Viruses in the family Reoviridae are characterized by the ability to endogenously synthesize nascent RNA within the virus. However, the mechanisms for assembling their RNA genomes with transcriptional enzymes into a multilayered virion and for priming such a virion for transcription are poorly understood. By cryo-EM and novel asymmetric reconstruction, we determined the atomic structure of the transcription complex inside aquareoviruses (ARV) that are primed for infection. The transcription complex is anchored by the N-terminal segments of enclosing capsid proteins and contains an NTPase and a polymerase. The NTPase has a newly discovered domain that translocates the 5′ end of plus-sense RNA in segmented dsRNA genomes from the NTPase to polymerase VP2 when the virus changes from the inactive (quiescent) to the primed state. Conformation changes in capsid proteins and transcriptional complexes suggest a mechanism for relaying information from the outside to the inside of the virus during priming.
KEYWORDS: NTPases, RNA polymerases, aquareovirus, RNA structure, RNA virus, reovirus, transcriptional regulation, RNA, endogenous transcription, priming, cryo-EM
ABSTRACT
Reoviruses carry out genomic RNA transcription within intact viruses to synthesize plus-sense RNA strands, which are capped prior to their release as mRNA. The in situ structures of the transcriptional enzyme complex (TEC) containing the RNA-dependent RNA polymerase (RdRp) and NTPase are known for the single-layered reovirus cytoplasmic polyhedrosis virus (CPV), but not for multilayered reoviruses, such as aquareoviruses (ARV), which possess a primed stage that CPV lacks. Consequently, how the RNA genome and TEC respond to priming in reoviruses is unknown. Here, we determined the near-atomic-resolution asymmetric structure of ARV in the primed state by cryo-electron microscopy (cryo-EM), revealing the in situ structures of 11 TECs inside each capsid and their interactions with the 11 surrounding double-stranded RNA (dsRNA) genome segments and with the 120 enclosing capsid shell protein (CSP) VP3 subunits. The RdRp VP2 and the NTPase VP4 associate with each other and with capsid vertices; both bind RNA in multiple locations, including a novel C-terminal domain of VP4. Structural comparison between the primed and quiescent states showed translocation of the dsRNA end from the NTPase to the RdRp during priming. The RNA template channel was open in both states, suggesting that channel blocking is not a regulating mechanism between these states in ARV. Instead, the NTPase C-terminal domain appears to regulate RNA translocation between the quiescent and primed states. Taking the data together, dsRNA viruses appear to have adapted divergent mechanisms to regulate genome transcription while retaining similar mechanisms to coassemble their genome segments, TEC, and capsid proteins into infectious virions.
IMPORTANCE Viruses in the family Reoviridae are characterized by the ability to endogenously synthesize nascent RNA within the virus. However, the mechanisms for assembling their RNA genomes with transcriptional enzymes into a multilayered virion and for priming such a virion for transcription are poorly understood. By cryo-EM and novel asymmetric reconstruction, we determined the atomic structure of the transcription complex inside aquareoviruses (ARV) that are primed for infection. The transcription complex is anchored by the N-terminal segments of enclosing capsid proteins and contains an NTPase and a polymerase. The NTPase has a newly discovered domain that translocates the 5′ end of plus-sense RNA in segmented dsRNA genomes from the NTPase to polymerase VP2 when the virus changes from the inactive (quiescent) to the primed state. Conformation changes in capsid proteins and transcriptional complexes suggest a mechanism for relaying information from the outside to the inside of the virus during priming.
INTRODUCTION
Viruses, divided into 7 classes by the Baltimore classification system, have various genome replication strategies. RNA viruses (groups III, IV, and V) do not rely on host polymerases and instead carry their own RNA-dependent RNA polymerases (RdRp) for genome transcription and replication. However, as RNA regulation is alien to most eukaryotic cells, exposed RNA viral genomes are vulnerable to host antiviral defense mechanisms, such as RIG-I and MDA-5 in humans (1). While each virus family has evolved different strategies to avoid host antivirals, reoviruses (belonging to the family Reoviridae) possess the remarkable ability to transcribe their own genetic material inside sealed capsids with minimal host involvement, an ability known as endogenous transcription (2). By incorporating enzymatic functions vital for transcription inside themselves, reoviruses are transcriptionally self-sufficient. This unique characteristic allows reoviruses to successfully “hide” their genomes from host antivirals, allowing the viruses to infect a wide variety of animal hosts, but it also forces these relatively isolated nanomachines to find very different triggers to convert from the inactive to the infectious state.
Most reoviruses, e.g., aquareovirus (ARV) (3) and bluetongue virus (BTV) (4), conceal their genetic material beneath two or three layers of capsid. The medically significant rotavirus, which causes 215,000 deaths each year (5), is also multilayered. Cytoplasmic polyhedrosis virus (CPV), however, is a unique single-shelled member of the family (6–8). As the simplest reovirus, CPV has been extensively studied. From it, a basic turreted reovirus infection process has been elucidated, as follows. Upon interacting with a host cell, a quiescent virion infects it via endocytosis. Host cell factors, such as S-adenosyl methionine (SAM), bind the turret proteins, ultimately remodeling and expanding the virus to make its internal environment more conducive to genome transcription (9). Transcriptionally active virions use viral RdRp, possibly aided by the viral NTPase, to transcribe new plus-sense RNA strands, which are capped by the turret proteins prior to expulsion into the host cytosol. The capped transcript is translated by host ribosomes to synthesize viral proteins, which are assembled into virions inside cytosolic vesicles (known as inclusion bodies or viral factories). Ultimately, whole progeny virions are released, ready to infect new host cells.
As a turreted reovirus, ARV can be assumed to possess a capping mechanism similar to that of CPV, but not necessarily a similar overall life cycle. The purified CPV virion is highly infectious (10), but extra treatment is required to achieve high infectivity in ARV virions. This is because CPV lacks an external capsid layer, which in ARV consists of the penetration protein VP5 and the protection protein VP7. ARV VP5 is covered by VP7; removal of the protein allows ARV to transition from a transcriptionally inactive (quiescent) form to a maximally infectious form known as the infectious subvirion particle (ISVP) (11), which uses its newly exposed VP5 penetration proteins to escape the endocytic pathway and invade its host (12). ARV ISVP, which is primed for yet does not actively engage in genetic transcription, possesses significant surface level structural differences from quiescent and transcribing CPV (13) and even from transcribing mammalian orthoreovirus (ORV), with which it shares significant sequence homology (14). The fact that CPV lacks a primed state makes it an inadequate tool for studying its multishelled cousins, which may, like ARV, require more complicated structural changes or even protein removal to prepare the virus for transcription.
While most ARV capsid proteins have been resolved to near-atomic resolution by icosahedral reconstruction (12, 15), the structure and location of RdRp and NTPase (together known as the transcribing enzyme complex [TEC]) remain unknown, precluding a full description of the transcription mechanism for multishelled reoviruses. Here, we used cryo-electron microscopy (cryo-EM) and a novel classification protocol based on our recent asymmetric-reconstruction method (13) to obtain asymmetric reconstructions of the ARV grass carp reovirus (GCRV) before and after priming, revealing the in situ structures of 11 TECs inside each capsid and their interactions with the 11 surrounding double-stranded RNA (dsRNA) genome segments and the 120 enclosing capsid shell protein (CSP) VP3 subunits. Our de novo atomic model of the NTPase VP4 contains an additional C-terminal domain that both holds the dsRNA end in the quiescent state and translocates it to the RdRp VP2 in the primed state. Our results point to a highly divergent mechanism of genome transcription regulation and suggest a conserved coassembly model among members of the Reoviridae.
RESULTS
Eleven TECs resolved in the asymmetric reconstruction of the primed ARV.
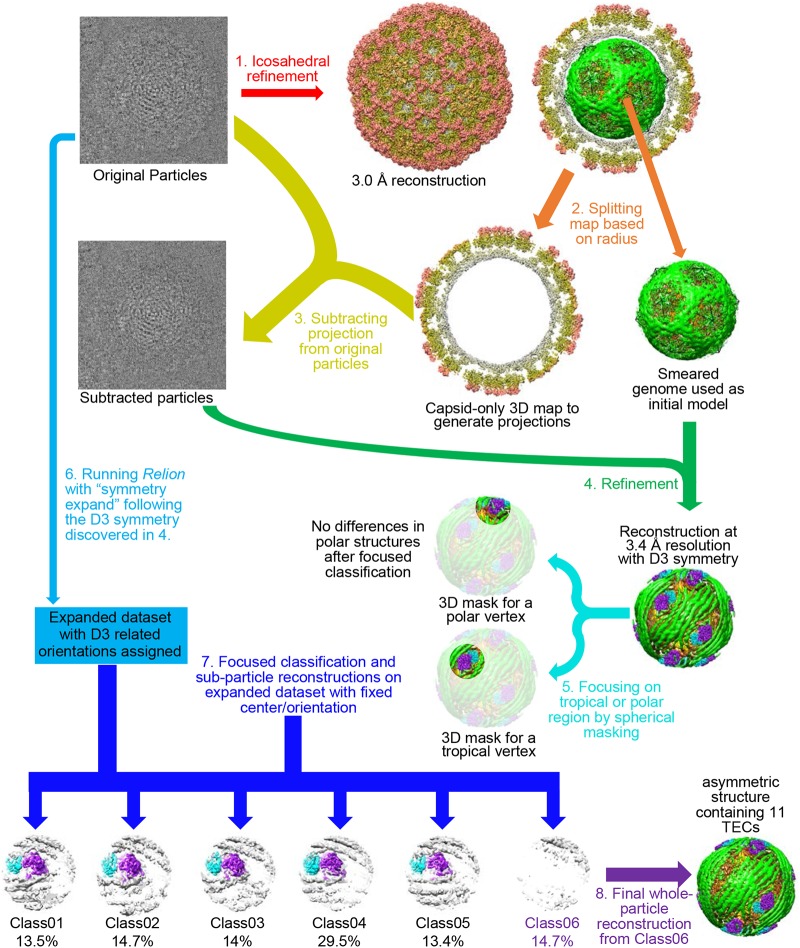
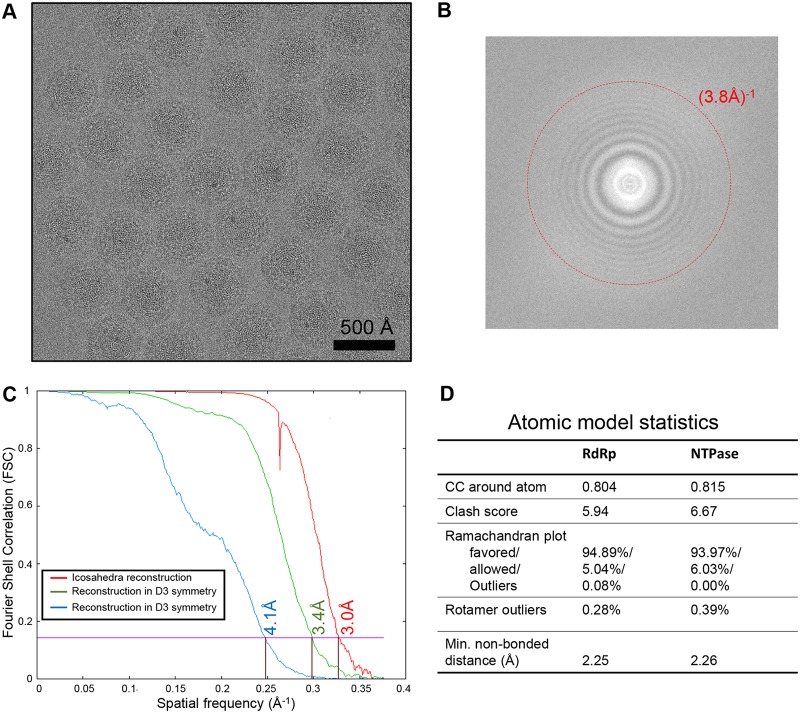
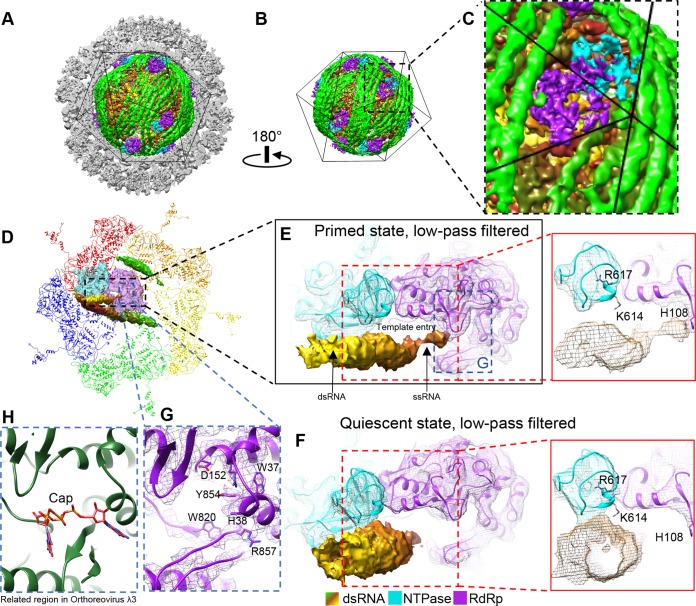
In order to resolve the structures of the TEC and genome inside primed-state ARV, we performed our asymmetric reconstruction by following a new protocol as described in Materials and Methods and outlined in Fig. 1. Grass carp reovirus ISVP was imaged using “superresolution” electron-counting technology to maximize the image contrast contributed by the internal genome and TEC (Fig. 2A and B). Compared to the workflow used to obtain the asymmetric structure of CPV (13), our new procedure contains two additional processes: symmetry expansion and focused classification (i.e., processes 6 and 7 in Fig. 1) under the framework of Relion (16). Three types of structures were obtained through this procedure: a capsid shell structure with icosahedral symmetry at 3.0-Å resolution, a structure exhibiting D3 symmetry (with a 3-fold rotation axis and a perpendicular 2-fold rotation axis) at 3.4-Å resolution, and a genuine asymmetric structure exhibiting pseudo-D3 symmetry at 4.1-Å resolution (Fig. 2C). Atomic models of TECs were built by imposing D3 symmetry onto density maps, and model statistics are reported (Fig. 2D). Our final asymmetric structure of primed-state ARV contains 11 TECs, 1 under each of the virion's 12 vertices, with only the northern tropical vertex lacking a TEC (Fig. 3 and 4A to D). The six TECs on the two poles are related to each other by D3 symmetry, and the remaining five TECs (tropical TECs) are related by pseudo-D3 symmetry.
FIG 1.
Asymmetric cryo-EM refinement/classification workflow for primed ARV showing ordered genome and 11 associated TECs in each virus. Arrows of the same color represent the same process applied to various data. Text in black describes the properties of the data. Colored text describes the sequential data-processing steps. 3D, three dimensional.
FIG 2.
Raw data, cryo-EM reconstruction, and model validation. (A and B) Cryo-EM micrograph of primed ARV particles (A) and Fourier spectrum of a representative micrograph showing the visibility of Thon rings (B). (C) Fourier shell correlation (FSC) curves showing the masked icosahedral reconstruction (red), unmasked reconstruction under D3 symmetry (green), and unmasked reconstruction without applying any symmetry (blue). (D) Model quality validation. CC, cross correlation.
FIG 3.
Density slices perpendicular to the pseudo-3-fold axis of the asymmetric reconstruction of the primed ARV. Note that the pseudo-D3 symmetry breaks at the slice framed in red (northern tropic).
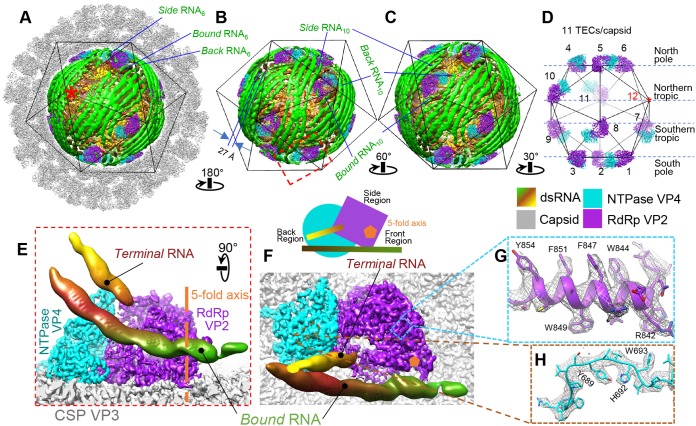
FIG 4.
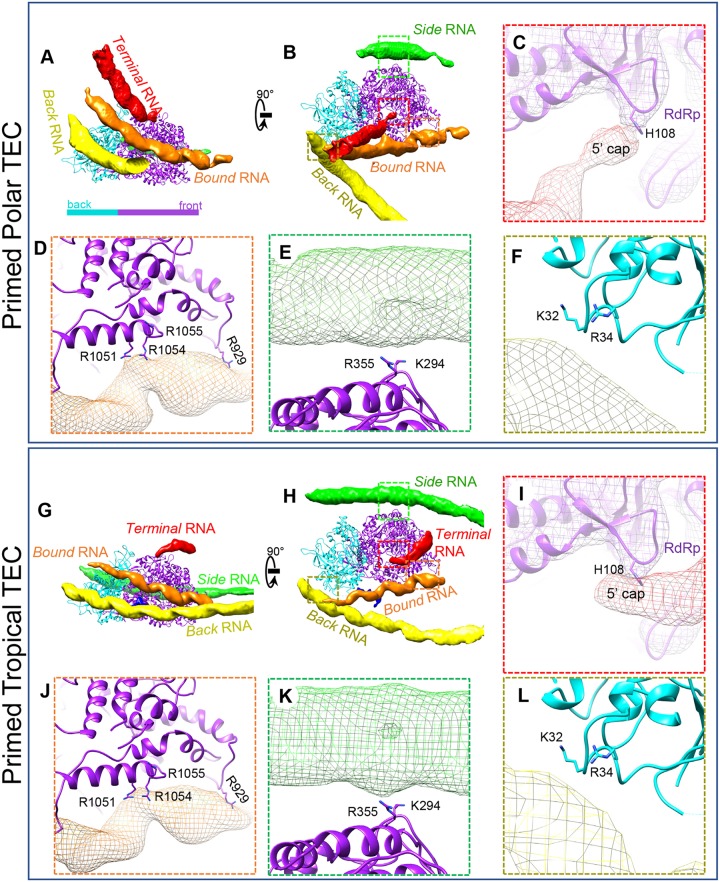
Asymmetric cryo-EM reconstruction of the primed ARV showing the ordered genome and 11 associated TECs in each virion. (A to D) Surface representations of the cryo-EM density map showing the full particle (A), the genome and TECs (B and C), and the TECs alone (D). The asterisk marks the vertex lacking a TEC. The three segments of RNA under vertices 6 and 10 are labeled “bound,” “back,” and “side” RNA. (E and F) Two orthogonal views of the region boxed in red in panel B. For clarity, all but two of the surrounding RNA densities, labeled bound and terminal RNA, were removed. The 5-fold axis is labeled; a cartoon diagram shows the definition of front, back, and side regions based on TEC geometry. (G and H) Superposition of our atomic model (color) on the density (gray mesh) extracted from the boxed regions in panel F. Color keys for the figure are the same throughout and are shown under panel D.
Each TEC is a heterodimer of two protein subunits. The protein closest to the capsid's 5-fold axis is the RdRp VP2, and the protein further away from the 5-fold axis is the NTPase VP4 (Fig. 4E and F). To facilitate subsequent structural description, we designated the three TEC sides away from the 5-fold axis the front, back, and side regions (Fig. 4F). Our in situ TEC structures reveal sufficient high-resolution features, such as clearly visible side chains, to support de novo atomic modeling for both RdRp and NTPase (Fig. 4G and H).
The dsRNA genome is tightly packed inside the capsid surrounding TECs (Fig. 3 and 4A to C). Each dsRNA duplex shows long persistence and is separated from its neighbors by an average of 27 Å (Fig. 4B). Several dsRNA strands form stabilizing interactions with each TEC, allowing visualization of their major and minor grooves, just as they do for the stem-loops of single-stranded RNA (ssRNA) viruses (17–19). Three major dsRNA densities interact with the TEC and are labeled “bound,” “side,” and “back” RNA based on their locations relative to the above-described sides of the TEC (Fig. 4A to C), with a fourth RNA, labeled “terminal RNA,” closely approaching the upper region of the TEC (Fig. 4E and F). This terminal RNA elongates directly toward the TEC, differing from other interacting RNAs.
RdRp VP2 has novel ligand interactions and an open template channel.
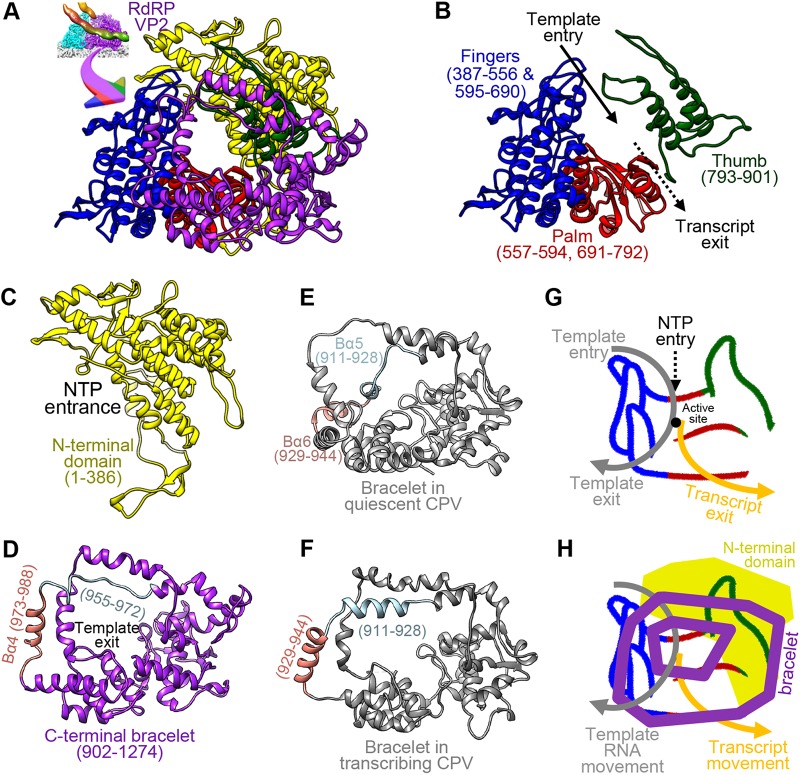
The RdRp VP2 contains 1,274 residues organized into three domains: the N-terminal (amino acids [aa] 1 to 386) (NTD), core, and C-terminal bracelet (aa 902 to 1274) domains. The core domain is sandwiched between the N-terminal and C-terminal bracelet domains and can be further divided into thumb (aa 793 to 901), finger (aa 387 to 556 and 595 to 690), and palm (aa 557 to 594 and 691 to 792) subdomains, following previously established terminology (20) (Fig. 5A to D). The domain arrangement of ARV RdRp follows that of ORV λ3 (21) and CPV RdRp (13, 22, 23). The finger and thumb subdomains perform transcript elongation and proofreading, whereas the palm catalyzes phosphodiester bond formation between new nucleoside triphosphates (NTPs) and the growing strand via D591, D740, and D741, which are highly conserved within the Reoviridae polymerases (see Fig. 7) (13, 21). Similar to ORV λ3 (21), ARV RdRp possesses four channels: the template entry, NTP entry, template exit, and transcript exit channels. The template exit channel penetrates the bracelet domain, as it does in other reoviruses (13, 24), whereas the NTP entry channel opens near the N-terminal domain (21, 24) (Fig. 5C, D, G, and H). All four channels intersect at the active site of the palm subdomain, which is unoccupied in our structure of the primed-state ARV RdRp. In notable contrast to the quiescent-CPV RdRp, no structures in the primed-ARV RdRp bracelet domain occlude the template exit channel or the active site. In the former, the same sites are blocked by the helix-loop structures formed by aa 911 to 928 and aa 929 to 944, respectively (Fig. 5D and F). Thus, all four channels in RdRp are open in ARV's primed state, and the protein's conformation resembles elongation state ORV RdRp (21).
FIG 5.
Structure of ARV RdRp VP2 with bound RNA and comparison of RdRp bracelet domains in ARV and CPV. (A to D) Ribbon diagrams of the atomic model of ARV RdRp VP2 with domains shown together (A) or separated. The RdRp core domain (B), comprising the finger, palm, and thumb subdomains, is sandwiched between the N-terminal domain (C) and the C-terminal bracelet domain (D). (E and F) The segment blocking the template exit in the CPV RdRp bracelet domain undergoes large conformational changes between the quiescent (E) and transcribing (F) states (13). The equivalent segment in ARV RdRp is similarly colored in panel D. (G and H) Cartoons showing the hand metaphor of polymerase core structure, with color coding following that in panel A. The polygons represent N- and C-terminal domains. (G) Four channels intersecting at the active site are labeled. (H) The N-terminal domain and bracelet are displayed on back and front planes, respectively.
FIG 7.
Sequence alignments of ARV, ORV, and CPV RdRps (A) and NTPases (B) (46).
NTPase VP4 has a unique C-terminal domain.
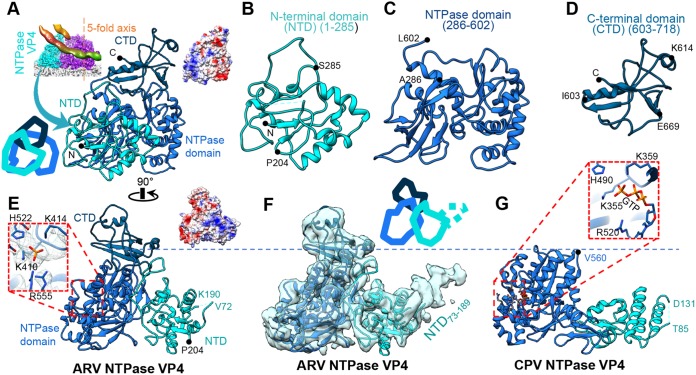
ARV's NTPase VP4 contains 728 residues arranged into three domains: an NTD (aa 1 to 285), an NTPase domain (aa 286 to 602), and a C-terminal domain (CTD) (aa 603 to 718; residues 718 to 728 are flexible) (Fig. 6A to D and 7B). VP4 contacts the inner capsid at the base of its NTD and NTPase domains, which fold around the end of the RdRp opposite to the capsid's 5-fold axis.
FIG 6.
Structure of NTPase VP4. (A to E) Ribbon diagrams of the atomic model of VP4 shown in two orthogonal views with its three domains in different colors (A and E) or in larger views as separated individual domains (B to D). (F and G) Comparison of NTPases from ARV (F) and CPV (G) (13) demonstrating the existence of an extra CTD (aa 603 to 718) in ARV. The atomic model is superimposed on a low-pass-filtered density map (semitransparent surface) showing the flexible (and thus unmodeled) NTD73–189 subdomain (F) in contrast to the shorter flexible segment NTD86–130 in CPV (G). The boxed region in panel E contains a phosphate group, whose location is equivalent to the third phosphate in GTP identified in the CPV NTPase in the boxed region in panel G. Cartoon diagrams were added to panels A and F to show the position of each domain. Surface charge diagrams were added to panels A and E in the same views.
The surface of VP4's NTD is highly positively charged (Fig. 6A, B, and E). The domain contains a protruding density that is visible only at low resolution and likely corresponds to its unmodeled residues 73 to 189, referred to as the NTD73–189 subdomain (Fig. 6F). Within NTD73–189 is a positively charged motif (aa 137 to 143 [RRVAAGR]) that potentially interacts with nearby RNA backbones. Our observation that NTPase VP4 interacts with RNA is consistent with a previous study showing that the homologous ORV μ2 is also an RNA-interacting protein (25). Located at the surface of ARV NTPase VP4 is a conserved residue, P204, that may be involved in filamentous inclusion body formation and microtubule-associated virion assembly, just as it is in the homologous ORV μ2 (14, 26, 27).
Unlike the N-terminal domain, the NTPase domain is mostly negatively charged (Fig. 6C). Structurally, it resembles the corresponding domain in CPV's NTPase VP4 (13) and binds an additional density at the conserved NTP binding site close to the key residues (K410 and K414, as previously predicted based on the homologous ORV μ2 [14]) in the primed state (Fig. 6E). Thus, we propose that ARV VP4 is also an NTPase. However, this additional density is too small to fit a full NTP molecule, and no NTP was added to the phosphate-buffered saline (PBS) buffer to obtain the viral sample. As observed, this small density could potentially be a single phosphate group whose position corresponds to the γ-phosphate group of a bound NTP.
Relative to CPV's NTPase, ARV's NTPase possesses an additional CTD (aa 603 to 718) (Fig. 6D and F). This domain has a mixed surface charge distribution with two positively charged residues (K614 and R617) forming a positively charged region at the far surface of VP4, close to the terminal RNA (Fig. 4E, 6D, and 8C). This region is also close to the template entry channel of RdRp, suggesting that the domain may be involved in template RNA regularization between different viral states.
FIG 8.
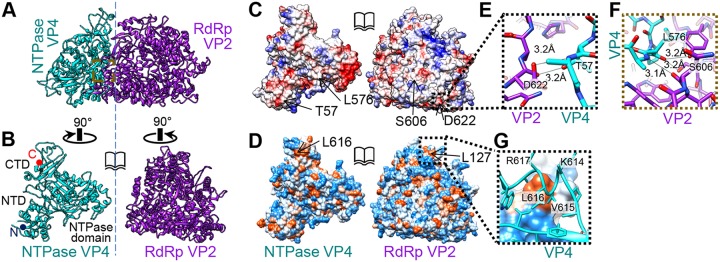
Interactions between RdRp and NTPase. (A) NTPase interacts with RdRp through RdRp's finger subdomain, near the template entrance. (B) NTPase and RdRp rotated 90° in opposite directions to show the triangle-shaped interface. (C) Same view as in panel B, showing positive (blue), neutral (white), and negative (red) Coulomb potentials. (D) Same view as in panel B, showing high hydrophobicity (orange) and high hydrophilicity (blue). (E) NTD-finger interaction in the region boxed in panel C. (F) NTPase domain-finger interaction in the region boxed in panel A. (G) CTD-NTD interaction in the region boxed in panel D.
Overall, NTPase VP4's proximity to RdRp VP2 and its affinity for binding cofactors crucial for RNA transcription (e.g., NTP and the RNA genome) away from the RdRp suggest that the protein is a key mediator in the polymerase's, and therefore the virion's, conversion from the transcriptionally inactive (quiescent) state to the primed state.
Interactions between RdRp and NTPase.
NTPase VP4 mostly interacts with RdRp through its finger subdomain (Fig. 8A), similar to the in situ structure of CPV (13). The shared interface is triangular and occupies a surface area of roughly 1,200 Å2 (Fig. 8B). Although both RdRp and NTPase are highly charged (Fig. 8C), the two proteins predominantly interact through hydrogen bonds and hydrophobic interactions. We found three strong interactions between the two proteins: (i) hydrogen bonds between the VP4 NTD (near aa 57) and the RdRp finger subdomain (near aa 622) (Fig. 8E), (ii) hydrogen bonds between the VP4 NTPase domain (aa 575 to 577) and the RdRp finger subdomain (aa 416, 601, and 606) (Fig. 8F), and (iii) hydrophobic interactions between the VP4 CTD (aa 615 and 618) and the RdRp N-terminal domain (near aa 127) (Fig. 8D and G). The last is close to the positively charged residues (K614 and R617) in the CTD (Fig. 8G).
CSP VP3's N termini anchor TECs and neighboring CSPs.
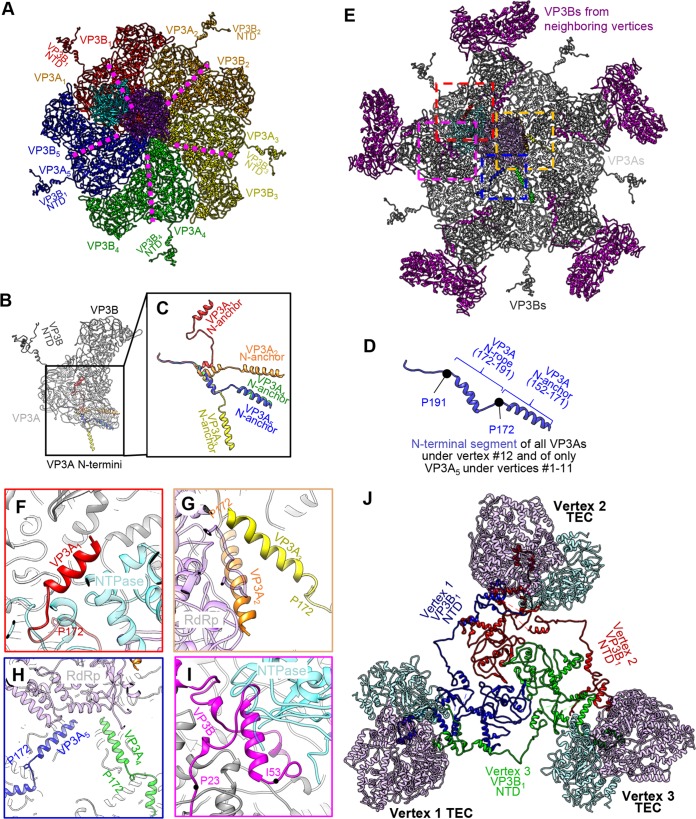
One hundred twenty copies of the 1,214-residue-long CSP VP3 form the innermost capsid shell with icosahedral symmetry T=2. Ten wedge-shaped VP3 monomers form one vertex in the icosahedron; the 5-fold pore at its center lines up with the TEC's RNA exit channel for direct transcript capping and release through turret protein VP1. Icosahedral reconstruction revealed two different conformers of VP3, termed VP3A and VP3B (3, 15). VP3A conformers form the star that houses the 5-fold pore, while VP3B conformers wedge themselves between the legs of the star to fill out the pentagonal shape. In our asymmetric reconstruction, we could further differentiate VP3A-VP3B dimers by their position in the vertex relative to the TEC: looking from the virion center, the VP31 dimer (containing VP3A1 and VP3B1) is mostly positioned behind the TEC, with the VP32 dimer through the VP35 dimer following in a clockwise direction (Fig. 9A). Under this convention, the VP31 to VP33 dimers form the primary seat upon which the TEC is positioned; in contrast, the VP34 dimer and the VP35 dimer make minimal contact with the TEC. Several fragments of VP3 were not resolved in the previous icosahedral reconstruction, most notably the first 187 residues of VP3A and aa 501 to 522 of VP3B (12). Our asymmetric reconstruction fully resolves VP3B, as well as residues 152 to 191 of VP3A. The latter residues were found to take at least 6 conformations depending on their positions relative to the TEC (Fig. 9B and C).
FIG 9.
The N-terminal segment of VP3 forms an adaptive anchor to interact with the TEC. (A) Inside view of a polar vertex showing the position of a TEC relative to five VP3A subunits (VP3A1 through VP3A5) and five VP3B subunits (VP3B1 through VP3B5). (B to D) Superposition of the five VP3 dimers from panel A showing dramatic conformational differences among the five subunits, VP3A1 through VP3A5, in the N-terminal fragment (colored in panels B and C). In the vertex without a TEC (i.e., position 12 in Fig. 4D), this fragment in the five VP3A subunits has a conformation (D) similar to that of the VP3A5 subunit (blue in panel C). (E) Same as panel A, with subunits colored as in panel B and five VP3B subunits from neighboring vertices added. (F to I) Enlarged views of the regions boxed in similar colors in panel E. (I) Note that the N-terminal domain of a VP3B of a neighboring vertex also interacts with this TEC. (J) Three polar TECs and their associated VP3B1 N-terminal segments (aa 15 to 518).
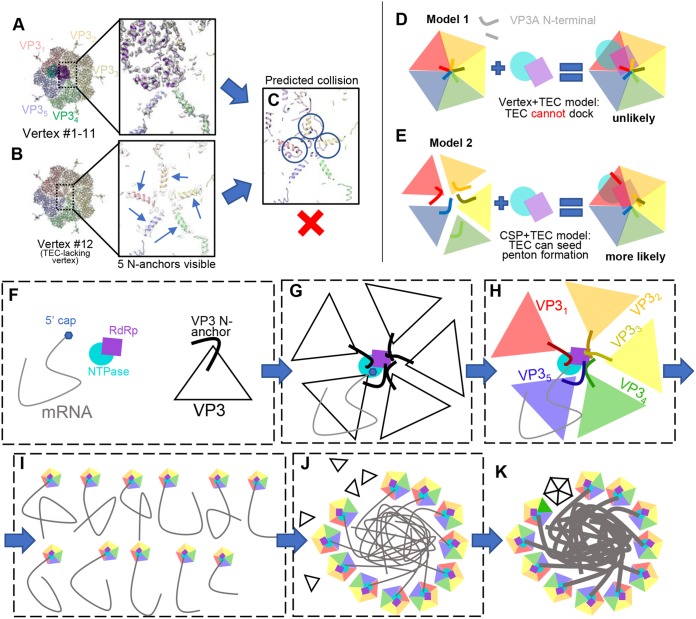
For VP3A, the newly modeled N-terminal residues include an N-terminal helix (N anchor) (aa 152 to 171) joined to the rest of the molecule by a varying rope-like fragment (N rope) (aa 172 to 191) (Fig. 9D). While the N anchor is folded into an α-helix in all VP3A conformers, the N rope can fold into a helix (VP3A4 and VP3A5) or partially unfold into extended loops (VP3A1 to VP3A3). Significant differences in the rope conformations of VP3A1, VP3A2, and VP3A3 allow their otherwise similar N anchors to reach and interact with NTPase, RdRp, and RdRp, respectively (Fig. 9B, C, and E to G). The N anchors of VP3A1 and VP3A2 are positioned similarly to the two “N-terminal helices” in the CPV TEC structure, but no N rope-like features were observed in CPV VP1 (13). In VP3A4 and VP3A5, the N rope is folded into a helix and the N anchor helix does not interact with the TEC (Fig. 9E and H). Notably, the three VP3A subunits whose N anchors contact the TEC (VP3A1 to VP3A3) are also the ones upon which the TEC is situated. Notably, the newly modeled residues of VP3A subunits at the vertex lacking a TEC (i.e., vertex 12) adopt much simpler conformations. Their N anchors are structurally identical to those of VP3A5 at the other 11 TEC-associated vertices. This uniform N anchor conformation creates steric hindrance that prevents the docking of a TEC at this vertex (see Fig. 15A to C).
FIG 15.
TEC-lacking vertex 12 and implications for the assembly mechanism. (A and B) Density map and models at vertices with (A) and without (B) a TEC. Note that in the icosahedral reconstruction, no N anchors of VP3A were visible due to averaging. (C) Direct model superposition of the TEC in panel A on five VP3As in panel B showing collision of TECs with N anchors (circled). (D and E) Two proposed models of assembly. (D) In model 1, vertices assemble first, followed by attachment of a TEC. The TECs would collide with assembled N anchors. (E) In model 2, the TEC seeds VP3A assembly, where VP3A N anchors can adapt the observed asymmetric pattern. (F to K) “Wool ball” model of virus assembly. (F) Building blocks of ARV assembly. TECs interact with mRNA and VP3, drawing the latter to their ideal positions (G), leading to properly folded N anchors around the TEC (H). The 11 genomic RNA segments, each with a vertex (I), coalesce to form a wool ball, bringing VP3s in close proximity to accelerate assembly (J). (K) Vertex 12 seals the capsid, while RdRp replicates a complementary strand from the plus-sense mRNA template in a confined environment.
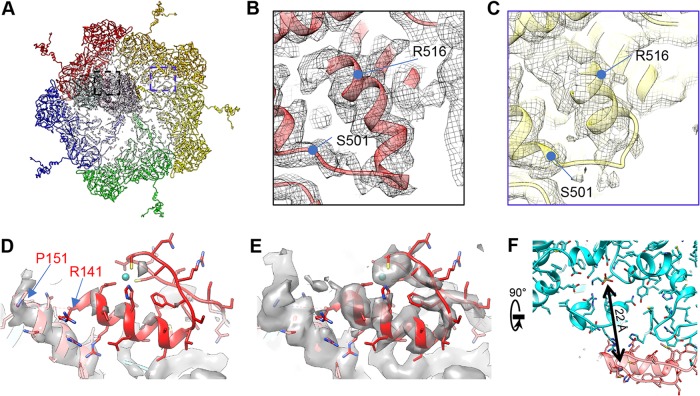
For all the VP3A conformers, the remainder of the N terminus (aa 1 to 147) is relatively flexible, and no strong densities could be found in a high-resolution map. However, at a lower display threshold (1 σ) (Fig. 10E), we found a zinc finger structure belonging to VP3A1 (aa 115 to 141) that binds to VP4 with a conformation similar to the zinc finger structure reported in VP3B conformers (15) (see Fig. 14M and N).
FIG 10.
Asymmetric feature found in primed ARV structure. (A to C) Structural comparisons showing plug-helix conformational change (VP3B aa 500 to 525). (A) A polar vertex region. (B and C) Boxed regions from panel A showing one plug-helix region interacting with a TEC (B) and one not interacting with a TEC (C). The plug-helix interacting with a TEC is less flexible. (D and E) Flexible zinc finger structure. The density maps are shown at 2 σ (D) and 1 σ (E), with the “hide dust” function off. The zinc finger at VP3A1 is much weaker than other modeled regions. The locations of P151 and R141 are labeled. (F) The VP3A1 Zn finger interacts with the TP-binding motif in VP4. The distance between the Zn atom and the γ-phosphate P atom is 22 Å.
FIG 14.
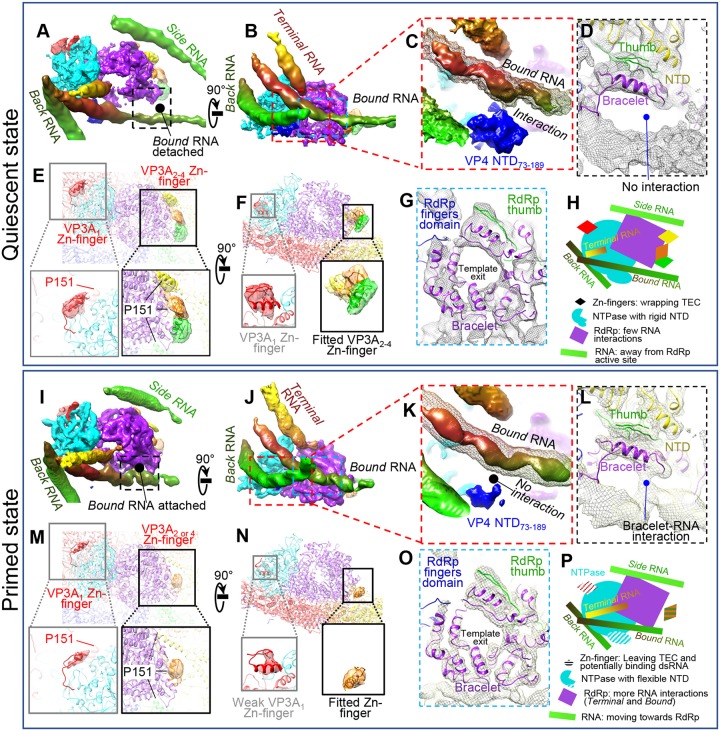
Comparison between quiescent and primed ARV shows similarities and differences in RNA, TEC, and CSP. (A to H) Structural details of a TEC under a polar vertex in the quiescent state. (A and B) Two orthogonal views showing four RNAs interacting with TECs. (C) Enlargement of the area boxed in red in panel B showing interaction of NTPase NTD73–189 with bound RNA (mesh). The solid density is at a higher display threshold than the mesh. (D) Enlargement of the boxed area in panel A showing detachment of the bound RNA from the RdRp in the quiescent state. (E and F) Magnified vertices showing only CSP N-terminal densities, with four structured densities (VP3A1 to VP3A4 Zn fingers, aa 117 to 141) labeled. P151 locations in VP3A1 to VP3A4 conformers are labeled to show the close proximity between N anchors and the corresponding Zn fingers. (G) Enlargement of the area boxed in blue in panel B showing the bracelet domain. (H) Cartoon of the relative positions of RNA and TEC. (I to P) Same view as in panels A to H for primed-state structures. Key differences from the quiescent state are as follows: bound RNA attaches to the RdRp bracelet domain (A, D, G, and L), the NTD73–189 domain in NTPase becomes more flexible (C and K), and the VP3A Zn finger becomes more flexible (E, F, M, and N). Unlike CPV, no large conformation changes in the bracelet domain could be found (G and O). (P) Cartoon of TEC and RNA structures in the primed state. All the map segmentations are based on the Chimera map segmentation function. No atomic model was used to generate the segmentation.
Unlike in VP3A, much more of the N terminus is modeled in VP3B. Residues 15 to 190 in all five VP3B subunits form an extended, largely loop-like structure that inserts itself underneath a flexible loop (aa 172 to 184) of a VP3B in an adjacent vertex and can even interact with the adjacent vertex's TEC (Fig. 9I). Because the first 200 residues of VP3 generally extend to great lengths to bind distant proteins, we renamed this region of both VP3 conformers the “daisy chain” domain.
Residues 500 to 525 form the tip of the apical domain of VP3, which in VP3A conformers forms the center of the vertex. In VP3B, the same residues, unmodeled in previous icosahedral reconstructions (12, 15), are now resolved as a loop linked to a short helix. This structure is best resolved in the VP3B conformer interacting with the TEC (i.e., VP3B1), where it is wedged between the RdRp VP2 and the NTPase VP4 (Fig. 10B).
TEC interactions with RNA.
Our in situ structure of the TEC reveals several RNA binding features. While our asymmetric reconstruction shows that the structures of 11 TECs are the same, the surrounding RNA can adopt different conformations (Fig. 11A, B, G, and H). Based on the specific residues the RNA strands interact with, we can classify them into RdRp-binding RNA (terminal, bound, and side) and NTPase-binding RNA (back).
FIG 11.
TEC interaction with RNA. Shown are structural comparisons of polar TECs (A to F) and tropical TECs (G to L) in a primed particle. (A and B) Two orthogonal views of polar TECs and four interacting RNA segments: terminal RNA, bound RNA, side RNA, and back RNA. (C to F) The boxed regions in panel B showing labeled residues interacting with terminal, bound, side, and back RNAs, respectively. (G to L) Same view as in panels A to F but on primed tropical TECs. RNA-interacting residues on TECs are conserved between polar TECs and tropical TECs.
For RdRp-binding RNA, we found that the terminal RNA‘s conformation depended on the position of its bound TEC. Comparison between polar and tropical TECs showed that the terminal RNAs approached RdRp from different directions but shared the same skinny geometry and binding site at H108 (Fig. 11A, C, G, and I). This “thin RNA binding to H108” feature is universal for all 11 TECs (6 polar and 5 tropical). Based on this, we propose that it is the plus-strand's 5′ end that we observed binding the TEC in our asymmetric reconstruction. Other RdRp-binding RNA strands are less influenced by the TEC's location. The bound RNA is anchored to the RdRp in two locations in the C-terminal bracelet domain. R929 (analogous to CPV's K997) anchors one segment of the bound RNA (Fig. 11D and J), while another RNA binding region downstream of R929, involving arginine residues 1051, 1054, and 1055, binds another segment (Fig. 11D and J). K294 and R355 of the N-terminal domain anchor the side RNA (Fig. 11E and K).
Back RNA is an NTPase-binding RNA in the primed state that interacts with both K32 and R34 on the positively charged NTD (Fig. 11F and L). This RNA is more curved near polar TECs. The positively charged NTD73–189 subdomain is close to both back and bound RNAs, but no observation showed that NTD73–189 can anchor RNA in the primed state.
With the exception of R929 (K997 in CPV), these RNA binding features have not been reported in previous asymmetric reconstructions of CPV (13, 23).
Priming introduces changes in RNA binding and protein structure.
The primed ARV described above was produced by removing the outermost protein, VP7, from the intact virion (3, 15). To investigate how the internal components of ARV—the genomic RNA and its associated TECs—respond to this external change, we also obtained an asymmetric reconstruction of quiescent-state ARV by reprocessing our old virion images with our new asymmetric-reconstruction protocol (Fig. 12 and 13A to C). Externally, the asymmetrically reconstructed quiescent ARV resembles the previous icosahedral reconstruction (15), but internally, it resembles our new primed-state ARV in that it contains 11 TECs in each capsid, again with the northern tropical vertex lacking a TEC (Fig. 12). The reported resolution near the TEC is 6 Å, after averaging all the TECs.
FIG 12.
Density slices perpendicular to the pseudo-3-fold axis of the asymmetric reconstruction of the quiescent ARV. Note that the pseudo-D3 symmetry breaks at the slice boxed in red (northern tropic).
FIG 13.
Reconstruction of quiescent ARV and binding changes in the terminal RNA during the priming process. (A and B) Asymmetric reconstruction of quiescent ARV; the TEC locations resemble those in primed ARV. (C) Magnified view of the boxed region in panel B to show a tropical TEC. (D to F) Structure comparison of RNA template entrances in the primed (D and E) and quiescent (F) states. The view in panel D is the same as that in Fig. 9A, except that the density maps of the terminal, side, and bound RNAs and the TEC are shown as shaded surfaces and as wire frames superimposed on the atomic models, respectively. The densities were low-pass filtered to facilitate comparison with the quiescent-ARV reconstruction. The terminal RNA bound to the CTD of NTPase in the quiescent state (F) detaches from the NTPase to bind H108 of the RdRp in the primed state (E). (G and H) Comparison of the cap-binding sites in primed ARV (G) and in the ORV λ3 crystal structure (PDB 1N1H) (H). Note that in primed ARV, the RNA 5′ cap is not bound to the cap-binding site.
The most significant conformational change during the priming process is the movement of the terminal RNA. NTPase's C-terminal domain is unique to ARV, as indicated by comparison with CPV's NTPase (Fig. 6E and G). For polar TECs in the quiescent state, K614 and R617 in this domain appear to anchor the terminal RNA so that its tip points toward the RNA template entry channel (Fig. 13F). This dsRNA is released from the NTPase in the primed state, and the 5′ end of its plus-sense strand extends to bind the key residue H108 in the N-terminal domain of RdRp VP2 (Fig. 11C and I and 13D and E). The cap density ends at H108, leaving the nearby cap-binding site unoccupied (Fig. 13G and H). Although this contrasts with previous findings that initiation state orthoreovirus RdRp λ3's cap-binding site directly binds the plus-sense RNA cap so that the minus-sense strand can enter the polymerase for transcription (21), it is possible that the binding of a single RNA base by H108 is an intermediate step in ARV's priming process. H108 may need to fully anchor the plus-sense ssRNA cap before passing it to the cap-binding site on the thumb subdomain about 10 Å behind it. Although no helicase-related motif could be found near the template entry channel to unwind dsRNA, the 3′ end of the minus-sense (template) ssRNA strand is positioned closer to the RdRp active site in the primed state. Thus, the movement and binding of the terminal RNA's plus-sense strand's cap seems to facilitate the minus-sense strand's entry into the active site as a template.
The bound RNA also undergoes large conformational changes during the priming process. In the quiescent state, the bound RNA does not interact with either R929 or the arginine-rich region of the RdRp bracelet domain as it does in the primed state (Fig. 14A and D). Instead, it is anchored to the NTPase NTD73–189 subdomain (Fig. 14C). During priming, the bound RNA detaches from the NTD73–189 subdomain and attaches to the RdRp's bracelet domain (Fig. 14K and L). Thus, it moves in the same direction as the terminal RNA: from NTPase to RdRp.
Inner ARV proteins also change during the transition from quiescent to primed states. The NTPase NTD73–189 subdomain is more structured in the quiescent state than in the primed state (Fig. 14C and K); its greater flexibility in the latter state can be attributed to the removal of constraints imposed on it by the bound RNA. Quiescent inner capsid proteins, specifically the N terminus zinc finger of VP3A, are also more structured. We can identify four structured regions near the VP3A1 to VP3A4 N anchor domains, the VP3A1 to VP3A4 Zn fingers, which, like the corresponding N anchors, are strongly associated with NTPase, RdRp, RdRp, and RdRp, respectively (Fig. 14E and F). Based on the positions of these Zn fingers, which bind the TEC in quiescent ARV, we identified another density in the filtered map of primed-state ARV (Fig. 14M and N, orange), which is located near but not overlapping the newly identified VP3A2 to VPA3A4 Zn fingers. Based on the distance between P151 and this density, it more likely contains the VP3A2 or VP3A4 Zn finger, as VP3A3's N anchor is relatively far away. Regardless, the four VP3A Zn fingers have more defined positions and features in the quiescent state. Greater freedom in the Zn finger residues in the primed state may correlate with greater flexibility in the neighboring TEC and RNA. This observation suggests that VP3 may help coordinate conformational changes between the outer-layer capsid, RNA, and TEC, most likely through directly interacting with all three elements.
Despite significant conformational changes in the ARV CSP and NTPase, we did not observe any similarly significant conformational changes in RdRp. In particular, residues 955 to 988 (roughly equivalent to aa 911 to 928 and aa 929 to 944 in CPV RdRp) of the C-terminal bracelet do not refold to occlude the template exit channel as they do in CPV (Fig. 5D to F and 14G and O) (13). Thus, both the domain and template exit channels remain open at all times in ARV, and premature transcription is blocked by a different mechanism, most likely through VP3, VP4, or both proteins in conjunction.
DISCUSSION
In this study, we found that the ARV NTPase VP4 possesses an extra C-terminal domain that is nonexistent in the CPV NTPase VP4. Two positively charged residues in this extra domain anchor the tip of the terminal RNA strand outside RdRp VP2's template entry channel in the quiescent state (Fig. 13F). This makes the positioning of C-terminal occlusion helices in the active site and in front of the RNA template exit channel, as seen in CPV RdRp, unnecessary in ARV. Both the bound and terminal RNA strands move from the NTPase to the RdRp when the virus transitions from the quiescent to the primed state. Thus, ARV NTPase may serve to bind RNA segments during viral assembly and to prevent premature transcription in the quiescent state, allowing RdRp to focus on its primary function of synthesizing RNA transcript in the transcribing state. From these structural observations, the primed state can be distinguished from the quiescent and transcribing states in that internally stored transcription factors (e.g., genomic dsRNA) are released from their anchoring structures and able to enter, but have not already entered, the viral polymerase. Primed virions may perhaps be prevented from entering the transcribing state by incomplete viral remodeling (i.e., further structural changes are needed to fully expand the capsid to allow sufficient space for transcription) or insufficient externally supplied cofactors (e.g., host NTP and SAM).
Because little external (host-provided) energy is needed to trigger the primed state, transitioning from the quiescent to the primed state must represent a natural transition from a higher-energy to a lower-energy state. Thus, the quiescent state in ARV is metastable. Similar local energy minimum states achieved prior to cell entry have been reported in many other viruses, such as poliovirus (28) and influenza virus (29). In order to transition out of this local minimum into the lower-energy primed state and thus “prime” the virion, the energy barrier around the quiescent state must be overcome. We believe the observed increased flexibility in viral proteins in the primed state can facilitate this process. Greater flexibility in RNA-contacting proteins allows greater freedom of movement in the RNA, which can potentially disturb short-range attractive interactions between positively charged residues and the RNA backbone. This can explain terminal RNA's detachment from K614 and R617 in polar VP4 (Fig. 13E and F). Movements introduced by protein flexibility can also allow the establishment of new interactions between positively charged residues and the RNA backbone, such as that observed between R1051, R1054, and R1055 and the RNA genome in the primed state (Fig. 11D and J and 14D and L). Conformational changes in the RNA genome were not addressed in previous studies, as conformational changes typically pertain to proteins. Here, however, given the more significant observed RNA conformational changes relative to observed protein conformations, it seems that few new protein-protein interactions are formed in the primed state and that proteins are unlikely to power these large RNA changes. It is more likely that the flexible protein structure releases RNA so that it can extend to occupy locations further away from the viral center (backbone charge driven) and/or to establish new interactions with other viral proteins (interaction driven). This mechanism is more efficient when backbone charges are maximized (stronger repulsion) and the genome has long persistence lengths (rigid genome). Both factors appear more strongly in viruses with a dsRNA genome. While we observed at best minimal conformational changes in the RdRp and the CTD of VP4, we found that the terminal RNA does extend toward RdRp and interacts with H108 in the primed state of ARV. This observation strongly supports the assumption that energy related to RNA can contribute to gene translocation and state transition. A similar theory has long been accepted for dsDNA viruses, such as phages (30).
The Zn fingers of VP3 may be multifunctional. In addition to stabilizing the capsid in VP3B conformers, they may also play a role in genome regulation because each finger in VP3A1 through VP3A4 shows greater flexibility in the primed state. A 10-residue-long linker (aa 141 to 151) separates the N anchor and the Zn finger; thus, the detached Zn finger can potentially interact with surrounding dsRNA. The fact that VP3A1's Zn finger density is visible and well defined but weaker than that of the other portions of VP3A1 suggest that this zinc finger is not bound to every VP4 and can potentially switch between TEC-binding and TEC-detaching states. The spatial proximity of the VP4 NTPase motif and the VP3A1 Zn finger suggests that the latter's detachment may potentially change the activity of the former (Fig. 10F). Zn finger detachment in VP3A2 through VP3A4 can also change RdRp activity through new interactions with RNA. These Zn fingers, which in ARV contact all the other major elements of the virion, are highly conserved in the family Reoviridae (31, 32) and can, in conjunction with the rest of the VP3 daisy chain domain, work as “sensors” to “inform” the TEC of the state of the dsRNA genome and the external environment.
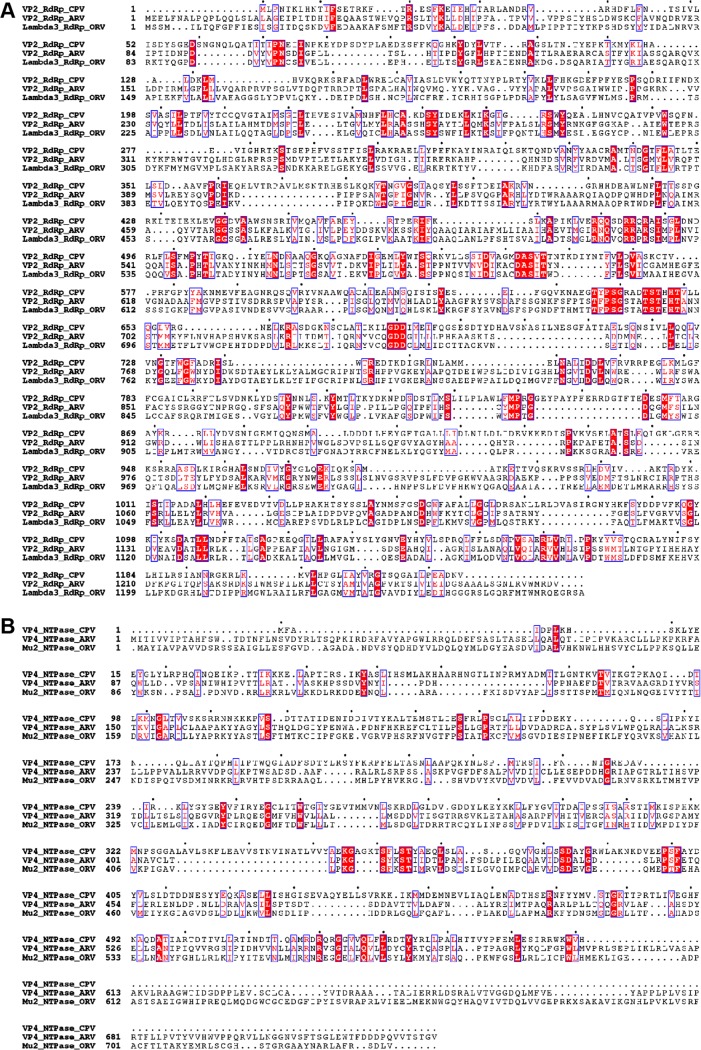
Structural variation among members of the Reoviridae appears to be the rule rather than the exception—as one might expect from RNA viruses, given their more rapid mutation and evolution rate relative to DNA viruses. Some members, such as rotavirus and BTV, lack an NTPase protein in the TEC (33), whereas others, such as CPV and ARV, have one. However, the number of domains each member's NTPase contains also varies, with the current study revealing that ARV's NTPase VP4 contains a new domain that binds to genomic RNA, which CPV's NTPase lacks. The specific locations of the NTPase's various functions can also vary. Sequence alignment between ORV μ2 and ARV VP4 showed that, although they have 26% identity overall, only the NTPase domain is highly conserved (Fig. 7B). In ARV VP4's CTD, the two key residues for RNA association are K614 and R617. No positively charged residues can be found in the corresponding positions of ORV μ2; however, ORV μ2 contains a significant peptide insertion (PRKXSAKAVIKG) in a location equivalent to ARV VP4's E669 (labeled in Fig. 6D), which is in turn located close to the finger subdomain and template entry of RdRp. In ORV, this highly charged peptide insertion may serve to anchor a dsRNA end, similar to K614 and R617 in ARV VP4. This variation between ORV and ARV shows that even close evolutionary relatives can have distinct NTPase structures and differing ways of regulating RNA. This variation holds true even within a single virion: the RNA regulation methods of polar TECs and tropical TECs are different, especially for terminal RNA (Fig. 11A, C, G, and I).
The numerous TEC interactions with the CSP VP3 N-terminal segments and RNA provide evidence for a novel coassembly model of dsRNA viruses. For many dsRNA viruses, expression of CSP alone leads to the formation of core-like particles (CLPs) (34–36), indicating that the constraints to form a capsid with the proper size and shape are fully encoded within CSP-CSP interactions. This notion is consistent with our observation of a TEC-lacking vertex in ARV virions (Fig. 15B). However, in the presence of the TEC, CSP binds to the TEC with higher affinity than to itself, as coexpression of polymerase and other capsid proteins leads to the formation of CLPs with TECs under all capsid vertices (35, 36). Most importantly, viral particles under native conditions (27, 37) or directly purified from infected cells typically contain RNA genomes, but empty particles can be obtained via special treatments, presumably due to loss of the genome through broken vertices (38). These observations can now be explained by the extensive interactions each TEC has with numerous CSPs (e.g., the extended N-terminal daisy chains of three VP3A and two VP3B molecules [Fig. 9E to J] and the inner surfaces of two VP3A and one VP3B molecules [Fig. 9A]) and the genomic RNA (Fig. 4, 11, 13, and 14). As the number of dsRNA segments of ARV matches the number (i.e., 11) of observed TECs inside each virion (Fig. 4A to D)—just as it does in CPV (i.e., 10 segments and 10 TECs) (13)—it is likely that each TEC associates with one and only one segment of the dsRNA genome, perhaps by recognizing terminal RNA (Fig. 15F to I). These genome-TEC-CSP interactions bring multiple copies of CSP into close proximity to significantly increase their local concentrations in the cytoplasm, improving their chances of finding their interacting interfaces and consequently increasing the yield of infectious virions containing both TEC and genome (Fig. 15E). This mechanism is preferable in the assembly process because an independently assembled CSP penton's flexible N terminus (e.g., residues 1 to 151) can create steric hindrance for the attachment of a TEC (Fig. 15D). In such a coassembly model, each TEC binds one genome segment, and interactions among the 11 segments lead to the formation of a “wool ball” (Fig. 15I and J). Tethered to the TECs in this wool ball through their N anchors, CSPs are brought together in approximately correct quantities and at approximately correct distances to coalesce snugly around the genome. As the ability to form a capsid vertex is entirely encoded within the CSP, the formation of a vertex bereft of TEC (i.e., vertex 12 in Fig. 15B) around the wool ball will happen naturally, ultimately leading to the formation of a complete infectious virion (Fig. 15K).
MATERIALS AND METHODS
Sample preparation and cryo-EM imaging. Primed GCRV, an ARV, was prepared as previously described (12). Each 2.5-μl sample of purified quiescent and primed GCRV virions was applied to a 400-mesh Quantifoil grid (R 1.2/1.3) from Quantifoil Micro Tools and vitrified in a Vitrobot Mk IV (Thermo Fisher Scientific) after blotting (blot time, 7 s; 100% humidity and 22°C). The cryo-EM samples were imaged in a Titan Krios electron microscope (Thermo Fisher Scientific) operated at 300-kV acceleration voltage and recorded on a Quantum LS energy-filtering direct electron-detecting camera (Gatan Inc.) operated in superresolution mode with a 20-eV slit width around the zero-loss peak. Data collection was facilitated by the Leginion automatic microscopy package (39). The total dosage was 56 e−/Å2, and the dosage used for refinement and reconstruction was 25 e−/Å2. The targeted defocus was 1.5 μm to 2.5 μm. The calibrated pixel size was 1.33 Å/pixel. In total, 5,810 movies were collected.
Single-particle reconstructions.
ISVPs were selected from aligned and averaged movies with UCSF MotionCorr (40). A total of 94,412 ISVPs were initially selected. The particles were extracted and initially subjected to icosahedral refinement with RELION (16), resulting in a 2.9-Å-resolution map. A symmetry relaxation method was applied to the refined data set as previously described (13) to obtain a 3.4-Å-resolution map of TECs. However, this method failed to resolve the true asymmetric structure (11 TECs). Consequently, the refinement was trapped in a local minimum with pseudo-D3 symmetry, restricting our ability to classify vertices to only 2 groups—6 polar and 6 tropical—in CPV. To precisely locate the unoccupied vertex, a local spherical mask was applied underneath different vertices to focus on areas of interest under the vertices (namely, TECs under vertices) and to minimize the strong noise introduced by the dsRNA genome in the liquid crystalline state. Each particle was expanded with 6 duplicates related to D3 symmetry, and exhaustive classification was conducted on the duplicated particles with symmetry-related orientations. We found that focusing classification on the northern tropical vertex resolved a class showing a vertex without a TEC, whereas all other tropical vertices were occupied with identical TECs. The above-described method allowed us to reconstruct GCRV ISVPs in the primed state with 11 TECs.
The structure for ARV in its quiescent state was determined by applying the same method to a previously published GCRV virion data set (15). The resolution near the reconstructed TEC was 6 Å.
Atomic model building and refinement.
The RdRp VP2, the CSP VP3, and the NTPase VP4 were modeled in Coot (41). VP2 modeling was guided by a homology model generated with Phyre2 (42) based on ORV λ3 (Protein Data Bank [PDB] accession no. 1N1H). VP3 was modeled by combining its previous atomic structure as determined by icosahedral reconstruction (13) with novel asymmetric regions built de novo using Coot. By docking the previously published model of the CSP VP3 (PDB accession no. 3IYL, chains X and Y) into our density map and comparing modeled residues with the full protein sequence, we pinpointed areas of unmodeled density that could be attributed to the missing regions of the protein. These unmodeled regions were filled using the Baton Build function in Coot and combined with the previous model. VP4, a new protein, was modeled entirely de novo using Baton Build. A string of polyalanines was placed in the density map using C-α bumps and landmark residues (such as Tyr and Trp) as a visual guide; this polyalanine chain was subsequently mutated to the proper sequence using Coot's Mutate Residue Range tool prior to local (Coot) and global (PHENIX) refinement. Poorly fitting residues were manually adjusted using the Rotate/Translate, Rotamer Fit, and Real Space Refine Zone tools. Initial models were further refined in PHENIX (43). The final quality of our models was evaluated based on model geometry, EMRinger score (45), fit to the density map, and agreement with the Ramachandran plot. Figures were generated using Chimera (44).
Accession number(s).
Newly determined data have been deposited in the Protein Data Bank (PDB) under number 6M99 and in the Electron Microscopy Data Bank (EMDB) under number EMD-9050.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (AI094386, GM071940, and DE025567). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111, and U24GM116792) and NSF (DBI-1338135 and DMR-1548924). K.D. was supported by a Whitcome Fellowship.
We thank Peng Ge for assistance in atomic modeling of the NTPase, for stimulating discussions during functional interpretation, and for proofreading the paper. We also thank former members of the Zhou group, including Xing Zhang, Qin Fang, and Lingpeng Cheng, for their contributions while working on the ARV project at UCLA.
REFERENCES
- 1.Barral PM, Sarkar D, Su ZZ, Barber GN, DeSalle R, Racaniello VR, Fisher PB. 2009. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol Therapeut 124:219–234. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estes MK. 1990. Rotaviruses and their replication. Virology 141:1353–1404. [Google Scholar]
- 3.Cheng LP, Fang Q, Shah S, Atanasov IC, Zhou ZH. 2008. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol 382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy P. 2007. Orbiviruses, p 1976–2000. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization-Coordinated Global Rotavirus Surveillance Network. 2016. Global, regional, and national estimates of rotavirus mortality in children < 5 years of age, 2000-2013. Clin Infect Dis 62:S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill CL, Booth TF, Prasad BV, Grimes JM, Mertens PP, Sutton GC, Stuart DI. 1999. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol 6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Zhang J, Yu X, Lu X, Zhang Q, Jakana J, Chen DH, Zhang X, Zhou ZH. 1999. Visualization of protein-RNA interactions in cytoplasmic polyhedrosis virus. J Virol 73:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou ZH. 2008. Cypovirus, p 27–43. In Patton JT. (ed), Segmented double-stranded RNA viruses: structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 9.Yu XK, Jiang JS, Sun JC, Zhou ZH. 2015. A putative ATPase mediates RNA transcription and capping in a dsRNA virus. Elife 4:e07901. doi: 10.7554/eLife.07901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne CC, Harrap KA. 1977. Cytoplasmic polyhedrosis viruses, p 105–129. In Maramorosch K. (ed), The atlas of insect and plant viruses. Academic Press, New York, NY. [Google Scholar]
- 11.Nason EL, Samal SK, Prasad BVV. 2000. Trypsin-induced structural transformation in aquareovirus. J Virol 74:6546–6555. doi: 10.1128/JVI.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 2010. 3.3 Angstrom cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Ding K, Yu XK, Chang W, Sun JC, Zhou ZH. 2015. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527:531. doi: 10.1038/nature15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Tao Y, Reinisch KM, Harrison SC, Nibert ML. 2004. Orthoreovirus and Aquareovirus core proteins: conserved enzymatic surfaces, but not protein-protein interfaces. Virus Res 101:15–28. doi: 10.1016/j.virusres.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Cheng LP, Zhu J, Hui WH, Zhang XK, Honig B, Fang Q, Zhou ZH. 2010. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol 397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui ZC, Gorzelnik KV, Chang JY, Langlais C, Jakana J, Young R, Zhang JJ. 2017. Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis. Proc Natl Acad Sci U S A 114:11697–11702. doi: 10.1073/pnas.1707102114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai XH, Li ZH, Lai M, Shu S, Du YS, Zhou ZH, Sun R. 2017. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 541:112. doi: 10.1038/nature20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ. 2016. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun 7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Sattar AKMA, Wang CC, Karam JD, Konigsberg WH, Steitz TA. 1997. Crystal structure of a Pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087–1099. doi: 10.1016/S0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 21.Tao YZ, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage; structural studies of reovirus polymerase lambda 3. Cell 111:733–745. doi: 10.1016/S0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 22.Li XW, Zhou NY, Chen WY, Zhu B, Wang XR, Xu B, Wang JW, Liu HR, Cheng LP. 2017. Near-atomic resolution structure determination of a cypovirus capsid and polymerase complex using cryo-EM at 200 kV. J Mol Biol 429:79–87. doi: 10.1016/j.jmb.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Liu HR, Cheng LP. 2015. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 24.Estrozi LF, Settembre EC, Goret G, McClain B, Zhang X, Chen JZ, Grigorieff N, Harrison SC. 2013. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol 425:124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brentano L, Noah DL, Brown EG, Sherry B. 1998. The reovirus protein mu 2, encoded by the M1 gene, is an RNA-binding protein. J Virol 72:8354–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker JSL, Broering TJ, Kim J, Higgins DE, Nibert ML. 2002. Reovirus core protein mu 2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol 76:4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah PNM, Stanifer ML, Hohn K, Engel U, Haselmann U, Bartenschlager R, Krausslich HG, Krijnse-Locker J, Boulant S. 21 July 2017. Genome packaging of reovirus is mediated by the scaffolding property of the microtubule network. Cell Microbiol 19. doi: 10.1111/cmi.12765. [DOI] [PubMed] [Google Scholar]
- 28.Hogle JM. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol 56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr CM, Chaudhry C, Kim PS. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci U S A 94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stent G. 1964. Molecular biology of bacterial viruses. R Soc Health J 84:151. [Google Scholar]
- 31.Reinisch KM, Nibert M, Harrison SC. 2000. Structure of the reovirus core at 3.6 angstrom resolution. Nature 404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 32.Yu XK, Ge P, Jiang JS, Atanasov I, Zhou ZH. 2011. Atomic model of CPV reveals the mechanism used by this single-shelled virus to economically carry out functions conserved in multishelled reoviruses. Structure 19:652–661. doi: 10.1016/j.str.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Magen T, Spencer E, Patton JT. 2007. An ATPase activity associated with the rotavirus phosphoprotein NSP5. Virology 369:389–399. doi: 10.1016/j.virol.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki N, Hagiwara K, Naitow H, Higashi T, Cheng RH, Tsukihara T, Nakagawa A, Omura T. 2005. Transcapsidation and the conserved interactions of two major structural proteins of a pair of phytoreoviruses confirm the mechanism of assembly of the outer capsid layer. J Mol Biol 345:229–237. doi: 10.1016/j.jmb.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 35.Nason EL, Rothagel R, Mukherjee SK, Kar AK, Forzan M, Prasad BVV, Roy P. 2004. Interactions between the inner and outer capsids of bluetongue virus. J Virol 78:8059–8067. doi: 10.1128/JVI.78.15.8059-8067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Sun J, Atanasov I, Ryazantsev S, Zhou ZH. 2011. Electron tomography reveals polyhedrin binding and existence of both empty and full cytoplasmic polyhedrosis virus particles inside infectious polyhedra. J Virol 85:6077–6081. doi: 10.1128/JVI.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Yu XK, Lu Y, Zhang JQ, Zhou ZH. 2002. Molecular interactions and viral stability revealed by structural analyses of chemically treated cypovirus capsids. Virology 298:45–52. doi: 10.1006/viro.2002.1473. [DOI] [PubMed] [Google Scholar]
- 39.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li XM, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng YF. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera; a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Barad BA, Echols N, Wang RY-R, Cheng Y, DiMaio F, Adams PD, Fraser JS. 2015. EMRinger: side chain directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]