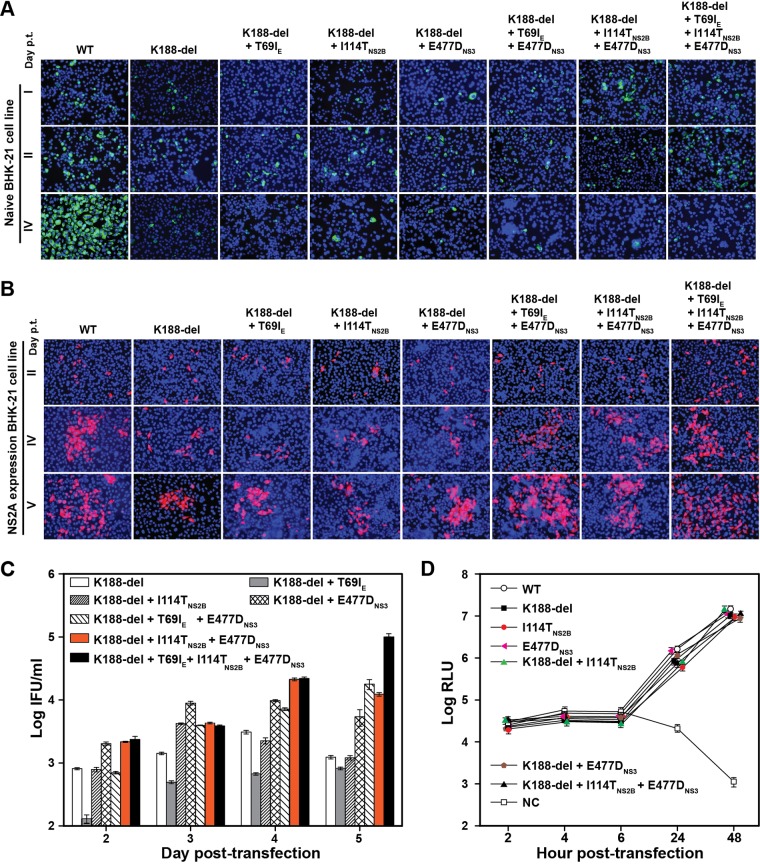
In the presence of K188-del, addition of selection II mutations (individually or in combination) did not rescue the defect of virion assembly in naive BHK-21 cells, as indicated by lack of increase in E-positive cells after RNA transfection (Fig. 8A). No infectious PIVNS2A was detected after incubating new BHK-21 cells with the culture fluids collected from the cells shown in Fig. 8A (data not shown). In contrast, in NS2A-expressing cells, addition of all three adaptive mutations to K188-del (K188-del plus T69IE plus I114TNS2B plus E477DNS3) significantly improved PIVNS2A titers, although addition of one or two mutations had various effects on PIVNS2A production (Fig. 8B and C). Finally, using a transient luciferase replicon, we showed that selection II mutations, alone or in combination with K188-del, did not affect viral RNA synthesis (Fig. 8D). Collectively, the results demonstrated that selection II mutations were responsible for the improvement of PIVNS2A production in the NS2A-expressing cells.
FIG 8.
Effects of selection II mutations on viral replication in the presence of the K188-del mutation. (A) IFA of naive BHK-21 cells transfected with genome-length RNA of the WT or mutants (K188-del alone or in combination with various selection II mutations). E-positive cells (green) were stained with MAb 4G2. (B) IFA of NS2A-expressing BHK-21 cells transfected with WT or mutant genome-length RNA. Viral E protein was stained with MAb 4G2 (red). (C) Quantification of PIVNS2A titers generated from NS2A-expressing BHK-21 cells transfected with the indicated genome-length RNAs. Mean values and standard deviations from the results of three independent experiments are shown. (D) Luciferase replicon analysis. WT or mutant luciferase replicon RNAs containing single or multiple selection II mutations were electroporated into naive BHK-21 cells. Luciferase activities were measured at the indicated time points. NC, negative control using an NS4B lethal mutant, K143A. Each data point shows the average result from three independent experiments, and the error bars represent standard deviations.
EM analysis of PIVNS2A production.
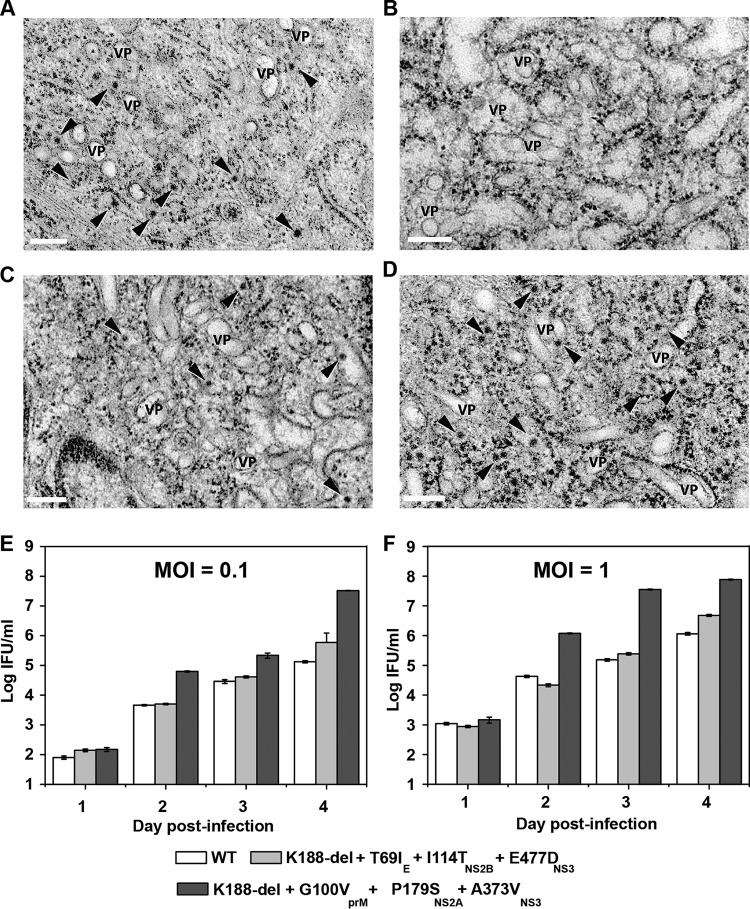
Transmission electron microscopy (EM) was used to examine virus production. As shown in Fig. 9A to D, infection of naive or NS2A-expressing BHK-21 cells with WT DENV-2 or selection I PIVNS2A caused rearrangement of the ER membrane and induction of vesicle packets (VP), indicating that both the WT virus and PIVNS2A could establish efficient replication in cells. However, progeny virions (indicated by arrowheads) were observed only in the WT-virus-infected naive cells (Fig. 9A), WT-virus-infected NS2A-expressing cells (Fig. 9C), and PIVNS2A-infected NS2A-expressing cells (Fig. 9D). We also observed more virions at 96 h postinfection (p.i.) than at 72 h p.i. in the PIVNS2A-infected NS2A-expressing cells (data not shown). It should also be noted that the appearance of virus particles was delayed in the PIVNS2A-infected NS2A-expressing cells (72 h p.i.) compared to infections with the WT virus in naive or NS2A-expressing cells (48 h p.i.). In contrast, no virions were detected in the PIVNS2A-infected cells without NS2A expression at any time point (Fig. 9B).
FIG 9.
Characterization of PIVNS2A replication. (A to D) Electron microscopic analysis of naive and NS2A-expressing BHK-21 cells infected with WT DENV-2 and PIVNS2A. (A) Naive BHK-21 cells infected with WT virus at 48 h postinfection. (B) Naive BHK-21 cells infected with PIVNS2A P30 (selection I) at 48 h postinfection. (C) NS2A-expressing BHK-21 cells infected with WT virus at 48 h postinfection. (D) NS2A-expressing BHK-21 cells infected with PIVNS2A P30 (selection I) at 72 h. Scale bars, 200 nm. The arrowheads indicate virus particles. VP, vesicle packets. (E and F) Growth kinetics of the WT, selection I recombinant PIVNS2A (K188-del plus G100VprM plus P179SNS2A plus A373VNS3), and selection II recombinant PIVNS2A (K188-del plus T69IE plus I144TNS2B plus E477DNS3) on NS2A-expressing BHK-21 cells at MOI of 0.1 (E) and 1 (F).
Growth kinetics of PIVNS2As in NS2A-expressing BHK-21 cells.
We characterized the growth kinetics of recombinant PIVNS2As representative of selections I and II in NS2A-expressing cells. At both multiplicities of infection (MOI) of 0.1 and 1, selection I recombinant PIVNS2A (K188-del plus G100VprM plus P179SNS2A plus A373VNS3) yielded the highest titer of >1 × 107 IFU, followed by selection II recombinant PIVNS2A (K188-del plus T69IE plus I144TNS2B plus E477DNS3) and WT virus (Fig. 9E and F). The results demonstrate that a high titer of PIVNS2A could be achieved through adaptive mutations.
NS2A-NS3 interaction.
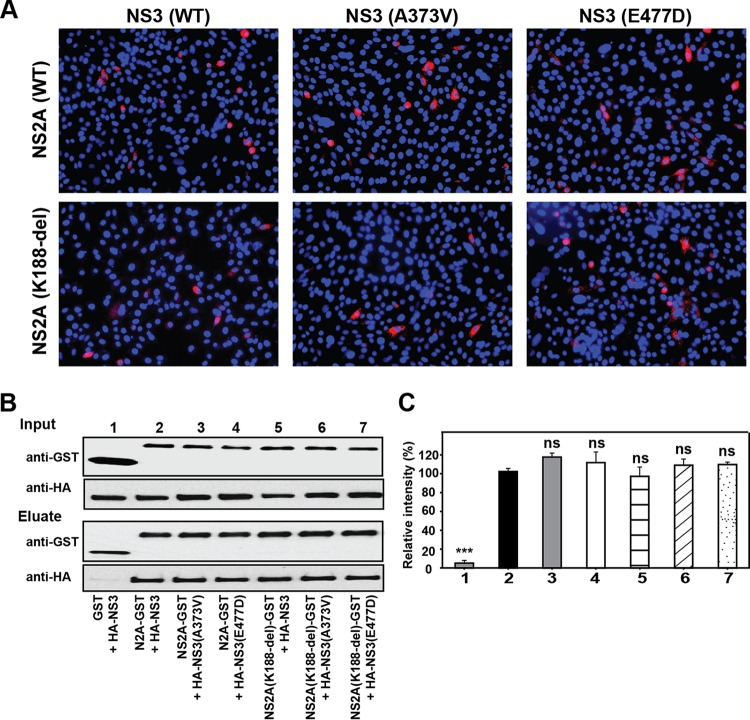
Our results showed that (i) PIVNS2As from four independent selections consistently accumulated mutations in the NS3 gene, whereas only one out of the four independent selections exhibited the NS2B I114T mutation (Fig. 4C); (ii) addition of the NS3 mutation alone to K188-del (K188-del plus A373VNS3 and K188-del plus E477DNS3) enhanced PIVNS2A production in the NS2A-expressing cells (Fig. 6C and 8C). These observations prompted us to examine the interaction between the NS2A and NS3 proteins. We initially probed the NS2A-NS3 interaction using an in situ proximity ligation assay (PLA). A PLA could detect two molecules in <40-nm proximity in cells (25). As a control, positive PLA signals were observed in BHK-21 cells that were cotransfected with plasmids expressing NS3 and NS2B, a known pair of interacting proteins (Fig. 10A). Coexpression of WT NS2A and NS3 revealed PLA-positive signals, whereas expression of NS2A or NS3 alone did not yield any positive signals (Fig. 10A). Mutations in NS2A (K188-del) and NS3 (A373V and E447D) did not change the positive PLA signals (Fig. 11A).
FIG 10.
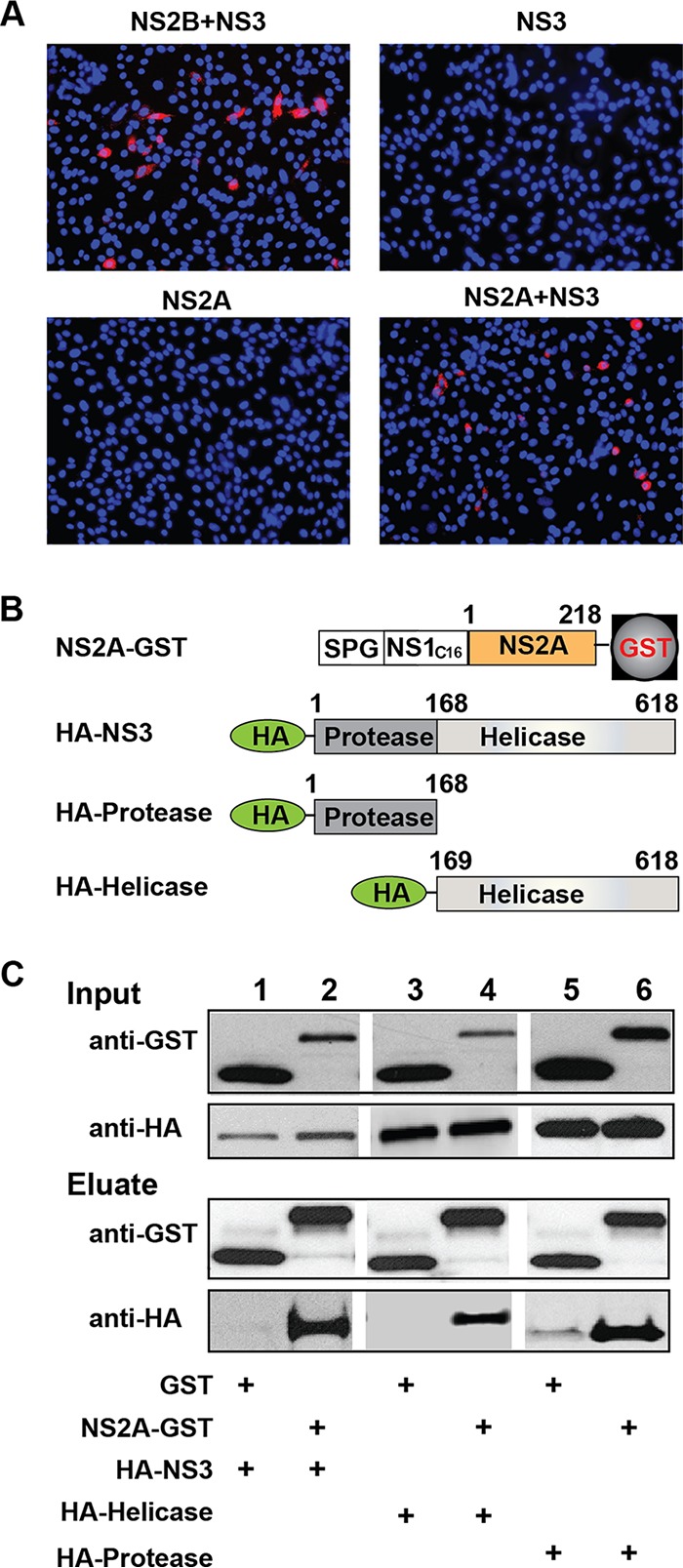
Characterization of NS2A-NS3 interaction. (A) Duo-link analysis. A Duo-link assay was performed to probe NS2A-NS3 interaction. BHK-21 cells were transfected with plasmids encoding C-terminally myc-tagged NS3 and/or C-terminally HA-tagged NS2A. As a positive control, cells were also cotransfected with plasmids expressing C-terminally HA-tagged NS2B and C-terminally myc-tagged NS3. Positive fluorescence signals are indicated in red. (B) Schematic diagram of constructs for co-IP assay. NS2A was fused with an N-terminal leader sequence comprising a signal peptide from Gaussia luciferase (SPG), the last 16 amino acids of NS1 (C16), and a C-terminal GST tag. NS3, the protease domain, and the helicase domain were fused with an N-terminal HA tag. (C) Co-IP analysis. Co-IP was performed to probe the NS2A-NS3 interaction. Two micrograms of rabbit anti-GST antibody was used to pull down GST and its fusion proteins. Mouse MAb against GST and mouse MAb against HA were used to detect GST- and HA-tagged proteins, respectively, in cell lysates and eluates.
FIG 11.
NS2A and NS3 mutations did not alter the proximity and interaction between NS2A and NS3. (A) Duo-link analysis. A Duo-link assay was performed to examine the effects of NS2A and NS3 mutations on the intracellular proximity of NS2A and NS3 proteins. BHK-21 cells were cotransfected with two plasmids: one plasmid encoding C-terminally myc-tagged WT or mutant NS3 (A373V or E477D) and another plasmid encoding C-terminally HA-tagged WT or K188-del NS2A. Positive fluorescence signals are shown in red. (B) Co-IP of NS2A and NS3 proteins. HEK 293T cells were cotransfected with two plasmids: one encoding GST or GST-tagged NS2A (WT or K188-del) and the other encoding HA-tagged NS3 (WT, A373V, or E477D). (C) Densitometry analysis. The intensity of each band from the immunoblot in panel B was quantified with ImageJ software. The relative pulldown efficiencies were calculated as follows: [(a/b)/(c/d)] × 100%, where a is the intensity of HA-NS3 (WT or mutant), b is the intensity of GST or NS2A-GST (WT or K188-del), c is the intensity of the WT HA-NS3 in lane 2, and d is the intensity of the WT NS2A-GST in lane 2. The means and standard deviations from the results of three independent experiments are shown. Statistical analysis was performed using an unpaired Student t test. ***, P < 0.001 (extremely significant); ns, nonsignificant (P > 0.05).
Next, we validated the NS2A-NS3 interaction using coimmunoprecipitation (co-IP). After cotransfection of HEK-293T cells with plasmids expressing NS2A and NS3, the two proteins were coimmunoprecipitated (Fig. 10B and C). In agreement with the PLA results, mutations in NS2A (K188-del) and NS3 (A373V and E447D) did not affect co-IP efficiency (Fig. 11B and C). Since NS3 contains protease and helicase domains, we tested whether NS2A binds to individual NS3 domains. The results showed that NS2A could pull down both protease and helicase domains (Fig. 10B and C). Collectively, the results indicate that (i) NS2A interacts with both NS3 protease and helicase domains and (ii) mutations in NS2A (K188-del) or NS3 (A373V and E447D) do not affect the NS2A-NS3 interaction.
Efficacy of PIVNS2A in mice.
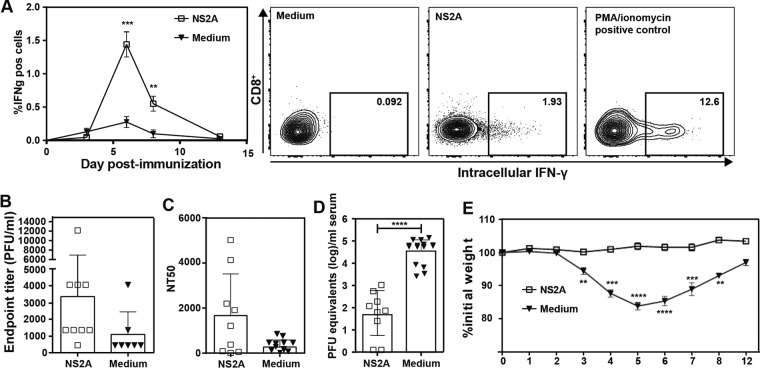
We used CD11c-cre × IFNARfl/fl mice (see Materials and Methods), which lack alpha/beta interferon (IFN-α/β) receptor only in CD11c-expressing cells, to test the efficacy of PIVNS2As as a potential vaccine (26). These CD11c-cre × IFNARfl/fl mice were able to generate a protective immune response for nonreplicative vaccines, whereas AG129 or IFNAR mice did not produce a robust immune response to the same nonreplicative vaccines (26). The mice were immunized once subcutaneously with 106 IFU PIVNS2As or placebo (medium containing fetal calf serum [FCS]). The immunized mice generated significant CD8+ T cell responses (Fig. 12A), DENV-specific binding antibodies, and neutralizing antibodies compared to the placebo-treated mice (Fig. 12B and C). Notably, the antibody responses exhibited large variation among different individuals, possibly due to the low infection rate or low immunogenicity of PIVNS2A. Further experiments are needed to measure viral RNA synthesis at the injection site and in draining lymph nodes at early times postinfection to determine whether differences in viral infection/replication efficiency (and thus antigen) account for the variation in antibody responses. Nevertheless, the immunized mice were protected after a challenge with DENV-2 strain D2Y98P, which caused substantial disease in the CD11c-cre × IFNARfl/fl mice (Fig. 12D and E). Peak viremia (measured by reverse transcription-quantitative PCR [RT-qPCR]) was significantly reduced (Fig. 12D), and the mice did not show any disease, as illustrated by their stable body weight compared to the placebo-treated mice, which lost up to 20% of their body weight after infection (Fig. 12E).
FIG 12.
In vivo efficacy of PIVNS2A. Mice (CD11c-cre × IFNARfl/fl) were immunized with 106 IFU PIVNS2As (i.e., K188-del plus G100VprM plus P179SNS2A plus A373VNS3 produced as described for Fig. 9F) or 10% FCS medium subcutaneously. (A) CD8+ T cell response in the blood was monitored on days 3, 6, 8, and 13. Representative flow cytometry graphs from day 6 are shown on the right. (B) IgG antibody titers after immunization. At 1 month postimmunization, blood was drawn prior to challenge with virulent DENV-2 (strain D2Y98P). The IgG antibody titers against DENV-2 were measured by ELISA. (C) NT50. Neutralizing antibodies against DENV-2 were measured using U937–DC-SIGN cells as target cells. (D) Viremia on day 3 after challenge. After 1 month of immunization, mice were challenged with 107 PFU D2Y98P virus. On day 3 postchallenge, peak viremia was measured by real-time RT-PCR. (E) Weights of mice over the course of 12 days postchallenge. The data are presented as means ± standard deviations. Statistical analysis was performed using Student's t test. **, P < 0.01 (highly significant); ***, P < 0.001 (extremely significant); ****, P < 0.0001. Nonsignificant differences are not indicated. The data are derived from the results of two or three independent experiments with totals of 9 to 15 mice.
DISCUSSION
Vaccine represents the most effective means to prevent and control infectious diseases. The goal of this study was to prove the concept of using NS2A-mediated virion assembly-defective virus for dengue vaccine development. We showed that an NS2A K188-del mutation did not significantly affect viral RNA replication but completely abolished virion assembly (Fig. 1B and 2). The assembly could be rescued by trans-complementation with exogenous WT NS2A protein. The efficiency of trans-complementation could be significantly improved by adaptive mutations in viral structural and nonstructural genes, leading to high-yield PIVNS2A production with titers of >107 IFU/ml. Mice immunized with the PIVNS2As generated robust humoral and cellular responses and were protected from virulent WT-virus challenge. The concept of using single-round infectious PIVs as a vaccine approach has been previously reported for a number of flaviviruses (15, 16). However, all previous PIVs were prepared through trans-complementation of subgenomic RNA with a capsid gene deletion in cells expressing exogenous capsid protein. Cells infected with subgenomic PIVs efficiently secreted empty virus-like particles consisting of prM-E proteins (15). Compared with the subgenomic PIVs, the current PIVNS2A contains intact structural proteins and should resemble authentic WT virus. Kummerer and Rice previously showed that the YFV NS2A K190S mutation (equivalent to DENV-2 NS2A K188 in this study) abolished virion assembly but did not affect the formation of empty virus-like particles in the PIVNS2A-infected cells (18), suggesting that YFV NS2A specifically functions in mediating genomic RNA into virion formation.
Both structural and nonstructural proteins elicit antibodies in patients (27), among which antibodies against E protein are critical for protection against flavivirus infection. Antibodies against DENV NS1 were recently shown to block NS1-mediated vascular leakage (28, 29). The roles of antibodies against other nonstructural proteins are not well understood. Besides antibodies, CD8+ T cell-mediated immunity was shown to protect mice from DENV infection (30). Among the 10 viral proteins, NS3 and NS5 harbor major epitopes that are targeted by the CD8+ T cells (31). It remains to be determined why the efficacies of live attenuated tetravalent dengue vaccine (Dengvaxia) differed in four serotypes (3–5). Since Dengvaxia contains only DENV prM and E genes in the backbone of YFV-17D, no DENV-specific CD8+ T cell immunity to nonstructural proteins can be established (32). This weakness is eliminated in our PIVNS2A vaccine platform, since all structural and nonstructural proteins are expressed during PIVNS2A infection. Since PIVNS2A infection is limited to a single round, the PIVNS2A vaccine is safer than the live attenuated vaccine.
Reversion of PIVNS2A to fully replicative WT virus was a safety concern. We found that the current PIVNS2A is remarkably stable. No WT virus was recovered after continuous culturing of PIVNS2A on NS2A-expressing cells for over 4 months (Fig. 4). Instead, several adaptive mutations were accumulated in the K188-del genomic RNA, leading to an improved PIVNS2A titer of >107 FIU/ml. Furthermore, no WT reversion virus was recovered because no infectious virus was detected after PIVNS2A was inoculated in mice (data not shown). Finally, continuous culturing of PIVNS2A-infected naive BHK-21 cells for five rounds (each round for 3 days) did not yield any infectious virus (data not shown). These in vitro and in vivo results indicate the safety of PIVNS2A as a vaccine candidate. To further minimize the safety concern, other attenuating changes, such as methyltransferase mutations (33–35), could be added to the K188-del genomic RNA.
Flavivirus assembly is a highly regulated process requiring incorporation of nucleocapsid into an enveloped membrane formed by the prM and E proteins. Accumulating evidence indicates that flavivirus assembly is modulated by multiple nonstructural proteins, including NS1 (17), NS2A (18, 19, 22, 36, 37), NS2B (20), and NS3 (21). In the current study, we identified adaptive mutations in both structural and nonstructural genes after continuous culturing of PIVNS2A on NS2A-expressing cells (Fig. 4C). All four independent selections accumulated NS3 mutations. In the NS2A-expressing cells, the NS3 mutation alone moderately improved the PIVNS2A yield and synergistically increased PIVNS2A production when combined with other mutations; in contrast, on naive BHK-21 cells, the NS3 mutations showed negligible effects on viral replication (Fig. 6 and 8). These data clearly underscore the role of NS3 in viral assembly. In agreement with our results, the interplay between NS2A and NS3 in YFV assembly has been well documented: (i) mutations in the helicase domain of NS3 (D343 to A, G, or V) could compensate for the defect in virion production caused by an NS2A mutation (Q189S) (18), (ii) the function of NS3 in virion assembly was independent of its enzymatic activities (21), and (iii) NS3 D343G mutation could rescue the virion assembly defects of an NS2A triple mutant (R22A/K23A/R24A) (36).
Vossmann et al. recently reported NS2A-NS3 interaction during YFV assembly (36). In line with the YFV result, we also detected NS2A-NS3 interaction in DENV. We extended the finding by showing that both protease and helicase domains of DENV-2 NS3 contributed to the NS2A-NS3 interaction. None of the NS2A and NS3 mutations identified in this study affected NS2A-NS3 interaction (Fig. 10 and 11), suggesting that other interactions (with viral and host proteins) are required for flavivirus assembly.
We observed that, in the absence of NS2A K188-del, mutations in structural protein (G100VprM or T69IE) alone increased virion production on the NS2A-expressing cells (Fig. 5F and 7F). Conversely, in the presence of NS2A K188-del, these structural-gene mutations alone reduced PIVNS2A production on the NS2A-expressing cells (Fig. 6C and 8C); however, they synergized with other mutations in nonstructural genes to increase PIVNS2A production (Fig. 6C and 8C). Taken together, the results suggest a fine-tuning network between structural and nonstructural proteins to regulate multiple events during virion assembly, such as transition from replication to package, condensing genomic RNA, rearrangement of capsid, and nucleocapsid encapsulation. The NS2A protein could play a critical role in orchestrating these events during virion formation.
In summary, we have proved the concept of using DENV PIVNS2A as a vaccine approach. We have also demonstrated the feasibility of generating a high titer of such PIVNS2A for vaccine production. The PIVNS2A vaccine approach could be applied to other flaviviruses. Mechanistically, we have identified distinct mutations in both structural and nonstructural genes that participate in DENV-2 assembly. These adaptive mutations have provided entry points to unravel the molecular details of flavivirus assembly. The technology described in this study should be applicable to vaccine development for other viral pathogens.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
All media and antibiotics were purchased from Life Technologies. Naive BHK-21 and C6/36 cells were purchased from the American Type Culture Collection (ATCC), Bethesda, MD. Naive BHK-21 cells were maintained in a high-glucose Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT) and 1% penicillin-streptomycin. C6/36 cells were grown in RPMI 1640 medium containing 10% FBS and 1% penicillin-streptomycin. NS2A-expressing BHK-21 cells were maintained in high-glucose DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1 mg/ml G418 (23). U937 cells expressing dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN) were obtained by lentiviral transfection and subsequent cell sorting (26); the cells were maintained in RPMI 1640 medium supplemented with 5% or 10% FBS.
DENV-2 strain New Guinea C (GenBank accession number AF038403) was generated from an infectious cDNA clone (pACYC-NGC FL) (24). DENV-2 strain D2Y98P was propagated in C6/36 cells and used for challenge experiments (38).
The following antibodies were used in this study: mouse monoclonal antibody (MAb) against DENV-2 E protein 4G2 (ATCC), mouse MAb against myc tag (Sigma), rabbit polyclonal antibody (PAb) against hemagglutinin (HA) tag (Sigma), mouse MAb against glutathione S-transferase (GST) (Abcam), rabbit PAb against GST (Abcam), and goat anti-mouse IgG conjugated with Alexa Fluor 488 or with Alexa Fluor 568 (Life Technologies).
Mice.
Female or male 6- to 8-week-old mice were used. LoxP-flanked ifnar1 (ifnar1fl/fl) (39) animals were bred with mice that express Cre recombinase specifically in CD11c+ dendritic cells (CD11c-Cre). The mice were kindly provided by Ulrich Kalinke (Institute for Experimental Infection Research, Twincore, Center for Experimental and Clinical Infection Research, Hannover, Germany). All the mice were bred and kept under specific-pathogen-free conditions at the Biomedical Resource Centre, Singapore. The mouse experiments were conducted according to the rules and guidelines of the Agri-Food and Veterinary Authority (AVA) and the National Advisory Committee for Laboratory Animal Research (NACLAR), Singapore. The experiments were reviewed and approved by the Institutional Review Board of the Biological Resource Center, Singapore (IACUC protocol 100566). Mice were immunized with 106 IFU PIVNS2As (containing recombinant K188-del plus G100VprM plus P179SNS2A plus A373VNS3 mutations produced as described for Fig. 9F) or 10% FCS medium subcutaneously and analyzed for T cell and antibody responses as previously reported (26).
Plasmid construction.
Standard molecular biology procedures were performed for all plasmid constructions. Two shuttle plasmids were used to generate prM, E, NS2A, NS2B, and NS3 mutations: pACYC-B (containing the T7 promoter followed by nucleotides 1 to 5426 of the DENV-2 genome) and pTA-E (containing viral genome from nucleotide positions 5427 to 10723, followed by the hepatitis delta virus ribozyme sequence [HDVr]). Mutations were individually introduced into the subclone pACYC-B (for prM, E, NS2A, and NS2B mutations) or pTA-E (for NS3 mutations) by site-directed mutagenesis using corresponding primer pairs. The DNA fragments containing individual mutations were cloned into pACYC NGC FL (23) through two pairs of unique restriction sites: SacII (upstream of the T7 promoter) and XhoI (nucleotide position 5426 of the viral genome), and XhoI and ClaI (downstream of the HDVr sequence). To engineer NS2A, NS2B, and NS3 mutations into the pACYC NGC replicon clone, the DNA was digested with NheI (nucleotide position 2545 of the viral genome) and XhoI or XhoI and ClaI from the pACYC NGC FL clones containing corresponding mutations and cloned into the plasmid pACYC NGC replicon through the same restriction enzymes.
The pXJ vector (40), which contains a cytomegalovirus (CMV) promoter and multiple restriction enzyme sites (EcoRI, NotI, BamHI, XhoI, and KpnI), was used to construct the plasmids that transiently expressed NS2A and NS3 proteins. To ensure the correct topology of NS2A expressed in the ER, a leader sequence that contained the signal peptide of Gaussia luciferase (SPG) and the last 16 amino acids of NS1 (NS1C16) were fused in frame to the N terminus of NS2A by overlap PCR (41, 42). An HA tag was fused in frame to the C terminus of the NS2A gene. The resulting PCR product containing the leader sequence-NS1C16-NS2A-HA tag was cloned into the pXJ plasmid through EcoRI and BamHI restriction enzyme sites, resulting in pXJ-SPG-C16-NS2A-HA. Alternatively, GST was fused to the C terminus of NS2A, resulting in plasmid pXJ-SPG-C16-NS2A-GST. For constructs expressing the C-terminally myc-tagged NS3, N-terminally HA-tagged NS3, protease domain (the N-terminal 168 amino acids of NS3), or helicase domain (the C-terminal 450 amino acids of NS3), the corresponding DNA fragments were amplified from pACYC NGC FL using PCR; the PCR products were cloned into the pXJ vector at NotI and KpnI restriction enzyme sites. All the constructs were validated by DNA sequencing and restriction enzyme digestion. Primer sequences are available upon request.
Plasmid transfection, RNA transcription, and RNA transfection.
Plasmids were transfected using an X-tremeGene 9 DNA transfection reagent (Roche) according to the manufacturer's instructions. DENV-2 genome-length or replicon RNAs were in vitro transcribed using a T7 mMessage mMachine kit (Ambion, Austin, TX) from cDNA plasmids prelinearized by XbaI. The RNA transcripts (10 μg) were electroporated into BHK-21 cells or NS2A-expressing BHK-21 cells following a protocol described previously (22).
Co-IP.
HEK 293T cells in a 10-cm dish were transfected with the indicated plasmids. At 48 h p.t., the cells were lysed in 1 ml IP buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 0.5% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace), and EDTA-free protease inhibitor cocktail [Roche]) by rotating at 4°C for 1 h. The lysates were clarified by centrifugation at 15,000 rpm at 4°C for 30 min and subjected to co-IP using protein G-conjugated magnetic beads (Millipore) according to the manufacturer's instructions. Briefly, immune complexes were formed at 4°C overnight by mixing 450 μl of cell lysates with anti-GST mouse MAb (2 μg) in a 500-μl reaction system containing 400 mM sodium chloride and 1 mg/ml bovine serum albumin (BSA). Subsequently, the complexes were precipitated with protein G-conjugated magnetic beads, followed by 4 washes with phosphate-buffered saline (PBS) containing 0.1% Tween 20. Finally, the bound proteins were eluted with 50 μl 4× lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) supplemented with 100 mM dithiothreitol (DTT) and heated at 70°C for 10 min. Ten microliters of eluates were loaded on 4 to 20% Mini-Protean TGX gels (Bio-Rad) and analyzed by SDS-PAGE and Western blotting as previously described (22).
Immunofluorescence assay (IFA).
Cells were grown in an 8-well Lab-Tek chamber slide (Thermo Fisher Scientific). At the indicated time points, the cells were fixed in PBS supplemented with 4% paraformaldehyde at room temperature for 20 to 30 min and permeabilized with 0.1% Triton X-100 in PBS at room temperature for 10 min. After 1 h of incubation in a blocking buffer containing 1% FBS in PBS, the cells were incubated with primary antibody for 1 h in blocking buffer, followed by three washes with PBS. The cells were then incubated with secondary antibody for 1 h in blocking buffer, after which the cells were washed as described above. The cells were mounted in mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories, Inc.). Fluorescence images were taken using a Leica DM4000 B system. Images were merged using Adobe Photoshop CS3 software.
Virus production.
Naive BHK-21 cells or NS2A-expressing BHK-21 cells were transfected with genome-length RNA as described above. The transfected cells were seeded in one T-75 flask (8 × 106 cells in 15 ml DMEM supplemented with 10% FBS). After incubation at 37°C for 24 h, the cells were cultured in DMEM with 2% FBS at 30°C for another 4 days. Every 24 h p.t., 500 μl culture fluids was collected and stored at −80°C. On day 5 p.t., all the culture fluids were centrifuged at 4°C at 500 × g for 5 min, aliquoted, and stored at −80°C. The extracellular viral titer was quantified by plaque assay or IFA on naive BHK-21 cells.
In situ PLA.
PLA was performed using a Duo-link in situ kit (Olink Bioscience) according to the manufacturer's instructions. Briefly, 5 × 104 BHK-21 cells were seeded into each well of an 8-well chamber slide. The next day, the cells were cotransfected with pXJ-SPG-C16-NS2A-HA (WT or NS2A K188-del) and pXJ-NS3-myc (WT or NS3 mutants). Cells transfected with pXJ-SPG-C16-NS2A-HA or pXJ-NS3-myc alone were set as negative controls. At 48 h p.t., the cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking in PBS containing 1% FBS and 0.05% Tween 20 for 1 h, the cells were simultaneously incubated with two different primary antibodies (mouse anti-myc MAb and rabbit anti-HA PAb) for 1 h at room temperature. Subsequent steps (including incubation with PLA probes, ligation, amplification, and preparation for imaging) were performed according to the manufacturer's instructions. Images were acquired using a Leica DM4000 B microscope system armed with 20× objectives.
Electron microscopy.
Naive BHK-21 or NS2A-expressing BHK-21 cells were infected with WT DENV-2 or PIVNS2A. At the indicated time points, the infected cells were fixed with 3% (wt/vol) glutaraldehyde in 0.1 M cacodylate buffer for 2 h at room temperature. The cells were then washed several times in 0.1 M cacodylate buffer, followed by fixation with 1% OsO4 in 0.1 M cacodylate buffer for 1 h. After washing the cells in 0.1 M cacodylate buffer, specimens were dehydrated in graded acetones for 10 to 20 min each. Subsequently, samples were infiltrated with Epon resin and polymerized in molds for 2 days at 60°C. Thin sections (50 nm) were cut on a Leica UC7 ultramicrotome using a Diatome diamond knife and collected on Formvar- and carbon-coated copper mesh grids. Before viewing in a TF30 transmission electron microscope, the cells were poststained with 2% aqueous uranyl acetate (UA) and Reynold's lead citrate.
Flow cytometry-based neutralization assay.
The flow cytometry-based neutralization assay was performed as described previously (34). Briefly, 4G2- or heat-inactivated serum/plasma was serially diluted, and a constant amount of virus was added. The antibody-virus mixture was incubated at 37°C for 30 min and then added to U937-DC cells. After incubation overnight, the infected cells were harvested, washed in PBS, and fixed and permeabilized with Cytofix/Cytoperm (BD). The percentage of infected cells was quantified by flow cytometry detecting intracellular E protein with Alexa Fluor 647-labeled 4G2 antibody. The data were analyzed using GraphPad Prism software for the calculation of the 50% neutralization titer (NT50).
Immunization and infection of mice.
All mice were immunized via the subcutaneous (s.c.) route with 106 IFU of PIVNS2As or 10% FCS. The mice were challenged with 1 × 107 PFU of a highly virulent DENV-2 strain (D2Y98P) via the intraperitoneal route (38, 43). The peak viremia on day 3 postchallenge was measured using RT-PCR, and the weights of the mice were monitored daily.
IgG ELISA.
Ninety-six-well polystyrene plates were coated with polyethylene glycol (PEG)-concentrated, UV-inactivated DENV. The plates were incubated overnight at 4°C. Before use, the plates were washed three times in PBS (pH 7.2) containing 0.05% Tween 20 (PBS-T). After washing, sera were diluted 1:50 in PBS and heat inactivated for 1 h at 55°C, and 3-fold serial dilutions were added to the wells. The plates were incubated for 1 h at room temperature, followed by three washes with PBS-T. Peroxidase-conjugated rabbit anti-mouse IgG in PBS was added, followed by 1 h of incubation at room temperature and three additional washes with PBS-T. Tetramethylbenzidine (TMB) was added as the enzyme substrate. The reaction was stopped with 1 M HCl, and the optical densities were read at 450 nm using an automatic enzyme-linked immunosorbent assay (ELISA) plate reader. Endpoint titers were defined as the lowest dilution of plasma in which binding was 2-fold greater than the mean binding observed with the negative controls.
T cell restimulation.
Mice were immunized with 106 IFU of PIVNS2As or 10% FCS. Five drops of blood were collected in 3 ml of fluorescence-activated cell sorter (FACS) buffer. Cells were collected by centrifugation, and red blood cells were lysed. Single-cell suspensions were stimulated with DENV-2-specific peptides NS3-198, NS3-237, and NS4B-96 for 12 h (34, 44), together with brefeldin for the last 5 h of stimulation, and the cells were subsequently stained for the surface markers anti-mouse CD4 and CD8. The cells were then fixed and stained intracellularly for IFN-γ expression. The cells were acquired on a FACSCanto (Becton Dickinson).
Statistical analysis.
Statistical tests were performed with GraphPad Prism software using Student's t test.
ACKNOWLEDGMENTS
We thank Eric Hanssen for his assistance with the electronic microscope experiment at the Advanced Microscopy facility, Bio21 Institute, University of Melbourne. Xuping Xie received a postdoctoral fellowship from Novartis Institutes for BioMedical Research. Jing Zou received a postdoctoral fellowship from the McLaughlin Endowment at the University of Texas Medical Branch. The Shi laboratory was supported by a University of Texas STARs Award, a Kleberg Foundation Award, UTMB CTSA UL1TR-001439, and NIH grant AI127744.
C.S., X.X., J.Z., R.Z., and R.A. performed the experiments. C.S., X.X., B.Z., R.Z., J.M., K.F., and P.-Y.S. interpreted the data and wrote the manuscript.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB, O'Rourke EJ. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med 146:201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 4.Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramirez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortes Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F, CYD15 Study Group. 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- 5.Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon IK, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A, CYD15 Study Group. 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 6.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group. 2015. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 373:1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead SS. 2016. Development of TV003/TV005, a single dose, highly immunogenic live attenuated dengue vaccine; what makes this vaccine different from the Sanofi-Pasteur CYD vaccine? Expert Rev Vaccines 15:509–517. doi: 10.1586/14760584.2016.1115727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vannice KS, Durbin A, Hombach J. 2016. Status of vaccine research and development of vaccines for dengue. Vaccine 34:2934–2938. doi: 10.1016/j.vaccine.2015.12.073. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach BD, Murray CJ, Thiel H-J, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. 2010. Virus-like particles in vaccine development. Expert Rev Vaccines 9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 11.Pincus S, Mason PW, Konishi E, Fonseca BA, Shope RE, Rice CM, Paoletti E. 1992. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology 187:290–297. doi: 10.1016/0042-6822(92)90317-I. [DOI] [PubMed] [Google Scholar]
- 12.Khromykh AA, Varnavski AN, Westaway EG. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J Virol 72:5967–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qing M, Liu W, Yuan Z, Gu F, Shi PY. 2010. A high-throughput assay using dengue-1 virus-like particles for drug discovery. Antiviral Res 86:163–171. doi: 10.1016/j.antiviral.2010.02.313. [DOI] [PubMed] [Google Scholar]
- 14.Gehrke R, Ecker M, Aberle SW, Allison SL, Heinz FX, Mandl CW. 2003. Incorporation of tick-borne encephalitis virus replicons into virus-like particles by a packaging cell line. J Virol 77:8924–8933. doi: 10.1128/JVI.77.16.8924-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason PW, Shustov AV, Frolov I. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351:432–443. doi: 10.1016/j.virol.2006.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rumyantsev AA, Goncalvez AP, Giel-Moloney M, Catalan J, Liu Y, Gao QS, Almond J, Kleanthous H, Pugachev KV. 2013. Single-dose vaccine against tick-borne encephalitis. Proc Natl Acad Sci U S A 110:13103–13108. doi: 10.1073/pnas.1306245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaturro P, Cortese M, Chatel-Chaix L, Fischl W, Bartenschlager R. 2015. Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog 11:e1005277. doi: 10.1371/journal.ppat.1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol 76:4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J Virol 82:4731–4741. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XD, Deng CL, Ye HQ, Zhang HL, Zhang QY, Chen DD, Zhang PT, Shi PY, Yuan ZM, Zhang B. 2016. Transmembrane domains of NS2B contribute to both viral RNA replication and particle formation in Japanese encephalitis virus. J Virol 90:5735–5749. doi: 10.1128/JVI.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol 82:3342–3352. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Zou J, Puttikhunt C, Yuan Z, Shi PY. 2015. Two distinct sets of NS2A molecules are responsible for dengue virus RNA synthesis and virion assembly. J Virol 89:1298–1313. doi: 10.1128/JVI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou G, Chen YL, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi PY. 2011. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem 286:14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 26.Zust R, Toh YX, Valdes I, Cerny D, Heinrich J, Hermida L, Marcos E, Guillen G, Kalinke U, Shi PY, Fink K. 2014. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: implications for a new mouse model to test dengue vaccines. J Virol 88:7276–7285. doi: 10.1128/JVI.03827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdes K, Alvarez M, Pupo M, Vazquez S, Rodriguez R, Guzman MG. 2000. Human dengue antibodies against structural and nonstructural proteins. Clin Diagn Lab Immunol 7:856–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med 7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 29.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, Hume DA, Stacey KJ, Young PR. 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med 7:304ra142. doi: 10.1126/scitranslmed.aaa3863. [DOI] [PubMed] [Google Scholar]
- 30.Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. 2009. A protective role for dengue virus-specific CD8+ T cells. J Immunol 182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, Teo GH, Gan VC, Lye DC, Leo YS, Hanson BJ, Smith KG, Bertoletti A, Kemeny DM, MacAry PA. 2013. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol 87:2693–2706. doi: 10.1128/JVI.02675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guirakhoo F, Arroyo J, Pugachev KV, Miller C, Zhang ZX, Weltzin R, Georgakopoulos K, Catalan J, Ocran S, Soike K, Ratterree M, Monath TP. 2001. Construction, safety, and immunogenicity in nonhuman primates of a chimeric yellow fever-dengue virus tetravalent vaccine. J Virol 75:7290–7304. doi: 10.1128/JVI.75.16.7290-7304.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li SH, Dong H, Li XF, Xie X, Zhao H, Deng YQ, Wang XY, Ye Q, Zhu SY, Wang HJ, Zhang B, Leng QB, Zuest R, Qin ED, Qin CF, Shi PY. 2013. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J Virol 87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zust R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K. 2013. Rational design of a live attenuated dengue vaccine: 2′-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard K, Shi P-Y, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vossmann S, Wieseler J, Kerber R, Kummerer BM. 2015. A basic cluster in the N terminus of yellow fever virus NS2A contributes to infectious particle production. J Virol 89:4951–4965. doi: 10.1128/JVI.03351-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu RH, Tsai MH, Chao DY, Yueh A. 2015. Scanning mutagenesis studies reveal a potential intramolecular interaction within the C-terminal half of dengue virus NS2A involved in viral RNA replication and virus assembly and secretion. J Virol 89:4281–4295. doi: 10.1128/JVI.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant D, Tan GK, Qing M, Ng JK, Yip A, Zou G, Xie X, Yuan Z, Schreiber MJ, Schul W, Shi PY, Alonso S. 2011. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J Virol 85:7775–7787. doi: 10.1128/JVI.00665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. 2006. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood 108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- 40.Lee CM, Xie X, Zou J, Li SH, Lee MY, Dong H, Qin CF, Kang C, Shi PY. 2015. Determinants of dengue virus NS4A protein oligomerization. J Virol 89:6171–6183. doi: 10.1128/JVI.00546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Pethel M, Falgout B, Lai CJ. 1992. Mutational analysis of the octapeptide sequence motif at the NS1-NS2A cleavage junction of dengue type 4 virus. J Virol 66:7225–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan GK, Ng JK, Trasti SL, Schul W, Yip G, Alonso S. 2010. A non mouse-adapted dengue virus strain as a new model of severe dengue infection in AG129 mice. PLoS Negl Trop Dis 4:e672. doi: 10.1371/journal.pntd.0000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, Shresta S. 2010. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol 185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou J, Xie X, Lee T, Chandrasekaran R, Reynaud A, Yap L, Wang QY, Dong H, Kang C, Yuan Z, Lescar J, Shi PY. 2014. Dimerization of flavivirus NS4B protein. J Virol 88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shustov AV, Frolov I. 2010. Efficient, trans-complementing packaging systems for chimeric, pseudoinfectious dengue 2/yellow fever viruses. Virology 400:8–17. doi: 10.1016/j.virol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]