EBV is a well-established B cell-tropic virus. However, we have recently shown that the EBV type 2 (EBV-2) strain also infects primary T cells in culture and in healthy Kenyan children. This finding suggests that EBV-2, unlike the well-studied EBV-1 strain, utilizes the T cell compartment to persist. As EBV is human specific, studies to understand the role of T cells in EBV-2 persistence require an in vivo model. Thus, we developed an EBV-2 humanized mouse model, utilizing immunodeficient mice engrafted with human cord blood CD34+ stem cells. Characterization of the EBV-2-infected humanized mice established that both T cells and B cells are infected by EBV-2 and that the majority of infected mice develop a B cell lymphoma resembling diffuse large B cell lymphoma. This new in vivo model can be utilized for studies to enhance our understanding of how EBV-2 infection of T cells contributes to persistence and lymphomagenesis.
KEYWORDS: B lymphocytes, B cell lymphoma, Epstein-Barr virus, T lymphocytes, humanized mouse model
ABSTRACT
Epstein-Barr virus (EBV) has been classified into two strains, EBV type 1 (EBV-1) and EBV type 2 (EBV-2) based on genetic variances and differences in transforming capacity. EBV-1 readily transforms B cells in culture while EBV-2 is poorly transforming. The differing abilities to immortalize B cells in vitro suggest that in vivo these viruses likely use alternative approaches to establish latency. Indeed, we recently reported that EBV-2 has a unique cell tropism for T cells, infecting T cells in culture and in healthy Kenyan infants, strongly suggesting that EBV-2 infection of T cells is a natural part of the EBV-2 life cycle. However, limitations of human studies hamper further investigation into how EBV-2 utilizes T cells. Therefore, BALB/c Rag2null IL2rγnull SIRPα humanized mice were utilized to develop an EBV-2 in vivo model. Infection of humanized mice with EBV-2 led to infection of both T and B cells, unlike infection with EBV-1, in which only B cells were infected. Gene expression analysis demonstrated that EBV-2 established a latency III infection with evidence of ongoing viral reactivation in both B and T cells. Importantly, EBV-2-infected mice developed tumors resembling diffuse large B cell lymphoma (DLBCL). These lymphomas had morphological features comparable to those of EBV-1-induced DLBCLs, developed at similar rates with equivalent frequencies, and expressed a latency III gene profile. Thus, despite the impaired ability of EBV-2 to immortalize B cells in vitro, EBV-2 efficiently induces lymphomagenesis in humanized mice. Further research utilizing this model will enhance our understanding of EBV-2 biology, the consequence of EBV infection of T cells, and the capacity of EBV-2 to drive lymphomagenesis.
IMPORTANCE EBV is a well-established B cell-tropic virus. However, we have recently shown that the EBV type 2 (EBV-2) strain also infects primary T cells in culture and in healthy Kenyan children. This finding suggests that EBV-2, unlike the well-studied EBV-1 strain, utilizes the T cell compartment to persist. As EBV is human specific, studies to understand the role of T cells in EBV-2 persistence require an in vivo model. Thus, we developed an EBV-2 humanized mouse model, utilizing immunodeficient mice engrafted with human cord blood CD34+ stem cells. Characterization of the EBV-2-infected humanized mice established that both T cells and B cells are infected by EBV-2 and that the majority of infected mice develop a B cell lymphoma resembling diffuse large B cell lymphoma. This new in vivo model can be utilized for studies to enhance our understanding of how EBV-2 infection of T cells contributes to persistence and lymphomagenesis.
INTRODUCTION
Epstein-Barr virus (EBV) is a well-established B cell-tropic virus able to transform B cells in vitro and drive lymphomagenesis in vivo. However, this virus is also associated with T cell lymphomas. The oncogenic potential of EBV in T cells is demonstrated by the strong association of EBV with a subset of T cell and NK cell lymphomas (1–3) and hydroa vacciniforme-like lymphomas (4). EBV is also associated with T cell lymphoproliferative disorders (LPDs) such as secondary hemophagocytic lymphohistiocytosis (5), hydroa vacciniforme (6), and chronic active EBV (7). Our understanding of the role that EBV plays in these T cell diseases has been hampered by the inability of EBV-1 to infect T cells in experimental settings.
EBV has been classified into two major strains which are referred to as EBV type 1 (EBV-1) and EBV type 2 (EBV-2) based on genetic differences in the latent genes encoding Epstein-Barr nuclear antigen 2 (EBNA-2), EBNA-3a, and EBNA-3c (8–12). These two strains also have functional differences in their transforming capacities. EBV-1 readily transforms B cells in culture, leading to the outgrowth of immortalized lymphoblastoid cell lines (LCL), while EBV-2 is poorly transforming (10, 13, 14). The paradox of EBV-2 is that although infection of B cells in culture does not readily yield LCL, EBV-2 is equally detected in endemic Burkitt's lymphoma (BL) (12, 15, 16), suggesting that the failure to transform B cells in vitro is not representative of EBV-2's oncogenic capability in vivo. In addition, the differing abilities of the two EBV strains to immortalize B cells in vitro are thought to be a model for how EBV establishes latency in vivo (17), suggesting that in vivo the EBV strains use alternative methods to establish latency.
Along these lines, we recently reported that EBV-2, but not EBV-1, readily infects and establishes a latent infection in mature human CD3+ (hCD3+) T cells in vitro (14). Infection with EBV-2 resulted in latent gene expression in T cells and induced activation and proliferation in culture. We have also found that EBV-2 infects T cells in healthy infants (18), strongly indicating that EBV-2 infection of T cells is not an artifact of cell culture but likely a natural part of the EBV-2 life cycle. Because EBV is strictly a human pathogen, it is challenging to study primary infections. Thus, it is currently unclear whether EBV-2 utilizes the T cell compartment to establish latency and/or long-term persistence.
Distinct patterns of EBV latent gene expression are observed in both healthy hosts and in different EBV-associated LPDs (19). EBV-encoded RNAs (EBERs), small nontranscribed, highly expressed RNAs, are found in all EBV latently infected cells and are thus useful for their detection (20). Following primary infection, EBV establishes a growth latency program (also referred to as latency III) in naive B cells, where all EBV latent genes are expressed (e.g., EBNA-1, -2, -3a, -3b, -3c, EBNA leader protein [EBNALP], and latent membrane protein 1 [LMP-1] and LMP-2) (21). This growth program is also seen in B cell LPDs that occur in immunodeficient hosts (22). Similar to EBV-1 in B cells, EBV-2 also expresses the growth program following primary infection of T cells ex vivo (14). Notably, this was the first observation of the growth program in cells of non-B cell origin. A second pattern of latent gene expression is termed latency II in which only EBNA-1, LMP-1, and LMP-2 are expressed. The latency II gene expression profile is observed in germinal center B cells following primary infection (17) and in a subset of Hodgkin's lymphomas (23), nasopharyngeal carcinoma (24), and T/NK cell lymphomas (25). Latency I is restricted to EBNA-1 only and found in memory B cells and in Burkitt's lymphoma (13, 26).
The use of hematopoietic mouse models for studying EBV infection and EBV-driven lymphomagenesis has been well documented (reviewed in reference 27). Early studies utilized a model with the engraftment of peripheral blood lymphocytes (PBL) in severe combined immunodeficiency (SCID) mice (reviewed in reference 28). However, this model had significant limitations due to a number of factors, including the mouse strain used (e.g., SCID mice) and the source of the human cells (e.g., mature PBL). Advancements in engineering greater levels of immunodeficiency in the recipient mice and the use of human CD34+ hematopoietic stem cells (HSCs) to reconstitute the human immune system have led to more robust reconstitution and the development of functional human lymphocytes (reviewed in references 29, 30, and 31). The contributions of different EBV latent and lytic proteins in B cell lymphomagenesis have been studied using these advanced humanized mouse (hu-mouse) models (32–37). Development of diffuse large B cell lymphomas (DLBCL) and LPD following EBV infection has been reported but with variable frequencies (38, 39). The EBV hu-mouse models to date have utilized EBV-1 strains that have well-defined B cell tropism; however, no EBV-2 models have been reported in the literature.
The BALB/c Rag2null IL2rγnull humanized mouse (BRG hu-mouse) model transplanted with cord blood (CB)-derived CD34+ HSCs results in the development of mature human B cells and T cells (40, 41). Both CD4+ and CD8+ T cell subsets are observed in BRG hu-mice along with normal thymic CD4/CD8 expression patterns. The BRG strain was further modified with the introduction of the NOD-derived Sirpa allele (referred to from this point as the BRGS strain), which improves human chimerism (42, 43). A significant advantage of the BRGS hu-mouse model is the combination of B cell maturation and robust IgG production as well as significant thymic engraftment and T cell generation without the need for human thymus cotransplantation, as required in other models (44).
In this study, we used BRGS hu-mice to address two questions: (i) whether, in contrast to EBV-1, EBV-2 can infect T cells in humanized mice, and (ii) whether, similar to EBV-1, EBV-2 is pathogenic in humanized mice. We report here that EBV-2 infection of BRGS hu-mice resulted in infection of T cells as well as B cells and also induces lymphomagenesis, providing an in vivo model to study the role of T cells in EBV-2 persistence and pathogenesis.
RESULTS
EBV type 2 infects CD3+ T cells in humanized mice.
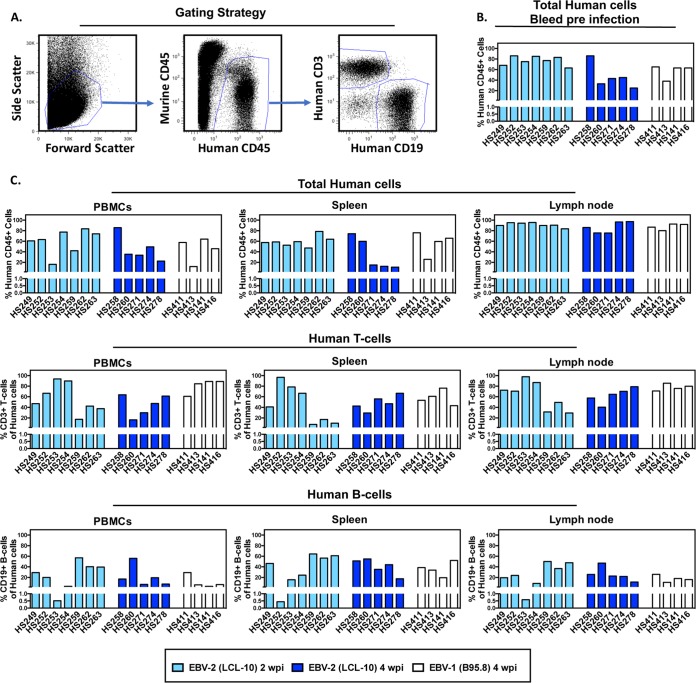
Because we observed that EBV-2 can infect CD3+ T cells both ex vivo (14) and in healthy Kenyan children (18), we hypothesized that EBV-2 could also infect T cells in hematopoietic engrafted humanized mice. To address this question, BRGS mice transplanted at birth with CD34+ HSCs isolated from human umbilical CB (here referred to as hu-mice) were used. Prior to infection with EBV, adult hu-mice were bled, and the percentage of human CD45+ (hCD45+) lymphocytes was determined by flow cytometry to confirm successful hematopoietic engraftment. In hu-mice used for infection, the percentage of human CD45+ cells in peripheral blood ranged from 25% to 86% (Fig. 1A and B). Notably, human chimerism in hu-mice is consistently variable. In addition to differences in stem cell donors, a fluctuating emergence of B cells followed by increased T cell frequencies is common in hu-mice, yet each hu-mouse has its own unique time course, resulting in variable B and T cell numbers among these mice (40). Hu-mice were infected intravenously (i.v.) with 1 × 108 genome copies of EBV-2. For comparison, hu-mice were also infected with the EBV-1 strain (1 × 108 genome copies) or were mock infected with an equivalent volume of phosphate-buffered saline (PBS). Infection of human lymphocytes was analyzed in spleen, lymph nodes (LN), and peripheral blood mononuclear cells (PBMC) at 2 and 4 weeks postinfection (wpi) for EBV-2 and at 4 wpi for EBV-1. At the time of sacrifice, flow cytometry was again used to validate the chimerism of human hematopoietic cells, including T and B cell subsets. All hu-mice had ≥75% hCD45+ cells in LN and between 11 and 86% hCD45+ cells in the spleens and PBMC. Human CD3+ T cells and hCD19+ B cells could be detected in all infected hu-mice with variable ranges in the percentages of these cells in the PBMC, spleen, and LN (Fig. 1C and Table 1). The percentages of hCD45+ chimerism and of the T and B cell subsets among the human cells were not significantly different among the three groups, with the exception of a decreased T cell frequency in the PBMC of EBV-2-infected mice compared to that in EBV-1-infected mice after 4 wpi (Fig. 1C and Table 1).
FIG 1.
Human lymphocyte engraftment levels of BRGS mice pre- and postinfection with EBV. To develop an EBV-2 hu-mouse model, BRGS mice transplanted with CD34+ HSCs isolated from human umbilical CB (hu-mice) were used. (A) CD34+ CB-engrafted BRGS mice were analyzed by flow cytometry with antibodies to murine CD45 and to human CD45, CD3, and CD19 prior to infection and at the time of euthanasia to determine human lymphocyte engraftment levels. (B) Prior to infection, blood samples from hu-mice were analyzed by flow cytometry to confirm engraftment of human lymphocytes. Human chimerism was calculated for each mouse, identified by number on the x axis, and is shown as the percentage of human hematopoietic cells in the total population (human and mouse) of hematopoietic cells [(percentage of hCD45 cells/percentage of hCD45 + mCD45 cells) × 100]. (C) Following euthanasia of EBV-infected hu-mice, the spleen, LN, and blood were harvested. A proportion of each sample was used for flow cytometric analysis of human lymphocyte subsets. Human hematopoietic chimerism was calculated as the percentage of human hematopoietic cells in the total population (human and mouse) of hematopoietic cells [(percentage of hCD45 cells/percentage of hCD45 + mCD45 cells) × 100]. Human B cell (hCD45+/hCD19+) and T cell (hCD45+/hCD3+) subsets were calculated as a percentage of total human CD45+ cells.
TABLE 1.
EBV-infected BRGS hu-mice
| Time point and virus or inoculuma | Mouse | Tumor (tissue) | Total no. of hCD45+ cells |
% hCD3+ T cells |
% hCD19+ B cells |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spleen | LN | Spleen | LN | PBMC | Spleen | LN | PBMC | |||
| 2 wpi | ||||||||||
| LCL-10 | HS249 | No | 4.77E+06 | 2.24E+06 | 40.80 | 72.30 | 47.00 | 46.4 | 19.5 | 29.1 |
| HS252 | No | 1.15E+07 | 7.00E+06 | 96.60 | 70.60 | 66.50 | 0.44 | 23.8 | 20.0 | |
| HS253 | No | 5.00E+06 | 3.35E+06 | 78.50 | 97.80 | 93.70 | 15.6 | 0.58 | 0.51 | |
| HS254 | No | 9.18E+06 | 3.57E+06 | 66.90 | 86.9 | 89.80 | 24.2 | 8.68 | 3.3 | |
| HS259 | No | 1.92E+07 | 1.38E+06 | 7.75 | 31.40 | 17.40 | 64.3 | 50.2 | 57.1 | |
| HS262 | No | 2.88E+07 | 3.31E+06 | 17.10 | 49.30 | 42.40 | 56.5 | 36.7 | 40.3 | |
| HS263 | No | 1.37E+07 | 1.16E+06 | 10.50 | 29.30 | 37.40 | 61.1 | 47.8 | 39.6 | |
| PBS | HS255 | No | 1.95E+06 | 4.41E+06 | 65.30 | 82.00 | 76.80 | 25.6 | 14.5 | 8.61 |
| 4 wpi | ||||||||||
| LCL-10 | HS258 | Yes (spleen) | 3.65E+07 | 1.41E+06 | 42.5 | 57.7 | 63.8 | 51.3 | 25.7 | 17.0 |
| HS260 | Yes (spleen) | 8.50E+07 | 1.30E+06 | 29.4 | 40.3 | 16.4 | 54.9 | 47.1 | 56.0 | |
| HS271 | No | 4.23E+06 | 3.01E+06 | 55.9 | 64.7 | 29.8 | 35.3 | 22.7 | 6.87 | |
| HS274 | No | 2.69E+06 | 2.46E+06 | 46.8 | 70.3 | 47.3 | 44.1 | 21.9 | 19.4 | |
| HS278 | No | 8.7E+05 | 2.02E+06 | 66.5 | 79 | 61.3 | 17.5 | 11.3 | 7.5 | |
| B95.8 | HS411 | Yes (spleen) | 4.22E+07 | 8.58E+06 | 53.4 | 70.9 | 60.7 | 38.5 | 25.9 | 29.2 |
| HS413 | No | 6.67E+06 | 5.13E+06 | 60.7 | 85.5 | 84.5 | 33.8 | 10.7 | 6.06 | |
| HS414 | Yes (spleen) | 1.63E+07 | 1.63E+07 | 76.3 | 75.7 | 88.8 | 19.5 | 18 | 3.12 | |
| HS416 | Yes (spleen) | 2.01E+07 | 9.74E+06 | 43.2 | 79.9 | 88.8 | 52.1 | 16 | 6.51 | |
LCL-10 and B95.8 are EBV-2 and EBV-1 strains, respectively.
To determine whether EBV-2 could infect T cells in hu-mice, the hCD3+ T cell and non-T cell fractions were isolated by magnetic bead separation from spleens, LN, and PBMC of hu-mice at 2 and 4 wpi. DNA was extracted, and quantitative PCR (qPCR) was used to detect and quantify EBV DNA. The purity of the CD3+ T cell fractions was evaluated by flow cytometry (Fig. 2A) (see Materials and Methods), and they consisted of >85% human CD3+ T cells with <1.6% contamination by human CD19+ B cells. At 2 wpi and 4 wpi, EBV DNA was detected in the T cell fractions isolated from LN and PBMC in all EBV-2-infected hu-mice (Fig. 2B). Only 4 of 7 hu-mouse spleen T cell fractions were EBV-2 positive (EBV-2+) at 2 wpi, but by 4 wpi, EBV-2 was detected in the T cell fractions isolated from spleens of every infected hu-mouse. When we calculated the number of EBV copies per 106 cells in each positive cell fraction, the EBV viral load ranged from 1.62 log EBV copies to 6.42 log copies per 1 × 106 cells (Fig. 2B). At 2 wpi significantly higher viral loads were detected in the T cell fractions isolated from LN than from the spleen (P = 0.0312); however, by 4 wpi there were no significant differences in viral loads detected in the T cell fractions isolated from the spleen, LN, and PBMC. To estimate the efficiency of T cell infection in hu-mice, we calculated the approximate frequency of infection, assuming one EBV genome per T cell. The frequencies ranged between 1:104 and 1:508 at 2 wpi and between 1:42 and 1:223 at 4 wpi, with the highest frequencies in the LN at both 2 and 4 wpi.
FIG 2.
EBV type 2 infects T cells in BRGS humanized mice. Hu-mice were infected via i.v. injection with 1 × 108 genome copies of EBV-2 (LCL-10) or EBV-1 (B95.8). At 2 and 4 wpi, human T cell and non-T cell fractions were isolated from spleen, LN, and PBMC via magnetic columns, and DNA was subsequently isolated. (A) Postsorting purity analysis for murine CD45− human CD3 and human CD19 cells was performed on all T cell and non-T cell fractions. Representative flow cytometry plots for hu-mouse HS274 are shown. (B and C) Multiplex qPCR for EBV genome (BALF5 gene) was performed to determine the log EBV copy number per 1 × 106 cells for each cellular fraction. Two groups of EBV-2-infected hu-mice (n = 7) and one PBS control mouse were analyzed at 2 wpi, and two groups of EBV-2-infected mice (n = 5) were analyzed at 4 wpi (B). One group of EBV-1-infected hu-mice (n = 4) was analyzed at 4 wpi (C). Dashed lines indicate the limits of detection of the BALF5 PCR. ND, not detected; NA, not acquired.
We also evaluated EBV infection of the non-T cell fractions harvested from the spleen, LN, and PBMC of these same mice. Due to limited cell numbers obtained from individual mice, it was not feasible to isolate CD19+ B cells directly although they were present in all tissues examined (Fig. 1C and Table 1). Thus, we utilized the B cell-containing non-T cell fractions to monitor infection levels in the B cell compartment. Evaluation of the non-T cell fractions indicated that CD19+ B cells were present in a range from 10.6% to 50.8%, with contamination of <1% human CD3+ T cells (Fig. 2A) (see Materials and Methods). EBV-2 was also detected in the majority of non-T cell fractions isolated from PBMC, spleen, and LN at both 2 and 4 wpi (Fig. 2B). The mean viral loads of EBV-2 in T cells and B cells in different organs were not significantly different. For various experimental groups, PBS-treated hu-mice were included as a control (n = 8); the data of a representative mock-infected hu-mouse (HS255) were included in Table 1. At the time mice were euthanized, PBS-treated hu-mice were screened by PCR for infection, and EBV was not detected in any of these mice (data not shown). Together, these data suggest that EBV-2 was able to persist in both the B and T cell compartments for at least 4 weeks.
EBV type 1 does not infect CD3+ T cells in humanized mice.
Previous studies have not detected EBV-1 infection of T cells in an NOD SCID IL2Rγnull (NSG) humanized mouse model (34). To determine if EBV-1 could infect T cells in the BRGS hu-mouse model, we evaluated EBV load in both the T cell and non-T cell fractions of EBV-1-infected hu-mice at 4 wpi. EBV-1 was detected only in the non-T cell fractions of PBMC, spleens, and LN (PBMC median, 2.67 log copies/1 × 106 human cells; spleen median, 3.37 log copies/1 × 106 cells; LN median, 3.58 log copies/1 × 106 cells) (Fig. 2C). EBV-1 was not detected in the CD3+ T cell fraction in any of the mice tested, confirming that infection of T cells is unique to EBV-2 in the BRGS hu-mouse model. Notably, similar to what was observed in the EBV-2-infected mice, three of the EBV-1-infected mice developed visible tumors in their spleens by 4 wpi (Table 1), suggesting that both EBV types are oncogenic in the hu-mouse model. Collectively, these data demonstrate that EBV-2, but not EBV-1, infects T cells in the BRGS hu-mouse model.
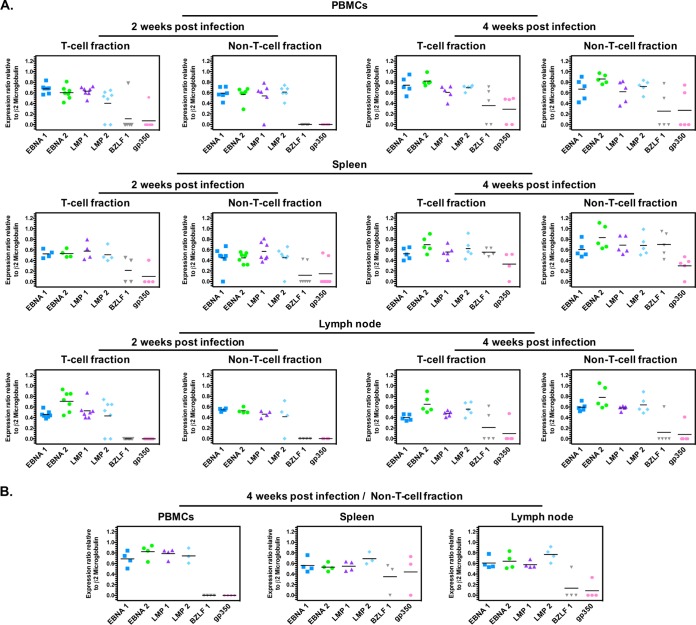
EBV type 2 establishes a latency III infection in T and B cells in humanized mice.
In previously described EBV-1 hu-mouse models reconstituted with human CD34+ HSCs, EBV-infected cells expressed the latency III program (34, 38, 45–48). Lytic infection could also be detected in the EBV-infected hu-mice (34, 38, 46–48). To determine the latency program in EBV-2-infected mice, specifically in T cell compared to non-T cell fractions, EBV gene expression was analyzed in all cell fractions that were positive for both EBV genome and the housekeeping human β2-microglobulin transcripts. The EBV latency gene transcripts LMP-1, LMP-2, EBNA-1, and EBNA-2 and the EBV lytic transactivator gene BZLF1 and late lytic gene gp350 transcripts were measured using a reverse transcription-PCR (RT-PCR) assay and quantified relative to human β2-microglobulin transcript levels (Fig. 3). At 2 and 4 wpi, all four latency gene transcripts were detected in the majority of blood, spleen, and LN samples isolated from EBV-2-infected hu-mice (Fig. 3A), characteristic of the latency III expression pattern. Both T and non-T cell fractions at 2 and 4 wpi were variably positive for BZLF1 and gp350, indicative of lytic reactivation. Of note, at 4 wpi, BZLF1 and gp350 were detected consistently in both the T cell and non-T cell fractions of EBV-2-infected hu-mice. These results demonstrate that EBV-2 established a latency III infection, with evidence of ongoing viral reactivation in both the B and T cell compartments in the BRGS hu-mouse model.
FIG 3.
EBV type 2 gene expression pattern in humanized mouse cell fractions. Purified RNA was isolated from all T cell and non-T cell fractions harvested at 2 and 4 wpi. For all cell fractions that were positive for EBV genome, RNA was reverse transcribed, and biplex PCRs were carried out for EBV genes EBNA-1, EBNA-2, LMP-1, LMP-2, BZLF1, gp350, and human β2-microglobulin. (A) EBV-2-infected mice. Cell fractions were analyzed as follows at 2 wpi: T cell fraction in PBMC, n = 7; non-T cell fraction in PBMC, n = 6; T cell fraction in spleen, n = 4; non-T cell fraction in spleen, n = 7; T cell fraction in LN, n = 7; non-T cell fraction in LN, n = 4. At 4 wpi, n = 5 for all cell fractions. (B) EBV-1-infected mice. At 4 wpi, n = 4 for all non-T cell fractions. The EBV-2 LCL-10 lymphoblastoid cell line was used as a positive control for all latent amplicons, and the EBV-2 Jijoye Burkitt's lymphoma cell line induced to reactivate was used for the lytic amplicon.
Because the infection of hu-mice with EBV-1 has not been characterized, we evaluated EBV gene expression in the non-T cell fractions isolated from EBV-1-infected mice at 4 wpi (Fig. 3B). Consistent with previous reports of a latency III pattern observed in EBV-1 hu-mouse models (34, 38, 46–48), we observed a latency III pattern of gene transcripts in the non-T cell fractions of EBV-1-infected hu-mice. Sporadic BZLF1 and gp350 transcript-positive samples were also observed for non-T cell fractions in EBV-1-infected hu-mice.
Human chimerism and activation status of human T and B cells in EBV type 2-infected mice.
Once we had determined that EBV-2 efficiently infects hu-mice, we further characterized the immune system of these infected hu-mice to detect differences in T cell responses to EBV-2 infection. To this end, we analyzed the human lymphocyte subsets of hu-mice pre- and postinfection with EBV-2 in comparison to those of the PBS-injected littermate controls. We performed this analysis at 2 wpi as this would allow time for the virus to efficiently establish infection. Prior to infection, the hu-mice were bled to determine human T and B cell chimerism (as shown in Fig. 1A) and distributed into experimental groups to have similar ranges of human hematopoietic chimerism (Fig. 4A, left). At 2 wpi spleens, LN, and PBMC were likewise evaluated for human T and B cell chimerism. While the LN and spleens showed similar levels of chimerism between the infected and control hu-mice (data not shown), there was a notable increase in human chimerism in the blood of infected mice over the 2 weeks on an individual mouse basis and using the preblood data as baseline (Fig. 4A, right). This change reflected an overall increase in the percentage of human cells although the frequencies of CD3+ T cells and CD19+ B cells within the hCD45+ blood population did not significantly differ between infected and control mice (Fig. 4A, right).
FIG 4.
Human lymphocyte subsets in EBV type 2-infected versus uninfected humanized mice. (A) Equivalent ranges of human hematopoietic (hCD45+), T (hCD3+), and B (hCD19+) cell chimerism were found in blood of hu-mice prior to infection with EBV-2 or PBS treatment (left). The percent change in human CD45+, T, and B cell chimerism in PBMC of PBS-treated or EBV-2-infected mice is defined as follows (calculated, respectively, for each cell type): (percentage of hCD45, CD3, or CD19 cells at 2 wpi − the percentage of hCD45, CD3, or CD19 cells prebleed)/(percentage of hCD45, CD3, or CD19 cells prebleed) (right). The dashed line represents no change (0%) in chimerism from preinfection to 2 wpi. (B) Representative flow plots and the percentages of naive (CD45RA−), effector memory (CD45RO+ CCR7−), and activated (HLA-DR+) cells among CD4+ and CD8+ T cells, as indicated, in the LN, spleen, or blood (PBMC) of uninfected (PBS) or EBV-2-infected humanized BRGS mice. (C) Representative flow plots and the percentages of CD69+ or CD5 proteins in CD4+ and CD8+ T cells from indicated tissues of control or EBV-2-infected hu-mice. MFI, mean fluorescence intensity. (D) The percentages of B cells that were CD69+ and CD5+ from indicated tissues of PBS-treated or EBV-2-infected hu-mice. Each dot represents an individual mouse from a total of 9 EBV-2-infected and 8 PBS-injected mice from two independent experiments. Bars are means ± standard errors of the means. *, P < 0.05; **, P < 0.01.
In our evaluation of the phenotype of T cells in the EBV-2-infected mice in comparison to that of the PBS controls, we saw no differences among the CD4+ or CD8+ T cell subsets in frequencies of naive (CD45RA+), effector memory (CD45RO+ CCR7−), central memory (CD45RO+ CCR7+), or activated T cells, as defined by HLA-DR+ (Fig. 2B and data not shown). On the other hand, the percentage of CD69+ cells was significantly decreased in the CD4+ and CD8+ human T cell and CD19+ human B cell subsets of infected hu-mice but only in the LN tissue (Fig. 4C and D, left panels). To increase our detection of T cells, we costained tissue samples with anti-human CD3 and CD5 antibodies (Abs). Using this approach, we noticed a significant increase in the level of CD5 expression by both CD4+ and CD8+ human T cells in the PBMC and by CD8+ T cells in the spleen of EBV-2-infected hu-mice relative to levels in noninfected hu-mice (Fig. 4C, right panels). No significant difference in the percentages of CD5+ cells was observed among the CD19+ CD20+ B cells from EBV-2-infected and PBS-treated mice (Fig. 4D, right panel). In summary, we were able to observe some differences in EBV-2-infected mice compared to PBS control mice, including increased human chimerism in the blood, decreased CD69 expression in LN, and increased CD5 expression on human T cells, which suggest that EBV-2 infection modulated the activity of the human immune system in the hu-mouse model.
EBV type 2- and EBV type 1-infected humanized mice develop B cell lymphoma.
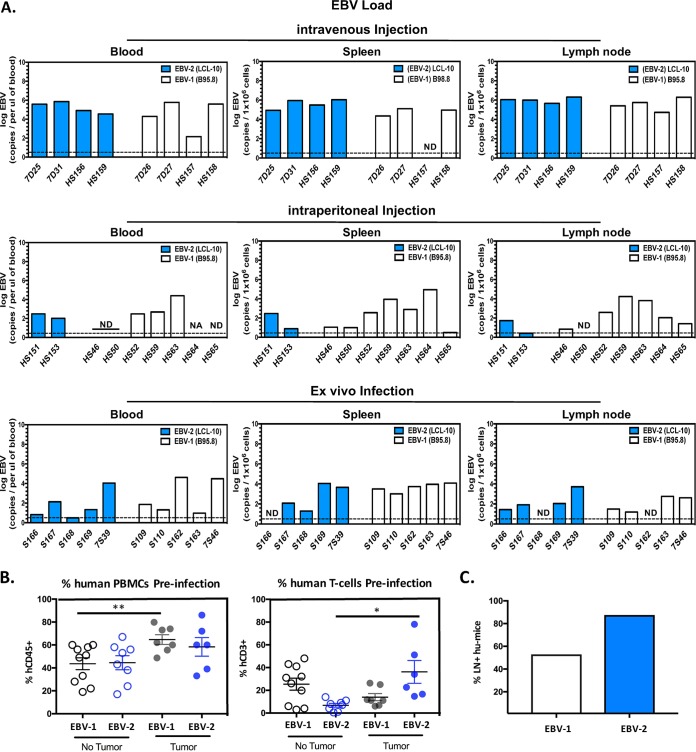
EBV-2 infection of B cells in culture results in limited outgrowth of transformed B cells (15) although EBV-2 is associated with Burkitt lymphoma in people (15, 16). We observed evidence of lymphoma development in both EBV-1- and EBV-2-infected hu-mice at 4 wpi, indicating EBV-2 transformation and oncogenic ability in hu-mice. To determine the pathogenic capacity of the EBV types in the BRGS hu-mouse model, hu-mice were infected with either EBV-1 or EBV-2 and monitored for signs of illness. For these experiments, multiple routes of infection were tested, including direct i.v. and intraperitoneal (i.p.) infection of hu-mice, as well as i.v. injection of ex vivo-infected CB mononuclear cells into nonhumanized BRGS mice. EBV-1 has been shown to induce lymphoma development in a number of hu-mouse models; thus, groups of hu-mice were infected with EBV-1 (n = 16) as a control. In these mice, 75% developed tumors (i.v. infection, 3 out of 4 tumors detected; i.p. infection, 4 out of 7 tumors detected; ex vivo infection, 5 out of 5 tumors detected) between 5 and 9 wpi in the spleen, LN, liver, and/or kidney (Table 2), consistent with previous observations in EBV-1 hu-mouse models (35, 46, 49, 50). The route of infection did not seem to have an effect on the frequency of tumor development in EBV-1-infected mice.
TABLE 2.
Frequency of EBV-2- and EBV-1-infected BRGS hu-mice that developed tumors
| Virus (strain) and mousea | Infection route | Age at time of infection (wk) | Time point (wpi) | Tumor [tissue(s)] | % hCD45+ cells preinfection | Total no. of hCD45+ cells |
% hCD45+ cells |
% hCD3+ cells |
% hCD19+ cells |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen | LN | Spleen | LN | Blood | Spleen | LN | Blood | Spleen | LN | Blood | ||||||
| LCL-10 | ||||||||||||||||
| 7D25 | i.v. | 19 | 6.4 | Yes (spleen, LN) | 59.94 | 1.52E+08 | 2.18E+07 | 69 | 21.90 | 32.9 | 37.3 | 5.02 | 24.7 | 61.4 | 89 | 73.6 |
| 7D31 | i.v. | 19 | 5.5 | Yes (spleen, LN) | 72.31 | 4.12E+07 | 7.75E+07 | 59 | 45.60 | 28.5 | 12.8 | 3.53 | 2.93 | 83.7 | 89.4 | 93.6 |
| HS156 | i.v. | 21 | 6.5 | Yes (spleen, LN, kidney) | 38.87 | 3.97E+07 | 1.55E+07 | 58 | 33.60 | 9.8 | 37.4 | 22.5 | 13.6 | 59.8 | 71.7 | 85.2 |
| HS159 | i.v. | 21 | 8 | Yes (spleen, peritoneal cavity) | 59.81 | 9.42E+05 | 9.42E+05 | 6.00 | 34.60 | 51.4 | 41.9 | 42 | 15 | 49.7 | 45.6 | 75.5 |
| HS151 | i.p. | 16 | 12 | Yes (peritoneal cavity) | 93 | 1.2E+07 | 1.9E+07 | 29.6 | 90 | 40.7 | 68.6 | 90.4 | 89.0 | 29.9 | 8.3 | 6.7 |
| HS153 | i.p. | 16 | 12 | No | 94 | 7.70E+06 | 4.7E+06 | 66.7 | 82.8 | 60.1 | 53.7 | 78.8 | 64.7 | 41.1 | 18.7 | 11.6 |
| S166 | ex vivo | 16 | 8 | No | NAb | 4.03E+06 | 6.31E+05 | 29.65 | 52.6 | 10.4 | 97.7 | 92.2 | 91.9 | 0.06 | 0.05 | 3.4 |
| S167 | ex vivo | 16 | 8 | Yes (spleen) | NA | 4.25E+06 | 5.65E+05 | 35.38 | 35.3 | 40.2 | 93.8 | 95.9 | 94.4 | 3.8 | 0.06 | 1.5 |
| S168 | ex vivo | 16 | 8 | No | NA | 2.41E+06 | 1.54E+04 | 27.34 | 0.96 | 35.0 | 98.3 | 71.6 | 96.8 | 0.06 | 4.5 | 1.3 |
| S169 | ex vivo | 16 | 8 | Yes (spleen, liver, peritoneal cavity) | NA | 3.34E+07 | 2.47E+06 | 29.55 | 41.9 | 72.0 | 98.1 | 85.4 | 89.8 | 0.3 | 5.7 | 9.1 |
| 7S39 | ex vivo | 17 | 6 | Yes (spleen, LN) | NA | 2.84E+07 | 2.86E+06 | 41.35 | 51.1 | 65.1 | 95.5 | 59.5 | 91.4 | 3.6 | 39.8 | 1.0 |
| B95.8 | ||||||||||||||||
| 7D26 | i.v. | 19 | 6.5 | Yes (LN) | 55.54 | 5.00E+08 | 1.70E+08 | 58 | 35.4 | 79.1 | 56.3 | 8.14 | 5.44 | 38.5 | 86.9 | 91.8 |
| 7D27 | i.v. | 19 | 5 | Yes (spleen) | 61.01 | 1.70E+08 | 4.83E+06 | 63 | 65.90 | 25.2 | 13 | 20.7 | 7.23 | 76.6 | 74.2 | 88.4 |
| HS157 | i.v. | 21 | 8 | No | 58.97 | 1.39E+05 | 1.12E+05 | 0.40 | 4.60 | 0.6 | 32.5 | 56.4 | 16.4 | 12.4 | 8.7 | 13.9 |
| HS158 | i.v. | 21 | 8 | Yes (spleen, LN, liver, kidney) | 48.73 | 1,31E+08 | 1.34E+07 | 74 | 58.50 | 45.3 | 43.4 | 28.5 | 9.5 | 37 | 67.3 | 81.2 |
| HS46 | i.p. | 20 | 11 | No | 28 | 4.13E+06 | 2.85E+06 | 45.94 | 57.00 | ND | 94.5 | 89.90 | NA | 3.2 | 8.7 | NA |
| HS50 | i.p. | 20 | 11 | No | 49 | 5.81E+06 | 8.06E+05 | 11.75 | 27.80 | ND | 95.5 | 94.60 | NA | 1.7 | 4.5 | NA |
| HS52 | i.p. | 20 | 11 | Yes (spleen, LN) | 60 | 1.39E+07 | 2.83E+06 | 69.49 | 49.60 | ND | 89.7 | 90.70 | NA | 8.8 | 9.7 | NA |
| HS59 | i.p. | 20 | 10 | Yes (spleen, liver, kidney) | 64 | 8.22E+07 | 4.02E+04 | 80.90 | 2.68 | ND | 60.9 | 11.00 | NA | 20.2 | 46.9 | NA |
| HS63 | i.p. | 12 | 7 | Yes (spleen, liver) | 62 | 4.09E+07 | 7.50E+06 | 58.36 | 43.10 | ND | 25.0 | 39.40 | NA | 72.0 | 54.8 | NA |
| HS64 | i.p. | 12 | 7 | Yes (spleen, liver) | 74 | 6.97E+06 | 5.40E+06 | 24.30 | 75.00 | ND | 44.4 | 18.00 | NA | 51.0 | 76.7 | NA |
| HS65 | i.p. | 12 | 10 | No | 58 | 6.19E+06 | 3.09E+06 | 38.20 | 61.80 | ND | 90.3 | 92.60 | NA | 9.0 | 4.5 | NA |
| S109 | ex vivo | 12 | 7 | Yes (kidney, liver) | NA | 2.52E+07 | 6.96E+06 | 44.49 | 26.1 | ND | 60.2 | 30.3 | NA | 33.3 | 57.9 | NA |
| S110 | ex vivo | 12 | 5 | Yes (spleen, kidney, liver) | NA | 1.90E+07 | 239E+06 | 8.48 | 34.6 | ND | 15.5 | 15.5 | NA | 77.4 | 74.5 | NA |
| S162 | ex vivo | 18 | 7 | Yes (Spleen) | NA | 3.59E+07 | 1.74E+05 | 66.49 | 4.6 | 34.7 | 75.5 | 51.6 | 18.4 | 20.4 | 24.2 | 76.1 |
| S163 | ex vivo | 18 | 6 | Yes (Spleen, LN) | NA | 7.36E+07 | 9.40E+05 | 53.39 | 17.1 | 18.3 | 57.8 | 60.6 | 43.1 | 36.8 | 32.1 | 50.1 |
| 7S46 | ex vivo | 17 | 6 | Yes (Spleen, LN) | NA | 1.56E+08 | 1.43E+06 | 69.72 | 59.7 | 14.9 | 38.5 | 60.9 | 20 | 59.2 | 37 | 75.1 |
LCL-10 and B95.8 are EBV-2 and EBV-1 strains, respectively.
NA, not attained.
Of the mice infected with EBV-2 (n = 11), 8 out of 11 (72.7%) developed tumors (i.v. infection, 4 out of 4 tumors detected; i.p. infection, 1 out of 2 tumors detected; ex vivo infection, 3 out of 5 tumors detected). Similar to what was observed in the EBV-1-infected BRGS mice, tumors were observed in the spleen, LN, kidneys, peritoneal cavity, and liver (Table 2). Of note, the i.v. route of infection appeared most efficient as mice infected with EBV-2 using this route became visibly ill, with signs of malaise and ruffled fur, between 5 and 8 wpi, and 100% of infected mice developed tumors. Comparatively, signs of illness were delayed to 8 and 12 wpi in the majority of hu-mice infected with EBV-2 using the i.p. and ex vivo routes of infection, with 50% and 60% of these mice developing tumors, respectively (Table 2). At harvest, >2-fold-higher EBV-2 loads were detected in all tissues examined from i.v.-infected hu-mice than in those from i.p.- and ex vivo-infected mice (Fig. 5A). Of note, for EBV-2- or EBV-1-infected mice that did not develop tumors, this was not due to failure of the virus to establish infection, as all mice included in the study were EBV positive by PCR in the blood, spleen, and/or LN at the time of harvest (Fig. 5A). A higher percentage of hCD45+ lymphocytes preinfection corresponded with an increased likelihood that EBV-1- and EBV-2-infected mice would develop tumors. However, for the EBV-2-infected mice that developed tumors, the percentage of human T cells preinfection was significantly higher than that of EBV-2 mice that did not develop tumors (Fig. 5B). Interestingly, the majority of EBV-2-infected hu-mice developed LN (Fig. 5C), which does not occur in all hu-mice and requires high human T cell chimerism (40, 42). Collectively, these observations suggest that T cells contribute to lymphoma development in EBV-2-infected mice. Importantly, EBV-2-infected (72.7%) and EBV-1-infected (75.0%) hu-mice developed tumors at comparable frequencies, suggesting that, in contrast to the impaired ability of EBV-2 to immortalize cells in vitro, EBV-2 in vivo infection efficiently induces development of lymphomas with a frequency comparable to that of EBV-1. Notably, both EBV-1 and EBV-2 tumor cells expanded when cultured in vitro and were maintained in culture for up to 2 months, further demonstrating the in vivo transformation capacity of EBV-2.
FIG 5.
Viral loads and immune parameters of EBV type 1- and 2-infected BRGS humanized mice with and without tumor development. Hu-mice were infected utilizing i.v. or i.p. injection with 1 × 108 genome copies of EBV-1 (B95.8 virus) or EBV-2 (LCL-10 virus). A subset of BRGS mice were also infected via the ex vivo route of infection: CD34+ depleted CB cells were infected in vitro with 10 genome copies of EBV-1 or EBV-2 per cell, and at 12 h postinfection 5 × 106 CB cells were i.v. injected into adult nonhumanized BRGS mice. Mice were euthanized when signs of illness were observed (5 to 8 wpi) or at 8 and 12 wpi if signs of illness were not observed. Following euthanasia, spleen, LN, and blood were harvested, and DNA was isolated. (A) Multiplex qPCR for EBV genome (BALF5 gene) was performed to determine the log EBV copy number per 1 × 106 cells for tissues or log EBV copies per microliter of blood. Dashed lines indicate limits of detection of the BALF5 PCR. ND, not detected; NA, not acquired. (B) Tumors developed in humanized mice that had higher human chimerism in blood, as determined by flow cytometry 1 to 6 weeks preinfection (left) or with higher human T cell chimerism, specifically for EBV-2-infected hu-mice, also determined by flow cytometry of PBMC at 1 to 6 weeks prior to infection (right). Each dot represents data from an individual hu-mouse infected with EBV-1 (B95.8) or EBV-2 (LCL-10), and data are segregated according to tumor development in mice, as indicated. Bars are means ± standard errors of the means. *, P < 0.05; **, P < 0.01. (C) Increased frequency of hu-mice with lymph node development among EBV-2-infected hu-mice relative to levels in EBV-1-infected hu-mice.
Characterization of B cell lymphomas in EBV-infected humanized mice.
Tumors from both EBV-1- and EBV-2-infected hu-mice were first analyzed to confirm EBV infection utilizing in situ hybridization (ISH) for EBER. EBV-1- and EBV-2-associated tumors were found to be positive for EBER (Fig. 6A). Additionally, RT-PCR analysis of tumors associated with EBV-1 (n = 3) and EBV-2 (n = 3) showed expression of EBNA-1, EBNA-2, LMP-1, and LMP-2, consistent with the latency III program of EBV gene expression. Transcripts of the EBV lytic transactivator gene BZLF1, indicative of a lytic infection, were also detected in some tumor samples (Fig. 6B). Hematoxylin and eosin (H&E) staining revealed that all tumors showed the histologic morphology of diffuse large B cell lymphoma (DLBCL). Specifically, tumors showed similar morphological features with diffuse infiltration of high-grade lymphoma cells in the tissue. The lymphoma cells were large and had round or slightly irregular nuclei with vesicular chromatin and frequent prominent nucleoli. The cytoplasm was moderate to abundant and showed pale pink staining. Frequent atypical mitotic figures and apoptotic cells were noted. Large areas of coagulating necrosis were commonly present (Fig. 6C). Immunohistochemistry (IHC) analysis established the presence of both CD20+ and CD20− cells in these tumors (Fig. 6D). CD20 is downregulated when B cells differentiate into plasmablasts (51). Tumors were negative for the plasma cell terminal differentiation marker CD138 (Fig. 6D), suggestive of a plasmablastic phenotype (52). In the majority of other EBV-1 hu-mouse models, infected mice developed DLBCL with type III EBV latency and limited detection of the lytic protein BZLF1 (36, 37, 53), consistent with what was observed in EBV-1- and EBV-2-infected hu-mice. For EBER ISH staining, previously confirmed EBV-positive B cell lymphoma tumor tissue was used as a positive control, and EBV-negative B cell lymphoma tumor tissue was used as a negative control; an isotype control and a CD138+ germinal center control were used for IHC staining (Fig. 6E). Collectively, these data demonstrate that both EBV types induced a B cell lymphoproliferative disease resembling DLBCL with immunologic features of plasmablasts, as observed in some cases of EBV+ DLBCL of the elderly (54).
FIG 6.
EBV types 1 and 2 induce diffuse large B cell lymphomas in BRGS humanized mice. Tumors harvested from EBV-infected hu-mice were analyzed utilizing a variety of techniques to characterize EBV-positive lymphomas. (A) Tumors isolated from the spleen and kidney of EBV-1- and EBV-2-infected hu-mice were examined via EBER in situ hybridization (ISH) to detect Epstein-Barr virus (EBV)-encoded small RNA-1 and -2 (EBER-1/2). Digoxigenin-labeled probe (Dako) was utilized to detect EBER-1/2, and nuclear fast red was used as a counterstain. (B) Purified RNA was isolated from tumors and reverse transcribed, and biplex PCRs were carried out for EBV genes EBNA-1, EBNA-2, LMP-1, LMP-2, BZLF1, and human β2-microglobulin. The EBV-2 LCL-10 lymphoblastoid cell line was used as a positive control for all latent amplicons, and the EBV-2 Jijoye Burkitt's lymphoma cell line induced to reactivate was used for the lytic amplicon. (C) Tumors isolated from the spleen and kidney of EBV-1- and EBV-2-infected hu-mice were examined by H&E staining to assess the types of tumors in each animal. Representative H&E-stained EBV-1- and EBV-2-associated tumors isolated from the spleen or kidney are shown. (D) CD20 and CD138 antibody staining of tumor sections isolated from the spleen and kidney of EBV-1- and EBV-2-infected hu-mice. DAB stain was used to visualize positive cells, which appear brown in the images. (E) EBV-positive and -negative tumor tissues were used for EBER ISH staining controls, and an isotype control and CD138+ germinal center control were used for IHC staining. All images were acquired with a Leica DM500 microscope using the 10× and 100×objectives, as indicated.
EBV type 2 lymphomas have a plasmablast phenotype and exhibit monoclonal B cell expansions.
To further characterize the lymphomas that developed in the hu-mice following EBV-1 or EBV-2 infection, we analyzed by flow cytometry digested tumors and lymph tissue that visibly contained tumors. We consistently detected a large population of CD19+ CD20− B cells in lymphoid tissues and tumors from both EBV-1- and EBV-2-infected mice but not in non-tumor-bearing and uninfected control mice (Fig. 7A). In fact, we could observe these CD19+ CD20− cells in blood of hu-mice up to 2 weeks prior to onset of tumor symptoms (data not shown). The CD19+ CD20− B cell phenotype is associated with plasmablasts and plasma cells. To further characterize these B cell subsets in infected hu-mice, we analyzed the CD19+ CD20+ and CD19+ CD20− cells for Ig clonality via intracellular staining for Ig(κ) and Ig(λ) light chains (LCs). In normal human lymphoid tissue, Ig(κ)- and Ig(λ)-expressing B cells exist in roughly equivalent proportions (Fig. 7B), and the majority of B cells express intermediate levels of Ig LC. Only plasmablasts or plasma cells express high levels of intracellular Ig. In the EBV-associated tissues with tumors, we found an increased percentage of CD19+ CD20− cells (Fig. 7A) with increased intracellular Ig LC expression (Fig. 7B and C). Importantly, in the majority of infected mice (Table 2), the tumors appeared monoclonal, with a predominant Ig(λ) population, although in some hu-mice we observed monoclonal Ig(κ) B cells and polyclonal populations (Fig. 7B and C).
FIG 7.
Clonality and immunophenotype of EBV type 1- and 2-associated tumors. (A) CD20 expression on CD19+ (hCD45+ CD19+ gate) cells in spleen and LN from uninfected or EBV-2-infected humanized mouse, as indicated. (B) Expression (intracellular plus extracellular) of Ig(κ) and Ig(λ) light chains in CD19+ CD20+ or CD19+ CD20− B cells, as indicated, from uninfected LN or from spleen or LN cells from EBV-2-infected hu-mouse. The upper right graph illustrates the gating for Ig(κ) (y axis) or Ig(λ) (x axis) follicular (FO) or plasmablast (PB) populations that was used for all flow plots. (C) Ig light chain expression in tumor-associated tissues from EBV-1- or EBV-2-infected hu-mice. The columns show the percentages of plasmablasts (CD19+ CD20−) that are Ig(λ)+ (intracellular [IC] plus extracellular [EC]). (D) CD10, CD21, and CD38 expression of CD19+ CD20+ and CD19+ CD20− cells, as indicated, from EBV-2 tumor-associated tissue. Blue lines are included as positive controls and represent expression of CD10 on immature splenic B cells, of CD21 on mature LN B cells, and of CD38 on mature splenic B cells from a normal hu-mouse. The filled gray lines represent negative controls and the expression of the relevant protein on mCD45+ cells. Data are representative of at least 3 mice infected with EBV-2 and are similar to phenotypes observed in lymph tissue from tumor-bearing EBV-1-infected mice.
We also characterized the lymphomas found in the spleen for expression of CD5, CD10, CD19, CD21, CD27, CD38, and IgD proteins, markers that identify distinct B cell subsets. Single-cell suspensions were analyzed by flow cytometry. The B cell tumors were CD5− CD10− CD19lo CD21− CD27+ CD38high IgD− (Fig. 7D and 8; also data not shown), a phenotype consistent with plasmablasts (55). Interestingly, comparison of the EBV-1- and EBV-2-associated tumors revealed remarkable similarities in the tumor immunophenotypes, including not only a plasmablastic phenotype but also infiltration of human T cells and downregulation of the EBV receptor CD21 on B cells (Fig. 7D and 8). Collectively, these data demonstrate that both EBV strains induced a B cell lymphoproliferative disease resembling DLBCL.
FIG 8.
Side-by-side comparison of EBV type 1 and 2 tumor immunophenotypes. Flow cytometric analysis was performed on tumors isolated from the spleen of an EBV-2- and EBV-1-infected hu-mouse. Tumor single-cell suspensions were initially gated on for human lymphocytes (murine CD45−) and assessed for the percentages of T cells (CD3+) and B cells (CD19+). Human CD19+ B cells were further analyzed for expression of surface IgD, CD38, CD27, CD20, CD10, and CD21.
DISCUSSION
Since the first description of the second strain of EBV, the biology of EBV-2 has represented a paradox. Unlike the worldwide distribution of EBV-1, EBV-2 has a more restricted geographic prevalence (56). EBV-2 differs genotypically from EBV-1 in key latency genes as well as in two lytic genes (8–12). EBV-2 does not readily establish immortalized B cell lines while the capacity to immortalize B cells is a prominent feature of EBV-1 (8, 57). And, in contrast to EBV-1, we found that EBV-2 readily infects resting primary T cells in vitro and establishes a persistent infection of T cells of healthy Kenyan children (14, 18). In this paper, we used a humanized mouse model to understand the unique biology of EBV-2. Consistent with both our in vitro and in vivo observations (14, 18), EBV-2, but not EBV-1, established latency in T cells in hu-mice. Surprisingly, in spite of the poor in vitro B cell-immortalizing ability of EBV-2, we observed B cell lymphoproliferations characteristic of DLBCL in EBV-2-infected hu-mice. These results suggest that infection of T cells by EBV-2 is part of the normal biology of this virus.
Consistent with the observations of Ma et al., who reported that T cells are not infected by EBV-1 in NSG hu-mice (34), we also did not observe EBV-1 infection of T cells in the hu-mice. However, EBV-2 infected both T and B cells in hu-mice. Interestingly, the viral loads are comparable to those observed in Kenyan infants' T cell fractions when calculated as the EBV copy number/cell (18). The viral loads in the T and B cells in the hu-mice were also comparable and indicative of a robust infection. The EBV-2-infected T cells in hu-mice expressed a latency III program in both peripheral circulating lymphocytes and secondary lymphoid tissue (e.g., lymph nodes and spleen). This program is characteristic of the pattern of viral latent gene expression in immortalized B cells in vitro and in B cell LPD in immunocompromised patients in vivo. The latency III transcription program, also termed the growth program, is also thought to occur in lymphoid tissue following primary EBV infection of B cells (17). That we are also observing this is in EBV-infected T cells suggests that this is a common lymphotropic program and not unique to B cells.
Transcripts of the lytic transactivator gene BZLF1 were also detected in a number of the T cell and non-T cell fractions, indicative of a productive lytic infection or early latent infection of newly infected cells. Transcripts of one late lytic gene, gp350, were also detected in many of the T cell and non-T cell fractions, providing further support for productive lytic infection occurring in EBV-2-infected hu-mice. We did not further evaluate EBV-2-infected hu-mice for production of infectious virus, so we cannot rule out a potential abortive infection. Importantly, no BZLF1 transcripts were detected following infection of T cells in vitro (14), pointing to a striking difference between in vitro and in vivo infection of T cells by EBV-2. Previous hu-mouse model studies utilizing BZLF1 mutant viruses suggested that lytic EBV infection was essential for lymphoma development (33, 34), and given that EBV-2 induced B cell lymphomas in the hu-mice, this is consistent with those observations.
Important questions in hu-mice concern the effects of EBV-2 T cell infection and whether the mice mount an immune response to EBV-2. We detected limited alterations in the EBV-2-infected T cells compared to the T cells of PBS-injected control hu-mice. However, our observations of a significant decrease of the CD69 protein in LN are consistent with trends of lower HLA-DR (also an activation marker) and CCR7 expression, which, similar to CD69, retains lymphocytes in LN. Furthermore, after T cell activation levels of CD5 are known to increase (58) on T cells in the blood and spleen, suggesting an exit of activated T cells from the LN at 2 weeks following EBV-2 infection. Along these lines, another interesting effect of EBV-2 infection was increased human chimerism in the blood. Perhaps the combination of EBV-2-infected T and B cells proliferating and activated T cells exiting the LN and proliferating in response to the infection itself collectively contributes to the increased human chimerism in the blood. Additionally, EBV-2-infected hu-mice developed LN at an increased frequency relative to infection with EBV-1. It is well established in many models of humanized mice that LN development is delayed, typically occurring 3 to 4 months following human engraftment, and that the development of LN does not occur in all hu-mice and is dependent on high human T cell chimerism (40, 42). Thus, the ability of EBV-2 to infect T cells could perhaps have an intriguing correlation with the increased human chimerism and LN development following infection in hu-mice.
Given that EBV-2 poorly immortalizes B cells in culture, it was a reasonable hypothesis that in hu-mice, EBV-2 infection would not result in B cell lymphomas. However, hu-mice infected with EBV-2 developed B cell lymphomas resembling DLBCL, as determined by both pathology analysis and flow cytometric phenotype. The tumors that developed in our EBV-2-infected mice were typically a mixed B cell population, with a polyclonal repertoire evident among the CD20+ B cells and a more monoclonal population evident in the CD20− population. In most of the tumor-associated tissues there was an expanded CD20− monoclonal population, as defined by LC isotype. The apparent contrast between EBV-2's poor immortalizing capacity in vitro compared to EBV-2's lymphomagenic capacity in vivo raises as yet unanswered questions as to how the in vivo environment supports this ability. One intriguing possibility is that infection of T cells by EBV-2 supports the transformation of B cells in vivo. Notably, other investigators have demonstrated that in EBV-1 hu-mouse models, CD4 T cells contribute to viral persistence and lymphomagenesis (36, 48, 59). Although T cells are not infected by EBV-1, these studies suggest that CD4 T cells contribute to EBV-1 persistence and lymphomagenesis by providing CD40 signaling when LMP-1 is no longer expressed (36, 48). Further studies using the EBV-2 hu-mouse model described here are required to determine the role that infected T cells play in EBV-2 persistence and lymphomagenesis.
These EBV-2 B cell lymphomas were morphologically similar to EBV-1-induced DLBCL and arose with similar kinetics and frequencies as well as similar EBV latency III gene expression profiles. The lymphomas induced in EBV-infected hu-mice are similar to DLBCLs observed in elderly humans with respect to histology, plasmablastic phenotype, expression of the latency III program, and monoclonality (reviewed in reference 60). EBV-1 and EBV-2 are detected with equal frequency in Burkitt's lymphoma (12, 15, 16) and in HIV-positive non-Hodgkin lymphomas (61, 62), so there is evidence for EBV-2 to drive B cell lymphomagenesis.
EBV is also associated with malignancies and diseases that originate from T cells, including NK/T cell lymphomas (1), hemophagocytic lymphohistiocytosis (5), hydroa vacciniforme (6), and chronic active EBV (7, 63). The etiology of these T cell diseases remains unknown, and previously only xenograft mouse models have been utilized to reproduce features of EBV-associated T cell diseases (39). The ability to readily infect T cells with EBV-2 in the BRGS hu-mouse model could allow the elucidation of EBV's role in the etiology of these EBV-associated T cell lymphomas and LPD. However, we did not observe any signs of T cell lymphoproliferative disease in the EBV-2-infected hu-mice. Relatedly, we have previously performed long-term in vitro cultures of EBV-2-infected primary T cells. EBV-2-infected T cell cultures consistently survived out to 9 wpi, with detectable EBV latent gene transcripts detectable through at least 5 wpi, demonstrating that infection is maintained in the T cells (C. B. Coleman and R. Rochford, unpublished observations). Yet while EBV-2 substantially prolongs the survival of primary T cells in vitro, we have yet to generate an immortalized T cell line. This suggests that, unlike infection of B cells, EBV is unable to independently immortalize T cells and that additional genetic hits may be required for the development of EBV-associated T cell lymphoproliferative disorders. It is possible that the rapid development of B cell lymphomas in the EBV-2-infected hu-mice did not allow sufficient time for the development of T cell lymphoproliferative disorders or that other genetic hits are needed.
While there was minor contamination of the isolated T cell fractions, several lines of evidence support the T cell infection. Importantly, the isolated T cell fractions had only minor contaminations of murine cells and human B cells; the levels of human B cell contamination of the T cell fractions were 0.00 to 1.59% and <8% for murine cells (not a target for EBV). Additionally, at 2 wpi there were T cell fractions isolated from the LN that were positive for EBV while the corresponding non-T cell fraction, which contains B cells, was negative for EBV. And T cell fractions isolated form B95.8-infected hu-mice, which had very similar percentages of contaminating B cells (0.2 to 1.48%), were all EBV negative. Further, viral loads in the T cell and non-T cell fractions of LCL-10-infected mice were similar; thus, although the non-T cell fractions contained only 10.6 to 50.8% B cells, the minor B cell contamination of the T cell fractions would not account for the viral loads that were detected in the T cells fractions. Further, our results are consistent with our previous studies showing EBV-2 infection of T cells in vitro (14) and in healthy infants (18). Because of the limited cell numbers isolated following EBV-2 infection, we were not able to confirm if EBV-2 infected CD4 or CD8 T cells.
Additional factors must be considered in evaluating EBV pathogenicity in hu-mouse models. For example, different EBV-1 strains have been shown to cause various EBV-associated diseases in these hu-mouse models. As demonstrated here and by others, infection with the EBV-1 B95.8 isolate resulted in development of a B cell lymphoma resembling DLBCL (33, 34, 46). In our model, we found that both the EBV-1 (B95.8) and EBV-2 (LCL-10) strains induced B cell lymphomas with striking similarities. In both strains the DLBCL tumors had a plasmablast phenotype, and the lymphoproliferative disease appeared to begin as a polyclonal process with outgrowth of monoclonal tumors. In fact, in individual mice we found multiple tumors with distinct monoclonal populations [Ig(κ) or Ig(λ)] based on tumor location (data not shown). On the other hand, infection of hu-mice with the Akata EBV-1 strain has been shown to induce B cell lymphoproliferative disease (38), arthritis (64), or hemophagocytic lymphohistiocytosis (HLH) (65); however, the factors that dictate which disease state occurs are currently unclear. In this study, we used an EBV-2 strain (LCL-10) derived from a Kenyan EBV-2 isolate. Interestingly, we found that the EBV-2 strain isolated from the Jijoye BL cell line was incapable of establishing infection in our hu-mouse model (data not shown). The association of the two EBV types with specific human diseases is not apparent, but there seem to be clear differences in individual EBV strains in hu-mouse models, beyond those specific to the two EBV types, that affect the pathogenicity of EBV.
Using our human chimerism analysis of PBMC prior to infection as a baseline to assess efficacy of tumor development, we found a clear correlation with higher human chimerism and tumor development in both strains but more notably with EBV-1 infection. Consistent with the reduced tumor incidence in the hu-mice with lower human chimerism preinfection, hu-mice with <30% hCD45+ cells preinfection were often not productively infected, or viral loads were below our PCR limit of detection of 4 genome copies. Importantly, by measuring the frequency of human T cells among the hCD45+ cells in the blood preinfection, we noted that mice with a higher frequency of T cell chimerism had a higher incidence of B cell tumor development by EBV-2. In contrast, this correlation was not observed in EBV-1-infected mice that developed lymphomas. This supports the hypothesis that infection of T cells by EBV-2 supports the transformation of B cells. The increase of CD8+ T cells in the LN and spleens of mice with tumors could either be an expansion of EBV+ T cells or the mounting of an anti-tumor response by the human T cells.
We have shown here that EBV-2 infection of hu-mice results in establishment of a latency III gene program in T and B cells and the subsequent development of B cell lymphomas resembling DLBCL. Thus, the EBV-2 BRGS hu-mouse model described here provides an important in vivo model to study the role of T cells in EBV-2 persistence and pathogenesis.
MATERIALS AND METHODS
BRGS humanized mice.
For this study, we utilized BALB/c Rag2null Il2rγnull SirpaNOD (BRGS) mice that are immunodeficient, devoid of murine T, B, and NK cells, and highly permissive to xenograft transplantation as previously described (42, 43). CB CD34+ cells were prepared using a CD34-positive selection kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and expanded in culture as previously described (40). Approximately 1.0 × 105 to 4.0 × 105 in vitro-expanded CD34+ cells were injected intravenously (i.v.) (typically) or intraphepatically (i.h.) (less frequently) into 1- to 3-day-old BRGS mice that were previously irradiated with 300 rad. For ex vivo infection of BRGS mice, the CD34− population of CB mononuclear cells was infected in vitro and the following morning injected i.v. into tail veins of lightly irradiated (300 rad) adult BRGS mice. Human umbilical CB samples, rejected due to low volume or other reasons, were obtained from the University of Colorado Cord Blood Bank at ClinImmune Labs (Aurora, CO).
Investigators in this study were blinded from donor identities, and the studies were performed in compliance with University of Colorado Institutional Review Boards. Hu-mice were housed and maintained on a diet enriched with Septra under specific-pathogen-free and biosafety level 2 conditions at the University of Colorado, Denver, Anschutz Medical Center (UCD-AMC) vivarium (Aurora, CO). Animal procedures were approved by the UCD-AMC Institutional Animal Care and Use Committee.
Evaluation of human chimerism and immunophenotype by flow cytometry.
PBMC from hu-mice between 8 and 18 weeks of age were analyzed by flow cytometry with antibodies (Abs) to murine CD45 (mCD45), human CD45 (hCD45), hCD3, hCD5, hCD8, hCD19, and hCD20 at least twice to confirm engraftment of human B and T cells. Human chimerism is defined as the percentage of human (hCD45+) hematopoietic cells in the total population (human and mouse) of hematopoietic (hCD45+ and mCD45+) cells. Abs used in flow cytometric analyses were as described previously (40, 41, 43) and purchased from BioLegend (San Diego, CA). Cells were incubated with Abs in staining buffer (PBS, 1% bovine serum albumin [BSA], 0.1% sodium azide) for 15 min at 4°C and washed two times with the same buffer. Samples were collected on a cyan analyzer at the flow cytometry core at the UCD Cancer Center, and analyses were performed with FlowJo software, as described previously (40).
Production of EBV stocks and infection of hu-mice.
The EBV cell lines B95.8 and LCL-10 were used in this study for production of virus. The B95.8 cell line has previously been typed and found to carry EBV-1 (66), and the LCL-10 cell line carries type 2 EBV (67). EBV stocks were generated as previously described (14). Virus stocks were quantified following DNase treatment by quantitative PCR (qPCR), as described below. Hu-mice (with proven human hematopoietic chimerism) were directly infected via i.p. or i.v. injection with 1 × 108 genome copies of EBV-1 (B95.8 virus stock) or EBV-2 (LCL-10 virus stock) in a total volume of 500 μl or 100 μl, respectively. Alternatively, mice were indirectly infected (ex vivo method) as follows. CD34+ cell-depleted cord blood mononuclear cells (5 × 106) were infected ex vivo with 10 genome copies per cell of EBV-1 (B95.8 virus stock) or EBV-2 (LCL-10 virus stock), incubated for 12 h, and transferred i.v. into intact BRGS mice. Control mice were i.v. injected with 100 μl of PBS. Following infection, mice were monitored daily for signs of malaise and ruffled fur as indicators of illness. Mice were euthanized when signs of illness were observed or at indicated time points.
Analysis of human lymphocyte subsets in EBV-infected mice.
Following euthanasia of EBV-infected or control hu-mice, the spleen, LN, and blood were harvested. Tissues were homogenized to single-cell suspensions, and PBMC were isolated from whole blood by Ficoll-Paque purification. For experiments evaluating which cell fractions were infected by EBV or the incidence of tumor development in the BRGS hu-mouse model, a portion of each sample was used for flow cytometric analysis of human lymphocyte subsets (anti-mCD45, -hCD45, -hCD3, and -hCD19). The remaining cells were used to assess EBV load and gene expression patterns in each compartment, as described below. Mice with <1 × 106 total human lymphocytes in the spleen and LN were excluded due to insufficient numbers from studies in which T cell and non-T cell fractions were analyzed. For experiments evaluating the incidence of tumor development in the BRGS hu-mouse model, mice in which virus was undetectable in the PBMC, spleen, and LN at the time of harvest, utilizing the PCR for EBV copy number described below, were excluded from the study. For experiments comparing lymphocyte subsets and activation status in EBV-2-infected versus PBS control mice at 2 wpi, samples were stained, as described above, for hCD45, hCD3, hCD4, hCD8, hCD19, hCD20, hCD45RA, hCD45RO, hCCR7/CD197, hCD69, hCD5, and hHLA-DR and analyzed via flow cytometric analysis.
Tumor tissue analysis.
Following euthanasia of EBV-infected hu-mice, observed tumors were harvested, and a portion was formalin fixed. The remaining portion of the tumors was homogenized to obtain single-cell suspensions. Paraffin-embedded sections and tumor cell suspensions were then analyzed utilizing a variety of techniques to characterize EBV-positive lymphomas.
(i) H&E stain.
Tumors from hu-mice infected with EBV-1 (4 tumors) and EBV-2 (5 tumors) were stained with hematoxylin and eosin (H&E) and analyzed by a pathologist to classify tumor types.
(ii) EBER in situ hybridization (ISH).
Digoxigenin-labeled probe (Ventana Medical Systems, Inc., Tucson, AZ), with nuclear fast red as a counterstain, was utilized to detect Epstein-Barr virus (EBV)-encoded small RNA-1 and -2 (EBER-1/2) for EBV infection. The assay was performed on a Ventana Benchmark XT platform with an iViewBlue detection kit according to the manufacturer's instructions (Ventana Medical Systems, Inc., Tucson, AZ).
(iii) Immunohistochemistry.
Following deparaffinization and rehydration, tissue sections were blocked with 10% BSA for 30 min in a humidity chamber; the primary Ab was incubated overnight in the humidity chamber at 4°C, followed by secondary Ab added for 30 min to the humidity chamber at room temperature (RT). Abs and dilutions were as follows: anti-CD20 (1:200, clone MEM-269; Thermo Fisher), anti-CD138 (1:100, clone DL-101, BioLegend), and goat anti-mouse IgG/IgM (1:500; Thermo Fisher). Subsequently, slides were stained with diaminobenzidine tetrachloride (DAB) (ImmPACT DAB; Vector Laboratories, Inc.), and counterstaining was done with Vector Hematoxylin QS (Vector Laboratories). A nonspecific binding control, matched for concentration and isotype, was used for each slide. All images were acquired with a Leica DM500 scope utilizing 10× and 100× objectives.
(iv) Flow cytometric analysis.
Tumor cell suspensions were analyzed by flow cytometric analysis to determine the tumor cell phenotype and clonality of the tumors. Tumor cells were stained, as described above, with antibodies to cell surface proteins, including mCD45, hCD45, hCD3, hCD19, hCD20, hIgD, hCD38, hCD27, hCD10, and hCD21. For evaluation of intracellular immunoglobulin (Ig) light chains, following extracellular staining cells were fixed with 2% formaldehyde for either 5 min at 37°C or 20 min at RT. Fixed cells were washed two times in staining buffer and then incubated with saponin permeabilization buffer (0.5% BSA, 0.5% saponin in PBS) for 15 min at RT. Cells were subsequently stained with Abs for Ig(κ) or Ig(λ) for 30 min in the dark at RT, followed by five washes in staining or saponin buffer.
Isolation of T cell and non-T cell fractions.
T cell and non-T cell fractions were isolated from the PBMC, LN, and spleens of EBV-2-infected hu-mice at 2 wpi and 4 wpi and from EBV-1-infected hu-mice at 4 wpi. First, human cells were enriched from the spleens of hu-mice by negative depletion of mouse cells using mCD45 magnetic columns (Miltenyi Biotec, Bergisch Gladbach, Germany). This enrichment was not performed for PBMC and LN. For all tissues, human CD3+ T cell and non-T cell fractions were isolated using a human Pan T Cell Isolation kit (Miltenyi-Biotec, Bergisch Gladbach, Germany). Purity analysis of cell fractions was assessed with anti-mCD45, -hCD3, and -hCD19 Ab staining. Purity of human T cell and non-T cell fractions was calculated on mCD45− cells. Notably, mCD45+ cells accounted for <8% of all cell fractions analyzed. The purity of T cell fractions ranged from 87.4% to 92.2%, with a contamination of 0.00% to 1.59% human CD19+ B cells. The percentage of human CD19+ B cells in the non-T cell fraction ranged between 15.2% and 48.1% in PBMC, 25.8% and 50.8% in spleen, and 10.6% and 21.7% in LN, with a contamination of 0.00% to 0.65% human CD3+ T cells. DNA and RNA were extracted from cell fractions using an AllPrep DNA/RNA kit (Qiagen, Venio, Netherlands).
PCR for EBV copy numbers.
EBV DNA levels were quantified as described previously using primers and probes designed to detect a 70-bp region of the EBV BALF5 gene and the human β-actin gene as a control for DNA input (68, 69). PCRs were run using an iCycler thermocycler equipped with an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA).
RT-PCR for EBV latent gene transcripts.
Purified RNA was reverse transcribed using a QuantiTect reverse transcription kit (Qiagen, Venio, Netherlands). Primers and probes were obtained as previously described (70), and biplex PCR was performed for EBV transcripts and human β2-microglobulin using iTaq Universal Probes mix (Bio-Rad, Hercules, CA). RNA isolated from either the LCL-10 or Jijoye BL line was used as a positive control for EBV-2 transcripts.
ACKNOWLEDGMENTS
This study was supported by NIAID R21 A1122670 (R.R. and R.P.), NIAID R01 A1124474 (R.P.), and the University of Colorado Anschutz Medical Campus (AMC) Cancer Center.
We thank the University of Colorado AMC Vivarium for assistance with mouse husbandry and the histology core for assisting with preparation of tumor tissues. We are very grateful to all members of their laboratories for numerous useful discussions. We are grateful to ClinImmune Labs (University of Colorado) for providing human umbilical CB samples. We thank Hélène Strick-Marchand (Institut Pasteur, Paris, France) for providing the BRGS mouse strain.
C.B.C., J.L., L.A.S., and N.A.S. performed the experiments; Z.P. interpreted the histological findings; B.H. and Z.P. analyzed tumor immunophenotype and characterization. B.M.F. contributed to humanization of BRGS mice. C.B.C., J.L., R.P., and R.R. designed the study.
We have no conflicts of interest to declare.
REFERENCES
- 1.Park S, Ko YH. 2014. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorders. J Dermatol 41:29–39. doi: 10.1111/1346-8138.12322. [DOI] [PubMed] [Google Scholar]
- 2.Gru AA, Haverkos BH, Freud AG, Hastings J, Nowacki NB, Barrionuevo C, Vigil CE, Rochford R, Natkunam Y, Baiocchi RA, Porcu P. 2015. The Epstein-Barr virus (EBV) in T cell and NK cell lymphomas: time for a reassessment. Curr Hematol Malig Rep 10:456–467. doi: 10.1007/s11899-015-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haverkos BM, Gru AA, Geyer SM, Bingman AK, Hemminger JA, Mishra A, Wong HK, Pancholi P, Freud AG, Caligiuri MA, Baiocchi RA, Porcu P. 2016. Increased levels of plasma Epstein-Barr virus DNA identify a poor-risk subset of patients with advanced stage cutaneous T-cell lymphoma. Clin Lymphoma Myeloma Leuk 16(Suppl):S181–S190.e4. doi: 10.1016/j.clml.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintanilla-Martinez L, Ridaura C, Nagl F, Sáez-de-Ocariz M, Durán-McKinster C, Ruiz-Maldonado R, Alderete G, Grube P, Lome-Maldonado C, Bonzheim I, Fend F. 2013. Hydroa vacciniforme-like lymphoma: a chronic EBV+ lymphoproliferative disorder with risk to develop a systemic lymphoma. Blood 122:3101–3110. doi: 10.1182/blood-2013-05-502203. [DOI] [PubMed] [Google Scholar]
- 5.Kasahara Y, Yachie A. 2002. Cell type specific infection of Epstein-Barr virus (EBV) in EBV-associated hemophagocytic lymphohistiocytosis and chronic active EBV infection. Crit Rev Oncol Hematol 44:283–294. doi: 10.1016/S1040-8428(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 6.Iwatsuki K, Satoh M, Yamamoto T, Oono T, Morizane S, Ohtsuka M, Xu ZG, Suzuki D, Tsuji K. 2006. Pathogenic link between hydroa vacciniforme and Epstein-Barr virus-associated hematologic disorders. Arch Dermatol 142:587–595. doi: 10.1001/archderm.142.5.587. [DOI] [PubMed] [Google Scholar]
- 7.Fujiwara S, Kimura H, Imadome K, Arai A, Kodama E, Morio T, Shimizu N, Wakiguchi H. 2014. Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr Int 56:159–166. doi: 10.1111/ped.12314. [DOI] [PubMed] [Google Scholar]
- 8.Rowe M, Young LS, Cadwallader K, Petti L, Kieff E, Rickinson AB. 1989. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 63:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sculley TB, Apolloni A, Stumm R, Moss DJ, Mueller-Lantczh N, Misko IS, Cooper DA. 1989. Expression of Epstein-Barr virus nuclear antigens 3, 4, and 6 are altered in cell lines containing B-type virus. Virology 171:401–408. doi: 10.1016/0042-6822(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 10.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol 64:4084–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology 350:164–170. doi: 10.1016/j.virol.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. 2015. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol 89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe M, Rowe DT, Gregory CD, Young LS, Farrell PJ, Rupani H, Rickinson AB. 1987. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J 6:2743–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman CB, Wohlford EM, Smith NA, King CA, Ritchie JA, Baresel PC, Kimura H, Rochford R. 2015. Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic cytokines. J Virol 89:2301–2312. doi: 10.1128/JVI.03001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young LS, Yao QY, Rooney CM, Sculley TB, Moss DJ, Rupani H, Laux G, Bornkamm GW, Rickinson AB. 1987. New type B isolates of Epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol 68:2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- 16.Kaymaz Y, Oduor CI, Yu H, Otieno JA, Ong'echa JM, Moormann AM, Bailey JA. 2017. Comprehensive transcriptome and mutational profiling of endemic Burkitt lymphoma reveals EBV type-specific differences. Mol Cancer Res 15:563–576. doi: 10.1158/1541-7786.MCR-16-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babcock GJ, Hochberg D, Thorley-Lawson DA. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497–506. doi: 10.1016/S1074-7613(00)00049-2. [DOI] [PubMed] [Google Scholar]
- 18.Coleman CB, Daud II, Ogolla SO, Ritchie JA, Smith NA, Sumba PO, Dent AE, Rochford R. 2017. Epstein-Barr virus type 2 infects T cells in healthy Kenyan children. J Infect Dis 216:670–677. doi: 10.1093/infdis/jix363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattle SB, Farrell PJ. 2006. The role of Epstein-Barr virus in cancer. Expert Opin Biol Ther 6:1193–1205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 20.Hislop AD, Taylor GS, Sauce D, Rickinson AB. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol 25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 21.Thorley-Lawson DA, Hawkins JB, Tracy SI, Shapiro M. 2013. The pathogenesis of Epstein-Barr virus persistent infection. Curr Opin Virol 3:227–232. doi: 10.1016/j.coviro.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha A, Robertson ES. 2011. Epstein-Barr virus-associated B cell lymphomas: pathogenesis and clinical outcomes. Clin Cancer Res 17:3056–3063. doi: 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallesen G, Hamilton-Dutoit SJ, Rowe M, Young LS. 1991. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet 337:320–322. doi: 10.1016/0140-6736(91)90943-J. [DOI] [PubMed] [Google Scholar]
- 24.Brooks L, Yao QY, Rickinson AB, Young LS. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol 66:2689–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox CP, Shannon-Lowe C, Rowe M. 2011. Deciphering the role of Epstein-Barr virus in the pathogenesis of T and NK cell lymphoproliferations. Herpesviridae 2:8. doi: 10.1186/2042-4280-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E. 1991. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci U S A 88:6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Münz C. 2017. Humanized mouse models for Epstein-Barr virus infection. Curr Opin Virol 25:113–118. doi: 10.1016/j.coviro.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Rochford R, Mosier DE. 1995. Differential Epstein-Barr virus gene expression in B cell subsets recovered from lymphomas in SCID mice after transplantation of human peripheral blood lymphocytes. J Virol 69:150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito R, Takahashi T, Katano I, Ito M. 2012. Current advances in humanized mouse models. Cell Mol Immunol 9:208–214. doi: 10.1038/cmi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh NC, Kenney LL, Jangalwe S, Aryee K-E, Greiner DL, Brehm MA, Shultz LD. 2017. Humanized mouse models of clinical disease. Annu Rev Pathol 12:187–215. doi: 10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theocharides APA, Rongvaux A, Fritsch K, Flavell RA, Manz MG. 2016. Humanized hemato-lymphoid system mice. Haematologica 101:5–19. doi: 10.3324/haematol.2014.115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McHugh D, Caduff N, Barros MHM, Rämer PC, Raykova A, Murer A, Landtwing V, Quast I, Styles CT, Spohn M, Fowotade A, Delecluse H-J, Papoudou-Bai A, Lee Y-M, Kim J-M, Middeldorp J, Schulz TF, Cesarman E, Zbinden A, Capaul R, White RE, Allday MJ, Niedobitek G, Blackbourn DJ, Grundhoff A, Münz C. 2017. Persistent KSHV infection increases EBV-associated tumor formation in vivo via enhanced EBV lytic gene expression. Cell Host Microbe 22:61–73.e7. doi: 10.1016/j.chom.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Ma S-D, Yu X, Mertz JE, Gumperz JE, Reinheim E, Zhou Y, Tang W, Burlingham WJ, Gulley ML, Kenney SC. 2012. An Epstein-Barr virus (EBV) mutant with enhanced BZLF1 expression causes lymphomas with abortive lytic EBV infection in a humanized mouse model. J Virol 86:7976–7987. doi: 10.1128/JVI.00770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma S-D, Hegde S, Young KH, Sullivan R, Rajesh D, Zhou Y, Jankowska-Gan E, Burlingham WJ, Sun X, Gulley ML, Tang W, Gumperz JE, Kenney SC. 2011. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J Virol 85:165–177. doi: 10.1128/JVI.01512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White RE, Rämer PC, Naresh KN, Meixlsperger S, Pinaud L, Rooney C, Savoldo B, Coutinho R, Bödör C, Gribben J, Ibrahim HA, Bower M, Nourse JP, Gandhi MK, Middeldorp J, Cader FZ, Murray P, Münz C, Allday MJ. 2012. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest 122:1487–1502. doi: 10.1172/JCI58092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma S-D, Xu X, Plowshay J, Ranheim EA, Burlingham WJ, Jensen JL, Asimakopoulos F, Tang W, Gulley ML, Cesarman E, Gumperz JE, Kenney SC. 2015. LMP1-deficient Epstein-Barr virus mutant requires T cells for lymphomagenesis. J Clin Invest 125:304–315. doi: 10.1172/JCI76357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma S-D, Tsai M-H, Romero-Masters JC, Ranheim EA, Huebner SM, Bristol JA, Delecluse H-J, Kenney SC. 2017. Latent membrane protein 1 (LMP1) and LMP2A collaborate to promote Epstein-Barr virus-induced B cell lymphomas in a cord blood-humanized mouse model but are not essential. J Virol 91:e01928p-16. doi: 10.1128/JVI.01928-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yajima M, Imadome K-I, Nakagawa A, Watanabe S, Terashima K, Nakamura H, Ito M, Shimizu N, Honda M, Yamamoto N, Fujiwara S. 2008. A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses. J Infect Dis 198:673–682. doi: 10.1086/590502. [DOI] [PubMed] [Google Scholar]
- 39.Fujiwara S, Imadome K-I, Takei M. 2015. Modeling EBV infection and pathogenesis in new-generation humanized mice. Exp Mol Med 47:e135. doi: 10.1038/emm.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang J, Kelly M, Freed BM, McCarter MD, Kedl RM, Torres RM, Pelanda R. 2013. Studies of lymphocyte reconstitution in a humanized mouse model reveal a requirement of T cells for human B cell maturation. J Immunol 190:2090–2101. doi: 10.4049/jimmunol.1202810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang J, Weiss N, Freed BM, Torres RM, Pelanda R. 2011. Generation of hematopoietic humanized mice in the newborn BALB/c-Rag2null Il2rγnull mouse model: a multivariable optimization approach. Clin Immunol 140:102–116. doi: 10.1016/j.clim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legrand N, Huntington ND, Nagasawa M, Bakker AQ, Schotte R, Strick-Marchand H, de Geus SJ, Pouw SM, Böhne M, Voordouw A, Weijer K, Di Santo JP, Spits H. 2011. Functional CD47/signal regulatory protein alpha (SIRPα) interaction is required for optimal human T- and natural killer- (NK) cell homeostasis in vivo. Proc Natl Acad Sci U S A 108:13224–13229. doi: 10.1073/pnas.1101398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang J, Zhang B, Kelly M, Peterson JN, Barbee J, Freed BM, Di Santo JP, Matsuda JL, Torres RM, Pelanda R. 2017. Replacing mouse BAFF with human BAFF does not improve B cell maturation in hematopoietic humanized mice. Blood Adv 1:2729–2741. doi: 10.1182/bloodadvances.2017010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wege AK, Melkus MW, Denton PW, Estes JD, Garcia JV. 2008. Functional and phenotypic characterization of the humanized BLT mouse model. Curr Top Microbiol Immunol 324:149–165. [DOI] [PubMed] [Google Scholar]
- 45.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, Wege AK, Haase AT, Garcia JV. 2006. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med 12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 46.Strowig T, Gurer C, Ploss A, Liu Y-F, Arrey F, Sashihara J, Koo G, Rice CM, Young JW, Chadburn A, Cohen JI, Münz C. 2009. Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J Exp Med 206:1423–1434. doi: 10.1084/jem.20081720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cocco M, Bellan C, Tussiwand R, Corti D, Traggiai E, Lazzi S, Mannucci S, Bronz L, Palummo N, Ginanneschi C, Tosi P, Lanzavecchia A, Manz MG, Leoncini L. 2008. CD34+ cord blood cell-transplanted Rag2−/− γc−/− mice as a model for Epstein-Barr virus infection. Am J Pathol 173:1369–1378. doi: 10.2353/ajpath.2008.071186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heuts F, Rottenberg ME, Salamon D, Rasul E, Adori M, Klein G, Klein E, Nagy N. 2014. T cells modulate Epstein-Barr virus latency phenotypes during infection of humanized mice. J Virol 88:3235–3245. doi: 10.1128/JVI.02885-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chijioke O, Müller A, Feederle R, Barros MHM, Krieg C, Emmel V, Marcenaro E, Leung CS, Antsiferova O, Landtwing V, Bossart W, Moretta A, Hassan R, Boyman O, Niedobitek G, Delecluse H-J, Capaul R, Münz C. 2013. human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep 5:1489–1498. doi: 10.1016/j.celrep.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antsiferova O, Müller A, Rämer PC, Chijioke O, Chatterjee B, Raykova A, Planas R, Sospedra M, Shumilov A, Tsai M-H, Delecluse H-J, Münz C. 2014. Adoptive transfer of EBV specific CD8+ T cell clones can transiently control EBV infection in humanized mice. PLoS Pathog 10:e1004333. doi: 10.1371/journal.ppat.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jego G, Bataille R, Pellat-Deceunynck C. 2001. Interleukin-6 is a growth factor for nonmalignant human plasmablasts. Blood 97:1817–1822. doi: 10.1182/blood.V97.6.1817. [DOI] [PubMed] [Google Scholar]
- 52.Qian Y, Wei C, Lee FE-H, Campbell J, Halliley J, Lee JA, Cai J, Kong M, Sadat E, Thomson E, Dunn P, Seegmiller AC, Karandikar NJ, Tipton C, Mosmann T, Sanz I, Scheuermann RH. 2010. Elucidation of seventeen human peripheral blood B cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin Cytom 78(Suppl 1):S69–S82. doi: 10.1002/cyto.b.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma S-D, Xu X, Jones R, Delecluse H-J, Zumwalde NA, Sharma A, Gumperz JE, Kenney SC. 2016. PD-1/CTLA-4 blockade inhibits Epstein-Barr virus-induced lymphoma growth in a cord blood humanized-mouse model. PLoS Pathog 12:e1005642. doi: 10.1371/journal.ppat.1005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsi ED, Lorsbach RB, Fend F, Dogan A. 2011. Plasmablastic lymphoma and related disorders. Am J Clin Pathol 136:183–194. doi: 10.1309/AJCPV1I2QWKZKNJH. [DOI] [PubMed] [Google Scholar]
- 55.Kaminski DA, Wei C, Qian Y, Rosenberg AF, Sanz I. 2012. Advances in human B cell phenotypic profiling. Front Immunol 3:302. doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. 2009. The extent of genetic diversity of Epstein-Barr Virus and its geographic and disease patterns: a need for reappraisal. Virus Res 143:209–221. doi: 10.1016/j.virusres.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rickinson AB, Young LS, Rowe M. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol 61:1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Domingues RG, Lago-Baldaia I, Pereira-Castro I, Fachini JM, Oliveira L, Drpic D, Lopes N, Henriques T, Neilson JR, Carmo AM, Moreira A. 2016. CD5 expression is regulated during human T cell activation by alternative polyadenylation, PTBP1 and miR-204. Eur J Immunol 46:1490–1503. doi: 10.1002/eji.201545663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johannessen I, Asghar M, Crawford DH. 2000. Essential role for T cells in human B cell lymphoproliferative disease development in severe combined immunodeficient mice. Br J Haematol 109:600–610. doi: 10.1046/j.1365-2141.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- 60.Shannon-Lowe C, Rickinson AB, Bell AI. 2017. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci 372:20160271. doi: 10.1098/rstb.2016.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyle M, Sewell W, Sculley T, Apolloni A, Turner J, Swanson C, Penny R, Cooper D. 1991. Subtypes of Epstein-Barr virus in human immunodeficiency virus-associated non-Hodgkin lymphoma. Blood 78:3004–3011. [PubMed] [Google Scholar]
- 62.Borisch B, Finke J, Hennig I, Delacrétaz F, Schneider J, Heitz PU, Laissue JA. 1992. Distribution and localization of Epstein-Barr virus subtypes A and B in AIDS-related lymphomas and lymphatic tissue of HIV-positive patients. J Pathol 168:229–236. doi: 10.1002/path.1711680212. [DOI] [PubMed] [Google Scholar]
- 63.Cohen JI, Jaffe ES, Dale JK, Pittaluga S, Heslop HE, Rooney CM, Gottschalk S, Bollard CM, Rao VK, Marques A, Burbelo PD, Turk S-P, Fulton R, Wayne AS, Little RF, Cairo MS, El-Mallawany NK, Fowler D, Sportes C, Bishop MR, Wilson W, Straus SE. 2011. Characterization and treatment of chronic active Epstein-Barr virus disease: a 28-year experience in the United States. Blood 117:5835–5849. doi: 10.1182/blood-2010-11-316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuwana Y, Takei M, Yajima M, Imadome K-I, Inomata H, Shiozaki M, Ikumi N, Nozaki T, Shiraiwa H, Kitamura N, Takeuchi J, Sawada S, Yamamoto N, Shimizu N, Ito M, Fujiwara S. 2011. Epstein-Barr virus induces erosive arthritis in humanized mice. PLoS One 6:e26630. doi: 10.1371/journal.pone.0026630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sato K, Misawa N, Nie C, Satou Y, Iwakiri D, Matsuoka M, Takahashi R, Kuzushima K, Ito M, Takada K, Koyanagi Y. 2011. A novel animal model of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood 117:5663–5673. doi: 10.1182/blood-2010-09-305979. [DOI] [PubMed] [Google Scholar]
- 66.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A 81:7632–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simbiri KO, Smith NA, Otieno R, Wohlford EEM, Daud II, Odada SP, Middleton F, Rochford R. 2015. Epstein-Barr virus genetic variation in lymphoblastoid cell lines derived from Kenyan pediatric population. PLoS One 10:e0125420. doi: 10.1371/journal.pone.0125420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moormann AM, Chelimo K, Sumba OP, Lutzke ML, Ploutz-Snyder R, Newton D, Kazura J, Rochford R. 2005. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis 191:1233–1238. doi: 10.1086/428910. [DOI] [PubMed] [Google Scholar]
- 69.Moormann AM, Chelimo K, Sumba PO, Tisch DJ, Rochford R, Kazura JW. 2007. Exposure to holoendemic malaria results in suppression of Epstein-Barr virus-specific T cell immunosurveillance in Kenyan children. J Infect Dis 195:799–808. doi: 10.1086/511984. [DOI] [PubMed] [Google Scholar]
- 70.Kubota N, Wada K, Ito Y, Shimoyama Y, Nakamura S, Nishiyama Y, Kimura H. 2008. One-step multiplex real-time PCR assay to analyse the latency patterns of Epstein-Barr virus infection. J Virol Methods 147:26–36. doi: 10.1016/j.jviromet.2007.08.012. [DOI] [PubMed] [Google Scholar]