CCHFV is an important human pathogen that is both endemic and emerging throughout Asia, Africa, and Europe. A common feature of acute disease is liver injury ranging from mild to fulminant hepatic failure. The processes through which CCHFV induces severe liver injury are unclear, mostly due to the limitations of existing small-animal systems. The only small-animal model in which CCHFV consistently produces severe liver damage is mice lacking IFN-I signaling. In this study, we used antibody-mediated blockade of IFN-I signaling in mice to study CCHFV liver pathogenesis in various transgenic mouse systems. We found that liver injury did not depend on cytotoxic immune cells and observed extensive activation of death receptor signaling pathways in the liver during acute disease. Furthermore, acute CCHFV infection resulted in a nearly complete loss of Kupffer cells. Our model system provides insight into both the molecular and the cellular features of CCHFV hepatic injury.
KEYWORDS: Crimean-Congo hemorrhagic fever virus, Kupffer cells, Nairoviridae, death receptor signaling, hepatic injury, pathogenesis
ABSTRACT
Crimean-Congo hemorrhagic fever virus (CCHFV) can cause severe hepatic injury in humans. However, the mechanism(s) causing this damage is poorly characterized. CCHFV produces an acute disease, including liver damage, in mice lacking type I interferon (IFN-I) signaling due to either STAT-1 gene deletion or disruption of the IFN-I receptor 1 gene. Here, we explored CCHFV-induced liver pathogenesis in mice using an antibody to disrupt IFN-I signaling. When IFN-I blockade was induced within 24 h postexposure to CCHFV, mice developed severe disease with greater than 95% mortality by 6 days postexposure. In addition, we observed increased proinflammatory cytokines, chemoattractants, and liver enzymes in these mice. Extensive liver damage was evident by 4 days postexposure and was characterized by hepatocyte necrosis and the loss of CLEC4F-positive Kupffer cells. Similar experiments in CCHFV-exposed NOD-SCID-γ (NSG), Rag2-deficient, and perforin-deficient mice also demonstrated liver injury, suggesting that cytotoxic immune cells are dispensable for hepatic damage. Some apoptotic liver cells contained viral RNA, while other apoptotic liver cells were negative, suggesting that cell death occurred by both intrinsic and extrinsic mechanisms. Protein and transcriptional analysis of livers revealed that activation of tumor necrosis factor superfamily members occurred by day 4 postexposure, implicating these molecules as factors in liver cell death. These data provide insights into CCHFV-induced hepatic injury and demonstrate the utility of antibody-mediated IFN-I blockade in the study of CCHFV pathogenesis in mice.
IMPORTANCE CCHFV is an important human pathogen that is both endemic and emerging throughout Asia, Africa, and Europe. A common feature of acute disease is liver injury ranging from mild to fulminant hepatic failure. The processes through which CCHFV induces severe liver injury are unclear, mostly due to the limitations of existing small-animal systems. The only small-animal model in which CCHFV consistently produces severe liver damage is mice lacking IFN-I signaling. In this study, we used antibody-mediated blockade of IFN-I signaling in mice to study CCHFV liver pathogenesis in various transgenic mouse systems. We found that liver injury did not depend on cytotoxic immune cells and observed extensive activation of death receptor signaling pathways in the liver during acute disease. Furthermore, acute CCHFV infection resulted in a nearly complete loss of Kupffer cells. Our model system provides insight into both the molecular and the cellular features of CCHFV hepatic injury.
INTRODUCTION
Crimean-Congo hemorrhagic fever virus (CCHFV) is an enveloped virus and a member of the genus Orthonairovirus in the Nairoviridae family (for reviews, see references 1 to 3). CCHFV infects a large number of wild and domesticated mammalian species, including giraffes, buffaloes, zebras, bovines, and ovines, in addition to some avian species, such as ostriches. However, infection in these animals is generally asymptomatic, at most producing a prolonged (>5-day) viremia (4, 5). In marked contrast, CCHFV infection in humans can lead to an acute and potentially life-threatening disease termed Crimean-Congo hemorrhagic fever (CCHF) (2, 6, 7). CCHFV is naturally spread through the bites of ixodid ticks, primarily those of the genus Hyalomma. In addition, human infections can also result from occupational or ceremonial exposure to infected animals during the slaughter of livestock, such as ostriches, cattle, and sheep (3, 8). Nosocomial infections also place hospital staff at significant risk (7, 9). The rate of mortality from CCHF ranges from 3 to 30% and is suspected to depend on multiple factors, including the viral strain, the speed of diagnosis, and access to emergency health care (2). There are currently no licensed vaccines or therapeutics to prevent or treat CCHFV infections, although ribavirin may provide some therapeutic benefit (10).
Hepatic injury is a salient feature of CCHF and a significant factor contributing to morbidity and mortality (6, 7, 11–13). Infection in humans leads to a spectrum of liver damage that can range from mild injury to fulminant hepatic failure (6, 7). The prevalence of liver injury has led to the clinical characterization of disease severity to be partially based on elevated liver enzymes (11, 12). A limited number of autopsy findings have explored the liver damage caused by CCHFV (6, 13, 14). In general, liver injury is characterized by Kupffer cell hyperplasia, hepatocellular necrosis, and the formation of councilman bodies. Within livers, CCHFV antigen can be detected in hepatocytes and nonparenchymal liver cells, including Kupffer cells and endothelial cells. Based on histopathological evidence, it has been speculated that CCHFV replication within hepatocytes directly leads to the loss of these cells, thereby resulting in organ dysfunction (13). This has been supported by in vitro data showing that CCHFV induces endoplasmic reticulum stress, which leads to apoptosis in the Huh7 hepatocyte-like cell line (15). However, CCHFV infection in humans induces robust expression of inflammatory cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6), and severe disease correlates with higher levels of these molecules (16–18). TNF-α is a member of the tumor necrosis factor (TNF) superfamily of death receptors/ligands, which also includes Fas (APO-1/CD95) and TNF-related apoptosis-inducing ligand (TRAIL) (19). TNF superfamily death ligands/receptors can be potent mediators of hepatic damage during infectious and noninfectious liver insults. The involvement of these molecules in CCHFV-mediated liver pathology is less clear. Additionally, higher levels of NK cells and cytotoxic T cells (CTLs) have been reported in fatal cases (20, 21), potentially implicating these cells as contributors to liver damage. Overall, the molecular and cellular mechanisms of CCHFV-mediated liver injury in vivo remain poorly characterized and largely unexplored outside epidemiological studies.
Severe disease models for CCHFV have been developed in mice (22–24). This work revealed that CCHFV produces acute disease only in mice lacking functional type I interferon (IFN-I) signaling either through STAT-1 deficiency (22) or through deletion of the type I interferon receptor (23, 24). These murine infection models recapitulate the CCHFV-mediated hepatic injury observed in humans with elevated liver enzymes and marked liver pathology (25). Liver pathology correlates with the presence of CCHFV antigen, which can be detected in hepatocytes, Kupffer cells, endothelial cells, and stellate cells. Consistent with human disease, infected mice also have high levels of inflammatory systemic cytokine activity, including TNF-α and IL-6 activity. Thus, mouse models can be used to provide insight into the pathogenic processes that lead to the liver injury and mortality incurred by CCHFV infection.
We and others have used antibody-mediated IFN-I blockade to study severe disease caused by IFN-I-hypersensitive viruses, including Zika virus (26), West Nile virus (27), and vesicular stomatitis virus (28) in mice. The IFN-I antibody blockade system has the advantage of allowing disruption of IFN-I in any murine system, enabling exploration of virus immunobiology and pathogenesis in various transgenic mice without the need for additional crossbreeding. Previously, we used this system to develop a severe-disease model for CCHFV (29). In that study, mice treated with antibody to block IFN-I signaling and infected with the prototypical laboratory CCHFV strain IbAr 10200 developed severe CCHFV disease with kinetics similar to those in IFN-I receptor knockout (IFNAR−/−) mice. Here, we expand upon this work and report on a refined antibody-mediated IFN-I blockade model using CCHFV strain Afg09-2990, which was isolated from a fatal human case (7, 30) and which was found to produce severe disease in IFNAR−/− mice (31). Using this model system, we investigated the molecular and cellular factors contributing to CCHFV-mediated liver pathology. Our findings provide novel insight into CCHFV-mediated liver pathogenesis. Additionally, our work presents a powerful platform for the investigation of CCHFV-mediated pathology and medical countermeasure (MCM) evaluation in mice.
RESULTS
CCHFV strain Afg09-2990 produces a lethal infection in mice following antibody-mediated IFN-I blockade.
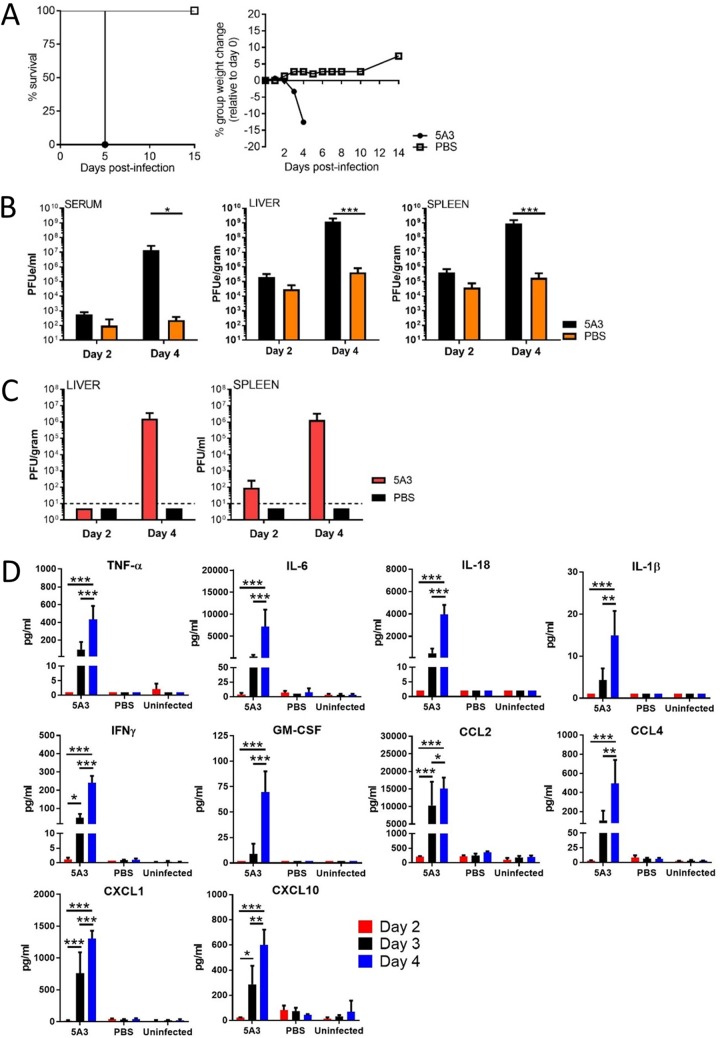
We previously used the prototypical laboratory CCHFV strain IbAr 10200, isolated from a tick, for our infection model (29). Here, we investigated whether CCHFV strain Afg09-2990, isolated from a fatal human case (7, 30), could also produce acute disease in mice when IFN-I was disrupted using the anti-IFNAR1 monoclonal antibody MAR1-5A3 (5A3). Groups of eight C57BL/6J (BL6) mice were infected with 100 PFU of CCHFV, and at 24 h after exposure, they were injected with phosphate-buffered saline (PBS) only (no IFN-I blockade) or 5A3 in PBS (IFN-I blocked). On day 3, 5A3-treated mice began to lose weight, and all mice succumbed to disease by day 5 (Fig. 1A). In contrast, PBS-treated, infected mice did not show signs of disease and did not lose weight. We examined the viral load on days 2 and 4 in a different group of mice. On day 2, low levels of viral RNA were detected by reverse transcription-quantitative PCR (RT-qPCR) in the sera, livers, and spleens of mice in both groups, with mean values in 5A3-treated mice trending higher (but not significantly) than those in the PBS group (Fig. 1B). By day 4, there were markedly increased levels of viral RNA detected in the sera, livers, and spleens of 5A3-treated mice. In contrast, viral RNA abundances in mice with intact IFN-I signaling were comparatively lower than those in blocked mice and only slightly increased from the day 2 values. The differences in the viral load on day 4 between 5A3-treated and PBS-treated mice were significant in serum, liver, and spleen (two-way analysis of variance [ANOVA], P < 0.05). Infectious virus was present in the livers of all three 5A3-treated mice on day 4, with a mean titer of 8.6 × 105 PFU/gram (Fig. 1C). Viable virus could be isolated from the spleens of 1/3 infected 5A3-treated mice on day 2 and 2/3 mice on day 4, with mean titers of 96 PFU/gram and 1.4 × 105 PFU/gram, respectively. No infectious virus was isolated from infected PBS-treated mice. These findings indicate that viral replication was markedly higher in the 5A3-treated mice.
FIG 1.
Lethal CCHFV infection in mice treated with 5A3. (A) BL6 mice (n = 8/group) were infected and at 24 h postinfection were treated with 5A3. Survival and group weights were monitored for 15 days and plotted using Prism software. (B) Mice were infected with CCHFV in the presence or absence of 5A3, and the viral RNA titers in serum, liver, and spleen were examined on days 2 and 4 by qRT-PCR (n = 3/group). Mean titers ± SD for the PFU equivalents (PFUe) were graphed. Statistical significance was determined by two-way ANOVA. *, P < 0.05; ***, P < 0.005. (C) The numbers of PFU of virus were determined in the liver and spleen on days 2 and 4 by plaque assay. Mean titers as the number of PFU per gram of tissue ± SD (n = 3/group) were graphed. The black dashed lines mark the limit of assay detection. (D) Inflammatory cytokines and chemoattractants were measured from serum (n = 3/group) on days 2, 3, and 4. Statistical significance was determined by two-way ANOVA. *, P < 0.01; **, P < 0.005; ***, P < 0.001.
Cytokine and monocyte chemokine activity was evaluated on days 2, 3, and 4 in uninfected mice and in CCHFV-infected mice with or without IFN-I disruption. TNF-α, IL-6, IL-18, IL-1β, gamma interferon (IFN-γ), and granulocyte-macrophage colony-stimulating factor (GM-CSF) activity was detected at high levels only in 5A3-treated mice starting on day 3, and this activity increased by significant levels (two-way ANOVA, P < 0.05) on day 4 (Fig. 1D). Similarly, the chemoattractants CCL2, CCL4, CXCL1, and CXCL10 were detectable only in 5A3-treated animals beginning on day 3 and increased significantly by day 4 (two-way ANOVA, P < 0.05) (Fig. 1D). These data indicate, similarly to the data from our previous study with strain IbAr 10200 (29), that strain Afg09-2990 produces a lethal infection in mice when IFN-I is transiently blocked by 5A3.
Mice infected with CCHFV in the presence of IFN-I antibody blockade develop hepatic injury.
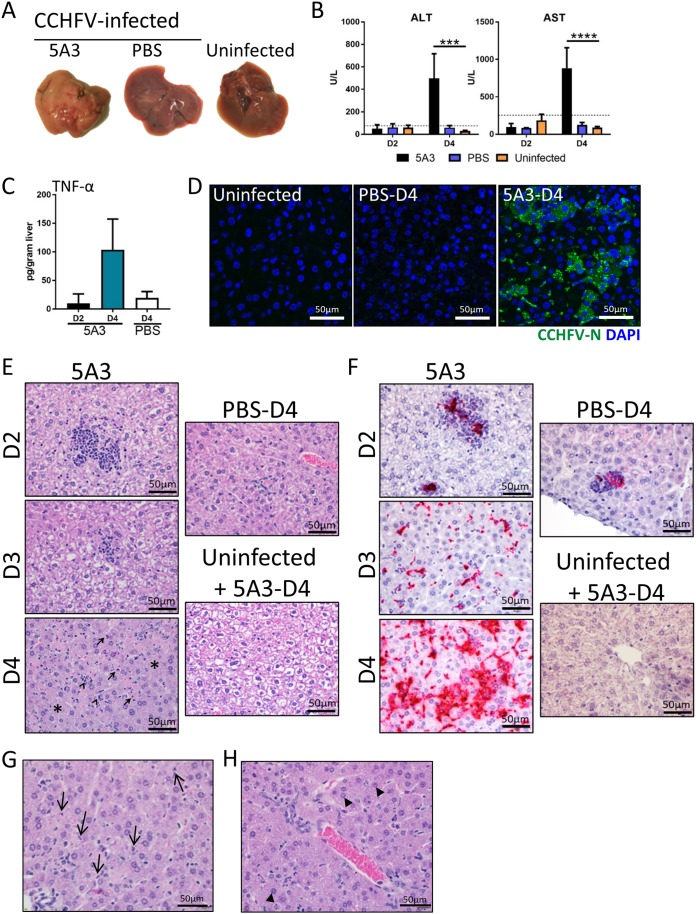
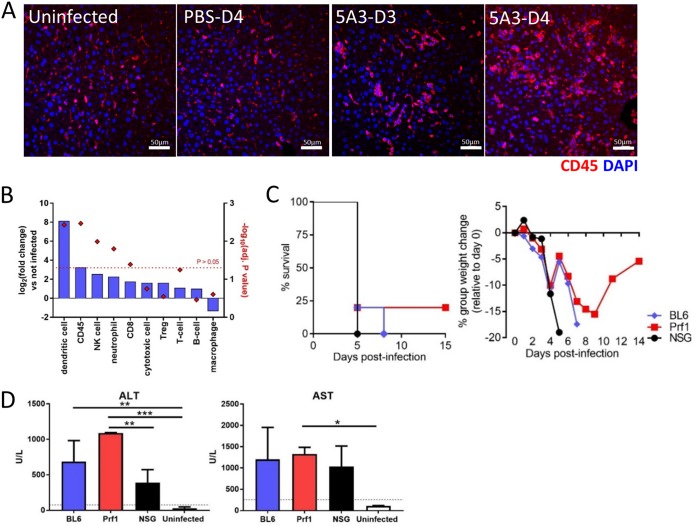
We next investigated the impact of CCHFV on the liver in 5A3-treated, infected BL6 mice compared to PBS-treated, infected mice and uninfected mice. No gross differences in liver appearance were detected on day 2, regardless of treatment. By day 4, livers from 5A3-treated, infected mice were uniformly pale in appearance in comparison to those of infected PBS-treated mice and uninfected mice, indicative of liver damage (Fig. 2A). Consistent with the gross pathology, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) liver enzymes were significantly elevated in 5A3-treated mice compared to PBS-treated, infected mice and uninfected mice (two-way ANOVA, P < 0.05) on day 4 but not on day 2 (Fig. 2B). ALT and AST values in infected PBS-treated mice were not elevated over those in uninfected mice. 5A3 treatment of uninfected mice did not increase ALT or AST levels above those in untreated and uninfected mice (data not shown). Elevated levels of TNF-α were detected by enzyme-linked immunosorbent assay (ELISA) in the livers of infected 5A3-treated mice on day 4 compared to those on day 2 and day 4 in PBS-treated, infected mice (Fig. 2C). However, these differences were not significant (one-way ANOVA, P > 0.05). Viral nucleocapsid (N) protein could be detected, but only in the livers of 5A3-treated mice, indicating direct infection of the liver (Fig. 2D).
FIG 2.
CCHFV-mediated pathogenesis in mouse liver. (A) Gross pathology of livers from CCHFV-infected mice treated or untreated (PBS) with 5A3 or uninfected mice. Livers were harvested on day 4 and photographed. (B) Liver enzymes from mouse serum (n = 3/group) taken on days 2 and 4. Uninfected mice were included as a baseline. Dotted lines mark the normal high levels for healthy mice. Statistical significance was determined by two-way ANOVA. ***, P < 0.001; ****, P < 0.0001. (C) TNF-α cytokine levels in liver homogenates in 5A3-treated (day 2 [D2] or day 4 [D4]) or PBS-treated (day 4) infected mice (n = 3/group) were determined by ELISA and graphed as the number of picograms per gram of tissue. (D) N (green) was detected in day 4 liver samples by immunofluorescence assay. Nuclei were stained with DAPI (blue). (E) Representative H&E staining of livers from mice infected with CCHFV in the presence or absence of an IFN-I blockade and harvested on days 2, 3, and 4. Uninfected, 5A3-treated mice serve as a negative control. 5A3-treated mice showed a progressive liver deterioration which was not observed in infected control mice (PBS) or uninfected mice. Arrows indicate necrotic cells, and arrowheads point to neutrophils. The asterisks mark areas of widespread degeneration. (F) Representative ISH staining showing the presence of CCHFV RNA (red) in the livers of mice taken on days 2, 3, and 4. Cells were counterstained with hematoxylin. (G) H&E-stained liver section from a CCHFV-infected mouse treated with 5A3 on day 4. Arrows point to necrotic cellular debris within sinusoids. (H) Liver section from a 5A3-treated mouse on day 4 showing oval cell hyperplasia (arrowheads).
Histopathological changes in livers from mice infected with CCHFV in the presence or absence of IFN-I signaling blockade were evaluated on days 2, 3, and 4 (3 mice per time point). Infected 5A3-treated mice developed hepatic lesions, marked by inflammation and hepatocellular necrosis, as early as day 2 (Fig. 2E). By days 3 and 4, widespread inflammation with hepatocellular degeneration and necrosis was observed in all animals (Fig. 2E). This was accompanied by Kupffer cell hypertrophy, evident in the sinusoids (Fig. 2G), and occasional periportal oval cell hyperplasia (Fig. 2H). Infected, PBS-treated mice developed similar, minimal hepatic lesions as early as day 2, but no significant progression occurred on day 3 or day 4 (Fig. 2E). Consistent with our previous findings that 5A3 treatment alone does not increase inflammation in mice (26, 32), 5A3 treatment of uninfected mice did not result in increased liver inflammation compared to that in untreated mice (Fig. 2E and data not shown). Hepatic lesions in CCHFV-infected mice correlated with the presence of CCHFV RNA (Fig. 2F). Early in the course of disease, CCHFV RNA was present only within inflammatory foci in the liver and Kupffer cells. However, by day 3 and, to a greater extent, day 4, CCHFV RNA was detected in Kupffer cells, hepatocytes, and endothelial cells. In PBS-treated, infected mice, CCHFV RNA was detected in only a small number of inflammatory foci at all time points (Fig. 2F). This corresponded with the presence of viral N protein (Fig. 2D). No in situ hybridization (ISH) signal was detected in 5A3-treated and uninfected mice. These findings demonstrate that CCHFV induces severe hepatic injury in wild-type BL6 mice when IFN-I signaling is disrupted.
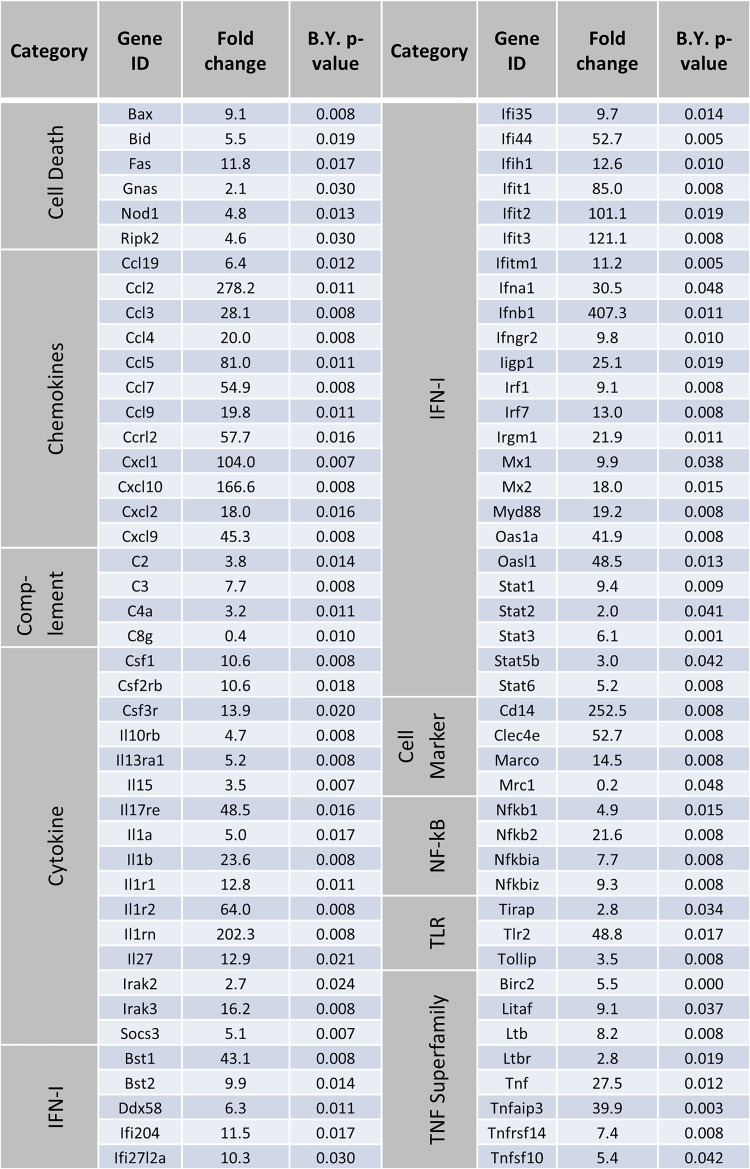
Transcriptional analysis of CCHFV-infected livers.
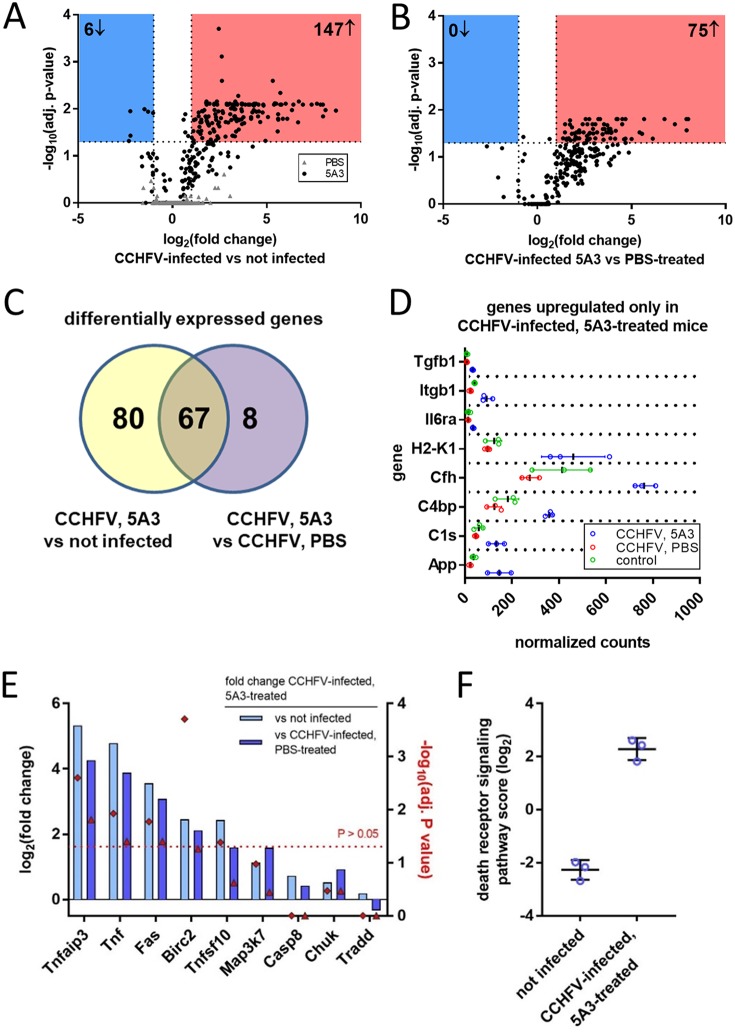
Using the NanoString nCounter platform and total RNA isolated from mouse liver homogenates, we performed targeted gene expression profiling of 687 genes related to host immunology and inflammatory processes. We compared gene expression in the livers of CCHFV-infected mice treated with 5A3 (IFN-I blockade) to that in the livers of uninfected mice at day 4 postexposure. Between CCHFV-infected 5A3-treated mice and uninfected mice, expression of 147 genes significantly increased and that of 6 genes significantly decreased (Fig. 3A and Table 1). This included extensive upregulation of several chemokine genes, including Ccl2, Cxcl1, Cxcl10, and Cxcl9, consistent with their presence in serum (Fig. 1D). Large increases in the expression of genes associated with myeloid-lineage cells, including Cd14, Marco, Clec5a, and Clec4e, were also observed. The six genes that significantly decreased included Mrc1, which is involved in anti-inflammatory macrophage responses, in addition to Dpp4 and Mme, which are involved in cellular metabolism. No significant changes were observed between PBS-treated, infected mice (in which IFN-I was active) and uninfected mice (Fig. 3A). We also evaluated gene expression changes between infected mice with and without IFN-I disruption and found 75 genes significantly increased in expression, but no transcripts were significantly decreased (Fig. 3B). We plotted the shared and unique genes that significantly increased between 5A3-treated, infected mice and uninfected mice or between 5A3-treated, infected mice and PBS-treated, infected animals (Fig. 3C). Eighty genes that were increased between 5A3-treated, infected mice and uninfected animals were not significantly increased in PBS-treated, infected mice (Fig. 3C). These genes were generally involved in pathogen sensing, such as Ifna1, Ifnb1, Myd88, Dxd58 (RIG-I), and Ifih1 (MDA-5). Eight genes were uniquely increased between infected mice with and without IFN-I activity (Fig. 3C and D).
FIG 3.
Transcriptional activation in CCHFV-infected livers. (A) Transcriptional activation in liver homogenates from mice (n = 3/group) infected with CCHFV with (black circles) and without (gray triangles) IFN-I blockade compared to uninfected animals. Changes in global gene expression levels are indicated by log2 fold changes (x axis) versus the adjusted (adj.) log10 P value (y axis). The dashed line indicates statistical significance. (B) Transcription activation in infected mice with and without IFN-I blockade. Data were graphed as described in the legend to panel A. (C) Venn diagram showing the number of overlapping and unique gene changes in panel A versus panel B. (D) Genes increased only in CCHFV-infected, 5A3-treated mice versus infected, PBS-treated mice. (E) Log2 fold change (left y axis) in genes associated with the death receptor pathways between 5A3-treated mice and untreated or uninfected animals. The right y axis indicates the significance for each group, indicated by a red diamond. The red dashed line denotes the limits of significance (P > 0.05). (F) Death receptor activation score for uninfected mice versus mice infected in the presence of IFN-I blockade. Death receptor activation is based on the presence of transcripts in panel E.
TABLE 1.
Gene expression changes in 5A3-treated, infected mice versus uninfected micea
B.Y., Benjamini-Yekutieli.
Sixty-seven transcripts were significantly increased between acutely infected mice and PBS-treated, infected mice or between acutely infected mice and uninfected mice (Fig. 3C). This included significant increases in expression of members of the TNF superfamily of death receptors and ligands, including Tnf, Fas, and, to a lesser extent, Tfnsf10 (TRAIL) (Fig. 3E and Table 1). Activation of Tnf and Fas in the livers was confirmed using ISH (data not shown). Several other death receptor pathway transcripts were significantly increased in infected mice with IFN-I blockade (Fig. 3E). A qualitative aggregate score of death receptor activation further indicated extensive activation of this pathway in acutely infected mice versus uninfected animals (Fig. 3F). Together, these data indicate that, in addition to multiple genes involved in pathogen recognition and inflammation, death receptor pathways are activated in the livers of acutely infected, IFN-I-blockaded mice at 4 days postexposure.
Investigating the role of cytotoxic immune cells in CCHFV-mediated pathogenesis.
CCHFV infection in the presence of IFN-I blockade resulted in an increased number of CD45+ cells within the liver, as indicated by IHC (Fig. 4A). No increases in CD45+ cells were apparent in uninfected or PBS-treated, infected mice. These findings were verified using specific gene markers for immune cells, which indicated a statistically significant increase in the CD45+ cell population (Fig. 4B). Based on immune cell transcript analysis, we observed significant increases of NK cells, CD8+ T cells (CTLs), neutrophils, and dendritic cells in the livers of infected mice with disrupted IFN-I signaling. A slight, but not significant, decrease in macrophages was detected in infected 5A3-treated mice compared to uninfected mice.
FIG 4.
Role of cytotoxic immune cells in CCHFV pathogenesis. (A) Liver sections from days 3 and 4 (5A3-treated mice) or day 4 (PBS-treated, infected mice and uninfected mice) were evaluated for CD45 (red) by immunofluorescence assay. Nuclei were stained with DAPI (blue). (B) NanoString detected log2 fold changes (left y axis) in the indicated liver immune cell populations in 5A3-treated, infected mice versus uninfected animals. The right y axis indicates the significance for each group, indicated by a red diamond. The red dotted line denotes the limits of significance (P > 0.05). Treg, regulatory T cells. (C) BL6, NSG, and Prf1−/− mice (n = 8/group) were infected with CCHFV and injected with 5A3 (i.p.) at 24 h postinfection. Survival and weight were monitored. (D) Liver enzyme levels in mouse serum (n = 3/group) taken on day 4 were determined as described in the legend to Fig. 2D. The black dotted lines mark the upper limit of normal expression in healthy mice. Statistical significance was determined by one-way ANOVA. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Because cytotoxic immune cells can contribute to pathogenesis by damaging infected hepatocytes and other liver cells, we evaluated the contribution of cytotoxic immune cells to CCHFV-mediated pathogenesis using perforin 1 (Prf1)-deficient and NOD-SCID-γ (NSG) mice. Prf1−/− mice lack potent cytotoxic immune activity (33); NSG mice lack mature T and B cells, and NK cell activity is severely reduced (34). Groups of eight BL6, NSG, and Prf1−/− mice were infected with CCHFV, and at 24 h after infection, mice were treated with 5A3. All infected mice lost weight starting on day 2 (Fig. 4C). By day 5, all NSG mice and most BL6 and Prf1−/− animals met euthanasia criteria. The single surviving BL6 and Prf1−/− animals continued to lose weight, and the BL6 mouse met euthanasia criteria by day 8. There were no significant survival differences between BL6 and NSG mice (log-rank test, P = 0.36). A single Prf1−/− mouse survived, but this survival was not statistically significantly different from that for BL6 mice (log-rank test, P = 0.54). The weight of this survivor began to increase after day 9 and by the end of the study (day 14) returned to a level approximately 5% less than the group starting weight. Liver enzyme levels in all infected groups were compared to those in uninfected BL6 control mice on day 4 (Fig. 4D). Baseline liver enzyme levels in NSG mice were similar to those in BL6 mice (35). The levels of ALT in all infected mice were significantly increased over those in control animals (one-way ANOVA, P < 0.05). While AST values were elevated in infected mice versus control mice, only the values for Prf1−/− mice were significantly different (one-way ANOVA, P < 0.05). AST and ALT levels in NSG and Prf1−/− mice were not significantly different from those in infected BL6 mice (one-way ANOVA, P < 0.05). Thus, in the absence of IFN-I signaling, mice deficient in cytotoxic immune cell activity succumbed to CCHFV-mediated disease with kinetics similar to those in wild-type mice with concomitant increases in liver enzymes.
Rag2-dependent lymphocytes are not essential for lethal infection but are required for virus control.
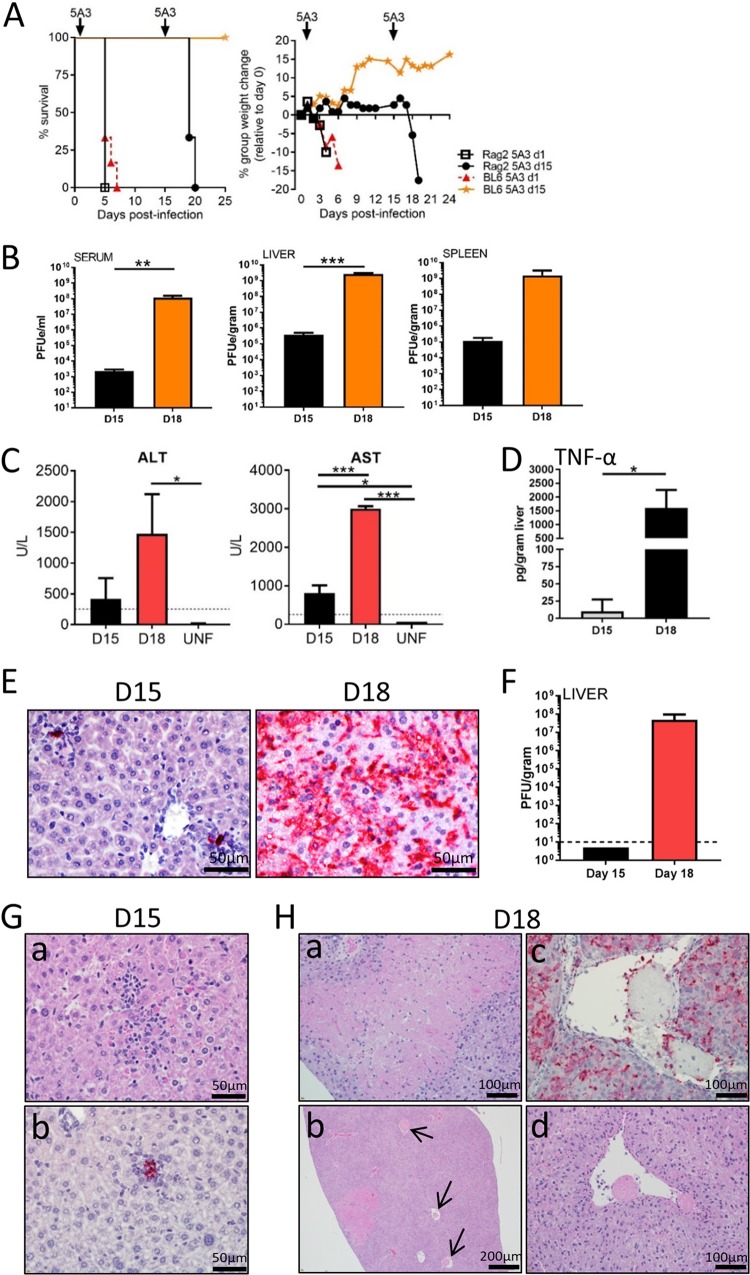
To further determine whether CTLs played a key role in CCHFV liver injury, the ability of CCHFV to induce acute disease and liver pathogenesis was evaluated in Rag2−/− mice, which are devoid of mature T cells, NKT cells, and B cells (36). Groups of six BL6 or Rag2−/− mice were infected with CCFHV (Fig. 5A). One group each of BL6 and Rag2−/− mice was treated with 5A3 at 24 h postchallenge. Infected 5A3-treated Rag2−/− mice and BL6 mice lost weight starting on day 2, with no differences in the kinetics or magnitudes being detected between the groups. The mean times to death (MTDs) between BL6 and Rag2−/− mice were not statistically different (log-rank test, P > 0.05). CCHFV-infected Rag2−/− mice and BL6 mice with intact IFN-I signaling did not lose weight or show signs of disease, even after 2 weeks. Therefore, we investigated if virus was cleared from Rag2−/− mice independently of the adaptive immune system. To this end, antibody-mediated IFN-I blockade was induced at 15 days postinfection in both Rag2−/− and BL6 mice (Fig. 5A). Subsequent to 5A3 treatment, Rag2−/− mice began to lose weight on day 17 (3 days after IFN-I disruption), and all mice reached euthanasia criteria by day 20. In sharp contrast, BL6 mice showed no signs of disease when IFN-I was blocked on day 15.
FIG 5.
CCHFV infection of Rag2-deficient mice. (A) BL6 or Rag2−/− mice (n = 6 to 8/group) were infected with CCHFV on day 0. On day +1 (d1) or day +15 (d15), one group each of BL6 and Rag2−/− mice was injected i.p. with 5A3. Survival and group weights were monitored for 25 days postinfection. Results for BL6 mice treated on day +15 (orange stars) were from a separate study. (B) qRT-PCR to detect virus was performed using serum, liver, and spleen from CCHFV-infected mice (n = 3/group) taken on day 15 (D15) and day 18 (D18). Values are graphed as the mean number of PFUe per milliliter of serum or per gram of tissue ± SD. Three mice were used per sample. Statistical significance was determined by one-way ANOVA. **, P < 0.005; ***, P < 0.0005. (C) ALT and AST values in CCHFV-infected Rag2−/− mice (n = 3/group) were determined on day 15 and day 18 and graphed as mean values ± SDs. The black dotted lines denote the upper limit of normal for mice for each enzyme. Two uninfected (UNF) Rag2−/− mice were used as a control for AST and ALT levels. Levels are shown as means ± SDs. Statistical significance was determined by one-way ANOVA. *, P < 0.05; ***, P < 0.0005. (D) TNF-α protein levels were determined as described in the legend to Fig. 2 from day 15 and day 18 liver homogenates. Statistical significance was determined by one-way ANOVA. *, P < 0.05. (E) Representative ISH staining on Rag2−/− mouse livers on day 15 and day 18 (after 5A3 treatment). (F) The numbers of PFU of virus on days 15 and 18 were determined as described in the legend to Fig. 1. The dashed line marks the limit of detection. (G) H&E (a) and ISH (b) staining of samples from Rag2−/− mice taken on day 15 postinfection. CCHFV RNA-positive areas correspond to areas of inflammation. (H) H&E (a, b, d) and ISH (c) staining of samples from Rag2−/− mice taken on day 18 (4 days after 5A3 treatment). Note areas of widespread necrosis (a) and fibrin deposits (b, c, and d). Arrows point to fibrin occlusions in vesicles (b).
In a second experiment, Rag2−/− mice were infected and on day 15 three animals were euthanized, while a second group of three mice was treated with 5A3 on day 15 and euthanized on day 18. Mice from both groups were analyzed for viral titers, cytokine expression, liver enzymes, and liver pathology. A low level of viremia was detectable on day 15, and this increased significantly by 5 logs following IFN-I blockade (one-way ANOVA, P < 0.05) (Fig. 5B). Viral RNA was detected in liver and spleen on day 15, and RNA abundances increased following 5A3 treatment, but only the increase in the liver was significant (one-way ANOVA, P < 0.05) (Fig. 5B). On both day 15 and day 18, ALT and AST values were elevated compared to those in uninfected Rag2−/− mice (Fig. 5C). The day 15 AST values and the day 18 AST and ALT values in infected mice were significantly higher than those in uninfected Rag2−/− mice (one-way ANOVA, P < 0.05). TNF-α protein could be detected in liver homogenates for a single Rag2−/− mouse on day 15 but was present in all three mice on day 18, with a mean value of 1,606 pg/gram of liver (Fig. 5D).
Livers from Rag2−/− mice were examined for histopathological changes and the presence of virus by ISH on day 15 and day 18. CCHFV RNA was detected in only a few foci on day 15, but RNA was detected throughout the liver following 5A3 treatment on day 18 (Fig. 5E). Viable virus was detected only in day 18 liver samples, with a mean titer of 5.0 × 107 PFU/gram (Fig. 5F). On day 15, all livers exhibited multiple small foci of hepatocellular necrosis with inflammation that consisted primarily of histiocytes and corresponded with the presence of viral RNA (Fig. 5G). Kupffer cell hypertrophy and oval cell hyperplasia were also present on all livers. Together, these findings indicate that a chronic or chronic active hepatitis occurred in mice lacking Rag2 on day 15. Livers harvested on day 18 exhibited a more severe pathology (Fig. 5H). This included extensive hepatic necrosis (both lytic and coagulative) with infiltration by macrophages and neutrophils and widespread necrotic cellular (karyorrhectic) debris. Animals also had fibrin thrombi occluding vessels, indicative of disseminated intravascular coagulation (DIC) in areas of coagulative necrosis (infarcts). Kupffer cell hypertrophy was also present in the animals on day 18. Thus, in Rag2−/− mice with intact IFN-I signaling, some liver inflammation and chronic hepatitis occur. However, administration of 5A3 leads to marked deterioration of the liver structure, similar to the findings for wild-type mice. These experiments indicate that Rag2-dependent functions (i.e., adaptive immunity) are required for virus control. Additionally, these data demonstrate that CTLs and NKT cells are dispensable for CCHFV-mediated liver pathology.
CCHFV infection results in the death of infected and uninfected cells in the liver.
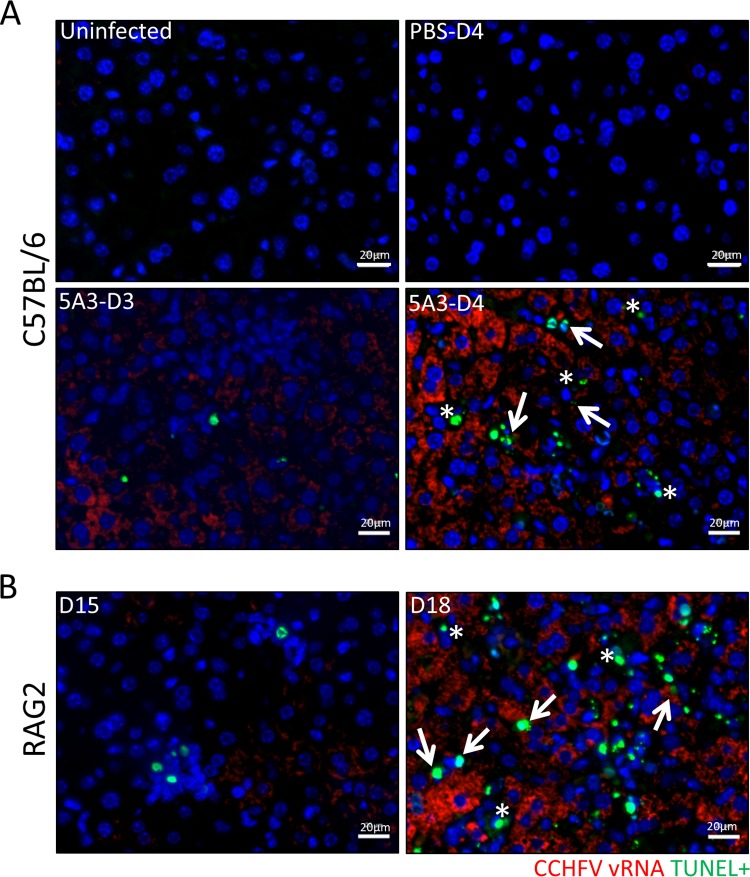
We next explored if CCHFV was present in sites of liver cell death. A fluorescent in situ hybridization (FISH) staining protocol designed to identify CCHFV RNA-expressing cells that was compatible with terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was developed. Some TUNEL-positive cells were present in infected BL6 mice on day 3, but these numbers increased on day 4 (Fig. 6A). In contrast, no TUNEL-positive cells were detected in uninfected mice or infected mice with intact IFN-I signaling. In Rag2−/− mice, some TUNEL staining was observed on day 15, prior to 5A3 treatment, but this staining increased subsequent to IFN-I blockade (Fig. 6B). For both BL6 and Rag2−/− mice, TUNEL staining could be detected in viral RNA-positive cells. However, TUNEL-positive cells lacking viral RNA were also detected. These findings suggest that CCHFV infection induces cell death within infected livers, but cell death does not always associate with the presence of viral RNA.
FIG 6.
Cell death in infected and uninfected liver cells. TUNEL and CCHFV RNA FISH costaining of CCHFV-infected and uninfected livers from BL6 (A) or Rag2−/− (B) mice is shown. TUNEL (green) and CCHFV RNA FISH (red) staining was performed as indicted in the Materials and Methods. Arrows point to CCHFV RNA-positive/TUNEL-positive cells, and asterisks denote TUNEL-positive cells lacking viral RNA. Nuclei were stained with DAPI (blue).
Loss of Kupffer cells in CCFHV-infected livers.
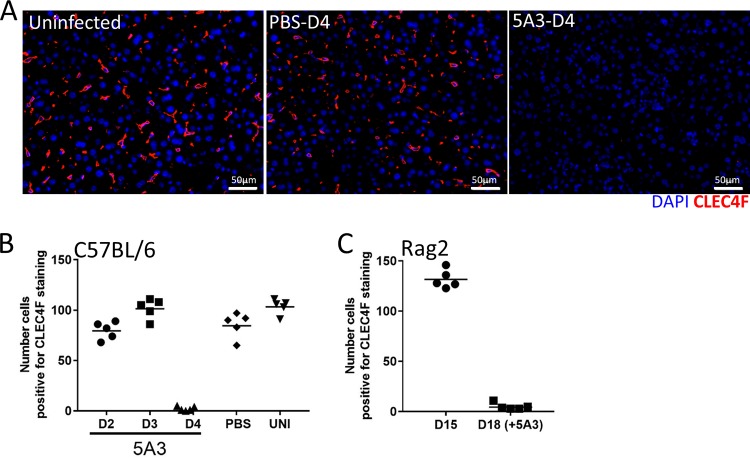
Histological findings indicated that cell populations resembling Kupffer cells within the sinusoids were necrotic and reduced in numbers in infected mice (Fig. 2G). Therefore, we directly investigated if Kupffer cell populations were decreasing by staining liver sections for CLEC4F, a marker highly specific for this cell type (37, 38). Kupffer cells were detected in uninfected livers and in the livers of infected mice with intact IFN-I signaling on day 4. However, mice infected with CCHFV in the presence of IFN-I blockade had a sharp reduction in the CLEC4F marker on day 4 (Fig. 7A). The CLEC4F population appeared to be lost abruptly after day 3 in 5A3-treated mice, as the numbers of CLEC4F cells were similar to those in control mice on days 2 and 3 (Fig. 7B). Similar results were observed in liver sections of Rag2-deficient mice on day 18 (Fig. 7C). We confirmed the loss of Kupffer cells by also staining the livers for F4/80, which is expressed at high levels on these populations (38). F4/80 staining was similar to that of CLEC4F (data not shown). These findings suggest that Kupffer cells are abruptly lost during CCHFV infection, but only following acute disease.
FIG 7.
Loss of CLEC4F-positive cell populations in infected liver. (A) Liver sections from infected (5A3-treated or controls) or uninfected mice were stained for the presence of Kupffer cells using a CLEC4F antibody (red). Nuclei were stained with DAPI (blue). (B and C) Quantification of CLEC4F-positive cells in liver sections from infected BL6 mice (B) and Rag2−/− mice (C). CLEC4F-positive cells were counted in 10 random fields for each group, and mean values were plotted as a dot plot. UNI, uninfected.
DISCUSSION
CCHFV-mediated liver pathogenesis.
A predominant model of CCHFV-induced liver injury suggests that liver cell death results directly from CCHFV replication and is termed intrinsic cell death. This model is based mostly on histopathological examination of a limited number of infected human livers (6, 13, 14) but is supported by in vitro data showing that CCHFV replication alone leads to apoptosis in the hepatocyte-like cell line Huh7 (15). Our findings also support this model, as we observed the presence of TUNEL-positive staining in cells also positive for viral RNA. One caveat to a conclusion that virus causes intrinsic cell death in vivo is the possibility that virally infected cells are being destroyed by cytotoxic immune cells, particularly CD8+ T cells targeting cognate antigen, or by NK cells. The ability of cytotoxic immune cells to induce hepatic injury is well described. For example, cytotoxic immune cell targeting of hepatitis B virus- or lymphocytic choriomeningitis virus-infected cells leads to hepatocyte loss and contributes to hepatitis (39–41). Indeed, larger increases in cytotoxic immune cell populations, including CD8+ T cells and NK cells, occur in fatal human cases of CCHFV (20, 21). We found transcripts for both NK and CD8+ T cells present in CCHFV-infected livers, and this timing coincided with the liver cell death. However, our results indicate that cytotoxic immune activity does not play a significant role in liver injury in CCHFV-infected mice. CCHFV-mediated liver pathogenesis in Rag2-deficient mice was indistinguishable from that in BL6 mice, excluding a major role of cytotoxic T cells (and B cells and NKT cells) in this process. Similarly, infection in NSG mice, which also lack mature T cells and B cells and have significantly impaired NK activity, resulted in severe disease with hepatic injury. Prf1-deficient mice also developed liver injury. Moreover, mice deficient in cytotoxic immune activity succumbed to disease with kinetics similar to those in wild-type mice. The dispensability of NK cells and T cells for CCHFV-induced hepatic injury supports a conclusion that CCHFV replication can produce intrinsic liver cell death in vivo.
However, we also identified TUNEL-positive liver cells that appeared to lack viral RNA. This finding supports a model whereby cell death during CCHFV infection can also be driven by extrinsic factors that do not require viral replication (but that are independent of cytotoxic immune activity). While we cannot rule out the possibility that viral RNA was not detected in these dying cells due to degradation, we found that extrinsic factors capable of causing extrinsic cell death were active in infected livers. Our study showed that the genes for both Fas (APO-1/CD95) and TNF-α are upregulated within CCHFV-infected livers, with 12- and 28-fold increases in gene expression, respectively. This activation correlated with high levels of TNF-α protein in both the liver and serum. Additionally, gene transcripts associated with death receptor pathways were significantly elevated in animals with acute disease. Members of the TNF superfamily of death receptors/ligands, particularly TNF-α and the Fas ligand (FasL), are major contributors to extrinsic liver cell death, and aberrant expression of these factors following infectious and noninfectious insults can lead to hepatic damage (19). Some data suggest that death receptor pathways contribute to cell loss during CCHFV infection (15, 42). One study found that the neutralization of TNF-α partially blocked apoptosis in CCHFV-infected SW-13 cells (42). In CCHFV-infected humans, higher serum levels of TNF-α and other inflammatory cytokines correlated with fatal cases (16–18). Despite these findings, the influence of the death receptor molecules on CCHFV pathogenesis and, specifically, the loss of liver parenchymal and nonparenchymal cells have not been investigated in vitro or in vivo. Given its reoccurring presence in severe CCHFV disease in animals and humans and the known negative impact that aberrant levels of TNF-α can have on liver tissue, it is tempting to speculate that it is a major contributor to CCHFV-mediated liver pathology. However, it will be important to rule out the involvement of other factors, such as infiltrating neutrophils, which we also detected in infected livers. Neutrophils can interact directly with distressed hepatocytes and facilitate their death via respirator burst and granule-effector molecules, such as neutrophil elastase (43). Studies are under way to determine whether an extrinsic (death receptor-mediated) or intrinsic mechanism(s) (viral replication) is the main factor contributing to liver cell loss.
CCHFV interaction with Kupffer cells.
Macrophages are important primary targets of CCHFV (13, 44, 45). Kupffer cells are the largest population of tissue-resident macrophages (46) and are cellular targets of CCHFV in humans (13) and in mice (25 and this study). Our data are in agreement with conclusions based on human histopathology analysis that suggest that Kupffer cells are the first liver cells to be infected, with infection of nearby hepatocytes and other nonparenchymal cells occurring secondarily. In fact, in Rag2−/− mice, Kupffer cells appear to support CCHFV infection even after 15 days and in the presence of intact IFN-I signaling. Despite supporting CCHFV replication early during infection, our findings reveal that the Kupffer cell population is destroyed late during disease. This was indicated by the lack of CLEC4F and F4/80 staining in livers on day 4 in both wild-type mice and Rag2-deficient animals. This loss occurred abruptly, as normal levels of cells were present on day 3 and absent on day 4. It is not evident from human data if these cells are similarly lost because such analysis requires specifically staining for the Kupffer cell population. Whether the loss of Kupffer cells is beneficial or detrimental to the host remains to be determined. During hepatitis B virus infection, Kupffer cells play important roles in removing infected and apoptotic hepatocytes, preventing the subsequent release of danger response molecules, such as HMGB1, thus reducing inflammation (47). In response to extensive inflammation, Kupffer cells can also function in an anti-inflammatory role (i.e., M2 phenotype) (48) and reduce the inflammatory responses of infiltrating monocyte-derived macrophages, which can produce high levels of inflammatory cytokines (49). Indeed, we suspect that monocyte-derived macrophages (MDM) are an important source of inflammatory cytokine production in CCHFV-infected livers. We base this on the fact that while Kupffer cells can be an important source of inflammatory cytokines (46), the levels of these molecules (transcripts and protein) increase at a time when Kupffer cells (and other liver-resident cells) are lost. While the transcript data suggested a slight decrease in macrophages in acutely infected mice, this reflected a loss of Kupffer cells, which express CD163 (50), the genetic marker used to detected macrophages. Other genetic signatures of MDMs, including MACRO, CD14, CLEC5A, and CLEC4E transcripts, increased in infected livers on day 4. These markers are generally, but not exclusively, associated with macrophages (51–53). Our findings warrant a deeper investigation aimed at determining if Kupffer cells are critical for initial CCHFV liver infiltration and how the loss of these cells may impact liver pathogenesis by disrupting important mechanisms of homeostatic control.
Antibody-mediated IFN-I blockade as a tool to study CCHFV pathogenesis and immunobiology.
Antibody-mediated IFN-I blockade has been shown to be extremely useful in the development of severe-disease mouse models for many different IFN-I-sensitive viruses (26–28). In addition to their usefulness in severe-disease models, antibody blockade of IFN-I signaling was shown to increase viral clearance and control of a persistent lymphocytic choriomeningitis virus infection by altering the immune response (54, 55). As demonstrated in the current study, the use of antibody-mediated IFN-I blockade is a powerful and convenient platform for studying CCHFV pathogenesis in various mouse models. IFN-I blockade produced similar disease in multiple mouse strains with an MTD of approximately 5 days after signaling was prevented. Our findings confirm that CCFHV strain Afg09-2990 produces a lethal infection in mice when IFN-I is blocked either by genetic means (31) or by antibody (this study) and that the disease induced by strain Afg09-2990 is indistinguishable from that induced by strain IbAr 10200. CCHFV strains are genetically diverse with seven distinct clades (56), and some antigenic heterogeneity exists between strains (57, 58). Accordingly, the ability to produce severe disease in a small-animal model with diverse strains will be important in evaluating the ability of MCMs to protect against heterologous CCHFV isolates.
The choice of murine models in the evaluation of CCHFV vaccine protective efficacy is important. Kortekaas et al. recently reported that despite the development of anti-CCHFV glycoprotein antibody responses in STAT-1 knockout mice, the protein vaccine failed to protect, likely due to the hypersensitivity of these mice to the virus (59). We initially used the antibody IFN-I blockade system to evaluate the protective efficacy of an M segment-based DNA vaccine (29) because it allowed vaccine immunogenicity to be evaluated in the presence of an active IFN-I system with signaling blocked only during CCHFV challenge. In that study, IFN-I was blocked prior to CCHFV challenge. Here we refined this model, demonstrating that an acute infection ensues when IFN-I is blocked in mice with functional adaptive immunity up to 24 h postexposure. Such dynamic control of IFN-I signaling will be particularly useful for vaccine studies, as delaying IFN-I blockade postinfection permits evaluation of CCHFV-targeting vaccine immune responses in an immune-intact animal prior to challenge and limits the impact of IFN-I disruption on anamnestic immune responses following challenge.
We also identified a unique CCHFV infection model using Rag2-deficient mice. These findings reiterate the importance of IFN-I in protecting even highly immunocompromised mice from acute disease. They also demonstrate that adaptive immunity is required to fully control and clear infection. While infection in Rag2-deficient mice did not produce outward signs of disease, such as cachexia, the animals developed hepatitis. How long CCHFV persists in the Rag2−/− animals or if the mice eventually succumb to disease is currently being investigated. Regardless, this model will help identify what adaptive immune populations are needed to control CCHFV. Furthermore, it may allow the identification of cells, such as Kupffer cells, which can support CCHFV replication in the presence of active IFN-I signaling. This is an important area of study, because in nature humans are the only known host in which CCHFV causes severe disease, but the virus can produce an extended viremia in various animals (5). The mechanism(s) and cell populations involved in allowing the extended viral replication without disease are unknown.
The role of IFN-I signaling in acute CCHFV disease in humans.
While rodent models for CCHFV disease demonstrate that IFN-I has a predominant role in preventing severe disease, the extent to which deficiencies in IFN-I signaling mediate disease severity in humans is less apparent. Some human data suggest that Toll-like receptor 8/9 (TLR8/9) or TLR3 polymorphisms are associated with acute CCHFV disease, potentially linking the dynamics of IFN-I signaling to human disease severity (60, 61). The authors speculated that certain TLR polymorphisms may limit IFN-I activation in response to CCHFV. This conclusion is supported by findings unrelated to CCHFV indicating that TLR3 polymorphisms can functionally impair TLR3 receptor activation, thus limiting IFN-I signaling (62). Indeed, polymorphisms in pathogen-sensing networks, including many of the TLRs, can influence host susceptibility to viral infection in general (63). More studies are needed to resolve whether IFN-I signaling deficiencies similar to those in mice are key factors influencing human susceptibility to acute CCHFV disease.
Summary.
Here, we developed a refined model of CCHF in mice using antibody-mediated blockade of IFN-I and used it to explore CCHFV-induced liver pathogenesis. Our salient findings include determining that cytotoxic immune cells, while present during infection, are dispensable for acute disease. Furthermore, we found that not all dying liver cells are infected with CCHFV, suggesting that extrinsic factors, such as death receptor signaling pathways, which are active in infected mice, may contribute to liver cell loss. The antibody-mediated IFN-I blockade model will be a powerful platform for future CCHFV pathogenesis studies.
MATERIALS AND METHODS
Viruses and cells.
Huh7 and SW-13 cells were propagated in Dulbecco's modified Eagle medium with Earle's salts (DMEM) (Corning) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco), 10 U/ml penicillin and 10 μg/ml streptomycin (Gibco), 1% 100 mM sodium pyruvate (Sigma), 1% 200 mM l-glutamine (HyClone), and 1% 1 M HEPES (Gibco). CCHFV strain Afg09-2990 (30), courtesy of the Bernard Nocht Institute for Tropical Medicine, was used for all experiments. This virus was passaged three times in Vero cells, followed by two passages in Huh7 cells. The virus was collected from clarified cell culture supernatants and stored at −80°C. All CCHFV work was performed in biosafety level 4/animal biosafety level 4 containment at the United States Army Medical Research Institute of Infectious Diseases.
Mice.
C57BL/6J (BL6), Rag2-deficient [BL6(Cg)-Rag2tm1.1Cgn/J], NOD-SCID-γ (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ), and perforin-deficient (C57BL/6-Prf1tm1Sdz/J) mice (6 to 8 weeks old) were obtained from The Jackson Laboratory. Mice were challenged with 100 PFU of CCHFV by intraperitoneal (i.p.) injection of virus diluted in a total volume of 0.2 ml PBS. Mice were injected by the intraperitoneal (i.p.) route with 2.5 mg of anti-IFNAR1 (MAR1-5A3 [5A3]; Leinco Technologies, Inc.) diluted in 0.4 ml of PBS at 24 h postinfection.
Ethics statement.
All animal research was conducted under a United States Army Medical Research Institute of Infectious Diseases Institutional Animal Care and Use Committee (IACUC)-approved protocol in compliance with the Animal Welfare Act, PHS Policy, and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to the principles stated in the Guide for the Care and Use of Laboratory Animals of the National Research Council (64).
RT-qPCR.
Mouse serum samples were inactivated using a 3:1 ratio of the TRIzol LS reagent (Thermo Fisher Scientific). Liver and spleen tissues were homogenized in 750 μl complete DMEM using a TissueLyser II homogenizer (Qiagen), and supernatants were inactivated using a 3:1 ratio of the TRIzol LS reagent and placed at −80°C. Total nucleic acid was purified using an EZ1 virus minikit (v.2.0; Qiagen) on an EZ1 Advanced XL robot (Qiagen) according to the manufacturer's recommendations. The viral load was determined in triplicate using a real-time RT-PCR assay specific to CCHFV as previously described (65). Challenge stock virus was used to generate the standard curve, allowing calculation of the number of PFU equivalents (PFUe) in the tested samples.
Plaque assay.
Liver and spleen homogenates were diluted 1:10 in DMEM, and subsequently, 100 μl of sample was adsorbed to confluent SW-13 cell monolayers in 6-well plates for 1 h in a 37°C and 5% CO2 incubator and rocked at ∼15-min intervals. Following adsorption, a 2-ml solid overlay (Earle's basal minimal essential medium [EBME], 0.5% [wt/vol] SeaKem ME agarose [Lonza], 5% [vol/vol] heat-inactivated FBS [Gibco], 4% 200 mM l-glutamine [Gibco]) was added to each well. The plates were incubated for 3 days in a 37°C and 5% CO2 incubator and stained with 2 ml of a solid overlay mixture containing 5% 33-mg/ml neutral red solution (Gibco). Cells were incubated for an additional 24 h in a 37°C and 5% CO2 incubator before plaque counting. Virus titers were calculated and are given as the number of PFU per gram of tissue.
TNF-α ELISA.
TNF-α levels in liver homogenates (25 μl) were detected using a commercially available ELISA (R&D Systems, Minneapolis, MN) following the manufacturer's protocol.
Liver enzymes.
Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in serum from CCHFV-infected animals were measured using a Piccolo Xpress analyzer (Abaxis) and a General Chemistry 13 panel (Abaxis) following the manufacturer's protocol.
Histology.
A necropsy was performed on the liver. Tissues were immersed in 10% (vol/vol) neutral buffered formalin for 30 to 70 days. Tissues were then trimmed and processed according to standard protocols (66). Histology sections were cut at 5 to 6 μm on a rotary microtome, mounted onto glass slides, and stained with hematoxylin and eosin (H&E). Examination of the tissue was performed by a board-certified veterinary pathologist.
Cytokine and chemokine analysis.
Serum cytokine and chemokine analysis was performed using a magnetic bead-based Plex mouse panel (Thermo Fisher Scientific) targeting the indicated molecules. Twenty-five microliters of serum per mouse per time point was used. Plates were analyzed on a Magpix system (Millipore Sigma) and quantitated against standard curves using MILLIPLEX analyst software.
NanoString gene expression analysis.
Total RNA samples were analyzed using the nCounter mouse immunology (v.1) and nCounter mouse inflammation (v.2) panels. Probe set-target RNA hybridization reactions were performed according to the manufacturer's protocol (NanoString). For each hybridization reaction, 100 ng total RNA or any quantity that was present in a 5-μl aliquot of purified RNA, if the amount was less than 100 ng, was used. Purified probe set-target RNA complexes from each reaction were processed and immobilized on nCounter cartridges using an nCounter Max preparation station, and transcripts were quantified on a digital analyzer (Gen, v.2). Data from each NanoString panel were first processed independently using nSolver (v.4.0) software (NanoString) as follows: following quality control checks on the individual RCC files, raw counts across samples were normalized to the geometric mean counts for spiked synthetic DNA-positive controls present in the hybridization reactions to mitigate platform-associated sources of variation. No background subtraction or thresholding was performed at this stage. Candidate reference genes were selected using the nCounter advanced analysis (nCAA) module (v.2.0.115), which implements the geNorm algorithm for downselection (67). Starting with a set of candidate reference genes (14 or 6 genes for the Immunology [v.2] and Inflammation [v.1] panels, respectively), the algorithm identified the top five most stable genes for each panel (for the Immunology [v.2] panel, Sdha, Hprt, Gapdh, Polr2a, and Tbp; for the Inflammation [v.1] panel, Cltc, Gusb, Hprt, Pgk1, and Gapdh). For each sample, normalization was performed by dividing the counts for each gene by the geometric mean for the five selected reference genes. These two normalized data sets were then combined in nSolver as a multi-RLF merge experiment and then input into the nCAA module for differential expression, gene set, and biological pathway analysis. The threshold for differential expression was a log2 fold change of >1 and a Benjamini-Yekutieli-adjusted P value of <0.05. Immune cell detection in the liver was determined using NanoStrain software.
TUNEL staining.
Formalin-fixed paraffin-embedded (FFPE) liver tissue sections were deparaffinized in Xyless II buffer (LabChem) 2 times for 10 min each time and rehydrated using an ethanol gradient (100%, 95%, and 70%). Apoptotic cells were detected using a DeadEnd fluorometric TUNEL assay (Promega) according to the manufacturer's protocol.
ISH.
CCHFV was detected in infected liver samples by in situ hybridization (ISH) probes targeting the M segment (GenBank accession number HM452306.1) of CCHFV strain Afg09-2990 (nucleotide positions 631 to 2702; Advanced Cell Diagnostics; Newark, CA). Formalin-fixed paraffin-embedded (FFPE) liver sections were deparaffinized and peroxidase blocked. Sections were then incubated with ISH probes at 40°C in a hybridization oven for 2 h. After rinsing, the ISH signal was amplified using the kit-provided preamplifier and amplifier conjugated to alkaline phosphatase and incubated with a Fast Red substrate solution for 10 min at room temperature. Sections were then stained with hematoxylin, air dried, and mounted.
Fluorescent in situ hybridization.
A probe set consisting of 48 individual probes was designed to detect CCHFV S segment viral RNA. Individual probes were tagged with a 3′ T (C6-biotin) motif and generated by LGC Biosearch, Inc. Tissue sections were processed as described above and were then treated with 20 μg/ml of proteinase K for 10 min at room temperature and washed with PBS. The samples were then washed in water and boiled in antigen-unmasking solution (Vector Laboratories) for 10 min. Endogenous biotin was blocked using a biotin-blocking kit according to the manufacturer's instructions (Thermo Fisher Scientific). Samples were incubated in formamide wash buffer (2× saline-sodium citrate [SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 10% formamide in nuclease-free H2O) for 5 min. CCHFV-specific probes diluted (125 nM) in formamide hybridization buffer (10% dextran sulfate [wt/vol], 2× SSC, 10% formamide in nuclease-free H2O) were added, and the slides were incubated at 37°C overnight. On the following day, samples were washed in formamide wash buffer 2 times for 30 min each time at 37°C. Biotin-labeled probes were detected using a tyramide labeling kit according to the manufacturer's protocol (Thermo Fisher Scientific).
Immunostaining.
FFPE tissue sections were deparaffinized using xylene and a series of ethanol washes. After 0.1% Sudan black B (Sigma) or TrueBlack (Biotium) treatment to eliminate the autofluorescence background, the sections were heated in citrate buffer (pH 6.0) for 15 min to reverse formaldehyde cross-linking. Rinsed sections were blocked with PBS containing 5% normal goat serum overnight at 4°C. For CCHFV detection, samples were incubated with a polyclonal rabbit anti-CCHFV N protein antibody (1:500; IBT) or monoclonal rat anti-mouse CD45 antibody (1:200; BD Pharmingen) for 2 h at room temperature. For CLEC4F detection, samples were incubated with a polyclonal rabbit anti-CLEC4F antibody (1:50; Thermo Fisher Scientific) overnight at 4°C. Following primary antibody incubation, sections that had been washed with PBS were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody or Alexa Fluor 561-conjugated goat anti-mouse IgG antibody (1:200) for 1 h at room temperature and mounted onto slides using Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories).
Microscopy.
Images were acquired using a Zeiss LSM 700 system, a Zeiss LSM 880 confocal system, or an Olympus BX46 microscope. Images were processed using Zeiss Zen confocal software, CellSens software, or ImageJ software.
Statistical analysis.
Comparisons of RT-qPCR virus titers, weight loss, cytokine expression, and blood chemistry parameters were performed using one-way or two-way ANOVA with the Bonferroni correction. Student's t tests were used to compare NanoString transcriptomic data. Survival analyses among mouse study cohorts were made using the log-rank test. In all tests, significance was determined as a P value of <0.05. All analyses were performed using GraphPad Prism (v.6) software.
ACKNOWLEDGMENTS
We thank Rebecca Brocato for reviewing the manuscript, Matthew Voorhees for assistance with cytokine analysis, and the Veterinary Medicine Division for technical assistance.
This project was funded by a grant awarded to J.W.G. from the Military Infectious Disease Research Program.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
We report that we have no conflicts of interest.
REFERENCES
- 1.Whitehouse CA. 2004. Crimean-Congo hemorrhagic fever. Antiviral Res 64:145–160. [DOI] [PubMed] [Google Scholar]
- 2.Ergonul O. 2006. Crimean-Congo haemorrhagic fever. Lancet Infect Dis 6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. 2013. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd AJ, Leman PA, Swanepoel R. 1989. Viremia and antibody response of small African and laboratory animals to Crimean-Congo hemorrhagic fever virus infection. Am J Trop Med Hyg 40:541–547. doi: 10.4269/ajtmh.1989.40.541. [DOI] [PubMed] [Google Scholar]
- 5.Spengler JR, Estrada-Pena A, Garrison AR, Schmaljohn C, Spiropoulou CF, Bergeron E, Bente DA. 2016. A chronological review of experimental infection studies of the role of wild animals and livestock in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus. Antiviral Res 135:31–47. doi: 10.1016/j.antiviral.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negredo A, de la Calle-Prieto F, Palencia-Herrejon E, Mora-Rillo M, Astray-Mochales J, Sanchez-Seco MP, Bermejo Lopez E, Menarguez J, Fernandez-Cruz A, Sanchez-Artola B, Keough-Delgado E, Ramirez de Arellano E, Lasala F, Milla J, Fraile JL, Ordobas Gavin M, Martinez de la Gandara A, Lopez Perez L, Diaz-Diaz D, Lopez-Garcia MA, Delgado-Jimenez P, Martin-Quiros A, Trigo E, Figueira JC, Manzanares J, Rodriguez-Baena E, Garcia-Comas L, Rodriguez-Fraga O, Garcia-Arenzana N, Fernandez-Diaz MV, Cornejo VM, Emmerich P, Schmidt-Chanasit J, Arribas JR, Crimean Congo Hemorrhagic Fever@Madrid Working Group. 2017. Autochthonous Crimean-Congo hemorrhagic fever in Spain. N Engl J Med 377:154–161. doi: 10.1056/NEJMoa1615162. [DOI] [PubMed] [Google Scholar]
- 7.Conger NG, Paolino KM, Osborn EC, Rusnak JM, Gunther S, Pool J, Rollin PE, Allan PF, Schmidt-Chanasit J, Rieger T, Kortepeter MG. 2015. Health care response to CCHF in US soldier and nosocomial transmission to health care providers, Germany, 2009. Emerg Infect Dis 21:23–31. doi: 10.3201/eid2101.141413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanepoel R, Leman PA, Burt FJ, Jardine J, Verwoerd DJ, Capua I, Bruckner GK, Burger WP. 1998. Experimental infection of ostriches with Crimean-Congo haemorrhagic fever virus. Epidemiol Infect 121:427–432. doi: 10.1017/S0950268898001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burney MI, Ghafoor A, Saleen M, Webb PA, Casals J. 1980. Nosocomial outbreak of viral hemorrhagic fever caused by Crimean hemorrhagic fever-Congo virus in Pakistan, January 1976. Am J Trop Med Hyg 29:941–947. doi: 10.4269/ajtmh.1980.29.941. [DOI] [PubMed] [Google Scholar]
- 10.Ergonul O, Celikbas A, Dokuzoguz B, Eren S, Baykam N, Esener H. 2004. Characteristics of patients with Crimean-Congo hemorrhagic fever in a recent outbreak in Turkey and impact of oral ribavirin therapy. Clin Infect Dis 39:284–287. doi: 10.1086/422000. [DOI] [PubMed] [Google Scholar]
- 11.Ergonul O, Celikbas A, Baykam N, Eren S, Dokuzoguz B. 2006. Analysis of risk-factors among patients with Crimean-Congo haemorrhagic fever virus infection: severity criteria revisited. Clin Microbiol Infect 12:551–554. doi: 10.1111/j.1469-0691.2006.01445.x. [DOI] [PubMed] [Google Scholar]
- 12.Swanepoel R, Gill DE, Shepherd AJ, Leman PA, Mynhardt JH, Harvey S. 1989. The clinical pathology of Crimean-Congo hemorrhagic fever. Rev Infect Dis 11(Suppl 4):S794–S800. doi: 10.1093/clinids/11.Supplement_4.S794. [DOI] [PubMed] [Google Scholar]
- 13.Burt FJ, Swanepoel R, Shieh WJ, Smith JF, Leman PA, Greer PW, Coffield LM, Rollin PE, Ksiazek TG, Peters CJ, Zaki SR. 1997. Immunohistochemical and in situ localization of Crimean-Congo hemorrhagic fever (CCHF) virus in human tissues and implications for CCHF pathogenesis. Arch Pathol Lab Med 121:839–846. [PubMed] [Google Scholar]
- 14.Baskerville A, Satti A, Murphy FA, Simpson DI. 1981. Congo-Crimean haemorrhagic fever in Dubai: histopathological studies. J Clin Pathol 34:871–874. doi: 10.1136/jcp.34.8.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues R, Paranhos-Baccala G, Vernet G, Peyrefitte CN. 2012. Crimean-Congo hemorrhagic fever virus-infected hepatocytes induce ER-stress and apoptosis crosstalk. PLoS One 7:e29712. doi: 10.1371/journal.pone.0029712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papa A, Bino S, Velo E, Harxhi A, Kota M, Antoniadis A. 2006. Cytokine levels in Crimean-Congo hemorrhagic fever. J Clin Virol 36:272–276. doi: 10.1016/j.jcv.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Ergonul O, Tuncbilek S, Baykam N, Celikbas A, Dokuzoguz B. 2006. Evaluation of serum levels of interleukin (IL)-6, IL-10, and tumor necrosis factor-alpha in patients with Crimean-Congo hemorrhagic fever. J Infect Dis 193:941–944. doi: 10.1086/500836. [DOI] [PubMed] [Google Scholar]
- 18.Saksida A, Duh D, Wraber B, Dedushaj I, Ahmeti S, Avsic-Zupanc T. 2010. Interacting roles of immune mechanisms and viral load in the pathogenesis of Crimean-Congo hemorrhagic fever. Clin Vaccine Immunol 17:1086–1093. doi: 10.1128/CVI.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin XM, Ding WX. 2003. Death receptor activation-induced hepatocyte apoptosis and liver injury. Curr Mol Med 3:491–508. doi: 10.2174/1566524033479555. [DOI] [PubMed] [Google Scholar]
- 20.Akinci E, Yilmaz M, Bodur H, Onguru P, Bayazit FN, Erbay A, Ozet G. 2009. Analysis of lymphocyte subgroups in Crimean-Congo hemorrhagic fever. Int J Infect Dis 13:560–563. doi: 10.1016/j.ijid.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Yilmaz M, Aydin K, Akdogan E, Sucu N, Sonmez M, Omay SB, Koksal I. 2008. Peripheral blood natural killer cells in Crimean-Congo hemorrhagic fever. J Clin Virol 42:415–417. doi: 10.1016/j.jcv.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Bente DA, Alimonti JB, Shieh WJ, Camus G, Stroher U, Zaki S, Jones SM. 2010. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. J Virol 84:11089–11100. doi: 10.1128/JVI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zivcec M, Metcalfe MG, Albarino CG, Guerrero LW, Pegan SD, Spiropoulou CF, Bergeron E. 2015. Assessment of inhibitors of pathogenic Crimean-Congo hemorrhagic fever virus strains using virus-like particles. PLoS Negl Trop Dis 9:e0004259. doi: 10.1371/journal.pntd.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bereczky S, Lindegren G, Karlberg H, Akerstrom S, Klingstrom J, Mirazimi A. 2010. Crimean-Congo hemorrhagic fever virus infection is lethal for adult type I interferon receptor-knockout mice. J Gen Virol 91:1473–1477. doi: 10.1099/vir.0.019034-0. [DOI] [PubMed] [Google Scholar]
- 25.Zivcec M, Safronetz D, Scott D, Robertson S, Ebihara H, Feldmann H. 2013. Lethal Crimean-Congo hemorrhagic fever virus infection in interferon alpha/beta receptor knockout mice is associated with high viral loads, proinflammatory responses, and coagulopathy. J Infect Dis 207:1909–1921. doi: 10.1093/infdis/jit061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith DR, Hollidge B, Daye S, Zeng X, Blancett C, Kuszpit K, Bocan T, Koehler JW, Coyne S, Minogue T, Kenny T, Chi X, Yim S, Miller L, Schmaljohn C, Bavari S, Golden JW. 2017. Neuropathogenesis of Zika virus in a highly susceptible immunocompetent mouse model after antibody blockade of type I interferon. PLoS Negl Trop Dis 11:e0005296. doi: 10.1371/journal.pntd.0005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto AK, Daffis S, Brien JD, Gainey MD, Yokoyama WM, Sheehan KC, Murphy KM, Schreiber RD, Diamond MS. 2011. A temporal role of type I interferon signaling in CD8+ T cell maturation during acute West Nile virus infection. PLoS Pathog 7:e1002407. doi: 10.1371/journal.ppat.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheehan KC, Lai KS, Dunn GP, Bruce AT, Diamond MS, Heutel JD, Dungo-Arthur C, Carrero JA, White JM, Hertzog PJ, Schreiber RD. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res 26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 29.Garrison AR, Shoemaker CJ, Golden JW, Fitzpatrick CJ, Suschak JJ, Richards MJ, Badger CV, Six CM, Martin JD, Hannaman D, Zivcec M, Bergeron E, Koehler JW, Schmaljohn CS. 2017. A DNA vaccine for Crimean-Congo hemorrhagic fever protects against disease and death in two lethal mouse models. PLoS Negl Trop Dis 11:e0005908. doi: 10.1371/journal.pntd.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olschlager S, Gabriel M, Schmidt-Chanasit J, Meyer M, Osborn E, Conger NG, Allan PF, Gunther S. 2011. Complete sequence and phylogenetic characterisation of Crimean-Congo hemorrhagic fever virus from Afghanistan. J Clin Virol 50:90–92. doi: 10.1016/j.jcv.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Oestereich L, Rieger T, Neumann M, Bernreuther C, Lehmann M, Krasemann S, Wurr S, Emmerich P, de Lamballerie X, Olschlager S, Gunther S. 2014. Evaluation of antiviral efficacy of ribavirin, arbidol, and T-705 (favipiravir) in a mouse model for Crimean-Congo hemorrhagic fever. PLoS Negl Trop Dis 8:e2804. doi: 10.1371/journal.pntd.0002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuszpit K, Hollidge BS, Zeng X, Stafford RG, Daye S, Zhang X, Basuli F, Golden JW, Swenson RE, Smith DR, Bocan TM. 2018. [18F]DPA-714 PET imaging reveals global neuroinflammation in Zika virus-infected mice. Mol Imaging Biol 20:275–283. doi: 10.1007/s11307-017-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, Podack ER, Zinkernagel RM, Hengartner H. 1994. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 34.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, Greiner DL, Handgretinger R. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 35.Hilger N, Glaser J, Muller C, Halbich C, Muller A, Schwertassek U, Lehmann J, Ruschpler P, Lange F, Boldt A, Stahl L, Sack U, Oelkrug C, Emmrich F, Fricke S. 2016. Attenuation of graft-versus-host-disease in NOD scid IL-2Rgamma(−/−) (NSG) mice by ex vivo modulation of human CD4(+) T cells. Cytometry A 89:803–815. doi: 10.1002/cyto.a.22930. [DOI] [PubMed] [Google Scholar]
- 36.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68:855–867. doi: 10.1016/0092-8674(92)90029-C. [DOI] [PubMed] [Google Scholar]
- 37.Haltiwanger RS, Lehrman MA, Eckhardt AE, Hill RL. 1986. The distribution and localization of the fucose-binding lectin in rat tissues and the identification of a high affinity form of the mannose/N-acetylglucosamine-binding lectin in rat liver. J Biol Chem 261:7433–7439. [PubMed] [Google Scholar]
- 38.Yang CY, Chen JB, Tsai TF, Tsai YC, Tsai CY, Liang PH, Hsu TL, Wu CY, Netea MG, Wong CH, Hsieh SL. 2013. CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in alpha-galactosylceramide presentation in liver. PLoS One 8:e65070. doi: 10.1371/journal.pone.0065070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Q, Zhu YY, Chen J, Ye YB, Li JY, Liu YR, Hu ML, Zheng YC, Jiang JJ. 2015. Activated natural killer cells accelerate liver damage in patients with chronic hepatitis B virus infection. Clin Exp Immunol 180:499–508. doi: 10.1111/cei.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gossmann J, Lohler J, Utermohlen O, Lehmann-Grube F. 1995. Murine hepatitis caused by lymphocytic choriomeningitis virus. II. Cells involved in pathogenesis. Lab Invest 72:559–570. [PubMed] [Google Scholar]
- 41.Iannacone M, Sitia G, Isogawa M, Marchese P, Castro MG, Lowenstein PR, Chisari FV, Ruggeri ZM, Guidotti LG. 2005. Platelets mediate cytotoxic T lymphocyte-induced liver damage. Nat Med 11:1167–1169. doi: 10.1038/nm1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlberg H, Tan YJ, Mirazimi A. 2015. Crimean-Congo haemorrhagic fever replication interplays with regulation mechanisms of apoptosis. J Gen Virol 96:538–546. doi: 10.1099/jgv.0.000011. [DOI] [PubMed] [Google Scholar]
- 43.Ramaiah SK, Jaeschke H. 2007. Role of neutrophils in the pathogenesis of acute inflammatory liver injury. Toxicol Pathol 35:757–766. doi: 10.1080/01926230701584163. [DOI] [PubMed] [Google Scholar]
- 44.Zivcec M, Scholte FE, Spiropoulou CF, Spengler JR, Bergeron E. 2016. Molecular insights into Crimean-Congo hemorrhagic fever virus. Viruses 8:106. doi: 10.3390/v8040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peyrefitte CN, Perret M, Garcia S, Rodrigues R, Bagnaud A, Lacote S, Crance JM, Vernet G, Garin D, Bouloy M, Paranhos-Baccala G. 2010. Differential activation profiles of Crimean-Congo hemorrhagic fever virus- and Dugbe virus-infected antigen-presenting cells. J Gen Virol 91:189–198. doi: 10.1099/vir.0.015701-0. [DOI] [PubMed] [Google Scholar]
- 46.Bilzer M, Roggel F, Gerbes AL. 2006. Role of Kupffer cells in host defense and liver disease. Liver Int 26:1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 47.Sitia G, Iannacone M, Aiolfi R, Isogawa M, van Rooijen N, Scozzesi C, Bianchi ME, von Andrian UH, Chisari FV, Guidotti LG. 2011. Kupffer cells hasten resolution of liver immunopathology in mouse models of viral hepatitis. PLoS Pathog 7:e1002061. doi: 10.1371/journal.ppat.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon LJ, Barnes M, Tang H, Pritchard MT, Nagy LE. 2013. Kupffer cells in the liver. Comp Physiol 3:785–797. doi: 10.1002/cphy.c120026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arango Duque G, Descoteaux A. 2014. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5:491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiraoka A, Horiike N, Akbar SM, Michitaka K, Matsuyama T, Onji M. 2005. Expression of CD163 in the liver of patients with viral hepatitis. Pathol Res Pract 201:379–384. doi: 10.1016/j.prp.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Bowdish DM, Sakamoto K, Kim MJ, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG. 2009. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog 5:e1000474. doi: 10.1371/journal.ppat.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakker AB, Baker E, Sutherland GR, Phillips JH, Lanier LL. 1999. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc Natl Acad Sci U S A 96:9792–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. 2009. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med 206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks DG. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han N, Rayner S. 2011. Epidemiology and mutational analysis of global strains of Crimean-Congo haemorrhagic fever virus. Virol Sin 26:229–244. doi: 10.1007/s12250-011-3211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zivcec M, Guerrero LIW, Albarino CG, Bergeron E, Nichol ST, Spiropoulou CF. 2017. Identification of broadly neutralizing monoclonal antibodies against Crimean-Congo hemorrhagic fever virus. Antiviral Res 146:112–120. doi: 10.1016/j.antiviral.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmed AA, McFalls JM, Hoffmann C, Filone CM, Stewart SM, Paragas J, Khodjaev S, Shermukhamedova D, Schmaljohn CS, Doms RW, Bertolotti-Ciarlet A. 2005. Presence of broadly reactive and group-specific neutralizing epitopes on newly described isolates of Crimean-Congo hemorrhagic fever virus. J Gen Virol 86:3327–3336. doi: 10.1099/vir.0.81175-0. [DOI] [PubMed] [Google Scholar]
- 59.Kortekaas J, Vloet RP, McAuley AJ, Shen X, Bosch BJ, de Vries L, Moormann RJ, Bente DA. 2015. Crimean-Congo hemorrhagic fever virus subunit vaccines induce high levels of neutralizing antibodies but no protection in STAT1 knockout mice. Vector Borne Zoonotic Dis 15:759–764. doi: 10.1089/vbz.2015.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engin A, Arslan S, Kizildag S, Ozturk H, Elaldi N, Dokmetas I, Bakir M. 2010. Toll-like receptor 8 and 9 polymorphisms in Crimean-Congo hemorrhagic fever. Microbes Infect 12:1071–1078. doi: 10.1016/j.micinf.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 61.Engin A, Arslan S, Ozbilum N, Bakir M. 2016. Is there any relationship between Toll-like receptor 3 c.1377C/T and -7C/A polymorphisms and susceptibility to Crimean Congo hemorrhagic fever? J Med Virol 88:1690–1696. doi: 10.1002/jmv.24519. [DOI] [PubMed] [Google Scholar]
- 62.Pandey S, Mittal B, Srivastava M, Singh S, Srivastava K, Lal P, Mittal RD. 2011. Evaluation of Toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in cervical cancer susceptibility. Mol Biol Rep 38:4715–4721. doi: 10.1007/s11033-010-0607-z. [DOI] [PubMed] [Google Scholar]
- 63.Medvedev AE. 2013. Toll-like receptor polymorphisms, inflammatory and infectious diseases, allergies, and cancer. J Interferon Cytokine Res 33:467–484. doi: 10.1089/jir.2012.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Council NR. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 65.Koehler JW, Delp KL, Hall AT, Olschner SP, Kearney BJ, Garrison AR, Altamura LA, Rossi CA, Minogue TD. 2018. Sequence optimized real-time reverse transcription polymerase chain reaction assay for detection of Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg 98:211–215. doi: 10.4269/ajtmh.17-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prophet EB, Mills B, Arrington JB, Sobin LH. 1992. Laboratory methods for histotechnology. Armed Forces Institute of Pathology, Washington, DC. [Google Scholar]
- 67.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]