According to the World Health Organization, approximately 20 million HEV infections occur annually, resulting in 3.3 million cases of hepatitis E and >44,000 deaths. The lack of an efficient animal model that can mimic the full-spectrum of infection outcomes hinders our ability to delineate the mechanism of HEV pathogenesis. Here, we successfully generated immunoglobulin heavy chain JH−/− knockout gnotobiotic pigs using CRISPR/Cas9 technology, established a novel JH−/− knockout and wild-type gnotobiotic pig model for HEV, and systematically determined the dynamic of acute HEV infection in gnotobiotic pigs. It was demonstrated that knockout of the Ig heavy chain in pigs decreased the level of HEV replication. Infected wild-type and JH−/− knockout gnotobiotic piglets developed more pronounced HEV-specific lesions than other studies using conventional pigs, and the infected JH−/− knockout pigs had significantly enlarged livers. The availability of this novel model will facilitate future studies of HEV pathogenicity.
KEYWORDS: hepatitis E virus, HEV, gnotobiotic pig, Ig heavy chain knockout, B cell depletion
ABSTRACT
Hepatitis E virus (HEV), the causative agent of hepatitis E, is an important but incompletely understood pathogen causing high mortality during pregnancy and leading to chronic hepatitis in immunocompromised individuals. The underlying mechanisms leading to hepatic damage remain unknown; however, the humoral immune response is implicated. In this study, immunoglobulin (Ig) heavy chain JH−/− knockout gnotobiotic pigs were generated using CRISPR/Cas9 technology to deplete the B-lymphocyte population, resulting in an inability to generate a humoral immune response to genotype 3 HEV infection. Compared to wild-type gnotobiotic piglets, the frequencies of B lymphocytes in the Ig heavy chain JH−/− knockouts were significantly lower, despite similar levels of other innate and adaptive T-lymphocyte cell populations. The dynamic of acute HEV infection was subsequently determined in heavy chain JH−/− knockout and wild-type gnotobiotic pigs. The data showed that wild-type piglets had higher viral RNA loads in feces and sera compared to the JH−/− knockout pigs, suggesting that the Ig heavy chain JH−/− knockout in pigs actually decreased the level of HEV replication. Both HEV-infected wild-type and JH−/− knockout gnotobiotic piglets developed more pronounced lymphoplasmacytic hepatitis and hepatocellular necrosis lesions than other studies with conventional pigs. The HEV-infected JH−/− knockout pigs also had significantly enlarged livers both grossly and as a ratio of liver/body weight compared to phosphate-buffered saline-inoculated groups. This novel gnotobiotic pig model will aid in future studies into HEV pathogenicity, an aspect which has thus far been difficult to reproduce in the available animal model systems.
IMPORTANCE According to the World Health Organization, approximately 20 million HEV infections occur annually, resulting in 3.3 million cases of hepatitis E and >44,000 deaths. The lack of an efficient animal model that can mimic the full-spectrum of infection outcomes hinders our ability to delineate the mechanism of HEV pathogenesis. Here, we successfully generated immunoglobulin heavy chain JH−/− knockout gnotobiotic pigs using CRISPR/Cas9 technology, established a novel JH−/− knockout and wild-type gnotobiotic pig model for HEV, and systematically determined the dynamic of acute HEV infection in gnotobiotic pigs. It was demonstrated that knockout of the Ig heavy chain in pigs decreased the level of HEV replication. Infected wild-type and JH−/− knockout gnotobiotic piglets developed more pronounced HEV-specific lesions than other studies using conventional pigs, and the infected JH−/− knockout pigs had significantly enlarged livers. The availability of this novel model will facilitate future studies of HEV pathogenicity.
INTRODUCTION
Hepatitis E, caused by hepatitis E virus (HEV), is typically an acute icteric disease of worldwide importance. Transmission of HEV occurs by the fecal-oral route, which often results in large explosive waterborne outbreaks in developing countries and sporadic foodborne cases in industrialized countries, including the United States (1–4). The mortality rate for HEV infection ranges from 0.4 to 2% in immunocompetent healthy individuals (5); however, infected pregnant women experience a significantly elevated level of mortality, up to 28 to 30% (6, 7). While hepatitis E is generally recognized as a self-limiting acute disease, immunosuppressed individuals such as solid organ transplant recipients (8), individuals with concurrent HIV infections (9), and patients with lymphoma or leukemia tend to progress to a state of chronicity (10) with cirrhotic disease and elevated mortality (11). The underlying mechanisms of disease severity and hepatic damage experienced by these populations are currently not understood, nor do we possess an adequate animal model for addressing the current knowledge gaps.
HEV is a single-stranded, positive-sense RNA virus classified in the family Hepeviridae (12), which consists of two genera: Orthohepevirus and Piscihepevirus. Within the genus Orthohepevirus, four species are recognized. Species Orthohepevirus A includes all HEV strains that are known to infect humans and numerous other mammalian species. At least eight distinct genotypes have been identified thus far within species Orthohepevirus A: genotypes 1 to 4 are known to infect humans (1, 12, 13) with genotypes 1 and 2 affecting only humans, whereas genotypes 3 and 4 affect humans and several other animal species such as domestic pigs (14), deer (15), and rabbits (16, 17). Genotype 5 and 6 HEVs infect wild boars (18), genotype 7 HEV infects dromedary camels (19) and possibly humans (20), and genotype 8 HEV infects the Bactrian camel (19). Species Orthohepevirus B consists of HEV strains that infect avian species (21–23), Orthohepevirus C infects rodents (24, 25), and Orthohepevirus D infects bats (26). The genus Piscihepeirus includes the sole strain of HEV infecting cutthroat trout (27).
Pigs are a major animal reservoir for HEV and a major source of zoonotic infections in humans (4). As the natural host for genotypes 3 and 4 HEV infections in humans (13, 28), the pig model has been used to study HEV biology and cross-species infections (13, 29). However, the typical outbred conventional pig experimentally infected with HEV does not develop the level of pathogenicity and progression of disease seen in immunocompromised and pregnant populations (30). Infection with HEV in conventional pigs are in general clinically asymptomatic with only mild to moderate hepatic changes observed (31). The typical course of HEV infection includes fecal shedding of HEV RNA in infected individuals at 1 week postinfection (wpi), which can persist for up to 8 wpi with a peak in viral titer at approximately 4 wpi (32, 33), a viremic phase lasting 1 to 2 weeks, followed by clearance of the virus at 8 to 9 wpi with the development of IgG anti-HEV at 2 to 4 wpi (34). Replication of HEV occurs primarily in the gastrointestinal tract (31, 35), with only limited levels of virus replication in hepatocytes. As a result, direct viral effects within hepatic tissue is limited. Consequently, the humoral immune response has long been thought to exacerbate the hepatic disease process as an immune-mediated event, leading to the development of the observed liver lesions (36). Likewise, in nonhuman primates (37, 38) and chickens (39) experimentally infected with HEV, hepatic lesions and alterations in serum levels of liver enzymes often correspond to the appearance of HEV antibodies, further suggesting that anti-HEV IgG may play a role in the development of hepatic lesions.
Here, we report the successful establishment of an immunoglobulin (Ig) heavy chain knockout JH−/− gnotobiotic piglet model that better mimics the course of acute HEV infection observed in humans. The dynamic of acute HEV infection was systematically determined in both Ig heavy chain knockout and wild-type gnotobiotic piglets experimentally infected with a genotype 3 human HEV. The presence and magnitude of viremia and fecal viral shedding, IgG anti-HEV antibody response to infection, immune correlates of infection, magnitude of infection and presence of viral RNA in extrahepatic sites, and liver pathology associated with HEV infection were determined.
RESULTS
Successful development of Ig heavy chain JH−/− knockout gnotobiotic piglets.
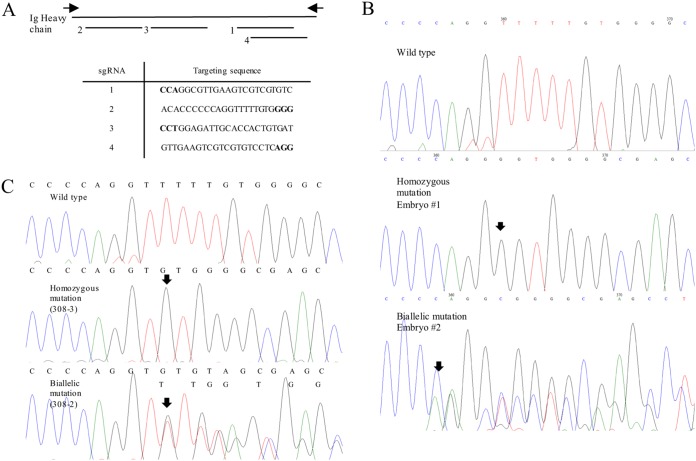
In order to delineate the differentiating characteristics between Ig heavy chain JH−/− knockout and wild-type gnotobiotic piglets, CRISPR/Cas9 technology was utilized to alter the region designated as the Ig heavy chain leading to the generation of “JH−/− knockout” piglets (Table 1). Similarly, as demonstrated in our previous studies (40–42), no wild-type allele was observed in any of the genotyped embryos (Table 2 and Fig. 1), suggesting that the approach is effective in producing Ig heavy chain knockout pigs. Pigs carrying the Ig heavy chain knockout phenotype were produced by transferring CRISPR/Cas9 injected embryos into estrus matched surrogate sows. Included in this study was a total of 21 live piglets and one stillborn that were born from 6 surrogate dams (Tables 3 and 4). Genotyping results indicated that all Ig heavy chain knockout pigs carried the modified Ig heavy chain region (Fig. 1 and Table 2). All wild-type gnotobiotic piglets were derived by embryo transfer of in vitro-fertilized embryos carrying identical parental line as the Ig heavy chain knockout pigs. Gnotobiotic piglets were retrieved by hysterectomy of pregnant sows with the removal of the entire uterus under sterile conditions and revived and housed in individual sterile isolators where all procedures occurred for the duration of the study.
TABLE 1.
Experimental infection of wild-type and immunoglobulin JH−/− knockout gnotobiotic piglets with a genotype 3 human strain (US2) of hepatitis E virus
| Group | No. of pigletsa | Phenotype of piglets | Inoculab | No. of piglets undergoing necropsy at 4 wpi |
|---|---|---|---|---|
| 1 | 5 | JH knockout | PBS | 5 |
| 2 | 6 | JH knockout | G3 (US2) HEV | 6 |
| 3 | 4 | Wild-type | PBS | 4 |
| 4 | 5 | Wild-type | G3 (US2) HEV | 6 |
Piglets were retrieved by hysterectomy at 111 to 113 days of gestation and maintained germfree in sterile isolators for the duration of the study. Variation in piglet numbers per groups was due to the efficiency in retrieval of live piglets following embryo transfer.
Piglets were inoculated with US2 strain of HEV or mock infected (PBS) at 8 days of age via ear-vein injection.
TABLE 2.
Efficacy of CRISPR/Cas9 to introduce targeted modifications in Ig heavy chain locus during embryogenesis
| Parameter | Observation |
|---|---|
| Gene | Ig heavy chain |
| Total no. injected | 132 |
| No. (%) of blastocysts on day 7 | 24 (18.2) |
| No. of animals genotyped | 15 |
| Genotype (no. of pigs)a | |
| Homozygous | 3 |
| Biallelic | 12 |
| Mosaic | 0 |
| Heterozygous | 0 |
| WT | 0 |
Single embryos at the blastocyst stage were used for genotyping. No wild-type (WT) allele was found.
FIG 1.
Ig heavy chain targeting CRISPR sequences and representative types of Ig heavy chain mutations in this study. (A) Disruption of Ig heavy chain in pigs by CRISPR/Cas9 with a total of four target sites designed using a web-based program. Boldface letters indicate the protospacer adjacent motif (PAM) sequence of target sites. Black arrows indicate primers used to amplify the target region. (B) Types of genetic mutations of the Ig heavy chain in single blastocyst. The sequencing results indicated either homozygous or biallelic mutation of Ig heavy chain in single embryos with no wild-type alleles observed. (C) Representative genotype of Ig heavy chain knockout pigs. Arrows indicate sites of mutations.
TABLE 3.
Phenotyping of Ig heavy-chain-modified piglets produced using CRISPR/Cas9 technology and retrieved by hysterectomy
| Surrogate ID | Modification | No. of embryos transferred into a surrogate | Stage of embryo (day) | Pregnancy | No. of pigletsa |
|---|---|---|---|---|---|
| 308 | Ig heavy chain | 82 | 6 | Y | 3 piglets and 1 stillborn |
| 68-8 | Ig heavy chain | 74 | 6 | Y | 5 piglets |
| 61-12 | Ig heavy chain | 89 | 5 | Y | 4 piglets |
| 29-1 | Ig heavy chain | 81 | 6 | Y | 3 piglets |
| 61-13 | Ig heavy chain | 80 | 6 | Y | 4 piglets |
| Y465 | WT | 83 | 6 | Y | 8 piglets and 3 stillborn |
| 70-3 | WT | 89 | 5 and 6 | Y | 8 piglets |
| 55-09 | WT | 68 | 6 and 7 | Y | 2 piglets |
| 70-04 | WT | 105 | 5 | Y | 2 piglets |
A total of 21 piglets were delivered from five surrogate gilts carrying the immunoglobulin heavy chain knockout JH−/− phenotype. The piglets were maintained in gnotobiotic status in sterile isolators for the duration of the experimental period. WT, wild type.
TABLE 4.
Genotyping the Ig heavy chain modified piglets
| Pig | Mutation | Genotypea | Length (bp) |
|---|---|---|---|
| WT | None | GCACACCCCCCAGGTTTTTGTGGGGCGAGCCTGGAGATTGCACCACTGTGATTACTATGCTATGGATCTCTGGGGCCCAGGCGTTGAAGTCGTCGTGTCCTCAGGTAAGAACGGCCC | |
| 308-1 | Homozygous | GCACACCCCCCAGGTTTTTAGTGGGGCGAGCCT | 1 |
| 308-2 | Biallelic mutation | GCACACCCCCCAGGTTTTTTGTGGGGCGAGCCTGGAG | 1 |
| GCACACCCCCCAGGT - - - - - - GGGGCGAGCCTGGAG | 6 | ||
| 308-3 | Homozygous | GCACACCCCCCAGGT - - - - - - - GTGGGGCGAGCCTGGAG | 4 |
| 68-8-1 | Biallelic mutation | GCACACCCCCCAGGT - - - - GTGGGGCGAGCCTGGAG | 4 |
| GCACACCCCCCAGGTT - - - - | 282 | ||
| 68-8-2 | Homozygous | GCACACCCCCCAGGT - - - - GTGGGGCGAGCCTGGAG | 4 |
| 68-8-3 | Biallelic mutation | GCACACCCCCCAGGTTTTTTTTGTGGGGCGAGCCTGGAG | 3 |
| GCACACCCCCCAGGTTTTTTGTGGGGCGAGCCTGGAG | 1 | ||
| 68-8-4 | Biallelic mutation | GCACACCCCCCAGG - - - - - GTGGGGCGAGCCTGGAG | 5 |
| GCACACCCCCCAGGTTTTTTGTGGGGCGAGCCTGGAG | 1 | ||
| 68-8-5 | Biallelic mutation | GCACACCCCCCAGGTTTTTTGTGGGGCGAGCCTGGAG | 1 |
| GCACACCCCCCAGG - - - - - - GGCGAGCCTGGAGATTGCAC | 9 | ||
| 61-12-1 | Homozygous | GCACACCCCCCAGGCCACACTCACCCCTCCAGGGGCGAGCCTGGAG | 14 |
| 61-12-2 | Homozygous | TCCCAGCCCTGCGGCCGAGGG - - - - - - GTGGGGCGAGCCTGGAG | 59 |
| 61-12-3 | Homozygous | GCACACCCCCCAGG - - - - - - GGGGGCGAGCCTGGAG | 6, 1 |
| 61-12-4 | Biallelic mutation | GCACACCCCCCAGGTTTTTTGTGGGGCGAGCCTGGAG | 1 |
| GGGAAGTCAGGAGGGAGC - - - - - - AGATTGCACCAC | 107 | ||
| 29-1-1 | Homozygous | GCACACCCCCCAGGT - - - - GTGGGGCGAGCCT | 4 |
| 29-1-2 | Homozygous | GCACACCCCCCA - - - - - - GGGGCGAGCCT | 9 |
| 29-1-3 | Biallelic mutation | GCACACCCCCCA - - - - - - GGGGCGAGCCT | 9 |
| GCACACCCCCCAGGTTTTTGTGGGGGCGAGCCT | 1 | ||
| 61-13-1 | Biallelic mutation | GCACACCCCCCAGGTTTTT - - - - - - CTCAGGT | 80 |
| GCACACCCCCCAG - - - - - - GTGGGGCGAGCCTGGAGATTGCACCATGTGATTACTATGCTATGGGATCTCTGGGGCCCAGGCGTTGAAGTCGCTCGTGTCCT - - - GTAAGAACGGCCC | 9, 2, 2 | ||
| 61-13-2 | Biallelic mutation | GCACACCCCCCAGGTTTT - - - - - - - TCGTGTCCTCAGGTAAGAA CGGCCC | 74 |
| GCACACCCCC - - - - - - CAGGTAAGAACGGCCC | 91 | ||
| 61-13-3 | Homozygous | GCACACCCCCCAGGTTTTTGT - - - - - - CGTGTCCTCAGGT | 72 |
| 61-13-4 | Homozygous | GCACACCCCCCAGGTTTTT - - - - - - CTCAGGT | 80 |
Genotype of each knockout piglet with mutations on each allele individually or on both alleles. The number of base pairs for each indicated sequence is indicated in column 4. Underlining indicates targeting sequence. Boldface letters indicate insertion or change in nucleotides, and a hyphen (“-”) indicates the deletion of a nucleotide. All piglets carried mutation on the Ig heavy chain.
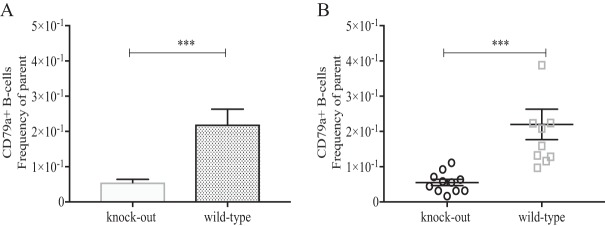
The sterility of the surgical setup, recovery chamber, and individual isolators was determined and confirmed by swabbing of the respective equipment and streaking on blood agar plates for any bacterial growth. The sterility of the gnotobiotic piglets was determined through the verification of a lack of bacterial growth (on both blood agar plates and Luria broth) from fecal swab samples throughout the course of the experiment. The JH−/− Ig heavy chain knockout gnotobiotic piglets were characterized and exhibited significantly reduced numbers of CD79a+ B lymphocytes (Fig. 2A and B) in the peripheral blood mononuclear cell (PBMC) populations, despite having normal levels of CD3+ CD4+ lymphocytes and no alteration in CD16+ natural killer cell populations.
FIG 2.
B-lymphocyte cell counts in peripheral blood samples of wild-type and immunoglobulin JH knockout gnotobiotic piglets experimentally infected with HEV. PBMCs were collected from each piglet at 28 dpi at necropsy. Cells were gated based on CD3 and quantified for the intracellular marker CD79a+ as a measure of the total B-cell population in the peripheral blood. Asterisks indicate statistical significance between designated groups determined by Tukey's t test with a P value of <0.001.
Lower fecal viral RNA loads in HEV-infected JH−/− gnotobiotic piglets than in wild-type piglets.
In general, HEV-infected conventional pigs shed virus in the feces by 1 wpi, with peak fecal viral shedding at 4 wpi and virus clearance by 7 to 8 wpi (33). In the Ig JH−/− gnotobiotic piglets infected with US-2 HEV in this study, viral RNAs were detected in feces as early as 4 days postinoculation (dpi) with all piglets testing positive by 7 dpi and remained positive through necropsy at 28 dpi (Table 5). Likewise, the wild-type gnotobiotic piglets had detectable viral RNA in the feces from 7 dpi, with all piglets testing positive by 9 dpi (Table 5), and remained so until necropsy at 28 dpi.
TABLE 5.
Detection of HEV RNA in the serum (fecal) samples of wild-type and Ig JH−/− knockout gnotobiotic piglets experimentally infected with the US2 strain of human HEV
| Group | Inoculum | Piglet phenotype | No. of positive samples/total no. tested at various times dpia: |
||||
|---|---|---|---|---|---|---|---|
| 0 dpi | 7 dpi | 14 dpi | 21 dpi | 28 dpi | |||
| 1 | PBS | Knockout | 0/5 (0/5) | 0/5 (0/5) | 0/5 (0/5) | 0/5 (0/5) | 0/5 (0/5) |
| 2 | HEV | Knockout | 0/6 (0/6) | 4/6 (6/6) | 5/6 (6/6) | 4/6 (6/6) | 5/6 (6/6) |
| 3 | PBS | Wild type | 0/4 (0/4) | 0/4 (0/4) | 0/4 (0/4) | 0/4 (0/4) | 0/4 (0/4) |
| 4 | HEV | Wild type | 0/6 (0/6) | 1/6 (3/6) | 4/6 (6/6) | 3/6 (6/6) | 3/6 (6/6) |
dpi, days postinoculation. HEV RNA was detected via strain-specific nested RT-PCR and qPCR. The amplified PCR-positive products were sequenced to confirm the identity as the US2 strain of HEV.
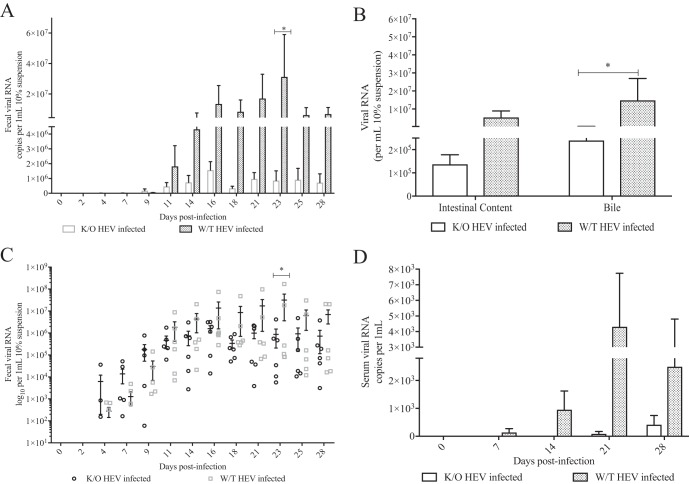
Quantification of HEV RNA loads in fecal samples (three times per week), weekly serum samples, intestinal content, bile, and hepatic, as well as extrahepatic tissues, including thymus, duodenum, jejunum, ileum, large intestine, gallbladder, spleen, liver, brain and spinal cord, and mesenteric lymph node tissues, was determined by quantitative reverse transcription-PCR (qRT-PCR). The infected Ig JH−/− gnotobiotic piglets reached peak viral RNA loads in the feces at 16 dpi, whereas the viral RNA loads in infected wild-type gnotobiotic piglets increased until 23 dpi (Fig. 3A and C). Viral RNA loads in both groups tapered but remained positive throughout the experimental period in accordance with the typical course of HEV infection. Neither group experienced a significant decrease in viral RNA loads by the 4-wpi necropsy time point. The fecal viral RNA loads were significantly higher in the HEV-infected wild-type gnotobiotic pigs than the JH−/− knockout gnotobiotic pigs (P < 0.05) (Fig. 3A and C) at 23 dpi. The infected wild-type pigs also had numerically larger amounts of fecal viral RNA loads at other time points compared to the JH−/− knockout pigs, although the difference was not statistically significant at other time points. Likewise, the viral RNA loads in the intestinal content were also higher in the infected wild-type piglets than JH−/− knockout pigs; although, the difference was not significant (Fig. 3B). Overall, the JH−/− knockout pigs exhibited a reduced level of fecal HEV RNA shedding in the first 4 wpi compared to wild-type pigs; however, a significant difference was observed only at 23 dpi due to individual pig variations in both groups (Fig. 3A and C).
FIG 3.
Quantification of HEV RNA loads in serum, fecal, and bile samples of wild-type and JH knockout gnotobiotic piglets experimentally infected with HEV. Viral RNAs were extracted from fecal samples three times weekly (A), intestinal contents and bile at 28 dpi (B), as a scatterplot of fecal viral RNA, with each symbol indicating the value for an individual piglet (C), and serum samples at 0, 7, 14, 21, and 28 dpi (D) for the quantification of HEV RNA copy numbers by qRT-PCR. The data are expressed as means ± the SEM. The detection limit is 10 viral genomic copies, which corresponds to 400 copies per 1-ml sample or per gram of tissue. Titers below the reported detection limit were considered negative. Asterisks indicate statistical significance between designated groups as determined by two-way ANOVA and a P value of <0.05.
Lower serum viral RNA loads and fewer incidence of viremia in HEV-infected JH−/− piglets compared to wild-type piglets under gnotobiotic condition.
In the HEV-infected JH−/− knockout pigs, when tested by a nested RT-PCR assay, 4/6 pigs became viremic by 14 dpi, and 3/6 remained positive at necropsy at 28 dpi (Table 5). However, when tested by a quantitative qRT-PCR assay, which is less sensitive than nested RT-PCR and has a detection limit of 400 copies/μl sample, only two piglets in the JH−/− knockout pig group were positive for HEV RNA in the serum, and none were positive until 3 wpi (Fig. 3D). In the infected wild-type pig group, viremia was detected as early as at 7 dpi, with 4/6 pigs being positive at 14 dpi and 3/6 pigs remaining positive at necropsy at 28 dpi (Table 5). When tested by the qRT-PCR assay for the sera from the infected wild-type pig group, the results were similar to those obtained by the nested RT-PCR assay: 1/6 positive at 7 dpi (Fig. 3D) and 4/6 positive at 14 dpi and remaining positive by 28 dpi. The infected wild-type pigs had numerically larger amounts of viral RNA loads in the serum than the JH−/− knockout pigs, although the difference was not significant (Fig. 3D). Overall, the data suggest that the level of viral RNA loads in serum is reduced in the JH−/− knockout pigs, with fewer positive piglets compared to the wild-type pigs.
Both the IgM and IgG anti-HEV responses in the JH knockout and wild-type pigs were analyzed by enzyme-linked immunosorbent assay (ELISA). Anti-HEV antibody responses are typically observed during acute HEV infections by approximately 4 to 5 wpi in humans and conventional pigs (5, 32, 33). In this study, seroconversion to HEV antibodies was not detected in gnotobiotic pigs by the necropsy time point at 28 dpi (data not shown).
No difference in the amount of viral RNAs in extrahepatic tissues between HEV-infected JH−/− knockout pigs and wild-type pigs.
During the acute phase of HEV infection in pigs, viral RNA is typically detectable in a variety of hepatic and extrahepatic tissues due to the circulating virus in the blood during the viremic stage. A qRT-PCR assay was used to quantify the amount of HEV RNA in samples of liver and various extrahepatic tissues, including thymus, duodenum, jejunum, ileum, large intestine, gallbladder, spleen, brain and spinal cord, and mesenteric lymph node tissues. HEV RNA was present in all tissues for the majority of piglets in both groups except in the thymus and mesenteric lymph node. All thymus samples had very low viral RNA titers in only 2/6 JH−/− knockout piglets and 0/6 wild-type pigs. Mesenteric lymph node samples were positive at low levels in 3/6 JH−/− knockout piglets, but in all wild-type piglets (data not shown). In the bile sample, the amount of HEV RNA was significantly higher (P = 0.0169) in wild-type pigs than in JH−/− knockout pigs (Fig. 3B), although there was no difference in the viral RNA loads in other extrahepatic tissues between the JH−/− knockout and wild-type groups (data not shown).
Elevation of serum level of liver enzyme γ-glutamyl transferase (GGT), but not other enzymes, in HEV-infected JH−/− knockout gnotobiotic pigs at 3 wpi.
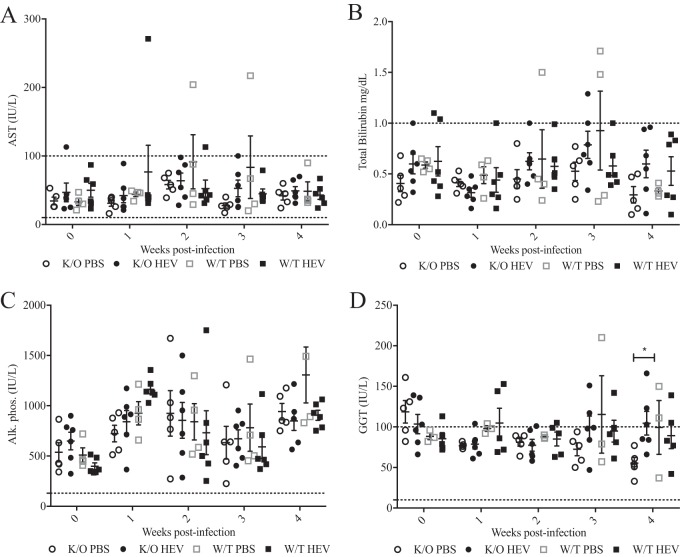
The serum levels of liver enzymes, including aspartate aminotransferase (AST) (Fig. 4A), total bilirubin (Fig. 4B), and alkaline phosphatase (Fig. 4C), were assessed and found to be similar among the mock-infected JH−/− knockout, HEV-infected JH−/− knockout, mock-infected wild-type, and HEV-infected wild-type piglets. Total bilirubin appeared to be different between HEV-infected JH−/− knockout pigs and mock-infected JH−/− knockout piglets at 2 to 4 wpi; however, the differences were not statistically significant (Fig. 4B). The serum levels of GGT were found to be statistically elevated at 3 wpi (Fig. 4D) in HEV-infected JH−/− knockout gnotobiotic pigs compared to the JH−/− knockout phosphate-buffered saline (PBS) group. There were abnormally elevated levels of GGT (10 to 100 IU/liter, considered to be within normal limits), AST (10 to 100 IU/liter), and total bilirubin (<1.0 mg/dl) in serum samples from all groups at various time points. In addition, all serum samples for all piglets in all groups had elevated alkaline phosphatase (>100 IU/liter) from 0 to 4 wpi (36 days of age) (Fig. 4C).
FIG 4.
Circulating levels of liver enzymes in serum samples collected weekly from HEV-infected wild-type and HEV-infected JH−/− knockout gnotobiotic piglets. Serum was collected weekly from each piglet beginning with 0 wpi and continuing until necropsy at 4 wpi. Liver enzymes, including AST (A), total bilirubin (B), alkaline phosphatase (C), and GGT (D), were measured at the Iowa State University Veterinary Diagnostic Lab. Normal limits were provided with the analysis based on the laboratory's standards for each specific test, which are indicated on each scatterplot graph as a dotted line. Each sample indicates an individual piglet for each time point and are expressed as means ± the SEM. Analysis was completed using two-way ANOVA (*, P < 0.05).
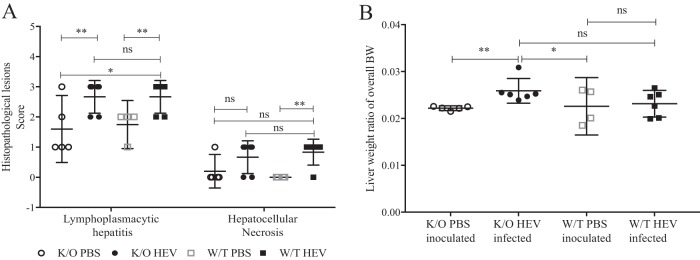
Histological liver lesions characterized by lymphoplasmacytic hepatitis and hepatocellular necrosis in both HEV-infected JH−/− knockout and wild-type pigs.
At necropsy at 28 dpi, the histological lesions in the liver of infected pigs were characterized as lymphoplasmacytic hepatitis and hepatocellular necrosis (Fig. 5A). The average lesion score of lymphoplasmacytic hepatitis in HEV-infected JH−/− knockout pigs was significantly higher (P = 0.0111) than that in PBS-inoculated JH−/− knockout pigs. Similarly, the average lymphoplasmacytic hepatitis lesion score was also higher (P < 0.05) in HEV-infected wild-type pigs than in PBS-inoculated wild-type pigs (Fig. 5A). The hepatocellular necrosis lesion score in HEV-infected wild-type pigs was also significantly higher (P < 0.05) than that in PBS-inoculated wild-type pigs, but there was no difference in lymphoplasmacytic hepatitis or hepatocellular necrosis lesion score between HEV-infected wild-type pigs and HEV-infected JH−/− knockout pigs (Fig. 5A).
FIG 5.
Histopathological lesions in wild-type and JH knockout gnotobiotic piglets experimentally infected with HEV. (A) Histopathological lesions in the liver, including lymphoplasmacytic hepatitis and hepatocellular necrosis. Lymphoplasmacytic hepatitis was scored as follows: 0, no inflammation; 1, 1 to 2 focal lymphoplasmacytic infiltrates/10 hepatic lobules; 2, 3 to 5 focal infiltrates/10 hepatic lobules; 3, 6 to 10 focal infiltrates/10 hepatic lobules; and 4, >10 focal infiltrates/10 hepatic lobules. Hepatocellular necrosis was characterized by the presence of individual hepatocytes with an eosinophilic cytoplasm with or without fragmented or absent nuclei. (B) Liver/body weight ratio. The liver weights were evaluated as a ratio of the overall body weight. Asterisks indicate statistical significance as determined by two-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Higher liver/body weight ratio in HEV-infected JH−/− knockout pigs compared to PBS-inoculated controls.
The liver and body weights of each pig were measured at necropsy and the liver/body weight ratio was calculated to determine whether the livers were enlarged. The average liver/body weight ratio in HEV-infected JH−/− knockout pigs was significantly higher than that of the PBS-inoculated JH−/− knockout (P < 0.01) or wild-type pigs (P < 0.05) (Fig. 5B), indicating that the presence of hepatic lesions and inflammation in this group led to larger livers than in the PBS-inoculated group. There was no significant difference in liver/body weight ratio between the wild-type pig infected and mock-infected groups (Fig. 5B).
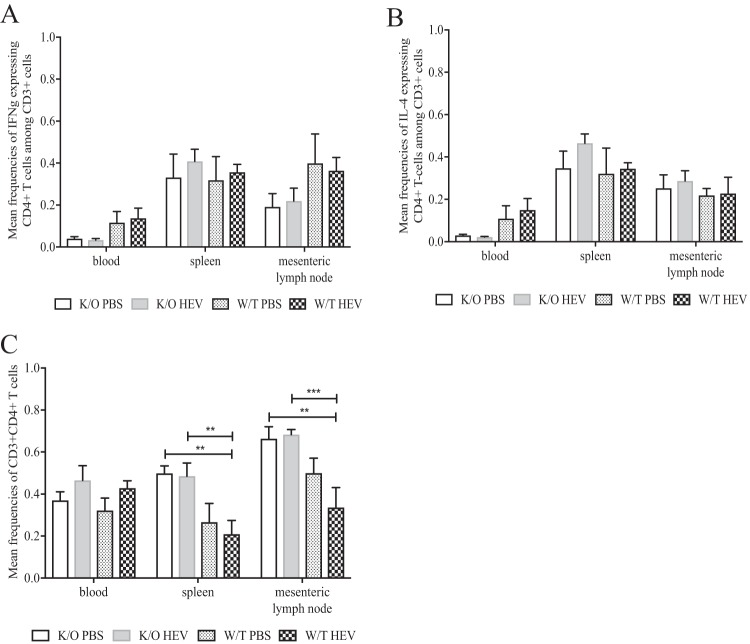
IFN-γ- and IL-4-specific CD4+ T-cell responses were not significantly altered in HEV-infected JH−/− knockout piglets compared to HEV-infected wild-type pigs and mock-infected groups at 28 dpi.
PBMCs from heparinized plasma and mononuclear cells (MNCs) from the spleen and mesenteric lymph nodes were isolated and stimulated with HEV-specific recombinant capsid antigen, followed by staining and flow cytometry analysis (Fig. 6). HEV-specific T-cell (Th1) responses were analyzed for the frequency of gamma interferon (IFN-γ) and interleukin-4 (IL-4) expression and compared between mock-infected JH−/− knockout, HEV-infected JH−/− knockout, mock-infected wild-type, and HEV-infected wild-type groups. Despite an apparent numerically lower level of expression of IFN-γ (Fig. 6A) in the PBMC T-cell population for all groups, the observed difference was not statistically significant. Expression of IL-4 (Fig. 6B) appeared to be numerically elevated in both mock-infected wild-type and HEV-infected groups compared to the mock-infected and HEV-infected JH−/− knockout groups, again the difference was not statistically significant. In addition, the total CD3+ CD4+ T cells (Th1) in the spleens and mesenteric lymph nodes were significantly elevated in both the mock-infected and the HEV-infected JH−/− knockout groups compared to the wild-type-infected groups (P < 0.01) (Fig. 6C). The difference was discernible between both JH−/− knockout pig groups and the wild-type-infected group at P < 0.01; while, the difference between JH−/− infected and wild-type infected was more significant at a P of <0.001.
FIG 6.
Frequencies of IFN-γ and IL-4 intracellular cytokines in mononuclear cells isolated from spleen and mesenteric lymph node tissues and PBMCs from the peripheral blood of wild-type and JH knockout piglets experimentally infected with HEV. PBMCs and MNCs were collected from each piglet at 28 dpi at necropsy. Cells were gated on CD3 and stained for extracellular and intracellular markers CD4+ and IFN-γ and IL-4, respectively, after stimulation with HEV-specific capsid protein. The mean frequencies of IFN-γ (A), IL-4 (B), and CD4+ (C) are indicated and expressed as means ± the SEM. All frequencies were determined by flow cytometry with 100,000 events per sample (*, P < 0.05).
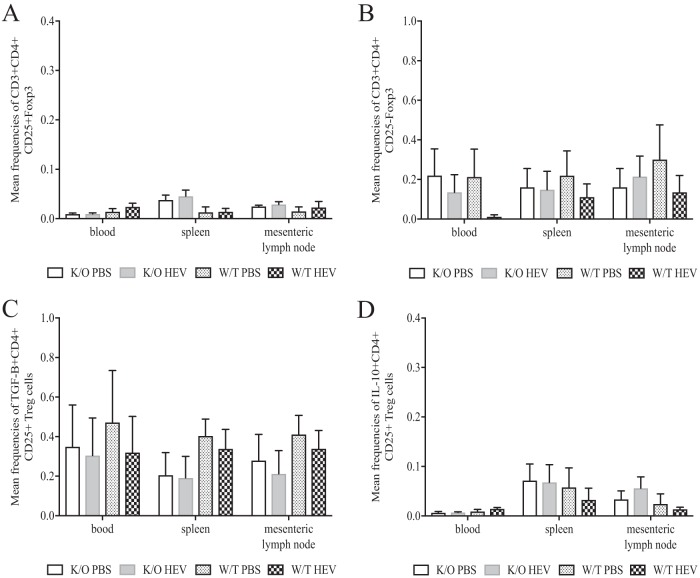
IL-10- and TGF-β-producing CD4+ CD25+ Foxp3+ and CD4+ CD25− Foxp3+ Treg cell populations were not significantly altered in HEV-infected JH−/− knockout piglets compared to HEV-infected wild-type pigs and mock-infected groups at 28 dpi.
To examine the influence of HEV infection on the functionality of T-regulatory (Treg) cell subsets, we determined the frequencies of IL-10 and TGF-β expression as intracellular cytokines in PBMCs from heparinized plasma and MNCs from both splenic and mesenteric lymph node preparations. The mean frequencies of CD4+ CD25+ and CD4+ CD25− Treg cell subsets (Fig. 7A and B) were similar among all groups in the blood, spleen, and mesenteric lymph nodes. Likewise, the total Foxp3+ Treg cells in the blood, spleen, and mesenteric lymph nodes were not significantly altered between mock-infected and HEV-infected wild-type or JH−/− knockout groups of piglets (Fig. 7B). There was no discernible difference in the frequency of transforming growth factor β (TGF-β; Fig. 7C) expression in any of the aforementioned groups. Likewise, the IL-10 expression (Fig. 7D) was not significantly altered based on pig phenotype (wild type versus JH−/− knockout) or infection status (HEV infected versus mock infected). There was an overall lower level of expression of IL-10+ CD4+ CD25+ Treg cells in the blood versus MNCs isolated from spleen or lymph nodes, but this difference was consistent in all four groups regardless of infection or pig phenotype status (Fig. 7D).
FIG 7.
Frequencies of TGF-β and IL-10 intracellular cytokines in mononuclear cells isolated from spleen and mesenteric lymph node tissues and PBMCs from the peripheral blood of wild-type and JH knockout piglets experimentally infected with HEV. PBMCs and MNCs were collected from each piglet at 28 dpi at necropsy. Cells were gated on CD3 and stained for extracellular and intracellular markers CD4+, CD25+, Foxp3, and TGF-β and IL-10, respectively, after stimulation with HEV-specific capsid protein. The mean frequencies of CD25+ Foxp3+ (A), CD25− Foxp3+ (B), TGF-β (C), and IL-10 (D) are indicated and expressed as means ± the SEM. All frequencies were determined by flow cytometry with 100,000 events per sample.
DISCUSSION
HEV is an important but understudied pathogen with the potential to cause significant mortality in immunocompromised populations such as organ transplant recipients (8), those with concurrent systemic immunosuppressive diseases such as lymphoma (10) or HIV/AIDS (43, 44), and in HEV-infected pregnant women who are burdened with a higher mortality rate reaching up to 28 to 30% (5). In this study, we successfully established a novel gnotobiotic pig model for HEV infection and systematically determined the dynamics of acute HEV infection in wild-type and Ig heavy chain JH−/− knockout gnotobiotic piglets. We also attempted to investigate the role of immunoglobulin heavy-chain JH in HEV pathogenesis and acute virus infection.
Pigs are the natural host for genotype 3 and 4 HEVs (13, 45, 46), which cause the majority of sporadic, cluster, and chronic cases of hepatitis E in humans, mostly from industrialized countries (4, 43). As an animal model, aside from nonhuman primates, pigs match humans (47) more closely than any other animals in terms of physiological characteristics and immunological parameters and responses. The gnotobiotic piglet model is extremely attractive especially for studying viral pathogenicity and immune responses, since the animals are raised in isolated sterile conditions with no interference from other infectious agents. In this case, infection with HEV was the only experimental manipulation in the piglets; therefore, the obtained results are solely attributable and specific to HEV. In addition, it has been shown that gnotobiotic piglets are more susceptible to virus infection (48, 49) and tend to develop more progressive disease with discernible lesions. Lacking maternal antibodies at the time of infection may allow for the increased pathogenesis of HEV and afford the opportunity to utilize a model that more closely matches symptomatic human infection. This is important in the study of HEV infection because the conventional pig model develops only mild-to-moderate hepatitis lesions and clears the infection asymptomatically (33, 36).
The application of the CRISRP/Cas9 system to produce Ig JH−/− knockout piglets significantly impacts the production of large animal models for biomedical research and in this case, allowed for the identification and isolation of each arm of the immune system in response to HEV infection. The CRISPR/Cas9 system allows for effective site-specific genome modification or targeted modifications during embryogenesis; therefore, bypassing the need for somatic cell nuclear transfer, which is associated with developmental defects in animal production. In a previous study, we generated RAG2/IL2RG double knockout pigs using direct injection of CRISRP/Cas9 system (41), thus eliminating the breeding step in generating these pigs. The efficiency of this approach was high enough (100%) to place these pigs into a human norovirus challenge study without genotyping. Similarly, in this HEV infection study, all JH−/− knockout genetically modified pigs were generated using direct injection of CRISPR/Cas9 into developing embryos without reversion to wild-type genotyping. In addition, none of the Ig heavy chain knockout piglets carried a mosaic genotype, which can be a shortcoming of the direct injection of the CRISPR/Cas9 system. These results suggest that the CRISPR/Cas9 system is a valid approach to use for animal production without having to establish a breeding program or herd for such use. Large animal models, namely, pigs, closely recapitulate the clinical signs of disease in human patients (50). However, use of the pig model in biomedical research is limited due to the cost of housing, housing requirements, and relatively prolonged gestation period. Using CRISPR/Cas9 system in large animal production may help reduce these costs through increased efficiency, as well as opening up the opportunity to develop specific knockout models in order to assess various immunological parameters.
During the acute phase of HEV infection, fecal virus shedding is typically apparent by 1 wpi and continues for approximately 3 to 4 weeks (32, 33, 51). By this time, the development of anti-HEV IgG is able to clear the infection, usually resulting in an asymptomatic course of infection devoid of clinical signs in both immunocompetent humans and pigs. The viremic phase typically lasts 1 to 3 weeks (34), and the acute infection is considered to peak at 4 wpi. In the present study, it was important to demonstrate that this novel gnotobiotic pig model mimics the typical course of HEV infection in humans since this is the first reported use of gnotobiotic piglets for the study of HEV. To evaluate the gross and microscopic lesions in the liver attributable to HEV infection, the infected animals had to be euthanized at 28 days postinfection, at which time the pathological liver lesions usually peak. In all HEV-inoculated gnotobiotic piglets, HEV was detectable in the feces during the first week postinfection and remained positive with relatively high viral RNA levels until necropsy at 28 dpi (Table 5, Fig. 3A and C). Fecal virus shedding appeared much earlier at 2 dpi in JH−/− knockout gnotobiotic piglets than in wild-type piglets (7 dpi). Interestingly, the viral RNA loads in fecal samples, intestinal content collected at necropsy, and serum samples were generally higher with significant differences observed at 23 dpi in the HEV-infected wild-type gnotobiotic piglets than in the HEV-infected JH−/− knockout gnotobiotic piglets. Furthermore, the incidence of viremia and viral RNA loads in sera were also higher in HEV-infected wild-type piglets than in HEV-infected JH−/− knockout gnotobiotic piglets. It is possible that the decreased level of HEV replication in JH−/− knockout gnotobiotic piglets is attributable to the lack of Ig heavy chain in the infected pigs. Unfortunately, due to the limited scope of the study, which is to study the infection dynamic and pathogenicity during acute infection, the animal experiment was terminated during the time of peak pathological liver lesions prior to the appearance of anti-HEV antibodies in pigs, which typically occurs at 4 to 5 weeks postinfection. The lack of detectable anti-HEV antibodies in infected JH−/− knockout and wild-type pigs at all time points validates the gnotobiotic pig model in that there are no maternal effects carried over after birth since piglets were retrieved via hysterectomy. Specifically, these piglets were naive to colostrum and were maintained on sterile whole cow's milk. Future studies are warranted to extend the infection period beyond the 4-week acute infection in order to definitively determine the role that the Ig heavy chain plays in susceptibility to virus infection and clearance. It is likely that the differences in virus replication level observed during the 4 weeks postinfection between the HEV-infected JH−/− knockout and wild-type groups will be greatly exacerbated with a prolonged experimental period, thus highlighting the importance of the humoral immune response to HEV infection.
HEV infection in immunocompetent humans and outbred conventional pigs generally causes only mild-to-moderate hepatic lesions without clinical disease (2). In this study, both wild-type and JH−/− knockout gnotobiotic piglets experimentally infected with HEV developed more pronounced lymphoplasmacytic hepatitis and hepatocellular necrosis lesions than other studies using conventional pigs (Fig. 5A). All HEV-infected gnotobiotic piglets recorded a histopathologic score of 2, which corresponds to three to five focal infiltrates per ten liver lobules or a score of 3 corresponding to six to ten focal infiltrates per ten liver lobules (33). Pigs with an assessed score of 3 also had hepatocellular necrosis or apoptosis, which was indicative of HEV-specific liver lesions. In addition, the HEV-infected JH−/− knockout pigs had significantly enlarged livers both grossly and as a liver/body weight ratio (Fig. 5B) compared to the PBS-inoculated group. Therefore, it appears that the HEV-infected gnotobiotic pigs induced more severe histological and gross liver pathology and inflammation than the conventional pigs. This presented as expected since gnotobiotic pigs are more sensitive to virus infection and are considered an excellent model for pathogenicity studies of viral infections (48, 49). Therefore, the gnotobiotic pig model may aid in future studies of HEV pathogenesis and disease, an aspect which has thus far been difficult to reproduce in the available animal model systems.
Serum levels of liver enzymes during HEV infections tend to elevate at the time of peak virus infection (30, 52, 53). In this study, we showed that the serum levels of liver enzymes, with the exception of GGT (Fig. 4A to D), were not good indicators for liver damage, nor do they assess overall liver health in the HEV-infected gnotobiotic piglets. Alkaline phosphatase was dramatically, yet inconsequentially elevated (Fig. 4C) in these piglets and likely due to the chosen feed source, as well as the young age of the gnotobiotic piglets. It is likely that this enzyme will return to normal values after approximately 60 days of age. Furthermore, feeding sterile boxed cow's milk in order to retain their sterility have had an adverse effect on the alkaline phosphatase levels, rendering it an insignificant biomarker in the gnotobiotic piglet model (54). Total bilirubin (Fig. 4B) and AST (Fig. 4A) were observed to be at similar levels in both the PBS mock-infected and the HEV-infected groups. All animals presented with elevated values, albeit with no discernible trend or significance. The serum level of the liver enzyme GGT (Fig. 4D) was significantly elevated in HEV-infected JH−/− knockout gnotobiotic pigs at 23 dpi, which may indicate liver damage in HEV-infected JH−/− knockout pigs, since the predominant source of GGT is the biliary epithelium, and significantly higher HEV RNA loads were observed in bile (P = 0.0169) in wild-type versus JH−/− knockout pigs.
At necropsy, neither the Ig heavy chain JH−/− knockout piglets nor the wild-type piglets had developed the antibodies necessary for clearance of HEV infection. By infecting gnotobiotic piglets at an early age (8 days) and completing the study by 35 days of age, the piglets are less immunocompetent than conventional pigs, which may result in a reduced humoral immune response and play a role in the level of HEV replication and pathogenesis. However, gnotobiotic piglets have been widely used to study other enteric viruses such as rotavirus infection dynamics and respond with a strong antibody response following infection by 28 dpi (55, 56). Therefore, the lack of antibodies is less likely influenced by the age of piglet, but rather by the typical time course for HEV infection. It has long been suspected that liver damage during HEV infection is a result of an immune-mediated (especially antibody-mediated) event and not due to the result of direct virus replication in hepatocytes (37, 38). Studies in nonhuman primates (38) demonstrated a correlation between liver lesions, elevated levels of liver enzymes, and symptomatic hepatic disease with the appearance of anti-HEV antibodies. Therefore, a longer term gnotobiotic pig infection study is needed to determine the significance of IgG anti-HEV antibodies in control of virus infection and pathogenesis and to determine the immune response of gnotobiotic piglets infected with HEV at an early age. Unfortunately, gnotobiotic piglets do have a rather limited life span due to the diet (sterile milk) and housing (plastic sterile bubble) requirements. The presence of liver lesions in the present study prior to an antibody response suggests that the liver damage is not solely mediated by the humoral immune response and requires further validation.
Cytokines play important roles regulating the immune response during virus infection but are currently not well understood in the context of HEV infection. During the acute HEV infection (up to 28 dpi) in the gnotobiotic pigs, we systematically determined the Treg, Th1, and Th2 cytokine responses in PBMC and splenic and mesenteric lymph node MNC populations. When gated on CD3+, CD4+ Th1 cell frequencies in flow cytometric analyses were significantly elevated in the mock-infected and HEV-infected Ig heavy chain JH−/− knockout groups compared to wild-type piglets (Fig. 6C). However, the IFN-γ+ Th cells, which are the critical effector memory T cells (57) used in pigs to mount cell-mediated immune responses, were not significantly different between any of the groups. The frequency of expression of IFN-γ+ T cells appeared to be numerically lower in PBMCs and to a lesser degree in lymph node MNCs in both JH−/− knockout groups compared to the wild-type pig groups (Fig. 6A), but not in the MNC populations isolated from the spleen. Although the difference was not significant, it points to a trend in a somewhat dampened cell-mediated immune response in JH−/− knockout pigs compared to the wild-type piglet. In addition to the Th responses, Treg cells are also important in maintaining the balance between clearing virus infection and minimizing immune-mediated pathology during infection (58, 59). In this study, there was no difference in Treg cell populations (CD25+ or CD25−) between mock-infected and HEV-infected groups (Fig. 7A and B), regardless of the phenotypes of the pigs. Similarly, the Treg-specific cytokines TGF-β and IL-10, typically expressed as a functional determination of the Treg cell population (Fig. 7C and D), were not significantly different among any of the groups. Therefore, while Treg cells may play a role in viral clearance or mitigation of immune-mediated pathology, this was not evident during the first 28 dpi of HEV infection in gnotobiotic pigs.
Overall, the results from this study showed a weak cell-mediated immune response in both the JH−/− knockout pigs and wild-type pigs at the presumptive peak virus infection at 28 dpi. Since this was the first attempt at HEV infection of gnotobiotic piglets with a limited study scope focusing on acute virus infection, the full course of infection from virus inoculation to clearance has not been evaluated. We demonstrated that Ig JH−/− knockout phenotype pigs exhibit a reduction in the mean frequencies of B lymphocytes while maintaining other cell populations, including T cells and NK cells. However, this study did not reach a time point when the wild-type pigs produced HEV-specific antibodies, which is a limitation. Therefore, a prolonged follow-up study is needed to fully discern the differences in immune responses between these JH−/− knockout and wild-type animals. Nevertheless, a discernible difference was observed with respect to liver lesions in HEV-infected gnotobiotic groups. We also noted a difference in the liver size both grossly and as a ratio of liver/body weight ratio in the HEV-infected JH−/− knockout group and a significant difference in viral RNA loads in bile at 28 dpi. The HEV infection dynamic data in gnotobiotic piglets from this study will help in the design of future experiments using a gnotobiotic pig model. Clearly, the JH−/− knockout gnotobiotic pig model for HEV requires further refinement to determine whether anti-HEV antibodies play a role in pathogenicity and virus clearance, and the presence of liver damage prior to the onset of an antibody response requires investigation.
MATERIALS AND METHODS
Generation of Ig JH−/− knockout gnotobiotic pigs by CRISPR/Cas9 technology.
To disrupt the Ig heavy chain, a total of four target sites were designed using a web-based program (http://www.crispr-cas.org/p/resources.html) (Fig. 1A and Table 6). To minimize potential off-targeting events, target sequences were blasted against the whole pig genome and those carrying 17 or more identities with other loci in the pig genome were excluded. The four sgRNAs, containing a target sequence and tracker RNA, were synthesized by in vitro transcription by using a MEGAshortscript kit (Ambion) with a double-stranded DNA as a template (IDT). Cas9 mRNA was generated as described in our previous studies (40–42).
TABLE 6.
Primers and double-stranded DNA used for the development of Ig heavy chain knockout piglets
| Name | Sequence (5′–3′) |
|---|---|
| Ig heavy chain G-block 1 (dsDNA) | TTAATACGACTCACTATAGGCGTTGAAGTCGTCGTGTCGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTGTTTTAGA |
| Ig heavy chain G-block 2 (dsDNA) | TTAATACGACTCACTATAGGACACCCCCCAGGTTTTTGTGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTGTTTTAGA |
| Ig heavy chain G-block 3 (dsDNA) | TTAATACGACTCACTATAGGAGATTGCACCACTGTGATGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTGTTTTAGA |
| Ig heavy chain G-block 4 (dsDNA) | TTAATACGACTCACTATAGGGTTGAAGTCGTCGTGTCCTCGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTGTTTTAGA |
| T7_Ig heavy chain 1F | TTAATACGACTCACTATAGGCGTTGAAGTCGTCGTGTC |
| T7_Ig heavy chain 2F | TTAATACGACTCACTATAGGACACCCCCCAGGTTTTTGTG |
| T7_Ig heavy chain 3F | TTAATACGACTCACTATAGGAGATTGCACCACTGTGAT |
| T7_Ig heavy chain 4F | TTAATACGACTCACTATAGGGTTGAAGTCGTCGTGTCCTC |
| T7_sgRNA R | AAAAGCACCGACTCGGTGCC |
| Cas9 F | TAATACGACTCACTATAGGGAGAATGGACTATAAGGACCACGAC |
| Cas9 R | GCGAGCTCTAGGAATTCTTAC |
| Genomic primer F | GACACTTTGGAGGTCAGGAACGGGAG |
| Genomic primer R | CTTCTCTCTCCGACATGGCTCTTTCAGAC |
Pig oocytes were obtained either from ovaries collected at a local abattoir or purchased commercially (DeSoto, Inc.). To collect oocytes from ovaries, medium-sized follicles were aspirated using an 18-gauge needle attached to a 10-ml syringe. Cumulus cell-oocyte complexes (COC) were washed twice with prewarmed TL-HEPES medium and COCs with a multiple layer of cumulus cells, stimulated to mature in vitro in a TCM-199-based maturation medium containing 0.5 IU/ml follicle-stimulating hormone (FSH), 0.5 IU/ml luteinizing hormone (LH), 0.82 mM cysteine, 3.02 mM glucose, 0.91 mM sodium pyruvate, and 10 ng/ml epidermal growth factor for 42 to 44 h at 38.5°C and 5% CO2 incubator. After maturation, COCs were denuded by 0.1% hyaluronidase. Oocytes were then extruded and the first polar body was used for in vitro fertilization (IVF). Groups of 30 oocytes were placed in 50-μl droplets of IVF medium (modified Tris-buffered medium with 113.1 mM NaCl, 3 mM KCl, 7.5 mM CaCl2, 11 mM glucose, 20 mM Tris, 2 mM caffeine, 5 mM sodium pyruvate, and 2 mg/ml bovine serum albumin [BSA]) and covered with mineral oil. Extended semen was washed with PBS at 720 relative centrifugal force for 3 min three times. After the wash, the sperm pellet was resuspended with IVF media. Subsequently, 50 μl of diluted sperm (2.5 × 105 sperm/ml) was added into mTBM drops that contained oocytes. The gametes were coincubated for 5 h at 38.5°C and 5% CO2. Presumable zygotes were washed three times with Porcine Zygote Medium 3 (PZM-3) (60) and then placed in PZM-3 and incubated at 38.5°C in 5% CO2 and 5% O2.
Two hours after IVF, sgRNAs and Cas9 mRNA were introduced into presumptive zygotes to disrupt Ig heavy chain via microinjection. Each zygote received 10 ng/μl of sgRNAs and 20 ng/μl of Cas9 mRNA by direct injection into the cytoplasm using a FemtoJet microinjector (Eppendorf, Hamburg, Germany). Microinjection was conducted in manipulation medium (TCM199 with 0.6 mM NaHCO3, 2.9 mM HEPES, 30 mM NaCl, 10 ng/ml gentamicin, and 3 mg/ml BSA) and covered with mineral oil on the heated stage of a Nikon inverted microscope (Nikon Corporation, Tokyo, Japan). Injected zygotes were washed with PZM-3 and then cultured in PZM-3 until further analyses. For embryo transfer, embryos were cultured in PZM-3 in the presence of 10 ng/ml granulocyte-macrophage colony-stimulating factor (61).
To confirm mutations of the Ig heavy chain region, PCR verification was performed with the primers listed in Table 6. Genomic DNA was extracted from an individual blastocyst by incubating blastocysts in embryo lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 8.5], 0.5% Nonidet P40, 0.5% Tween 20, and 200 μg/ml proteinase K) for 30 min at 65°C, followed by 10 min at 95°C. Genomic DNA was extracted from the tissue of each piglet using PureLink Genomic DNA kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The predicted mutation sites were amplified by using Platinum Taq DNA polymerase (Thermo Fisher Scientific). PCR was conducted using the following conditions: initial denaturing at 95°C for 2 min, denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min for 34 cycles, final extension at 72°C for 5 min, and holding at 4°C. The PCR products were either directly sequenced or inserted into a cloning vector (TOPO; Invitrogen) to confirm the mutations introduced by CRISPR/Cas9 system.
At day 5 to 6 post-IVF, blastocysts and embryos carrying over 16 cells were transferred into surrogate gilts. The embryos were surgically transferred into the oviduct of the gilts. Pregnancy was determined by ultrasound examination at day 30 to 35 of gestation. Sows impregnated with wild-type or knockout embryos, respectively, underwent hysterectomy at 111 to 113 days of gestation for the retrieval of gnotobiotic piglets. Piglets were further maintained in plastic sterile microbiological isolators (“bubbles”) for the duration of the study. All animals were delivered and housed at the Virginia Polytechnic Institute and State University (Blacksburg, VA) in accordance with the approved procedures of the Institutional Animal Care and Use Committee (IACUC). Twenty-one gnotobiotic piglets were used, corresponding to six litters (IACUC no. 13-127-CVM).
Virus inoculum.
A genotype 3 strain of human HEV (US-2) originally isolated from a patient in the United States was used in this study (62). The infectious stock of the US-2 HEV was prepared as a 10% fecal suspension in 0.01 M PBS using the feces of a pig experimentally infected with the US-2 strain of human HEV from a previous study (34). The fecal suspension was further purified to produce an endotoxin-free virus preparation using cellufine sulfate (JNC Corporation, Tokyo, Japan) as the binding substrate for column purification and elution into PBS. The genomic equivalent (GE) titer of the final viral stock was determined by HEV-specific quantitative qRT-PCR with a GE titer of ∼4.5 × 106 per ml of fecal suspension.
Experimental infection of wild-type and JH−/− knockout gnotobiotic piglets with a genotype 3 strain of human HEV.
The experimental design for the HEV infection of gnotobiotic pigs is detailed in Table 1. The gnotobiotic piglets were housed in the sterile isolators for 7 days before virus inoculation. On day 8, a total of 6 JH−/− knockout gnotobiotic piglets were intravenously inoculated with approximately 9 × 106 GE titer of US-2 HEV infectious stock, and 5 other JH−/− knockout piglets were similarly mock inoculated with PBS (Table 1). As controls, a total of 5 wild-type gnotobiotic pigs were intravenously inoculated with approximately 9 × 106 GE titer of US-2 HEV infectious stock, and another 4 wild-type gnotobiotic pigs were similarly inoculated with PBS buffer.
After inoculation, the gnotobiotic piglets were monitored daily for a total of 4 wpi. Blood samples were collected from each pig weekly, and fecal samples were collected three times per week. At 4 wpi, all gnotobiotic piglets were humanely euthanized and necropsied. Blood samples were collected for serum and PBMC isolation prior to euthanasia. The total body and liver weights were measured. During necropsy, tissue samples, including spinal cord, brain, lung, liver, kidney, spleen, duodenum, jejunum, ileum, large intestine, and mesenteric lymph node tissues, were collected from each pig. A portion of the tissue samples were fixed in 10% formalin to process for routine histological examination of pathological lesions, and another portion of the tissues samples were immediately frozen on dry ice and further stored at −80°C for detection and quantification of HEV RNA by qRT-PCR (Table 5). In addition, samples of bile and intestinal content were also collected and stored at −80°C for the detection and quantification of HEV RNA by RT-PCR and qRT-PCR (Fig. 3B). Furthermore, sections of spleen and mesenteric lymph node were sterilely collected in RPMI 1640 medium and stored on wet ice for immediate isolation of MNCs.
Histological examination of liver lesions.
Evaluation of gross and histological lesion were conducted by a board-certified veterinary pathologist in a blinded fashion. The formalin-fixed tissues were trimmed and routinely processed for histological examination. The lesion scoring criteria, previously described (33), were as follows: score 0, no inflammation; score 1, 1 to 2 focal lymphoplasmacytic infiltrates per 10 hepatic lobules; score 2, 2 to 5 focal infiltrates/10 hepatic lobules; score 3, 6 to 10 focal infiltrates/10 lobules; and score 4, >10 focal infiltrates/10 hepatic lobules (Fig. 5A).
Detection of HEV RNA by a nested RT-PCR.
To detect HEV RNA in weekly fecal samples, total RNAs were extracted with TRIzol LS reagent (Invitrogen) from 200 μl of each 10% fecal suspension. The total RNAs were resuspended in 30 μl of DNase-, RNase-, and proteinase-free water (Eppendorf, Inc.). Synthesis of cDNA and first-round PCR amplification were performed using Superscript III one-step reverse transcriptase PCR kit (Invitrogen) with primers specific for the US-2 strain of HEV: forward primer US202:1198F22 (5′-ATCGCCCTGACACTGTTCAATC-3′) and reverse primer US202:2064R25 (5′-AGGAATTAATTAAGACTCCCGGGTT-3′). The cDNA synthesis was carried out at 55°C for 30 min, followed by PCR amplification with an initial denaturing incubation at 94°C for 2 min and 40 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 68°C for 1 min, with a final incubation at 68°C for 5 min. The second round nested RT-PCR was completed using Hi-Fidelity Taq polymerase (Invitrogen) using 5 μl of first-round PCR product as the template and the same primers (US202:1198F22 and US202:2064R25) in a reaction as described previously (25, 34, 47). The amplified PCR products were visualized by gel electrophoresis on 1% agarose in Tris-acetate-EDTA buffer (TAE; Thermo Fisher Scientific). An expected PCR product band of 895 bp was extracted and purified from the gel using QG buffer (Qiagen) and sequenced to verify the authenticity of the amplified product at the Genomic Sequencing Center at the Biocomplexity Institute of Virginia Tech.
Quantification of HEV RNA by qRT-PCR.
The amounts of viral RNAs in weekly serum and fecal swab samples, as well as in a panel of tissue samples collected at necropsy, were quantified by qRT-PCR using HEV-specific primers and probe. Briefly, fecal swab materials and intestinal content were suspended in 10% sterile PBS (wt/vol). Serum and bile samples were diluted in 10% sterile PBS (vol/vol). One gram of tissue samples (liver, spleen, jejunum, and lymph node) were homogenized in 1 ml of TRIzol LS reagent (Invitrogen) to prepare a 10% tissue suspension. Briefly, each 1-g sample was placed in 1 ml of TRIzol for 5 min at room temperature, homogenized using individual sterile gentleMACS dissociator tubes (Miltenyi Biotec), and centrifuged at 4°C for 5 min. Samples were separated into 1.5-ml microcentrifuge tubes for the addition of 200 μl of chloroform per 1-ml sample. Samples were incubated at room temperature for 10 min and centrifuged at 12,000 rpm for 15 min at 4°C for phase separation, and supernatants were added to an equivalent amount of 70% ethanol. Total RNAs were extracted using RNeasy microkit columns (Qiagen) from 10% serum, bile, fecal swab materials, intestinal contents, and tissue suspension in 70% ethanol using a standard Qiagen protocol. The amounts of HEV RNA were quantified using the SensiFAST Probe No-ROX One-Step kit (Bioline, USA, Inc.) with the forward primer JVHEVF (5′-GGTGGTTTCTGGGGTGAC-3′), the reverse primer JVHEVR (5′-AGGGGTTGGTTGGATGAA-3′), and the hybridization probe JVHEVP (5′-TGATTCTCAGCCCTTCGC-3′) according to a protocol described previously (63). The qRT-PCR assays were performed in a CFX96 real-time PCR machine (Bio-Rad Laboratories). The standard curve was produced with purified HEV genomic RNAs, and the thermal cycling conditions were as follows: 45°C for 10 min (reverse transcription) and 95°C for 2 min (initial denaturation), followed by 95°C for 5 s (denaturation) and 60°C for 20 s (PCR amplification) for 40 cycles. The detection limit of the qRT-PCR assay was reported earlier as 10 viral genomic copies, which corresponds to approximately 400 copies of viral RNA per 1-ml sample or per gram of tissue (63, 64). Samples with GE titers below the detection limit were considered negative.
ELISA to detect IgG anti-HEV in pigs.
Weekly serum samples were tested for IgG anti-HEV antibodies by an ELISA as described previously (14, 34, 47). Briefly, a truncated recombinant HEV capsid protein containing the immunodominant region of HEV ORF2 (amino acids 452 to 617) was used as the antigen (GenWay Biotech, Inc., San Diego, CA) and plated at 6 μg/10 ml of carbonate coating buffer to each well. The serum samples were tested at a 1:100 dilution in blocking buffer. Preimmune and hyperimmune anti-HEV positive pig sera were included as negative and positive controls, respectively. Horseradish peroxidase-conjugated goat anti-swine IgG (Sigma) was used as the secondary antibody at 1:2,000 in blocking buffer. The plate was developed using ABTS substrate and absorbance was measured at 405 nm on a Promega GloMax Discover plate reader.
Determination of serum levels of liver enzymes in pigs.
A panel of liver enzymes, including aspartate aminotransferase (AST), gamma-glutamyl transferase (GGT), alkaline phosphatase, and total bilirubin, were measured in weekly serum samples using established protocols and standard values from the Veterinary Diagnostic Lab at Iowa State University College of Veterinary Medicine (Ames, IA).
Isolation of PBMCs from peripheral blood and MNCs from spleen and lymph node tissues.
Blood was collected from each pig for PBMC isolation prior to euthanasia and isolated using density gradient centrifugation with Ficoll-Paque Premium (GE Healthcare). Buffy coats were resuspended in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Viability counts of PBMCs were performed with 1:1 trypan blue as the stain and a Cellometer (Nexelom). Cells were stored at a density of 107/ml in FBS with 5% dimethyl sulfoxide at −80°C overnight before moving to liquid nitrogen for storage prior to staining. Samples of spleen and MLN tissues were collected in RPMI 1640 medium supplemented with gentamicin, ampicillin, and HEPES on the day of euthanasia and processed for the isolation of MNCs as described previously (65, 66). Briefly, the spleen tissues were minced and transferred through a 40-μm-pore-size cell strainer and pelleted by centrifugation. A density-gradient isolation procedure was followed using Percoll in order to collect the interface containing MNCs. The isolated cells were resuspended in enriched RPMI 1640 medium supplemented with 8% FBS, gentamicin, ampicillin, 2 mM l-glutamine, 100 μM nonessential amino acids, 1 mM sodium pyruvate, and 1:1,000 2-mercaptoethanol. For the mesenteric lymph nodes, the tissue samples were incubated for 30 min at 37°C on a horizontal shaker in RPMI medium supplemented with 8% FBS, gentamicin, ampicillin, HEPES, collagenase D (1 mg/ml), and DNase I (100 U/ml), prior to transferring the suspension to a cell strainer and collection by centrifugation. Cells were counted with 1:1 trypan blue using a Cellometer and stored as described above.
Intracellular cytokine staining and flow cytometry analyses.
Splenic and mesenteric lymph node MNCs and blood PBMCs were thawed at room temperature; resuspended in RPMI 1640 medium with 10% FBS, ampicillin, and gentamicin; and plated at a concentration of 2 × 106 cells/100 μl/well of a 96-well V-bottom tissue-culture plate. Two wells were prepared for each sample using three separate plates for each of the three staining protocols. An unstimulated preparation of RPMI 1640 medium with 10% FBS, ampicillin, gentamicin, and 0.2 μg of anti-human CD49 costimulatory monoclonal antibody (MAb) was applied to one well for each sample per plate at 100 μl per well. A stimulation preparation of RPMI 1640 medium with 10% FBS, ampicillin, gentamicin, 0.2 μg of CD49 MAb, and 1 μl of JPT Pepmix (purified recombinant HEV ORF2 antigen) was applied to one well for each sample per plate at 100 μl per well. A cell stimulation mixture (eBioscience, Inc.) was used as a positive stimulation control. Cells were stimulated for 12 h at 37°C. Brefeldin A (GolgiPlug; BD Biosciences) was added to each well at 0.2 μl/well, followed by further incubation at 37°C for 5 h. The plates were stored overnight and protected from light at 4°C prior to antibody staining. PBMCs and MNCs were washed with PBS once, resuspended in cold PBS with 3% FBS (fluorescence-activated cell sorting buffer), and stained with one of the following three panels of fluorochrome-conjugated antibodies according to manufacturer's recommendations and procedures described previously (45, 48, 49).
In panel 1, PBMCs (2 × 106 cells per well) were sequentially stained with the following MAb sets diluted in cold PBS with 3% FBS at 4°C for 15 min per incubation period: Spectral Red-conjugated mouse (IgG1) anti-pig CD3ε (clone PPT3; Southern Biotech) and phycoerythrin (PE)-conjugated mouse (IgG1) anti-pig CD16 (AbD Serotec), followed by permeabilization with BD Cytofix/Cytoperm buffer (BD Pharmingen) at 4°C for 15 min and three washes with BD Perm/Wash buffer (BD Pharmingen). Intracellular markers were stained with mouse IgG2b anti-human CD79a (clone 11D10; Vector Laboratories), followed by PE-Cy7-goat anti-mouse IgG2b (Southern Biotech) and fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse IgG2b (BioLegend) at 4°C for 15 min. Appropriate isotype-matched control antibodies were included as background staining controls (Southern Biotech, BD Biosciences, eBioscience, and Affymetrix eBioscience). The frequencies of CD16+ and CD79a+ cells were expressed as percentages of parental CD3gated cells.
In panel 2, in vitro HEV-specific antigen-stimulated PBMCs and splenic and mesenteric lymph node MNCs were stained with antibodies to determine the frequencies of CD3+ CD4+ Foxp3+ Treg cells, CD25+ activation status, and the expression of regulatory cytokines IL-10 and TGF-β in these cells (45, 48). PBMCs, spleen-derived MNCs, and mesenteric lymph node-derived MNCs (2 × 106 cells/well) were stained at 4°C for 15 min with FITC-conjugated mouse (IgG2b) anti-pig CD4a, SPRD-conjugated mouse (IgG2a) anti-pig CD3, and mouse (IgG1) anti-pig CD25 (IgG1, clone K231.3B2; AbD Serotec), followed by APC-conjugated rat (IgG1) anti-mouse (clone X56; BD Pharmingen). After staining for the extracellular markers, the cells were permeabilized with the Foxp3-staining buffer set (eBioscience) at 4°C for 15 min. Intracellular staining was completed with PE-Cy7-conjugated rat (IgG2) anti-mouse/rat Foxp3 (cloneFJK-16S; eBioscience), biotin mouse (IgG1) anti-pig IL-10 (clone 945a; Cell Sciences), and PE-conjugated mouse (IgG1) anti-human TGF-β1 (clone 9016; R&D Systems) at 4°C for 15 min. The appropriated isotype-matched control antibodies and PE-conjugated mouse (IgG1) isotype control (clone P3.6.2.8.2; eBioscience) were used as background staining controls. The frequencies of CD4+ Foxp3+, CD25+, IL-10+, and TGF-β+ cells were expressed as percentages of parental CD3 gated cells.
In panel 3, in vitro HEV-specific antigen-stimulated PBMCs and spleen-derived and mesenteric lymph node-derived MNCs were stained with antibodies to determine CD3+ CD4+ cells expressing IL-4 and IFN-γ regulatory cytokines. PBMCs and MNCs were stained at 4°C for 15 min with FITC-conjugated mouse (IgG2b) anti-pig CD4a (clone 74-12-4; BD Pharmingen) and SPRD-conjugated (IgG1) anti-pig CD3ε, followed by permeabilization with BD Cytofix/Cytoperm buffer (BD Pharmingen) at 4°C for 15 min. PBMCs and MNCs were washed three times with BD Perm/Wash buffer (BD Pharmingen) and subsequently stained with PE-conjugated mouse (IgG1) anti-pig IFN-γ (clone P2G10; BD Pharmingen) and Brilliant Violet 421 anti-human IL-4 antibody (IgG1 clone MP4-25D2; BioLegend). The appropriate isotype-matched control antibodies were used as background staining controls (BioLegend, BD Biosciences, and Southern Biotech).
For all three staining panels, at least 100,000 events were acquired on FACSCalibur flow cytometer (BD Biosciences), and the data were analyzed using FloJo V10 software (Tree Star). The absolute numbers of B, Treg, and NK cells were based on the frequencies of cells in each sample, respectively.
Statistical analyses.
All data were statistically analyzed using GraphPad Prism 7.0 (GraphPad Software, Inc.). The differences between the mean values of two treatment groups were analyzed with two-way analysis of variance (ANOVA) or a two-tailed unpaired Student t test and Tukey multiple-comparison tools. The presence or absence of hepatic infiltrates was analyzed by Fisher exact test. A P value of <0.05 was considered statistically significant for all analyses. All graphs are reported as means ± the standard errors of the mean (SEM).
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (R01 AI074667 and R01 AI050611). D.M.Y. is supported by a Ruth L. Kirschstein National Research Service Award Institutional Research Training Grant (T32OD010430).
We thank Melissa Makris for technical assistance in flow cytometry; Karen Hall, Kimberly Allen, Peter Jobst, and the Teaching and Research Animal Care Support Staff (TRACCS) for their support in the animal study; and Lijuan Yuan's laboratory for their support in the animal study.
REFERENCES
- 1.Yugo DM, Meng XJ. 2013. Hepatitis E virus: foodborne, waterborne, and zoonotic transmission. Int J Environ Res Public Health 10:4507–4533. doi: 10.3390/ijerph10104507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson S, Purcell R. 2003. Hepatitis E virus. Rev Med Virol 13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 3.Dalton H, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 4.Meng X-J. 2011. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res 161:23–30. doi: 10.1016/j.virusres.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X-J. 2013. Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis 33:41–49. doi: 10.1055/s-0033-1338113. [DOI] [PubMed] [Google Scholar]
- 6.Navaneethan U. 2009. Seroprevalence of hepatitis E infection in pregnancy: more questions than answers. Indian J Med Res 130:677–679. [PubMed] [Google Scholar]
- 7.Navaneethan U, Al Mohajer M, Shata MT. 2008. Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int 28:1190–1199. doi: 10.1111/j.1478-3231.2008.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haagsma EB, van den Berg AP, Porte RJ, Benne CA, Vennema H, Reimerink JH, Koopmans MP. 2008. Chronic hepatitis E virus infection in liver transplant recipients. Liver Transpl 14:547–553. doi: 10.1002/lt.21480. [DOI] [PubMed] [Google Scholar]
- 9.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med 361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 10.Kamar N, Garrouste C, Haagsma EB, Garrigue V, Pischke S, Chauvet C, Dumortier J, Cannesson A, Cassuto-Viguier E, Thervet E, Conti F, Lebray P, Dalton HR, Santella R, Kanaan N, Essig M, Mousson C, Radenne S, Roque-Afonso AM, Izopet J, Rostaing L. 2011. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140:1481–1489. doi: 10.1053/j.gastro.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 11.Singh A, Seth R, Gupta A, Shalimar, Nayak B, Acharya SK, Das P. 2016. Chronic hepatitis E: an emerging disease in an immunocompromised host. Gastroenterol Rep (Oxf) 2016:gow024. doi: 10.1093/gastro/gow024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purdy MA, Harrison TJ, Jameel S, Meng XJ, Okamoto H, Van der Poel WHM, Smith DB, ICTV Report Committee. 2017. ICTV virus taxonomy profile: Hepeviridae. J Gen Virol 98:2645–2646. doi: 10.1099/jgv.0.000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng XJ. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol 140:256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng XJ, Purcell R, Halbur P, Lehman J, Webb D, Tsareva T, Haynes J, Thacker B, Emerson S. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonoda H, Abe M, Sugimoto T, Sato Y, Bando M, Fukui E, Mizuo H, Takahashi M, Nishizawa T, Okamoto H. 2004. Prevalence of hepatitis E virus (HEV) infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J Clin Microbiol 42:5371–5374. doi: 10.1128/JCM.42.11.5371-5374.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18:1274–1281. doi: 10.3201/eid1808.120057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- 18.de Deus N, Peralta B, Pina S, Allepuz A, Mateu E, Vidal D, Ruiz-Fons F, Martín M, Gortázar C, Segalés J. 2008. Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet Microbiol 129:163–170. doi: 10.1016/j.vetmic.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Sridhar S, Teng JLL, Chiu TH, Lau SKP, Woo PCY. 2017. Hepatitis E virus genotypes and evolution: emergence of camel hepatitis E variants. Int J Mol Sci 18:E869. doi: 10.3390/ijms18040869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee GH, Tan BH, Teo EC, Lim SG, Dan YY, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, Purdy MA, Teo CG. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150:355–357. doi: 10.1053/j.gastro.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 21.Payne CJ, Ellis TM, Plant SL, Gregory AR, Wilcox GE. 1999. Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet Microbiol 68:119–125. doi: 10.1016/S0378-1135(99)00067-X. [DOI] [PubMed] [Google Scholar]
- 22.Billam P, Huang F, Sun Z, Pierson F, Duncan R, Elvinger F, Guenette D, Toth T, Meng XJ. 2005. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J Virol 79:3429–3437. doi: 10.1128/JVI.79.6.3429-3437.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun ZF, Larsen CT, Huang FF, Billam P, Pierson FW, Toth TE, Meng XJ. 2004. Generation and infectivity titration of an infectious stock of avian hepatitis E virus (HEV) in chickens and cross-species infection of turkeys with avian HEV. J Clin Microbiol 42:2658–2662. doi: 10.1128/JCM.42.6.2658-2662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johne R, Heckel G, Plenge-Bönig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich R. 2011. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16:1452–1455. doi: 10.3201/eid1609.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johne R, Plenge-Bönig A, Hess M, Ulrich R, Reetz J, Schielke A. 2010. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol 91:750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- 26.Drexler J, Seelen A, Corman V, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose S, Adu-Sarkodie Y, Oppong S, Kalko E, Osterman A, Rasche A, Adam A, Müller M, Ulrich R, Leroy E, Lukashev A, Drosten C. 2012. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86:9134–9147. doi: 10.1128/JVI.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158:116–123. doi: 10.1016/j.virusres.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Kasorndorkbua C, Guenette D, Huang F, Thomas P, Meng XJ, Halbur P. 2004. Routes of transmission of swine hepatitis E virus in pigs. J Clin Microbiol 42:5047–5052. doi: 10.1128/JCM.42.11.5047-5052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao D, Huang Y-W, Meng X-J. 2010. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84:13040–13044. doi: 10.1128/JVI.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yugo DM, Cossaboom CM, Meng XJ. 2014. Naturally occurring animal models of human hepatitis E virus infection. ILAR J 55:187–199. doi: 10.1093/ilar/ilu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams TP, Kasorndorkbua C, Halbur PG, Haqshenas G, Guenette DK, Toth TE, Meng XJ. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J Clin Microbiol 39:3040–3046. doi: 10.1128/JCM.39.9.3040-3046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell RH, Emerson SU. 2001. Hepatitis E virus, 4th ed, p 3051–3061 Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 33.Halbur PG, Kasorndorkbua C, Gilbert C, Guenette D, Potters MB, Purcell RH, Emerson SU, Toth TE, Meng XJ. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol 39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng XJ, Halbur P, Shapiro M, Govindarajan S, Bruna J, Mushahwar I, Purcell R, Emerson S. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol 72:9714–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Billam P, Pierson F, Li W, LeRoith T, Duncan R, Meng X. 2008. Development and validation of a negative-strand-specific reverse transcription-PCR assay for detection of a chicken strain of hepatitis E virus: identification of nonliver replication sites. J Clin Microbiol 46:2630–2634. doi: 10.1128/JCM.00536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emerson S, Purcell R. 2007. Hepatitis E. Pediatr Infect Dis J 26:1147–1148. doi: 10.1097/INF.0b013e31815dd7c2. [DOI] [PubMed] [Google Scholar]
- 37.Tsarev SA, Emerson SU, Tsareva TS, Yarbough PO, Lewis M, Govindarajan S, Reyes GR, Shapiro M, Purcell RH. 1993. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J Infect Dis 167:1302–1306. doi: 10.1093/infdis/167.6.1302. [DOI] [PubMed] [Google Scholar]
- 38.Tsarev SA, Tsareva TS, Emerson SU, Rippy MK, Zack P, Shapiro M, Purcell RH. 1995. Experimental hepatitis E in pregnant rhesus monkeys: failure to transmit hepatitis E virus (HEV) to offspring and evidence of naturally acquired antibodies to HEV. J Infect Dis 172:31–37. doi: 10.1093/infdis/172.1.31. [DOI] [PubMed] [Google Scholar]
- 39.Billam P, LeRoith T, Pudupakam RS, Pierson FW, Duncan RB, Meng XJ. 2009. Comparative pathogenesis in specific-pathogen-free chickens of two strains of avian hepatitis E virus recovered from a chicken with Hepatitis-Splenomegaly syndrome and from a clinically healthy chicken. Vet Microbiol 139:253–261. doi: 10.1016/j.vetmic.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei S, Ryu J, Wen K, Twitchell E, Bui T, Ramesh A, Weiss M, Li G, Samuel H, Clark-Deener S, Jiang X, Lee K, Yuan L. 2016. Increased and prolonged human norovirus infection in RAG2/IL2RG-deficient gnotobiotic pigs with severe combined immunodeficiency. Sci Rep 6:25222. doi: 10.1038/srep25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu J, Lee K. 2017. CRISPR/Cas9-mediated gene targeting during embryogenesis in swine. Methods Mol Biol 1605:231–244. doi: 10.1007/978-1-4939-6988-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JT, Cho B, Ryu J, Ray C, Lee EJ, Yun YJ, Ahn S, Lee J, Ji DY, Jue N, Clark-Deener S, Lee K, Park KW. 2016. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod Biol Endocrinol 14:74. doi: 10.1186/s12958-016-0206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalton H, Bendall RP, Rashid M, Ellis V, Ramnarace R, Stableforth W, Headdon W, Abbott R, McLaugfhlin C, Froment E, Hall KJ, Thatcher P, Henley WE. 2011. Host risk factors and autochthonous hepatitis E infection. Eur J Gastroenterol Hepatol 23:1200–1205. doi: 10.1097/MEG.0b013e32834ca4da. [DOI] [PubMed] [Google Scholar]
- 44.Dalton HR, Fellows HJ, Stableforth W, Joseph M, Thurairajah PH, Warshow U, Hazeldine S, Remnarace R, Ijaz S, Hussaini SH, Bendall RP. 2007. The role of hepatitis E virus testing in drug-induced liver injury. Aliment Pharmacol Ther 26:1429–1435. doi: 10.1111/j.1365-2036.2007.03504.x. [DOI] [PubMed] [Google Scholar]
- 45.Cao D, Cao QM, Subramaniam S, Yugo DM, Heffron CL, Rogers AJ, Kenney SP, Tian D, Matzinger SR, Overend C, Catanzaro N, LeRoith T, Wang H, Pineyro P, Lindstrom N, Clark-Deener S, Yuan L, Meng XJ. 2017. Pig model mimicking chronic hepatitis E virus infection in immunocompromised patients to assess immune correlates during chronicity. Proc Natl Acad Sci U S A 114:6914–6923. doi: 10.1073/pnas.1705446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanford BJ, Dryman BA, Huang YW, Feagins AR, Leroith T, Meng XJ. 2011. Prior infection of pigs with a genotype 3 swine hepatitis E virus (HEV) protects against subsequent challenges with homologous and heterologous genotypes 3 and 4 human HEV. Virus Res 159:17–22. doi: 10.1016/j.virusres.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X, Halbur P, Haynes J, Tsareva T, Bruna J, Royer R, Purcell R, Emerson S. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol 143:1405–1415. doi: 10.1007/s007050050384. [DOI] [PubMed] [Google Scholar]
- 48.Wen K, Li G, Yang X, Bui T, Bai M, Liu F, Kocher J, Yuan L. 2012. CD4+ CD25− FoxP3+ regulatory cells are the predominant responding regulatory T cells after human rotavirus infection or vaccination in gnotobiotic pigs. Immunology 137:160–171. doi: 10.1111/j.1365-2567.2012.03617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan L, Wen K, Azevedo MS, Gonzalez AM, Zhang W, Saif LJ. 2008. Virus-specific intestinal IFN-γ-producing T cell responses induced by human rotavirus infection and vaccines are correlated with protection against rotavirus diarrhea in gnotobiotic pigs. Vaccine 26:3322–3331. doi: 10.1016/j.vaccine.2008.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers CS, Hao Y, Rokhlina T, Samuel M, Stoltz DA, Li Y, Petroff E, Vermeer DW, Kabel AC, Yan Z, Spate L, Wax D, Murphy CN, Rieke A, Whitworth K, Linville ML, Korte SW, Engelhardt JF, Welsh MJ, Prather RS. 2008. Production of CFTR-null and CFTR-DeltaF508 heterozygous pigs by adeno-associated virus-mediated gene targeting and somatic cell nuclear transfer. J Clin Invest 118:1571–1577. doi: 10.1172/JCI34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell R, Emerson S. 2008. Hepatitis E: an emerging awareness of an old disease. J Hepatol 48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Aggarwal R, Kamili S, Spelbring J, Krawczynski K. 2001. Experimental studies on subclinical hepatitis E virus infection in cynomolgus macaques. J Infect Dis 184:1380–1385. doi: 10.1086/324376. [DOI] [PubMed] [Google Scholar]
- 53.Krawczynski K, Meng X-J, Rybczynska J. 2011. Pathogenetic elements of hepatitis E and animal models of HEV infection. Virus Res 161:78–83. doi: 10.1016/j.virusres.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Butterworth PJ. 1983. Alkaline phosphatase. Biochemistry of mammalian alkaline phosphatases. Cell Biochem Funct 1:66–70. doi: 10.1002/cbf.290010202. [DOI] [PubMed] [Google Scholar]
- 55.Kim YB. 1975. Developmental immunity in the piglet. Birth Defects Orig Artic Ser 11:549–557. [PubMed] [Google Scholar]
- 56.Saif LJ, Ward LA, Yuan L, Rosen BI, To TL. 1996. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch Virol Suppl 12:153–161. [DOI] [PubMed] [Google Scholar]
- 57.Wack A, Openshaw P, O'Garra A. 2011. Contribution of cytokines to pathology and protection in virus infection. Curr Opin Virol 1:184–195. doi: 10.1016/j.coviro.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 58.Boer MC, Joosten SA, Ottenhoff TH. 2015. Regulatory T cells at the interface between human host and pathogens in infectious diseases and vaccination. Front Immunol 6:217. doi: 10.3389/fimmu.2015.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ndure J, Flanagan KL. 2014. Targeting regulatory T cells to improve vaccine immunogenicity in early life. Front Microbiol 5:477. doi: 10.3389/fmicb.2014.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. 2002. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod 66:112–119. doi: 10.1095/biolreprod66.1.112. [DOI] [PubMed] [Google Scholar]
- 61.Lee K, Redel BK, Spate L, Teson J, Brown AN, Park KW, Walters E, Samuel M, Murphy CN, Prather RS. 2013. Piglets produced from cloned blastocysts cultured in vitro with GM-CSF. Mol Reprod Dev 80:145–154. doi: 10.1002/mrd.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erker JC, Desai SM, Schlauder GG, Dawson GJ, Mushahwar IK. 1999. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J Gen Virol 80(Pt 3):681–690. doi: 10.1099/0022-1317-80-3-681. [DOI] [PubMed] [Google Scholar]
- 63.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Mokhtari C, Marchadier E, Haim-Boukobza S, Jeblaoui A, Tesse S, Savary J, Roque-Afonso AM. 2013. Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection. J Clin Virol 58:36–40. doi: 10.1016/j.jcv.2013.06.038. [DOI] [PubMed] [Google Scholar]
- 65.Subramaniam S, Pineyro P, Tian D, Overend C, Yugo DM, Matzinger SR, Rogers AJ, Haac ME, Cao Q, Heffron CL, Catanzaro N, Kenney SP, Huang YW, Opriessnig T, Meng XJ. 2014. In vivo targeting of porcine reproductive and respiratory syndrome virus antigen through porcine DC-SIGN to dendritic cells elicits antigen-specific CD4T cell immunity in pigs. Vaccine 32:6768–6775. doi: 10.1016/j.vaccine.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Yuan L, Ward LA, Rosen BI, To TL, Saif LJ. 1996. Systematic and intestinal antibody-secreting cell responses and correlates of protective immunity to human rotavirus in a gnotobiotic pig model of disease. J Virol 70:3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]